Abstract
The functions of human natural killer (NK) cells are controlled by diverse families of antigen receptors. Prominent among these are the killer cell immunoglobulin-like receptors (KIR), a family of genes clustered in one of the most variable regions of the human genome. Within this review we discuss the vast polymorphism of the KIR gene complex which rivals that of the human leucocyte antigen (HLA) complex. There are several aspects to this polymorphism. Initially there is presence/absence of individual KIR genes, with four of these genes, termed framework genes, being present in all individuals tested to date, except on those very occasional instances when the gene has been deleted. Within each gene, alleles are present at different frequencies. We provide details of a new website that enables convenient searching for data on KIR gene, allele and genotype frequencies in different populations and show how these frequencies vary in different worldwide populations and the high probability of individuals differing in their KIR repertoire when both gene and allele polymorphism is considered. The KIR genes present in an individual may be classified into A and/or B haplotypes, which respectively have a more inhibitory role or a more activating role on the function of the NK cell. Family studies have been used to ascertain the make-up of these haplotypes, inclusion of allele typing enabling determination of whether one or two copies of a particular gene is present. In addition to genetic diversification the KIR gene complex shows differences at the functional level with different alleles having different protein expression levels and different avidity with their HLA ligand.
Keywords: genotypes, haplotypes, HLA ligands, killer cell immunoglobulin-like receptors, natural killer cells, transplantation, website
Introduction
Human natural killer (NK) cells are bone marrow-derived lymphocytes that share a common progenitor with T cells, do not express antigen-specific cell surface receptors and comprise 10–15% of all circulating lymphocytes. Owing to their early production of cytokines and chemokines and their ability to lyse target cells without prior sensitization (hence the term ‘natural killer’ cells), NK cells are crucial components of the innate immune system, providing a first line of defence against infectious agents.1 The NK cells were discovered as a result of their ability to kill certain tumour cell lines that expressed little or no major histocompatibility complex (MHC) class I molecules.2 This led to the ‘missing-self’ hypothesis, which formulated that NK cells recognize and, thereafter, eliminate cells that fail to express self-MHC molecules.
The cytolytic activity of human NK cells is modulated by the interaction of inhibitory and activatory membrane receptors, expressed on their surface, with MHC class I antigens expressed by host cells. The receptors belong to two distinct families, the C-type lectins-like group (CD94: NKG2) mapping to chromosome 12q1.3–13.4 and the immunoglobulin-like super family consisting of the killer cell immunoglobulin-like receptors (KIR), leucocyte immunoglobulin-like receptors and the leucocyte-associated immunoglobulin-like receptors mapping to chromosome 19q13.4.3–5 Whereas the other gene families are believed to have limited polymorphism, KIRs show extensive polymorphism. The genes encoding the KIR receptors are clustered in one of the most variable regions of the human genome in terms of both gene content and sequence polymorphism. This extensive variability generates a repertoire of NK cells in which KIR are expressed at the cell surface in a combinatorial fashion. Interactions between KIR and their appropriate ligands on target cells result in the production of positive or negative signals, which regulate NK cell function.6,7 Interestingly, the human leucocyte antigen (HLA) ligands for KIR genes are highly polymorphic whereas those for CD94-NKG2 are not. Variation in KIR is the result of gene and allele content, giving rise to haplotype diversity and leading to a staggering number of different genotypes. Genotype is defined as the repertoire of KIR genes present in an individual. This diversity is compounded by functional diversity (variegated expression, ligand-binding specificity and inhibitory strength). A few years ago a clearer picture emerged of the genomic organization of the KIR8,9 and the extent of KIR diversity within the human population,10,11 leading to a search for potential consequences for human disease, infection and outcomes in stem cell transplantation.12–14
Nomenclature
To date, 15 distinct KIR gene loci (including two pseudogenes KIR2DP1 and KIR3DP1) have been identified, which vary with respect to their presence or absence on different KIR haplotypes, creating considerable diversity in the number of KIR genotypes observed in the population. Some confusion arises with the number of KIR genes that are mentioned in publications. The distinction between what are individual genes and what are alleles of the same gene has not always been clear. This is compounded by the fact that genes with separate names, KIR3DL1 and KIR3DS1 are now taken as allelic. Similarly 2DL2 and 2DL3 are also allelic and so some publications may refer to 17 KIR genes. This has been noted by the nomenclature committee who although they still name alleles as either KIR3DL1 or KIR3DS1, use a non-coinciding numbering system for these alleles.15 However, this does not happen for KIR2DL2/2DL3. In the present review we refer to these genes as 2DL2/3 and 3DL1/S1. Each KIR gene encodes either an inhibitory or an activating KIR, except KIR3DL1/S1, which encodes one or the other depending on which allele is present, and KIR2DL4, which shares structural features with both inhibitory and activating KIR.16
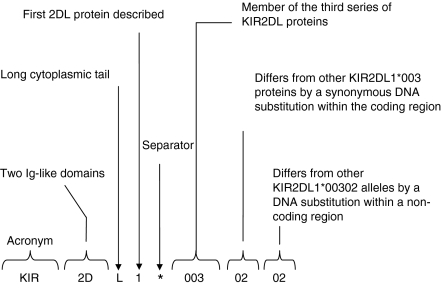
The names given to the KIR genes by a subcommittee of the World Health Organization Nomenclature Committee for Factors of the HLA System, are based on the structures of the molecules they encode (Fig. 1).15 The KIR genes have either two or three extracellular immunoglobulin domains, called 2D or 3D and either a long (L) or a short (S) intracellular domain. The first digit following the KIR acronym corresponds to the number of immunoglobulin-like domains in the molecule and the ‘D’ denotes ‘domain’. The D is followed by either an ‘L’, indicating a ‘Long’ cytoplasmic tail, (these proteins have inhibitory function), or ‘S’ indicating a ‘Short’ cytoplasmic tail, (these proteins have activating function), or a ‘P’ for ‘pseudogene’. The final digit indicates the number of the gene encoding a protein with this structure. Where two or more genes have very similar structures and have very similar sequences, they may be given the same number but distinguished by a final letter, for example, the KIR2DL5A and KIR2DL5B genes.17 KIR alleles are named in a similar fashion to alleles of the HLA system (Fig. 1). Hence, the first three digits distinguish alleles differing in exon sequences that lead to non-synonymous changes. (The HLA nomenclature is on the point of being changed to allow for the expansion in the number of alleles). The next two digits indicate alleles that differ in exon sequences leading to synonymous changes and the last two digits are used for those alleles that only differ in an intron, promoter or other non-coding region.
Figure 1.
Nomenclature of killer cell immunoglobulin-like receptors (KIR) genes and alleles.
