Abstract
It is known that head and neck squamous cell carcinomas (HNSCC) originating from different anatomic locations can exhibit varying behavior that is not predictable by histopathology of the primary tumor. Using a microarray containing 27,323 cDNA clones, we generated sets of gene expression profiles for 36 HNSCC primary tumors (12 oral cavity, 12 oropharynx, and 12 larynx/hypopharynx). From these datasets, we ranked genes according to their ability to differentiate between patients whose disease progressed within a 24 month period (aggressive phenotype) and those that did not (non-aggressive phenotype) based on levels of gene expression. A merging of datasets from the three sites revealed that only a fraction of identified genes were shared between any two sites. This contrasted greatly with the significant overlap (approximately 50%) in down-regulated genes identified in tumor/normal comparisons using cases both from oropharynx and larynx/hypopharynx. From these data, we conclude that HNSCC tumors originating from different anatomic sites share consistent changes in gene expression when comparing primary tumors to normal adjacent mucosa; these common changes most likely reflect alterations required for tumor development. In contrast, once a tumor has developed, tumor-host interactions at the different anatomic sites are likely responsible for the site-specific signatures associated with aggressive versus non-aggressive disease. Predictions of outcome based on gene expression profiling are therefore heavily influenced by the anatomic site of the primary tumor.
Keywords: Microarray, Squamous cell carcinoma, Head and neck cancer, Prognosis
Introduction
Head and neck squamous cell carcinomas (HNSCC) constitute an anatomically heterogeneous group of cancers. These arise from all mucosal sites within the head and neck, primarily the oral cavity, oropharynx, hypopharynx, larynx and nasopharynx. Over the years, clinical observations have demonstrated numerous differences in HNSCC from various head and neck sites. Lindberg systematically assessed the frequency of lymph node metastases in patients with T1 tumors at presentation; the frequencies ranged from 8% in patients with T1 soft palate cancers to 71% in patients with T1 tonsil cancers [1]. Lindberg also noted that the rate of lymph node metastases may be directly associated with tumor T stage at some sites (e.g. floor of mouth), whereas for other sites (e.g. tongue base), increasing tumor T stage does not impact the rate of lymph node metastases. Survival rates also differed between groups, with laryngeal carcinomas generally being associated with a better outcome than oral and oropharyngeal carcinomas. In 2003, the death rate per 100,000 population for laryngeal versus oral/oropharyngeal carcinomas was 2.36 versus 4.06, respectively [2]. An analysis of the Surveillance, Epidemiology, and End-Results (SEER) data for HNSCC demonstrates that this is not due to clinical stage at presentation [3]. Analysis of the SEER registry data demonstrated that site-specific survival differences remain across stages; defined as local, regional, and distant. This site-specific difference may also be seen in the varied treatment approaches. While surgery and radiation therapy were once the only primary treatments offered, head and neck sites such as the larynx and oropharynx have seen a paradigm shift to chemotherapy and radiotherapy [4], while oral cavity tumors are still treated predominantly with surgery [3]. There is also a stage related difference in therapy, with early stage lesions often managed with single modality treatment and advanced stage disease treated with multimodality therapy.
The causes for site-specific differences in tumor behavior remain enigmatic. Beyond anatomic factors which impact symptomatology and lymphatic drainage, are there additional site-specific factors that impact outcome? Smoking status affects clinical and molecular characteristics of head and neck cancers, with non-smokers having a greater percentage of oral cavity tumors than smokers, who had higher rates of laryngeal, pharyngeal and floor of mouth cancers, as well as chromosomal losses [5]. Interestingly, however, survival was not significantly different among these groups. Epstein Barr Virus (EBV) has long been associated with nasopharyngeal cancers, and other data to supporting this concept are emerging from Human Papilloma Virus (HPV) related studies [6]. For instance, in addition to differences in HPV involvement of separate anatomic sites, with common involvement of the oropharynx, it is becoming apparent that HPV related carcinomas have an improved outcome compared to HPV negative carcinomas when adjusted for anatomic site and clinicopathologic stage [7, 8]. Additionally, the heterogeneity of oncogene alteration among head and neck tumors supports such differences [9]. Differences in immune host response, tumor adhesion properties, invasiveness modulators, are possible pathways that may differ with tumor anatomic site. Many global gene profiling investigations of HNSCC study patient cohorts with tumors grouped from different anatomic sites, or restrict investigations to one anatomic site [10]. This approach does not allow for direct comparison of expression profiles between different anatomic sites.
We hypothesize that there are cellular and molecular distinctions between HNSCC at different anatomic sites that further impact biologic potential, and that these distinctions are present at the time of initial tumor treatment. This study evaluates site-specific differences through global gene expression profiling. This is part of a long-term goal to identify site-specific prognostic biomarkers that can be used at diagnosis to add prognostic information, thereby guiding therapeutic decisions. In order to discover prognostically significant site-specific signatures, we compared 6 clinically aggressive carcinomas with 6 non-aggressive carcinomas for each of the three sites studied (oropharynx, oral cavity and larynx/hypopharynx). The definition of “clinically aggressive” used was tumor progression at 24 months, regardless of treatment. We compared the data collected from these three sites in order to ascertain whether differences in gene expression that are predictive of disease progression are common across sites, or whether these differences are specific to a given anatomic location.
Materials and Methods
Patient Cohort
Patients recruited for this study were treated for histologically confirmed head and neck squamous cell carcinoma (HNSCC) at Montefiore Medical Center in the Bronx, a region with high incidence of HNSCC. All patients consented to participation under protocols approved by the Institutional Review Boards. Only patients undergoing primary therapy with curative intent were included in the study. Patients were treated by primary chemoradiotherapy or primary surgery +/− adjuvant radiotherapy as deemed clinically appropriate. Treatment start date was defined as the date of surgery, if appropriate, or the date the non-surgical therapy started. Disease progression was defined as the date of the first pathologically documented locoregional persistence and/or recurrence and/or distant metastasis. Persistence was defined as the presence of pathologically documented carcinoma less than six months after initial treatment with curative intent.
