While nosocomial infections by Staphylococcus epidermidis have gained much attention, this skin colonizer has apparently not evolved to cause disease, but maintain the commonly benign relationship with its host. Accordingly, S. epidermidis does not produce aggressive virulence determinants. Rather, factors that normally sustain the commensal lifestyle of S. epidermidis seem to rise to additional benefit during infection. Furthermore, we are beginning to comprehend the roles of S. epidermidis in balancing the epithelial microflora and serving as a reservoir of resistance genes. In this review, the molecular basis of the commensal and infectious lifestyles of S. epidermidis will be discussed.
Whereas previously only regarded as an innocuous commensal microorganism on the human skin, Staphylococcus epidermidis is nowadays seen as an important opportunistic pathogen. It is now the most frequent cause of nosocomial infections, at a rate about as high as that due to its more virulent cousin Staphylococcus aureus1. In particular, S. epidermidis represents the most common source of infections on indwelling medical devices. This likely stems from the fact that S. epidermidis is a permanent and ubiquitous colonizer of human skin, and the resulting high probability of device contamination during insertion2. While S. epidermidis infections only rarely develop into life-threatening diseases, their frequency and the fact that they are extremely difficult to treat represent a serious burden for the public health system. The costs related to vascular catheter-related bloodstream infections caused by S. epidermidis amount to an estimated $ 2 billion annually in the United States alone3–5. Treatment is complicated by specific antibiotic resistance genes and the formation of biofilms, multicellular agglomerations that have intrinsic resistance to antibiotics and mechanisms of host defense3. Furthermore, recent investigation has identified specific molecular determinants facilitating S. epidermidis immune evasion and ability to cause chronic disease. Interestingly, many of these determinants are believed to have original functions in the non-infectious lifestyle of this microorganism, emphasizing the accidental nature of S. epidermidis infections. A better understanding of S. epidermidis physiology not only during infection, but also in its commensal status is urgently needed to evaluate therapeutic strategies against S. epidermidis.
S. epidermidis – the species
Staphylococci are common bacterial colonizers of the skin and mucous membranes of humans and other mammals4. S. epidermidis in particular is the most frequently isolated species from human epithelia. It colonizes predominantly the axillae, head, and nares5. Analysis of the S. epidermidis genome indicated that the species is well equipped with genes assumed to provide protection from the harsh conditions encountered in its natural habitat9, 10. For example, to cope with extremes of salt concentration and osmotic pressure, S. epidermidis has eight sodium ion/proton exchangers and six transport systems for osmoprotectants9.
S. epidermidis belongs to the group of coagulase-negative staphylococci (CoNS), which is distinguished from coagulase-positive staphylococci such as S. aureus by lacking the enzyme coagulase. The species shows a high degree of diversity with 74 identified sequence types (STs)6. Most isolates belong to clonal complex (CC) 2, which comprises the most frequently isolated ST2. Possibly, the successful spread of ST2 may be due to the fact that all ST2 isolates contain IS256 insertion sequences and ica genes7, two factors found correlated with S. epidermidis invasiveness13–16. In addition, most ST2 isolates show in vitro capacity to form biofilms7. Genome information is available for two strains of S. epidermidis: the biofilm-negative ATCC122288 and the biofilm-positive clinical isolate RP62A9. Of note, no genome sequence is available yet for an isolate of the most frequently found and potentially most invasive ST2.
An opportunistic pathogen
As part of the human epithelial microflora, S. epidermidis usually has a benign relationship with its host. Furthermore, it has been proposed that S. epidermidis may have a probiotic function by preventing colonization of more pathogenic bacteria such as S. aureus17. However, there is no clear evidence indicating that S. epidermidis secretes factors that impact colonization of other microorganisms in vivo.
In contrast to the relatively scarce information on the non-infectious lifestyle of S. epidermidis, S. epidermidis infections and mechanisms by which S. epidermidis promotes disease have gained much interest. Among CoNS, S. epidermidis clearly causes the greatest number of infections2, 9. In clinical microbiology, CoNS are often not further specified, as the major interest is in making a distinction between S. aureus and other staphylococci. However, based on reports that have performed species identification1, 5, one can assume that the vast majority of non-specified CoNS infections are due to S. epidermidis. Particularly, S. epidermidis represents the most frequent causative agent involved with infections of any type of indwelling medical devices, such as peripheral or central intravenous catheters (CVCs)9. These infections usually commence with the introduction of bacteria from the skin of the patient or that of health care personnel during device insertion and have increased in number most likely owing to the increased use of such devices1, 18. S. epidermidis now accounts for at least 22% of bloodstream infections in intensive care unit patients in the USA, which occur in at least 4–5/1000 CVC insertions1, 18. In addition to the abundance of S. epidermidis on the skin, this high frequency is likely due to elaborate mechanisms to colonize catheter surfaces, which will be discussed later in this article. Furthermore, S. epidermidis may be involved in prosthetic joint, vascular graft, surgical site, central nervous system shunt, and cardiac device infections9. Last but not least, second only to S. aureus, S. epidermidis causes ~ 13% of prosthetic valve endocarditis (PVE) infections, with a high rate of intracardiac abscesses (38%) and 24% mortality10. However, PVE and other serious complications are rare among S. epidermidis infections, which altogether may be characterized as predominantly subacute and chronic.
The fact that S. epidermidis usually does not cause severe infections raises the interesting question why it is advantageous for this species – as opposed to its virulent cousin S. aureus – to maintain a low level of virulence. Massey et al. have developed a mathematical model outlining that for a strain with a high level of asymptomatic transmission such as S. epidermidis, avirulent strains out-compete virulent strains – in contrast to S. aureus, for which asymptomatic transmission is low and virulent strains out-compete avirulent strains11. This model is based on the assumption that S. epidermidis is more readily transmissible than S. aureus. The authors explain this by (i) the widespread colonization of S. epidermidis on human epithelia, while S. aureus almost exclusively colonizes the nares, (ii) colonization of all humans with S. epidermidis in contrast to S. aureus, which is only found in some individuals, and (iii) specific genetic factors involved in colonization and bacterial interference, such as cross-inhibiting quorum-sensing signals (Box 1). However, while quorum-sensing interference favors at least one subtype of S. epidermidis over S. aureus in vitro21, 22, there is no evidence that it plays a role in vivo17.
The staphylococcal accessory gene regulator (agr) quorum-sensing system and cross-inhibition by agr autoinducing peptides (AIPs)
Quorum-sensing in staphylococci is accomplished by the agr system, which consists of an AIP precursor peptide maturation and export enzyme (AgrB) and a two-component signal transduction system (AgrC, AgrA)75. Quorum-sensing-controlled target genes of agr are regulated directly by the DNA-binding protein AgrA or via the regulatory RNAIII139, 140. AIPs (or pheromones) are 7 to 9 amino acids in length and have a conserved cysteine residue, whose sulfhydryl group reacts with the C-terminal carboxy group to form a thiolactone that is essential for activity141, 142. Binding of the AIP to AgrC stimulates AgrC to auto-phosphorylate, which in turn leads to phosphorylation and activation of AgrA. AgrA activates the P2 promoter controlling expression of agrBDCA, thereby closing the quorum-sensing circuit. It also activates the P3 promoter that drives expression of RNAIII and the embedded PSM, δ-toxin (hld).
In general, AIPs of self activate, whereas AIPs of non-self (different species or subgroups) inhibit the agr response, unless the groups are closely related (e.g. S. aureus agr types I and IV)138, 143. The S. epidermidis agr type I is by far the most frequently isolated type from infections. The AIP of S. epidermidis agr type I inhibits all S. aureus agr types except for the rare type IV, while only S. aureus type IV inhibits S. epidermidis type I76. Interference by quorum-sensing cross-inhibition between S. aureus and S. epidermidis seems therefore to be in favor of S. epidermidis, but it is not known whether this plays a role during colonization in vivo.
In accordance with the low virulence potential of S. epidermidis and the Massey et al. model, the following paragraphs will show that S. epidermidis is well equipped with determinants promoting persistence, such as immune evasion molecules, rather than those which aggressively attack the host, such as toxins.
Evasion of host defenses
Pathogen survival in the human body requires evasion of host defenses. While a limited subset of host defense mechanisms such as antimicrobial peptides (AMPs) are present on the human skin12, S. epidermidis has to cope with multiple additional mechanisms of host defense after penetration through the epithelial barrier. The innate immune system reacts first and in a rather non-specific way to any invading microorganism, including S. epidermidis. For example, as a key part of innate host defense, neutrophils ingest bacteria and kill them using reactive oxygen species and AMPs13. S. epidermidis has several mechanisms to evade being ingested and killed by neutrophils, as outlined below.
The role of the specific, acquired immune response to S. epidermidis infection is less well understood. The fact that our immune system has difficulties clearing long-lasting S. epidermidis infections despite production of antibodies against S. epidermidis proteins25 indicates that acquired host defense may not be very efficient against S. epidermidis. This may be due in part to S. epidermidis exopolymers that protect from antibody recognition. Furthermore, our immune system may have evolved not to react in a strong manner to prevalent colonizing bacteria.