Ligands
The HLA class I molecules act as ligands for some of the KIR genes. The alleles of the HLA-C locus can be distinguished into two groups of ligands (C1 and C2) by the amino acid present at position 80 of the molecule with approximately 50% of alleles being in each group. HLA-C group 1 with asparagine at position 80 provides the ligand for KIR2DL2 and KIR2DL3, whereas HLA-C group 2 with lysine at position 80 provides the ligand for KIR2DL1. Recently it has been shown that whereas KIR2DL1 has only interaction with HLA-C2 group, KIR2DL2, and to a weaker extent KIR2DL3, also bind to HLA-C2 group.18KIR3DL1 has specificity for the HLA-Bw4 epitope at residues 77–83, present on some HLA-A molecules in addition to many of the HLA-B alleles as each HLA-B allele has a Bw4 or Bw6 epitope. KIR3DL2 has as its ligand HLA-A3 and HLA-A11 allele families but only when certain virally derived peptides are loaded and HLA-G is the ligand for KIR2DL4. As all individuals will carry an HLA-C allele, HLA-C may be more important in the regulation of NK cells.
Methods to detect KIR genes
As many of the laboratories interested in typing for presence/absence of KIR genes were histocompatibility laboratories the tendency was to use methods familiar to the laboratory, i.e. sequence-specific primers (SSP)19–22 and sequence-specific oligonucleotide probes (SSOP).23 However, these methods are not able to determine the number of gene copies present. Allele typing is limited and has been performed in only a few laboratories. Continuous discovery of new alleles and the difficulties inherent because of similarity in sequences, even between alleles of different genes, requires constant revision of the SSP and SSOP typing systems. As a result, some laboratories have resorted to sequencing for allele determination whereas others have used mass spectrometry or real-time reverse transcription–polymerase chain reaction, which not only could prove useful for allele determination but also for determining copy number of either gene or allele.24–26 Recently, we reported a KIR allele discrimination method using a high-resolution melting technique, which bypassed the primer design restrictions imposed in SSP systems and allowed identification of alleles that had previously given ambiguous typing results by SSOP.27
Website and KIR genotypes
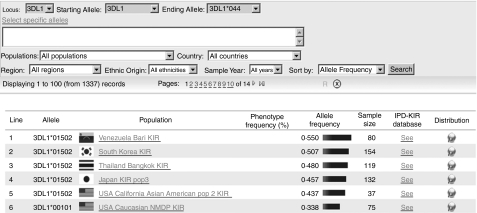
A website initially set up to contain data on frequencies of HLA alleles in global populations has been extended to include KIR frequency data. The website http://www.allelefrequencies.net is freely available and contains at present KIR data from 172 populations (19 640 individuals).28 Most of the data are taken from publications and reference to the publication and demographic details of the populations are given on the website. The data are available in two formats; KIR gene or allele frequencies (Fig. 2) and KIR genotypes (i.e. presence or absence of KIR genes) (Fig. 3). Phenotypic frequencies (number of individuals in a population having that gene or allele) are given as percentages and allele frequencies are given in three decimal format. Also available on the website are KIR typing results, including allele typing, of 84 International Histocompatibility Workshop (IHW) cell lines and 12 Centre d’Etude du Polymorphisme Humain (CEPH) families from the 13th IHW.
Figure 2.
Killer cell immunoglobulin-like receptors (KIR) allele frequency search.
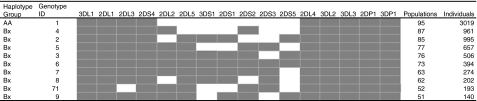
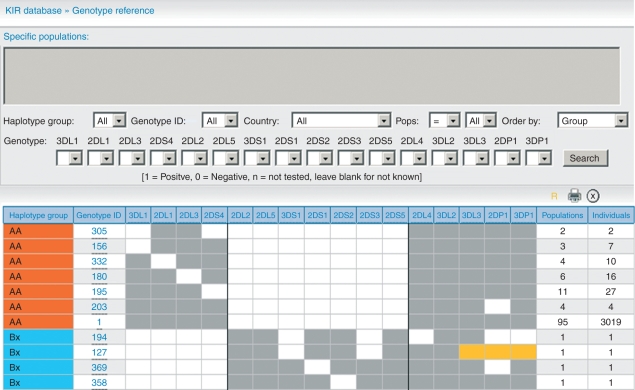
Figure 3.
Most common killer cell immunoglobulin-like receptors (KIR) genotypes reported in http://www.allelefrequencies.net.
The reader is referred to this website, which is regularly updated and contains different methods of sorting data. This review contains a brief summary of the data therein; 355 different genotypes have been reported in 10 040 individuals from 95 populations. Figure 3 shows the most common genotypes. The genotypes have been labelled as AA or Bx where x can be either an A or B haplotype. This is because of the difficulty, without family studies, of distinguishing in the presence of a B haplotype whether the other haplotype is A or B. Table 1 shows distribution of genotypes by geographic region. Only two genotypes occurred in all 10 geographic regions and only one genotype occurred in all populations. Ten genotypes are common, being reported in more than 50 of the 95 populations and representing 7341 (73·1%) of the total of individuals tested, whereas 178 genotypes only occurred in one population, 166 of these in only one individual (Table 2).
Table 1.
Distribution of killer cell immunoglobulin-like receptors (KIR) genotypes by geographic region
| Region | Populations | Genotypes | Individuals |
|---|---|---|---|
| Asia | 21 | 157 | 2076 |
| Australia | 1 | 6 | 42 |
| East Europe | 6 | 134 | 1067 |
| Middle East | 8 | 105 | 1028 |
| North Africa | 1 | 22 | 67 |
| North America | 5 | 66 | 599 |
| Pacific | 8 | 86 | 387 |
| South and Central America | 21 | 136 | 2046 |
| Sub-Saharan Africa | 6 | 55 | 407 |
| Western Europe | 18 | 136 | 2321 |
| Total | 95 | 10 040 |
Table 2.
Distribution of killer cell immunoglobulin-like receptors (KIR) genotypes by population
| Genotypes | Populations | Individuals |
|---|---|---|
| 178 | 1 | 200 |
| 41 | 2 | 116 |
| 33 | 3 | 154 |
| 13 | 4 | 82 |
| 5 | 5 | 34 |
| 39 | 6–10 | 597 |
| 36 | 11–50 | 1516 |
| 10 | > 50 | 7341 |
| Total 355 | 10 040 |
Haplotypes
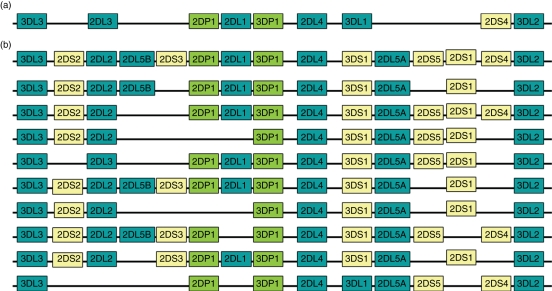
Genotypes can be resolved into two broad haplotypes termed A and B based on KIR gene content and this grouping is referred to in many analyses. A 24-kilobase band is present in group B and absent in group A using HindIII digestion and Southern blot analyses.19 The basis of each A or B haplotype consists of four framework genes, found, with very few exceptions, to be present in all individual tested to date: KIR2DL4, KIR3DL2, KIR3DL3 and KIR3DP1. Duplication and deletion of genes have led to many different haplotypes, examples of which are shown in Fig. 4. Full haplotype-length sequencing has been performed for KIR haplotypes, showing the order of the genes on each haplotype to be KIR3DL3 at the centromeric end, KIR3DL2 at the telomeric end and KIR2DL4 in the middle.8,9,29 The A haplotype is generally non-variable in its gene organization, using up to eight genes: those of the framework and KIR2DL1, KIR2DL3, KIR2DS4 and KIR3DL1. Indeed, one genotype consisting of two identical A haplotypes with all eight genes, is present in all 95 populations with available genotyping data, a total of 3019 (30·1%) individuals (Fig. 5). Occasionally AA genotypes have one of the genes normally present on an A haplotype missing. The B haplotype is defined by the presence of one or more of the genes encoding activating KIRs, KIR2DS1/2/3/5, KIR3DS1 and the genes encoding inhibitory KIRs, KIR2DL5A/B and KIR2DL2. Hence, variability on the B haplotype is created mainly by the presence or absence of the genes and, to a lesser extent, by alleles whereas in the A haplotype it is very exceptional to have variability in gene content but there is much more allele variability. Corresponding to this is the fact that it is the inhibitory genes that, in the main, have more alleles than the activating genes. Of the 335 alleles reported to date, 243 are from the inhibitory genes, whereas 79 are from the activating genes.15,30 The remaining 13 alleles are contributed by the pseudogenes KIR2DP1 and KIR3DP1. It is not known if the B haplotype, with its many gene arrangements, does not require allele polymorphism or if natural selection has acted against variability at the allele level of these genes because of possible autoimmune destruction. It has been suggested that the activating KIR genes evolved from inhibitory KIR genes and are short-lived in comparison with the genes encoding the inhibitory KIR and so there may not have been enough time for polymorphism to develop.31
Figure 4.