We hypothesized that the gene signatures of biologically more aggressive carcinomas were present at the time of initial treatment. This study was not designed to examine changes in expression profiles during the course of disease, and we do not exclude the possibility that tumor expression profiles change over time. We also hypothesized that disease progression within 2 years bespoke greater tumor aggression than either no progression within 2 years, or the disease progression after 2 years. Therefore we defined aggressive versus nonaggressive disease as the presence or absence of disease-progression within 2 years, respectively. For the purpose of site-specific analyses, we selected samples from patients with aggressive disease versus nonaggressive disease (Table 1).
Table 1.
Overview of clinical follow-up for HNSCC patients
| HN# | Sub-Site | T | N | M | LRR, DM or DOD | Months to LRR, DM or DOD | Survival status | Follow-up months |
|---|---|---|---|---|---|---|---|---|
| Oral cavity aggressive | ||||||||
| 65 | Floor of mouth | 4 | 2c | 0 | LRR | 5 | ALIVE | 42 |
| 12 | Alveolar ridge | 4a | 1 | 0 | LRR + DM | 8 | DUCa | 10 |
| 134 | Anterior tongue | 2 | 2c | 0 | DM + DOD | 17 | DOD | 17 |
| 180 | Anterior tongue | 3 | 2b | 0 | LRR + DM | 5 | DOD | 12 |
| 53 | Anterior tongue | 4a | 1 | 0 | DM | 6 | DOD | 16 |
| 25 | Anterior tongue | 2 | 0 | 0 | LRR(PERS) | 2 | DOD | 5 |
| Oral cavity non-aggressive | ||||||||
| 67 | Alveolar ridge | 2 | 0 | 0 | 32 | ALIVE | 32 | |
| 77 | Alveolar ridge | 4 | 0 | 0 | 36 | ALIVE | 36 | |
| 137 | Alveolar ridge | 4a | 2c | 0 | 24 | ALIVE | 24 | |
| 119 | Retromolar trigone | 2 | 0 | 0 | 28 | ALIVE | 28 | |
| 147 | Anterior tongue | 1 | 0 | 0 | 24 | ALIVE | 24 | |
| 81 | Anterior tongue | 2 | 0 | 0 | 34 | ALIVE | 34 | |
| Oropharynx aggressive | ||||||||
| 128 | Tonsil | 1 | 3 | 0 | DM | 11 | ALIVE | 21 |
| 32 | Tonsil | 3 | 2b | 0 | DM | 17 | DOD | 25 |
| 36 | Base of tongue | 3 | 2b | 0 | DM | 8 | DOCa | 9 |
| 66 | Base of tongue | 4 | 2c | 0 | DOD | 4 | DOCa | 4 |
| 60 | Soft palate | 2 | 2c | 0 | LRR | 12 | ALIVE/DM | 42 |
| 68 | Oropharyngeal wall | 2 | 3 | 0 | LRR | 16 | DOD | 17 |
| Oropharynx non-aggressive | ||||||||
| 62 | Tonsil | 2 | 1 | X | 31 | ALIVE | 31 | |
| 26 | Soft palate | 4b | 2a | 0 | DOC/NED | 24 | DOC/NED | 24 |
| 63 | Base of tongue | 4a | 2c | 0 | 37 | ALIVE | 37 | |
| 64 | Tonsil | 2 | 2a | 0 | 41 | ALIVE | 41 | |
| 2 | Base of tongue | 3 | 1 | 0 | 59 | ALIVE | 59 | |
| 17 | Tonsil | 2 | 0 | 0 | 2nd PRIMARY | 55 | DOC(2nd PRIM) | 60 |
| Larynx/Hypopharynx aggressive | ||||||||
| 18 | Supraglottis—NOS | 3 | 2c | 0 | LRR | 16 | DOD | 46 |
| 7 | Glottis—TVC | 2 | 0 | 0 | DOD(PERS) | 22 | DOD | 22 |
| 97 | Supraglottis—NOS | 3 | 0 | 0 | DM | 10 | ALIVE | 30 |
| 84 | Supraglottis—NOS | 4a | 2c | 1 | LRR(PERS) | 4 | DOD | 5 |
| 39 | Pharyngeal wall | 4 | 0 | 0 | DM | 10 | DOD | 28 |
| 91 | Hypopharynx—NOS | 2 | 3 | 0 | DM | 24 | DOD | 32 |
| Larynx/Hypopharynx non-aggressive | ||||||||
| 21 | Supraglottis—NOS | 4a | 0 | 0 | 49 | ALIVE | 49 | |
| 28 | Supraglottis—NOS | 3 | 1 | 0 | 49 | ALIVE | 49 | |
| 48 | Supraglottis—NOS | 3 | 0 | 0 | 63 | ALIVE | 63 | |
| 56 | Supraglottis—NOS | 3 | 1 | 0 | DOC | 28 | DOC(LUNG CA) | 28 |
| 76 | Suprahyoid epiglottis | 2 | 0 | 0 | 37 | ALIVE | 37 | |
| 6 | Pyriform sinus | 4 | 1 | 0 | 48 | ALIVE | 48 | |
Abbreviations: DM, distant metastasis; LRR, loco-regional recurrence; PERS, persistent disease; DOD, died of disease; DOC, died of other causes; DUC, died of unknown causes
aCancer unresolved at time of death
Survival status (either death due to disease or death due to other causes or alive) is also presented in Table 1 for the sake of completeness. Death from other causes may reflect inherent tumor biology or other comorbidities unrelated to the primary cancer. However, this issue is not relevant here, as patients were classified as having either aggressive or nonaggressive disease based on their disease-progression status, not on their survival status.
HNSCC Samples
Tumor and adjacent mucosal samples were procured from either biopsies and/or resection specimens and snap-frozen in liquid nitrogen within 30 min of the procedure, and stored at −80°C until total RNA extraction. Internal controls were taken for all research specimens to confirm the histological nature of the tissues. An exception to this was for mucosal biopsies from patients who were treated with chemoradiation protocols after initial biopsies. Biopsies of normal, site-specific, squamous mucosa were usually harvested from contralateral corresponding regions. These mucosal biopsies were gratuitous from the point of view of standard of patient care. Therefore, as per the IRB-approved protocol, the sizes of these biopsies were minimized to prevent patient morbidity; submissions of internal histology controls were not always feasible. All carcinoma specimens were confirmed as containing at least 10% carcinoma cells. We did not further quantify the percentage of tumor cells per specimen, as we intend to immunohistochemically validate significant genes which will confirm the nature of the cells (carcinoma versus host stromal cells) elaborating putative biomarkers.
