Abstract
Colonization of the infant gut by microorganisms over the first year of life is crucial for development of a balanced immune response. Early alterations in the gastrointestinal microbiota of neonates has been linked with subsequent development of asthma and atopy in older children. Here we describe high-resolution culture-independent analysis of stool samples from 6-month old infants fed daily supplements of Lactobacillus casei subsp. Rhamnosus (LGG) or placebo in a double-blind, randomized Trial of Infant Probiotic Supplementation (TIPS). Bacterial community composition was examined using a high-density microarray, the 16S rRNA PhyloChip, and the microbial assemblages of infants with either high or low LGG abundance were compared. Communities with high abundance of LGG exhibited promotion of phylogenetically clustered taxa including a number of other known probiotic species, and were significantly more even in their distribution of community members. Ecologically, these aspects are characteristic of communities that are more resistant to perturbation and outgrowth of pathogens. PhyloChip analysis also permitted identification of taxa negatively correlated with LGG abundance that have previously been associated with atopy, as well as those positively correlated that may prove useful alternative targets for investigation as alternative probiotic species. From these findings we hypothesize that a key mechanism for the protective effect of LGG supplementation on subsequent development of allergic disease is through promotion of a stable, even, and functionally redundant infant gastrointestinal community.
Introduction
There is growing evidence that failure to develop a balanced immune response plays a key role in asthma and allergy development [1], [2], [3], and that environmental microbial exposure and host sampling of the developing gastrointestinal (GI) microbial community over the first year of life are crucial to immune response maturation [4], [5]. Culture-based approaches have suggested that the development of the GI microbiome is a progressive event beginning at birth and continuing until infants are weaned, with particular organisms acquired in distinct phases [6]. More recent, culture-independent studies have demonstrated that rather than a progressive colonization, the first year of life is characterized by fluctuating diversity of the microbial assemblage until convergence, with weaning, towards a GI community that more resembles that of an adult [7], [8], [9], [10]. As with adult GI bacterial consortia, inter-personal differences in GI microbial communities are evident in infants, particularly in the rate and stability of communities colonizing neonates [10], [11]. By 12 months old, the infant GI microbial community structure is relatively stable and the consortium largely resembles that of an adult, in which the Bacteroidetes and Firmicutes represent the two most dominant phyla [8], [9], [10].
A direct association has recently been demonstrated between the presence and abundance of specific microbial species in the GI tract of infants during the first 6 months of life and subsequent development of allergic disease at ages 1 and 2 [12], [13], demonstrating that early events in GI colonization precede development of allergic disease later in life. The first indication that a link existed between the GI microbiome and allergy was reported in the early 1980's in a study that described “dysbacteriosis” in infants with dermatological manifestations of food allergy, primarily due to low Bifidobacteria and Lactobacilli in combination with high numbers of species from the Enterobacteriaceae family [7]. Since then several studies have examined specific bacterial species in GI samples and demonstrated that their abundance correlated with atopy and asthma development [14], [15], [16], [17], [18]. A cross-sectional study of 1 year old infants in Estonia (low allergy prevalence) and Sweden (high allergy prevalence) demonstrated that more Estonian children possessed Lactobacilli and Eubacteria in their stool compared to Swedish children who were more likely to be colonized by Clostridium difficile [11]. A follow-up prospective study of stool samples from Estonian and Swedish children who were sampled over the first year of life and clinically followed up to 2 years of age, demonstrated that infants who developed allergy consistently exhibited lower levels of Bifidobacterial colonization compared to those that did not [16], [18]. Species-specific q-PCR analysis of the feces of 957 one-month-old infants in the KOALA birth cohort also demonstrated that a high abundance of Escherichia coli or C. difficile [12], [13] was associated with the development of eczema or atopy respectively [14].
The hygiene hypothesis suggests that a lack of microbial exposures during the crucial stages of immune maturation in infancy, results in immune modulation (Th2-biased response) that increases susceptibility to development of allergic disease [17]. Several studies have linked probiotic species with immunomodulation [14], [15], [16], [18], [19], [20], [21] and demonstrated their efficacy in protection against development of allergy and atopy. A randomized, controlled, double-blind study of 159 newborns, found that early feeding of Lactobacillus casei decreased the rate of atopic dermatitis at age two by 50% [22] and that this protective effect was sustained past infancy [23]. In animal models, probiotic supplementation has been shown to attenuate the respiratory inflammatory response [24] and reduce allergen-induced skin inflammation in sensitized mice [25]. Maassen and colleagues [26] demonstrated that of the eight Lactobacillus species used in their study of gut epithelial cytokine response, none induced local production of TGF-beta or IL-10, both of which are associated with a Th2 phenotype (a hallmark of atopic disease [27], [28], [29]). These observations are supported by other studies, which suggest that exposure to probiotics including Lactobacillus species, favors development of a Th1 phenotype and suppression of Th2 secreted cytokines [30].
The overall indication is that early events in gut microbial colonization play a key role in development of inflammatory disease at sites remote from the GI tract, and that a GI microbiome composed of beneficial bacterial species protects against allergy and atopic disease. Recent studies have demonstrated that the human gut is a densely populated, diverse bacterial microbiome [8], [9], [10], representing a complex host-microbial interaction. Therefore, it is likely that the species identified in previous studies may act as biomarkers for specific microbial consortia associated with, or protective against, allergic disease development. There is precedence for such a paradigm; several studies have demonstrated that bacterial community composition is dramatically altered in diseases such as obesity and periodontal disease [8], [31], [32] and that these changes are associated with altered consortium functionality [9].
While several studies have linked probiotics with improved clinical outcome, no study to date has examined the effect of supplementation on the overall GI microbiome to determine if the beneficial effects are due solely to high abundance of the species supplemented, or to a more global change in GI community structure. Here we describe culture-independent analysis of stool samples from 6-month-old infants at high risk for asthma development in the Trial of Infant Probiotic Supplementation (TIPS) study who were fed daily Lactobacillus casei subsp. rhamnosus (LGG) or placebo from birth to 6 months. Bacterial community composition was profiled using the 16S rRNA PhyloChip [33], [34], [35], a high-density, culture-independent microarray that can identify approximately 8,500 bacterial taxa (defined as groups of organisms that share at least 97% 16S rRNA sequence identity) in a single assay.