Biofilm formation
Biofilms are multicellular, surface-attached agglomerations of microorganisms. They have a characteristic physiology and architecture that form the basis of biofilm resistance to many antibiotics and mechanisms of host defense3. In accordance with this general notion, S. epidermidis shows significant, genome-wide adaptation to the biofilm mode of growth including down-regulation of basic cell processes such as nucleic acid, protein and cell wall biosyntheses14. These gene regulatory changes may explain limited activity of many antibiotics that target actively growing cells, such as penicillins15, aminoglycosides16, and quinolones17, against S. epidermidis biofilms.
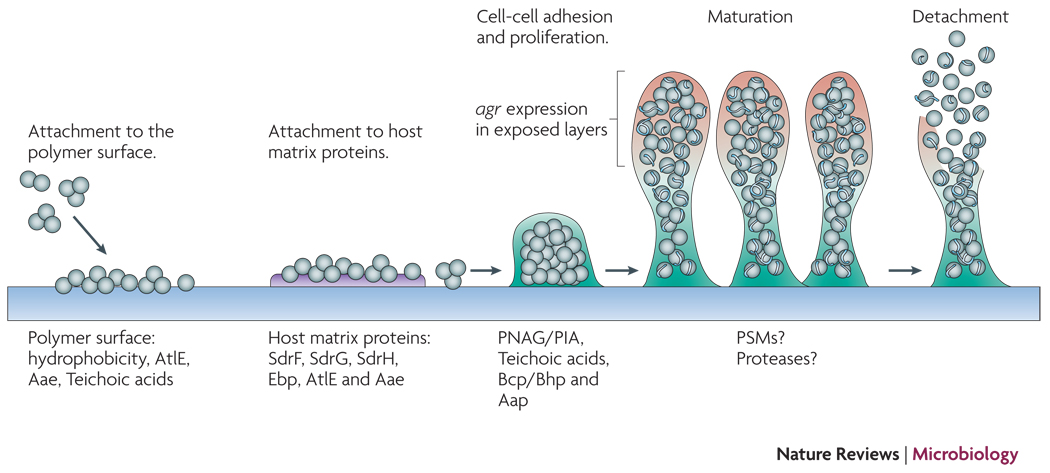
Biofilm formation proceeds via initial adhesion and subsequent aggregation into multicellular structures (Fig. 1). Thus, the development of a biofilm requires adhesive forces for the colonization of surfaces and the interaction of cells among each other. Disruptive forces are needed for the formation of fluid-filled channels that are important for nutrient delivery to all biofilm cells and give the mature biofilm its typical three-dimensional structure. Disruptive forces are also involved in the detachment of cell clusters from the biofilm, which limits biofilm expansion and may lead to the dissemination of infection18.
Figure 1. Biofilm development in S. epidermidis.
Attachment to uncoated material is mainly dependent on cell surface hydrophobicity, while dedicated surface proteins mediate adhesion to host matrix-covered devices. Afterwards, exopolysaccharide, specific proteins, and accessory macromolecules provide intercellular aggregation. Mechanisms of biofilm maturation, structuring, and detachment are poorly understood, but possibly involve quorum-sensing controlled expression of detergent-like peptides and proteolytic activity in exposed layers of the biofilm. Genome-wide gene expression is significantly different in the biofilm compared to the planktonic mode of growth and includes down-regulation of basic cell processes.
Adhesion to abiotic surfaces such as catheters is mainly governed by bacterial cell surface hydrophobicity19. Specific proteins that impact surface adhesion in S. epidermidis, such as the abundant surface protein AtlE20, a bifunctional adhesin/autolysin, and the Bap/Bhp protein21 likely contribute to the hydrophobic character of the cell surface.
In vivo, matrix proteins fast cover abiotic surfaces such as those of indwelling medical devices. S. epidermidis has a vast array of surface proteins called MSCRAMMs (Microbial surface components recognizing adhesive matrix molecules) (Tab. 1) with the potential to interact with matrix proteins. MSCRAMMs may be covalently bound to the bacterial surface by sortase A22, or via yet incompletely understood non-covalent interaction with, probably, surface polymers such as teichoic acids23 (Fig. 2). Binding activity to fibrinogen and collagen has been demonstrated for the covalently anchored proteins SdrG and SdrF36, 37, respectively, and for the non-covalently bound autolysins AtlE and Aae, which show a less specific interaction and may bind to fibrinogen, fibronectin, and vitronectin32, 38.
Tab. 1.
Virulence factors of S. epidermidis
| Virulence factor | Gene | Function | Reference |
|---|---|---|---|
| Immune evasion | |||
| Biofilm formation | |||
| Primary attachment | |||
| (To abiotic surfaces) | |||
| AtlE | atlE | Bifunctional autolysin/adhesin Abundant protein impacting surface hydrophobicity |
20 |
| Aae | aae | Bifunctional autolysin/adhesin | 77 |
| Teichoic acids | (multiple biosynthetic genes) |
In S. aureus Impact attachment (via binding of autolysins?) |
78 |
| (To matrix proteins) | |||
| SdrF | sdrF | Binds to collagen | 36, 44 |
| SdrG (Fbe) | sdrG | Binds to fibrinogen | 79 |
| SdrH | sdrH | Putative binding function only | 39 |
| Ebp | ebp | Binds to elastin | |
| AtlE/Aae | atlE/aae | Bind to multiple matrix proteins | 32, 38 |
| Intercellular aggregation | |||
| PNAG/PIA | icaADBC | Intercellular polysaccharide adhesin | 49, 53 |
| Biofilm-associated protein Bap/Bhp |
bap/bhp | Intercellular protein adhesin | 21 |
| Accumulation-associated protein Aap |
aap | Intercellular protein adhesin precursor (requires proteolytic processing for activity) |
71, 72 |
| Teichoic acids | Component of biofilm matrix | 80 | |
| Protective exopolymers | |||
| PNAG/PIA | icaADBC | Protects from IgG, AMPs, phagocytosis, complement |
96, 97 |
| PGA | capABCD | Protects from AMPs, phagocytosis | 49 |
| Resistance to AMPs | |||
| SepA protease | sepA | AMP degradation | 81 |
| Dlt, MprF, VraFG |
dltABCD mprF vraFG |
In analogy to S. aureus D-alanylation of teichoic acids (Dlt), lysylation of phospholipids (MprF), putative AMP export (VraFG) |
110–112 |
| Aps system |
apsR (graR), apsS (graS), apsX |
AMP sensor, regulator of AMP resistance mechanisms |
59 |
| Toxins | |||
| Phenol-soluble modulins (PSMs) |
psmα,psmδ, psmε, hld, psmβ1, psmβ2 |
Pro-inflammatory cytolysins | 26, 87, 105 |
| Exoenzymes | |||
| Proteases | |||
| Cysteine protease (SspB, Ecp)(S.aureus staphopain homologue) |
sspB | Unknown, tissue damage? | 82 |
| Metalloprotease/elastase (SepA) |
sepA | Tissue damage?, lipase maturation, AMP resistance |
81, 120, 145 |
| (S.aureus aureolysin homologue) |
|||
| Glutamylendopeptidase (GluSE, SspA, Esp), serine protease |
sspA | Degradation of fibrinogen and complement factor C5 |
82, 83 |
| (S. aureus V8 protease homologue) |
|||
| Lipases GehC, GehD | gehC, gehD | Persistence in fatty acid secretions? | 146–148 |
| Others | |||
| Staphyloferrins |
sfna locus (S.aureus staphyloferrin A) |
Siderophores (iron acquisition) | 149, 150 |
| SitABC | sitABC | Iron importer | 151 |
| Fatty acid modifying enzyme (FAME) |
unidentified | Detoxification of bactericidal fatty acids | 83 |
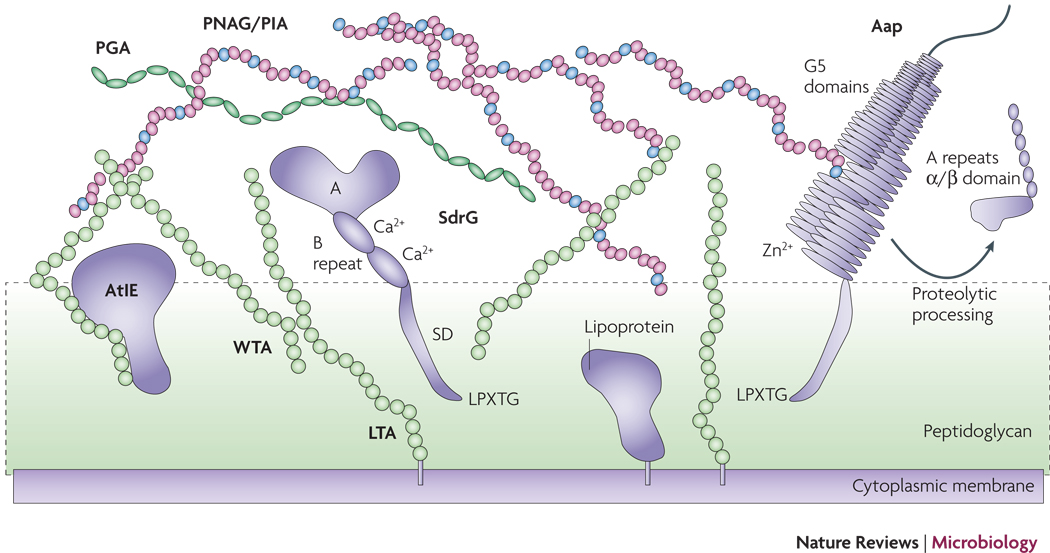
Figure 2. The S. epidermidis cell surface.
