Abstract
We describe a high-throughput screening system to detect interactions between leucocyte surface proteins, taking into account that these interactions are usually of very low affinity. The method involves producing the extracellular regions of leucocyte proteins with tags so that they can be bound to nanoparticles to provide an avid reagent to screen over an array of 36 similar proteins immobilized using the Proteon™ XPR36 with detection by surface plasmon resonance. The system was tested using established interactions that could be detected without spurious binding. The ability to detect new interactions was shown by identifying a new interaction between carcinoembryonic antigen-related cell adhesion molecule 1 and carcinoembryonic antigen-related cell adhesion molecule 8.
Keywords: carcinoembryonic antigen-related cell adhesion molecule, high-throughput screening, leucocyte-associated immunoglobulin-like receptor 1, leucocyte membrane protein, ProteOn, surface plasmon resonance
Introduction
The immune system needs to be highly regulated to obtain the right level of response for the time necessary at the required site and this is reflected by the complexity of proteins at the surfaces of leucocytes.1 While interacting partners have been found for many of these proteins, many ligands are still to be defined. With the ease of making recombinant proteins corresponding to extracellular regions there is considerable interest in screening for new interactions at the protein levels.2–6 Most of these interactions are of low affinity; for example, the T-cell receptor (TCR) binds to major histocompatibility complex (MHC)/peptide at μm affinity, CD8 to MHC class I protein KD > 100 μm, CD2 binds CD58 with a KD of 9–22 μm and CD4 to TCR is even weaker.7–10 To readily detect such interactions one requires multivalent reagents that are highly avid such as fusion proteins with immunoglobulin Fc or other alternative sequences to make proteins multivalent such as the cartilage oligomatrix protein, COMP, that makes pentamers or by immobilizing the reagent on particles.2,11,12 We have recently increased the sensitivity of screens using nanoparticles coated in recombinant proteins to recognize immobilized ligands using surface plasmon resonance (SPR) as a method of detection.10 The recombinant proteins comprising the extracellular domains of the studied molecule, rat CD4 domains 3 and 4 and a biotin tag, were expressed by transient expression in eukaryotic cells and biotinylated in vitro. The recombinant proteins were captured on avidin-coated nanoparticles to provide highly avid beads that had previously been used to recognize weak interactions on cells but were now applied to protein directly and detected by SPR.2,5 This gave very good signal-to-noise ratios even for interactions at the limit of detection with purified protein with KD approaching 1 mm for the monomeric interaction. In addition to its high sensitivity for screening, this method avoids the requirement to purify the proteins before applying to the array system, minimizing possible denaturation of the protein during the purification steps that may lead to non-specific binding. It is also effective in cost and time.
We now extend this method using the recently developed ProteOn™ XPR36 interaction array system (Bio-Rad Laboratory, Hercules, CA) that is an SPR biosensor with a multichannel module and interaction array sensor chip that allows for analysis of up to 36 protein interactions in a single injection step rather than four in the conventional method. We establish the technology using proven weak interactions and describe a new interaction within the carcinoembryonic antigen (CEA) family.
