Abstract
A series of substituted 6-arylquinazolin-4-amines were prepared and analyzed as inhibitors of Clk4. Synthesis, structure activity-relationships and the selectivity of a potent analogue against a panel of 402 kinases are presented. Inhibition of Clk4 by these agents at varied concentrations of assay substrates (ATP and receptor peptide) highly suggests that this chemotype is an ATP competitive inhibitor. Molecular docking provides further evidence that inhibition is the result of binding at the kinase hinge region. Selected compounds represent novel tools capable of potent and selective inhibition of Clk1, Clk4 and Dyrk1A.
Keywords: kinase inhibition, pre-mRNA splicing, Clk, Dyrk1A
The removal of intron sequences from genes occurs via the actions of the splicesome, a protein complex that removes intervening sequences at the nuclear pre-mRNA level to afford properly coded mRNA for translation.1,2 Many genes produce multiple mRNA isoforms through the actions of alternative splicing and, importantly, numerous human diseases are caused by improper splicing.3 Exogenous manipulation of the splicesome is theorized to be a powerful means to control gene translation and ultimately correct disease phenotypes via rectification of splicing abnormalities.
The modulation of kinases provides a powerful mechanism to control numerous aspects of cell function and offers the potential for the management of many diseases.4 Small molecule inhibitors of kinases (both selective and promiscuous) represent important biochemical tools for basic research and several kinase inhibitors have gained FDA approval as drugs.5,6 In 2002, Manning et al reviewed the human kinome and found 518 putative kinase genes7 providing a roadmap to explore the consequences of kinase modulation within cellular biology. The possible control of gene splicing through the modulation of kinase activity represents an intriguing concept.8 There are several reports of kinases that alter the function of the splicesome. Among these are the cdc2-like kinase (Clk) family.9 A major target of Clk kinases is the prominent family of serine- and arginine-rich (SR) splicing proteins10,11 which are involved in the assembly of the splicesome and are implicated in both constitutive and alternative splicing control and selection of splicing sites.12,13
The Clk family contains four characterized isoforms (Clk1, Clk2, Clk3 and Clk4). The Clks are capable of auto-phosphorylation (at serine, threonine and tyrosine residues) and phosphorylation of exogenous proteins (at serine and threonine residues). Members of the Clk family have been implicated in the regulation of alternative splicing of PKC II14, TF15, Tau16 and β-globin17 pre-mRNA. These studies suggest that small molecule modulation of the Clk family of kinases may represent an important mechanism for the control of mRNA splicing.
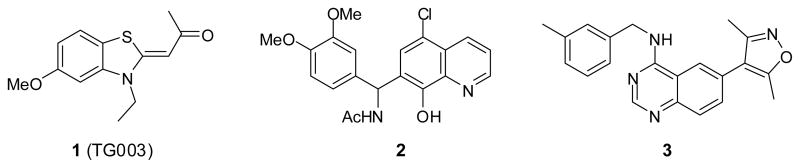
Hagiwara and coworkers have reported TG003 (1)(Figure 1) as a small molecule with low-nanomolar IC50 values versus Clk1 and Clk4.17 Additionally, Clk inhibitors are presented in patent reports from Sirtris Pharmaceuticals (no structures presented) and Chronogen Inc (a series of substituted quinolines related to structure 2).18,19 The report from Hagiwara and coworkers does not define the selectivity of TG003 beyond the Clk family and 4 additional kinases (PKA, PKC, SRPK1 and SRPK2) and the patents do not disclose details regarding structure-activity relationships nor selectivity. Thus, a need remains for small molecule probes of this important class of enzymes. Here, we report a novel class of quinazoline small molecules as potent and selective inhibitors of Clk1 and Clk4.
Figure 1.
Structures of known Clk inhibitors 1 (TG003), 2 and the lead structure 3.
We recently performed a high-throughput screen for small molecule modulators of Lamin A splicing (PubChem AID: 1487); details of this study will be reported elsewhere. Among the actives was a series of substituted quinazolines including 3 (Figure 1). The previously reported ability of quinazoline small molecules to modulate kinases through competitive inhibition at the ATP binding site suggested potential kinase stimulated influence of splicing. Given the association of the Clk family with splicing, we speculated that 3 might be a Clk inhibitor and this was confirmed via a commercial kinase profile.20 Based on these results, we examined selected in-house compound libraries for Clk4 inhibition via a bioluminescent, luciferase-based assay capable of visualizing substrate (ATP) depletion and product (ADP) formation.21 In primary screening, this biochemical assay revealed that 3 (Figure 1) possessed an IC50 value versus Clk4 of 316 nM.
