Summary
Historically, developmental stage and tissue-specific patterns of gene expression were assumed to be determined primarily by DNA regulatory sequences and their associated activators, while the general transcription machinery including core promoter recognition complexes, coactivators, and chromatin modifiers was held to be invariant. New evidence suggests that significant changes in these general transcription factors including, TFIID, BAF, and Mediator may facilitate global changes in cell-type-specific transcription.
Introduction
From the first discovery of sequence-specific DNA binding proteins to the recent findings that a small collection of transcription factors (Oct 4, Sox2, KLF3 and Myc) can reprogram fibroblasts into induced pluripotent stem cells (iPS), the seminal role of transactivators in driving specific transcriptional programs in metazoan organisms has been widely recognized (Takahashi and Yamanaka, 2006; Tjian, 1978). This large family of easily identifiable gene regulatory proteins represents between 5–10% of the coding capacity of animal and human genomes (Babu et al., 2004). Our ability to readily pick these transcription factors out of a line up of unknown genes stems from the early studies that defined their “modular” structural architecture comprised of well defined functional motifs that include DNA binding, activation, oligomerization and regulatory domains. Given that different members of this huge protein family are differentially expressed in distinct cell types that make up multi-cellular organisms, it seemed reasonable to posit that these regulatory factors would be the primary gate keepers of differential gene transcription.
Although the transactivators were the first to be isolated and characterized both biochemically and genetically - it was soon revealed that these key gene regulatory transcriptional activators must work in concert with a slew of other “basal factors” (TFIIA-H) as well as numerous co-activators to assemble RNA polymerase II (Pol II) into a pre-initiation complex (PIC) at the promoters of activated genes. Thus, over a 20 year period (1980–2000) there emerged a consensus that transcription activation would require these 3 distinct categories of transcription factors (transacting factors, co-activators, and basal factors) to work coordinately to assemble an active pre-initiation complex capable of triggering RNA synthesis by Pol II. (Fig 1).
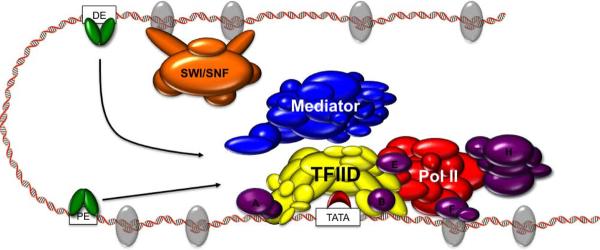
Figure 1. Model of Preinitiation Complex Assembly.
Sequence-specific activators (green) bind proximal (PE) and distal (DE) enhancer elements within the regulatory DNA of a gene and recruit TFIID (yellow) to the core promoter. These activators and/or TFIID in turn recruit chromatin remodeling complexes such as SWI/SNF (orange), coactivators including Mediator (MED, blue), and TFIIA and TFIIB (purple). The TFIID/TFIIA/TFIIB heterotrimer sequentially recruits TFIIE, TFIIF, PolII (red), and TFIIH (purple), allowing for promoter escape and productive transcriptional elongation.
Implicit in this view of how transcriptional regulation works is the notion that the “basal” or “core” transcriptional apparatus would be nearly universal in its role as a highly conserved “engine” that drives transcription while the sequence-specific transacting factors (activators and repressors) would serve as the master switches that would turn transcription of specific genes on or off in a cell and gene specific manner. This well defined division of labor between “activators” and the “core machinery” seemed eminently reasonable – especially since the components of the “core machinery” were so highly conserved from yeast to man. In contrast, sequence-specific transcription factors varied considerably from organism to organism and their expression reflected, indeed even defined, cell-to-cell differences. Thus, it did not seem necessary to invoke a potential role of the core machinery in regulating gene or cell-specific transcription.
Several nagging findings began to emerge in the 1980's that hinted perhaps this tidy model with well-defined functions for the 3 transcription factor families may not be entirely correct. The first clue that this picture was over simplified came from the finding that the core promoter recognition factor TATA binding protein (TBP) was a component of a large (1md) multi-subunit complex (≈15 subunits) named TFIID; the other subunits of the complex were named TBP associated factors (TAFs). Surprisingly, in addition to TBP, at least 2 different TAFs bind core promoter DNA with sequence-specificity. Along with this early conundrum was the later finding that core promoters in metazoans were composed of multiple composite cis-elements (TATA, INR, DPE, MTE, etc.) each recognized by distinct components of the TBP/TAF complex as well as other basal factors (Verrijzer et al., 1995). Most importantly, different genes bore very different combinations of core promoter elements suggesting that indeed, there may be many different flavors of core promoters – a rather different picture from the simple TATA vs. TATA-less world we had come to accept. Furthermore, different classes of genes and different core promoter architectures were found to recruit unique combinations of TBP and TAFs, suggesting that within a given cell TBP, TFIID, and other TAF containing complexes may regulate unique subsets of genes (Huisinga and Pugh, 2004; Kuras et al., 2000). Next came the finding of “cell type specific” subunits of the core promoter recognition complex TFIID – another clue that perhaps the so called basal apparatus that forms the heart of the PIC may exist in different cell-type-specific forms. Thus the discovery of an ovarian specific TAF4b in humans was followed by the identification of several other tissue specific TFIID TAF subunits in flies, mice and humans that compounded the mystery of core promoter recognition functions.