Two groups of killer cell immunoglobulin-like receptors (KIR) haplotypes (a and b).
Figure 5.
Killer cell immunoglobulin-like receptors (KIR) genotype search.
KIR3DP1 and KIR2DL4 divide the centromeric from the telomeric parts of the haplotype. Within each of these two regions there is extensive linkage disequilibrium (see also section on KIR alleles). For example a recent report has shown that in 27 global populations the average linkage disequilibrium is nearly complete (Cramer’s V statistic = 0·99) between centromeric B haplotype loci KIR2DL2 and KIR2DS2 and very strong (Cramer’s V statistic = 0·92) between the telomeric genes KIR3DS1 and KIR2DS1. However, much less linkage disequilibrium is found between centromeric and telomeric parts; for example Cramer’s V statistic = 0·1 for KIR2DL2 and KIR3DS1 (J. A. Hollenbach, A. Meenagh, C. Sleator et al., submitted).
In a previous report on 77 families in Northern Ireland (plus an additional 27 families added more recently) we examined KIR genes and alleles, making it possible to ascertain if an individual had one or two copies of the gene, although it was necessary to make some assumptions.32 Gene content was first ascertained23 and those genes present were allele typed.33–39 Of the 418 haplotypes of the parents of the 104 families (haplotype information was derived from three parents in one family), there were 122 different haplotypes, taking into account both genes and alleles. Of these, 48 were A and 74 were B. Sixty-six haplotypes only occurred on one occasion. In total, 230 (55%) of haplotypes were A and 188 (45%) were B. The percentage of individuals who were homozygous for the A haplotype was 32·3%, the percentage homozygous for the B haplotype was 12·1% and 55·6% of individuals had both A and B haplotypes. B haplotypes have previously been shown to be more prevalent in non-Caucasian populations such as Australia Aborigines and Asian Indians,40–43 whereas in Caucasian populations approximately 55% of the population will have A haplotypes and 30% have two A haplotypes.44 It is believed that populations with higher frequencies of B haplotypes will be those under strong pressure from infectious diseases. The addition of 27 new families to the haplotype study resulted in the definition of 19 new individual haplotypes, some of which occurred more than once. This would indicate that even in a small ethnically homogeneous population, the number of families (77 in the original report) needs to be greatly increased to cover all potential haplotype variation.
It is important to note that genes normally associated with the A haplotype can also be found on the B haplotype. These genes, KIR3DL1, KIR2DS4, KIR2DL1, KIR2DL3, were present on 102, 99, 113 and 52 of the 188 B haplotypes, respectively. Ninety-six B haplotypes had both KIR3DL1 and KIR2DS4.
The only activating gene, bar KIR2DL4, on the A haplotype is KIR2DS4. There are two versions of KIR2DS4, one with the full sequence and one with a short deletion. The deleted version has a 22-base-pair deletion in exon 5, which causes a frame shift leading to a stop codon in exon 745 and it is believed that this version is not expressed at the cell surface. The deleted version (KIR2DS4 alleles *003,004,006,007) is quite common, at 80% in the Northern Ireland population, nearly 60% of the population having only the deleted KIR2DS4. However, there is a trend for decreased frequency of the deleted version in those populations that are homozygous for the A haplotypes.46 Interestingly we found that 30 (62·5%) of the different A haplotypes and 155 (67·4%) of total A haplotypes contained both a deleted version of KIR2DS4 and a deleted version of KIR2DL4, (2DL4-9A). Consequently, in those individuals who have the genotype AA, 43·1% did not have an activating KIR, leading to 13·9% in the overall population not having an activating receptor.
The tightly clustered KIR genes possess a high level of sequence homology, which would facilitate unequal crossing over producing high levels of insertion, deletion and recombination of KIR loci resulting in shorter or longer haplotypes and facilitating rapid diversification of the KIR gene complex.47 Two studies identified two copies of both KIR2DL4 and KIR3DL1/S1 on one haplotype.48,49 Further work on this topic showed that 4·5% of Caucasian individuals had a recombinant allele of the pseudogene KIR3DP1 that associated strongly with gene duplications of KIR2DL4 and KIR3DL1/S1 and was possibly formed by recombination of KIR3DP1 and KIR2DL5A.50 The reciprocal haplotype lacking the KIR3DL1/S1 and KIR2DS4 was also found in an individual from Northern Ireland. Again emphasizing possible unequal recombination, we have reported a haplotype which has two alleles of KIR2DL5A.32
The haplotype with the framework genes KIR2DL4 and KIR3DL1/S1 deleted has been completely sequenced and showed to be comprised of five genes, KIR3DL3, KIR2DL3, KIR2DP1, a novel KIR2DL1/2DS1 gene and KIR3DL2.51 This novel gene is also reported in a haplotype in a CEPH family from Utah, which has only four complete KIR genes. In this haplotype it is present with another novel gene, KIR2DL3/2DP1 situated between the two framework genes KIR3DL3 and KIR3DL2.51 Screening for the two hybrid genes in different ethnic populations found the KIR2DL1/2DS1 hybrid gene in an African American and a Canadian individual and similar, though not identical, hybrid genes to the KIR2DL1/2DS1 and KIR2DL3/2DP1 genes, in other populations.51
Gene frequencies in different populations
Framework genes are present with very few exceptions in all individuals; the only published exceptions being for KIR2DL4: one CEPH family member,22 one from the Bubi population on Bioko Island Equatorial Guinea52 and two from South Asia.40 However, in our study on families we found two haplotypes, on different individuals, in which KIR2DL4 was not present.32 In addition, individuals have been reported to the website as being negative for KIR2DL4 (n = 1), KIR3DL2 (n = 13), KIR3DL3 (n = 10) and KIR3DP1 (n = 15). Some of these reports may be the result of inaccurate typing, which is also possible for some of the genotypes that only occur in one individual: we have taken all data published at face value but are actively pursuing ways of analysing the data to take accuracy into account. Other individuals negative for these genes may be the result of gene deletions, as mentioned in the previous section.
