Abstract
The activity of nuclear receptors is modulated by numerous coregulatory factors. Corepressors can either mediate the ability of nuclear receptors to repress transcription, or can inhibit transactivation by nuclear receptors. As we learn more about the mechanisms of transcriptional repression, the importance of repression by nuclear receptors in development and disease has become clear. The protein encoded by the mammalian Hairless (Hr) gene was shown to be a corepressor by virtue of its functional similarity to the well-established corepressors N-CoR and SMRT. Mutation of the Hr gene results in congenital hair loss in both mice and men. Investigation of Hairless function both in vitro and in mouse models in vivo has revealed a critical role in maintaining skin and hair by regulating the differentiation of epithelial stem cells, as well as a putative role in regulating gene expression via chromatin remodeling.
Introduction
Nuclear receptors regulate developmental and physiological processes by regulating the expression of specific target genes. Nuclear receptors typically activate transcription, some without the benefit of a ligand. Some nuclear receptors can repress transcription. A subset of nuclear receptors, including receptors for thyroid hormone, retinoic acid and vitamin D, repress transcription in the absence of their ligand [Jepsen and Rosenfeld, 2002; Mangelsdorf and Evans, 1995; Privalsky, 2004; Smith and O'Malley, 2004]. In general, less is known about the mechanisms of transcriptional repression, but a number of studies indicate the importance of repression by nuclear receptors in development and disease [Privalsky, 2004].
Repression by nuclear receptors occurs through their association with specific proteins that include corepressors. To date, corepressors are far less numerous than their counterparts, the coactivators. Corepressors include proteins that mediate basal repression by unliganded receptors (N-CoR, SMRT) and those that inhibit the activity of ligand-bound receptors (RIP140, L-CoR) [Cavailles et al., 1995; Fernandes et al., 2003; Steel et al., 2005].
The first nuclear receptor corepressors to be identified were Nuclear receptor Co-Repressor (N-CoR) and Silencing Mediator for Retinoic Acid and Thyroid hormone receptors (SMRT) [Chen and Evans, 1995; Horlein et al., 1995; Sande and Privalsky, 1996]. N-CoR and SMRT were originally shown to mediate repression by unliganded TR and RAR, and were subsequently shown to play a role in transcriptional repression by other factors, as well [Privalsky, 2004]. Both N-CoR and SMRT bind to retinoic acid receptors (RAR) and thyroid hormone receptors (TR) in the absence of ligand, and mutant receptors that lack interaction with N-CoR and SMRT no longer repress [Chen and Evans, 1995; Horlein et al., 1995].
Similar to coactivators, N-CoR and SMRT are part of large multiprotein complexes that function via associated enzymatic activities that modify chromatin structure, such as histone deacetylation. For N-CoR and SMRT, functional domains and interacting protein complexes have been characterized in great detail (reviewed in [Perissi and Rosenfeld, 2005; Privalsky, 2004]). N-CoR and SMRT are broadly expressed.
The protein encoded by the mammalian Hairless (Hr, formerly hr) gene was initially shown to be a nuclear receptor corepressor by virtue of its functional similarities to N-CoR and SMRT, including: 1) interaction with nuclear receptors (TR) in the absence of ligand, 2) mediating repression by unliganded TR, 3) multiple independent repression domains, 4) multiple receptor interaction domains comprised of conserved hydrophobic residues, and 5) interaction with histone deacetylases (HDACs) [Guenther et al., 2000; Horlein et al., 1995; Huang et al., 2000; Kao et al., 2000; Li et al., 1997a; Nagy et al., 1997; Ordentlich et al., 1999; Perissi et al., 1999; Wen et al., 2000; Xu et al., 2002]. HR can also inhibit transcription by both activated and orphan receptors.
The Hr gene is essential for proper skin function, as both humans and mice with mutations in Hr suffer from congenital hair loss and in some cases epidermal abnormalities [Ahmad et al., 1999; Panteleyev et al., 1998b]. This review will summarize the properties of HR protein function as a nuclear receptor corepressor, including its in vivo role in regulating epithelial stem cell differentiation.
History of Hairless
Research on Hairless began with the simple discovery of a mouse with no hair [Brooke, 1926; Sumner, 1924]. Studies of these hairless (Hrhr) mutant mice showed that initial hair growth is normal, but after the hair is shed it does not grow back. This observation implicated the Hr gene as an essential regulator of the hair cycle, the process by which hair follicles are maintained in adult mammals [Mann, 1971; Montagna et al., 1952; Orwin et al., 1967; Panteleyev et al., 1998a; Zarach et al., 2004]. Over 60 years later, the murine Hr gene was isolated by cloning the retroviral insertion site in the original Hrhr allele [Cachon-Gonzalez et al., 1994; Stoye et al., 1988]. The rat Hr gene was concurrently isolated as a gene that is regulated by thyroid hormone (TH) in neonatal rat brain [Thompson, 1996]. The identification of the rodent Hr genes [Cachon-Gonzalez et al., 1994; Stoye et al., 1988; Thompson, 1996] led to the identification of the human Hr gene, revealing that Hr is mutated in congenital hair loss disorders (alopecia universalis, papular atrichia) that phenotypically resemble the mouse mutants [Ahmad et al., 1998; Cichon et al., 1998].
Identifying the molecular function of HR
The rat Hr gene encodes a 130 kDa protein (HR) that at the time did not show significant homology to known structural or functional motifs, other than several cysteine residues proposed to form a zinc finger [Cachon-Gonzalez et al., 1994]. Thus, the primary sequence of HR did not provide insight into its function as a corepressor.
Identifying the function of the HR protein as a nuclear receptor corepressor was facilitated by the surprising observation that HR interacts with TR. Interaction with TR was discovered in a yeast two-hybrid screen for HR-interacting proteins [Thompson and Bottcher, 1997]. The interaction of HR with TR was demonstrated in vitro using multiple biochemical assays, and in vivo by coimmunoprecipitation with both overexpressed and endogenous proteins [Potter et al., 2001; Potter et al., 2002; Thompson and Bottcher, 1997]. Evidence that HR bound to TR more avidly in the absence of TH suggested that HR might function as a corepressor [Thompson and Bottcher, 1997].
The results of cotransfection assays demonstrated that HR can mediate repression by unliganded TR, while HR has little effect on transcriptional activation by TR [Potter et al., 2001]. Repression by HR requires receptor binding, as both deletion and point mutants of HR that lack TR-binding no longer repress [Potter et al., 2001]. HR can mediate repression by unliganded TR on different TH response elements (TREs) [Potter et al., 2001; Potter et al., 2002]. Notably, HR represses TR-mediated basal transcription most effectively in pituitary-derived cell lines, a cell type in which HR is endogenously expressed, and not in many other cell lines such as CV-1 and COS. This may reflect the presence of tissue-specific factors, or may be due to compensation by N-CoR and/or SMRT expression in most cells [Misiti et al., 1998].
Evidence that HR expression is induced by TH in brain and other tissues suggests that HR would not act as a corepressor for TR in vivo. However, HR could function via TR in tissues in which HR expression is not TH-dependent, such as skin and adult brain [Thompson, 1996]. In addition, recent work indicates that HR can suppress TR-mediated transcriptional activation at physiologic TH concentrations [Thompson and Beaudoin, 2006]. Thus, HR may modulate TR/TH signaling in addition to mediating basal repression.
HR functional domains
Repression domains
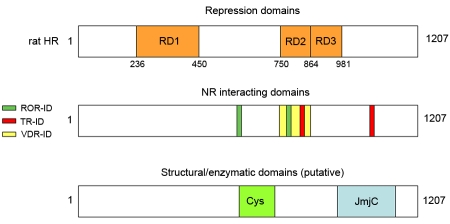
Analysis of HR deletion derivatives expressed as fusion proteins with the GAL4 DNA binding domain revealed three domains capable of mediating repression [Potter et al., 2001]. Repression domains include a single amino terminal domain (amino acids 236-450; RD1), and two carboxyl terminal domains (amino acids 750-864, RD2; amino acids 864-981, RD3) (Figure 1). The functional organization differs from N-CoR and SMRT, as RD2 overlaps with a receptor interaction domain (see below).
Figure 1. Schematic representation of rat HR structural and functional domains.
Repression domains (RD1, 236-450; RD2, 750-864; RD3, 864-981); TR-interacting domains (TR-ID1, 816-830; TR-ID2, 1026-1038); ROR-interacting domains, ROR-ID1, 586-590; ROR-ID2, 778-782); cysteine-rich domain, 587-712; JmjC domain, 964-1175. Note that rat HR is 1207 amino acids, mouse and human Hr initiate at an internal AUG (amino acid 27 in rat Hr) and are 1182 and 1189 amino acids, respectively.