The characterization of tissue-specific TAFs was closely followed by the discovery of various TBP-related factors (TRF1, 2, 3, etc.) some of which also revealed potential gene and cell-specific functions. By 2006, the idea that the core promoter recognition complex in metazoans may not be monolithic, invariant and universal for all promoters in an organism had gained some traction. However, the prevailing models nevertheless still largely assumed that the primary driver of cell specificity would lie squarely on the shoulders of the sequence-specific transcriptional activators and repressors. At the same time, the core promoter recognition machinery, although endowed with some degree of combinatorial specificity due to subunit exchanges, remained relegated to a supporting role in driving developmental cell fate and cellular differentiation. This notion was certainly bolstered by the advent of iPS cells induced by a tetrad of sequence-specific, cell-selective DNA binding transcription factors. This generally accepted enhancer/promoter centric view of transcriptional regulation suffered a refreshing reality check in 2007 when it was discovered that during skeletal muscle differentiation, not only are tissue-specific TAFs called into play but surprisingly, there was a concomitant near whole sale elimination of the prototypic, core promoter recognition complex TFIID including the loss of the iconic TBP subunit (Deato and Tjian, 2007). Remarkably, as cycling myoblasts are induced to differentiate into multi-nucleated myotubes – the so called universal core complex, TFIID – is dispatched both at the protein and mRNA levels leaving active transcription in muscle fibers to be directed by what appears to be a much pared down TRF3/TAF3 complex. Concomitant with this loss of TBP and TFIID during muscle differentiation was an equally unexpected loss of the CRSP/Med complex. Thus, at least in skeletal muscle formation, there appeared to be a rather dramatic re-casting of the core promoter recognition apparatus that apparently must accompany the well-documented induction of key myogenic transcription factors (MyoD, MyoG, etc).
Tissue-Specific TAFs
While a variety of TAF paralogues have been identified by bioinformatic analysis, the first concrete evidence of subunit diversity within TFIID came with the discovery of a substoichiometric TAF4 paralog in cultured B cells (Dikstein et al., 1996). Subsequently, TAF4b was found to be highly enriched in mouse testis and ovary and to be an intrinsic component of an altered ovary-specific TFIID (Freiman et al., 2001). TAF4b knockout females while viable are infertile as a result of deficient folliculogenesis, and knockout males show age dependent infertility resulting from a defect in spermatogenic maintenance (Falender et al., 2005). Subsequently, gene expression analysis revealed a requirement for TAF4b in regulating a subset of ovarian granulosa cell-specific genes and genes required for spermatogonial stem cell maintenance (Geles et al., 2006). Furthermore, TAF4b containing TFIID does not necessarily require unique tissue-specific activators, but rather induces the expression of and cooperates with ubiquitous general activators such as c-Jun to regulate cell-type-specific gene expression (Geles et al., 2006). More detailed developmental analysis confirmed that TAF4b is the primary transcriptional integrator of extracellular signals that specify the developmental and proliferative program of granulosa cells (Voronina et al., 2007). Although not explicitly required in B cells, TAF4b cooperates with known regulators of B cell specific transcription such as OCA-B and NF-κB, and may serve a redundant function to TAF4 in this context (Freiman et al., 2002; Wolstein et al., 2000; Yamit-Hezi and Dikstein, 1998). Promoter-specific differences in both basal and activated transcription by canonical and TAF4b containing TFIID were recapitulated in vitro, and these differences correlated with TAF4b target genes identified in vivo (Liu et al., 2008). The molecular mechanism of TAF4b-specific transcription has been further elucidated by electron microscopy and 3D particle reconstruction studies, which reveal distinct structural differences between canonical and TAF4b containing TFIID that likely explain their promoter-selective activities. Intriguingly, a TAF4b homologue is enriched in Xenopus ovaries but absent from lower metazoans, suggesting that unlike canonical TAFs, tissue-specific paralogs may have evolved to direct species-specific transcriptional programs (Xiao et al., 2006) The importance of tissue-specific TAFs was further validated by the identification of five testis restricted (tTAF) subunits in Drosophila, no hitter (paralog of TAF4), cannonball (paralog of TAF5), meiosis I arrest (paralog of TAF6), spermatocyte arrest (paralog of TAF8), and ryan express (paralog of TAF12) (Lin et al., 1996). Disrupting each of these proteins alters expression of testis specific genes, and together they form a stable complex, which is required for meiotic cell cycle progression and normal spermatid differentiation (Hiller et al., 2004; Hiller et al., 2001). Intriguingly, the tTAF complex is thought to mediate spermatid gene activation in part by displacing the repressive Polycomb complex and promoting its sequestration to the nucleolus (Chen et al., 2005); hence, a derepressive tTAF strategy may represent a novel cell-type-specific mechanism of TAF dependent coactivation. While tTAFs are restricted to Drosophila, a testis-linked paralog of TAF7 called TAF7l appears unique to vertebrates. High level expression of mouse TAF7l in late spermatocytes and haploid spermatids is coincident with a reduction in TAF7 expression, and as spermatogenesis proceeds TAF7l it relocalized to the nucleus where it associates with TBP and other TAFs to form an alternative TFIID-like complex (Pointud et al., 2003). Viable TAF7l knockout mice display altered patterns of spermatocyte gene expression and defects in spermiogenesis which lead to reduced fertility, consistent with a testis function (Cheng et al., 2007). Additional TAF paralogs, including TAF9b, while shown to associate with TFIID and promote alternative gene expression programs are just beginning to be characterized, and the full repertoire of tissue-specific TAFs remains to be discovered (Frontini et al., 2005).