Tissues (approximately 100 mg) were homogenized using a Brinkmann Model PT 10/35 Tissue Homogenizer in 1 ml Trizol reagent (Invitrogen). Chloroform (200 μl) was added to separate the solution into aqueous and organic phases, with RNA remaining in the aqueous phase. The aqueous phase was separated and RNA precipitated using 500 μl isopropanol. RNA pellets were washed with ice-cold 75% ethanol and dried. RNA was quantitated using a NanoDrop ND-1000 spectrophotometer. All RNA samples were then precipitated in ethanol and stored at −80°C until use.
T7 Linear Amplification, Fluorescent Labeling, and Hybridization to Microarrays
Approximately 5 μg of total RNA was used for T7 linear amplification. Linear amplification of primary tumor total RNA and subsequent fluorescent labeling of corresponding cDNA was carried out using the MessageAmp T7 linear amplification kit (Ambion) and cDNA labeling protocols developed at the AECOM Microarray Facility (http://microarray1k.aecom.yu.edu; [11]). In order to optimize differences in gene expression profiles among the individual primary tumors, we first created pools of total RNA for each anatomic site by pooling equal aliquots of total RNA from each of the 12 primary tumors representing that specific site. We then utilized a two-channel cDNA microarray containing 27,323 cDNA clones to compare gene expression between each HNSCC primary tumors (Cy5) and its corresponding site-specific RNA pool (Cy3). Therefore, the ratio of the fluorescence intensities of the two dyes represented a measure of differential gene expression between the individual primary tumor and its corresponding RNA pool. For corresponding tumor to normal comparisons, we compared differential gene expression between normal adjacent mucosa (Cy5) and primary HNSCC tumor for each patient (Cy3) using the same cDNA microarray platform. Hybridization to cDNA arrays was carried out overnight at 50°C in a buffer containing 30% formamide, 3× SSC, 0.75% SDS and 100 ng of human Cot-1 DNA. Following hybridization, slides were briefly washed with a solution of 1X SSC, 0.1% SDS, then washed for 20 min at room temperature in 0.2× SSC, 0.1% SDS and 20 min at room temperature in 0.1× SSC (without SDS). Slides were immediately dried and scanned using the GenePix 4000A microarray scanner. This software gives an integrated intensity per spot for each channel in addition to an integrated background count.
Data Processing, Normalization, and Supervised Clustering
For each spot on the microarray, we calculated the median intensity over the spot in the two fluorescence channels and from this subtracted the median of the background intensity. We computed an intensity dependent normalization factor for each microarray experiment by fitting a robust curve using the lowess function from the R statistical package [12]. Data designated to be of poor quality, or which did not achieve a signal to noise ratio of at least two-fold, were discarded from subsequent analysis. Statistical methods (an elemental t test) were used to generate a list of the genes that best discriminated between the two groups (aggressive versus non-aggressive phenotype). Unsupervised clustering of primary HNSCC tumor samples based on patterns of gene expression was carried out using Spearman Rank clustering. Samples with similar expression profiles were clustered using the Cluster program and the results visualized with TreeView [13]. For the purposes of overlapping datasets, independent cDNA clones originating from the same gene were treated as independent measurements due to the possibility of splice variants.
Network Generation and Associated Functional Analysis
To determine potential relationships between molecules that distinguish tumor behavior and particular cellular functions, we subjected each site-specific signature to analysis using Ingenuity Pathways Analysis (IPA) (Ingenuity® Systems, www.ingenuity.com). To generate networks, each site-specific signature containing gene identifiers and corresponding expression values was uploaded into the application. Each gene identifier was mapped to its corresponding gene object in the Ingenuity Pathways Knowledge Base. These genes, called focus genes, were overlaid onto a global molecular network developed from information contained in the Ingenuity Pathways Knowledge Base. Networks of these focus genes were then algorithmically generated based on their connectivity. Several analyses were performed yielding similar results including: analysis of individual tumors, analysis of the median expression of the six aggressive and six non aggressive tumors for each site, and analysis of median aggressive minus median non aggressive values for each gene within each site-specific signature. Because median expression values minimize potential outlier effects, we present for each site-specific signature a comparison of the highest scoring network derived from the median values of the six aggressive and six non aggressive tumors at each site. The IPA program used most of the molecules in each signature to produce potential interactive networks with functional analysis; usage of molecules in each signature was as follows: OC 146 total, 138 mapped, 107 network eligible, 101 functional pathway eligible; OP 66 total, 54 mapped, 41 network eligible, 39 functional pathway eligible; LH 77 total, 72 mapped, 47 network eligible, 44 functional pathway eligible. Networks with scores greater than 20 containing at least 10 functional molecules from the signature are presented in Figs. 5–7 and functions associated with those networks are described in “Results”.
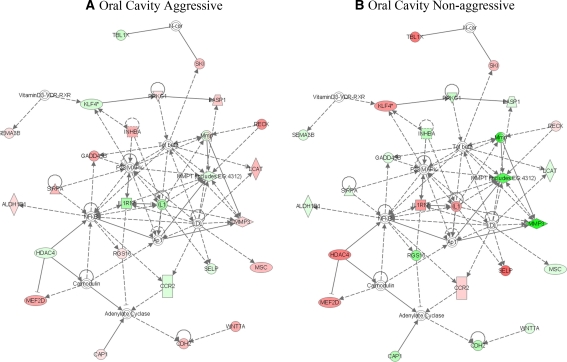
Fig. 5.