TR-interacting domains
Analysis of a series of HR deletion derivatives in multiple binding assays (far western, yeast two hybrid, coimmunoprecipitation) showed that HR interacts with TR via two carboxyl terminal regions (between amino acids 750-864 and 980-1084) (Figure 1), with the amino terminal region interacting with higher affinity [Potter et al., 2001]. Specific amino acids required for interaction were identified using site-directed mutagenesis, and essential residues were identified as those that inhibited or abolished interaction by yeast two hybrid and far western analyses [Potter et al., 2001]. Compiling essential residues in the two TR interaction domains (TR-ID1, 816-830; TR-ID2, 1026-1038) revealed a consensus motif for HR binding to TR (I/L-I-X-X-L/V-V) similar to those identified for N-CoR and SMRT binding to TR and RAR, including the L/I-X-X-I/V-I motif termed the CoRNR box [Cohen et al., 2001; Hu and Lazar, 1999; Nagy et al., 1999; Perissi et al., 1999; Webb et al., 2000] and the extended amphipathic α helix predicted and subsequently demonstrated to mediate corepressor: receptor interaction [Perissi et al., 1999; Xu et al., 2002]. The spacing of hydrophobic residues is conserved in the HR consensus, while the identity of hydrophobic residues is not, suggesting that the identity of specific residues determines the specificity of corepressor: receptor interaction. For example, HR does not bind to RAR, and SMRT shows higher affinity for binding to RAR than to TR [Potter et al., 2001; Privalsky, 2004].
JmjC domain
Subsequent to the initial identification of the Hr gene and predicted protein, a JmjC domain was recognized in the HR carboxyl terminus (amino acids 964-1175, rat HR) [Clissold and Ponting, 2001]. The JmjC domain is one of multiple conserved motifs originally identified in the Jumonji (JMJ) protein, a transcriptional repressor [Clissold and Ponting, 2001; Kim et al., 2003; Takeuchi et al., 1995; Toyoda et al., 2003]. The JmjC domain is conserved in a number of proteins, but until recently, had no known function [Clissold and Ponting, 2001; Takeuchi et al., 1999; Takeuchi et al., 2006; Takeuchi et al., 1995].
Recent work has shown that the JmjC domains in a number of proteins function as histone demethylases [Klose et al., 2006; Tsukada et al., 2006], including one (JMJD2A, also known as JHDM3A, KDM4A) that is found in a complex with N-CoR [Zhang et al., 2005]. Although the HR protein lacks conserved coregulator binding sites found in these proteins, preliminary results suggest that the JmjC domain in HR has a similar enzymatic activity with novel specificity (J. M. Sisk and C.C.T., unpublished observations). If so, part of the repression function of HR and other JmjC-domain containing proteins may lie in providing an additional component of the histone code for regulating transcriptional activity.
Interactions
Transcriptional repression often results from the association of corepressors with histone deacetylases (HDACs) [Burke and Baniahmad, 2000; Glass and Rosenfeld, 2000; Pazin and Kadonaga, 1997]. Multiple HDACs have been shown to interact with corepressors, and biochemical analyses have indicated that HDAC3 is found in an endogenous complex with corepressors [Guenther et al., 2001; Li et al., 2000a; Wen et al., 2000; Yoon et al., 2003]. HR interacts indirectly with several HDACS, including HDACs 1, 3 and 5 [Potter et al., 2001; Potter et al., 2002]. HR corepressor activity is reduced in the presence of an HDAC inhibitor (trichostatin A), suggesting that HDAC activity is partially responsible for repression by HR [Potter et al., 2002].
Biochemical studies have revealed the presence of N-CoR and SMRT in large multiprotein complexes that include HDACs, TBL1, TBLR1, as well as other proteins [Privalsky, 2004]. Although similar biochemical studies have not yet been performed with HR, based on its functional similarities, HR likely functions as part of a multiprotein complex.
HR is a corepressor for multiple nuclear receptors
HR interacts with RORα, an orphan nuclear receptor important for cerebellar development
Unlike N-CoR and SMRT, HR does not interact with retinoic acid receptor (RAR); however, HR does bind to RORα (Retinoic acid receptor-related Orphan Receptor α). RORα is a constitutively-active orphan nuclear receptor that is critical for cerebellar development [Dussault et al., 1998; Hamilton et al., 1996; Matysiak-Scholze and Nehls, 1997]. HR interacts with RORα, and inhibits transcriptional activation by all ROR isoforms (α, β, γ) [Moraitis et al., 2002]. Further analysis has shown that HR binding protects RORα from proteasome-mediated degradation [Moraitis and Giguere, 2003]. The domain of HR that interacts with ROR was mapped to a region with two motifs with the LxxLL consensus sequence for coactivator interaction [Heery et al., 1997; McInerney et al., 1998]. Surprisingly, mutation of both LxxLL motifs abolished the ability of HR to repress RORα activity, while mutation of TR-ID1 had no effect on activity [Moraitis et al., 2002]. Since coactivators interact with the AF-2 domain of nuclear receptors, the requirement for this domain was tested by changing the AF-2 domain of RARα (which does not interact with HR) to that of RORα. Remarkably, the specificity of HR corepressor action could be transferred to RARα by exchanging the AF-2 domain, a change of only 5 amino acids [Moraitis et al., 2002]. These results unexpectedly demonstrated that in the context of ROR, the repressive activity of HR is dependent on coactivator-type binding motifs.
HR interacts with vitamin D receptor, a nuclear receptor important for skin function
The interaction of HR with TR does not easily explain the prominent skin phenotype in Hr mutants, as although TH-deficient humans and mice do have some hair and skin defects, none are as striking as the complete hair loss and skin wrinkling observed in Hr mutant [Alonso and Rosenfield, 2003]. Targeted deletion of RXRα and β in mice results in a hair loss phenotype [Li et al., 2000b]. RXRs are heterodimeric partners for multiple nuclear receptors, and there is no evidence of direct interaction between HR and RXRs. However, HR does have multiple receptor-interacting domains, suggesting that HR may contact RXR in the context of a heterodimer with either TR or VDR.
Current evidence suggests that the receptor through which HR acts in the skin is the vitamin D receptor (VDR). HR interacts with VDR in multiple biochemical assays, including GST pulldown and coimmunoprecipitation [Hsieh et al., 2003]. Interaction of HR with VDR is weaker than interaction with TR and may require other proteins (C.C.T., unpublished observations). The region of VDR interaction with HR overlaps the sites responsible for interaction with TR and ROR (Figure 1); the specific amino acids required for interaction of HR with VDR have not been determined. In contrast to its role with TR, HR represses VDR activity both in the absence and presence of vitamin D [Hsieh et al., 2003]. HR can inhibit expression from both synthetic and naturally occurring VDR-responsive promoters, and in cultured keratinocytes, a cell type in which HR and VDR are normally expressed ([Hsieh et al., 2003] and G.M.J. Beaudoin and C.C.T., unpublished observations). Hr and VDR mRNAs colocalize in a subset of cells in the skin, indicating that the biochemically defined interaction of HR and VDR can occur in vivo [Hsieh et al., 2003]. Both mice and humans with mutations in VDR have a hair loss phenotype which resembles that of mice and humans with mutations in Hr [Li et al., 1997b; Yoshizawa et al., 1997]. Notably, although mutations in VDR cause hair loss, vitamin D deficiency does not result in hair loss either in mice or humans [Sakai et al., 2001]. This observation indicated that transcriptional activation by vitamin D-bound VDR is not essential for hair cycle regulation, and suggested that repression could be important for this function. Recent work supporting the role of repression by VDR in hair cycling has come from studies showing that mutant VDRs which do not activate transcription in response to vitamin D can rescue the hair loss phenotype of VDR null mice [Skorija et al., 2005]. The mutant VDRs retain the ability to bind HR and repress basal transcription, suggesting that repression by unliganded VDR via HR is essential for VDR function in the skin [Skorija et al., 2005].
Thus, multiple studies have shown that HR functions as a corepressor for nuclear receptors. By analogy with other corepressors, HR may possess additional functional properties. For example, it is not yet known whether HR can bind to antagonist-bound steroid receptors, and/or act as corepressors for other transcription factors [Jepsen and Rosenfeld, 2002; Privalsky, 2004]. The data summarized above is from rat HR, but the human HR protein also functions as a corepressor [Wang et al., 2007]. Thus, the function of Hr is conserved between mammalian species.
Hr expression
Hr mRNA is most abundantly expressed in the skin and brain. Within the skin, Hr mRNA is localized to specific cells of the epidermis and hair follicles. Early in postnatal development, Hr mRNA is detected in epidermis, then expression is downregulated as development proceeds. In hair follicles, Hr mRNA is present throughout the hair cycle (Figure 2, described below) [Panteleyev et al., 2000]. Surprisingly, analysis of HR protein expression showed that follicles actively growing hair do not contain detectable HR protein [Beaudoin et al., 2005]. The discordance between Hr mRNA expression and protein expression points to posttranscriptional or posttranslational control of HR protein expression and/or stability. Indeed, recent work has shown a novel mechanism of translational regulation residing in a conserved upstream open reading frame (ORF), supporting the idea that regulation of HR protein expression is critical for its role in regulating hair growth [Wen et al., 2009]. The expression and function of HR protein in the skin has been studied in detail and is described below.
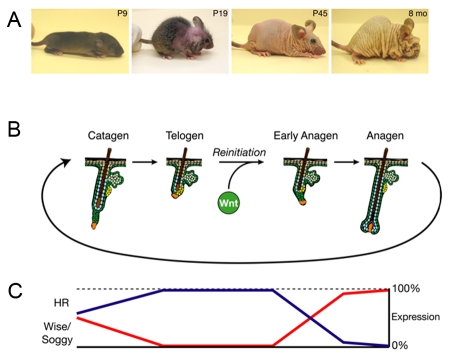
Figure 2. Model for HR action in hair regrowth.