Materials and methods
Construction, expression and purification of soluble recombinant proteins
Constructs for the recombinant proteins were prepared from complementary DNA clones from Geneservice Ltd (Cambridge, UK) by inserting the sequence for the extracellular domains into the pEF-BOS vector2,13 together with the sequence for domains 3 and 4 of rat CD4 (rCD4d3 + 4) and the sequence that allows biotinylation by biotin ligase BirA (Avidity LLC, Aurora, CO)2 near the C terminus. The boundary between the human carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) part and rCD4 was GLSPGSTSIT, for human CEACAM8 TVSDGAPSTSIT (CD4 linker is underlined).5 The pEF-BOS constructs were transfected into 3 × 106 HEK 293T cells in 75-cm2 flasks with polyethylenimine (PEI; linear MW ∼ 25 000, Polysciences, Inc., Washington, PA) at the DNA : PEI ratio of 1 : 10 by weight.14 The cells were incubated in serum-free X-vivo 10 medium (BioWhittaker, Walkersville, MD) for 5 days at 37°, and the supernatants were collected, concentrated and assayed by an inhibition enzyme-linked immunosorbent assay using OX68 monoclonal antibody (mAb; specific for the rCD4d3 + 4 tag) to measure the expression levels of recombinant proteins.11 The yield of the protein was 5–20 μg/ml. The protein was either captured on the chip surface via OX68 mAb or biotinylated, dialysed against phosphate-buffered saline (PBS) and coupled to avidin-coated yellow fluorescent beads (SPHERO™, 0·46 μm diameter; Spherotech Inc., Libertyville, IL) as described previously.2,6,15 Recombinant proteins were purified from concentrated transfection supernatant by OX68 antibody affinity chromatography. 2,15
Screening with multivalent nanoparticles with the ProteOn™ XPR36
The proteins to be tested were immobilized indirectly through the OX68 mAb that recognizes the rCD4d3 + 4 tag that had been coupled to the GLM chip of the ProteOn™ XPR36 protein interaction array system. Immobilization was performed by first injecting 150 μl of a 1 : 1 mixture of 0·2 m 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide (EDC) and 0·05 m sulpho-N-hydroxysuccinimide (sulpho-NHS) followed by the OX68 antibody (0·08 mg/ml in 10 mm sodium acetate, pH 5·0) (step 1 in Fig. 1) to obtain about 1000 Response Units (RU). Then, 1 m ethanolamine was applied to block the remaining active amine groups at surfaces and 10 mm glycine–HCl pH 2·5 was applied briefly to rinse off non-covalently bound protein. Thirty-six different recombinant proteins, including the control rCD4d3 + 4, were applied and captured onto the OX68 mAb, illustrated as step 2 in Fig. 1. This capturing step continued until binding reached saturation with all OX68 mAb binding sites occupied with the rCD4d3 + 4 tag on the recombinant proteins. This ensures that the rCD4d3 + 4 tag on the particles applied subsequently would not be detected and give rise to background binding. After capturing the recombinant proteins on the surface via rCD4d3 + 4/OX68 bridging on each channel, the EDC/NHS was applied again to covalently ‘fix’ the recombinant protein to the immobilized OX68 mAb followed by ethanolamine. Unbound protein was removed by 10 mm glycine–HCl pH 2·5. This fixing process functions by activating and subsequently deactivating the free amide and carboxyl groups in rCD4d3 + 4/OX68 complexes. In case this affected the activity of the recombinant protein, specific mAb recognizing the extracellular domain of the corresponding protein where available were applied at the end of each experiment to confirm the activity of the recombinant protein and ensure the maintenance of their correct conformation. No loss of antigenic activity was observed with six proteins tested with mAb.
Figure 1.
ProteOn™ XPR36 multichannel array system configuration for the measurement of real-time interactions over 36 spots simultaneously. The immobilization of monoclonal antibody OX68 against rCD4d3 + 4 (step 1), capturing recombinant protein with rCD4d3 + 4 tag (step 2) and the injection of multivalent nanoparticles in analyte flow (step 3) were operated sequentially throughout the process at 37°. The particles were coated with recombinant proteins anchored through biotin–avidin bridging.
To optimize the effective binding of multivalent nanoparticles to the immobilized proteins, fresh samples were prepared before each experiment. Ten microlitres of 0·1% wt/v avidin-coated particles in PBS were incubated with dialysed biotinylated recombinant proteins for 3 hr, followed by centrifuging (10 min at 13 000 rpm or 1500 g), removal of supernatant and resuspension in 250 μl HBS buffer (0·01 m HEPES, pH 7·4, 0·15 m NaCl, 3 mm ethylenediaminetetraacetic acid, 0·005% surfactant P20) with 1% bovine serum albumin (BSA). Ten microlitres of diluted particle suspension was then injected over the flow cells at 25 μl/min with HBS pH 7·4 as running buffer at 37° (step 3 in Fig. 1). The results shown are representative of at least three experiments.