Materials and Methods
Ethics Statement
The Committee on Human Research at UCSF approved all study protocols, and all parents provided written, informed consent.
Sample Collection
Stool samples from 6-month-old infants (n = 16) randomized in blocks of four to daily probiotic (Lactobacillus casei subsp. Rhamnosus (LGG; ATCC 53103; 1×109 CFU) or placebo supplementation in the TIPS trial were used for this study. Samples were collected on the day prior to, or the day of the 6 month clinical visit from diapers using a scoop attached to the lid of a sterile collection vessel, prior to storage at 4°C and hand delivery or overnight mail to the study team. Samples were immediately banked at −80°C upon receipt.
Sample Processing
Stool samples were thawed on ice prior to extraction of DNA using the UltraClean Fecal extraction kit according to the manufacturer's instructions (Mo Bio, CA). Universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACT T-3′; [36]) were used to amplify the 16S rRNA gene using 12 PCR reactions per sample performed across a gradient of annealing temperatures (48-58°C) to maximize diversity recovered. PCR reactions contained 0.02 U/µl Takara Ex Taq DNA Polymerase (Takara Bio Inc., Japan), 1× Takara buffer, 0.8 mM Takara dNTP mixture, 0.4 mg/ml bovine serum albumin (BSA) and 1.0 µM of each primer. PCR conditions were 1 cycle of 3 min at 95°C followed by 25 cycles of 95°C for 30 s, the gradient annealing temperature for 30 s, 72°C for 2 min and a final extension at 72°C for 10 min. A total of 100 ng of extracted DNA from the stool samples was used per PCR reaction. Amplified products from all 12 annealing temperatures were pooled, gel purified and processed for PhyloChip analysis as previously described [33], except that 500 ng of each amplicon was hybridized. Further details of the PhyloChip, its development and use are provided elsewhere [37], [38], [39], [40], [41].
Microarray Analysis
Data sets were conservatively filtered, with taxa determined as present if ≥90% of probes in a probe set (for an individual taxon) were positive. Changes in probe-set fluorescence intensity are equivalent to changes in taxon relative abundance between samples. For taxa determined to be present in at least one sample, PhyloChip probe-set fluorescence intensity data was log transformed prior to analysis using packages in the R statistical environment [42]. Hierarchical cluster analysis (HCA) was performed on a Bray-Curtis dissimilarity matrix generated from PhyloChip fluorescence intensity data using the vegan package [43], followed by average linkage clustering.
A two-tailed Welch's T-test was used to identify taxa that were significantly altered in relative abundance in specific groups and adjusted for false discovery using the qvalue package as previously described. Significance was assigned with a p-value ≤0.05, q-value of 0.057. The 16S rRNA sequences of significant taxa were used to construct a neighbor-joining with nearest-neighbor interchange tree using FastTree [44] which was annotated using the Interactive Tree of Life (http://itol.embl.de/; [45]).
Correlation analysis of each individual taxon abundance against that of LGG (OTU_ID 3821) was performed using the multtest [46] package available as part of the Bioconductor suite of analysis programs [47]. Q-values were again generated for all p-values; taxa with r±0.5, p<0.05, q<0.15 were considered significant.
Indices of Bacterial Community Phylogenetic Structure
Nearest-taxon index (NTI) and Net-relatedness index (NRI; [48], [49]) were calculated using the picante package [50]. A phylogenetic tree of representative sequences was constructed as described above and used together with taxon richness to calculate the mean phylogenetic distance (MPD) and mean nearest phylogenetic taxon distance (MNTD) using the phylogeny shuffle null model for each sample. MPD and MNTD values were used, as previously described [49], to calculate NRI and NTI values respectively for each sample. Inverse Simpson's diversity index [51] and Pielou's evenness [52] were calculated using the vegan package [43]. To accurately reflect community richness for this calculation, individual taxa were deemed to have an abundance of 0 if they did not meet the pf ≥0.9 criterion.
Q-PCR Validation of LGG Presence and Relative Abundance
Quantitative PCR (Q-PCR) was performed to validate the presence and relative abundance of L. casei using primers based on PhyloChip probe sequences (LcF 5′-CGCATGGTTCT TGGCTGAAA-3′ and LcR 5′-ACAACAGTTA CTCTGCCGAC-3′). A total of 10 ng of DNA per reaction was used in triplicate, 25 µl Q-PCR reactions at an annealing temperature of 55°C. Regression analysis of inverse cycle threshold values plotted against array fluorescence intensity was used to confirm relative abundance of L. casei reported by the array and concordance between the two independent molecular methods.