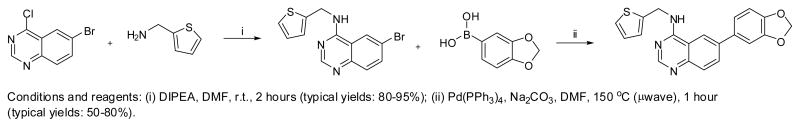
Based upon this result, we examined the SAR for this chemotype versus Clk4. Key to this effort was the synthetic preparation of these agents. In our hands, the most straightforward synthesis began with 6-bromo-4-chloroquinazoline (Scheme 1). Addition of primary and secondary amines (including aliphatic, benzylic, heteroaromatic and substituted analines) was accomplished in DMF with Hunig’s base at room temperature. The resulting 6-bromo-N-substituted-quinazolin-4-amines were then subjected to standard Suzuki-Miyaura couplings with various aryl boronic acids using tetrakis(triphenylphosphine)palladium(0) and sodium carbonate in DMF and heating using microwave irradiation. This general procedure22 resulted in the production of numerous N-substituted-6-arylquinazolin-4-amines including 4 which were purified via reverse-phase HPLC methods prior to testing in biological assays.
Scheme 1.
Conditions and reagents: (i) DIPEA, DMF, r.t., 2 hours (typical yields: 80–95%); (ii) Pd (PPh3)4, Na2CO3, DM (typical yields: 50–80%).
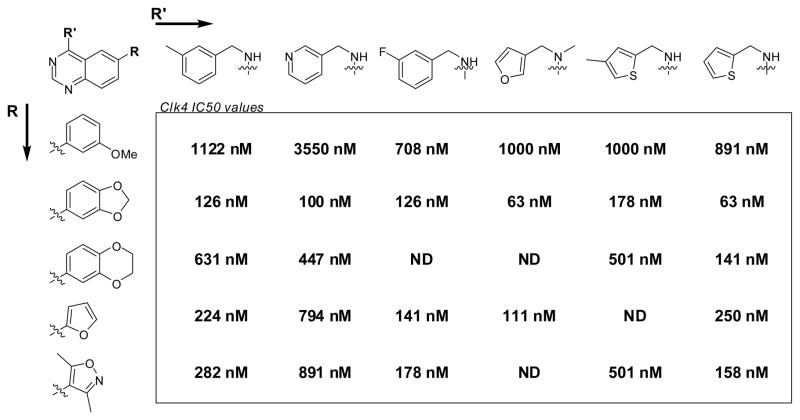
The primary phenotypic screen and the follow-up biochemical Clk4 assay were performed on the NIH Molecular Libraries Small Molecule Repository and several internal NCGC compound libraries. Each assay was performed using the quantitative high-throughput screening method whereby each molecular entity is screened in a dose-response format.23 This screening format provides an unprecedented level of detail with regards to nascent SAR from the primary screening data. The resulting analysis of both of the aforementioned screens provided several clues as to which substitution patterns were likely to result in active compounds. Substitutions at the 4 position of the quinazoline core scaffold were limited to substituted benzylic systems (for instance m-tolylmethanamine, pyridin-3-ylmethanamine and (3-fluorophenyl)methanamine)) and various heterocyclic methylamines (for instance 1-(furan-3-yl)-N-methylmethanamine, (4-methylthiophen-2-yl)methanamine and thiophen-2-ylmethanamine). Substitutions at the 6 position of the quinazoline core scaffold were limited to various aryl groups (for instance 3-methoxybenzene, benzo[d][1,3]dioxole, 2,3-dihydrobenzo[b][1,4]dioxine, furan, and 3,5-dimethylisoxazole). As SAR is often not additive, we chose to incorporate all of these substitutions in matrix format in hopes of identifying more potent Clk4 inhibitors and revealing as much SAR as possible. The results of this library are listed in Figure 2. The original lead 3 was found to possess an IC50 value of 282 nM (very similar to the IC50 value determined in the primary biochemical screen). Additionally, two novel small molecules were found with potencies of 63 nM including 6-(benzo[d][1,3]dioxol-5-yl)-N-(thiophen-2-ylmethyl)quinazolin-4-amine (4). The SAR revealed that the benzo[d][1,3]dioxole was consistently favored in the 6 position of the quinazoline ring. The SAR of the amine substitutions was more complex. The thiophen-2-ylmethanamine substitution was typically favored, but several structural combinations favored other moieties at this position.
Figure 2.
SAR results for matrix library of Clk4 inhibitors. Data represents the results from three separate experiments. All compounds achieved a maximum response greater than 90%.
