Abstract
According to a recent IARC Working Group report, alcohol consumption is causally related to an increased risk of cancer of the upper aerodigestive tract, liver, colorectum, and female breast (Lancet Oncol. 2007 8:292–3). Several lines of evidence indicate that acetaldehyde (AA), the first product of alcohol metabolism, plays a very important role in alcohol-related carcinogenesis, particularly in the esophagus. We previously proposed a model for alcohol-related carcinogenesis in which AA, generated from alcohol metabolism, reacts in cells to generate DNA lesions that form interstrand crosslinks (ICLs) (Nucleic Acids Res. 2005 33:3513–20). Since the Fanconi anemia-breast cancer associated (FANC-BRCA) DNA damage response network plays a crucial role in protecting cells against ICLs, in the present work we tested this hypothesis by exposing cells to AA and monitoring activation of this network. We found that AA exposure results in a concentration-dependent increase in FANCD2 monoubiquitination, which is dependent upon the FANC core complex. AA also stimulated BRCA1 phosphorylation at Ser1524 and increased the level of γH2AX, with both modifications occurring in a dose-dependent manner. However, AA did not detectably increase the levels of hyperphosphorylated RPA34, a marker of single-stranded DNA exposure at replication forks. These results provide the initial description of the AA-DNA damage response, which is qualitatively similar to the cellular response to mitomycin C, a known DNA crosslinking agent. We discuss the mechanistic implications of these results, as well as their possible relationship to alcohol-related carcinogenesis in different human tissues.
1. Introduction
It has been known for some time that the consumption of alcoholic beverages is related to an increased risk of cancers of the upper aerodigestive tract (UADT) and liver [1]. However, according to the most recent (2007) IARC Working Group, there is now sufficient evidence to conclude that ethanol (alcohol) consumption is causally related to an increased risk of cancer not only of the UADT and liver, but also of the colorectum and female breast [2]. These findings, particularly the risk of breast cancer, substantially expand the list of potential health risks from drinking alcohol.
There are a number of possible mechanisms by which alcohol consumption could increase cancer risk [3]. However, a variety of lines of evidence indicate that acetaldehyde (AA), generated by ethanol metabolism, plays a very important role in alcohol-related carcinogenesis, particularly in the UADT [1]. The strongest evidence comes from studies of individuals with a genetic deficiency in ALDH2, the enzyme responsible for the metabolism of AA to acetate [4,5]. Compared to individuals with fully active ALDH2, ALDH2-deficient individuals are at greatly increased risk of esophageal cancer when they consume equivalent amounts of alcohol [6,7].
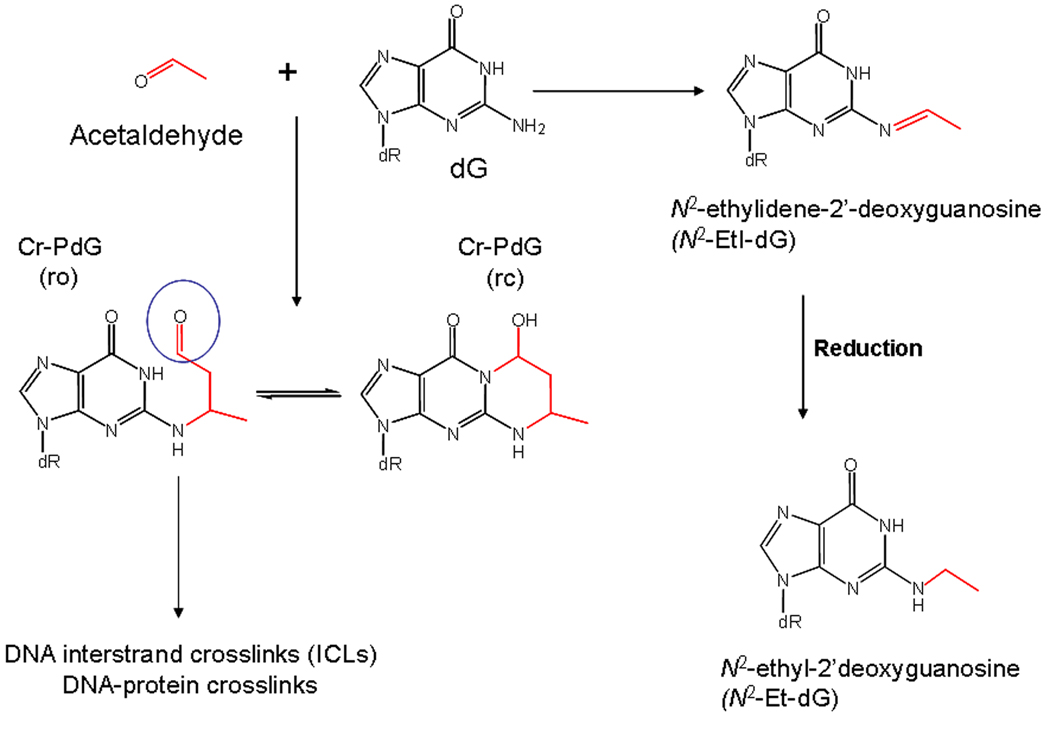