Results
Bacterial Community Composition
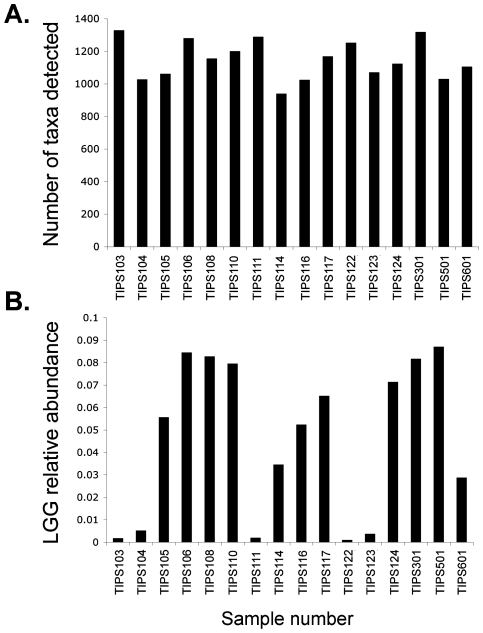
In the original TIPS trial, infants were randomized in groups of 4, therefore the cohort investigated in this study consisted of subjects fed LGG and those fed placebo. However, to protect the integrity of the TIPS trial, investigators remained blinded to the nature of the daily supplement. A total of 1,988 taxa, representing 46 different bacterial phyla were detected across all samples (a complete list of taxa detected is provided in Supplementary material, Table S1). This represents considerably greater diversity than previously reported in a clone library study of healthy infant stool samples [10], likely due to the ability of the array to sample in parallel and detect species that represent as little as 0.01% of the bacterial community [34]. Community richness (number of taxa present) was broadly similar across all samples studied (mean number of taxa per sample was 1146±125; Fig. 1A). This suggests that none of these infants had received antibiotics, (characteristically associated with a rapid, dramatic decline in bacterial community richness [53], [54]), proximal to the date of sample collection. Community richness ranged from 950 (TIPS 114) to 1,333 taxa (TIPS 103). Inter-subject microbiota variation has previously been described in a study of infant GI microbiota [10], suggesting that daily probiotic supplementation with LGG (1×109 CFU) did not result in dramatic domination of the communities by this species. Consortia were typically composed of members of the phyla Proteobacteria, Firmicutes, Actinobacteria and Bacteroidetes, which is in agreement with those detected in previous culture-independent studies of infant gastrointestinal microbiota [8], [9], [55]. At a more detailed phylogenetic level the greatest number of taxa detected belonged to the family Clostridiaceae, followed by the Enterobacteriaceae, Lachnospiraceae, Alteromonadaceae and Bacillaceae respectively.
Figure 1. A. Community Richness.
Bacterial community richness (number of taxa detected by 16S rRNA PhyloChip) in stool samples from 6 month old study subjects. B. LGG Abundance. LGG abundance (based on total fluorescence intensity) varies across subject samples.
Comparative analysis of LGG relative abundance in subject samples demonstrated that substantial differences existed across the cohort (Fig. 1B). To confirm differences in LGG abundance, independent Q-PCR analysis was performed on all samples with sufficient material (n = 11). Regression analysis demonstrated a significant correlation between the two independent molecular methods (r = 0.63; p<0.05), demonstrating concordance between the array and Q-PCR results and confirming the variation in relative abundance observed by the array in infant stool samples.
Effect of LGG Abundance on Bacterial Community Structure
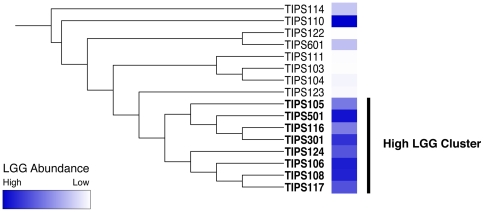
Hierarchical cluster analysis was performed on all samples using a distance matrix representing differences in PhyloChip taxon intensities. This demonstrated the existence of a clear cluster which contained the majority of samples with high LGG relative abundance (Fig. 2), suggesting that the presence of LGG in high abundance was associated with a specific community composition. To identify the organisms that characterized those communities, all samples were ranked by abundance of LGG and the five samples with the highest abundance were compared to the five with the lowest abundance using a two-tailed Welch's t-test. Following adjustment for false discovery a total of 682 taxa demonstrated significant differences between the two groups, all of which were more abundant in the high LGG samples (a complete list of taxa exhibiting significantly different abundance is provided in Supplementary Table, S2). The fact that the relative abundance of a number of taxa was significantly higher in the high LGG samples indicates that changes in community structure associated with high abundance LGG are not simply due to a dilution effect, but putatively to promotion of other members of the GI microbiota. Taxa of interest that were significantly more abundant in the high LGG samples included, amongst others, nine related Lactobacillaceae (L. crispatus [two strains], L. salivarius, L.sakei, L. manihotivorans, L. suntoryeus, L. kitasatonis, L. cypricasei and L. fuchuensis), and a member of the Bifidobacteriaceae (Bifidobacteriaceae genomosp. C1).
Figure 2. Hierarchical cluster analysis of infant stool samples.
Hierarchical cluster analysis reveals that LGG abundance is associated with specific bacterial community structures.
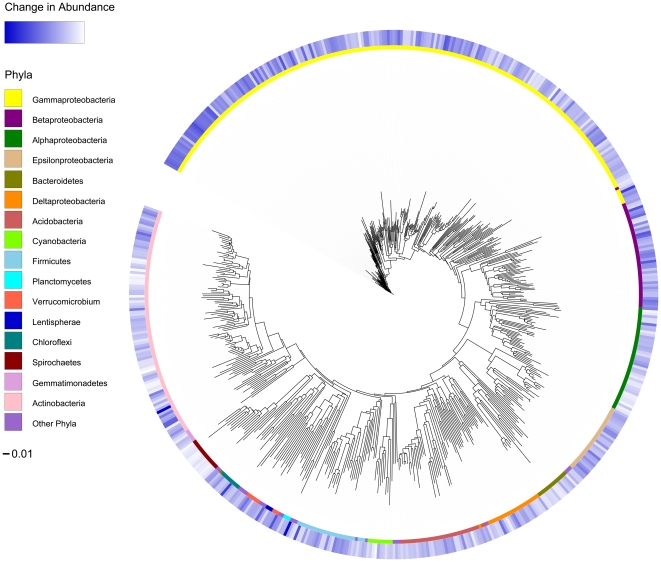
To determine if a high relative abundance of LGG impacted the abundance of phylogenetically related or, conversely phylogenetically distinct bacteria, community structure metrics Nearest Taxon Index (NTI) and Net Relatedness Index (NRI) were calculated (Table 1). Taxa promoted in the high LBB abundance communities had an NTI of 9.99 and NRI of 4.59, indicating that the species promoted in these communities were phylogenetically clustered at both the tips of the phylogenetic tree (NTI) and throughout the tree (NRI) relative to all bacteria detected. This clustering phenomenon is evident on a phylogenetic tree of all taxa that increased or decreased significantly in relative abundance; approximately 60% of the taxa that exhibited significantly higher abundance in the high abundance LGG communities belonged to the Proteobacteria and 37% of the total taxa were Gammaproteobacteria alone (Fig. 3). Various community metrics were also calculated for each individual sample and compared between high the LGG and low LGG samples by a two-tailed Welch's T-test (Table 1). Though the difference was small, Pielou's species evenness index was significantly higher in high LGG samples (p<0.041), demonstrating that communities with high abundance LGG were significantly more even than those with a low abundance of this species. Given the large size of the communities even this small, significant change in evenness is indicative of fundamental differences in the two communities. Species richness, community diversity (calculated using Inverse Simpson's index) Nearest Taxon Index (NTI) and the Net Relatedness Index (NRI) were not significantly different (p>0.05) between the two groups.