Network of oral cavity HNSCC discriminating aggressive and non aggressive tumors. The median values for aggressive (a) and non aggressive (b) oral cavity HNSCC were subjected to analysis by IPA. The highest scoring network for aggressive (48, 24 focus molecules) and non aggressive (48, 24 focus molecules) tumors are shown. Genes or gene products are represented as nodes, and the biological relationship between two nodes is represented as an edge (line). All edges are supported by at least 1 reference from the literature, from a textbook, or from canonical information stored in the Ingenuity Pathways Knowledge Base. The intensity of the node color indicates the degree of up- (red) or down- (green) regulation. Nodes are displayed using various shapes that represent the functional class of the gene product. Edges are displayed with various labels that describe the nature of the relationship between the nodes. A complete list of the shapes for each node, as well as the line types representing each edge, is available in the tutorial section at www.ingenuity.com
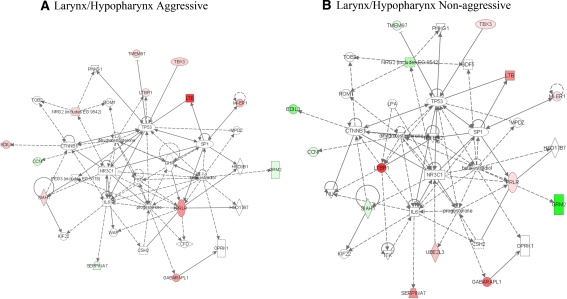
Fig. 7.
Network of laryngeal/hypopharyngeal HNSCC discriminating aggressive and non aggressive tumors. The median values for aggressive (a) and non aggressive (b) oral cavity HNSCC were subjected to analysis by IPA. The highest scoring network for aggressive (29, 14 focus molecules) and non aggressive (27, 13 focus molecules) tumors are shown. Colored symbols are focus molecules where red indicates increased expression and green represents decreased expression. A legend for the shapes and lines is available in the tutorial section at www.ingenuity.com
The Functional Analysis of a network identifies the biological functions and/or diseases that are most significant to the genes in the network. The network genes associated with biological functions and/or diseases in the Ingenuity Pathways Knowledge Base are considered for the analysis. Fischer’s exact test is used to calculate a P-value determining the probability that each biological function and/or disease assigned to that network is due to chance alone. The score takes into account the number of Network Eligible molecules in the network and its size, as well as the total number of Network Eligible molecules analyzed and the total number of molecules in Ingenuity’s knowledge base that could potentially be included in networks. The network Score is based on the hypergeometric distribution and is calculated with the right-tailed Fisher’s Exact Test. The score is the negative log of this P-value.
Results
We initially generated gene expression profiles of 45 primary HNSCC (all sites) from patients treated with curative intent (Fig. 1). Each gene expression profile was generated by comparing HNSCC total RNA against a Universal Human Reference (UHR) pool of RNA in a two color cDNA microarray experiment using a cDNA microarray containing 27,323 cDNA clones. To visualize the gene expression data, hierarchical clustering was performed using genes that satisfied stringent filtering criteria (SNR > 2, Benjamini-Hochberg corrected P < 0.05, and fold difference >2.0 or <0.5) and visualized using TreeView (Fig. 1). In our initial clustering of gene expression differences among the primary tumors, we found that there were larger differences in gene expression profiles between tumors defined by site of origin than there was with respect to the traditional clinical parameters (lymph node metastases at diagnosis, stage and tumor size), irrespective of tumor site. Assessment of tumor characteristics for the tumors across these three groups revealed strong separation along anatomic gradients with 80% (10/13) of laryngeal tumors falling in Group 1, 90% (9/10) of oral cavity tumors in Group 2, and 67% (12/18) of pharynx tumors in Group 3. However, no statistical associations were observed between cluster group and the other clinical characteristics evaluated. Therefore, an unsupervised hierarchical clustering of gene expression differences among primary tumors revealed larger differences in expression profiles between tumors from different anatomic sites than was seen with other clinical parameters (lymph node metastases at diagnosis, stage and tumor size)
Fig. 1.
Section of heat map for primary HNSCC tumors (N = 41); three principal groups (groups 1, 2 and 3) are identified. Genes whose expression is higher in the tumor sample relative to the reference sample are shown in red; those whose expression is lower than the reference sample are shown in green. Patient sample IDs are shown at the top of the figure; accession numbers for cDNA clones on the array are shown at the right. Anatomic site dominates unsupervised hierarchical clustering of gene expression profiles from HNSCC. Overall, assessment of tumor characteristics for the tumors across these three groups revealed strong separation along anatomic gradients with 80% (10/13) of laryngeal tumors falling in Group 1, 90% (9/10) of oral cavity tumors in Group 2, and 67% (12/18) of pharynx tumors in Group 3. P-value indicates the significance of associations between cluster groups and the each clinical characteristics. No statistical associations were observed between cluster group and the other clinical characteristics evaluated
To further investigate this site-specific phenomenon, we selected primary HNSCC tumor samples from oropharynx (N = 12), oral cavity (N = 12) and larynx/hypopharynx (N = 12) (Table 1). As mentioned, within each anatomic site population, we purposefully selected HNSCC samples from 6 patients with aggressive disease (HNSCC progression within 2 years) and 6 samples from patients with non-aggressive disease (no progression within 2 years).
Differential Gene Expression in HNSCC
Differences in gene expression profiles between aggressive and non-aggressive subgroups were evaluated in two ways. As part of our preliminary analysis of these patients, we compared pooled total RNA from the aggressive group to pooled total RNA from the non-aggressive group for each anatomic site using a microarray containing 27,323 cDNA clones (Fig. 2a). We also compared pooled total RNA from all 18 aggressive cancers to pooled total RNA from 18 non-aggressive cancers. In each experiment, the ratio of the fluorescence intensities of the two fluorescent dyes (Cy5-red and Cy3-green) therefore represented a measure of differential gene expression between the aggressive and non-aggressive phenotypes. In subsequent analyses of these patient samples, total RNA from each HNSCC sample was independently compared to a site-specific reference RNA pool composed of equal aliquots of RNA from each site-specific sample (oropharynx, larynx, or larynx/hypopharynx) (Fig. 2b).
Fig. 2.