AA generates multiple specific types of DNA adducts [8–11]. Of these, the most likely to be biologically important are the alpha-methyl-gamma-hydroxy-1,N2-propano-dG adducts (see Figure 1) [10]. Because these adducts can also be formed from crotonaldehyde [12], as well as the evidence that the generation of these adducts from biologically relevant acetaldehyde concentrations involves a crotonaldehyde intermediate [11], they will be referred to as CrPdG adducts. PdG adducts of this type can exist in two forms, with the additional ring either opened or closed (see Figure 1) [10]. In some sequence contexts, the ring-opened form can react with dG on the opposite strand of the DNA to form DNA interstrand crosslinks (ICLs) [13,14]. These lesions can also link to peptides in vitro [15], and are likely responsible for DNA-protein crosslinks resulting from AA [16,17]. Importantly, elevated levels of AA adducts including the CrPdGs were observed in white blood cells of ALDH2-deficient alcoholics relative to controls with fully active ALDH2 activity, consistent with a role for AA-related DNA adducts in the increased risk of cancer in these individuals [18].
Figure 1.
DNA adducts from AA. The direct adduct of DNA from AA is N2-EtI-dG, which can be reduced to the stable adduct N2-Et-dG. In the presence of polyamines, the CrPdG adducts can form from biologically relevant AA concentration through a crotonaldehyde intermediate [11]. The aldehyde moiety (circled) on the ring opened form of the CrPdG adduct can react with deoxyguanosine on the opposite strand to form an ICL, or with protein to form DNA-protein crosslinks. For further discussion, see [10,14].
The Fanconi anemia-BRCA (FANC-BRCA) DNA-damage response network plays an important role in protecting cells against replication blocking DNA lesions and ICLs [19,20]. Based on the chemical properties of the CrPdG adducts as described above, we previously proposed that the FANC-BRCA network would play a protective role against the genotoxic effects of these adducts, and by extension against alcohol related carcinogenesis [10]. One prediction of this hypothesis is that exposure of cells to AA should result in activation of the FANC-BRCA network. Therefore, in the present work, we sought to test this prediction, using AA concentrations comparable to in vivo exposure levels in the gastrointestinal tract following alcohol consumption.
Blood AA concentrations after alcohol drinking are low to undetectable in most people [21]. However, AA concentrations in the GI tract can be much higher, due to the metabolism of ethanol to AA by microorganisms [22]. Specifically, AA concentrations in human saliva after drinking ethanol can be 450 µM [23], or possibly higher depending upon ALDH2 genotype [24] and other factors [25,26]. Data for AA levels in the human colon after alcohol drinking are not available, but AA concentrations of up to 2.7 mM have been measured in the colon of rats following ethanol treatment [27]. In addition, recent data showing that Aldh2−/− mice showed a 10-fold increase in AA adducts in the stomach after drinking a liquid diet containing ethanol [28] further underscores the role of AA-derived DNA lesions in ethanol-related genotoxicity.