Table 1. Diversity and Phylogenetic Indices.
| Sample | Taxon Richness | Pielou's Evenness | Inverse Simpson's Index | Nearest Taxon Index | Net Relatedness Index |
| TIPS103 | 1333 | 0.9906 | 1185.09 | −1.50 | 2.26 |
| TIPS104 | 1014 | 0.9934 | 933.92 | −1.04 | −0.05 |
| TIPS105 | 1061 | 0.9932 | 974.79 | 1.25 | 1.39 |
| TIPS106 | 1285 | 0.9938 | 1187.24 | −0.02 | −5.30 |
| TIPS108 | 1158 | 0.9942 | 1076.34 | −0.30 | −0.33 |
| TIPS110 | 1203 | 0.9949 | 1129.03 | −0.64 | −0.25 |
| TIPS111 | 1278 | 0.9938 | 1183.64 | −2.40 | −0.67 |
| TIPS114 | 950 | 0.9875 | 814.61 | −2.77 | −2.01 |
| TIPS116 | 1032 | 0.9943 | 962.01 | 1.24 | 1.20 |
| TIPS117 | 1161 | 0.9935 | 1069.16 | −0.72 | −2.31 |
| TIPS122 | 1253 | 0.9911 | 1124.22 | −2.11 | −2.56 |
| TIPS123 | 1064 | 0.9926 | 970.82 | 0.16 | 1.94 |
| TIPS124 | 1117 | 0.9938 | 1033.53 | 0.82 | −2.12 |
| TIPS301 | 1319 | 0.9939 | 1223.26 | −1.74 | −1.30 |
| TIPS501 | 1020 | 0.9938 | 945.40 | 0.76 | 1.52 |
| TIPS601 | 1100 | 0.9932 | 1010.23 | −2.22 | −1.95 |
Samples in bold text were used as the examples of high LGG abundance and those in italics as examples of low LGG abundance in the t-test analysis.
Figure 3. Significant Differences in Abundance of Taxa.
Phylogenetic tree displaying taxa significantly increased in relative abundance in LGG dominated samples as a heatmap of fluorescence intensities in the outer ring. The inner ring displays the phylogenetic affiliation of each bacterial taxon at the level of class or higher. The scale bar indicates 0.01 nucleotide substitutions per base.
Correlation of LGG with Other Members of the Microbiota
We hypothesized that a proportion of the taxa that differentiated high LGG samples from other samples would be due to LGG-dependent interactions. Therefore we performed correlation analysis to identify the significant relationships that existed specifically between LGG and other taxa detected using all available samples. LGG was significantly correlated with 361 taxa (41% of which were also identified as significantly more abundant in high LGG samples; supplementary table S2). A total of 358 of these correlations were positive, 3 were negative. Positively correlated taxa included known probiotic species such as Lactobacillus fuchuensis and Bifidobacterium bifidum as well as several members of the Helicobacteraceae and a number of species that are known to produce antimicrobial compounds e.g. Streptomyces coelicolor. Interestingly S. coelicolor has been detected in the intestines of earthworms where it is particularly antagonistic to the anaerobic spore-forming bacteria [56]. Many of the positive relationships identified were with poorly characterized species, for example, the taxa most highly correlated with high LGG abundance were three Verrucomicrobia, two of which are unclassified and the third has the representative species Prosthecobacter dejongeii [57]. Characterized members of this recently described genus are fermenters and have been shown to encode a multitude of eukaryotic genes; their ancestors are hypothesized to have played an important role in the evolution of a proto-eukaryotic organism [58]. The three negative correlations revealed by this analysis were with Bacteroides uniformis, B. merdae and a swine intestinal clone classified as a member of the Lachnospiraceae.
Discussion
The protective effect of specific probiotic species against diseases such as atopy (reviewed in [59]), irritable bowel syndrome [60], [61], [62] and neonatal necrotizing enterocolitis [63] amongst others has previously been demonstrated in a number of clinical studies. However, the species-specific mechanism of protection conferred by feeding commensal organisms remains elusive. We took advantage of an on-going clinical study, the Trial of Infant Probiotic Supplementation (TIPS), to pose the question whether probiotic supplementation results in a species-specific increase in relative abundance that accounts for protection or if there is a global effect on the complex GI microbial consortium. In the latter case, we hypothesized that comprehensive analysis of these communities would identify the consortia that act in concert to protect against allergic disease development.
PhyloChip analysis of stool collected from 6-month old infants who had received daily probiotic or placebo supplements permitted high-resolution profiling of the microbial assemblages present in these samples. Although we remained blinded to the identity of which infants received probiotic supplementation, comparative analysis of LGG relative abundance amongst the samples demonstrated clear differences in the abundance of this species. While we cannot be sure that all of these infants received probiotic supplementation, the fact that substantial differences in LGG abundance existed amongst our subjects allowed us to focus our analysis efforts on the effect of LGG abundance (high or low) on GI community composition. Though it is likely that those with the highest abundance received supplements and those with the lowest did not, it is difficult to determine whether samples TIPS114 and TIPS601 represent supplemented infants that have not retained the LGG as efficiently as other infants in the study, or whether they have received placebo but have higher than average abundance of LGG from other dietary sources. Cluster analysis of these samples, supported the hypothesis that LGG abundance was associated with a distinct community composition. The majority of samples with high LGG abundance clustered together and formed a group distinct from those with low LGG abundance.