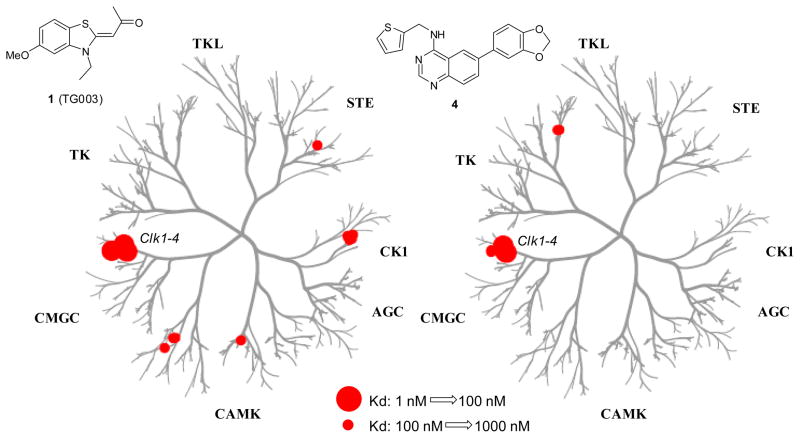
Prior to additional investigations into the SAR and optimization of this chemotype, it was important for us to gain an appreciation of the selectivity of these compounds towards the Clk family of kinases. Quinazoline based small molecules have been reported as potent inhibitors of numerous kinase families. Any small molecule Clk inhibitor that is additionally capable of promiscuous inhibition across the kinome will be of limited value as a tool compound for evaluating Clk biochemistry. To assess the selectivity of this general chemotype we choose to submit a representative reagent (analogue 4) across a commercial panel of kinases. In 2008, Ambit Biosciences reported a quantitative analysis of 38 known kinase inhibitors across a panel of 317 kinases.24 This commercially available panel contained 402 kinases at the time of submission and is based upon a competition binding assay of kinases fused to a proprietary tag. The data is first recorded as a % of kinase bound to an immobilized ligand in the presence and absence of the test reagent as compared to DMSO. Activities beyond a selected threshold are submitted for Kd determination. In addition to profiling 4 versus this panel, we submitted the reported small molecule Clk1/4 inhibitor TG003 (1) to generate a comparison between both agents. The results (tabulated in a dendrogram representation25) are shown in figure 3 and demonstrate that both agents are remarkably selective. TG003 (1) was determined to have Kd’s of 19 nM, 95 nM and 30 nM versus Clk1, Clk2 and Clk4, respectively. The Kd for TG003 (1) versus Clk3 was 3 μM. It was also found that TG003 had activity versus CSNK1D (150 nM), CSNK1E (300 nM), Dyrk1A (12 nM), Dyrk1B (130 nM), PIM1 (130 nM), PIM3 (280 nM) and Ysk4 (290 nM). The novel quinazoline 4 was found to have Kd’s of 37 nM, 50 nM and 27 nM versus Clk1, Clk4 and Dyrk1A, respectively. The only other locus of relevant activity (below 500 nM) was found for binding to the endothelial growth factor receptor (EGFR) (230 nM).
Figure 3.
Dendrogram representation of the selectivity profile for kinase binding by TG003 (1) and 4 within a panel of 402 kinases. Activity for 1: Clk1 = 19 nM, Clk2 = 95 nM, Clk3 = 3000 nM, Clk4 = 30 nM, CSNK1D = 150 nM, CSNK1E = 300 nM, CSNK1G2 = 270 nM, CSNK1G3 = 290 nM, Dyrk1A = 12 nM, Dyrk1B = 130 nM, PIM1 = 130 nM, PIM3 = 280 nM, Ysk4 = 290 nM. Activity for 4: Clk1 = 37 nM, Clk2 = 680 nM, Clk3 = 470 nM, Clk4 = 50 nM, Dyrk1A = 27 nM, Dyrk1B = 430 nM, EGFR = 230 nM. Note: 4 was active (>250 nM) versus numerous EGFR mutants.