TRF1/TRF2/TRF3
The extent of core promoter recognition complex diversity was expanded by the discovery of TBP-related factors or TRFs. The first TRF identified (TRF1), exhibits significant homology to TBP in its DNA binding domain and interacts with TFIIA, TFIIB and TATA box DNA; however considerable divergence exists throughout the rest of the protein which likely accounts for its regulatory diversity (Hansen et al., 1997). Intriguingly, TRF1 is unique to Drosophila where it is enriched in the nervous system and gonads and thus seems to have evolved to suit a specific purpose in these insect tissues. Polytene chromosome staining and genome wide promoter occupancy experiments suggest that TRF1 regulates a highly restricted set of genes relative to TBP, and that these loci are primarily associated with Pol III transcription (Isogai et al., 2007b). However, TRF1 has been isolated in association with both BRF and a larger complex containing novel proteins and been shown to direct both Pol II and Pol III transcription in vitro and in vivo much like TBP (Holmes and Tjian, 2000; Takada et al., 2000). Hence, TRF1 may represent an evolutionarily restricted alternative core promoter recognition complex that directs promoter-selective transcription by multiple polymerases in a subset of tissues.
TBP related factor 2 (TRF2, also called TLF, TLP, or TRP) though widely distributed from C. elegans to humans shows considerable diversity in its N and C-terminal domains both between species and when compared to other TBPs (Dantonel et al., 1999; Rabenstein et al., 1999). While TRF2 binds TFIIA and TFIIB and displays high conservation of the TBP DNA binding domain, it does not bind TATA box containing DNA and fails to complement TFIID in basal or activated in vitro transcription assays (Moore et al., 1999; Teichmann et al., 1999). Biochemically, TRF2 associates with the DRE-binding factor (DREF) and components of the NURF chromatin remodeling complex, and has been shown to selectively regulate the transcription of DRE-containing promoters of cell cycle and proliferation genes in cooperation with DREF and NURF (Hochheimer et al., 2002). Intriguingly, several genes have been shown to contain tandem transcriptional start sites whose promoters are specifically and differentially regulated by TRF2 and TFIID. Disruption experiments in C. elegans, Drosophila, and Xenopus demonstrated a requirement for TRF2 in early embryonic development, but also confirmed that TRF2 regulates a subset of developmental genes responsible for these phenotypes while being dispensable for the expression of others (Dantonel et al., 2000; Kaltenbach et al., 2000; Kopytova et al., 2006; Veenstra et al., 2000). Conversely, mouse TRF2 is highly enriched in the testis, and while knockout animals display spermiogenesis defects and altered patterns of testis-specific gene expression, they show no developmental defects, consistent with an evolutionarily divergent function in mammals (Martianov et al., 2001; Zhang et al., 2001). Recent genome wide localization of Drosophila TRF2 showed that it occupies over a thousand promoters most of which are not bound by TBP and are enriched for DREs but lack a canonical TATA box (Isogai et al., 2007a). Intriguingly, the Drosophila histone H1 promoter is specifically regulated by TRF2 while the remainder of the histone gene cluster is TFIID dependent, suggesting that differences in core promoter recognition complex activity may alter histone ratios. Hence, TRF2 exhibits evolutionarily diverse roles, and clearly extends the diversity of core promoter recognition complexes in its regulation of both tissue-specific and promoter-selective gene expression patterns. Additionally, the existence of separate TRF2 and TFIID dependent promoters within related gene clusters or in some cases single genes may allow a limited set of promoters and core promoter recognition complexes to produce dramatically varied expression patterns in response to signaling events or between tissues. This alternative regulation likely results from the association of TRF2 with novel transcription factors through its divergent N and C-terminal protein interaction domains and its recruitment to novel core promoter elements not occupied by TBP.
The most recently identified and perhaps most intriguing member of the TBP-related factor family is the vertebrate-specific TRF3 (also called TBP2). Initially identified based on homology, mammalian TRF3 shows near identity to TBP in its C-terminal region, which includes the TATA box, TFIIA and TFIIB binding domains, and considerable divergence from all known TBPs in its N-terminal region (Persengiev et al., 2003). Preliminary characterization showed TRF3 to be widely expressed in adult mammalian tissues though at varying levels relative to TBP, and to be primarily contained in a larger 150–200kDa complex. Many studies of TRF3 function have focused on oogenesis and early embryonic development. Murine TRF3 expression has been shown to increase rapidly during oocyte growth from primordial follicle formation until preovulation at the same time that TBP expression is effectively eliminated (Gazdag et al., 2007; Yang et al., 2006). However, these observational reports reach conflicting conclusions on the role of TRF3 in murine embryogenesis as one shows a steady increase in both TRF3 and TBP expression following fertilization while the other concludes that TRF3 remains silent as TBP expression is induced post fertilization; this conflict in immunofluorescence data will likely be resolved by more extensive studies and the use of additional approaches.