Analysis of the phylogenetic differences characteristic of samples with high LGG revealed a large number of taxa increased in relative abundance in these communities. These included a number of known beneficial species belonging to the Lactobacillaceae and Bifidobacteriaceae in addition to species that, through their secondary metabolite production could conceivably influence GI consortium composition. Community phylogenetic metrics (NTI and NRI) demonstrated that the promoted taxa were strongly phylogenetically related. This suggests functional redundancy within GI communities that possess LGG in high abundance. Ecologically this attribute is characteristic of a stable, resilient consortia, resistant to sub-population overgrowth that can reduce host fitness [64]. In addition, samples with high LGG abundance were significantly more even; initial evenness of microbial communities has been suggested to preserve the functional stability of an ecosystem [65]. In the case of human hosts, overgrowth by pathogens, such as Escherichia coli in 1-month old neonates has previously been shown to be associated with a higher risk of developing eczema and this risk is increased with greater numbers of this species [12]. These observations point to a protective mechanism by which high LGG abundance results in promotion of other protective species in a community that is functionally redundant, more even, and possibly resistant to pathogen overgrowth.
Given these data, it is tempting to suggest that perturbation of the GI community, due to antibiotic administration or viral infection early in life, may affect microbial colonization patterns and permit outgrowth of specific resistant members of the community that influence immune development. Putatively, this provides an explanation for the relationships demonstrated between these factors and subsequent allergic disease development [66], [67]. Certainly there is support in the literature for such a model; several recent studies have demonstrated that host susceptibility to enteric pathogens is influenced by the GI bacterial community composition [68], [69] and Hrncir et al showed a link between gut microbiota, diet and development of T reg cells (key to maintaining immunological homeostasis) in germ-free mice [70]. Given the extent of GI community diversity and the inherent inter-personal variability in consortium composition, a comprehensive analysis of the pattern of colonization over the first year of life in well defined groups is necessary to address this, and many other questions fundamental to understanding the complexity of GI microbial factors that impact development of allergic disease.
In addition to a large number of commonly isolated or identified gut microorganisms, the PhyloChip also identified a range of taxa that are not commonly associated with this environment (supplementary table 1). There are a number of possible explanations for this, firstly the PhyloChip is a highly sensitive technique, rather than detecting the most abundant taxa present in a sample, it detects taxa in parallel to as little as 0.01% of the total abundance. Secondly, the representative organism for a given taxon (Table S1) may not have been reported previously in the gut environment, but other, related organisms in that taxonomic group may have been identified. Other culture-independent studies using high-throughput sequencing have also detected unexpected organisms in specific host niches; Eckburg et al [71] noted the presence of a novel, deeply branching lineage related to the Cyanobacteria by high-throughput sequencing in mucosal tissue and fecal samples while Andersson et al also identified a diverse stomach microbiota which included Cyanobacteria and Chlamydia sequences [72]. The more frequent use of high-coverage culture-independent approaches and repeated detection of these species in specific niches, validates their presence and suggests a role for these unusual organisms in human health.
Mucosal recognition and discrimination between commensal and pathogenic species is key to the regulation of immune homeostasis. This study certainly suggests that the relationship between LGG abundance and the immune response is significantly more complex than is inferred from studies of single species in vitro. Supplemental analysis to determine which taxa exhibited a relationship specifically with LGG abundance demonstrated that 41% of taxa differentiating high LGG from low LGG samples also exhibited significant correlations with LGG. Proposed mechanisms by which probiotic species are believed to elicit a beneficial effect include direct antimicrobial activity, competitive colonization, stimulation of immune responses and inhibition of virulence gene or protein expression by pathogenic species [73]. In addition to these possible factors, LGG also appears to elicit a profound effect on GI community composition. The beneficial effect of this species appears not to be due solely to its high abundance, but to the global changes in the bacterial community composition of the infant gut when it is present in high abundance. It is clear that a large number of other taxa detected in this study that have not previously been associated with, but are now potentially implicated in protection against allergic disease development, merit further study. To fully understand the functional implications of probiotic supplementation, parallel microbial consortium and host response gene expression analyses in clearly defined groups of probiotic or placebo supplemented infants are necessary.
The overall health effects of probiotic supplementation are strain-specific [59]. Nevertheless, there are potential general implications from this LGG-focused study which demonstrate that high abundance of this species is associated with a dramatic change in GI microbial community composition, impacting the relative abundance of a large number of taxa previously associated with either an increased or decreased risk for the development of allergy and atopy. Recent findings demonstrate that the GI microbiome is a unique, personalized assemblage at the species level [74]. Therefore there is potential for multiple components of this consortium to confer a protective effect. The data presented here suggests that promotion of a phylogenetically even, functionally redundant infant GI community, composed of a multitude of probiotic species, rather than a community dominated by a single beneficial species, may represent a key factor in protection against allergic disease development.
Supporting Information
Total taxa detected by PhyloChip. A list of all taxa detected by the 16S rRNA PhyloChip in all samples.
(0.38 MB XLS)
Significantly correlated and T-tested taxa. A list of taxa that are significantly correlated, positively or negatively, with LGG together with those that demonstrate significant differences in abundance between high LGG (G1) and low LGG samples.