The genes encoding inhibitory KIR are nearly always present in populations at frequencies greater than 90%. The exceptions are those on the B haplotypes; KIR2DL2 and the KIR2DL5 genes, KIR2DL5A and KIR2DL5B. More detailed analysis can be performed on the website but in general it can be seen that it is the indigenous populations, especially Aborigines and Amerindians, who have outlying frequencies. For example, KIR2DL2, which is generally present at 40–60%, is absent in the Taiwan Taroko Atayal population, but present at 96% in the Papua New Guinea Nasioi. KIR3DL1 has generally very high frequencies (> 90%) with 10 populations having a frequency of 100%, but some populations have lower frequencies; Australia Aborigines (55%), Papua New Guinea Nasioi (59%), Brazil Amazon (65%).
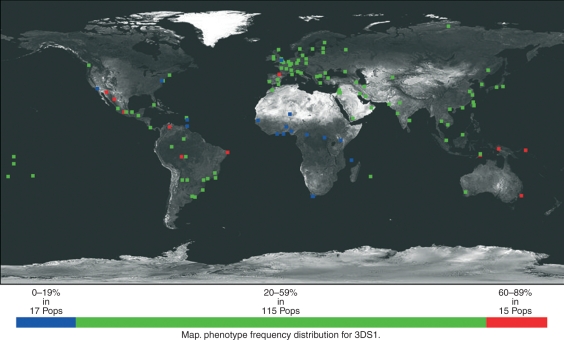
Activating KIR show much greater variation in their presence/absence in different populations. For example KIR2DS1 has four populations with greater than 80% frequency (Australia Aborigines, Brazil Amazon, Brazil Rodonia Province Karitiana and Papua New Guinea Nasioi) but three African populations with < 10%; Central Africa Republic Bagandu Biaka, Ghana and Nigeria Enugu Ibo. Similarly, KIR2DS2 has high frequencies (> 70%) in nine populations (e.g. Australia Aborigines, South Africa San and Xhosa and populations from India) but very low frequencies in Japan (8·5–16·0%), South Korea (16·9%) and China (17·3%). In some of the South American Amerindian populations KIR2DS3 is absent – Argentina Salta Wichis, Mexico Tarahumaras, Venezuela Bari and Venezuela Yucpa.53,54 The frequency of this gene is also low in Japan and China. The KIR2DS4 gene is present in seven populations at 100% – either from Africa or African Americans in USA. However, it has also low frequencies – Costa Rica (31%), Australia Aborigines (52%), Taiwan (59·4%). Selection against having KIR3DS1 has been reported in African populations25 with KIR3DS1 present in San (2·2%), Xhosa (4·0%), Nigeria (3·4% and 6·3%), Senegal (4·0%), Kenya (0·7%), Ghana (4·9%), Central Africa Republic Bagandu Biaka (2·9%). Global phenotype frequencies of KIR3DS1 are shown as an example of how the data can be represented (Fig. 6). Obviously there is a close inverted correspondence between the frequencies of KIR3DL1 and KIR3DS1 in an individual population. A very small percentage of individuals (0·34%) are negative for both KIR3DL1 and KIR3DS1.
Figure 6.
Global phenotype frequencies of 3DS1.
Such extensive diversity between modern populations may indicate that geographically distinct diseases have exerted recent, or perhaps ongoing, selection on KIR repertoires. The differences in frequencies therefore make the choice of controls for disease studies very important for all populations. We linked the published data by analysing all populations submitted to the website that had data for 13 KIR genes (excluding KIR2DP1 and KIR3DP1).55 The 56 populations analysed, using neighbour-joining dendrograms and correspondence analysis, grouped with a few exceptions according to a geographical gradient. Subsequently, we selected 38 of the 56 populations that we considered to be well defined in the anthropological sense. We found that based on KIR haplotype B genes (i.e. genes mainly encoding activating KIR) the populations were related to geography like a good anthropological marker such as HLA or Y chromosome. However, the results based on the KIR haplotype A (i.e. genes mainly encoding inhibitory KIR) did not show such a correlation.56
KIR alleles
There has been an increase in the number of known alleles from 87 in the first KIR nomenclature report in 2002 to 335 in the latest release on the IPD-KIR database, where the sequence of all KIR alleles is kept.30 This is a small increase compared with that in the HLA field but although many laboratories now type for KIR genes, notably for stem cell transplantation, very few laboratories type for KIR alleles. It is early days in the study of KIR alleles but one trusts that the finding of the new alleles can be independently confirmed (sequencing of alleles of the KIR genes is problematic because of similarities in the sequences of alleles from different genes and the size of the introns making it difficult to sequence from genomic DNA) and their possible clinical significance can be ascertained before we find ourselves in the same situation as for HLA alleles. There, 40% of HLA alleles have never been reported again after the report of their initial sequence in one individual.57
A report of allele frequency data in a Japanese population showed that for the KIR genes KIR2DL1, KIR2DL2/2DL3, KIR2DL4, KIR3DL1/S1, KIR3DL2 and KIR2DS4, one allele at each gene was at a very high-frequency (44–89%) compared with the next frequent allele.58 This is not the case in many other populations,55 emphasizing the conclusion reached by Parham and colleagues of the skewed distribution of KIR variants in the Japanese population, which reflected a distinct history of directional and balancing selection.58
Linkage disequilibrium has been reported between the alleles in a study examining the alleles of KIR2DL1, -2DL3, -3DL1 and -3DL2 in 34 families.59 Strong linkage disequilibrium existed between KIR2DL1 and -2DL3 alleles in the centromeric half and between KIR3DL1 and KIR3DL2 alleles in the telomeric half, but these two sets of pairs had little linkage disequilibrium between them and appeared to define the two halves of the KIR gene complex. This study was the first to show that in addition to gene content, diversity of KIR was the result of allele polymorphism and the combination of gene content and allele differences resulted in the vast majority of individuals having different KIR genotypes. A further study on individuals from North India determining only the alleles of KIR2DL1, -2DL3, -2DL5, -3DL1 and -3DL2 showed that all individuals had different KIR genotypes.43
In the Northern Ireland family study there were 188 (90%) different genotypes allowing for allele information. It is worth emphasizing that the Northern Ireland population is very homogeneous and drawn from a Caucasian population of 1·5 million, with very little immigration. Some alleles of the framework genes occurred more frequently on B haplotypes than A haplotypes Most notable of these was the occurrence of KIR2DL4*00501 on 43·6% of B but absent from A and KIR3DL2*007 on 43·6% of B but only 1·3% of A. In those genes that have been thought to be on A haplotypes (KIR2DL1, -2DL3, -3DL1, -2DS4) but that we found at a high occurrence on B haplotypes, there was little difference in the frequency of specific alleles on an A compared to a B haplotype, except the absence of KIR2DL1*00401 on A haplotypes, this allele being the most common allele of KIR2DL1 on B haplotypes at 27·7%. We have recently revisited the data obtained from the Northern Ireland family study and produced a list of those alleles in linkage disequilibrium in the centromeric region or the telomeric region (P. A. Gourraud, A. Meenagh, A. Cambon-Thomsen and D. Middleton, submitted). As expected, strong linkage disequilibrium between the KIR genes is driven by specific allelic associations in both regions. However, at the telomeric region KIR2DL4, KIR3DL1/S1 and KIR3DL2 have a particularly high number of alleles included in haplotypes in strong linkage disequilibrium, extending across relatively low linkage disequilibrium between pairwise loci. The data suggested that balancing between inhibitory and activating genes involves specific allele associations.