The role of TRF3 in early development has been most extensively investigated in Xenopus and zebrafish. Xenopus and zebrafish TRF3 bind TFIIA, TFIIB and TATA containing DNA, and Xenopus TRF3 supports TATA dependent transcription in egg extracts (Bartfai et al., 2004; Jallow et al., 2004). While highly expressed in the Xenopus oocyte, TRF3 is expressed at lower levels in the developing embryo and is recruited to a subset of genes not occupied by TBP in a time dependent manner. Importantly, TRF3 depletion causes defects in Xenopus gastrulation and blastopore closure and alters expression of many oocyte and embryo-specific genes, a subset of which also require TBP. Intriguingly, Xenopus TRF3 overexpression can partially rescue TBP knockdown phenotypes and gene expression patterns, consistent with a partial redundancy. Further genome wide analysis found that Xenopus TBP primarily regulates the synthesis of maternally deposited transcripts while a majority of embryonically transcribed genes require TRF2 or TRF3 (Jacobi et al., 2007). Additionally, TBP dependent Xenopus genes are widely conserved from yeast to humans while TRF3-specific genes are generally restricted to vertebrates. Xenopus knockdown and ChIP experiments demonstrated that many promoters while exclusively dependent on either TRF3 or TFIID are occupied by both, suggesting that differences in complex-specific transcription may occur at the level of activation rather than recruitment. Expression of zebrafish TRF3 increases steadily from fertilization to gastrulation after which it declines rapidly but remains detectable in all adult tissues and enriched in the ovary. As with Xenopus, knockdown of TRF3 in zebrafish inhibits normal gastrulation and results in mesodermal patterning defects. Intriguingly, both species show an enrichment of TRF3 in the ventral embryo consistent with a role in dorsoventral patterning. Additional knockdown studies in later stage zebrafish embryos demonstrate a specific requirement for TRF3 in the initiation of hematopoiesis by upregulation of the mespa gene, and partially define a lineage-specific transcriptional cascade in which TRF3 is the master regulator of this particular differentiation program (Hart et al., 2007). These findings were recently expanded to show that zebrafish TAF3 specifically interacts with the C-terminus of TRF3 but not TBP to form a TRF3/TAF3 complex which closely resembles that found in myotubes (Hart et al., 2009). Much like TRF3, zebrafish TAF3 is required for the induction of hematopoiesis and enriched at the promoter of mespa, an important early hematopoietic regulator. Thus, multiple studies in lower vertebrates revealed a requirement for TRF3 in early development and elucidated both redundant and overlapping functions with TBP in embryonic transcription. However, it is intriguing to note that the striking embryonic phenotypes of TRF2 depletion in Xenopus and zebrafish may not be conserved in the knockout mice and that the important embryonic functions of TRF3 in lower vertebrates likewise may not be fully preserved in mammals.
One of the first studies of murine TRF3 investigated the differentiation of C2C12 cells and found that while TBP along with several TAFs are degraded upon differentiation, at least some TRF3 in myotubes is retained and is found in a complex with TAF3 (Deato and Tjian, 2007). Depletion of TRF3 and TAF3 by RNAi suggested that this putative core promoter recognition complex may be required for myogenesis. Consistent with a requisite role in myotube differentiation, ChIP analysis found that TRF3/TAF3 occupies a key muscle-specific gene promoter. Further biochemical analysis of the complex suggested that TRF3/TAF3 is sufficient for modest activation of the Myogenin promoter in vitro by the myogenic transcription factors MyoD and E47 (Deato et al., 2008). Protein-protein interaction studies, along with additional in vitro transcription assays, revealed that MyoD can directly target TAF3, and that this interaction is necessary for in vitro activity. The recent report of a TRF3 knockout mouse extended its central role in oocyte maturation to mammals, as homozygous females display complete sterility due to oocyte arrest at the primordial follicle stage and eventual ovarian failure (Gazdag et al., 2009). Furthermore, TRF3 knockout oocytes show a reduction of PolII activity and the enrichment of chromatin marks normally associated with transcriptional activation, as well as altered patterns of oocyte-specific gene expression. Consistent with this, TRF3 is enriched at oocyte specific promoters in wild type animals. In contrast to the Xenopus and zebrafish knockouts, mice lacking TRF3 display no increase in embryonic lethality or apparent defects in early embryonic development, suggesting that like TRF2, TRF3 may have an evolutionarily unique function in mammals. It was also noted that these TRF3 KO mice did not display any obvious muscle developmental defect that might have been anticipated given its reported role in myogenesis of C2C12 in culture. However, previous KO studies of other key myogenic regulators including MyoD also failed to display any muscle phenotype until their functional role in myogenesis in vivo was revealed by wounding and tissue repair defects (Megeney et al., 1996). It is also possible that the apparent role of TRF3 in muscle gene regulation is masked by functionally overlapping factor(s) yet to be identified that can work in conjunction with TAF3 to help direct transcriptional programs in muscle that have not been revealed in the C2C12 studies. An example of such a compensatory rescue was seen in the case of the TAF4a KO which can largely be rescued by TAF4b (Mengus et al., 2005). Although both TRF2 and TRF3 predominantly show fertility defects (ie. in tissues that also express the highest levels of these 2 factors) an extensive range of expression studies indicate that both of the TBP-related factors are in fact widely expressed in varying levels in many mammalian cell types. We suspect that the potentially diversified functional roles of mammalian TRFs will remain unclear until more exhaustive studies in multiple cell types have been carried out. For example, the role of TRF3 in hematopoiesis was recently extended to mammals using an established mouse cell culture model. These authors also find that mouse TRF3 specifically binds TAF3, and demonstrate that this complex is enriched at the promoter of the mouse mespa homologue, Mesp1. During progenitor differentiation TRF3 is expressed at the onset of hematopoietic induction, and its siRNA mediated depletion blocks the expression of multiple hematopoietic markers including Mesp1 (Hart et al., 2009). It will be interesting for future studies to determine how widely TRF3 may be used either in association with TAF3 or as a component of as yet unidentified complexes. The association of TRF3 with vertebrate specific differentiation programs and its absence in lower organisms suggest that it may have specifically evolved to direct the greater complexity of cell-types and functions required by higher metazoans.