Proteins such as SdrG and Aap may be attached to the cell surface via sortase-catalyzed covalent anchoring. These proteins harbor a characteristic LPXTG motif at the C-terminus, of which the threonine residue is linked to peptidoglycan. Many autolysins such as AtlE are anchored non-covalently, likely via interaction with teichoic acids. Furthermore, lipoproteins are surface-attached via their fatty acid anchor that penetrates the cytoplasmic membrane. AtlE is a bifunctional adhesin/autolysin that contributres to biofilm formation by its surface hydrophobicity and to host matrix protein binding. SdrG is an example of the Sdr protein family of MSCRAMMs. It stretches the peptidoglycan layer by its SD repeat region and binds fibrinogen via its A region. The B repeats harbor a Ca2+ binding EF-hand domain. Aap proteins aggregate via Zn2+-dependent G5 domains and form fibrils that likely connect cells in the biofilm matrix. G5 domains also bind N-acetylglucosamine and may thus interact with the N-acetylglucosamine exopolysaccharide PNAG/PIA. PNAG/PIA is cationic and likely interacts with negatively charged surface polymers such as teichoic acids (lipoteichoic acids, LTA and wall teichoic acids, WTA) and poly-γ-glutamic acid (PGA).
The most intensively studied MSCRAMM of S. epidermidis is SdrG (Fbe), a fibrinogen-binding protein that belongs to the serine/aspartate (SD) repeat family. Three members of this family, SdrF, SdrG, and SdrH, are present in most strains of S. epidermidis39. SdrG has been described as necessary and sufficient to promote S. epidermidis adhesion to fibrinogen in vitro37, 40 and promotes CVC-associated infection in vivo24. SdrG binds to the thrombin cleavage site in the Bbeta chain of fibrinogen using a “dock, lock, and latch” mechanism42. This mechanism is believed to lead to a greatly stabilized MSCRAMM-ligand interaction. Emphasizing the importance of SdrG for S. epidermidis infection, expression of SdrG increases in an in vivo environment43 and antibodies to SdrG are present in human blood39. Recently, an important role during ventricular assist device driveline-related infection has also been demonstrated for SdrF25. In addition, several further S. epidermidis MSCRAMMs have been predicted and undergone preliminary characterization26, although their role in matrix protein binding and virulence is not yet understood.
After initial adhesion, biofilms develop via intercellular aggregation that is mediated by many different surface macromolecules. Among those, exopolysaccharide and some proteins appear be dedicated predominantly to the formation of the extracellular biofilm matrix. In addition, teichoic acids46, 47 and extracellular DNA originating from lysed cells27, may have accessory functions in aggregation, which are likely dependent on their polyanionic character (Fig. 1).
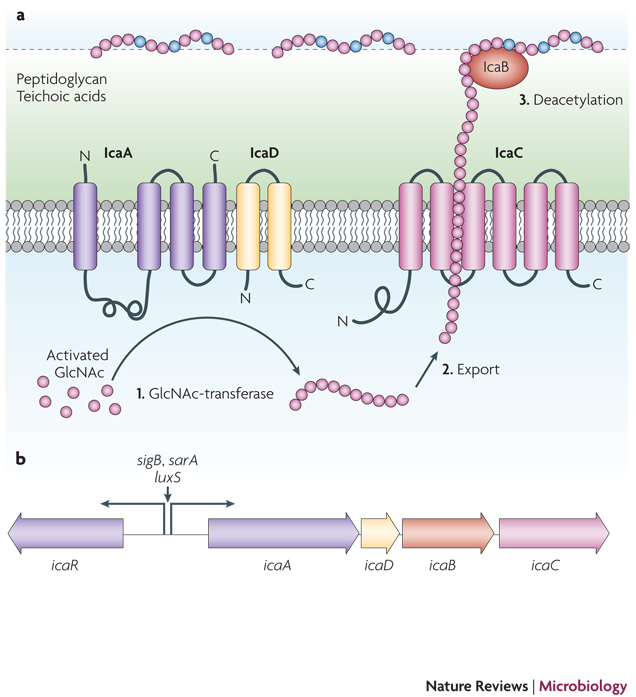
Many S. epidermidis strains produce a poly-N-acetylglucosamine (PNAG) homopolymer also named PIA (polysaccharide intercellular adhesin) that surrounds and connects S. epidermidis cells in a biofilm (Fig. 3)28. This polymer, which differs from other poly-N-acetylglucosamine polymers found in nature (such as chitin) by its β 1–6 linkage28, has recently also been detected in many other microorganisms including Yersinia pestis and Escherichia coli50, 51. Production of PNAG/PIA is crucial for biofilm formation in vitro52, 53 and has a significant impact on S. epidermidis biofilm-associated infection in most animal models 54–58. The biosynthesis of PNAG/PIA is accomplished by the gene products of the ica (intercellular adhesion) locus29. IcaA and IcaD produce a chain from activated N-acetylglucosamine monomers, whose elongation is dependent on the IcaC protein, likely due to the assumed exporter function of the latter30. Partial de-acetylation of the N-acetylglucosamine residues is accomplished by the cell surface-located enzyme IcaB after export31. De-acetylation introduces positive charges in the otherwise neutral polymer that are important for surface binding of PNAG/PIA and its multiple biological functions in biofilm formation and immune evasion discussed below31. Production of PNAG/PIA is subject to a variety of regulatory influences32, including by many global virulence regulators61–67, but excluding the quorum-sensing regulator agr33. While it is less well understood which environmental signals control PNAG/PIA expression, particularly in vivo, the complexity of regulation underpins the importance of PNAG/PIA for S. epidermidis pathophysiology.
Figure 3. The exopolysaccharide PNAG/PIA.
The immune evasion and biofilm aggregation exopolysaccharide PNAG/PIA, a partially de-acetylated β 1–6 linked N-acetylglucosamine homopolymer, is synthesized by the membrane-located N-acetylglucosamine transferase IcaA that needs the accessory IcaD membrane protein for activity. The growing PNAG/PIA chain is likely exported by the IcaC membrane protein. After export, the surface-located IcaB de-acetylase removes some of the N-acetyl groups, giving the polymer a cationic character that is essential for surface attachment. The Ica proteins are encoded in the ica gene locus, which contains the icaADBC operon and the icaR gene encoding a regulatory protein. Expression of the icaADBC operon is regulated either directly at the icaA promoter or via expression of IcaR by a series of global regulatory proteins. Furthermore, insertion and excision of the IS256 element may turn PNAG/PIA expression off and on.
More recently, it was recognized that PNAG/PIA is not absolutely essential for biofilm formation in all S. epidermidis strains, as biofilm formation has been demonstrated in strains lacking the ica genes34 and ica-negative S. epidermidis strains were isolated from biofilm-associated infection70. In some strains, biofilm formation may thus be mediated additionally or exclusively by specific surface proteins, namely Bap/Bhp21 and Aap35. The Aap protein requires proteolytic activation36 and zinc ions37 for its biofilm-promoting effect. Zn2+ is crucial for the modular association of so-called G5 tandem repeats37, which may underlie the formation of Aap-made fibril-like structures on the bacterial surface38 (Fig. 2). The same domains are known to interact with N-acetylglucosamine and thus potentially bind PIA/PNAG, forming a protein/polysaccharide biofilm network39. Based on the prevention of in vitro biofilm formation by a chelating agent, it has been suggested that biofilm formation in the strong biofilm forming strain S. epidermidis RP62A is solely dependent on Aap37. In support of this observation, monoclonal antibodies against Aap prevent biofilm formation in this strain40. However, this hypothesis is at variance with another report that did not find an impact of protein-mediated biofilm formation in the same strain41. Thus, the contribution of proteins to S. epidermidis biofilm formation and the involved mechanisms will certainly require intensive further investigation. In addition, the finding that biofilms solely made by proteins are not as robust as those with PNAG/PIA70 indicates that both proteins and exopolysaccharide participate in efficient S. epidermidis biofilm formation.