Results
Amplification of weak affinity by multivalent nanoparticles
A library of 36 proteins corresponding to leucocyte surface proteins with immunoglobulin superfamily (IgSF) domains were immobilized on the chip. The system was tested using the extremely weak interaction between mCD200 and mCD200RLa and mCD200RLc that have KD in the range of 0·2–5 mm, which compares with the CD200 and CD200R interaction that has a KD of around 3 μm, which is typical of most leucocyte interactions.11 In the screen of mCD200 nanoparticles over 36 proteins captured on the chip, clear specific binding occurred only to mCD200RLa and mCD200RLc demonstrating the sensitivity and specificity of the method as illustrated in Fig. 2, which shows clear binding for these two interactions compared with two negative traces (to rCD4d3 + 4 and hTOSO). It is noted that there is some drift in that the negative control binding after the bead addition is higher than before (see plateau after and before), but the positive binding is clearly distinguishable above this small change (Fig. 2). The particles were applied in the presence of 1% BSA to minimize non-specific binding. Other additives to reduce background binding were tested including a commercial ‘non-specific binding reducer’, trehalose (known for its activity in maintaining biological activity) and alginate, which is an ingredient of the GLM chip surface coat used to immobilize protein, but no significant effect was observed.
Figure 2.
Sensitive assay to detect low-affinity interactions using beads expressing multivalent ligands to immobilized proteins detected by ProteOn™ XPR36. Multivalent mCD200-coated nanoparticles injected at 25 μl/min at 37° over 36 spots coated with ∼ 1000 response units (RU) recombinant proteins. Six traces are shown: mCD200 nanoparticles gave clear binding to mCD200RLa and mCD200RLc but none to either mCD200RLb or mCD200RLe as expected or to the control proteins (rCD4d3 + 4 and TOSO), when only the signal due to the analyte itself was seen (no binding to the other 32 protein was observed – data not shown).
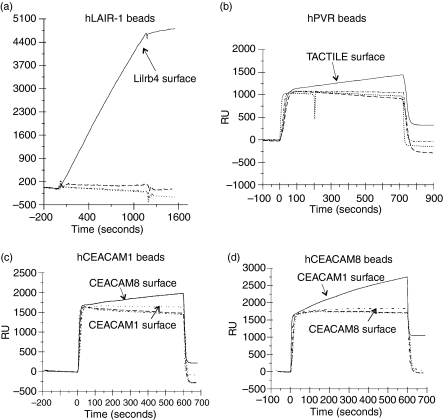
The recently described interaction between leucocyte-associated immunoglobulin-like receptor 1 (LAIR-1) and leucocyte immunoglobulin-like receptor, subfamily B, member 4 (Lilrb4)5 was also confirmed, as indicated in Fig. 3(a). LAIR-1 binds collagen with a KD of about 20 μm, but this assay also detected the much weaker binding to a second ligand, Lilrb4 whose KD is > 200 μm.5,16 There was no detectable binding to the other 35 proteins on the chip. In addition, the recently identified interaction between poliovirus receptor [poliovirus receptor (PVR), CD155, nectin and nectin-like (NECL)5] and TACTILE (CD96) has been confirmed, as indicated in Fig. 3(b).17,18
Figure 3.
Identification of specific interactions when coated nanoparticles were applied across the chip with 36 proteins immobilized (at least three interactions shown, as dashed or dotted lines). (a) Leucocyte-associated immunoglobulin-like receptor 1 (LAIR-1)-coated nanoparticles bound to leucocyte immunoglobulin-like receptor subfamily B member 4 (Lilrb4) proteins (solid line). (b) poliovirus receptor (PVR)-coated nanoparticles bound to TACTILE protein (solid line). (c) Carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1)-coated nanoparticles bound to CEACAM8 proteins (solid line) more strongly than to itself (dashed line) and the other proteins (dots and long dashes). (d) The reverse interaction where CEACAM8-coated nanoparticles bound to CEACAM1 protein (solid line) with trace binding to itself (dashed line) compared with other proteins (dots and long dashes).