General Co-activator Complexes
Chromatin remodeling complexes of the Swi/Snf family have also been shown to exhibit considerable subunit diversity; however the significance of specific complex subunit composition to developmental and cell-type-specific transcriptional programs remains unclear (Clapier and Cairns, 2009; Wang et al., 1996). The finding that specific subunits of the mammalian PBAF complex are enriched in cardiac progenitors and required for normal heart maturation provided the earliest support for the existence of cell-type-specific chromatin remodelers (Wang et al., 2004; Yan et al., 2005). Conversely, knockout studies of unique subunits of the related BAF-A and BAF-B complexes demonstrated overlapping deficiencies in ES cell pluripotency and lineage specific differentiation, while disruption of PBAF showed no specific ES cell phenotype, consistent with distinct developmental requirements for each complex (Gao et al., 2008; Huang et al., 2008; Yan et al., 2008). The first evidence of a requisite developmental stage specific switch in subunit composition came with the discovery that homologous BAF45 and BAF53 subunits must be exchanged during the transition from neuronal progenitors to postmitotic neurons (Lessard et al., 2007; Wu et al., 2007). Moreover, this exchange is now known to depend on microRNA mediated repression of the progenitor-specific subunits, adding another level of regulatory complexity to the neuronal transition (Yoo et al., 2009). Most recently, a novel esBAF complex was isolated from embryonic stem cells which has a unique subunit composition, is required for pluripotency and self-renewal, and is downregulated upon differentiation (Ho et al., 2009b). Further genome wide localization experiments demonstrated the existence of a cooperative transcriptional network between esBAF and known ES cell specific transcription factors which define self-renewal and pluripotency (Ho et al., 2009a).
Recent evidence has also hinted that the composition of the Mediator co-activator complex, thought to be universally required for transcription activation, may similarly be subject to regulation during differentiation. Careful fractionation of HeLa nuclear extracts yielded an altered Mediator complex, called CRSP/Med2, lacking two key subunits previously shown to be critical for nuclear hormone receptor (NHR)-dependent transcription (Taatjes and Tjian, 2004). This CRSP/Med2 complex while unable to support transcription directed by NHRs is sufficient to co-activate transcription by activators utilizing other Mediator subunits. Thus, combinatorial control of subunit composition could allow for cell-type and promoter-specific transcription dependent on distinct classes of activators. However, it remains to be seen whether these complexes are utilized in a cell-type-specific way. Strikingly, a recent study noted that, similar to TFIID, many subunits of the Mediator complex are degraded upon differentiation of the skeletal muscle cell culture model C2C12 cells to myotubes (Deato et al., 2008). These data support the notion that sub-complexes of Mediator, such as CRSP2/Med2, may be used in a cell-type-specific manner by degrading subunits to turn off large portions of a gene expression profile while retaining subunits necessary for the expression of a smaller, highly specific subset of genes. A more comprehensive analysis of the presence or absence of Mediator subunits in additional differentiation systems should provide clarification for the potential role of altered Mediator complexes in cell-type-specific transcription, much as it has in the case of altered TFIID.
Concluding remarks
A bevy of new data, some of which have been highlighted in this review, have begun to shake the premise that one-size-fits-all when dealing with the core transcription machinery. One of the most striking discoveries is the dramatic down-regulation and near complete destruction of the TFIID complex upon skeletal muscle formation from myoblast precursors. This unexpected finding opens the possibility that other tissues may similarly part with TFIID in favor of a more streamlined complex (Figure 2). It is tempting to speculate that distinct TAFs may be selectively retained in other specialized cell types in order to drive distinct sets of gene expression programs, reminiscent, though perhaps a more drastic version of the multiple cell-specific flavors of the BAF chromatin remodeling complexes.
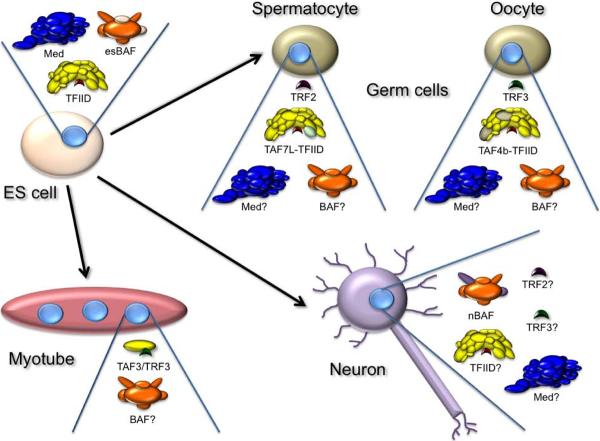
Figure 2. Model of Cell-Type-Specific Changes in General Transcription Factors.
ES cells and proliferative progenitors are believed to contain canonical TFIID consisting of TBP (red) and 12–15 associated TAFs (yellow), Mediator (blue), and ES BAF consisting of both common (orange) and ES cell-specific subunits (tan). Myotubes may replace TFIID with a novel complex of TRF (green) and TAF3 (yellow), and also downregulate Mediator and presumably retain a general BAF complex (orange). Neurons are known to contain a novel BAF complex with conserved (orange) and neuron specific subunits (purple). The disposition of Mediator and TFIID in most neuronal cell types is not known. Spermatocytes have elevated levels of TRF2 (purple) and of the TAF7 paralog TAF7l (lime) which may or may not be a component of an altered TFIID. Ovarian cells have elevated levels of TRF3 (green) and of an altered TFIID containing one or more subunits of the TAF4 paralog, TAF4b (tan). The subunit composition of Mediator and BAF complexes in germ cells is currently unknown.
One outstanding issue regarding the dramatic degradation of TBP observed in muscle fibers and also in multiple cell lines is the fate of tRNA and rRNA transcription. All three vertebrate RNA polymerases are thought to utilize TBP as part of a promoter recognition complex. Is it possible that various TBP-related factors, such as TRF2 or TRF3 like TRF1 in Drosophila, are able to substitute for TBP in Pol I and Pol III transcription? Presently there is no evidence either for or against this model in verebrates. Clearly more work is necessary to resolve this issue.