Biofilm detachment
In contrast to intercellular aggregation, S. epidermidis biofilm structuring and detachment are poorly understood. We know that biofilm detachment in S. epidermidis is controlled by the quorum-sensing system agr, as biofilms that are agr-dysfunctional produce thicker biofilms and have an obvious defect in detachment68, 78. In S. aureus a model has been proposed that involves agr expression at the exposed layers of a biofilm, promoting detachment of cell clusters from the biofilm surface, thereby controlling biofilm expansion42. Likewise, S. epidermidis agr activity is limited to the biofilm surface43, indicating a common staphylococcal mechanism of quorum-sensing-controlled biofilm detachment. Two detachment mechanisms have been proposed: enzymatic degradation of biofilm exopolymers and disruption of non-covalent interaction by detergent-like molecules (Fig. 1). With regard to enzymatic degradation of proteinaceous biofilm factors as suggested in S. aureus44, evidence for such a function of proteases in biofilm detachment in S. epidermidis has not been obtained. However, S. epidermidis produces a series of exoproteases with relatively low substrate specificity that may serve to degrade surface proteins81–83. As for degradation of biofilm exopolysaccharide, staphylococci do not appear to have a dedicated enzyme for PNAG/PIA hydrolysis in contrast to several other bacteria with PNAG/PIA production84, 85. Alternatively, detergent-like molecules may disrupt non-covalent such as electrostatic and hydrophobic interactions, as for example between the cationic PNAG/PIA and anionic surface polymers, or between hydrophobic parts of the bacterial surface. The short amphipathic phenol-soluble modulins (PSMs) that include the S. epidermidis δ-toxin have been proposed to have such a function45 (Fig. 4). Both S. epidermidis PSMs and exoproteases are strictly agr-regulated87, 88, lending support to the idea that they may be candidates for biofilm structuring activity.
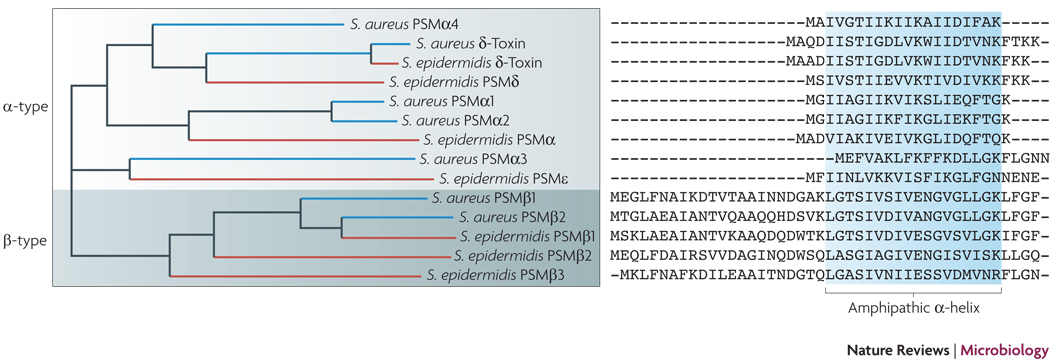
Figure 4. Phenol-soluble modulins.
Sequence alignment of S. epidermidis and S. aureus PSMs. PSMs serve as immune evasion molecules to their bacterial producer and, on the other hand, as PAMPs for pathogen recognition to the host. All PSMs contain an amphipathic α-helix and N-terminal N-formyl methionine, as they are secreted as the direct translational product without processing in an unknown manner. PSMs of the α-type are relatively short, ~ 20–25 amino acids. Particularly the S. aureus PSMα peptides 1 through 4 are strongly cytolytic. PSMs of the β-type are longer, ~ 45 amino acids, and do not have considerable cytolytic activity. Only the δ-toxin, an α-type PSM with moderate cytolytic activity, and the β-type PSMs are secreted by S. epidermidis in large amounts. Although part of the psmβ operon, the PSMβ3 peptide is not found in S. epidermidis culture filtrates for unknown reasons. The psmβ1 gene is duplicated in some strains of S. epidermidis.
In general, knowledge about the molecular mechanisms of biofilm formation and its regulation in S. epidermidis is almost exclusively based on in vitro research. The contribution to pathogenesis of some determinants such as PNAG/PIA54–58, AtlE46, Fbe (SdrG)24, SdrF25, and the regulators agr43, luxS47, and sigB90 has been demonstrated using animal models. Furthermore, there is evidence indicating that important biofilm factors are expressed in vivo58, 91. Nevertheless, there is an urgent need for more detailed in vivo research providing mechanistic insight into S. epidermidis biofilm-associated infection. A recently constructed bioluminescent strain of a biofilm-forming clinical isolate of S. epidermidis may be helpful in these endeavors48.
Protective exopolymers
S. epidermidis produces exopolymers, namely poly-γ-glutamic acid (PGA) and PNAG/PIA, that protect from important mechanisms of innate host defense. The pseudopeptide polymer PGA, which is synthesized by the gene products of the cap locus, is crucial for S. epidermidis resistance to neutrophil phagocytosis and AMPs, despite comparatively low production49. Except for Bacillus anthracis50, S. epidermidis is the only organism known so far in which PGA has a function in pathogenesis. Furthermore, PGA promotes growth of S. epidermidis at high salt concentrations and is induced under these conditions49. This is reminiscent of PGA production in many halophilic bacteria, where PGA is believed to contribute to osmotolerance51, and indicates a role of PGA during S. epidermidis colonization. Finally, expression of the cap genes appears to be increased during the biofilm mode of growth14. Interestingly, PGA is present in many CoNS, but absent from S. aureus9.
In addition to its role as part of the extracellular biofilm matrix, the already described exopolysaccharide PNAG/PIA has been found to protect S. epidermidis from neutrophil killing, complement deposition, immunoglobulins, and AMPs96, 97, and from Caenorhabditis elegans immune defenses in a nematode infection model98. The cationic PNAG/PIA protects from AMPs of cationic and anionic charge, indicating that its mechanism of action may not be limited to electrostatic repulsion of AMPs of the same charge52. It may thus also work by sequestering oppositely charged AMPs in a way similar to the proposed mechanism of protection from tobramycin by Pseudomonas aeruginosa alginate53.
Pathogen-associated molecular patterns
Pathogen-associated molecular patterns (PAMPs) are structures on the bacterial surface that the innate immune system recognizes as non-self via dedicated pathogen recognition receptors (PRRs), such as the Toll-like receptors (TLRs)52. Recognition of PAMPs acivates host defense mechanisms that include phagocytosis and cytokine release54. PAMPs such as lipoproteins and lipoteichoic acids are common in Gram-positive bacteria. Furthermore, there are reports suggesting that several additional molecules that are specific to S. epidermidis may stimulate innate host defense. For example, PNAG/PIA was reported to stimulate the Toll-like receptor 2 (TLR2)55. Recognition of PIA/PNAG by the human immune system would constitute an interesting example of the hide-and-seek interplay between pathogen and host, as a substance that S. epidermidis uses for immune evasion would trigger innate host defense mechanisms. However, this has not been confirmed using genetic deletion mutants, which is important to rule out that contaminating strongly pro-inflammatory substances such as lipoproteins were the basis of the observed effect, a situation that has led to frequent misidentification of alleged TLR2 stimulators102–104. Similarly, pro-inflammatory capacities of S. epidermidis PSMs56 have not yet been confirmed using synthetic peptides or gene deletion mutants. However, their similarity to S. aureus PSMs, for which activity has been confirmed57, indicates that the described pro-inflammatory effect of S. epidermidis PSMs is genuine; although activation of TLR2 by PSMs107 needs verification. Finally, an unusual short-chain pro-inflammatory lipoteichoic acid has been described in S. epidermidis58. However, the chemical characterization of the purified molecule does not indicate a teichoic acid-related polymer, and thus the identity of this molecule and the reported pro-inflammatory activity certainly require confirmation. Thus, there is a clear need for further characterization of S. epidermidis molecules that activate host defenses.