A) Onset of phenotype in Hr knockout mice. Note that initial hair growth is normal, but after the first hair cycle (around P17) the hair is shed and does not regrow. Hr-rhino and knockout mice get progressively more wrinkled with age. P9, postnatal day 9; P19, postnatal day 19; P45, postnatal day 45; 8 mo, 8 months. B) Schematic representation of hair cycle. Wnt signaling initiates the transition from rest (telogen) to growth (anagen). C) Summary of HR protein expression through the hair cycle in relation to Wise and Soggy mRNA expression. Regression (catagen) is characterized by the upregulation of HR protein and concurrent downregulation of Wise/Soggy mRNA. HR protein expression during rest (telogen) is predicted to repress Wise/Soggy mRNA expression, allowing Wnt activation. Subsequent to the reinitiation of hair growth (anagen), HR protein is downregulated while Wise/Soggy mRNA increases, resulting in a transient decrease in Wnt signaling required for proper hair growth. In Hr mutants, uncontrolled expression of Wise/Soggy and possibly other Wnt inhibitors prevents hair regrowth by suppressing Wnt signaling. Figure adapted from (Thompson et al., 2006).
In the brain, Hr mRNA and protein are broadly expressed, including in cerebellum, somatosensory cortex, hippocampus, dentate gyrus, retina and inner ear [Cachon-Gonzalez et al., 1999; Potter et al., 2002; Thompson, 1996]. In neonatal rodent brain, Hr mRNA expression is induced by TH, which is significant because of the well-established role of TH in brain development [Thompson, 1996; Thompson and Bottcher, 1997; Thompson and Potter, 2000]. Expression of Hr is also regulated by TH in other tissues, although notably not in skin or in adult brain [Thompson, 1996].
Hr mRNA is expressed at low levels in several other tissues. Hr mRNA was detected in a wide range of tissues as early as embryonic day 12.5 in mice [Cachon-Gonzalez et al., 1999]. Tissues with relatively high levels of Hr expression include oral, nasal, bladder and rectal epithelia, which are of interest based on the proposed function of HR protein in skin epithelial cells [Beaudoin et al., 2005]. Hr mRNA was also detected in embryonic cartilage and in embryonic and adult retina ([Cachon-Gonzalez et al., 1999]; J. M. Sisk and C.C.T., unpublished observations).
Notably, expression is detected in a number of tissues in which thus far no phenotype has been observed in Hr mutants. This may reflect redundancy with other corepressors, or regulation of protein expression. For example, the onset of the skin phenotype is correlated with detection of HR protein. Some phenotypic abnormalities have been observed. In the brain, abnormalities in neuronal morphology, inner ear and retinal cytoarchitecture have been observed [Cachon-Gonzalez et al., 1999; Garcia-Atares et al., 1998]. Alterations in the cerebellar cortex include changes in the size and shape of Purkinje cells, which may be related to HR interaction with RORα, as both RORα and TH play roles in Purkinje cell development and function [Dussault et al., 1998; Garcia-Atares et al., 1998; Hamilton et al., 1996; Matysiak-Scholze and Nehls, 1997; Steinmayr et al., 1998]. While the majority of patients with mutations in Hr have not been assessed for neural function, there are two cases of patients homozygous for a mutation in Hr that suffer from mental retardation [Aita et al., 2000; del Castillo et al., 1974]. In colon, Hrrh mice show hyperproliferation of the colon epithelium and an increased number of villi [Cachon-Gonzalez et al., 1999]. This reflects aspects of the skin phenotype and may be typical of other tissues with epithelial components in which HR is expressed.
Cellular mechanism and cellular biology
The HR protein is nuclear [Potter et al., 2002; Thompson and Bottcher, 1997]. In some cells, HR is found in subnuclear structures that contain both HDACs and corepressors known as matrix-associated deacetylase (MAD) bodies [Downes et al., 2000; Potter et al., 2001]. HR colocalizes with HDACs and other corepressors, including SMRT, HDAC3 and HDAC 5 [Potter et al., 2001]. Biochemical studies have revealed the presence of N-CoR and SMRT in large multiprotein complexes that include HDACs, TBL1, TBLR1, as well as other proteins (reviewed in [Privalsky, 2004]). Although similar biochemical studies have not been performed with HR, based on functional conservation, HR may be part of multiprotein complexes that include other corepressors.
The HR protein is phosphorylated in vivo, initially shown by immunoprecipitation of metabolically labeled cell extracts (G. B. Potter and C. C. T., unpublished observations). Proteomic profiling of phosphoproteins in human skin fibroblasts has identified two sites of phosphorylation in human HR (S416, S425) [Yang et al., 2006]. It is not known if phosphorylation is important for corepressor function as has been shown for SMRT [Hong and Privalsky, 2000].
To date, HR does not have a defined enzymatic activity although the presence of a conserved JmjC domain and preliminary results (C.C.T. and J.M. Sisk, unpublished observations) indicate that HR may have histone demethylase activity. As a corepressor, HR does not directly regulate target gene expression but does so via multiple nuclear receptors. Genes that are misregulated in the Hr null mouse model have been identified [Beaudoin et al., 2005; Zarach et al., 2004]. Further investigation is required to determine whether any or all of these genes are directly regulated by HR:nuclear receptor complexes.
Systemic biology and physiology
Based on the phenotype of Hr mutant mice and humans, Hr has an important role in the skin. The skin phenotype in all known murine Hr alleles is initially similar (Figure 2A). Animals appear normal for about the first two weeks after birth, after which they begin to lose their hair and eventually all hair is lost. In hairless (Hrhr) mutant mice, after the initial hair loss the skin remains relatively smooth. In rhino (Hrrh) mutant mice, once hair is lost the skin subsequently becomes wrinkled [Mann, 1971]. Hairless “knockout” mice (Hr-/-) made by targeted deletion have a more severe wrinkling phenotype [Zarach et al., 2004]. The severity of the phenotype is correlated with the severity of the deficit in Hr mRNA expression [Thompson and Beaudoin, 2006; Zarach et al., 2004].
Mammalian skin is comprised of three distinct structures, the overlying epidermis, hair follicles and sebaceous glands [Stenn and Paus, 2001]. All of these structures are affected in Hr mutant skin. The first phenotypic change detected in Hr mutant skin is the widening of the hair follicle near the epidermis [Mann, 1971; Zarach et al., 2004], forming as a consequence of aberrant cell proliferation and differentiation [Zarach et al., 2004]. The next alteration observed is that the lower part of the hair follicle disintegrates, and follicles do not reform as they would in wild type mice [Mann, 1971; Montagna et al., 1952; Orwin et al., 1967; Panteleyev et al., 1999]. Loss of hair follicles is complete, as markers expressed in regenerating hair follicles are not present [Panteleyev et al., 1998b; Zarach et al., 2004]. Cysts develop in the dermis [Mann, 1971; Montagna et al., 1952; Zarach et al., 2004], which increase in size with age, due in part to proliferation of the cells lining the cyst [Zarach et al., 2004]. The cysts have properties of sebaceous glands, as they contain lipids and express Scd1, but do not express adipocyte-specific markers [Bernerd et al., 1996; Mann, 1971; Zarach et al., 2004]. The cysts in Hr-/- skin contribute to the overall thickness of the skin, and the wrinkling phenotype.
How does a single gene/protein affect all of these processes in the skin? Although the epidermis, sebaceous glands, and hair follicles are distinct structures with characteristic properties, all are maintained by the same population of epithelial stem cells. The undifferentiated progenitors of all three components originate as stem cells found in a region of the follicle outer root sheath (ORS) known as the bulge [Alonso and Fuchs, 2003; Tumbar et al., 2004]. The current model for stem cell-mediated maintenance of the skin is that the bulge stem cells generate progenitor cells, and that production and maintenance of all three components occurs through the differentiation of progenitor cells into the mature cells that comprise each structure [Alonso and Fuchs, 2003].
Based on the increase in epidermal and sebaceous cells and decrease in hair cells in Hr-/- skin, my lab proposed a model in which the role of HR is to regulate epithelial stem cell differentiation, and that disruption of timing in Hr mutants leads to changes in cell fate favoring epidermis and sebaceous cells at the expense of the hair follicle [Zarach et al., 2004].
To test this model, we generated and characterized a gain of function mutant mouse in which the Hr cDNA was expressed in K14 positive cells (K14-rHr). Based on the analysis of K14-rHr skin, the model generally holds true ([Beaudoin et al., 2005] and unpublished observations). In K14-rHr epidermis, there is an increased number of undifferentiated and a reduced number of terminally differentiated keratinocytes, indicating a delay in epidermal differentiation. The hair of the K14-rHr transgenic mice is shorter than normal due to reduced production of each lineage of the hair bulb, and the hair cycle is accelerated. Sebaceous glands were not initially detected in K14-rHr skin, indicating delayed sebaceous gland differentiation. Thus, the K14-rHr transgenic phenotype is consistent with the model that HR promotes hair cell fate while suppressing progression into cells that populate the epidermis and sebaceous glands.
What about the most striking part of the phenotype, hair loss? Unlike the epidermis and sebaceous gland, the hair follicle is periodically destroyed and regenerated in a cyclic manner (Figure 2B) [Hardy, 1992; Stenn and Paus, 2001]. The hair cycle can be divided into distinct stages: anagen, catagen, and telogen. Active growth of hair occurs during anagen [Hardy, 1992; Stenn and Paus, 2001]. Once the hair has grown to an appropriate length, the hair follicle enters catagen, a period of regression in which the lower part of the hair bulb is destroyed by apoptosis [Lindner et al., 1997; Stenn and Paus, 2001]. After catagen is completed, the follicle enters a period of rest, telogen [Stenn and Paus, 2001]. Hair growth is reinitiated by reentry into anagen and the hair bulb is regenerated [Hardy, 1992; Stenn and Paus, 2001]. Signaling between the cells of the dermal papilla (mesenchymal cells) and bulge (epithelial cells), appears to be critical for re-growth [Jahoda et al., 1984; Oliver and Jahoda, 1988].