Likewise the identification of an ES cell-specific chromatin remodeling complex raises the interesting prospect that other co-activators may also have ES cell-specific counterparts. If this is indeed the case, the isolation and characterization of such ES cell-specific factors would have far reaching implications in our understanding of both ES cell pluripotency as well as the molecular pathway leading to the generation of iPS cells. Since the composition of the Mediator complex also appears to be regulated during myogenesis, it is conceivable that it too may have cell-type-specific versions in ES and differentiated cells. The presence of putative stem cell specific co-activators could provide one attractive explanation for at least part of the inefficiencies inherent in generating iPS cells from differentiated cells, since expression of ES cell-specific components would require reactivation of multiple transcriptional components not found in terminally differentiated cell types
It therefore seems likely that, the data highlighted in this review represents a potentially exciting new frontier in the study of cell-specific transcription, a field still in its relative infancy despite over two decades of study, and argues for continued effort to unravel the many remaining questions.
Acknowledgments
We wish to thank Gerald R. Crabtree and Jim Goodrich for critical reading of and helpful feedback on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Babu MM, Luscombe NM, Aravind L, Gerstein M, Teichmann SA. Structure and evolution of transcriptional regulatory networks. Curr Opin Struct Biol. 2004;14:283–291. doi: 10.1016/j.sbi.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Bartfai R, Balduf C, Hilton T, Rathmann Y, Hadzhiev Y, Tora L, Orban L, Muller F. TBP2, a vertebrate-specific member of the TBP family, is required in embryonic development of zebrafish. Curr Biol. 2004;14:593–598. doi: 10.1016/j.cub.2004.03.034. [DOI] [PubMed] [Google Scholar]
- Chen X, Hiller M, Sancak Y, Fuller MT. Tissue-specific TAFs counteract Polycomb to turn on terminal differentiation. Science. 2005;310:869–872. doi: 10.1126/science.1118101. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Buffone MG, Kouadio M, Goodheart M, Page DC, Gerton GL, Davidson I, Wang PJ. Abnormal sperm in mice lacking the Taf7l gene. Mol Cell Biol. 2007;27:2582–2589. doi: 10.1128/MCB.01722-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Dantonel JC, Quintin S, Lakatos L, Labouesse M, Tora L. TBP-like factor is required for embryonic RNA polymerase II transcription in C. elegans. Mol Cell. 2000;6:715–722. doi: 10.1016/s1097-2765(00)00069-1. [DOI] [PubMed] [Google Scholar]
- Dantonel JC, Wurtz JM, Poch O, Moras D, Tora L. The TBP-like factor: an alternative transcription factor in metazoa? Trends Biochem Sci. 1999;24:335–339. doi: 10.1016/s0968-0004(99)01436-x. [DOI] [PubMed] [Google Scholar]
- Deato MD, Marr MT, Sottero T, Inouye C, Hu P, Tjian R. MyoD targets TAF3/TRF3 to activate myogenin transcription. Mol Cell. 2008;32:96–105. doi: 10.1016/j.molcel.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deato MD, Tjian R. Switching of the core transcription machinery during myogenesis. Genes Dev. 2007;21:2137–2149. doi: 10.1101/gad.1583407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikstein R, Zhou S, Tjian R. Human TAFII 105 is a cell type-specific TFIID subunit related to hTAFII130. Cell. 1996;87:137–146. doi: 10.1016/s0092-8674(00)81330-6. [DOI] [PubMed] [Google Scholar]
- Falender AE, Freiman RN, Geles KG, Lo KC, Hwang K, Lamb DJ, Morris PL, Tjian R, Richards JS. Maintenance of spermatogenesis requires TAF4b, a gonad-specific subunit of TFIID. Genes Dev. 2005;19:794–803. doi: 10.1101/gad.1290105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freiman RN, Albright SR, Chu LE, Zheng S, Liang HE, Sha WC, Tjian R. Redundant role of tissue-selective TAF(II)105 in B lymphocytes. Mol Cell Biol. 2002;22:6564–6572. doi: 10.1128/MCB.22.18.6564-6572.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freiman RN, Albright SR, Zheng S, Sha WC, Hammer RE, Tjian R. Requirement of tissue-selective TBP-associated factor TAFII105 in ovarian development. Science. 2001;293:2084–2087. doi: 10.1126/science.1061935. [DOI] [PubMed] [Google Scholar]
- Frontini M, Soutoglou E, Argentini M, Bole-Feysot C, Jost B, Scheer E, Tora L. TAF9b (formerly TAF9L) is a bona fide TAF that has unique and overlapping roles with TAF9. Mol Cell Biol. 2005;25:4638–4649. doi: 10.1128/MCB.25.11.4638-4649.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Tate P, Hu P, Tjian R, Skarnes WC, Wang Z. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc Natl Acad Sci U S A. 2008;105:6656–6661. doi: 10.1073/pnas.0801802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdag E, Rajkovic A, Torres-Padilla ME, Tora L. Analysis of TATA-binding protein 2 (TBP2) and TBP expression suggests different roles for the two proteins in regulation of gene expression during oogenesis and early mouse development. Reproduction. 2007;134:51–62. doi: 10.1530/REP-06-0337. [DOI] [PubMed] [Google Scholar]