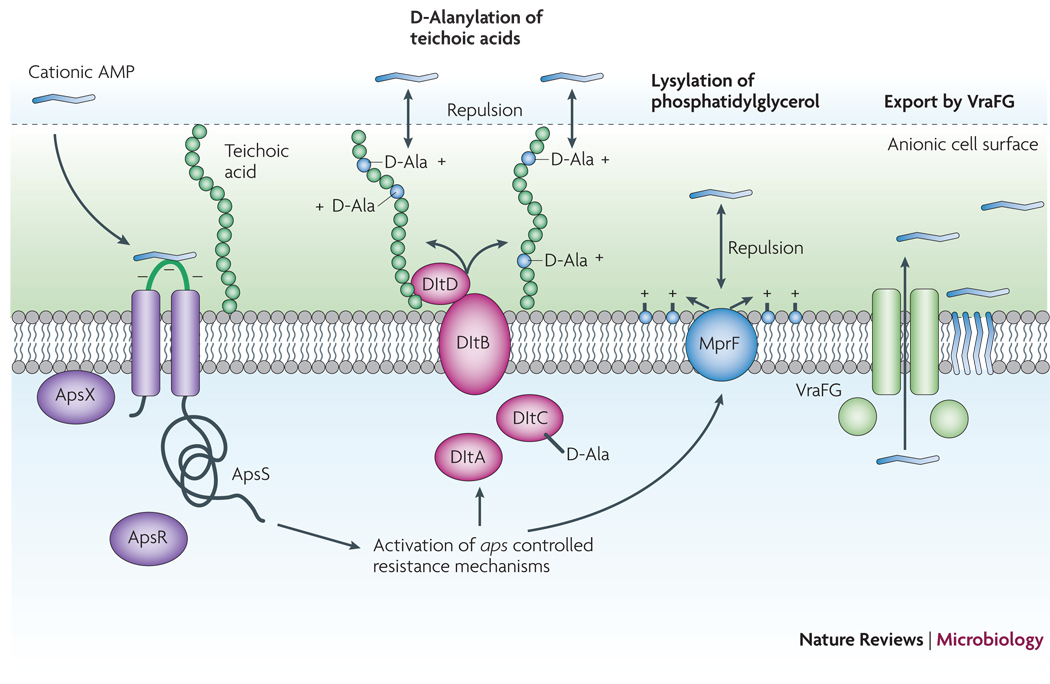
Sensing of antimicrobial peptides
In a way similar to the recognition of S. epidermidis PAMPs by the human immune system, S. epidermidis has mechanisms to sense the presence of harmful molecules produced by the host. Specifically, an AMP-sensing system has been identified, termed aps, that is activated by a variety of AMPs and triggers up-regulation of staphylococcal AMP-defensive systems59, including D-alanylation of teichoic acids60, lysylation of phospholipids by the MprF enzyme111, and the VraFG proteins61 (Fig. 5) The former two mechanisms decrease the anionic charge of the bacterial surface, thus preventing efficient attraction of cationic AMPs, while the latter likely function as an AMP exporter, removing AMPs from the cytoplasmic membrane. Thus, the Aps system – the first example of an AMP sensor in Gram-positive bacteria - has a function similar to the Gram-negative PhoP/PhoQ AMP sensor113, but is not evolutionarily related. Importantly, activation and protective response of the aps system is limited to cationic AMPs. Furthermore, the aps system represents an exceptional example of a 3-component sensor/regulator that contains an essential component of unknown function, ApsX, in addition to the classical components of a two-component system, the histidine kinase ApsS (GraS) and the response regulator protein, ApsR (GraR).
Figure 5. The Aps antimicrobial peptide sensor/regulator.
Cationic AMPs attach to the negatively charged bacterial surface and membrane by electrostatic interaction, a prerequisite for AMP antimicrobial activity, which is often based on pore formation in the bacterial cytoplasmic membrane. The S. epidermidis ApsS AMP sensor has one short extracellular loop with a high density of negatively charged amino acid residues that interacts with cationic AMPs. Transduction of this signal via ApsS and the accessory, essential ApsX, which has a yet unknown function, triggers expression of key AMP resistance mechanisms. The D-alanylation of teichoic acids, encoded by the products of the dlt operon, and lysylation of phosphatidylglycerol, catalyzed by the MprF enzyme, result in a decreased negative charge of the cell surface and membrane, respectively, leading to decreased attraction, or repulsion, of cationic AMPs. The VraFG ABC transporter also promotes resistance to AMPs and likely functions as an AMP exporter.
Toxins
In S. aureus and many other bacteria, toxins are the most important contributors to aggressive virulence. In contrast to the vast toxin repertoire of S. aureus, S. epidermidis toxin production is mostly limited to PSMs. While strain-specific production of enterotoxins has been described114, 115, S. epidermidis is not generally accepted as an enterotoxin producer. In contrast, all except naturally agr dysfunctional S. epidermidis strains produce PSMs68, 87 (Fig. 4). These already mentioned peptides characteristically are short, amphipathic, and α-helical and have pro-inflammatory and sometimes cytolytic function. S. epidermidis δ-toxin (also called PSMγ), a 24-amino acid peptide that differs from its S. aureus homologue only in one amino acid position, has been suggested to be involved in necrotizing enterocolitis in neonates62. Some S. epidermidis PSMs are related to S. aureus PSMs that have pronounced capacity to lyse human neutrophils57. However, the PSM production pattern in S. epidermidis shows strong production solely of the moderately cytolytic δ-toxin and non-cytolytic β-type PSMs14. Thus, the PSM production pattern, in addition to the general absence of highly aggressive toxins in S. epidermidis, contrasts the high cytolytic potential of S. aureus. This underpins the Massey et al.11 model proposing an evolutionary advantage for low aggressiveness of S. epidermidis.
Colonization and pathogenesis
Several studies have attempted to identify determinants that distinguish S. epidermidis strains that may cause infection from those that live on the skin. These studies focused on putative virulence determinants or used genome-wide approaches such as comparative genomic hybridization14–16, 117. Two main putative determinants of S. epidermidis invasiveness were identified in these studies: the ica genes encoding production of PNAG/PIA and the insertion element IS256. IS256 is believed to contribute to genetic adaptation that may play a role during infection63. For example, it may serve to abolish production of PNAG/PIA or function of the agr global virulence regulator by inserting into the ica or agr loci, respectively78, 118. As for PNAG/PIA, correlation with invasiveness may be due to the roles of this exopolymer in biofilm formation and immune evasion. In addition, results from a human colonization model indicate that ica-negative strains may even have a selective advantage over ica-positive strains on the skin64. However, there is also evidence suggesting that when corrected for clonal relatedness, there may not be any differences between commensal and infectious strains 117.
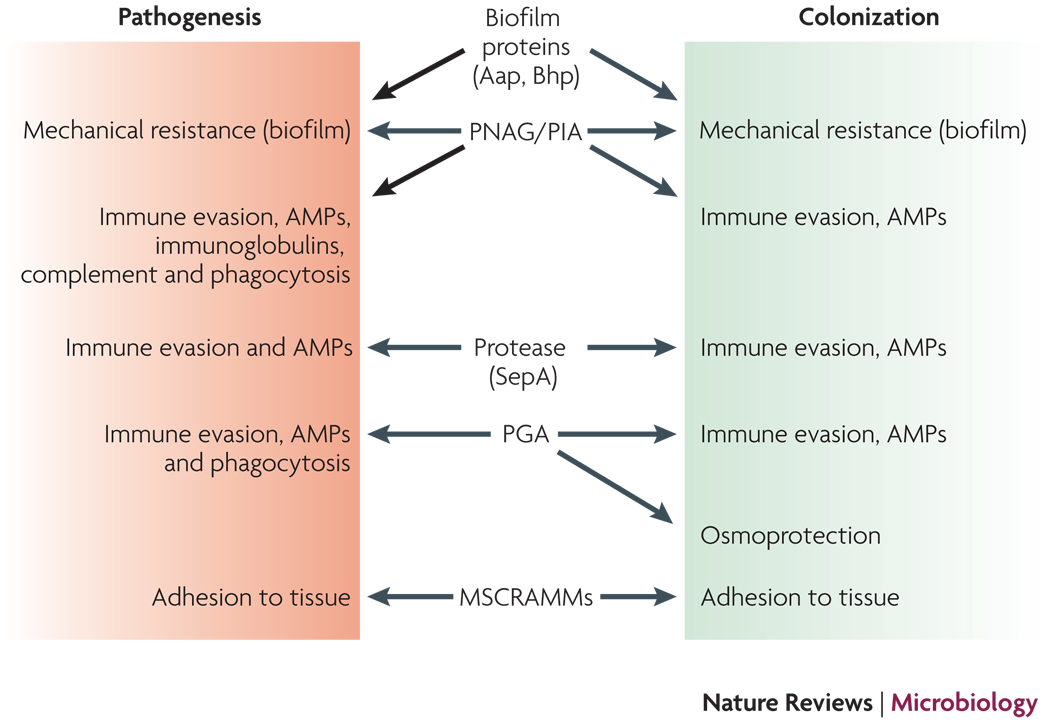
Several lines of evidence indicate that most “virulence factors” of S. epidermidis have original roles in the commensal lifestyle of S. epidermidis (Fig. 6). The roles of PNAG/PIA, PGA, and the SepA protease in protecting from AMPs indicate a key role of these polymers also during life on the skin60, 93, 120, where AMPs are a major determinant of innate host defense. Furthermore, intercellular adhesion by PIA and biofilm-related proteins can be assumed to be vital in an environment such as the skin, where there is considerable mechanical stress for the bacteria. Finally, the role of PGA in osmotolerance49 suggests an original function of this polymer in the non-infectious lifestyle of S. epidermidis. Moreover, there is no clear evidence indicating differences between infectious and commensal strains for the multitude of S. epidermidis MSCRAMMs, indicating that these proteins are valuable during both infection and colonization. This makes sense, as adhesion to host tissue is considered imperative during both these lifestyles. Together, this suggests that S. epidermidis should be regarded as an “accidental” pathogen, whose clinical importance stems less from a dedicated infectious lifestyle, but rather from (i) the frequency of contamination events and (ii) mechanisms such as adhesion and immune evasion that are beneficial for the bacteria during both colonization and chronic infection.