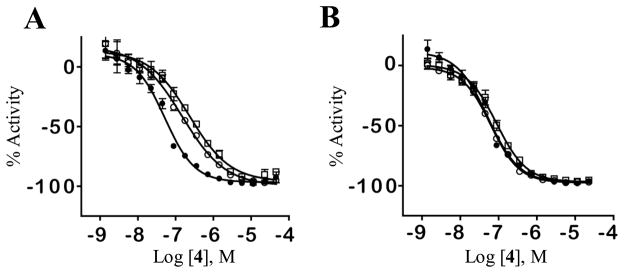
Based upon the potency and selectivity for 4 we next aimed to understand the binding mechanism of this chemotype at Clk1 and Clk4. We explored the binding modality by examining the inhibitory capacity of 4 in settings that varied both compound and substrate concentrations. The results are shown in figure 4. The dose response curve of 4 in the presence of 3 different ATP concentrations demonstrates a loss in potency when ATP levels rise (Figure 4A). Conversely, the % activity of 4 in the presence of varying concentrations of the peptide substrate has no affect on the compound potency (Figure 4B). Additionally, while maintaining 4 at a constant concentration (70 nM), an examination of the dose response of ATP demonstrated a sharp decline in enzyme inhibition at high ATP concentrations (>200 μM) while an increase in the dose of the peptide did not effect the potency of 4 (data not shown).
Figure 4.
A. Inhibitory dose response of 4 in the presence of three different ATP concentrations [1 μM (filled circles), 50 μM (empty circles), 100 μM (empty squares)]. B. Inhibitory dose response of 4 in the presence of three different peptide concentrations [50 μM (filled circles), 100 μM (empty circles), 200 μM (empty squares)].
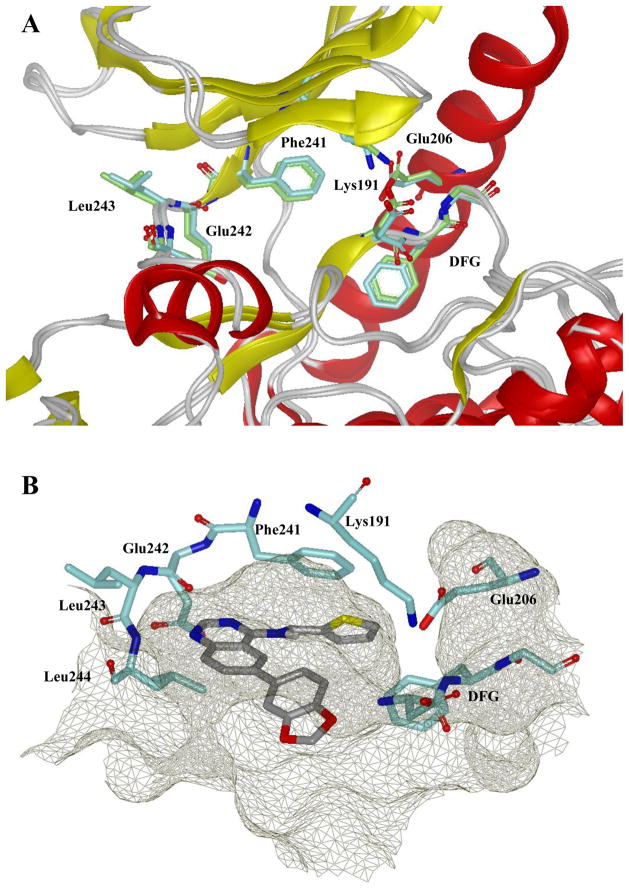
Following confirmation that this chemotype inhibits Clk4 via an ATP competitive mechanism, it was of interest to explore docking of 4 at a Clk kinase. As previously mentioned, quinazoline based small molecules have precedence as kinase inhibitors.26 Among these reagents is the clinically approved drug erlotinib (Tarceva ®).27 Erlotinib is currently indicated for treatment of non-small cell lung and pancreatic cancer and its actions are mediated through inhibition of the EGFR tyrosine kinase.28 In 2002, Stamos et al reported the structure of erlotinib bound to the ATP binding domain of EGFR (PDB code: 1M17).29 We theorized that the relationship of the 4-anilinoquinazoline structure of erlotinib to our newly discovered Clk family inhibitors may provide significant insight into the binding modality and mechanism of action for this class of compounds. There are no published X-ray structures of Clk4. There are structures of Clk1 (PDB code: 1Z57) and Clk3 (PDB code: 2EU9). Clk1 and Clk4 are highly homologous enzymes (>85% sequence identity) while Clk2 and Clk3 also share a high degree of sequence homology (>70% sequence identity). Based upon this, we utilized the X-ray structure of Clk1 as the template to derive a homology model of Clk4 (Figure 5A) using MOE molecular modeling software.30 Molecular docking was performed on 4 within the ATP binding domain of Clk1 and Clk4 to achieve an optimal binding pose using FRED31 (Figure 5B). In the crystal structure of erlotinib and EGFR, the N1 of the quinalozine heterocycle makes a critical H-bond to an amide NH of the hinge region of the ATP binding pocket. This interaction is mimicked within our docking analysis of 4 at Clk1 and Clk4. The thiophen-2-ylmethanamine moiety is oriented to fill an open pocket formed by the gatekeeper Phe241 (Phe239 in Clk4) while the benzo[d][1,3]dioxole extends toward the solvent exposed face of the hinge region. While this model partially accounts for various aspects of the SAR found to date, more extensive exploration of this binding modality is required to fully understand the potency and remarkable selectivity associated with this chemotype.