- Gazdag E, Santenard A, Ziegler-Birling C, Altobelli G, Poch O, Tora L, Torres-Padilla ME. TBP2 is essential for germ cell development by regulating transcription and chromatin condensation in the oocyte. Genes Dev. 2009;23:2210–2223. doi: 10.1101/gad.535209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geles KG, Freiman RN, Liu WL, Zheng S, Voronina E, Tjian R. Cell-type-selective induction of c-jun by TAF4b directs ovarian-specific transcription networks. Proc Natl Acad Sci U S A. 2006;103:2594–2599. doi: 10.1073/pnas.0510764103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen SK, Takada S, Jacobson RH, Lis JT, Tjian R. Transcription properties of a cell type-specific TATA-binding protein, TRF. Cell. 1997;91:71–83. doi: 10.1016/s0092-8674(01)80010-6. [DOI] [PubMed] [Google Scholar]
- Hart DO, Raha T, Lawson ND, Green MR. Initiation of zebrafish haematopoiesis by the TATA-box-binding protein-related factor Trf3. Nature. 2007;450:1082–1085. doi: 10.1038/nature06349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart DO, Santra MK, Raha T, Green MR. Selective interaction between Trf3 and Taf3 required for early development and hematopoiesis. Dev Dyn. 2009;238:2540–2549. doi: 10.1002/dvdy.22083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M, Chen X, Pringle MJ, Suchorolski M, Sancak Y, Viswanathan S, Bolival B, Lin TY, Marino S, Fuller MT. Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development. 2004;131:5297–5308. doi: 10.1242/dev.01314. [DOI] [PubMed] [Google Scholar]
- Hiller MA, Lin TY, Wood C, Fuller MT. Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev. 2001;15:1021–1030. doi: 10.1101/gad.869101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci U S A. 2009a;106:5187–5191. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A. 2009b;106:5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochheimer A, Zhou S, Zheng S, Holmes MC, Tjian R. TRF2 associates with DREF and directs promoter-selective gene expression in Drosophila. Nature. 2002;420:439–445. doi: 10.1038/nature01167. [DOI] [PubMed] [Google Scholar]
- Holmes MC, Tjian R. Promoter-selective properties of the TBP-related factor TRF1. Science. 2000;288:867–870. doi: 10.1126/science.288.5467.867. [DOI] [PubMed] [Google Scholar]
- Huang X, Gao X, Diaz-Trelles R, Ruiz-Lozano P, Wang Z. Coronary development is regulated by ATP-dependent SWI/SNF chromatin remodeling component BAF180. Dev Biol. 2008;319:258–266. doi: 10.1016/j.ydbio.2008.04.020. [DOI] [PubMed] [Google Scholar]
- Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- Isogai Y, Keles S, Prestel M, Hochheimer A, Tjian R. Transcription of histone gene cluster by differential core-promoter factors. Genes Dev. 2007a;21:2936–2949. doi: 10.1101/gad.1608807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai Y, Takada S, Tjian R, Keles S. Novel TRF1/BRF target genes revealed by genome-wide analysis of Drosophila Pol III transcription. EMBO J. 2007b;26:79–89. doi: 10.1038/sj.emboj.7601448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobi UG, Akkers RC, Pierson ES, Weeks DL, Dagle JM, Veenstra GJ. TBP paralogs accommodate metazoan- and vertebrate-specific developmental gene regulation. EMBO J. 2007;26:3900–3909. doi: 10.1038/sj.emboj.7601822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallow Z, Jacobi UG, Weeks DL, Dawid IB, Veenstra GJ. Specialized and redundant roles of TBP and a vertebrate-specific TBP paralog in embryonic gene regulation in Xenopus. Proc Natl Acad Sci U S A. 2004;101:13525–13530. doi: 10.1073/pnas.0405536101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenbach L, Horner MA, Rothman JH, Mango SE. The TBP-like factor CeTLF is required to activate RNA polymerase II transcription during C. elegans embryogenesis. Mol Cell. 2000;6:705–713. doi: 10.1016/s1097-2765(00)00068-x. [DOI] [PubMed] [Google Scholar]
- Kopytova DV, Krasnov AN, Kopantceva MR, Nabirochkina EN, Nikolenko JV, Maksimenko O, Kurshakova MM, Lebedeva LA, Yerokhin MM, Simonova OB, et al. Two isoforms of Drosophila TRF2 are involved in embryonic development, premeiotic chromatin condensation, and proper differentiation of germ cells of both sexes. Mol Cell Biol. 2006;26:7492–7505. doi: 10.1128/MCB.00349-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras L, Kosa P, Mencia M, Struhl K. TAF-Containing and TAF-independent forms of transcriptionally active TBP in vivo. Science. 2000;288:1244–1248. doi: 10.1126/science.288.5469.1244. [DOI] [PubMed] [Google Scholar]
- Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TY, Viswanathan S, Wood C, Wilson PG, Wolf N, Fuller MT. Coordinate developmental control of the meiotic cell cycle and spermatid differentiation in Drosophila males. Development. 1996;122:1331–1341. doi: 10.1242/dev.122.4.1331. [DOI] [PubMed] [Google Scholar]
- Liu WL, Coleman RA, Grob P, King DS, Florens L, Washburn MP, Geles KG, Yang JL, Ramey V, Nogales E, Tjian R. Structural changes in TAF4b-TFIID correlate with promoter selectivity. Mol Cell. 2008;29:81–91. doi: 10.1016/j.molcel.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianov I, Fimia GM, Dierich A, Parvinen M, Sassone-Corsi P, Davidson I. Late arrest of spermiogenesis and germ cell apoptosis in mice lacking the TBP-like TLF/TRF2 gene. Mol Cell. 2001;7:509–515. doi: 10.1016/s1097-2765(01)00198-8. [DOI] [PubMed] [Google Scholar]
- Megeney LA, Kablar B, Garrett K, Anderson JE, Rudnicki MA. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 1996;10:1173–1183. doi: 10.1101/gad.10.10.1173. [DOI] [PubMed] [Google Scholar]