Figure 6. S. epidermidis as a commensal and infectious microorganism.
Determinants that are believed to contribute to both S. epidermidis colonization and pathogenesis are shown. In animal models, only the roles of PNAG/PIA, PGA, and the MSCRAMM SdrG in infection have been demonstrated. Other roles are based on in vitro experiments and environmental challenges during colonization and infection. Not shown are regulators such as agr or sigB that control many of the depicted determinants and may thus also have important functions during both S. epidermidis lifestyles.
Antibiotic resistance and prophylaxis
Specific antibiotic resistance genes are widespread in S. epidermidis. Most notably, resistance to methicillin as an antibiotic of first choice against staphylococcal infections is at 75–90% among hospital isolates of S. epidermidis, which is even higher than the corresponding rate for S. aureus (40–60%)65. High-level resistance to methicillin is encoded on mobile genetic elements (MGEs), namely the staphylococcal cassette chromosome mec (SCCmec), which contains the mecA gene encoding a penicillin-binding protein, PBP2a, with decreased affinity for methicillin compared to other PBPs66. In S. epidermidis, 10 different SCCmec structures were identified, with the short SCCmec type IV element67 being the most abundant (36%)68. SCCmec type IV poses a particular problem, as it does not impose a fitness cost to its host and may thus spread without selective antibiotic pressure125. Interestingly, closely related strains may carry different SCCmec types, indicating frequent loss and acquisition of SCCmec elements by S. epidermidis68.
In addition to methicillin resistance, S. epidermidis strains have acquired resistance to several other antibiotics, including rifamycin, flouroquinolones, gentamicin, tetracycline, chloramphenicol, erythromycin, clindamycin, and sulfonamides9. Very rarely, there is resistance to streptogramins, linezolid, and tigecycline. Most antibiotic resistance genes are plasmid-encoded and more often found in methicillin-resistant than methicillin-susceptible strains69. This likely has no molecular reasons, but is due to the fact that both resistance to methicillin and other antibiotics is frequent among endemic nosocomial strains. Despite widespread resistance to methicillin and other antibiotics, 80% of S. epidermidis-infected catheters may still be treated with antibiotics such as vancomycin without catheter removal70. However, intermediate resistance to vancomycin has been described71. Additionally, staphylococcal biofilm formation significantly decreases the activity of vancomycin and other antibiotics129– 131.
The frequency of antibiotic resistance in S. epidermidis reflects antibiotic overuse. Furthermore, the ubiquity of S. epidermidis as a human commensal microorganism renders this bacterium an optimal carrier and reservoir for antibiotic resistance genes, particularly those that do not inflict a major fitness cost to the bacteria, such as SCCmec elements. Accordingly, there is evidence suggesting that methicillin resistance cassettes were transferred from S. epidermidis to S. aureus 124, 132. Especially the acquisition of SCCmec type IV by community-associated methicillin-resistant S. aureus (CA-MRSA)67 may have had an enormous impact on public health. It enabled the combination of methicillin resistance at no cost for fitness paired with exceptional virulence, which is the main molecular basis of the epidemic caused by CA-MRSA72. In addition, there is recent evidence indicating that CA-MRSA acquired other MGEs from S. epidermidis by horizontal gene transfer that may be important for efficient colonization 73. These findings emphasize an important role of S. epidermidis in human disease by providing a “reservoir” function for the transfer of genetic elements to enhance pathogenic success of S. aureus.
Together, these considerations highlight the need for prophylactic measures against S. epidermidis infections. Vaccination and decolonization, often discussed for other pathogens including S. aureus, appear not appropriate for S. epidermidis. First, there is no anti-staphylococcal vaccine and several lines of evidence indicate that it may be very difficult to use traditional active immunization for staphylococci135, 136. Second, eradication of S. epidermidis as a common part of the human microflora may not only be difficult to achieve owing to the fact that re-colonization from other individuals will be fast; it may also turn out to be counterproductive as it may allow potentially more harmful microorganisms to take the place of S. epidermidis. Thus, it is commonly agreed upon that the best way to deal with S. epidermidis infections is by prevention, which includes sterilization of medical equipment and body parts of patients and health care personnel in possible contact with indwelling medical devices during surgery9.
Unidirectional horizontal gene transfer?
Interestingly, while S. epidermidis appears to frequently transfer MGEs to S. aureus132, 134, it does not contain toxin genes, although acquisition of toxin genes from S. aureus using a similar mechanism would seem easy. The recent investigation of CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) sequences, short repeats that are involved in preventing uptake of conjugative elements such as phages and conjugative plasmids, may provide an explanation of why the transfer of MGEs between S. epidermidis and S. aureus is unidirectional74. These sequences have only been found in S. epidermidis, in one of the two genome-sequenced strains9, but in none of the many S. aureus genomes that are known. While CRISPR-mediated prevention of MGE uptake in S. epidermidis clearly needs to be further evaluated, this mechanism may represent a molecular cause for the absence of a highly diverse toxin repertoire and the resulting lack of aggressive virulence in S. epidermidis.
Outlook
To evaluate potential novel strategies to combat S. epidermidis infections, we need to better understand the relationship between the commensal and infectious lifestyles of this bacterium. To that end, we should more thoroughly investigate determinants that ensure survival of S. epidermidis in its natural habitat, which most notably will include the development and use of skin colonization models. Furthermore, the interaction of S. epidermidis with other bacteria and the reservoir function of S. epidermidis for genes that may be transferred to S. aureus will need to be elucidated in more detail. For several of these tasks, it would be helpful to determine the genome sequence of additional S. epidermidis strains, particularly those of ST2 that appear to be most widely distributed among infectious isolates. Finally, the molecular mechanisms influencing biofilm-associated infection of S. epidermidis will need to be explored using in vivo approaches.
Acknowledgments
This work was supported by the Intramural Research program of the National Institute of Allergy and Infectious Diseases, NIH.
Glossary
- Biofilms
Multicellular agglomerations of microorganisms on a surface with a characteristic three-dimensional structure and physiology.
- Quorum-sensing
Cell density-dependent gene regulation in bacteria. Quorum-sensing systems in Gram-positive bacteria commonly contain peptide-based secreted signals and a membrane-located sensor. The staphylococcal quorum-sensing system is termed agr (accessory gene regulator) and controls a series of genes involved in metabolism and virulence.
- Pseudopeptide
A peptide formed by peptide bonds via carboxyl groups other than the α-carboxyl group.
- Innate host defense
Part of the immune system that provides first line of defense, fast response to invading microorganisms, based on recognition of pathogen-associated molecular patterns (PAMPs). Consists mainly of phagocytes, platelets, and secreted antimicrobial peptides (AMPs).
- PAMPs
or pathogen-associated molecular patterns. Surface structures on pathogens that the innate immune system recognizes as non-self and which trigger activation of innate host defense commonly by binding to toll-like receptors (TLRs).
- Acquired host defense
Part of the immune system that depends on antigen-dependent clonal expansion of T and B cells after antigen presentation by professional antigen-presenting cells. Provides long-term humoral (e.g., antibody-based) and cell-mediated immunity, but is delayed in response.
- Neutrophils
short for neutrophil granulocytes (or polymorphonuclear leukocytes - PMNs), the most abundant leukocytes in human blood. Neutrophils are the primary cells in charge of eliminating invading microorganisms by uptake and subsequent killing via reactive oxygen species and antimicrobial proteins and peptides.
- Antimicrobial peptides
or AMPs, peptides with antimicrobial activity, such as defensins, cathelicidins, etc., secreted for example by epithelial cells or into neutrophil phagosomes.
- Sortase
Enzyme that covalently links secreted bacterial surface proteins to peptidoglycan. Most of these proteins are substrates of sortase A and are characterized by an LPXTG amino acid motif at their C-terminus.
- Teichoic acids
Anionic cell envelope glycopolymer in Gram-positive bacteria composed of many identical sugar-phosphate repeating units. May be linked to peptidoglycan (wall teichoic acids) or the cytoplasmic membrane via a lipid anchor (lipoteichoic acids).
- Two-component system
or TCS, bacterial sensory system composed of a membrane-located sensor (histidine kinase) and a cytoplasmic DNA-binding regulatory protein (response regulator), whose autophosphorylation-dependent activation is triggered by an extracellular signal.
- Enterotoxin
a protein toxin released by a microorganism into the intestine.
- Methicillin
Penicillin derivative that is resistant to penicillinase (an enzyme widespread in staphylococci providing resistance to penicillin).
- Mobile genetic elements
or MGEs, DNA such as plasmids or transposons that may be exchanged between bacteria by horizontal gene transfer, and which often carry virulence or antibiotic resistance genes.