Figure 5.
A. Ribbon representation of the catalytic clefts in the Clk1 and Clk4 kinase domain. Clk4 kinase domain is a homology model derived from the X-ray structure of Clk1 (PDB code: 1Z57). Protein kinase structural elements are labeled and the key residues are colored: Clk1 in cyan, Clk4 in green. This figure was prepared with the program VIDA (OpenEye Scientific Software). B. Docking model of 4 in the Clk1 catalytic cleft. The binding pocket is depicted by molecular surface in mesh grey and the hydrogen bonds are labeled as green dotted lines. This figure was prepared with the program VIDA (OpenEye Scientific Software).
An interesting aspect of these agents is the inhibitory activity of this chemotype versus Dyrk1A. The Dyrk1A [dual specificity tyrosine (Y)-phosphorylation-regulated kinase 1A] gene is located on chromosome 21 and is known to be highly expressed in CNS tissues. Dyrk1A knock-out mice are embryonic lethal and transgenic mice overexpressing Dyrk1A display learning and memory deficiencies.32,33 The Dyrk1A gene is located on the Down’s syndrome (DS) critical region of chromosome 21 and trisomy-driven overexpression in DS patients has been demonstrated.34 Further evidence has been presented that hyperphosphorylation of Tau by Dyrk1A is a causative factor in the early onset of Alzheimer disease in DS patients.35 To date, a class of pyrazolidine-3,5-diones discovered via in silico screening have been reported as Dyrk1A inhibitors.36,37 The most potent of these agents were demonstrated to have an IC50 value of 0.6 μM and selectivity versus a panel of 15 selected kinases highly suggested that these agents were fairly promiscuous. The binding of 4 to Dyrk1A with a potency of 27 nM suggests that 4 and related 6-arylquinazolin-4-amines may represent important new tool compounds for exploration of Dyrk1A biochemistry. We have confirmed that 4 and related analogues are potent inhibitors of Dyrk1A (data not shown). Interestingly, Dyrk1A has been implicated as an important modulator of pre-mRNA splicing via several molecular interactions including the phosphorylation of the SR protein cyclin L2.38
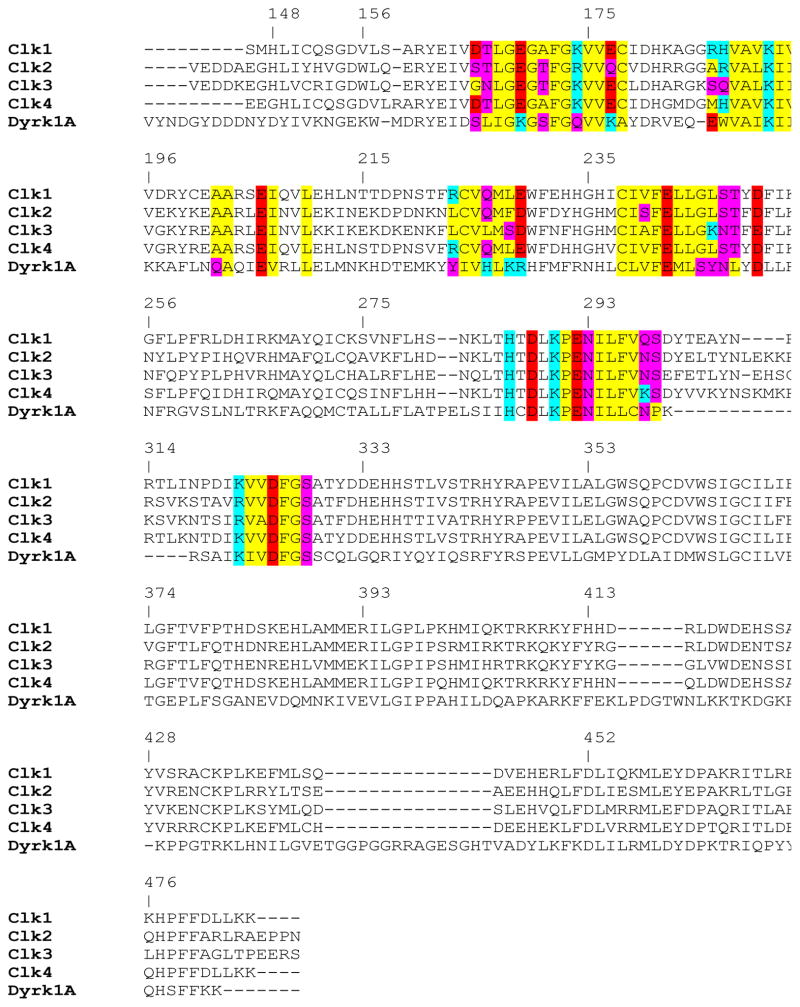
The fact that both 4 and TG003 were highly selective for the Clk family and Dyrk1A leads to questions regarding the relationship between these two enzyme classes. Clk and Dyrk are both members of the CMCG branch of the kinome, however, Dyrk1A and Clk1 are only 32.8% homologous. A sequence comparison is provided in Figure 6. While each kinase retains several key amino acids residues that seem to be fundamental to forming the ATP binding domain (including Glu206, Lys191 and matched hydrophobic residues at positions 243 and 244) there are significant differences that likely confer divergent structural aspects between the Clks and Dryk1A. A concerted effort to correlate compound SAR at each enzyme will be required to better understand the relationship between these kinases.