- Mengus G, Fadloun A, Kobi D, Thibault C, Perletti L, Michel I, Davidson I. TAF4 inactivation in embryonic fibroblasts activates TGF beta signalling and autocrine growth. EMBO J. 2005;24:2753–2767. doi: 10.1038/sj.emboj.7600748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PA, Ozer J, Salunek M, Jan G, Zerby D, Campbell S, Lieberman PM. A human TATA binding protein-related protein with altered DNA binding specificity inhibits transcription from multiple promoters and activators. Mol Cell Biol. 1999;19:7610–7620. doi: 10.1128/mcb.19.11.7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persengiev SP, Zhu X, Dixit BL, Maston GA, Kittler EL, Green MR. TRF3, a TATA-box-binding protein-related factor, is vertebrate-specific and widely expressed. Proc Natl Acad Sci U S A. 2003;100:14887–14891. doi: 10.1073/pnas.2036440100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pointud JC, Mengus G, Brancorsini S, Monaco L, Parvinen M, Sassone-Corsi P, Davidson I. The intracellular localisation of TAF7L, a paralogue of transcription factor TFIID subunit TAF7, is developmentally regulated during male germ-cell differentiation. J Cell Sci. 2003;116:1847–1858. doi: 10.1242/jcs.00391. [DOI] [PubMed] [Google Scholar]
- Rabenstein MD, Zhou S, Lis JT, Tjian R. TATA box-binding protein (TBP)-related factor 2 (TRF2), a third member of the TBP family. Proc Natl Acad Sci U S A. 1999;96:4791–4796. doi: 10.1073/pnas.96.9.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taatjes DJ, Tjian R. Structure and function of CRSP/Med2; a promoter-selective transcriptional coactivator complex. Mol Cell. 2004;14:675–683. doi: 10.1016/j.molcel.2004.05.014. [DOI] [PubMed] [Google Scholar]
- Takada S, Lis JT, Zhou S, Tjian R. A TRF1:BRF complex directs Drosophila RNA polymerase III transcription. Cell. 2000;101:459–469. doi: 10.1016/s0092-8674(00)80857-0. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Teichmann M, Wang Z, Martinez E, Tjernberg A, Zhang D, Vollmer F, Chait BT, Roeder RG. Human TATA-binding protein-related factor-2 (hTRF2) stably associates with hTFIIA in HeLa cells. Proc Natl Acad Sci U S A. 1999;96:13720–13725. doi: 10.1073/pnas.96.24.13720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjian R. The binding site on SV40 DNA for a T antigen-related protein. Cell. 1978;13:165–179. doi: 10.1016/0092-8674(78)90147-2. [DOI] [PubMed] [Google Scholar]
- Veenstra GJ, Weeks DL, Wolffe AP. Distinct roles for TBP and TBP-like factor in early embryonic gene transcription in Xenopus. Science. 2000;290:2312–2315. doi: 10.1126/science.290.5500.2312. [DOI] [PubMed] [Google Scholar]
- Verrijzer CP, Chen JL, Yokomori K, Tjian R. Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell. 1995;81:1115–1125. doi: 10.1016/s0092-8674(05)80016-9. [DOI] [PubMed] [Google Scholar]
- Voronina E, Lovasco LA, Gyuris A, Baumgartner RA, Parlow AF, Freiman RN. Ovarian granulosa cell survival and proliferation requires the gonad-selective TFIID subunit TAF4b. Dev Biol. 2007;303:715–726. doi: 10.1016/j.ydbio.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhai W, Richardson JA, Olson EN, Meneses JJ, Firpo MT, Kang C, Skarnes WC, Tjian R. Polybromo protein BAF180 functions in mammalian cardiac chamber maturation. Genes Dev. 2004;18:3106–3116. doi: 10.1101/gad.1238104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolstein O, Silkov A, Revach M, Dikstein R. Specific interaction of TAFII105 with OCA-B is involved in activation of octamer-dependent transcription. J Biol Chem. 2000;275:16459–16465. doi: 10.1074/jbc.275.22.16459. [DOI] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Olave IA, Qiu Z, Ghosh A, Graef IA, Crabtree GR. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56:94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- Xiao L, Kim M, DeJong J. Developmental and cell type-specific regulation of core promoter transcription factors in germ cells of frogs and mice. Gene Expr Patterns. 2006;6:409–419. doi: 10.1016/j.modgep.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Yamit-Hezi A, Dikstein R. TAFII105 mediates activation of anti-apoptotic genes by NF-kappaB. EMBO J. 1998;17:5161–5169. doi: 10.1093/emboj/17.17.5161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Cui K, Murray DM, Ling C, Xue Y, Gerstein A, Parsons R, Zhao K, Wang W. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev. 2005;19:1662–1667. doi: 10.1101/gad.1323805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Wang Z, Sharova L, Sharov AA, Ling C, Piao Y, Aiba K, Matoba R, Wang W, Ko MS. BAF250B-associated SWI/SNF chromatin-remodeling complex is required to maintain undifferentiated mouse embryonic stem cells. Stem Cells. 2008;26:1155–1165. doi: 10.1634/stemcells.2007-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Cao J, Huang L, Fang HY, Sheng HZ. Regulated expression of TATA-binding protein-related factor 3 (TRF3) during early embryogenesis. Cell Res. 2006;16:610–621. doi: 10.1038/sj.cr.7310064. [DOI] [PubMed] [Google Scholar]
- Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Penttila TL, Morris PL, Teichmann M, Roeder RG. Spermiogenesis deficiency in mice lacking the Trf2 gene. Science. 2001;292:1153–1155. doi: 10.1126/science.1059188. [DOI] [PubMed] [Google Scholar]