The temporal and spatial localization of the HR protein in hair follicles suggested that HR functions in both follicle regression and hair regrowth [Beaudoin et al., 2005]. Within hair follicles, HR protein is detected in the outer root sheath (ORS), which includes the bulge region. HR protein is initially detected as follicles start to regress (catagen), which coincides with the onset of phenotypic alterations in Hr mutant mice [Panteleyev et al., 1999; Zarach et al., 2004]. HR expression is maintained through the rest phase (telogen) and during the transition to hair regrowth. Once the hair follicle has regenerated, HR protein is undetectable in follicles actively growing hair (anagen) [Beaudoin et al., 2005]. Evidence that HR plays an active role in reinitiating hair growth came from studies in which the hair re-growth defect in Hr-/- mice was rescued by expression of Hr in K14-positive cells. Mice that express HR only in K14-positive cells (“transgenic rescue”) lose their hair, but regenerate hair follicles and produce normal hair [Beaudoin et al., 2005]. Thus, HR is both necessary and sufficient to reinitiate hair growth.
Transcriptional regulation by HR in vivo
Since HR is a corepressor, the defects observed in Hr mutant skin presumably arise from altered gene expression, specifically upregulation of gene expression. Putative target genes were identified using microarray analysis to compare gene expression between normal and Hr-/- skin at an age prior to observable phenotypic changes (P12). Microarray analysis revealed significant (> 2-fold) upregulation of several genes in Hr-/- skin with most of the identified genes playing roles in epidermal differentiation [Zarach et al., 2004]. These genes include filaggrin, loricrin, keratin 10, caspase-14, keratinocyte differentiation associated protein, and calmodulin-4) [Chien et al., 2002; Eckhart et al., 2000; Hwang and Morasso, 2003; Koshizuka et al., 2001; Lippens et al., 2000; Oomizu et al., 2000].
Misregulation of these genes likely underlies the initial epidermal phenotype, as in situ hybridization showed that transcriptional changes were localized to the upper part of the hair follicle, the site at which the phenotype is initially manifested [Zarach et al., 2004]. Genes regulated by HR with a role in reactivation of hair growth were identified as genes (identified from the P12 microarray) that were 1) upregulated in Hr-/- skin and 2) downregulated (relative to Hr-/-) in transgenic rescue skin, at the time hair follicles regenerate (P24) using Northern analysis [Beaudoin et al., 2005]. Genes that met these criteria included Wise (Wnt modulator in surface ectoderm), a protein that modulates Wnt signaling [Itasaki et al., 2003], and Soggy, a protein related to Dickkopfs, which inhibit Wnt signaling [Krupnik et al., 1999; Logan and Nusse, 2004].
The identification of putative inhibitors of Wnt signaling suggested that HR might be regulating this important signaling pathway. Indeed, expression of an endogenous Wnt target gene, Axin2, is correlated with the Hr genotype; Axin2 is expressed in wild type and transgenic rescue skin, and is not detected in Hr knockout skin [Beaudoin et al., 2005].
A number of studies have shown that Wnt signaling (like HR function) is critical for hair follicle regeneration [Huelsken et al., 2001; Lo Celso et al., 2004; Merrill et al., 2001; Niemann et al., 2002; Van Mater et al., 2003]. For example, blocking Wnt signaling by either loss of β-catenin expression or expression of a mutant Lef1 blocks reinitiation of hair growth [Huelsken et al., 2001; Merrill et al., 2001; Niemann et al., 2002]. Wnt signaling is sufficient for follicle regeneration, as inducing nuclear translocation of β-catenin in a transgenic mouse model induces hair regrowth [Lo Celso et al., 2004; Van Mater et al., 2003]. Transient Wnt signaling appears to be critical, as constitutive expression of a stabilized β-catenin induces skin tumors in mice and humans [Chan et al., 1999; Gat et al., 1998; Lo Celso et al., 2004; Van Mater et al., 2003].
The established role of Wnt signaling in hair regrowth suggested that HR may regulate hair follicle regeneration by repressing the expression of one or more Wnt inhibitors (Wise, Soggy), thereby allowing Wnt activation. Multiple lines of evidence support this hypothesis. First, Wise inhibits Wnt-induced reporter gene expression, including activation by Wnt10b, one of two Wnts that are proposed regulators of hair regrowth [Beaudoin et al., 2005; Reddy et al., 2001]. Second, expression of HR protein and Wise and Soggy mRNAs is inversely correlated in vivo, and HR suppresses expression of the Wise promoter in vitro [Beaudoin et al., 2005]. Third, nuclear HR protein colocalizes with expression of a Wnt-responsive reporter gene in the skin of TOP-Gal transgenic mice [Beaudoin et al., 2005; DasGupta and Fuchs, 1999], and no reporter gene expression is detected in the skin of TOP-Gal/Hr-/- mice (J. M. Sisk and C.C.T., unpublished observations).
These data support a model for HR action in the skin in which HR regulates hair follicle regeneration by promoting Wnt signaling (Figure 2). The HR protein promotes Wnt/β-catenin activation, and thus hair growth initiation, by repressing expression of soluble Wnt inhibitors at the proper time in the hair cycle. This model includes a means of inducing transient Wnt signaling, via the decrease in HR protein expression and concomitant increase in Wise expression subsequent to the reinitiation of hair growth. This model also specifies a molecular mechanism to explain the failure of hair follicles to regenerate in Hr mutant skin: inhibition of Wnt signaling caused by the persistent expression of Wnt inhibitors.
The function of HR in the epidermis and sebaceous glands may be through Wnt signaling as well, as HR expression in K14-positive cells also prevents the wrinkling phenotype. This suggests that HR expression also rescues the epidermal and sebaceous phenotypes, and is consistent with the idea that HR is regulating epithelial stem cell differentiation via the modulation of Wnt signaling. Unlike reinitiation of hair growth, hair morphogenesis does not seem to be sensitive to loss of Hr expression; an alternative factor may regulate Wnt signaling during morphogenesis.
The regulation of Wnt signaling by HR in the hair follicle is likely to be one component of a complex interplay of signaling pathways that regulate hair regrowth. Other pathways that have already been implicated in hair cycling include Sonic hedgehog and BMP signaling [Botchkarev and Sharov, 2004; Callahan and Oro, 2001]. Notably, Wise was independently identified as an inhibitor of BMP signaling (Sostdc1, Ectodin, USAG-1) [Balemans and Van Hul, 2002; Laurikkala et al., 2003; Yanagita et al., 2004] raising the possibility that Wise may also influence BMP signaling in the context of follicle regeneration. In addition, HR may regulate the expression of additional factors that modulate such pathways.
Disease
The clinical importance of Hr function was demonstrated when the human ortholog of Hr was identified as the gene mutated in two rare human diseases, alopecia universalis and atrichia with papular lesions [Ahmad et al., 1998; Cichon et al., 1998; Sprecher et al., 1998]. It was subsequently recognized that many cases of alopecia universalis may be misdiagnosed cases of papular atrichia, with the development of papular rash a phenotypic variation [Sprecher et al., 1999a; Sprecher et al., 1999b]. As in mice, humans with these diseases initially have normal hair, but within a few years the hair falls out and in general does not regrow [Ahmad et al., 1998; Cichon et al., 1998; Sprecher et al., 1998]. Both human diseases exhibit histological changes similar to those observed in Hr mutant mice [Ahmad et al., 1998; Cichon et al., 1998; Sprecher et al., 1998].
Recently, an autosomal dominant hair loss disorder, Marie Unna hereditary hypotrichosis, was mapped to the Hr locus, not to the coding region but to a conserved upstream ORF [Wen et al., 2009]. This elegant work showed that the ORF controlled the level of HR protein expression by a translational mechanism, reinforcing the idea that HR protein levels are critical for modulating hair growth.
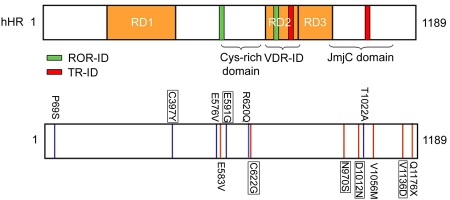
As in mouse, there are multiple mutant human Hr alleles, which include nonsense and missense mutations. Unlike in mice, the human phenotype is not correlated with the severity of the mutation. While nonsense mutations are presumably loss-of-function, a number of missense mutations have been tested to assess corepressor function (Figure 3).
Figure 3. Mutant Hr alleles from human patients lack corepressor activity.