Determination of alleles is also useful for positioning of KIR genes on a haplotype. Recently KIR2DS3*00103 has been shown to map to the centromeric side, and KIR2DS3*002 and KIR2DS3*003N to the telomeric sides of the haplotype.60KIR2DS5*002 was also shown to map to the same telomeric position as KIR2D3*002/003N, implying that these alleles belong to a single locus. We have extrapolated this work to our family data by determining the KIR2DS3 and KIR2DS5 alleles. KIR2DS3 was present on 67 (16%) of the 418 haplotypes. None of the four haplotypes positive for KIR2DS3*002 or KIR2DS3*003N had KIR2DS5, whereas in 53 haplotypes positive for KIR2DS3*00103, KIR2DS5*002 was present in 17, KIR2DS5*002 being the only KIR2DS5 allele found in the Northern Ireland population.39 Ten haplotypes that had two copies of KIR2DS3 (*00103 and *002) were negative for KIR2DS5, It would therefore appear that KIR2DS3 alleles *002 and *003N are allelic to KIR2DS5*002 and KIR2DS3*00103 forms a separate gene, emphasizing that we have still much to learn of the generic make-up of KIR.
Diversity in expression and interaction with HLA ligand
A further level of diversity is provided by interaction of KIR and its HLA ligands and variation in expression of KIR genes on the NK cell. This topic and how NK cells are licensed by interaction with their HLA ligand has been covered in much greater depth in a recent review,61 but is worth mentioning briefly in the present context.
Evidence of co-evolution is suggested by disease studies62,63 and population genetics.25,64 An inverse correlation exists in populations between the frequencies of the KIR A haplotype and the HLA-C2 group reducing the frequencies of potential pre-eclampsia pregnancies in which an increased prevalence of the AA genotype when the fetus carried the HLA-C2 group has been reported.65 Global studies on KIR3DL1/S1 diversity showed that positive selection was focused to the residues that interact with HLA and strong negative correlations between KIR3DS1 and its presumed HLA-Bw 4 ligand existed.25,64 In the latter study, the tendency was for inhibitory KIR to have positive correlations and activating KIR to have negative correlations, respectively, with their ligands. The one exception was the negative correlation between KIR2DL2 and HLA-C1 group which has been confirmed in data from 27 global populations from six broad ethnic groups with a corresponding positive correlation of KIR2DL3 with the HLA-C1 group ligand.32 However this study could not confirm the correlation between KIR3DL1/S1 and HLA-Bw4. In a recent study examining the relationship between KIRs and their HLA ligands in Europe, evidence in favour of co-evolution was shown. In southern European populations higher frequencies of activating KIR and those ligands associated with greater inhibition (HLA-C2 group and HLA-Bw4) were found, whereas in north and north-west Europe a lower frequency of activating receptors was accompanied by ligands associated with less inhibition.66 Consequently, a balance seems to have been struck to control high activation when needed and to allow more activation when the receptors are not as abundant.
Expression of KIR receptors is also influenced by the presence of HLA ligand. Individuals with KIR2DL1 or KIR3DL1 had greater numbers of NK cells expressing these genes if the HLA-C2 group or HLA-Bw4 ligands were, respectively, present in the individual.58 Furthermore, the effect of the ligand on its specific KIR diminished with the number of additional KIR that also had their ligand present, suggesting co-operation between receptor and ligand pairs.
The extensive sequence polymorphism of KIR genes gives rise to peculiar expression features67 and protein variants with differential binding affinity for HLA ligand.68 Promoter polymorphisms are obvious modifiers of transcription, which in the case of KIR genes can change methylation patterns.69 Whereas KIR2DL4 is expressed on all NK cells, other KIRs are only expressed on some NK cells because of patterns of KIR gene methylation.70,71 The KIR gene promoters are polymorphic and display significant structural and functional differences.72 Polymorphisms within the coding regions can also alter expression. For example, single-base polymorphisms in extracellular domains lead to intracellular sequestration in some alleles of KIR3DL1,73KIR2DL274 and KIR2DS3.75 We have previously mentioned frameshift deletions that cause premature stop codons, giving rise to truncated KIR proteins lacking transmembrane or cytoplasmic domains and to generation of soluble rather than membrane-anchored proteins.46,76 Interestingly some of the KIR alleles with some of these patterns are not uncommon: KIR2DS4*003 (46%), KIR3DL1*004 (35%).32 Indeed, KIR3DL1*004 has been shown to be the most protective allele against disease progression in human immunodeficiency virus (HIV) infection when present with the HLA-Bw4 ligand.77 Variation in the number of NK cells expressing a KIR3DL1 allele has been shown to correlate with binding of specific alleles to the KIR3DL1-specific monoclonal antibody Dx9, leading to a definition of high, low and no binders.58,78 These differences appear to be important in human disease as KIR3DL1 alleles with high expression gave stronger protection than those with low expression in HIV, except the previously mentioned KIR3DL1*004.77
The presence of the HLA-Bw4 epitope on an HLA-B allele delivers a stronger inhibitory signal resulting in better protection against NK cell-mediated cytolysis than if present on an HLA-A allele.79 However, this varies with the allele79 and with which amino acid is present at position 80 of the HLA-Bw4 epitope80 and with the KIR3DL1 allele.67
The expansive polymorphism of the KIR gene complex has been described. Whether this allows individuals to respond differently to specific viral infections remains to be determined, but it is possible that the diversity is the result of natural selection by pathogens. The different population frequency distribution from these studies indicates that KIR genes and alleles have been through rapid diversification and may have been under selection because of functional significance. Indeed, there is little conservation of KIR genes between species and only three KIR genes (KIR2DL4, KIR2DS4, KIR2DL5) have been preserved through hominid evolution.81 The diversification is thought to be more rapid for KIR genes than HLA, as HLA genes in humans and chimpanzees are more similar in sequence than their KIR counterparts.7,82 Even the CD94-NKG2 receptors are much more similar in chimpanzees and humans than KIR. Knowing the many associations of the MHC class I molecules in disease, this diversity of KIR has been sought in many diseases. However, it is imperative that knowledge from functional studies be acquired to ascertain the immunological relevance of the statistical associations found between KIR and several diseases.
Disclosures
None.