Figure 6.
Multiple sequence alignment of the catalytic domain of protein kinase for all four human Clk isozymes (Clk1, Clk2, Clk3 and Clk4) and human Dyrk1A. The amino acid residues that are within 10Å of the ATP binding site are highlighted: red for negatively charged, cyan for positively charged, yellow for hydrophobic and purple for hydrophilic. The numbering of amino acid residues is taken from Clk1 crystal structure (PDB code: 1Z57). Multiple sequence alignment was prepared by MOE molecular modeling software.
In conclusion, we report a novel class of Clk inhibitors based upon a core 6-arylquinazolin-4-amine scaffold. Selected agents were screened versus Clk4 to gain an appreciation of this chemotypes SAR and selected agents were found to inhibit this enzyme with potencies below 100 nM. One agent (analogue 4) was profiled against a panel of over 400 kinases and found to be remarkably selective for Clk1, Clk4 and Dyrk1A. The only other reported inhibitor of the Clk family [TG003 (1)] was also profiled and found to bind selectively to Clk1, Clk2, Clk4 and Dyrk1A. Analysis of the mechanism of action highly suggests that this chemotype inhibits Clk4 via competition with ATP binding. Molecular modeling also suggests that 4 and related agents inhibit the Clk isozymes through binding at the ATP binding domain. These agents provide useful tools for the study of Clk1, Clk4 and Dyrk1A and their respective roles in pre-mRNA splicing. Efforts to expand on the SAR of this chemotype in hopes of finding small molecules with divergent SAR for each isozyme of the Clk family and Dyrk1A are currently underway.
Acknowledgments
We thank Ms. Allison Mandich for critical reading of this manuscript. We thank Mr. Dac-Trung Nguyen for generation of the dendrogram representations of kinase activity. This research was supported by the Molecular Libraries Initiative of the National Institutes of Health Roadmap for Medical Research and the Intramural Research Program of the National Human Genome Research Institute at the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Krämer A. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 2.Black DL. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 3.Faustino NA, Cooper TA. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 4.Cohen P. Nature Rev Drug Discovery. 2002;1:309–315. doi: 10.1038/nrd773. [DOI] [PubMed] [Google Scholar]
- 5.Capdeville R, Buchdunger E, Zimmermann J, Matter A. Nature Rev Drug Discovery. 2002;1:493–502. doi: 10.1038/nrd839. [DOI] [PubMed] [Google Scholar]
- 6.Wilhelm S, Carter C, Lynch M, Lowinger T, Dumas J, Smith RA, Schwartz B, Simantov R, Kelley S. Nature Rev Drug Discovery. 2006;5:835–844. doi: 10.1038/nrd2130. [DOI] [PubMed] [Google Scholar]
- 7.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 8.Hagiwara M. Biochemica et Biophysica Acta. 2005;1754:324–331. doi: 10.1016/j.bbapap.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 9.Hanes J, von der Kammer H, Klaudiny J, Scheit KH. J Mol Biol. 1994;244:665–672. doi: 10.1006/jmbi.1994.1763. [DOI] [PubMed] [Google Scholar]
- 10.Nayler O, Stamm S, Ullrich A. Biochem J. 1997;326:693–700. doi: 10.1042/bj3260693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prasad J, Manley JL. Mol Cell Biol. 2003;23:4139–4149. doi: 10.1128/MCB.23.12.4139-4149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manley JL, Tacke R. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- 13.Zahler AM, Lane WS, Stolk JA, Roth MB. Genes Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- 14.Jiang K, Patel NA, Watson JE, Apostolatos H, Kleiman E, Hanson O, Hagiwara M, Cooper DR. Endocrinology. 2009;150:2087–2097. doi: 10.1210/en.2008-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwertz H, Tolley ND, Foulks JM, Denis MM, Risenmay BW, Buerke M, Tilley RE, Rondina MT, Harris EM, Kraiss LW, Mackman N, Zimmerman GA, Weyrich AS. J Exp Med. 2006;203:2433–2440. doi: 10.1084/jem.20061302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glatz DC, Rujescu D, Tang Y, Berendt FJ, Hartmann AM, Faltraco F, Rosenberg C, Hulette C, Jellinger K, Hampel H, Riederer P, Möller HJ, Andreadis A, Henkel K, Stamm S. J Neurochem. 2006;96:635–644. doi: 10.1111/j.1471-4159.2005.03552.x. [DOI] [PubMed] [Google Scholar]