Design of separate experiments to measure differential gene expression between six aggressive and six non-aggressive HNSCC primary tumors for each anatomic site (oropharynx, oral cavity and larynx/hypopharynx. (a) Pooled total RNA from six aggressive HNSCC primary tumors (A1 to A6) was compared to pooled total RNA from six non-aggressive HNSCC primary tumors (N1 to N6) on a single microarray experiment. (b) In the second analysis, RNA from each HNSCC primary tumors (A1 through N6) was individually compared to a pool of RNA represent all primary tumors from that site, for a total of 12 microarray experiments
With respect to differential gene expression comparing pooled total RNA from aggressive and non-aggressive subgroups, 36 HNSCC patients (18 aggressive subgroup, 18 non-aggressive subgroup), a total of 130 genes (59 up-regulated, 71 down-regulated) were identified as differentially expressed between patients with aggressive versus non-aggressive disease. This represented less than 0.5% of the total genes evaluated using this microarray (Fig. 3a). Of note, the number of observed gene expression differences between aggressive and non-aggressive subgroups increased 2 to 3 fold when differences were examined on an anatomic site-by-site basis. For example, when analyzing the oropharynx cases only, we identified a total of 392 genes (192 up-regulated, 200 down-regulated) with at least 3-fold difference in expression between the aggressive and non-aggressive subgroups. Similar numbers were obtained in comparisons of RNA pools from aggressive and non-aggressive subgroups of oral cavity SCC (488 genes: 206 up-regulated, 282 down-regulated) and laryngeal SCC (308 genes: 127 up-regulated, 181 down-regulated). These results revealed that many more differences in gene expression related to local recurrence could be detected when anatomic sites were examined independently.
Fig. 3.
(a) Overview of numbers of differentially expressed genes identified in first microarray experiments using pooled total RNA when comparing aggressive to non-aggressive HNSCC tumors. Comparisons included all 18 aggressive tumors against all 18 non-aggressive tumors, as well as the corresponding comparisons for each anatomic site. Breakdown of the (b) 59 up-regulated and (c) 71 down-regulated genes identified in the initial global comparison of all 36 cases, according to whether they are seen in a single or several of the corresponding site-specific comparisons. (d) Breakdown of the up-regulated and down-regulated genes identified in each site specific comparison of differentially expressed genes based on numbers seen in each of the other site-specific comparisons
Why did pooling of anatomic sites yield fewer changes in gene expression between aggressive and non aggressive tumors than analyzing the anatomic sites independently? We postulated two causes for these observations. First, when pooling anatomic sites, we might only be identifying those genes that are differentially expressed in aggressive versus non-aggressive phenotypes common to all three tumor sites. Alternatively and secondly, the genes identified may represent genes strongly expressed or repressed within a single site that are refractory to dilution because they have a sufficient difference in gene expression to be detected when all anatomic sites are pooled. In order to investigate these two postulates, we overlapped the 130 genes (59 up-regulated, 71 down-regulated) from the pooled comparison to those differences identified on a site-by-site basis. For the 59 up-regulated genes identified in our site-independent analysis, two-thirds of those genes (39 genes) were observed to be differentially expressed in only a single tumor site (Fig. 3b). Approximately one-quarter of the genes (15 genes) were seen in data from any two anatomic sites. And finally, only 5% of genes (3 genes) were seen in data from all three anatomic sites. These three genes were matrix metalloproteinase 10 (MMP10), the iron-binding protein lactotransferrin (LTF), and the serine protease inhibitor serpin A3 (ACT). For the 71 down-regulated genes identified in our site-independent analysis, the site-specific effect was even more pronounced (Fig. 3c). Seventy percent of those genes (49 genes) were observed to be differentially expressed in only a single tumor site. Twenty-eight percent of the genes (20 genes) were seen in data from any two anatomic sites. And finally, only a single gene was seen as down-regulated in data from all three anatomic sites. This gene was the inhibitor of Wnt proteins known as Wnt inhibitory factor 1 (WIF-1). Overall, the results demonstrated that genes identified in analysis pooling all three anatomic sites were, in fact, not observed across all anatomic sites, but were largely derived from dominant differences observed in a single anatomic site.
We next examined the set of genes for each anatomic site including oropharynx (392 genes), oral cavity (488 genes) and larynx (308 genes) that showed at least a 3-fold difference in expression between the aggressive and non-aggressive subgroups to determine if the gene expression differences distinguishing aggressive from non aggressive tumors were site specific. Of the 392 genes (192 up-regulated, 200 down-regulated) differentially expressed in the oropharynx dataset, 86% of these genes (336 genes: 168 up-regulated, 168 down-regulated) were observed exclusively in the oropharynx dataset (Fig. 3d). In contrast, seven percent of the genes (29 genes: 9 up-regulated, 20 down-regulated) were shared with the oral cavity dataset, and 6% of the genes (23 genes: 12 up-regulated, 11 down-regulated) were shared with the larynx dataset. Similar patterns of overlap were seen with differentially expressed genes identified from the other two anatomic sites. For example, within the oral cavity dataset, of the 488 differentially expressed genes (206 up-regulated, 282 down-regulated), 83% of these genes (406 genes: 165 up-regulated, 241 down-regulated) were observed exclusively in the oral cavity dataset. Only 10% of the genes (49 genes: 29 up-regulated, 20 down-regulated) were shared with the larynx dataset, and only 6% of the genes (29 genes: 9 up-regulated, 20 down-regulated) were shared with the oropharynx dataset. And finally, of the 308 differentially expressed genes (127 up-regulated, 181 down-regulated) in the larynx dataset, 75% of these genes (232 genes: 83 up-regulated, 149 down-regulated) were observed exclusively in the larynx dataset. Overall, these results reveal that differences in gene expression related to aggressiveness of HNSCC disease is highly site-specific, perhaps reflecting highly specific biological mechanisms of tumor aggressiveness that are heavily influenced by the anatomic site of the primary tumor.
Tumor Classification in HNSCC
One of the most clinically significant aspects of profiling the gene expression signatures of patient primary tumors is for the purpose of tumor classification and prognostic prediction. With this in mind, we individually profiled each HNSCC primary tumor sample by comparing gene expression between the primary tumor (Cy5-red) and its site-specific tumor RNA pool (Cy3-green). In order to isolate the most prognostically relevant genes, all genes were ranked according to their difference in expression levels between the aggressive and non-aggressive disease subtypes for each anatomic site using a student t-test to rank genes in our datasets for their ability to distinguish aggressive from non-aggressive phenotypes based on their level of expression. Using this approach for the 12 oropharyngeal SCC, we identified 66 genes with a P-value less than 0.1 (Fig. 4). Similarly, 176 genes could distinguish between aggressive and non-aggressive individually profiled oral cavity SCC. Finally, 95 genes could distinguish between aggressive and nonaggressive individually profiled laryngeal SCC. When these datasets were compared, there were virtually no common genes despite the liberal P-value cutoff. Only a single gene, coding for Kruppel-like factor 12 (KLF12) was common to both the oropharyngeal and laryngeal datasets. In both cases, lower expression of this gene was observed in the aggressive subgroup. Similarly, only a single gene, coding for hypothetical protein FLJ21272, was observed in both the oral cavity and laryngeal datasets. The results of these and the previous analyses with larger gene lists suggest that any classifiers of tumor behavior based on patterns of gene expression are heavily dependent on the anatomic site of the primary tumor.