Here we show that AA, at biologically relevant concentrations (0.1–1 mM), results in an increased FANCD2 monoubiquitination in human lymphoblastoid cells. The same concentrations also increased phosphorylation of the BRCA1 protein, and phosphorylation of histone H2AX. The implications of these findings for alcohol related carcinognesis in different human tissues are discussed.
2.Materials and Methods
2.1 Cells and Media
Lymphoblastoid cells from two apparently normal individuals (AG15793 and AG15997A) and from an XPA patient (GM02345) were obtained from the Coriell Cell Repository. Lymphoblastoid cells from an FA-G patient EUFA143 were originally provided by Dr. Alan D’Andrea to LHT. EUFA143 cells were complemented with a plasmid expressing the human FANCG gene, and the resulting cells referred to as EUFA143-T2 (hereafter called T2). These cells express physiological levels of FANCG protein and the survival of these cells after MMC treatment is indistinguishable from that of normal cells (see Suppl. Figure 1). Lymphoblastoid cells were maintained in culture using RPMI 1640 supplemented with 2 mM glutamine and 15% fetal bovine serum. For the chromosomal aberration assay, primary fibroblasts from a FA-A patient (GM00646) and the patient’s parent (GM02977) were obtained from the Coriell repository and grown in MEM plus 2 mM glutamine and 15% FBS.
2.2 Acetaldehyde exposure
AA is highly volatile, and has a low boiling point (20.2°C), which complicates the design of experiments. We exposed cells to AA for 24 hours using two different protocols. In both protocols, lymphoblasts were fed with regular media the day before experimental treatments. The day of the experiment, lymphoblasts were harvested from their growth flasks, counted and resuspended at a concentration of 1–2 × 107 / ml in media.
In the first protocol (sealed flask), media containing a 2× concentration of acetaldehyde was prepared on ice; then 10 mL was added to each T25 flask containing 107 cells in 10 ml of media without AA on ice. Caps were tightly closed, sealed with Parafilm and incubated at 37°C. In this condition, there is some head-space in the flask that acetaldehyde can potentially evaporate into; therefore, over time, the concentration of AA in the media that the cells are exposed to will become lower than the initial concentration in the media.
In the second protocol (full tube), 107 cells were exposed to AA in 15 mL conical test tubes which were filled with AA containing media. In this protocol, there is no headspace into which the AA can diffuse, so that the AA concentration remains essentially constant. Tubes were sealed tightly and wrapped with Parafilm before incubation in a tilted position at 37°C.
2.3 Extract Preparation
After the 24 hour incubation period, lymphoblasts were harvested by centrifugation and cell pellets were transferred to microfuge tubes using phosphate buffered saline and centrifuged again. Cells were lysed on ice for 30 min in a high-salt buffer [29] consisting of 20 mM Tris, pH 8.0, 600 mM NaCl, 1 mM EDTA, 0.5% Igepal (a detergent that is chemically indistinguishable from Nonidet P-40), 1 mM AEBSF (4-(2-Aminoethyl)benzenesulfonyl fluoride hydrochloride), 1 µM microcystin and protease inhibitors (Sigma). Lysates were then centrifuged at 16,000 × g for 20 min at 4°C, and the supernatant was transferred to a new tube. Protein concentration of the supernatant was determined using the Bio-Rad protein assay with BSA as the standard.
2.4 Lambda phosphatase treatment
To verify that the shift in BRCA1 migration after DNA damage was due to phosphorylation, approximately 50 µg of lysate protein was treated with 400 units of lambda phosphatase for 1 hr at 30°C. Control lysates were incubated for 1 hr at 30°C without enzyme. Prior to incubation with lambda phosphatase, the lysates were supplemented with 2 mM MnCl2. Reactions were stopped by the addition of SDS-PAGE buffer and boiling.