Identification of a new interaction between CEACAM1 and CEACAM8
This method has proved to be efficient in screening for potential interaction pairs so we tested a series of analytes over the 36-sample protein array to look for novel interactions concentrating on proteins with IgSF domains. New interactions within the CEACAM family were observed (Table 1). CEACAM1 (CD66a) particles bound CEACAM8 (CD66b) on the surface and the converse was also observed as illustrated by selected traces in solid lines (Fig. 3c,d). Both CEACAM1 and CEACAM8 interact homophilically, indicated as the dashed lines in Fig. 3(c,d), although the heterophilic binding gave a larger signal. The relationship between strength of binding and signal is not straightforward because the reagents themselves can interact minimizing their availability to the chip.19 The interaction between CEACAM1 and CEACAM8 was also verified in separate SPR experiments in a BIAcore where purified preparations of recombinant CEACAM1 and CEACAM8 proteins were passed over flow cells onto which CEACAM1, CEACAM8 and TACTILE (negative control) had been immobilized. The binding to each flow cell was measured and compared, as indicated in Fig. 4. At equilibrium CEACAM1 clearly bound more to CEACAM8 than to CEACAM1 (Fig. 4a) whereas CEACAM8 bound CEACAM1 with no detectable binding to CEACAM8 (Fig. 4b). In the latter (Fig. 4b) the CEACAM8 did elute from the CEACAM1 and this is probably because this protein preparation is multimeric, giving a high avidity interaction. These results showing an interaction between CEACAM1 and CEACAM8 in both orientations and using purified proteins are in agreement with the observations from the high-throughput screen using beads with the ProteOn™ where the CEACAM1–CEACAM8 interaction was first identified.
Table 1.
Thirty-two proteins immobilized on surfaces of the ProteOn™ array system, besides the four unlisted members of the mCD200R family (mCD200RLa, mCD200RLb, mCD200RLc and mCD200RLe), are listed in the first column and the proteins coated on the applied nanoparticles are listed in the first row
| CEACAM1 | CEACAM8 | TACTILE (CD96) | CD85 | CD85C | CD85E | CD85H | CD85K (LILRB4) | CRTAM | LAIR1 (CD305) | CD226 (DNAM) | CD200 | PVR (CD155, NECL5) | NECL1 | NECL2 | All other proteins | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TREM1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| TREM2 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| CD200 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| CEACAM1 (CD66) | ++ | +++ | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| CEACAM5 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| CEACAM6 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| CEACAM8 (CD67) | +++ | + | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| NCR3 (CD337) | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| TACTILE (CD96) | – | – | – | – | – | – | – | – | – | – | – | – | ++ | – | – | – |
| PVR (CD155, NECL5) | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| CD226 (DNAM) | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| NECL1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| NECL2 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| PSG (CD66F) | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| mGPNMB | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| CD85 | – | – | – | – | – | – | – | + | – | – | – | – | – | – | – | – |
| CD85C | – | – | – | – | – | – | – | + | – | – | – | – | – | – | – | – |
| CD85E | – | – | – | – | – | – | – | + | – | – | – | – | – | – | – | – |
| CD85H | – | – | – | – | – | – | – | + | – | – | – | – | – | – | – | – |
| CD85K (LILRB4) | – | – | – | – | – | – | – | + | – | +++ | – | – | – | – | – | – |
| CD83 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| CMRF35 (CD300C) | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| CRTAM | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| CD5 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| CD6 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| CD72 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| TOSO | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| LAIR1 (CD305) | – | – | – | – | – | – | – | +++ | – | – | – | – | – | – | – | – |
| ESAM | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| CD99 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| mMAIR | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| CD73 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
The responses are summarized: ‘–’: no specific binding; ‘+’: weak but specific binding; ‘+++’: strong and specific binding. (If not noted, all the proteins were constructed from human genes).
Figure 4.
Heterophilic interaction between carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) and CEACAM8 detected by surface plasmon resonance (BIAcore). Three flow cells were coated with biotinylated hCEACAM1/rCD4d3 + 4 [4600 response units (RU)], hCEACAM8/rCD4d3 + 4 (3000 RU), hTACTILE/rCD4d3 + 4 (3000 RU, negative control) which had been captured on the CM5 chip surface via anchoring through streptavidin (1500 RU) that had been immobilized previously. Purified proteins were injected at 10 μl/min at 25°. (a) hCEACAM1 (0·67 μm) gave specific binding to CEACAM8 (dashed line). (b) hCEACAM8 (0·67 μm) gave binding to CEACAM1 (solid line) but no detectable binding to CEACAM8. The results are representative of at least two separate experiments.