Fig. 4.
TreeView diagrams of genes identified in the second analysis based on the ability to distinguish aggressive from non-aggressive tumors (t-test, P < 0.01) for (a) oropharynx (66 genes), (b) oral cavity (176 genes), and (c) larynx/hypopharynx (95 genes). Each row in the TreeView diagram represents a single gene in the signature; each column represents a single HNSCC primary tumor. Red indicates increased expression of the gene in a tumor relative to the site-specific tumor pool; green indicates decreased expression. Genes for which no data are reported for a given tumor are indicated in grey. Division of the aggressive versus non-aggressive tumors for each site are indicated below each TreeView diagram
Potential Network Interactions and Functional Pathways in Site-Specific Signatures
Networks were generated for each signature using the Ingenuity Pathways Analysis program. In graphing the difference in median values, higher expression in aggressive disease is shown in red and lower expression in aggressive disease is shown in green. The signature molecules represent molecules whose change in expression correlates with aggressive or non aggressive disease. Thus the focus molecules (red or green) in the networks may be causal in a particular pathway or they may reflect changes in that pathway. With that in mind, the highest scoring networks and associated functions are presented for each site signature; the description given is intentionally brief to highlight possible key mechanisms that could explain aggressive behavior.
For oral cavity tumors, IPA constructed four networks with scores >20 incorporating at least 13 focus molecules and a diverse group of functions with some commonly associated with cancer including cellular movement, cell death and proliferation. The highest scoring networks are based on the median expression values for oral cavity aggressive tumors (score 48, focus molecules 24) (Fig. 5a) as compared to a similar network derived from non aggressive tumors (score 48, focus molecules 24) (Fig. 5b). A major function associated with this network is cellular movement; three matrix metalloproteinases are expressed at higher levels in aggressive tumors consistent with a more invasive phenotype. It is interesting that two molecules that would generally be downregulated in tumors (inhibin and GADD45B) are increased in aggressive tumors. In addition, IL1 and IL1 receptor antagonist are differentially expressed with respect to tumor behavior.
For oropharyngeal tumors, IPA constructed two networks with scores > 20 incorporating at least 11 focus molecules and a diverse group of functions with some commonly associated with cancer including post-translational modification, cellular development and cell death. The highest scoring networks are based on the median expression values for oropharyngeal aggressive tumors (score 41, focus molecules 18) (Fig. 6a) as compared to a similar network derived from non aggressive tumors (score 41, focus molecules 18) (Fig. 6b). A major function associated with this network is cellular signaling; several protein kinase C isoforms are down regulated in aggressive tumors as compared to increased expression in non aggressive tumors. There is also differential expression of cytokine/cytokine receptors with IL8 and FGF receptor 1 down regulated in aggressive tumors and interferon receptor 2 down regulated in non aggressive tumors. Cyclooxygenase 1 (PTGS1) is down regulated in aggressive tumors and upregulated in non aggressive tumors.
Fig. 6.
Network of oropharyngeal HNSCC discriminating aggressive and non aggressive tumors. The median values for aggressive (a) and non aggressive (b) oral cavity HNSCC were subjected to analysis by IPA. The highest scoring network for aggressive (41, 18 focus molecules) and non aggressive (41, 18 focus molecules) tumors are shown. Colored symbols are focus molecules where red indicates increased expression and green represents decreased expression. The intensity of each color reflects the expression of the gene relative to the tumor pool reference. A legend for the shapes and lines is available in the tutorial section at www.ingenuity.com
For laryngeal/hypopharyngeal tumors, IPA constructed three networks with scores >20 incorporating at least 13 focus molecules and a diverse group of functions with some commonly associated with cancer including cell cycle regulation, protein degradation and protein synthesis. The highest scoring networks are based on the median expression values for laryngeal/hypopharyngeal aggressive tumors (score 29, focus molecules 14) (Fig. 7a) as compared to a similar network derived from non aggressive tumors (score 27, focus molecules 13) (Fig. 7b). A major function associated with this network is cell proliferation; there is differential expression of several molecules associated with proliferation. For example, aggressive tumors express higher levels of neuregulin 2, prolactin receptor and TMEM97. Aggressive tumors also express higher levels of SIAH1, an E3 ligase that has been associated with response to hypoxia. While both aggressive and non aggressive tumors express mesodermal induction early response 1 homologue which may be associated with epithelial-mesenchymal transformation, aggressive tumors express higher levels of MI-ER1.
In reviewing all of the functions associated with the networks for the three site-specific signatures one can find cancer-related functions that are common to all signatures (e.g. cell death). However, the uniqueness of the signatures distinguishing aggressive from non aggressive tumors at each site is confirmed by the different networks and functions that dominate each site.