Naturally-occurring HR mutants tested for corepressor activity. Top, schematic representation of human HR protein (hHR) functional domains. Bottom, positions of mutated residues and altered amino acids. Blue indicates polymorphism, red indicates missense mutation. Mutants were tested for the ability to mediate repression by VDR and TR. Mutations in human HR were tested with VDR, mutants in rat HR (corresponding to the mutations found in human Hr alleles) were tested with TR (boxed). All proteins were expressed at equivalent levels except for P69S. All expressed proteins retain interaction with TR and VDR. In general, mutants lack corepressor activity, polymorphisms retain corepressor activity. Missense mutants (red): E583V (Paradisi et al., 2003); C622G/ C642G(rat) (Aita et al., 2000); N970S/N988S(rat) (Kruse et al., 1999); D1012N/D1030N(rat) (Klein et al., 2002); V1056M (Zlotogorski et al., 2002); V1136D/V1154D(rat) (Cichon et al., 1998), Q1176X (Henn et al., 2002). Polymorphisms (blue): P69S, C397Y/C422Y(rat), A576V, E591G/E611G(rat), R620Q, T1022A (Hillmer et al., 2002).
Surprisingly, mutations do not in general map to established functional domains, although several mutations map to the JmjC domain which has putative histone modifying activity. A set of HR mutants (amino acid changes associated with alopecia/atrichia) and polymorphisms (amino acid changes occurring in HR that are not associated with atrichia/alopecia) were tested for function with VDR [Wang et al., 2007]. This work showed that stably expressed missense mutants (except E583V) lose corepressor activity, while polymorphisms maintain corepressor activity [Wang et al., 2007]. Some polymorphisms (T1022A) do show partial activity. All mutants retain interaction with VDR, so the lack of corepressor function is not due to loss of receptor interaction. These investigators demonstrated that HR mutants show reduced interaction with HDAC1, suggesting that impaired HDAC interaction contributes to loss of corepressor function.
Supporting this idea, recent work has shown that a human HR isoform (Δ1072-1126) that arises from alternative splicing maintains interaction with VDR, but lacks both corepressor activity and HDAC1 binding [Malloy et al., 2009]. The deletion in this variant also removes part of the JmjC domain, so disruption of this domain may also contribute to loss of corepressor activity. A subset of HR mutants were tested for function with TR, and showed similar results; all mutants tested maintained interaction with TR but lost corepressor activity [Thompson et al., 2006]. The correlation between corepressor activity and mutant phenotype indicates that disruption of corepressor function underlies congenital hair loss disorders [Thompson et al., 2006; Wang et al., 2007].
Conclusions
The biological role of transcriptional activation by nuclear receptors is well established. Far less is known about the role of repression, but the study of corepressors is helping to elucidate the mechanisms and significance of repression by nuclear receptors. The study of HR over the past decade has contributed to this understanding.
HR corepressor function in vivo is essential for maintaining skin function. HR appears to be a member of a distinct class of corepressors that are spatially and temporally restricted, as Hr mRNA expression is restricted to specific tissues and/or cell types, and HR protein expression is further restricted temporally. The study of the biochemical and biological roles of HR has revealed its mechanism of action in regulating developmental processes in the skin, which will likely apply to other tissues, as well.
Acknowledgments
Work from the author’s lab at the Kennedy Krieger Institute was supported by the National Institutes of Health (NS41313). Special thanks to all lab members who contributed to this work, especially G. Beaudoin and J. Sisk. I am grateful to P. Coulombe, M. Haussler and their lab members for contributing their technical and intellectual expertise. Thanks to the reviewers for helpful suggestions, and to J. Sisk and C. McNally for help with artwork and figures.
Abbreviations
- HR
Hairless protein
- N-CoR
nuclear receptor corepressor
- ROR
retinoic acid receptor-related orphan receptor
- SMRT
silencing mediator of retinoic acid and thyroid hormone receptors
- TH
thyroid hormone
- TR
thyroid hormone receptor
- VDR
vitamin D receptor
References
- Ahmad W., Faiyaz ul Haque M., Brancolini V., Tsou H. C., ul Haque S., Lam H., Aita V. M., Owen J., deBlaquiere M., Frank J., Cserhalmi-Friedman P. B., Leask A., McGrath J. A., Peacocke M., Ahmad M., Ott J., Christiano A. M. Alopecia universalis associated with a mutation in the human hairless gene. Science. 1998;279:720–4. doi: 10.1126/science.279.5351.720. [DOI] [PubMed] [Google Scholar]
- Ahmad W., Panteleyev A. A., Christiano A. M. The molecular basis of congenital atrichia in humans and mice: mutations in the hairless gene. J Investig Dermatol Symp Proc. 1999;4:240–3. doi: 10.1038/sj.jidsp.5640220. [DOI] [PubMed] [Google Scholar]
- Aita V. M., Ahmad W., Panteleyev A. A., Kozlowska U., Kozlowska A., Gilliam T. C., Jablonska S., Christiano A. M. A novel missense mutation (C622G) in the zinc-finger domain of the human hairless gene associated with congenital atrichia with papular lesions. Exp Dermatol. 2000;9:157–62. doi: 10.1034/j.1600-0625.2000.009002157.x. [DOI] [PubMed] [Google Scholar]
- Alonso L. C., Rosenfield R. L. Molecular genetic and endocrine mechanisms of hair growth. Horm Res. 2003a;60:1–13. doi: 10.1159/000070821. [DOI] [PubMed] [Google Scholar]
- Alonso L., Fuchs E. Stem cells in the skin: waste not, Wnt not. Genes Dev. 2003b;17:1189–200. doi: 10.1101/gad.1086903. [DOI] [PubMed] [Google Scholar]
- Balemans W., Van Hul W. Extracellular regulation of BMP signaling in vertebrates: a cocktail of modulators. Dev Biol. 2002;250:231–50. [PubMed] [Google Scholar]
- Beaudoin G. M., 3rd, Sisk J. M., Coulombe P. A., Thompson C. C. Hairless triggers reactivation of hair growth by promoting Wnt signaling. Proc Natl Acad Sci U S A. 2005;102:14653–8. doi: 10.1073/pnas.0507609102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernerd F., Schweizer J., Demarchez M. Dermal cysts of the rhino mouse develop into unopened sebaceous glands. Arch Dermatol Res. 1996;288:586–95. doi: 10.1007/BF02505261. [DOI] [PubMed] [Google Scholar]
- Botchkarev V. A., Sharov A. A. BMP signaling in the control of skin development and hair follicle growth. Differentiation. 2004;72:512–26. doi: 10.1111/j.1432-0436.2004.07209005.x. [DOI] [PubMed] [Google Scholar]
- Brooke H. C. Hairless Mice. J Hered. 1926;17:173–174. [Google Scholar]
- Burke L. J., Baniahmad A. Co-repressors 2000. Faseb J. 2000;14:1876–88. doi: 10.1096/fj.99-0943rev. [DOI] [PubMed] [Google Scholar]
- Cachon-Gonzalez M. B., Fenner S., Coffin J. M., Moran C., Best S., Stoye J. P. Structure and expression of the hairless gene of mice. Proc Natl Acad Sci U S A. 1994;91:7717–21. doi: 10.1073/pnas.91.16.7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cachon-Gonzalez M. B., San-Jose I., Cano A., Vega J. A., Garcia N., Freeman T., Schimmang T., Stoye J. P. The hairless gene of the mouse: relationship of phenotypic effects with expression profile and genotype. Dev Dyn. 1999;216:113–26. doi: 10.1002/(SICI)1097-0177(199910)216:2<113::AID-DVDY3>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Callahan C. A., Oro A. E. Monstrous attempts at adnexogenesis: regulating hair follicle progenitors through Sonic hedgehog signaling. Curr Opin Genet Dev. 2001;11:541–6. doi: 10.1016/s0959-437x(00)00230-6. [DOI] [PubMed] [Google Scholar]
- Cavailles V., Dauvois S., L'Horset F., Lopez G., Hoare S., Kushner P. J., Parker M. G. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. Embo J. 1995;14:3741–51. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan E. F., Gat U., McNiff J. M., Fuchs E. A common human skin tumour is caused by activating mutations in β-catenin. Nat Genet. 1999;21:410–3. doi: 10.1038/7747. [DOI] [PubMed] [Google Scholar]
- Chen J. D., Evans R. M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–7. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- Chien A. J., Presland R. B., Kuechle M. K. Processing of native caspase-14 occurs at an atypical cleavage site in normal epidermal differentiation. Biochem Biophys Res Commun. 2002;296:911–7. doi: 10.1016/s0006-291x(02)02015-6. [DOI] [PubMed] [Google Scholar]