2.5 Western Blotting
For detection of FANCD2 and BRCA1, Western blots were prepared using 15–50 µg protein per lane on a 3–8% Tris-Acetate gel (Invitrogen). For γH2AX and RPA34 phosphorylation, 12% Bis-Tris (Invitrogen) gels were used. Separated proteins were transferred to nitrocellulose (ECL Hybond, GE Healthcare) using the NuPage transfer system (Invitrogen). Blots were stained with Amido Black to check protein loading and transfer prior to blocking in Tris buffered saline containing 0.1% Tween 20 (TBST) and 5% milk. Blots were probed with antibodies against FANCD2 (Novus), BRCA1 (Ab-1, Calbiochem), BRCA1 pSer 1524, pSer 1457, and pSer 1387 (Bethyl Laboratories), γH2AX (Millipore) or RPA34 (Cell Signaling). Antibody binding was visualized on X-ray film (Kodak XAR) using chemiluminescence (ECL Plus, GE Healthcare). Western blot X-ray films were scanned and analyzed using the NIH ImageJ program.
2.6 Chromosomal Aberrations
For the chromosomal aberration study, cells were exposed to 0.18 mM AA for 24 hours in tightly sealed flasks (same conditions as in [30]). Following exposure, cells were sent to the NCI Cytogenetics core facility for analysis of chromosomal aberrations by an investigator unaware of treatment conditions.
3. Results
3.1 AA Stimulates FANCD2 Monoubiquitination
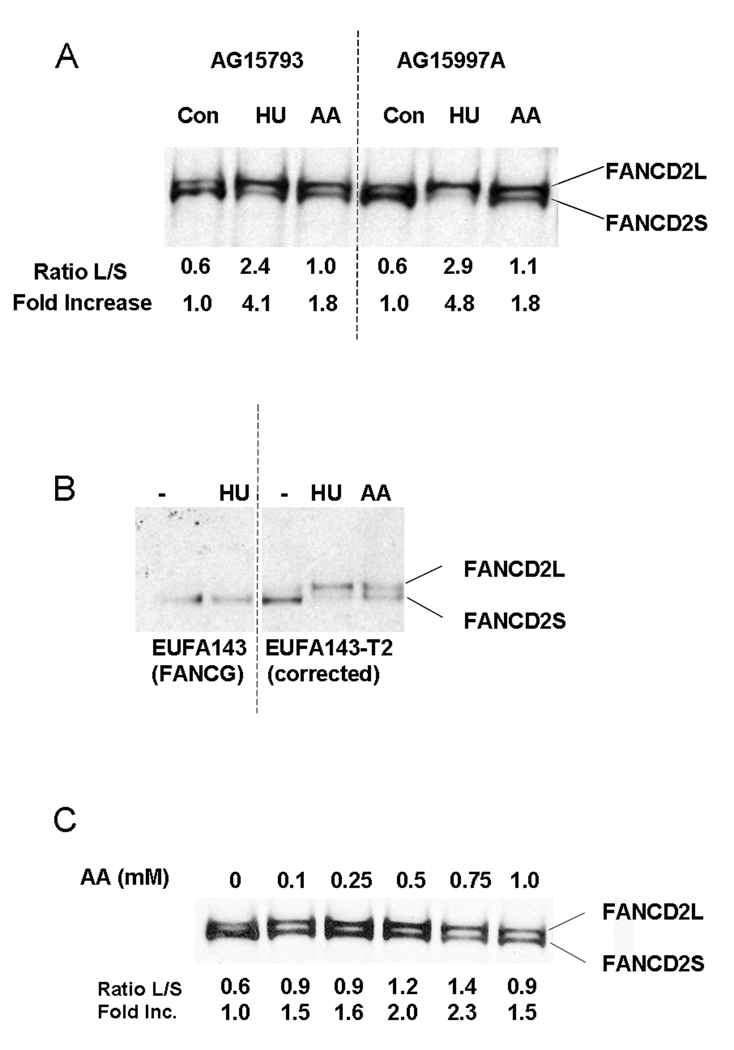
Based on published data for AA levels in the human and animal gastrointestinal tract following ethanol consumption (see Introduction), we chose an AA concentration of 1 mM for our initial experiments. We monitored activation of the FANC pathway after AA by assaying for FANCD2 monoubiquitination by Western blotting. Hydroxyurea (2 mM) was used as a positive control. As shown in Figure 2A, we found that 1 mM AA resulted in increased FANCD2 monoubiquitination, as represented by an increased ratio of the long form of FANCD2 (FANCD2-L) to the short form. In two different normal cell lines, the magnitude of induction was approx 1.8 fold.