(0.25 MB XLS)
Acknowledgments
We would like to thank I. Letunic for rapid implementation of the heatmap function in the International Tree of Life Project and N. Slusher for critical assessment of the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: SVL is supported by an American Lung Association Award (http://www.lungusa.org/), MDC and and HAB are supported by a National Institutes of Health (NIH) award (http://www.nih.gov/), HL080074 to MDC and HL074207 to HAB and ELB, and YLH by a Tobacco-Related Disease Research Program award (http://www.trdrp.org/). Part of this work was performed at Lawrence Berkeley National Laboratory under the auspices of the University of California under contract number DOE DE-AC02-05CH11231. This study was also supported in part by National Institutes of Health/National Center for Research Resources (NIH/NCRR) University of California San Francisco Clinical and Translational Science (UCSF-CTSI) Grant Number UL1 RR024131. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Yoo J, Tcheurekdjian H, Lynch SV, Cabana M, Boushey HA. Microbial manipulation of immune function for asthma prevention: inferences from clinical trials. Proc Am Thorac Soc. 2007;4:277–282. doi: 10.1513/pats.200702-033AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalliomaki M, Isolauri E. Pandemic of atopic diseases–a lack of microbial exposure in early infancy? Curr Drug Targets Infect Disord. 2002;2:193–199. doi: 10.2174/1568005023342452. [DOI] [PubMed] [Google Scholar]
- 3.Liu AH. Hygiene theory and allergy and asthma prevention. Paediatr Perinat Epidemiol. 2007;21(Suppl 3):2–7. doi: 10.1111/j.1365-3016.2007.00878.x. [DOI] [PubMed] [Google Scholar]
- 4.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rakoff-Nahoum S, Medzhitov R. Innate immune recognition of the indigenous microbial flora. Mucosal Immunol. 2008;1(Suppl 1):S10–14. doi: 10.1038/mi.2008.49. [DOI] [PubMed] [Google Scholar]
- 6.Copperstock MS. Human intestinal microflora in health and disease. In: Hentges DJ, editor. New York: Academic Press; 1983. pp. 77–99. [Google Scholar]
- 7.Kuvaeva IB, Orlova NG, Veselova OL, Kuznezova GG, Borovik TE. Microecology of the gastrointestinal tract and the immunological status under food allergy. Nahrung. 1984;28:689–693. doi: 10.1002/food.19840280645. [DOI] [PubMed] [Google Scholar]
- 8.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 10.Palmer C, Bik EM, Digiulio DB, Relman DA, Brown PO. Development of the Human Infant Intestinal Microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sepp E, Julge K, Vasar M, Naaber P, Bjorksten B, et al. Intestinal microflora of Estonian and Swedish infants. Acta Paediatr. 1997;86:956–961. doi: 10.1111/j.1651-2227.1997.tb15178.x. [DOI] [PubMed] [Google Scholar]
- 12.Penders J, Stobberingh EE, van den Brandt PA, Thijs C. The role of the intestinal microbiota in the development of atopic disorders. Allergy. 2007;62:1223–1236. doi: 10.1111/j.1398-9995.2007.01462.x. [DOI] [PubMed] [Google Scholar]
- 13.Penders J, Thijs C, van den Brandt PA, Kummeling I, Snijders B, et al. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study. Gut. 2007;56:661–667. doi: 10.1136/gut.2006.100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delcenserie V, Martel D, Lamoureux M, Amiot J, Boutin Y, et al. Immunomodulatory effects of probiotics in the intestinal tract. Curr Issues Mol Biol. 2008;10:37–54. [PubMed] [Google Scholar]
- 15.Galdeano CM, de Moreno de LeBlanc A, Vinderola G, Bonet ME, Perdigon G. Proposed model: mechanisms of immunomodulation induced by probiotic bacteria. Clin Vaccine Immunol. 2007;14:485–492. doi: 10.1128/CVI.00406-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nova E, Warnberg J, Gomez-Martinez S, Diaz LE, Romeo J, et al. Immunomodulatory effects of probiotics in different stages of life. Br J Nutr. 2007;98(Suppl 1):S90–95. doi: 10.1017/S0007114507832983. [DOI] [PubMed] [Google Scholar]
- 17.Strachan DP. Hay fever, hygiene, and household size. BMJ. 1989;299:1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Versalovic J. Probiotics: intestinal gatekeeping, immunomodulation, and hepatic injury. Hepatology. 2007;46:618–621. doi: 10.1002/hep.21916. [DOI] [PubMed] [Google Scholar]
- 19.Baken KA, Ezendam J, Gremmer ER, de Klerk A, Pennings JL, et al. Evaluation of immunomodulation by Lactobacillus casei Shirota: immune function, autoimmunity and gene expression. Int J Food Microbiol. 2006;112:8–18. doi: 10.1016/j.ijfoodmicro.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 20.Bell SG. Immunomodulation, Part V: probiotics. Neonatal Netw. 2007;26:57–60. doi: 10.1891/0730-0832.26.1.57. [DOI] [PubMed] [Google Scholar]
- 21.Gill H, Prasad J. Probiotics, immunomodulation, and health benefits. Adv Exp Med Biol. 2008;606:423–454. doi: 10.1007/978-0-387-74087-4_17. [DOI] [PubMed] [Google Scholar]
- 22.Kalliomaki M, Salminen S, Arvilommi H, Kero P, Koskinen P, et al. Probiotics in primary prevention of atopic disease: a randomised placebo-controlled trial. Lancet. 2001;357:1076–1079. doi: 10.1016/S0140-6736(00)04259-8. [DOI] [PubMed] [Google Scholar]
- 23.Kalliomaki M, Salminen S, Poussa T, Arvilommi H, Isolauri E. Probiotics and prevention of atopic disease: 4-year follow-up of a randomised placebo-controlled trial. Lancet. 2003;361:1869–1871. doi: 10.1016/S0140-6736(03)13490-3. [DOI] [PubMed] [Google Scholar]
- 24.Forsythe P, Inman MD, Bienenstock J. Oral treatment with live Lactobacillus reuteri inhibits the allergic airway response in mice. Am J Respir Crit Care Med. 2007;175:561–569. doi: 10.1164/rccm.200606-821OC. [DOI] [PubMed] [Google Scholar]
- 25.Park CW, Youn M, Jung YM, Kim H, Jeong Y, et al. New functional probiotic Lactobacillus sakei probio 65 alleviates atopic symptoms in the mouse. J Med Food. 2008;11:405–412. doi: 10.1089/jmf.2007.0144. [DOI] [PubMed] [Google Scholar]