- Cichon S., Anker M., Vogt I. R., Rohleder H., Putzstuck M., Hillmer A., Farooq S. A., Al-Dhafri K. S., Ahmad M., Haque S., Rietschel M., Propping P., Kruse R., Nothen M. M. Cloning, genomic organization, alternative transcripts and mutational analysis of the gene responsible for autosomal recessive universal congenital alopecia. Hum Mol Genet. 1998;7:1671–9. doi: 10.1093/hmg/7.11.1671. [DOI] [PubMed] [Google Scholar]
- Clissold P. M., Ponting C. P. JmjC: cupin metalloenzyme-like domains in jumonji, hairless and phospholipase A2beta. Trends Biochem Sci. 2001;26:7–9. doi: 10.1016/s0968-0004(00)01700-x. [DOI] [PubMed] [Google Scholar]
- Cohen R. N., Brzostek S., Kim B., Chorev M., Wondisford F. E., Hollenberg A. N. The specificity of interactions between nuclear hormone receptors and corepressors is mediated by distinct amino acid sequences within the interacting domains. Mol Endocrinol. 2001;15:1049–61. doi: 10.1210/mend.15.7.0669. [DOI] [PubMed] [Google Scholar]
- DasGupta R., Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development. 1999;126:4557–68. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- del Castillo V., Ruiz-Maldonado R., Carnevale A. Atrichia with papular lesions and mental retardation in two sisters. Int J Dermatol. 1974;13:261–5. doi: 10.1111/j.1365-4362.1974.tb05078.x. [DOI] [PubMed] [Google Scholar]
- Downes M., Ordentlich P., Kao H. Y., Alvarez J. G., Evans R. M. Identification of a nuclear domain with deacetylase activity. Proc Natl Acad Sci U S A. 2000;97:10330–5. doi: 10.1073/pnas.97.19.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussault I., Fawcett D., Matthyssen A., Bader J. A., Giguere V. Orphan nuclear receptor ROR α-deficient mice display the cerebellar defects of staggerer. Mech Dev. 1998;70:147–53. doi: 10.1016/s0925-4773(97)00187-1. [DOI] [PubMed] [Google Scholar]
- Eckhart L., Declercq W., Ban J., Rendl M., Lengauer B., Mayer C., Lippens S., Vandenabeele P., Tschachler E. Terminal differentiation of human keratinocytes and stratum corneum formation is associated with caspase-14 activation. J Invest Dermatol. 2000;115:1148–51. doi: 10.1046/j.1523-1747.2000.00205.x. [DOI] [PubMed] [Google Scholar]
- Fernandes I., Bastien Y., Wai T., Nygard K., Lin R., Cormier O., Lee H. S., Eng F., Bertos N. R., Pelletier N., Mader S., Han V. K., Yang X. J., White J. H. Ligand-dependent nuclear receptor corepressor LCoR functions by histone deacetylase-dependent and -independent mechanisms. Mol Cell. 2003;11:139–50. doi: 10.1016/s1097-2765(03)00014-5. [DOI] [PubMed] [Google Scholar]
- Garcia-Atares N., San Jose I., Cabo R., Vega J. A., Represa J. Changes in the cerebellar cortex of hairless Rhino-J mice (hr-rh-j) Neurosci Lett. 1998;256:13–6. doi: 10.1016/s0304-3940(98)00757-5. [DOI] [PubMed] [Google Scholar]
- Gat U., DasGupta R., Degenstein L., Fuchs E. De Novo hair follicle morphogenesis and hair tumors in mice expressing a truncated β-catenin in skin. Cell. 1998;95:605–14. doi: 10.1016/s0092-8674(00)81631-1. [DOI] [PubMed] [Google Scholar]
- Glass C. K., Rosenfeld M. G. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–41. [PubMed] [Google Scholar]
- Guenther M. G., Lane W. S., Fischle W., Verdin E., Lazar M. A., Shiekhattar R. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 2000;14:1048–57. [PMC free article] [PubMed] [Google Scholar]
- Guenther M. G., Barak O., Lazar M. A. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol. 2001;21:6091–101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton B. A., Frankel W. N., Kerrebrock A. W., Hawkins T. L., FitzHugh W., Kusumi K., Russell L. B., Mueller K. L., van Berkel V., Birren B. W., Kruglyak L., Lander E. S. Disruption of the nuclear hormone receptor RORalpha in staggerer mice. Nature. 1996;379:736–9. doi: 10.1038/379736a0. [DOI] [PubMed] [Google Scholar]
- Hardy M. H. The secret life of the hair follicle. Trends Genet. 1992;8:55–61. doi: 10.1016/0168-9525(92)90350-d. [DOI] [PubMed] [Google Scholar]
- Heery D. M., Kalkhoven E., Hoare S., Parker M. G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–6. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- Hong S. H., Privalsky M. L. The SMRT corepressor is regulated by a MEK-1 kinase pathway: inhibition of corepressor function is associated with SMRT phosphorylation and nuclear export. Mol Cell Biol. 2000;20:6612–25. doi: 10.1128/mcb.20.17.6612-6625.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlein A. J., Naar A. M., Heinzel T., Torchia J., Gloss B., Kurokawa R., Ryan A., Kamei Y., Soderstrom M., Glass C. K. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- Hsieh J. C., Sisk J. M., Jurutka P. W., Haussler C. A., Slater S. A., Haussler M. R., Thompson C. C. Physical and functional interaction between the vitamin D receptor and hairless corepressor, two proteins required for hair cycling. J Biol Chem. 2003;278:38665–74. doi: 10.1074/jbc.M304886200. [DOI] [PubMed] [Google Scholar]
- Huang E. Y., Zhang J., Miska E. A., Guenther M. G., Kouzarides T., Lazar M. A. Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes Dev. 2000;14:45–54. [PMC free article] [PubMed] [Google Scholar]
- Huelsken J., Vogel R., Erdmann B., Cotsarelis G., Birchmeier W. β-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell. 2001;105:533–45. doi: 10.1016/s0092-8674(01)00336-1. [DOI] [PubMed] [Google Scholar]
- Hu X., Lazar M. A. The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature. 1999;402:93–6. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- Hwang M., Morasso M. I. The novel murine Ca2+-binding protein, Scarf, is differentially expressed during epidermal differentiation. J Biol Chem. 2003;278:47827–33. doi: 10.1074/jbc.M306561200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itasaki N., Jones C. M., Mercurio S., Rowe A., Domingos P. M., Smith J. C., Krumlauf R. Wise, a context-dependent activator and inhibitor of Wnt signalling. Development. 2003;130:4295–305. doi: 10.1242/dev.00674. [DOI] [PubMed] [Google Scholar]
- Jahoda C. A., Horne K. A., Oliver R. F. Induction of hair growth by implantation of cultured dermal papilla cells. Nature. 1984;311:560–2. doi: 10.1038/311560a0. [DOI] [PubMed] [Google Scholar]
- Jepsen K., Rosenfeld M. G. Biological roles and mechanistic actions of co-repressor complexes. J Cell Sci. 2002;115:689–98. doi: 10.1242/jcs.115.4.689. [DOI] [PubMed] [Google Scholar]
- Kao H. Y., Downes M., Ordentlich P., Evans R. M. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev. 2000;14:55–66. [PMC free article] [PubMed] [Google Scholar]
- Kim T. G., Kraus J. C., Chen J., Lee Y. JUMONJI, a critical factor for cardiac development, functions as a transcriptional repressor. J Biol Chem. 2003;278:42247–55. doi: 10.1074/jbc.M307386200. [DOI] [PubMed] [Google Scholar]
- Klose R. J., Yamane K., Bae Y., Zhang D., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442:312–6. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- Koshizuka Y., Ikegawa S., Sano M., Nakamura K., Nakamura Y. Isolation of novel mouse genes associated with ectopic ossification by differential display method using ttw, a mouse model for ectopic ossification. Cytogenet Cell Genet. 2001;94:163–8. doi: 10.1159/000048809. [DOI] [PubMed] [Google Scholar]
- Krupnik V. E., Sharp J. D., Jiang C., Robison K., Chickering T. W., Amaravadi L., Brown D. E., Guyot D., Mays G., Leiby K., Chang B., Duong T., Goodearl A. D., Gearing D. P., Sokol S. Y., McCarthy S. A. Functional and structural diversity of the human Dickkopf gene family. Gene. 1999;238:301–13. doi: 10.1016/s0378-1119(99)00365-0. [DOI] [PubMed] [Google Scholar]
- Laurikkala J., Kassai Y., Pakkasjarvi L., Thesleff I., Itoh N. Identification of a secreted BMP antagonist, ectodin, integrating BMP, FGF, and SHH signals from the tooth enamel knot. Dev Biol. 2003;264:91–105. doi: 10.1016/j.ydbio.2003.08.011. [DOI] [PubMed] [Google Scholar]
- Li J., Wang J., Wang J., Nawaz Z., Liu J. M., Qin J., Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. Embo J. 2000a;19:4342–50. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Leo C., Schroen D. J., Chen J. D. Characterization of receptor interaction and transcriptional repression by the corepressor SMRT. Mol Endocrinol. 1997a;11:2025–37. doi: 10.1210/mend.11.13.0028. [DOI] [PubMed] [Google Scholar]
- Lindner G., Botchkarev V. A., Botchkareva N. V., Ling G., van der Veen C., Paus R. Analysis of apoptosis during hair follicle regression (catagen) Am J Pathol. 1997;151:1601–17. [PMC free article] [PubMed] [Google Scholar]
- Lippens S., Kockx M., Knaapen M., Mortier L., Polakowska R., Verheyen A., Garmyn M., Zwijsen A., Formstecher P., Huylebroeck D., Vandenabeele P., Declercq W. Epidermal differentiation does not involve the pro-apoptotic executioner caspases, but is associated with caspase-14 induction and processing. Cell Death Differ. 2000;7:1218–24. doi: 10.1038/sj.cdd.4400785. [DOI] [PubMed] [Google Scholar]