Technical considerations
The 36-sample chip provides a powerful way to screen for interactions between large numbers of proteins. As it is a six × six array, without individual fluidics to each sample, the immobilization step is relatively time consuming. In these experiments we had to ensure that all the OX68 (rCD4 mAb) was fully saturated with recombinant protein to prevent unbound OX68 binding the rCD4 part of the recombinant protein on the nanoparticles. We ensured saturation and then prevented dissociation by carrying out a cross-linking step. An alternative would be to use different tags on the proteins on the particles to those on the chip or to use biotin immobilization for both the chip and the bead – any unbound avidin sites can be blocked by free biotin, the binding of which is essentially irreversible. The absolute level of binding and hence sensitivity of detection probably depends on the orientation of individual proteins at surfaces.5 In this study PVR beads gave binding to TACTILE as expected (Fig. 3b) but the interaction was not seen in the opposite orientation (Table 1).17,18 This may have been because of the large size of the TACTILE or its extensive glycosylation. Interactions of the NECL proteins were also not observed in this screen and this may be a factor or it is possible that the protein constructs used were not active.18,20,21
Discussion
Novel weak interactions such as those between leucocyte surface proteins could be detected on a high throughput analysis using the new ProteOn™ array system with the previously established multivalent reagent approach.5 In this assay 36 proteins were immobilized on the chip surfaces via mAb or streptavidin followed by the injection of equivalent amounts (in moles) of proteins bound to multivalent nanoparticles to provide an avid array. Thirty-six interactions could be detected and compared simultaneously by SPR providing much higher input than that obtained previously by BIAcore analysis.4,5 We have validated this method by confirming previously described weak interactions such as that between LAIR-1/Lilrb4 and identifying a new interaction between CEACAM1/CEACAM8. The finding of an interaction between CEACAM8 and CEACAM1 suggests possible interplay between granulocytes (expressing CEACAM8) and epithelial cells and/or activated leucocytes (expressing CEACAM1).
Previous studies have demonstrated that CEACAM1 interacts homophilically,22 and this was confirmed in our biophysical approach as in Fig. 3(c). In addition we demonstrate that CEACAM1 also interacts heterophilically with CEACAM8, for which this is the first demonstration of a ligand. In some respects the CEACAM family resembles the CD2/signalling lymphocytic activation molecule family of IgSF receptors that interact either homophilically or to members within the family.23 CEACAM1 has been well studied because of its significant effects in cancer cell signalling.22 There are several variants in the protein and the outcome of the homophilic interaction depends on its concentration and the environment such as adaptor proteins or signalling processes.22 CEACAM8 is not expressed on as many cells as CEACAM1, but until now its binding partner was not known.24 Engagement of CEACAM8, but not CEACAM1, induced cellular adhesion, superoxide production and degranulation of eosinophils via Hck kinase.24 Like some other CEACAMs, CEACAM8 differs from CEACAM1 in that it is glycophosphatidylinositol-anchored, which results in CEACAM8’s dependence on other cell surface molecules, such as CD11, to transmit a signal through the membrane. In turn, the disruption of lipid rafts where CEACAM8 is localized or removal of the glycophosphatidylinositol anchor could inhibit adhesion and activation of eosinophils.24
The signalling mechanism may result from a co-operative complex composed of homodimers and heterodimers of CEACAM family members. Such interactions could also be present in the role of CEACAMs as receptors for hepatitis virus, Neisseria meningiditis and Neisseria gonorrhoeae and their association with other molecules such as integrins during cell migration and tumour lymphangiogenesis.25 Several members of the CEACAM (CD66) family, including CEACAM1 and CEACAM8, are expressed on the surface of human neutrophils and closely related with the regulation of adhesive activity of CD11/CD18, probably in a cooperative way. CEACAM1 (CD66a), CEACAM6 (CD66c) and pregnancy-specific glycoprotein (PSG) (CD66f) have been reported to interact homophilically and heterophilically among themselves. It is possible that CEACAM8 (CD66b) is also involved in such intrafamily interaction networks in regulating pathological processes, such as cancer, adhesion and viral infection.25,26 Cross-linking of CEACAM8 with mAb leads to activation of eosinophils and neutrophils and the identification of CEACAM1 as a ligand for CEACAM8 suggests that CEACAM1 may be involved in this type of function.24,27 The finding of new interactions such as between CEACAM1/CEACAM8 illustrate the potential of this high throughput screening method with multivalent reagents.