- 17.Muraki M, Ohkawara B, Hosoya T, Onogi H, Koizumi J, Koizumi T, Sumi K, Yomoda J-i, Murray MV, Kimura H, Furuichi K, Shibuya H, Drainer AR, Suzuki M, Hagiwara M. J Biol Chem. 2004;279:24246–24254. doi: 10.1074/jbc.M314298200. [DOI] [PubMed] [Google Scholar]
- 18.Perni, R. B.; Bemis, J.; Nunes, J. J.; Szczepankiewicz, B. G. WO 2009085226.
- 19.Hekimi, S.; McBride, K.; Hihi, A. K.; Kianicka, I.; Wang, Y.; Hayes, S. L.; Guimond, M.-P.; Sevigny, G.; Dumas, D.; Smith, J. WO 2008014602.
- 20.Reaction Biology Corporation. http://www.reactionbiology.com/pages/kinase.htm.
- 21.Tanega, C.; Shen, M.; Mott, B.; Thomas, C. J.; MacArthur, R.; Inglese, J.; Auld, D. S. manuscript in preparation.
- 22.Procedure for the preparation of 6-bromo-N-(thiophen-2-ylmethyl)quinazolin-4-amine: To a solution of 6-bromo-4-chloroquinazoline (0.3 g, 1.23 mmol) in DMF (8 mL)were added thiophen-2-ylmethanamine (0.139 g, 1.23 mmol) and Hunig’s base (0.21 mL, 1.23 mmol). The reaction mixture was stirred at rt for 2 h. Upon completion, the reaction mixture was diluted with EtOAc (100 mL) and washed with 10% KHSO4 (25 mL) and three times with 3N LiCl (3 × 30 mL). The organic layer was extracted, dried on MgSO4, filtered, and concentrated in vacuo. The residue was purified directly on silica column. Gradient elution with ethyl acetate (15→75%) in hexanes provided the title compound as a colorless solid: yield (0.39 g, 1.22 mmol, 99 %). Procedure for the preparation of 6-(benzo[d][1,3]dioxol-5-yl)-N-(thiophen-2-ylmethyl)quinazolin-4-amine (4): To a solution of 6-bromo-N-(thiophen-2-ylmethyl)quinazolin-4-amine (0.1 g, 0.31 mmol) in DMF (3 mL) were added benzo[d][1,3]dioxol-5-ylboronic acid (0.078 g, 0.47 mmol), sodium carbonate (0.066 g, 0.63 mmol), and tetrakis(triphenylphosphine)palladium(0) (0.036 g, 0.03 mmol). The reaction mixture was heated in a Biotage Initiator® microwave at 150 °C for 1 h. Upon completion, the reaction mixture was diluted with EtOAc (75 mL) and washed with NaHCO3 (50 mL), three times with 3N LiCl (3 × 30 mL), and brine (30 mL). The organic layer was collected, filtered through a pad of Celite, and concentrated in vacuo. The residue was purified directly on silica column. Gradient elution with ethyl acetate (5→65%) in hexanes provided 4 as a colorless solid: yield (0.065 g, 0.18 mmol, 58 %). 1H NMR (DMSO-d6) δ 4.97 (d, J = 5.53 Hz, 2H), 6.09 (s, 2H), 6.92–7.01 (m, 1H), 7.02–7.18 (m, 2H), 7.29–7.41 (m, 2H), 7.44 (s, 1H), 7.73 (d, J = 8.56 Hz, 1H), 8.08 (d, J = 8.46 Hz, 1H), 8.51 (d, J = 4.70 Hz, 2H), 8.99 (d, J = 4.89 Hz, 1H); 13C NMR (DMSO-d6) 54.88, 101.28, 107.14, 108.70, 115.04, 119.52, 120.54, 125.11, 125.79, 126.54, 127.99, 131.04, 133.21, 137.02, 142.18, 147.14, 148.11, 148.26, 154.75, 159.05. HRMS (ESI) m/z 362.0957 (M+H)+ (C20H16N3O2S requires 362.0958). Purity determination via LCMS was performed using an Agilent Diode Array Detector using a 3 minute gradient of 4% to 100% acetonitrile (containing 0.025% trifluoroacetic acid) in water (containing 0.05% trifluoroacetic acid) with a 4.5 minute run time at a flow rate of 1 mL/min and a 7 minute gradient of 4% to 100% acetonitrile (containing 0.025% trifluoroacetic acid) in water (containing 0.05% trifluoroacetic acid) with an 8 minute run time at a flow rate of 1 mL/min. 3 minute gradient retention time = 3.050; 7 minute gradient retention time = 4.531 min. Purity was determined to be > 95% in both methods.