References
- 1.CDC. National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2004, issued October 2004. Am J Infect Control. 2004;32:470–485. doi: 10.1016/S0196655304005425. [DOI] [PubMed] [Google Scholar]
- 2.Uckay I, et al. Foreign body infections due to Staphylococcus epidermidis. Ann Med. 2009;41:109–119. doi: 10.1080/07853890802337045. [DOI] [PubMed] [Google Scholar]
- 3.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 4.Kloos W, Schleifer KH. In: Bergey's Manual of Systematic Bacteriology. PHA S, S M, ME S, JG H, editors. Baltimore: Williams & Wilkins; 1986. [Google Scholar]
- 5.Kloos WE, Musselwhite MS. Distribution and persistence of Staphylococcus and Micrococcus species and other aerobic bacteria on human skin. Appl Microbiol. 1975;30:381–385. doi: 10.1128/am.30.3.381-395.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miragaia M, Thomas JC, Couto I, Enright MC, de Lencastre H. Inferring a population structure for Staphylococcus epidermidis from multilocus sequence typing data. J Bacteriol. 2007;189:2540–2552. doi: 10.1128/JB.01484-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li M, Wang X, Gao Q, Lu Y. Molecular characterization of Staphylococcus epidermidis strains isolated from a teaching hospital in Shanghai, China. J Med Microbiol. 2009;58:456–461. doi: 10.1099/jmm.0.007567-0. [DOI] [PubMed] [Google Scholar]
- 8.Zhang YQ, et al. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228) Mol Microbiol. 2003;49:1577–1593. doi: 10.1046/j.1365-2958.2003.03671.x. [DOI] [PubMed] [Google Scholar]
- 9.Rogers KL, Fey PD, Rupp ME. Coagulase-negative staphylococcal infections. Infect Dis Clin North Am. 2009;23:73–98. doi: 10.1016/j.idc.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 10.Chu VH, et al. Coagulase-negative staphylococcal prosthetic valve endocarditis--a contemporary update based on the International Collaboration on Endocarditis: prospective cohort study. Heart. 2009;95:570–576. doi: 10.1136/hrt.2008.152975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Massey RC, Horsburgh MJ, Lina G, Hook M, Recker M. The evolution and maintenance of virulence in Staphylococcus aureus: a role for host-to-host transmission? Nat Rev Microbiol. 2006;4:953–958. doi: 10.1038/nrmicro1551. [DOI] [PubMed] [Google Scholar]
- 12.Harder J, Schroder JM. Antimicrobial peptides in human skin. Chem Immunol Allergy. 2005;86:22–41. doi: 10.1159/000086650. [DOI] [PubMed] [Google Scholar]
- 13.Faurschou M, Borregaard N. Neutrophil granules and secretory vesicles in inflammation. Microbes Infect. 2003;5:1317–1327. doi: 10.1016/j.micinf.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 14.Yao Y, Sturdevant DE, Otto M. Genomewide analysis of gene expression in Staphylococcus epidermidis biofilms: insights into the pathophysiology of S. epidermidis biofilms and the role of phenol-soluble modulins in formation of biofilms. J Infect Dis. 2005;191:289–298. doi: 10.1086/426945. [DOI] [PubMed] [Google Scholar]
- 15.Khardori N, Yassien M, Wilson K. Tolerance of Staphylococcus epidermidis grown from indwelling vascular catheters to antimicrobial agents. J Ind Microbiol. 1995;15:148–151. doi: 10.1007/BF01569818. [DOI] [PubMed] [Google Scholar]
- 16.Duguid IG, Evans E, Brown MR, Gilbert P. Effect of biofilm culture upon the susceptibility of Staphylococcus epidermidis to tobramycin. J Antimicrob Chemother. 1992;30:803–810. doi: 10.1093/jac/30.6.803. [DOI] [PubMed] [Google Scholar]
- 17.Duguid IG, Evans E, Brown MR, Gilbert P. Growth-rate-independent killing by ciprofloxacin of biofilm-derived Staphylococcus epidermidis; evidence for cell-cycle dependency. J Antimicrob Chemother. 1992;30:791–802. doi: 10.1093/jac/30.6.791. [DOI] [PubMed] [Google Scholar]
- 18.O'Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 19.Vacheethasanee K, et al. Bacterial surface properties of clinically isolated Staphylococcus epidermidis strains determine adhesion on polyethylene. J Biomed Mater Res. 1998;42:425–432. doi: 10.1002/(sici)1097-4636(19981205)42:3<425::aid-jbm12>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 20.Heilmann C, Hussain M, Peters G, Gotz F. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol Microbiol. 1997;24:1013–1024. doi: 10.1046/j.1365-2958.1997.4101774.x. [DOI] [PubMed] [Google Scholar]
- 21.Tormo MA, Knecht E, Gotz F, Lasa I, Penades JR. Bap-dependent biofilm formation by pathogenic species of Staphylococcus: evidence of horizontal gene transfer? Microbiology. 2005;151:2465–2475. doi: 10.1099/mic.0.27865-0. [DOI] [PubMed] [Google Scholar]
- 22.Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 23.Navarre WW, Schneewind O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev. 1999;63:174–229. doi: 10.1128/mmbr.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo B, Zhao X, Shi Y, Zhu D, Zhang Y. Pathogenic implication of a fibrinogen-binding protein of Staphylococcus epidermidis in a rat model of intravascular-catheter-associated infection. Infect Immun. 2007;75:2991–2995. doi: 10.1128/IAI.01741-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arrecubieta C, et al. SdrF, a Staphylococcus epidermidis Surface Protein, Contributes to the Initiation of Ventricular Assist Device Driveline-Related Infections. PLoS Pathog. 2009;5:e1000411. doi: 10.1371/journal.ppat.1000411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowden MG, et al. Identification and preliminary characterization of cell-wall-anchored proteins of Staphylococcus epidermidis. Microbiology. 2005;151:1453–1464. doi: 10.1099/mic.0.27534-0. [DOI] [PubMed] [Google Scholar]
- 27.Rice KC, et al. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci U S A. 2007;104:8113–8118. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mack D, et al. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J Bacteriol. 1996;178:175–183. doi: 10.1128/jb.178.1.175-183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heilmann C, et al. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol. 1996;20:1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x. [DOI] [PubMed] [Google Scholar]
- 30.Gerke C, Kraft A, Sussmuth R, Schweitzer O, Gotz F. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J Biol Chem. 1998;273:18586–18593. doi: 10.1074/jbc.273.29.18586. [DOI] [PubMed] [Google Scholar]
- 31.Vuong C, et al. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J Biol Chem. 2004;279:54881–54886. doi: 10.1074/jbc.M411374200. [DOI] [PubMed] [Google Scholar]
- 32.O'Gara JP. ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol Lett. 2007;270:179–188. doi: 10.1111/j.1574-6968.2007.00688.x. [DOI] [PubMed] [Google Scholar]
- 33.Vuong C, Gerke C, Somerville GA, Fischer ER, Otto M. Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J Infect Dis. 2003;188:706–718. doi: 10.1086/377239. [DOI] [PubMed] [Google Scholar]
- 34.Kogan G, Sadovskaya I, Chaignon P, Chokr A, Jabbouri S. Biofilms of clinical strains of Staphylococcus that do not contain polysaccharide intercellular adhesin. FEMS Microbiol Lett. 2006;255:11–16. doi: 10.1111/j.1574-6968.2005.00043.x. [DOI] [PubMed] [Google Scholar]
- 35.Hussain M, Herrmann M, von Eiff C, Perdreau-Remington F, Peters G. A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect Immun. 1997;65:519–524. doi: 10.1128/iai.65.2.519-524.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohde H, et al. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol Microbiol. 2005;55:1883–1895. doi: 10.1111/j.1365-2958.2005.04515.x. [DOI] [PubMed] [Google Scholar]
- 37.Conrady DG, et al. A zinc-dependent adhesion module is responsible for intercellular adhesion in staphylococcal biofilms. Proc Natl Acad Sci U S A. 2008;105:19456–19461. doi: 10.1073/pnas.0807717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Banner MA, et al. Localized tufts of fibrils on Staphylococcus epidermidis NCTC 11047 are comprised of the accumulation-associated protein. J Bacteriol. 2007;189:2793–2804. doi: 10.1128/JB.00952-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bateman A, Holden MT, Yeats C. The G5 domain: a potential N-acetylglucosamine recognition domain involved in biofilm formation. Bioinformatics. 2005;21:1301–1303. doi: 10.1093/bioinformatics/bti206. [DOI] [PubMed] [Google Scholar]
- 40.Sun D, Accavitti MA, Bryers JD. Inhibition of biofilm formation by monoclonal antibodies against Staphylococcus epidermidis RP62A accumulation-associated protein. Clin Diagn Lab Immunol. 2005;12:93–100. doi: 10.1128/CDLI.12.1.93-100.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chaignon P, et al. Susceptibility of staphylococcal biofilms to enzymatic treatments depends on their chemical composition. Appl Microbiol Biotechnol. 2007;75:125–132. doi: 10.1007/s00253-006-0790-y. [DOI] [PubMed] [Google Scholar]