- 26.Maassen CB, van Holten-Neelen C, Balk F, den Bak-Glashouwer MJ, Leer RJ, et al. Strain-dependent induction of cytokine profiles in the gut by orally administered Lactobacillus strains. Vaccine. 2000;18:2613–2623. doi: 10.1016/s0264-410x(99)00378-3. [DOI] [PubMed] [Google Scholar]
- 27.Ball TM, Castro-Rodriguez JA, Griffith KA, Holberg CJ, Martinez FD, et al. Siblings, day-care attendance, and the risk of asthma and wheezing during childhood. N Engl J Med. 2000;343:538–543. doi: 10.1056/NEJM200008243430803. [DOI] [PubMed] [Google Scholar]
- 28.Gereda JE, Leung DY, Thatayatikom A, Streib JE, Price MR, et al. Relation between house-dust endotoxin exposure, type 1 T-cell development, and allergen sensitisation in infants at high risk of asthma. Lancet. 2000;355:1680–1683. doi: 10.1016/s0140-6736(00)02239-x. [DOI] [PubMed] [Google Scholar]
- 29.Prescott SL, Macaubas C, Holt BJ, Smallacombe TB, Loh R, et al. Transplacental priming of the human immune system to environmental allergens: universal skewing of initial T cell responses toward the Th2 cytokine profile. J Immunol. 1998;160:4730–4737. [PubMed] [Google Scholar]
- 30.Ghadimi D, Folster-Holst R, de Vrese M, Winkler P, Heller KJ, et al. Effects of probiotic bacteria and their genomic DNA on TH1/TH2-cytokine production by peripheral blood mononuclear cells (PBMCs) of healthy and allergic subjects. Immunobiology. 2008;213:677–692. doi: 10.1016/j.imbio.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 31.Kumar PS, Leys EJ, Bryk JM, Martinez FJ, Moeschberger ML, et al. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J Clin Microbiol. 2006;44:3665–3673. doi: 10.1128/JCM.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakamoto M, Huang Y, Ohnishi M, Umeda M, Ishikawa I, et al. Changes in oral microbial profiles after periodontal treatment as determined by molecular analysis of 16S rRNA genes. J Med Microbiol. 2004;53:563–571. doi: 10.1099/jmm.0.45576-0. [DOI] [PubMed] [Google Scholar]
- 33.Brodie EL, DeSantis TZ, Joyner DC, Baek SM, Larsen JT, et al. Application of a High-Density Oligonucleotide Microarray Approach To Study Bacterial Population Dynamics during Uranium Reduction and Reoxidation. Appl Environ Microbiol. 2006;72:6288–6298. doi: 10.1128/AEM.00246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brodie EL, DeSantis TZ, Parker JP, Zubietta IX, Piceno YM, et al. Urban aerosols harbor diverse and dynamic bacterial populations. Proc Natl Acad Sci U S A. 2007;104:299–304. doi: 10.1073/pnas.0608255104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeSantis TZ, Brodie EL, Moberg JP, Zubieta IX, Piceno YM, et al. High-density universal 16S rRNA microarray analysis reveals broader diversity than typical clone library when sampling the environment. Microb Ecol. 2007;53:371–383. doi: 10.1007/s00248-006-9134-9. [DOI] [PubMed] [Google Scholar]
- 36.Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York: John Wiley & Sons; 1991. pp. 115–175. [Google Scholar]
- 37.Yergeau E, Schoondermark-Stolk SA, Brodie EL, Dejean S, DeSantis TZ, et al. Environmental microarray analyses of Antarctic soil microbial communities. ISME J. 2009;3:340–351. doi: 10.1038/ismej.2008.111. [DOI] [PubMed] [Google Scholar]
- 38.Sunagawa S, Desantis TZ, Piceno YM, Brodie EL, Desalvo MK, et al. Bacterial diversity and White Plague Disease-associated community changes in the Caribbean coral Montastraea faveolata. ISME J. 2009. [DOI] [PubMed]
- 39.Shawkey MD, Firestone MK, Brodie EL, Beissinger SR. Avian incubation inhibits growth and diversification of bacterial assemblages on eggs. PLoS One. 2009;4:e4522. doi: 10.1371/journal.pone.0004522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sagaram US, DeAngelis KM, Trivedi P, Andersen GL, Lu SE, et al. Bacterial Diversity Analysis of Huanglongbing Pathogen-Infected Citrus, Using PhyloChip Arrays and 16S rRNA Gene Clone Library Sequencing. Applied and Environmental Microbiology. 2009;75:1566–1574. doi: 10.1128/AEM.02404-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeAngelis KM, Brodie EL, DeSantis TZ, Andersen GL, Lindow SE, et al. Selective progressive response of soil microbial community to wild oat roots. ISME J. 2009;3:168–178. doi: 10.1038/ismej.2008.103. [DOI] [PubMed] [Google Scholar]
- 42.Team RDC. Vienna: R Foundation for Statistical Computing; 2008. R: A language and environment for statistical computing. [Google Scholar]
- 43.Oksanen J, Kindt R, Legendre P, O'Hara B, Simpson GL, et al. vegan: Community Ecology Package. R package version 1.16-1 2008 [Google Scholar]
- 44.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 46.Pollard KS, Ge Y, Taylor S, Dudoit S multtest: Resampling-based multiple hypothesis testing. R package version 1.20.0 [Google Scholar]
- 47.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Webb CO, Losos JB, Agrawal AA. Integrating phylogenies into community ecology. Ecology. 2006;87:S1–S2. [Google Scholar]
- 49.Webb CO, Ackerly DD, McPeek MA, Donoghue MJ. Phylogenies and community ecology. Annual Review of Ecology and Systematics. 2002;33:475–505. [Google Scholar]
- 50.Kembel S, Ackerly D, Blomberg S, Cowan P, Helmus M, et al. picante: Tools for Integrating Phylogenies and Ecology. R package version. 2008;0.4-0 doi: 10.1093/bioinformatics/btq166. [DOI] [PubMed] [Google Scholar]
- 51.Hill MO. Diversity and evenness: a unifying notation and its consequences. Ecology. 1973;54:427–432. [Google Scholar]
- 52.Pielou EC. Measurement of Diversity in Different Types of Biological Collections. Journal of Theoretical Biology. 1966;13:131–&. [Google Scholar]
- 53.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Flanagan JL, Brodie EL, Weng L, Lynch SV, Garcia O, et al. Loss of Bacterial Diversity during Antibiotic Treatment of Intubated Patients Colonized with Pseudomonas aeruginosa. J Clin Microbiol. 2007;45:1954–1962. doi: 10.1128/JCM.02187-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edwards CA, Bohlen PJ. Biology and Ecology of Earthworms: Chapman and Hall. 1996.