Figure 2.
AA exposure stimulated FANCD2 monoubiquitination. A. Two normal lymphoblastoid cell lines were exposed to 1 mM AA or 2 mM HU using the sealed flask protocol for 24 hours. Whole cell extracts were prepared for Western blotting and probed with anti-FANCD2. The positions of the long (L) and short (S) forms of FANCD2 are indicated. The ratio of the L and S forms of FANCD2 were determined using the NIH ImageJ program and are shown under each lane. To calculate the fold increase, the L/S ratio of the untreated culture was set to 1 and the L/S ratio of the other samples were normalized to that value. B. Lymphoblasts from a FA-G patient (EUFA143) and from EUFA143 cells containing a human FANCG gene T2 were exposed to AA or HU (filled flask protocol) as indicated. No FANCD2-L band was detected in EUFA143 cells after HU or AA (data not shown). C. Normal lymphoblasts (AG15793) were exposed to different concentrations of AA for 24 hours as indicated, using the sealed flask protocol. L/S ratios and fold increase were determined as described for panel A.
To verify that FANCD2-L we detect does in fact represent FANCD2 monoubiquitination, we tested cells from a FA-G patient, EUFA143 cells, as well as EUFA143 cells expressing human FANCG (EUFA143-T2 cells). FANCD2 monoubiquitination requires the Fanconi core complex, which includes FANCG, and therefore should be absent from the patient cells. Consistent with this expectation, FANCD2-L was not observed in EUFA143 cells, either with or without HU (Figure 2B) or AA (not shown). However, in EUFA143-T2 cells, FANCD2-L was induced by both HU and AA (Figure 2B). These results verify that the FANCD2-L we detect represents monoubiquitinated FANCD2.
We then carried out a dose response test over the range of 0.1–1 mM AA (Figure 2C). Interestingly, an effect of AA could be detected even at 0.1 mM, although the effect was maximal at 0.75 mM. Substantial toxicity was observed when higher concentrations of AA (2–10 mM) were used (data not shown).
An earlier study showed that exposure of human lymphocytes from a single Fanconi anemia patient to AA resulted in increased levels of chromosomal aberrations (CA) [30]. However, this brief report did not contain any clinical or diagnostic information about the FA patient whose complementation group was not defined. Therefore, to confirm and extend this earlier study, and to verify the protective role of the FANC pathway in protecting against AA genotoxicity, we exposed primary fibroblasts from a FA-A patient as well as cells from the patients’ parent to AA (0.18 mM), the same concentration as used by Obe et al [30]. Cells were scored for CA by an investigator blinded to the identity of the cell lines.
As shown in Table 1, AA increased the number of cells with CA in both the FA-A and parental control lines. However, the magnitude of the AA-induced increase was significantly greater in the FA-A cells than the normal cells, consistent with the earlier results of Obe et al [30].
Table 1.
Chromosomal Aberrations from AA in FA and Control Cells
| Treatment Groups | # of Cells | ||
|---|---|---|---|
| Total | Normal Chromosomes | Abnormal Chromosomes | |
| Control | 100 | 89 | 11 |
| Control + AA | 100 | 77 | 23 a |
| FA-A | 100 | 86 | 14 |
| FA-A + AA | 84 | 53 | 31 b,c |
significantly different from control without ACD, P=0.0234
significantly different from FA-A without ACD, P = 0.00032
significantly different from control with ACD, P = 0.0391
P values based on Chi-square test
FA-A cells - GM00646
Control cells - GM02977 (parent of GM646).