References
- 1.Caligiuri MA. Human natural killer cells. Blood. 2008;112:461–9. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ljunggren HG, Karre K. In search of the ‘missing self’. MHC molecules and NK cell recognition. Immunol Today. 1990;11:237–44. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 3.Liu WR, Kim J, Nwankwo C, Ashworth LK, Arm JP. Genomic organisation of the human leukocyte immunoglobulin-like receptors within the leukocyte receptor complex on chromosome 19q13.4. Immunogenetics. 2000;51:659–69. doi: 10.1007/s002510000183. [DOI] [PubMed] [Google Scholar]
- 4.Trowsdale J, Barten R, Haude A, et al. The genomic context of natural killer receptor extended gene families. Immunol Rev. 2001;181:20–38. doi: 10.1034/j.1600-065x.2001.1810102.x. [DOI] [PubMed] [Google Scholar]
- 5.Wende H, Colonna M, Ziegler A, Volz A. Organisation of the leukocyte receptor cluster (LRC) on human chromosome 19q13.4.A. Mamm Genome. 1999;10:154–60. doi: 10.1007/s003359900961. [DOI] [PubMed] [Google Scholar]
- 6.Moretta L, Biassoni R, Bottino C, Mingari MC, Moratta A. Human NK-cell receptors. Immunol Today. 2000;9:420–2. doi: 10.1016/s0167-5699(00)01673-x. [DOI] [PubMed] [Google Scholar]
- 7.Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol. 2002;20:217–51. doi: 10.1146/annurev.immunol.20.092501.134942. [DOI] [PubMed] [Google Scholar]
- 8.Martin AM, Freitas EM, Witt CS, Christiansen FT. The genomic organization and evolution of the natural killer immunoglobulin-like receptor (KIR) gene cluster. Immunogenetics. 2000;51:268–80. doi: 10.1007/s002510050620. [DOI] [PubMed] [Google Scholar]
- 9.Wilson MJ, Torkar M, Haude A, et al. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci USA. 2000;97:4778–83. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Norman PJ, Parham P. Complex interactions: the interactions of human leukocyte antigen and killer cell immunoglobulin-like receptors. Semin Haematol. 2005;42:65–75. doi: 10.1053/j.seminhematol.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 11.Khakoo SI, Carrington M. KIR and disease: a model system or system of models? Immunol Rev. 2006;214:186–201. doi: 10.1111/j.1600-065X.2006.00459.x. [DOI] [PubMed] [Google Scholar]
- 12.Kulkarni S, Martin MP, Carrington M. The yin and yang of HLA and KIR in human disease. Semin Immunol. 2008;20:343–52. doi: 10.1016/j.smim.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costa P, Rusconi S, Fogli M, et al. Low expression of inhibitory natural killer receptors in CD8 cytotoxic T lymphocytes in long-term non-progressor HIV-1-infected patients. AIDS. 2003;17:257–60. doi: 10.1097/00002030-200301240-00017. [DOI] [PubMed] [Google Scholar]
- 14.Velardi A. Role of KIRs and KIR ligands in hematopoietic transplantation. Curr Opin Immunol. 2008;20:581–7. doi: 10.1016/j.coi.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 15.Marsh SGE, Parham P, Dupont B, et al. Killer-cell immunoglobulin-like receptor (KIR) nomenclature report 2002. Hum Immunol. 2003;64:648–54. doi: 10.1016/s0198-8859(03)00067-3. [DOI] [PubMed] [Google Scholar]
- 16.Rajagopalan S, Fu J, Long EO. Cutting edge: induction of IFN-γ production but not cytotoxicity by the killer cell Ig-like receptor KIR2DL4 (CD158d) in resting NK cells. J Immunol. 2001;167:1877–81. doi: 10.4049/jimmunol.167.4.1877. [DOI] [PubMed] [Google Scholar]
- 17.Gomez-Lozano N, Gardiner CM, Parham P, Vilches C. Some human KIR haplotypes contain two KIR2DL5 genes: KIR2DL5A and KIR2DL5B. Immunogenetics. 2002;54:314–9. doi: 10.1007/s00251-002-0476-2. [DOI] [PubMed] [Google Scholar]
- 18.Moesta AK, Norman PJ, Yawata M, et al. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL1 a stronger receptor for HLA-C than KIR2DL3. J Immunol. 2008;180:3969–79. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- 19.Uhrberg M, Valiante NM, Shum BP, et al. Human diversity in killer cell inhibitory receptor genes. Hum Immunol. 1997;7:753–63. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 20.Vilches C, Castano J, Gomez-Lozano N, Estefania E. Facilitation of KIR genotyping by a PCR-SSP method that amplifies short DNA fragments. Tissue Antigens. 2007;70:415–22. doi: 10.1111/j.1399-0039.2007.00923.x. [DOI] [PubMed] [Google Scholar]
- 21.Gomez-Lozano N, Vilches C. Genotyping of human killer-cell immunoglobulin-like receptor genes by polymerase chain reaction with sequence-specific primers: an update. Tissue Antigens. 2002;59:184–8. doi: 10.1034/j.1399-0039.2002.590302.x. [DOI] [PubMed] [Google Scholar]
- 22.Norman PJ, Cook MA, Carey BS, et al. SNP haplotypes and allele frequencies show evidence for disruptive and balancing selection in the human leukocyte receptor complex. Immunogenetics. 2004;56:225–37. doi: 10.1007/s00251-004-0674-1. [DOI] [PubMed] [Google Scholar]
- 23.Middleton D, Williams F, Halfpenny IA. KIR genes. Transpl Immunol. 2005;14:135–42. doi: 10.1016/j.trim.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 24.Cooley S, Trachtenberg E, Bergmann TL, et al. Donors with group B KIR haplotypes improve relapse-free survival after unrelated hematopoietic cell transplantation for acute myelogenous leukemia. Blood. 2009;113:726–32. doi: 10.1182/blood-2008-07-171926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Norman PJ, Abi-Rached L, Gendzekhadze K, et al. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat Genet. 2007;39:1092–9. doi: 10.1038/ng2111. [DOI] [PubMed] [Google Scholar]
- 26.Du Z, Sharm SK, Spellman S, Reed EF, Rajalingham R. KIR2DL5 alleles mark certain combination of activating KIR genes. Genes Immun. 2008;9:1–11. doi: 10.1038/gene.2008.39. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez A, McErlean C, Meenagh A, Shovlin T, Middleton D. Killer cell immunoglobulin-like receptor allele discrimination by high-resolution melting. Hum Immunol. 2009;70:858–63. doi: 10.1016/j.humimm.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 28.Middleton D, Menchaca L, Rood H, Komerofsky R. New Allele Frequency Database. Tissue Antigens. 2003;61:403–7. doi: 10.1034/j.1399-0039.2003.00062.x. http://www.allelefrequencies.net. [DOI] [PubMed] [Google Scholar]