- 57.Teske A, Hinrichs K-U, Edgcomb V, de Vera Gomez A, Kysela D, et al. Microbial Diversity of Hydrothermal Sediments in the Guaymas Basin: Evidence for Anaerobic Methanotrophic Communities. Appl Environ Microbiol. 2002;68:1994–2007. doi: 10.1128/AEM.68.4.1994-2007.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Staley JT, Bouzek H, Jenkins C. Eukaryotic signature proteins of Prosthecobacter dejongeii and Gemmata sp. Wa-1 as revealed by in silico analysis. FEMS Microbiol Lett. 2005;243:9–14. doi: 10.1016/j.femsle.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 59.Betsi GI, Papadavid E, Falagas ME. Probiotics for the treatment or prevention of atopic dermatitis: a review of the evidence from randomized controlled trials. Am J Clin Dermatol. 2008;9:93–103. doi: 10.2165/00128071-200809020-00002. [DOI] [PubMed] [Google Scholar]
- 60.Barbara G, Stanghellini V, Cremon C, De Giorgio R, Gargano L, et al. Probiotics and irritable bowel syndrome: rationale and clinical evidence for their use. J Clin Gastroenterol. 2008;42(Suppl 3 Pt 2):S214–217. doi: 10.1097/MCG.0b013e31817da129. [DOI] [PubMed] [Google Scholar]
- 61.Barrett JS, Canale KE, Gearry RB, Irving PM, Gibson PR. Probiotic effects on intestinal fermentation patterns in patients with irritable bowel syndrome. World J Gastroenterol. 2008;14:5020–5024. doi: 10.3748/wjg.14.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Williams E, Stimpson J, Wang D, Plummer S, Garaiova I, et al. Clinical trial: a multistrain probiotic preparation significantly reduces symptoms of irritable bowel syndrome in a double-blind placebo-controlled study. Aliment Pharmacol Ther. 2008 doi: 10.1111/j.1365-2036.2008.03848.x. [DOI] [PubMed] [Google Scholar]
- 63.Samanta M, Sarkar M, Ghosh P, Ghosh J, Sinha M, et al. Prophylactic probiotics for prevention of necrotizing enterocolitis in very low birth weight newborns. J Trop Pediatr. 2009;55:128–131. doi: 10.1093/tropej/fmn091. [DOI] [PubMed] [Google Scholar]
- 64.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 65.Wittebolle L, Marzorati M, Clement L, Balloi A, Daffonchio D, et al. Initial community evenness favours functionality under selective stress. Nature. 2009;458:623–626. doi: 10.1038/nature07840. [DOI] [PubMed] [Google Scholar]
- 66.Thomsen SF, Sluis SV, Stensballe LG, Posthuma D, Skytthe A, et al. Exploring The Association Between Severe Respiratory Syncytial Virus Infection and Asthma: a Registry-Based Twin Study. Am J Respir Crit Care Med. 2009 doi: 10.1164/rccm.200809-1471OC. [DOI] [PubMed] [Google Scholar]
- 67.Verhulst SL, Vael C, Beunckens C, Nelen V, Goossens H, et al. A longitudinal analysis on the association between antibiotic use, intestinal microflora, and wheezing during the first year of life. J Asthma. 2008;45:828–832. doi: 10.1080/02770900802339734. [DOI] [PubMed] [Google Scholar]
- 68.Lawley TD, Bouley DM, Hoy YE, Gerke C, Relman DA, et al. Host transmission of Salmonella enterica serovar Typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infect Immun. 2008;76:403–416. doi: 10.1128/IAI.01189-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sekirov I, Tam NM, Jogova M, Robertson ML, Li Y, et al. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect Immun. 2008;76:4726–4736. doi: 10.1128/IAI.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hrncir T, Stepankova R, Kozakova H, Hudcovic T, Tlaskalova-Hogenova H. Gut microbiota and lipopolysaccharide content of the diet influence development of regulatory T cells: studies in germ-free mice. BMC Immunol. 2008;9:65. doi: 10.1186/1471-2172-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, et al. Diversity of the Human Intestinal Microbial Flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Andersson AF, Lindberg M, Jakobsson H, Backhed F, Nyren Pal, et al. Comparative Analysis of Human Gut Microbiota by Barcoded Pyrosequencing. PLoS ONE. 2008;3:e2836. doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Corr SC, Hill C, Gahan CG. Chapter 1 understanding the mechanisms by which probiotics inhibit gastrointestinal pathogens. Adv Food Nutr Res. 2009;56:1–15. doi: 10.1016/S1043-4526(08)00601-3. [DOI] [PubMed] [Google Scholar]
- 74.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Total taxa detected by PhyloChip. A list of all taxa detected by the 16S rRNA PhyloChip in all samples.
(0.38 MB XLS)
Significantly correlated and T-tested taxa. A list of taxa that are significantly correlated, positively or negatively, with LGG together with those that demonstrate significant differences in abundance between high LGG (G1) and low LGG samples.
(0.25 MB XLS)