Acknowledgments
We are grateful to Debbie Hatherley and Marion H Brown for provision of reagents and to Marion H Brown for helpful comments. This work was supported by the Medical Research Council (G0500845 and G0400808).
Glossary
Abbreviations:
- CEACAM
carcinoembryonic antigen-related cell adhesion molecule
- GPI
glycophosphatidylinositol
- IgSF
immunoglobulin superfamily
- LAIR-1
leucocyte-associated immunoglobulin-like receptor 1
- Lilrb4
leucocyte immunoglobulin-like receptor subfamily B member 4
- SPR
surface plasmon resonance
Disclosures
The authors have no potential conflicts of interest.
References
- 1.Barclay AN. Membrane proteins with immunoglobulin-like domains – a master superfamily of interaction molecules. Semin Immunol. 2003;15:215–23. doi: 10.1016/s1044-5323(03)00047-2. [DOI] [PubMed] [Google Scholar]
- 2.Brown MH, Boles K, van der Merwe PA, Kumar V, Mathew PA, Barclay AN. 2B4, the natural killer and T cell immunoglobulin superfamily surface protein, is a ligand for CD48. J Exp Med. 1998;188:2083–90. doi: 10.1084/jem.188.11.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bushell KM, Sollner C, Schuster-Boeckler B, Bateman A, Wright GJ. Large-scale screening for novel low-affinity extracellular protein interactions. Genome Res. 2008;18:622–30. doi: 10.1101/gr.7187808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonzalez LC, Loyet KM, Calemine-Fenaux J, Chauhan V, Wranik B, Ouyang W, Eaton DL. A coreceptor interaction between the CD28 and TNF receptor family members B and T lymphocyte attenuator and herpesvirus entry mediator. Proc Natl Acad Sci USA. 2005;102:1116–21. doi: 10.1073/pnas.0409071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang L, Barclay AN. New assay to detect low-affinity interactions and characterization of leukocyte receptors for collagen including leukocyte-associated Ig-like receptor-1 (LAIR-1) Eur J Immunol. 2009;39:1167–75. doi: 10.1002/eji.200839188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Letarte M, Voulgaraki D, Hatherley D, Foster-Cuevas M, Saunders NJ, Barclay AN. Analysis of leukocyte membrane protein interactions using protein microarrays. BMC Biochem. 2005;6:2. doi: 10.1186/1471-2091-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Merwe PA, Bodian DL, Daenke S, Linsley P, Davis SJ. CD80 (B7-1) binds both CD28 and CTLA-4 with a low affinity and very fast kinetics. J Exp Med. 1997;185:393–403. doi: 10.1084/jem.185.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Merwe PA, Barclay AN, Mason DW, et al. Human cell-adhesion molecule CD2 binds CD58 (LFA-3) with a very low affinity and an extremely fast dissociation rate but does not bind CD48 or CD59. Biochemistry. 1994;33:10149–60. doi: 10.1021/bi00199a043. [DOI] [PubMed] [Google Scholar]
- 9.Gao GF, Jakobsen BK. Molecular interactions of coreceptor CD8 and MHC class I: the molecular basis for functional coordination with the T-cell receptor. Immunol Today. 2000;21:630–6. doi: 10.1016/s0167-5699(00)01750-3. [DOI] [PubMed] [Google Scholar]
- 10.van der Merwe PA, Barclay AN. Analysis of cell-adhesion molecule interactions using surface plasmon resonance. Curr Opin Immunol. 1996;8:257–61. doi: 10.1016/s0952-7915(96)80065-3. [DOI] [PubMed] [Google Scholar]
- 11.Hatherley D, Cherwinski HM, Moshref M, Barclay AN. Recombinant CD200 protein does not bind activating proteins closely related to CD200 receptor. J Immunol. 2005;175:2469–74. doi: 10.4049/jimmunol.175.4.2469. [DOI] [PubMed] [Google Scholar]