- 29.Horton R, Coggill P, Miretti MM, et al. The LRC haplotype project: a resource for killer immunoglobulin-like receptor-linked association studies. Tissue Antigens. 2006;68:450–6. doi: 10.1111/j.1399-0039.2006.00697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson J, Waller MJ, Stoehr P, Marsh SGE. IPD – the Immuno Polymorphism Database. Nucleic Acids Res. 2005;331:D523–6. doi: 10.1093/nar/gki032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abi-Rached L, Parham P. Natural selection drives recurrent formation of activating killer cell immunoglobulin-like receptor and Ly49 from inhibitory homologues. J Exp Med. 2005;201:1319–32. doi: 10.1084/jem.20042558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Middleton D, Meenagh A, Gourrand PA. KIR hapolotype content at the allele level in 77 Northern Irish families. Immunogenetics. 2007;59:145–58. doi: 10.1007/s00251-006-0181-7. [DOI] [PubMed] [Google Scholar]
- 33.Williams F, Meenagh A, Sleator C, Middleton D. Investigation of killer cell immunoglobulin-like receptor gene diversity: I KIR2DL4. Hum Immunol. 2004;65:31–8. doi: 10.1016/j.humimm.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 34.Maxwell LD, Williams F, Gilmore P, Meenagh A, Middleton D. Investigation of killer cell immunoglobulin-like receptor gene diversity: II KIR2DS4. Hum Immunol. 2004;65:613–21. doi: 10.1016/j.humimm.2004.02.028. [DOI] [PubMed] [Google Scholar]
- 35.Keaney L, Williams F, Meenagh A, Sleator C, Middleton D. Investigation of killer cell immunoglobulin-like receptor gene diversity: III KIR2DL3. Tissue Antigens. 2004;64:188–94. doi: 10.1111/j.1399-0039.2004.00263.x. [DOI] [PubMed] [Google Scholar]
- 36.Halfpenny IA, Middleton D, Barnett YA, Williams F. Investigation of killer cell immunoglobulin-like receptor gene diversity: IV KIR3DL1/S1. Hum Immunol. 2004;65:602–12. doi: 10.1016/j.humimm.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 37.Meenagh A, Williams F, Sleator C, Halfpenny IA, Middleton D. Investigation of killer cell immunoglobulin-like receptor gene diversity: V KIR3DL2. Tissue Antigens. 2004;64:226–34. doi: 10.1111/j.1399-0039.2004.00272.x. [DOI] [PubMed] [Google Scholar]
- 38.Meenagh A, Gonzalez A, Sleator C, McQuaid S, Middleton D. Investigation of killer cell immunoglobulin-like receptor gene diversity, KIR2DL1 and KIR2DS1. Tissue Antigens. 2008;72:383–91. doi: 10.1111/j.1399-0039.2008.01093.x. [DOI] [PubMed] [Google Scholar]
- 39.Gonzalez A, Meenagh A, Sleator C, Middleton D. Investigation of killer cell immunoglobulin-like receptor (KIR) gene diversity: KIR2DL2, KIR2DL5 and KIR2DS5. Tissue Antigens. 2008;72:11–20. doi: 10.1111/j.1399-0039.2008.01050.x. [DOI] [PubMed] [Google Scholar]
- 40.Norman PJ, Carrington CVF, Byng M, et al. Natural killer cell immunoglobulin-like receptor (KIR) locus profiles in African and South Asian populations. Genes Immun. 2002;3:86–95. doi: 10.1038/sj.gene.6363836. [DOI] [PubMed] [Google Scholar]
- 41.Rajalingam R, Du Z, Meenagh A, et al. Distinct diversity of KIR genes in three Southern Indian populations: comparison with world populations revealed a link between KIR gene content and pre-historic human migrations. Immunogenetics. 2008;60:207–17. doi: 10.1007/s00251-008-0286-2. [DOI] [PubMed] [Google Scholar]
- 42.Toneva M, Lepage V, Lafay G, et al. Genomic diversity of natural killer cell receptor genes in three populations. Tissue Antigens. 2001;57:358–62. doi: 10.1034/j.1399-0039.2001.057004358.x. [DOI] [PubMed] [Google Scholar]
- 43.Rajalingam R, Krausa P, Shilling HG, et al. Distinctive KIR and HLA diversity in a panel of North India Hindus. Immunogenetics. 2002;53:1009–19. doi: 10.1007/s00251-001-0425-5. [DOI] [PubMed] [Google Scholar]
- 44.Parham P. Immunogenetics of killer-cell immunoglobulin-like receptors. Tissue Antigens. 2003;62:194–200. doi: 10.1034/j.1399-0039.2003.00126.x. [DOI] [PubMed] [Google Scholar]
- 45.Maxwell LD, Wallace A, Middleton D, Curran MD. A common KIR2DS4 deletion variant in the human that predicts soluble KIR molecule analogous to the KIR1D molecule observed in the rhesus monkey. Tissue Antigens. 2002;60:254–8. doi: 10.1034/j.1399-0039.2002.600307.x. [DOI] [PubMed] [Google Scholar]
- 46.Middleton D, Gonzalez A, Gilmore PM. Studies on the expression of the deleted KIR2DS4*003 gene product and distribution of KIR2DS4 deleted and nondeleted version in different populations. Hum Immunol. 2007;68:128–34. doi: 10.1016/j.humimm.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 47.Barten R, Torkar M, Haude A, Trowsdale J, Wilson MJ. Divergent and convergent evolution of NK-cell receptors. Trends Immunol. 2001;22:52–7. doi: 10.1016/s1471-4906(00)01802-0. [DOI] [PubMed] [Google Scholar]
- 48.Williams F, Maxwell LD, Halfpenny IA, et al. Multiple copies of KIR 3DL/S1 and KIR 2DL4 genes identified in a number of individuals. Hum Immunol. 2003;64:729–32. doi: 10.1016/s0198-8859(03)00089-2. [DOI] [PubMed] [Google Scholar]
- 49.Martin MP, Bashirova A, Traherne J, Trowsdale J, Carrington M. Cutting edge: expansion of the KIR locus by unequal crossing over. J Immunol. 2003;171:2192–5. doi: 10.4049/jimmunol.171.5.2192. [DOI] [PubMed] [Google Scholar]
- 50.Gomez-Lozano N, Estefania E, Williams F, et al. The silent KIR3DP1 gene (CD158C) is transcribed and might encode a secreted receptor in a minority of humans, in whom the KIR3DP1, KIR2DL4 and KIR3DL1/KIR3DS1 genes are duplicated. Eur J Immunol. 2004;35:16–24. doi: 10.1002/eji.200425493. [DOI] [PubMed] [Google Scholar]
- 51.Traherne JA, Martin MP, Ward R, et al. In-line deletions create novel hybrid genes on extremely short KIR haplotypes. in press. [Google Scholar]
- 52.Gomez-Lozano N, de Pablo R, Puente S, Vilches C. Recognition of HLA-G by the NK cell receptor KIR2DL4 is not essential for human reproduction. Eur J Immunol. 2003;33:639–44. doi: 10.1002/eji.200323741. [DOI] [PubMed] [Google Scholar]
- 53.Flores AC, Marcos CY, Paladino N, et al. KIR genes polymorphism in Argentinean Caucasoid and Amerindian populations. Tissue Antigens. 2007;69:568–76. doi: 10.1111/j.1399-0039.2007.00824.x. [DOI] [PubMed] [Google Scholar]
- 54.Gendzekhadze K, Norman PJ, Abi-Rached L, Layrisse Z, Parham P. High KIR diversity in Amerindians is maintained using few gene content haplotypes. Immunogenetics. 2006;58:474–80. doi: 10.1007/s00251-006-0108-3. [DOI] [PubMed] [Google Scholar]