- 23.Inglese J, Auld DS, Jadhav A, Johnson RL, Simeonov A, Yasgar A, Zheng W, Austin CA. Proc Natl Acad Sci USA. 2006;103:11473–11478. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, Faraoni R, Floyd M, Hunt JP, Lockhart DJ, Milanov ZV, Morrison MJ, Pallares G, Patel HK, Pritchard S, Wodicka LM, Zarrinkar PP. Nat Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 25.Dendrogram representations were generated by an in-house visualization tool designated PhyloChem. Dendrogram clustering and apexes are based on the human phylogenetic kinase data. available at http://kinase.com/human/kinome.
- 26.Noble MEM, Endicott JA, Johnson LN. Science. 2004;303:1800–1804. doi: 10.1126/science.1095920. [DOI] [PubMed] [Google Scholar]
- 27.www.tarceva.com
- 28.Dowell J, Minna JD, Kirkpatrick P. Nature Rev Drug Discovery. 2005;4:13–14. doi: 10.1038/nrd1612. [DOI] [PubMed] [Google Scholar]
- 29.Stomos J, Sliwkowski MX, Eigenbrot C. J Biol Chem. 2002;277:46265–46272. doi: 10.1074/jbc.M207135200. [DOI] [PubMed] [Google Scholar]
- 30.MOE Molecular Operating Environment. Chemical Computing Group Inc; Montreal, Canada: 2008. Version 2008.10. [Google Scholar]
- 31.OpenEye Scientific Software, Inc. Santa Fe, NM: http://www.eyesopen.com/ [Google Scholar]
- 32.Fotaki V, Dierssen M, Alcantara S, Martinez S, Marti E, Casa C, Visa J, Soriano E, Estivill X, Arbones ML. Mol Cell Biol. 2002;22:6636–6647. doi: 10.1128/MCB.22.18.6636-6647.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahn KJ, Jeong HK, Choi HS, Ryoo SR, Kim YJ, Goo JS, Choi SY, Han JS, Ha I, Song WJ. Neurobiol Dis. 2006;22:463–472. doi: 10.1016/j.nbd.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 34.Dowjat WK, Adayev T, Kuchna I, Nowicki K, Palminiello S, Hwang YW, Wegiel J. Neurosci Lett. 2007;413:77–81. doi: 10.1016/j.neulet.2006.11.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ryoo SR, Jeong HK, Radnaabazar C, Yoo JJ, Cho HJ, Lee HW, Kim IS, Cheon YH, Ahn YS, Chung SH, Song WJ. J Biol Chem. 2007;282:34850–34857. doi: 10.1074/jbc.M707358200. [DOI] [PubMed] [Google Scholar]
- 36.Kim ND, Yoon J, Kim JH, Lee JT, Chon YS, Hwang MK, Ha I, Song WJ. Bioorg Med Chem Lett. 2006;16:3772–3776. doi: 10.1016/j.bmcl.2006.04.042. [DOI] [PubMed] [Google Scholar]
- 37.Koo KA, Kim ND, Chon YS, Jung MS, Lee BJ, Kim JH, Song WJ. Bioorg Med Chem Lett. 2009;19:2324–2328. doi: 10.1016/j.bmcl.2009.02.062. [DOI] [PubMed] [Google Scholar]
- 38.de Graff K, Hekerman P, Spelten O, Herrmann A, Packman LC, Büssow K, Müller-Newen G, Becker W. J Biol Chem. 2004;279:4612–4624. doi: 10.1074/jbc.M310794200. [DOI] [PubMed] [Google Scholar]