- 12.Voulgaraki D, Mitnacht-Kraus R, Letarte M, Foster-Cuevas M, Brown MH, Barclay AN. Multivalent recombinant proteins for probing functions of leucocyte surface proteins such as the CD200 receptor. Immunology. 2005;115:337–46. doi: 10.1111/j.1365-2567.2005.02161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci USA. 1995;92:7297–301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wright GJ, Puklavec MJ, Willis AC, Hoek RM, Sedgwick JD, Brown MH, Barclay AN. Lymphoid/neuronal cell surface OX2 glycoprotein recognizes a novel receptor on macrophages implicated in the control of their function. Immunity. 2000;13:233–42. doi: 10.1016/s1074-7613(00)00023-6. [DOI] [PubMed] [Google Scholar]
- 16.Lebbink RJ, de Ruiter T, Adelmeijer J, et al. Collagens are functional, high affinity ligands for the inhibitory immune receptor LAIR-1. J Exp Med. 2006;203:1419–25. doi: 10.1084/jem.20052554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baury B, Masson D, McDermott BM, Jr, et al. Identification of secreted CD155 isoforms. Biochem Biophys Res Commun. 2003;309:175–82. doi: 10.1016/s0006-291x(03)01560-2. [DOI] [PubMed] [Google Scholar]
- 18.Fuchs A, Cella M, Giurisato E, Shaw AS, Colonna M. Cutting edge: CD96 (tactile) promotes NK cell–target cell adhesion by interacting with the poliovirus receptor (CD155) J Immunol. 2004;172:3994–8. doi: 10.4049/jimmunol.172.7.3994. [DOI] [PubMed] [Google Scholar]
- 19.Mavaddat N, Mason DW, Atkinson PD, et al. Signaling lymphocytic activation molecule (CDw150) is homophilic but self-associates with very low affinity. J Biol Chem. 2000;275:28100–9. doi: 10.1074/jbc.M004117200. [DOI] [PubMed] [Google Scholar]
- 20.Galibert L, Diemer GS, Liu Z, et al. Nectin-like protein 2 defines a subset of T-cell zone dendritic cells and is a ligand for class-I-restricted T-cell-associated molecule. J Biol Chem. 2005;280:21955–64. doi: 10.1074/jbc.M502095200. [DOI] [PubMed] [Google Scholar]
- 21.Yu X, Harden K, Gonzalez LC, et al. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat Immunol. 2009;10:48–57. doi: 10.1038/ni.1674. [DOI] [PubMed] [Google Scholar]
- 22.Gray-Owen SD, Blumberg RS. CEACAM1: contact-dependent control of immunity. Nat Rev Immunol. 2006;6:433–46. doi: 10.1038/nri1864. [DOI] [PubMed] [Google Scholar]
- 23.Tangye SG, Phillips JH, Lanier LL. The CD2-subset of the Ig superfamily of cell surface molecules: receptor–ligand pairs expressed by NK cells and other immune cells. Semin Immunol. 2000;12:149–57. doi: 10.1006/smim.2000.0217. [DOI] [PubMed] [Google Scholar]
- 24.Yoon J, Terada A, Kita H. CD66b regulates adhesion and activation of human eosinophils. J Immunol. 2007;179:8454–62. doi: 10.4049/jimmunol.179.12.8454. [DOI] [PubMed] [Google Scholar]
- 25.Skubitz KM, Skubitz AP. Interdependency of CEACAM-1, -3, -6, and -8 induced human neutrophil adhesion to endothelial cells. J Transl Med. 2008;6:78. doi: 10.1186/1479-5876-6-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skubitz KM, Campbell KD, Skubitz AP. Synthetic peptides of CD66a stimulate neutrophil adhesion to endothelial cells. J Immunol. 2000;164:4257–64. doi: 10.4049/jimmunol.164.8.4257. [DOI] [PubMed] [Google Scholar]
- 27.Schroder AK, Uciechowski P, Fleischer D, Rink L. Crosslinking of CD66B on peripheral blood neutrophils mediates the release of interleukin-8 from intracellular storage. Hum Immunol. 2006;67:676–82. doi: 10.1016/j.humimm.2006.05.004. [DOI] [PubMed] [Google Scholar]