- Li M., Indra A. K., Warot X., Brocard J., Messaddeq N., Kato S., Metzger D., Chambon P. Skin abnormalities generated by temporally controlled RXRalpha mutations in mouse epidermis. Nature. 2000b;407:633–6. doi: 10.1038/35036595. [DOI] [PubMed] [Google Scholar]
- Li Y. C., Pirro A. E., Amling M., Delling G., Baron R., Bronson R., Demay M. B. Targeted ablation of the vitamin D receptor: an animal model of vitamin D-dependent rickets type II with alopecia. Proc Natl Acad Sci U S A. 1997b;94:9831–5. doi: 10.1073/pnas.94.18.9831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Celso C., Prowse D. M., Watt F. M. Transient activation of β-catenin signalling in adult mouse epidermis is sufficient to induce new hair follicles but continuous activation is required to maintain hair follicle tumours. Development. 2004;131:1787–99. doi: 10.1242/dev.01052. [DOI] [PubMed] [Google Scholar]
- Logan C. Y., Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- Malloy P. J., Wang J., Jensen K., Feldman D. Modulation of vitamin d receptor activity by the corepressor hairless: differential effects of hairless isoforms. Endocrinology. 2009;150:4950–7. doi: 10.1210/en.2009-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf D. J., Evans R. M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–50. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- Mann S. J. Hair loss and cyst formation in hairless and rhino mutant mice. Anat Rec. 1971;170:485–99. doi: 10.1002/ar.1091700409. [DOI] [PubMed] [Google Scholar]
- Matysiak-Scholze U., Nehls M. The structural integrity of ROR α isoforms is mutated in staggerer mice: cerebellar coexpression of ROR alpha1 and ROR alpha4. Genomics. 1997;43:78–84. doi: 10.1006/geno.1997.4757. [DOI] [PubMed] [Google Scholar]
- McInerney E. M., Rose D. W., Flynn S. E., Westin S., Mullen T. M., Krones A., Inostroza J., Torchia J., Nolte R. T., Assa-Munt N., Milburn M. V., Glass C. K., Rosenfeld M. G. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 1998;12:3357–68. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill B. J., Gat U., DasGupta R., Fuchs E. Tcf3 and Lef1 regulate lineage differentiation of multipotent stem cells in skin. Genes Dev. 2001;15:1688–705. doi: 10.1101/gad.891401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misiti S., Schomburg L., Yen P. M., Chin W. W. Expression and hormonal regulation of coactivator and corepressor genes. Endocrinology. 1998;139:2493–500. doi: 10.1210/endo.139.5.5971. [DOI] [PubMed] [Google Scholar]
- Montagna W., Chase H. B., Melaragno H. P. The skin of hairless mice. I. The formation of cysts and the distribution of lipids. J Invest Dermatol. 1952;19:83–94. doi: 10.1038/jid.1952.67. [DOI] [PubMed] [Google Scholar]
- Moraitis A. N., Giguere V., Thompson C. C. Novel mechanism of nuclear receptor corepressor interaction dictated by activation function 2 helix determinants. Mol Cell Biol. 2002;22:6831–41. doi: 10.1128/MCB.22.19.6831-6841.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraitis A. N., Giguere V. The co-repressor hairless protects RORalpha orphan nuclear receptor from proteasome-mediated degradation. J Biol Chem. 2003;278:52511–8. doi: 10.1074/jbc.M308152200. [DOI] [PubMed] [Google Scholar]
- Nagy L., Kao H. Y., Love J. D., Li C., Banayo E., Gooch J. T., Krishna V., Chatterjee K., Evans R. M., Schwabe J. W. Mechanism of corepressor binding and release from nuclear hormone receptors. Genes Dev. 1999;13:3209–16. doi: 10.1101/gad.13.24.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy L., Kao H. Y., Chakravarti D., Lin R. J., Hassig C. A., Ayer D. E., Schreiber S. L., Evans R. M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–80. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- Niemann C., Owens D. M., Hulsken J., Birchmeier W., Watt F. M. Expression of DeltaNLef1 in mouse epidermis results in differentiation of hair follicles into squamous epidermal cysts and formation of skin tumours. Development. 2002;129:95–109. doi: 10.1242/dev.129.1.95. [DOI] [PubMed] [Google Scholar]
- Oliver R. F., Jahoda C. A. Dermal-epidermal interactions. Clin Dermatol. 1988;6:74–82. doi: 10.1016/0738-081x(88)90069-7. [DOI] [PubMed] [Google Scholar]
- Oomizu S., Sahuc F., Asahina K., Inamatsu M., Matsuzaki T., Sasaki M., Obara M., Yoshizato K. Kdap, a novel gene associated with the stratification of the epithelium. Gene. 2000;256:19–27. doi: 10.1016/s0378-1119(00)00357-7. [DOI] [PubMed] [Google Scholar]
- Ordentlich P., Downes M., Xie W., Genin A., Spinner N. B., Evans R. M. Unique forms of human and mouse nuclear receptor corepressor SMRT. Proc Natl Acad Sci U S A. 1999;96:2639–44. doi: 10.1073/pnas.96.6.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orwin D. F., Chase H. B., Silver A. F. Catagen in the hairless house mouse. Am J Anat. 1967;121:489–507. doi: 10.1002/aja.1001210305. [DOI] [PubMed] [Google Scholar]
- Panteleyev A. A., Paus R., Ahmad W., Sundberg J. P., Christiano A. M. Molecular and functional aspects of the hairless (hr) gene in laboratory rodents and humans. Exp Dermatol. 1998a;7:249–67. doi: 10.1111/j.1600-0625.1998.tb00295.x-i1. [DOI] [PubMed] [Google Scholar]
- Panteleyev A. A., Ahmad W., Malashenko A. M., Ignatieva E. L., Paus R., Sundberg J. P., Christiano A. M. Molecular basis for the rhino Yurlovo (hr(rhY)) phenotype: severe skin abnormalities and female reproductive defects associated with an insertion in the hairless gene. Exp Dermatol. 1998b;7:281–8. doi: 10.1111/j.1600-0625.1998.tb00298.x-i1. [DOI] [PubMed] [Google Scholar]
- Panteleyev A. A., Paus R., Christiano A. M. Patterns of hairless (hr) gene expression in mouse hair follicle morphogenesis and cycling. Am J Pathol. 2000;157:1071–9. doi: 10.1016/S0002-9440(10)64621-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panteleyev A. A., Botchkareva N. V., Sundberg J. P., Christiano A. M., Paus R. The role of the hairless (hr) gene in the regulation of hair follicle catagen transformation. Am J Pathol. 1999;155:159–71. doi: 10.1016/S0002-9440(10)65110-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazin M. J., Kadonaga J. T. What's up and down with histone deacetylation and transcription? Cell. 1997;89:325–8. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- Perissi V., Rosenfeld M. G. Controlling nuclear receptors: the circular logic of cofactor cycles. Nat Rev Mol Cell Biol. 2005;6:542–54. doi: 10.1038/nrm1680. [DOI] [PubMed] [Google Scholar]
- Perissi V., Staszewski L. M., McInerney E. M., Kurokawa R., Krones A., Rose D. W., Lambert M. H., Milburn M. V., Glass C. K., Rosenfeld M. G. Molecular determinants of nuclear receptor-corepressor interaction. Genes Dev. 1999;13:3198–208. doi: 10.1101/gad.13.24.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter G. B., Beaudoin G. M., 3rd, DeRenzo C. L., Zarach J. M., Chen S. H., Thompson C. C. The hairless gene mutated in congenital hair loss disorders encodes a novel nuclear receptor corepressor. Genes Dev. 2001;15:2687–701. doi: 10.1101/gad.916701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter G. B., Zarach J. M., Sisk J. M., Thompson C. C. The thyroid hormone-regulated corepressor hairless associates with histone deacetylases in neonatal rat brain. Mol Endocrinol. 2002;16:2547–60. doi: 10.1210/me.2002-0115. [DOI] [PubMed] [Google Scholar]
- Privalsky M. L. The role of corepressors in transcriptional regulation by nuclear hormone receptors. Annu Rev Physiol. 2004;66:315–60. doi: 10.1146/annurev.physiol.66.032802.155556. [DOI] [PubMed] [Google Scholar]
- Reddy S., Andl T., Bagasra A., Lu M. M., Epstein D. J., Morrisey E. E., Millar S. E. Characterization of Wnt gene expression in developing and postnatal hair follicles and identification of Wnt5a as a target of Sonic hedgehog in hair follicle morphogenesis. Mech Dev. 2001;107:69–82. doi: 10.1016/s0925-4773(01)00452-x. [DOI] [PubMed] [Google Scholar]
- Sakai Y., Kishimoto J., Demay M. B. Metabolic and cellular analysis of alopecia in vitamin D receptor knockout mice. J Clin Invest. 2001;107:961–6. doi: 10.1172/JCI11676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sande S., Privalsky M. L. Identification of TRACs (T3 receptor-associating cofactors), a family of cofactors that associate with, and modulate the activity of, nuclear hormone receptors. Mol Endocrinol. 1996;10:813–25. doi: 10.1210/mend.10.7.8813722. [DOI] [PubMed] [Google Scholar]
- Skorija K., Cox M., Sisk J. M., Dowd D. R., MacDonald P. N., Thompson C. C., Demay M. B. Ligand-independent actions of the vitamin D receptor maintain hair follicle homeostasis. Mol Endocrinol. 2005;19:855–62. doi: 10.1210/me.2004-0415. [DOI] [PubMed] [Google Scholar]