Differential Gene Expression between Primary HNSCC Tumors and Normal Adjacent Mucosa
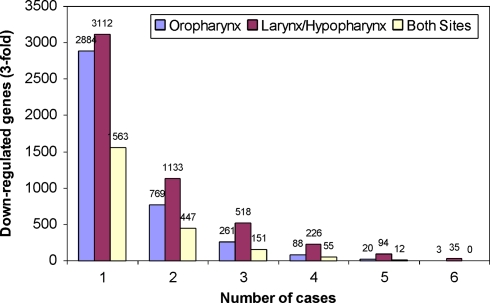
Given the site-specific nature of gene expression patterns and their ability to distinguish aggressive from non-aggressive disease, we decided to test whether or not a similar phenomenon was apparent when comparing primary tumors to corresponding normal adjacent mucosa from the same patient. This approach has historically been utilized in the identification of differentially expressed genes associated with carcinogenesis [11, 14, 15]. Drawing from the same patient population as for previous analyses, we selected 12 patients (6 OP cases, 6 LH cases) and utilized the same cDNA microarray technology to compare primary HNSCC tumor RNA (Cy3-green) to normal adjacent mucosa (Cy5-red) from the same patient. Using the 6 OP cases, we identified 261 genes that were down-regulated at least 3-fold in the primary tumor compared to the adjacent mucosa for at least 3 of the 6 OP patients (Fig. 8). A similar analysis identified 518 genes that were down-regulated at least 3-fold in the primary tumor compared to the adjacent mucosa for at least 3 of the 6 LH patients. We then overlapped these datasets in order to distinguish genes commonly down-regulated in both sites, compared to those specific to a given site. A total of 151 genes were identified as present in both datasets. This represented 58% of the genes identified in the OP dataset, and 29% of genes identified in the LH dataset of down-regulated genes. This contrasted greatly with what was observed in the previous analysis for aggressive versus non-aggressive disease, where only a single gene (KLF12) was common to both the oropharyngeal and laryngeal datasets (Fig. 4). Taken together, these data suggest that progression from normal mucosa to squamous cell carcinoma involves a common set of genes and, in contrast, local recurrence is dependent upon unique sets of genes involving the tumor-host microenvironment found at the various anatomic sites.
Fig. 8.
Numbers of down-regulated genes identified in comparisons of primary HNSCC to corresponding adjacent mucosa using the 27,323 element cDNA microarray. Bars indicate the number of genes down-regulated at least 3-fold in at least one patient through all six cases for a given anatomic site. Both oropharynx and larynx/hypopharynx cases are shown, as well as the number of genes that are found in both datasets for a given frequency
Discussion
Squamous cell carcinomas from various anatomic sites in the head and neck frequently behave differently, with different patterns of invasion and metastasis, treatment paradigms, and survival statistics. For instance, oral cavity tumors are primarily treated surgically, while adjacent oropharyngeal cancers are often treated with radiation or chemotherapy and radiation. Some authors evaluate tumor volume as a primary indicator of radioresponsiveness, although it does not hold true across all head and neck sites [16]. Proposed reasons for such clinical differences include lymphatic drainage patterns, mobility of the tissue treated, radiosensitivity, or undefined cellular or stromal differences resulting in unique tumor-stromal interactions in the various sites.
The data presented in this paper extend those well known clinical site-specific differences to differences in global gene expression patterns associated with aggressive disease. Different experimental designs were used to ask specific questions regarding the ability to identify signatures significantly associated with clinical outcome. The first question asked if aggressive and non aggressive tumor samples pooled across all three sites would generate the same discriminating genes when compared to gene sets produced by comparing aggressive and non aggressive tumor pools representing individual sites. When examining differences obtained by pooling RNA from sub-populations of tumors (aggressive versus non-aggressive phenotype), it was clear that more differences were detectable when each anatomic site was independently evaluated. Furthermore, the vast majority of genes identified in this manner were observed exclusively in one anatomic site. Only matrix metalloproteinase 10 (MMP10), the iron-binding protein lactotransferrin (LTF), and the serine protease inhibitor serpin A3 (SERPINA3) showed increased expression in the aggressive phenotypes independent of site. Recent work has established the increased expression of MMP10 in oral tongue squamous cell cancer, non-small cell lung cancer (NSCLC), and gastric cancer [17–19]. In the case of gastric cancer, levels of MMP10 were correlated with poor prognosis in advanced gastric cancer [19]. From the down-regulated genes, only a single gene (WIF-1) was seen as down-regulated in data from all three anatomic sites. Silencing of WIF-1 expression due to promoter hypermethylation is known to be an important mechanism underlying the activation of the Wnt signaling pathway in several solid tumors, including nasopharyngeal (NPC) and esophageal squamous cell (ESCC) carcinomas [20, 21].
Because the analysis of pooled tumor samples yielded the most discriminating results by analyzing each site separately, the next question asked if comparing individual tumor samples to a pool of site-specific tumor samples would yield a signature that would better discriminate aggressive from non aggressive tumor behavior. The concept behind this design is that all elements common to a tumor in a particular site would be eliminated and only those molecules distinguishing aggressive behavior would be revealed. When genes were ranked according to their ability to differentiate between the aggressive and non-aggressive disease subtypes for each anatomic site, we isolated molecular signatures of 66 genes, 176 genes and 95 genes for oropharyngeal, oral cavity, and laryngeal datasets, respectively. Of these three respective signatures, not a single gene was identified which was common to all three. Furthermore, only two genes coding for Kruppel-like factor 12 (KLF12) and hypothetical protein FLJ21272 were observed to be common to any two datasets out of the three. Loss of KLF12 in the aggressive phenotypes is intriguing, especially given its chromosomal location within 13q21, a common region of deletion in human cancers [22].
We subjected the three signatures to analysis with IPA to generate networks of interacting genes with potential functions associated with aggressive or non aggressive behavior. The genes within the signatures (focus molecules) may provide a function that is causally related to tumor behavior. Some of these genes were noted in the results section. Alternatively, the focus molecules may reflect changes in pathway activation where the pathway is causally related to tumor behavior and not the particular gene whose expression has changed. If we consider the networks presented for each anatomic site in Figs. 5–7, some overlap in nodal molecules (e.g. P38MAP kinase and NFκB) is present as expected. However, each site has nodes that appear to be significantly different such as: oral cavity—TGFβ, metalloproteinases, IL1 (Fig. 5); oropharynx—PKC, IL8, FGFR1 (Fig. 6); larynx/hypopharynx—IL6, TP53, PRLR, steroid hormones (Fig. 7). The differences in the discriminating molecules and the pathways activated at each site may reflect unique aspects of the tumor, the stroma or the tumor–stroma interaction. It should also be pointed out that such tumors can progress both genetically and behaviorally. However, our results suggest an initial dynamic reflected by our measurements that is associated with clinical outcome. This difference is in contrast to the large number of differentially expressed genes shared by multiple anatomic sites that are associated with the transformation from normal to tumor.