- 42.Yarwood JM, Bartels DJ, Volper EM, Greenberg EP. Quorum sensing in Staphylococcus aureus biofilms. J Bacteriol. 2004;186:1838–1850. doi: 10.1128/JB.186.6.1838-1850.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vuong C, Kocianova S, Yao Y, Carmody AB, Otto M. Increased colonization of indwelling medical devices by quorum-sensing mutants of Staphylococcus epidermidis in vivo. J Infect Dis. 2004;190:1498–1505. doi: 10.1086/424487. [DOI] [PubMed] [Google Scholar]
- 44.Boles BR, Horswill AR. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 2008;4:e1000052. doi: 10.1371/journal.ppat.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kong KF, Vuong C, Otto M. Staphylococcus quorum sensing in biofilm formation and infection. Int J Med Microbiol. 2006;296:133–139. doi: 10.1016/j.ijmm.2006.01.042. [DOI] [PubMed] [Google Scholar]
- 46.Rupp ME, Fey PD, Heilmann C, Gotz F. Characterization of the importance of Staphylococcus epidermidis autolysin and polysaccharide intercellular adhesin in the pathogenesis of intravascular catheter-associated infection in a rat model. J Infect Dis. 2001;183:1038–1042. doi: 10.1086/319279. [DOI] [PubMed] [Google Scholar]
- 47.Xu L, et al. Role of the luxS quorum-sensing system in biofilm formation and virulence of Staphylococcus epidermidis. Infect Immun. 2006;74:488–496. doi: 10.1128/IAI.74.1.488-496.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vuong C, Kocianova S, Yu J, Kadurugamuwa JL, Otto M. Development of real-time in vivo imaging of device-related Staphylococcus epidermidis infection in mice and influence of animal immune status on susceptibility to infection. J Infect Dis. 2008;198:258–261. doi: 10.1086/589307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kocianova S, et al. Key role of poly-gamma-DL-glutamic acid in immune evasion and virulence of Staphylococcus epidermidis. J Clin Invest. 2005;115:688–694. doi: 10.1172/JCI23523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Little SF, Ivins BE. Molecular pathogenesis of Bacillus anthracis infection. Microbes Infect. 1999;1:131–139. doi: 10.1016/s1286-4579(99)80004-5. [DOI] [PubMed] [Google Scholar]
- 51.Oppermann-Sanio FB, Steinbuchel A. Occurrence, functions and biosynthesis of polyamides in microorganisms and biotechnological production. Naturwissenschaften. 2002;89:11–22. doi: 10.1007/s00114-001-0280-0. [DOI] [PubMed] [Google Scholar]
- 52.Vuong C, et al. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell Microbiol. 2004;6:269–275. doi: 10.1046/j.1462-5822.2004.00367.x. [DOI] [PubMed] [Google Scholar]
- 53.Mah TF, et al. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature. 2003;426:306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- 54.Heine H, Ulmer AJ. Recognition of bacterial products by toll-like receptors. Chem Immunol Allergy. 2005;86:99–119. doi: 10.1159/000086654. [DOI] [PubMed] [Google Scholar]
- 55.Stevens NT, et al. Staphylococcus epidermidis Polysaccharide Intercellular Adhesin induces IL-8 expression in human astrocytes via a mechanism involving TLR2. Cell Microbiol. 2008 doi: 10.1111/j.1462-5822.2008.01264.x. [DOI] [PubMed] [Google Scholar]
- 56.Mehlin C, Headley CM, Klebanoff SJ. An inflammatory polypeptide complex from Staphylococcus epidermidis: isolation and characterization. J Exp Med. 1999;189:907–918. doi: 10.1084/jem.189.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang R, et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007;13:1510–1514. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- 58.Lambert PA, Worthington T, Tebbs SE, Elliott TS, Lipid S. a novel Staphylococcus epidermidis exocellular antigen with potential for the serodiagnosis of infections. FEMS Immunol Med Microbiol. 2000;29:195–202. doi: 10.1111/j.1574-695X.2000.tb01523.x. [DOI] [PubMed] [Google Scholar]
- 59.Li M, et al. Gram-positive three-component antimicrobial peptide-sensing system. Proc Natl Acad Sci U S A. 2007;104:9469–9474. doi: 10.1073/pnas.0702159104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peschel A, et al. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J Biol Chem. 1999;274:8405–8410. doi: 10.1074/jbc.274.13.8405. [DOI] [PubMed] [Google Scholar]
- 61.Li M, et al. The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol Microbiol. 2007;66:1136–1147. doi: 10.1111/j.1365-2958.2007.05986.x. [DOI] [PubMed] [Google Scholar]
- 62.Scheifele DW, Bjornson GL, Dyer RA, Dimmick JE. Delta-like toxin produced by coagulase-negative staphylococci is associated with neonatal necrotizing enterocolitis. Infect Immun. 1987;55:2268–2273. doi: 10.1128/iai.55.9.2268-2273.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ziebuhr W, et al. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol Microbiol. 1999;32:345–356. doi: 10.1046/j.1365-2958.1999.01353.x. [DOI] [PubMed] [Google Scholar]
- 64.Rogers KL, Rupp ME, Fey PD. The presence of icaADBC is detrimental to the colonization of human skin by Staphylococcus epidermidis. Appl Environ Microbiol. 2008;74:6155–6157. doi: 10.1128/AEM.01017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Diekema DJ, et al. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997–1999. Clin Infect Dis. 2001;32 Suppl 2:S114–S132. doi: 10.1086/320184. [DOI] [PubMed] [Google Scholar]
- 66.Hiramatsu K, Cui L, Kuroda M, Ito T. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 2001;9:486–493. doi: 10.1016/s0966-842x(01)02175-8. [DOI] [PubMed] [Google Scholar]
- 67.Ma XX, et al. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob Agents Chemother. 2002;46:1147–1152. doi: 10.1128/AAC.46.4.1147-1152.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miragaia M, Couto I, de Lencastre H. Genetic diversity among methicillin-resistant Staphylococcus epidermidis (MRSE) Microb Drug Resist. 2005;11:83–93. doi: 10.1089/mdr.2005.11.83. [DOI] [PubMed] [Google Scholar]
- 69.Miragaia M, et al. Molecular characterization of methicillin-resistant Staphylococcus epidermidis clones: evidence of geographic dissemination. J Clin Microbiol. 2002;40:430–438. doi: 10.1128/JCM.40.2.430-438.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Raad I, Hanna H, Maki D. Intravascular catheter-related infections: advances in diagnosis, prevention, and management. Lancet Infect Dis. 2007;7:645–657. doi: 10.1016/S1473-3099(07)70235-9. [DOI] [PubMed] [Google Scholar]
- 71.Schwalbe RS, Stapleton JT, Gilligan PH. Emergence of vancomycin resistance in coagulase-negative staphylococci. N Engl J Med. 1987;316:927–931. doi: 10.1056/NEJM198704093161507. [DOI] [PubMed] [Google Scholar]
- 72.Chambers HF. The changing epidemiology of Staphylococcus aureus? Emerg Infect Dis. 2001;7:178–182. doi: 10.3201/eid0702.010204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Diep BA, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 74.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 76.Otto M, Echner H, Voelter W, Gotz F. Pheromone cross-inhibition between Staphylococcus aureus and Staphylococcus epidermidis. Infect Immun. 2001;69:1957–1960. doi: 10.1128/IAI.69.3.1957-1960.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Heilmann C, et al. Identification and characterization of a novel autolysin (Aae) with adhesive properties from Staphylococcus epidermidis. Microbiology. 2003;149:2769–2778. doi: 10.1099/mic.0.26527-0. [DOI] [PubMed] [Google Scholar]
- 78.Gross M, Cramton SE, Gotz F, Peschel A. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect Immun. 2001;69:3423–3426. doi: 10.1128/IAI.69.5.3423-3426.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nilsson M, et al. A fibrinogen-binding protein of Staphylococcus epidermidis. Infect Immun. 1998;66:2666–2673. doi: 10.1128/iai.66.6.2666-2673.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sadovskaya I, Vinogradov E, Flahaut S, Kogan G, Jabbouri S. Extracellular carbohydrate-containing polymers of a model biofilm-producing strain, Staphylococcus epidermidis RP62A. Infect Immun. 2005;73:3007–3017. doi: 10.1128/IAI.73.5.3007-3017.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lai Y, et al. The human anionic antimicrobial peptide dermcidin induces proteolytic defence mechanisms in staphylococci. Mol Microbiol. 2007;63:497–506. doi: 10.1111/j.1365-2958.2006.05540.x. [DOI] [PubMed] [Google Scholar]
- 82.Dubin G, et al. Molecular cloning and biochemical characterisation of proteases from Staphylococcus epidermidis. Biol Chem. 2001;382:1575–1582. doi: 10.1515/BC.2001.192. [DOI] [PubMed] [Google Scholar]
- 83.Chamberlain NR, Brueggemann SA. Characterisation and expression of fatty acid modifying enzyme produced by Staphylococcus epidermidis. J Med Microbiol. 1997;46:693–697. doi: 10.1099/00222615-46-8-693. [DOI] [PubMed] [Google Scholar]