- Smith C. L., O'Malley B. W. Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr Rev. 2004;25:45–71. doi: 10.1210/er.2003-0023. [DOI] [PubMed] [Google Scholar]
- Sprecher E., Bergman R., Szargel R., Raz T., Labay V., Ramon M., Baruch-Gershoni R., Friedman-Birnbaum R., Cohen N. Atrichia with papular lesions maps to 8p in the region containing the human hairless gene. Am J Med Genet. 1998;80:546–50. doi: 10.1002/(sici)1096-8628(19981228)80:5<546::aid-ajmg28>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Sprecher E., Lestringant G. G., Szargel R., Bergman R., Labay V., Frossard P. M., Friedman-Birnbaum R., Cohen N. Atrichia with papular lesions resulting from a nonsense mutation within the human hairless gene. J Invest Dermatol. 1999b;113:687–90. doi: 10.1046/j.1523-1747.1999.00723.x. [DOI] [PubMed] [Google Scholar]
- Sprecher E., Bergman R., Szargel R., Friedman-Birnbaum R., Cohen N. Identification of a genetic defect in the hairless gene in atrichia with papular lesions: evidence for phenotypic heterogeneity among inherited atrichias. Am J Hum Genet. 1999a;64:1323–9. doi: 10.1086/302368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel J. H., White R., Parker M. G. Role of the RIP140 corepressor in ovulation and adipose biology. J Endocrinol. 2005;185:1–9. doi: 10.1677/joe.1.05896. [DOI] [PubMed] [Google Scholar]
- Steinmayr M., Andre E., Conquet F., Rondi-Reig L., Delhaye-Bouchaud N., Auclair N., Daniel H., Crepel F., Mariani J., Sotelo C., Becker-Andre M. staggerer phenotype in retinoid-related orphan receptor α-deficient mice. Proc Natl Acad Sci U S A. 1998;95:3960–5. doi: 10.1073/pnas.95.7.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenn K. S., Paus R. Controls of hair follicle cycling. Physiol Rev. 2001;81:449–494. doi: 10.1152/physrev.2001.81.1.449. [DOI] [PubMed] [Google Scholar]
- Stoye J. P., Fenner S., Greenoak G. E., Moran C., Coffin J. M. Role of endogenous retroviruses as mutagens: the hairless mutation of mice. Cell. 1988;54:383–91. doi: 10.1016/0092-8674(88)90201-2. [DOI] [PubMed] [Google Scholar]
- Sumner F. Hairless Mice. Journal of Heredity. 1924;15:475–481. [Google Scholar]
- Takeuchi T., Yamazaki Y., Katoh-Fukui Y., Tsuchiya R., Kondo S., Motoyama J., Higashinakagawa T. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Dev. 1995;9:1211–22. doi: 10.1101/gad.9.10.1211. [DOI] [PubMed] [Google Scholar]
- Takeuchi T., Kojima M., Nakajima K., Kondo S. jumonji gene is essential for the neurulation and cardiac development of mouse embryos with a C3H/He background. Mech Dev. 1999;86:29–38. doi: 10.1016/s0925-4773(99)00100-8. [DOI] [PubMed] [Google Scholar]
- Takeuchi T., Watanabe Y., Takano-Shimizu T., Kondo S. Roles of jumonji and jumonji family genes in chromatin regulation and development. Dev Dyn. 2006;235:2449–59. doi: 10.1002/dvdy.20851. [DOI] [PubMed] [Google Scholar]
- Thompson C. C., Beaudoin G. M. J. Advances in Developmental Biology. New York: Elsevier; 2006a. Hairless: A nuclear receptor corepressor essential for skin function. pp. 357–387. [Google Scholar]
- Thompson C. C., Sisk J. M., Beaudoin G. M., 3rd. Hairless and Wnt signaling: allies in epithelial stem cell differentiation. Cell Cycle. 2006b;5:1913–7. doi: 10.4161/cc.5.17.3189. [DOI] [PubMed] [Google Scholar]
- Thompson C. C., Bottcher M. C. The product of a thyroid hormone-responsive gene interacts with thyroid hormone receptors. Proc Natl Acad Sci U S A. 1997;94:8527–32. doi: 10.1073/pnas.94.16.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C. C., Potter G. B. Thyroid hormone action in neural development. Cereb Cortex. 2000;10:939–45. doi: 10.1093/cercor/10.10.939. [DOI] [PubMed] [Google Scholar]
- Thompson C. C. Thyroid hormone-responsive genes in developing cerebellum include a novel synaptotagmin and a hairless homolog. J Neurosci. 1996;16:7832–40. doi: 10.1523/JNEUROSCI.16-24-07832.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda M., Shirato H., Nakajima K., Kojima M., Takahashi M., Kubota M., Suzuki-Migishima R., Motegi Y., Yokoyama M., Takeuchi T. jumonji downregulates cardiac cell proliferation by repressing cyclin D1 expression. Dev Cell. 2003;5:85–97. doi: 10.1016/s1534-5807(03)00189-8. [DOI] [PubMed] [Google Scholar]
- Tsukada Y., Fang J., Erdjument-Bromage H., Warren M. E., Borchers C. H., Tempst P., Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–6. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- Tumbar T., Guasch G., Greco V., Blanpain C., Lowry W. E., Rendl M., Fuchs E. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–63. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Mater D., Kolligs F. T., Dlugosz A. A., Fearon E. R. Transient activation of β -catenin signaling in cutaneous keratinocytes is sufficient to trigger the active growth phase of the hair cycle in mice. Genes Dev. 2003;17:1219–24. doi: 10.1101/gad.1076103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Malloy P. J., Feldman D. Interactions of the vitamin D receptor with the corepressor hairless: analysis of hairless mutants in atrichia with papular lesions. J Biol Chem. 2007;282:25231–9. doi: 10.1074/jbc.M702939200. [DOI] [PubMed] [Google Scholar]
- Webb P., Anderson C. M., Valentine C., Nguyen P., Marimuthu A., West B. L., Baxter J. D., Kushner P. J. The nuclear receptor corepressor (N-CoR) contains three isoleucine motifs (I/LXXII) that serve as receptor interaction domains (IDs) Mol Endocrinol. 2000;14:1976–85. doi: 10.1210/mend.14.12.0566. [DOI] [PubMed] [Google Scholar]
- Wen Y., Liu Y., Xu Y., Zhao Y., Hua R., Wang K., Sun M., Li Y., Yang S., Zhang X. J., Kruse R., Cichon S., Betz R. C., Nothen M. M., van Steensel M. A., van Geel M., Steijlen P. M., Hohl D., Huber M., Dunnill G. S., Kennedy C., Messenger A., Munro C. S., Terrinoni A., Hovnanian A., Bodemer C., de Prost Y., Paller A. S., Irvine A. D., Sinclair R., Green J., Shang D., Liu Q., Luo Y., Jiang L., Chen H. D., Lo W. H., McLean W. H., He C. D., Zhang X. Loss-of-function mutations of an inhibitory upstream ORF in the human hairless transcript cause Marie Unna hereditary hypotrichosis. Nat Genet. 2009;41:228–33. doi: 10.1038/ng.276. [DOI] [PubMed] [Google Scholar]
- Wen Y. D., Perissi V., Staszewski L. M., Yang W. M., Krones A., Glass C. K., Rosenfeld M. G., Seto E. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc Natl Acad Sci U S A. 2000;97:7202–7. doi: 10.1073/pnas.97.13.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H. E., Stanley T. B., Montana V. G., Lambert M. H., Shearer B. G., Cobb J. E., McKee D. D., Galardi C. M., Plunket K. D., Nolte R. T., Parks D. J., Moore J. T., Kliewer S. A., Willson T. M., Stimmel J. B. Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARalpha. Nature. 2002;415:813–7. doi: 10.1038/415813a. [DOI] [PubMed] [Google Scholar]
- Yanagita M., Oka M., Watabe T., Iguchi H., Niida A., Takahashi S., Akiyama T., Miyazono K., Yanagisawa M., Sakurai T. USAG-1: a bone morphogenetic protein antagonist abundantly expressed in the kidney. Biochem Biophys Res Commun. 2004;316:490–500. doi: 10.1016/j.bbrc.2004.02.075. [DOI] [PubMed] [Google Scholar]
- Yang F., Stenoien D. L., Strittmatter E. F., Wang J., Ding L., Lipton M. S., Monroe M. E., Nicora C. D., Gristenko M. A., Tang K., Fang R., Adkins J. N., Camp D. G., 2nd, Chen D. J., Smith R. D. Phosphoproteome profiling of human skin fibroblast cells in response to low- and high-dose irradiation. J Proteome Res. 2006;5:1252–60. doi: 10.1021/pr060028v. [DOI] [PubMed] [Google Scholar]
- Yoon H. G., Chan D. W., Huang Z. Q., Li J., Fondell J. D., Qin J., Wong J. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. Embo J. 2003;22:1336–46. doi: 10.1093/emboj/cdg120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa T., Handa Y., Uematsu Y., Takeda S., Sekine K., Yoshihara Y., Kawakami T., Arioka K., Sato H., Uchiyama Y., Masushige S., Fukamizu A., Matsumoto T., Kato S. Mice lacking the vitamin D receptor exhibit impaired bone formation, uterine hypoplasia and growth retardation after weaning. Nat Genet. 1997;16:391–6. doi: 10.1038/ng0897-391. [DOI] [PubMed] [Google Scholar]
- Zarach J. M., Beaudoin G. M., 3rd, Coulombe P. A., Thompson C. C. The co-repressor hairless has a role in epithelial cell differentiation in the skin. Development. 2004;131:4189–200. doi: 10.1242/dev.01303. [DOI] [PubMed] [Google Scholar]
- Zhang D., Yoon H. G., Wong J. JMJD2A is a novel N-CoR-interacting protein and is involved in repression of the human transcription factor achaete scute-like homologue 2 (ASCL2/Hash2) Mol Cell Biol. 2005;25:6404–14. doi: 10.1128/MCB.25.15.6404-6414.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]