Overall, the results obtained by gene expression profiling revealed that differences in gene expression related to aggressiveness of HNSCC disease are highly site-specific. It is therefore plausible that specific biological mechanisms underlying tumor aggressiveness are heavily influenced by the anatomic site of the primary tumor, such that different mechanisms offer advantage only within the specific environment of a single anatomic site. While we have identified fundamental differences in gene expression distinguishing tumor behavior at each site, subsequent studies are required to elucidate the mechanism that each feature contributes to tumor behavior. In addition, the signatures themselves will need to be validated on a new set of patients to demonstrate usefulness.
Acknowledgments
The authors would like to thank Aldo Massimi and the Albert Einstein College of Medicine Microarray Facility for their assistance. This study was supported by a grant from the US National Cancer Institute, CA104402 (to TJB).
References
- 1.Lindberg R. Distribution of cervical lymph node metastases from squamous cell carcinoma of the upper respiratory digestive tracts. Cancer. 1972;29:1446–9. doi: 10.1002/1097-0142(197206)29:6<1446::AID-CNCR2820290604>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 3.Carvalho AL, Nishimoto IN, Califano JA, Kowalski LP. Trends in incidence and prognosis for head and neck cancer in the United States: a site-specific analysis of the SEER database. Int J Cancer. 2005;114:806–16. doi: 10.1002/ijc.20740. [DOI] [PubMed] [Google Scholar]
- 4.Forestiere AA, Goepfert H, Maor M, et al. Concurrent chemotherapy and radiotherapy for organ preservation in advanced laryngeal cancer. N Engl J Med. 2003;349:2091–8. doi: 10.1056/NEJMoa031317. [DOI] [PubMed] [Google Scholar]
- 5.Koch WM, Lango M, Sewell D, Zahurak M, Sidransky D. Head and neck cancer in nonsmokers: a distinct clinical and molecular entity. Laryngoscope. 1999;109:1544–51. doi: 10.1097/00005537-199910000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Sturgis EM, Wei Q, Spitz MR. Descriptive epidemiology and risk factors for head and neck cancer. Semin Oncol. 2004;31:726–33. doi: 10.1053/j.seminoncol.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 7.Smith EM, Wang D, Kim Y, et al. P16INK4a expression, human papillomavirus, and survival in head and neck cancer. 1. . Oral Oncol. 2008;44(2):133–42. doi: 10.1016/j.oraloncology.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 8.Li W, Thompson CH, O’Brien CJ, et al. Human papillomavirus positivity predicts favourable outcome for squamous carcinoma of the tonsil. Int J Cancer. 2003;106(4):553–8. doi: 10.1002/ijc.11261. [DOI] [PubMed] [Google Scholar]
- 9.Forastiere A, Koch W, Trotti A, Sidransky D. Head and neck cancer. N Engl J Med. 2001;345:1890–900. doi: 10.1056/NEJMra001375. [DOI] [PubMed] [Google Scholar]
- 10.Choi P, Chen C. Genetic expression profiles and biologic pathway alterations in head and neck squamous cell carcinoma. Cancer. 2005;104(6):1113–28. doi: 10.1002/cncr.21293. [DOI] [PubMed] [Google Scholar]
- 11.Belbin TJ, Singh B, Smith RV, et al. Molecular profiling of tumor progression in head and neck cancer. Arch Otolaryngol Head Neck Surg. 2005;131:10–8. doi: 10.1001/archotol.131.1.10. [DOI] [PubMed] [Google Scholar]
- 12.Dudoit S. Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. U. C. Berkeley Department of Statistics Technical Report #578. http://www.stat.berkeley.edu/tech-reports/578.ps.Z, 2000.
- 13.Eisen MB, Spellman PT, Brown PO, et al. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leethanakul C, Patel V, Gillespie J, et al. Distinct pattern of expression of differentiation and growth-related genes in squamous cell carcinoma of the head and neck revealed by the use of laser capture microdissection and cDNA arrays. Oncogene. 2000;19:3220–4. doi: 10.1038/sj.onc.1203703. [DOI] [PubMed] [Google Scholar]
- 15.Villaret DB, Wang T, Dillon D, et al. Identification of genes overexpressed in head and neck squamous cell carcinoma using a combination of complementary DNA subtraction and microarray analysis. Laryngoscope. 2000;110:374–81. doi: 10.1097/00005537-200003000-00008. [DOI] [PubMed] [Google Scholar]
- 16.Mukherji SK, Schmalfuss IM, Castelijns J, et al. Clinical applications of tumor volume measurements for predicting outcome in patients with squamous cell carcinoma of the upper aerodigestive tract. Am J Neuroradiol. 2004;25:1425–32. [PMC free article] [PubMed] [Google Scholar]
- 17.Ye H, Yu T, Temam S, et al. Transcriptomic dissection of tongue squamous cell carcinoma. BMC Genomics. 2008;9:69. doi: 10.1186/1471-2164-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin TS, Chiou SH, Wang LS, et al. Expression spectra of matrix metalloproteinases in metastatic non-small cell lung cancer. Oncol Rep. 2004;12:717–23. [PubMed] [Google Scholar]
- 19.Aung PP, Oue N, Mitani Y, et al. Systematic search for gastric cancer-specific genes based on SAGE data: melanoma inhibitory activity and matrix metalloproteinase-10 are novel prognostic factors in patients with gastric cancer. Oncogene. 2006;25:2546–57. doi: 10.1038/sj.onc.1209279. [DOI] [PubMed] [Google Scholar]
- 20.Wissmann C, Wild PJ, Kaiser S, et al. WIF1, a component of the Wnt pathway, is down-regulated in prostate, breast, lung, and bladder cancer. J Pathol. 2003;201:204–12. doi: 10.1002/path.1449. [DOI] [PubMed] [Google Scholar]
- 21.Chan SL, Cui Y, Hasselt A, et al. The tumor suppressor Wnt inhibitory factor 1 is frequently methylated in nasopharyngeal and esophageal carcinomas. Lab Invest. 2007;87:644–50. doi: 10.1038/labinvest.3700547. [DOI] [PubMed] [Google Scholar]
- 22.Chen C, Brabham WW, Stultz BG, et al. Defining a common region of deletion at 13q21 in human cancers. Genes Chromosomes Cancer. 2001;31:333–44. doi: 10.1002/gcc.1152. [DOI] [PubMed] [Google Scholar]