- 55.Middleton D, Meenagh A, Moscoso J, Arnaiz-Villena A. Killer immunoglobulin receptor gene and allele frequencies in caucasoid, oriental and black populations from different continents. Tissue Antigens. 2007;71:105–13. doi: 10.1111/j.1399-0039.2007.00973.x. [DOI] [PubMed] [Google Scholar]
- 56.Middleton D, Meenagh A, Serrano-Vela JI, Moscoso J, Arnaiz-Villena A. Different evolution of inhibitory and activating killer immunoglobulin receptors (KIR) in worldwide human populations. Open Immunol J. 2008;1:42–50. [Google Scholar]
- 57.Middleton D, Gonzalez F, Fernandez-Vina M, et al. A bioinformatics approach to ascertaining the rarity of HLA alleles. Tissue Antigens. 2009;74:480–5. doi: 10.1111/j.1399-0039.2009.01361.x. [DOI] [PubMed] [Google Scholar]
- 58.Yawata M, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med. 2006;203:633–45. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schilling HG, Guethlein LA, Cheng NW, et al. Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J Immunol. 2002;168:2307–15. doi: 10.4049/jimmunol.168.5.2307. [DOI] [PubMed] [Google Scholar]
- 60.Ordonez D, Meenagh A, Gomez-Lozano N, et al. Duplication, mutation and recombination of the human orphan gene KIR2DS3 contribute to the diversity of KIR haplotypes. Genes Immun. 2008;9:431–7. doi: 10.1038/gene.2008.34. [DOI] [PubMed] [Google Scholar]
- 61.Cheent K, Khakoo SI. Natural killer cells: integrating diversity with function. Immunol. 2009;126:449–57. doi: 10.1111/j.1365-2567.2009.03045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201–14. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 63.Carrington M, Martin MP. The impact of variation at the KIR gene cluster on human disease. Curr Top Microbiol Immunol. 2006;298:225–57. doi: 10.1007/3-540-27743-9_12. [DOI] [PubMed] [Google Scholar]
- 64.Single RM, Martin MP, Gao X, et al. Global diversity and evidence for coevolution of KIR and HLA. Nat Genet. 2007;39:1114–9. doi: 10.1038/ng2077. [DOI] [PubMed] [Google Scholar]
- 65.Hiby SE, Walker JJ, O’Shaughnessy KM, et al. Combinations of maternal KIR and fetal HLA-C genes influence the risk of preeclampsia and reproductive success. J Exp Med. 2004;200:957–65. doi: 10.1084/jem.20041214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guinan K, Cunningham R, Meenagh A, Megan D, Middleton D, Gardiner C. Receptor systems controlling natural killer cell functions are genetically stratified in Europe. Genes Immun. doi: 10.1038/gene.2009.60. In Press. [DOI] [PubMed] [Google Scholar]
- 67.Gardiner CM. Killer cell immunoglobulin-like receptors on NK cells: the how, where and why. Int J Immunogenet. 2008;35:1–8. doi: 10.1111/j.1744-313X.2007.00739.x. [DOI] [PubMed] [Google Scholar]
- 68.Carr WH, Pando MJ, Parham P. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J Immunol. 2005;175:5222–9. doi: 10.4049/jimmunol.175.8.5222. [DOI] [PubMed] [Google Scholar]
- 69.Uhrberg M. Shaping the human NK cell repertoire: an epigenetic glance at KIR gene regulation. Mol Immunol. 2005;42:471–5. doi: 10.1016/j.molimm.2004.07.029. [DOI] [PubMed] [Google Scholar]
- 70.Valiante NM, Uhrberg M, Shilling H, et al. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7:739–51. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- 71.Santourlidis S, Trompeter HI, Weinhold S, et al. Crucial role of DNA methylation in determination of clonally distributed killer cell Ig-like receptor expression patterns in NK cells. J Immunol. 2002;169:4253–9. doi: 10.4049/jimmunol.169.8.4253. [DOI] [PubMed] [Google Scholar]
- 72.Van Bergen J, Stewart CA, van Des Elsen PJ, Trowsdale J. Structural and functional differences between the promoters of independently expressed killer cell Ig-like receptors. Eur J Immunol. 2005;35:2191–9. doi: 10.1002/eji.200526201. [DOI] [PubMed] [Google Scholar]
- 73.Pando MJ, Gardiner CM, Gleimer M, McQueen KL, Parham P. The protein made from a common allele of KIR3DL1 (3DL1*004) is poorly expressed at cell surfaces due to substitution at positions 86 in Ig domain 0 and 182 in Ig domain 1. J Immunol. 2003;171:6640–9. doi: 10.4049/jimmunol.171.12.6640. [DOI] [PubMed] [Google Scholar]
- 74.Vandenbussche CJ, Dakshanamurthy S, Posch PE, Hurley CK. A single polymorphism disrupts the killer Ig-like receptor 2DL2/2DL3 D1 domain. J Immunol. 2006;177:5347–57. doi: 10.4049/jimmunol.177.8.5347. [DOI] [PubMed] [Google Scholar]
- 75.Vandenbussche CJ, Mulrooney TJ, Frazier WR, Dakshanamurthy S, Hurley CK. Dramatically reduced surface expression of NK cell receptor KIR 2DS3 is attributed to multiple residues throughout the molecule. Genes Immun. 2009;10:162–73. doi: 10.1038/gene.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Goodridge JP, Lathbury LJ, Steiner NK, et al. Three common alleles of KIR 2DL4 (CD158d) encode constitutively expressed, inducible and secreted receptors in NK cells. Eur J Immunol. 2007;37:199–211. doi: 10.1002/eji.200636316. [DOI] [PubMed] [Google Scholar]
- 77.Martin MP, Qi Y, Gao X, et al. Innate partnership of HLA-B and KIR2DL1 subtypes against HIV-I. Nat Genet. 2007;39:733–40. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gardiner CM, Guethlein LA, Schilling HG, et al. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J Immunol. 2001;166:2992–3001. doi: 10.4049/jimmunol.166.5.2992. [DOI] [PubMed] [Google Scholar]
- 79.Foley BA, De Santis D, Van Beelen E, et al. The reactivity of Bw4+ HLA-B and HLA-A alleles with KIR3DL1:implications for patient and donor suitability for haploidentical stem cell transplantations. Blood. 2008;112:435–43. doi: 10.1182/blood-2008-01-132902. [DOI] [PubMed] [Google Scholar]
- 80.Cella M, Longo A, Ferrara GB, Strominger JL, Colonna M. NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J Exp Med. 1994;180:1235–40. doi: 10.1084/jem.180.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moesta AK, Abi-Rached L, Norman PJ, Parham P. Chimpanzees use more varied receptors and ligands than humans for inhibitory killer cell Ig-like receptor recognition of MHC-C1 and MHC-C2 epitopes. J Immunol. 2009;182:3628–37. doi: 10.4049/jimmunol.0803401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Khakoo SI, Rajalingam R, Shum BP, et al. Rapid evolution of NK cell receptor systems demonstrated by comparison of chimpanzees and humans. Immunity. 2000;12:687–95. doi: 10.1016/s1074-7613(00)80219-8. [DOI] [PubMed] [Google Scholar]