3.1 AA Stimulates Phosphorylation of BRCA1
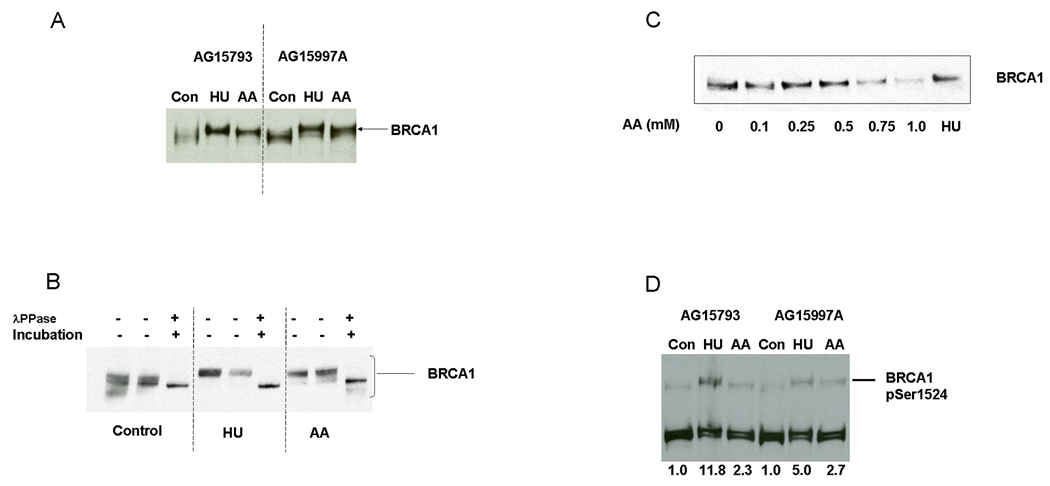
Previous work has shown that different DNA damaging agents stimulate phosphorylation of BRCA1, which can be detected by a reduced mobility of the protein under denaturing gel conditions. As shown in Figure 3A, exposure of cells to either HU or 1 mM AA resulted in a clear decrease in BRCA1 mobility (Figure 3A).
Figure 3.
AA treatment increases BRCA1 phosphorylation. Two normal lymphoblastoid cell lines were exposed to 1 mM AA or 2 mM HU for 24 hours using the sealed flask protocol. Whole cell extracts were probed with anti-BRCA1. Note the decreased mobility (upward shift) of BRCA1 after AA and HU treatment. B. Lysates from control, HU, or AA exposed AG15793 cells were either untreated, incubated without lambda phosphatase, or incubated with lambda phosphatase prior to denaturing and loading on the gel. C. Normal lymphoblasts (AG15793) were exposed to different concentrations of AA for 24 hours as indicated, using the sealed flask protocol. Note the progressive upward shift of the BRCA1 band, at AA concentrations of 0.5 mM and higher. D. Extracts from normal lymphoblastoid cell lines exposed to 1 mM AA or 2 mM HU using the sealed flask protocol were probed for BRCA1 phosphorylation at Ser1524. For quantification, the intensity of the pSer1524 signal was determined with ImageJ and normalized to the total FANCD2 signal (L + S) used here as a loading control.
To verify that the reduced mobility of BRCA1 was due to phosphorylation, as has been shown to be the case following other forms of DNA damage, we treated the lysates with lambda phosphatase prior to electrophoresis. As shown in 3B, the AA-stimulated mobility shift was reversed by phosphatase treatment, verifying that it does in fact represent phosphorylation. A dose response experiment (Figure 3C), showed that an upward shift of BRCA1 was detectable at 0.5 mM AA and higher concentrations.
3.2.1. AA stimulates BRCA1 phosphorylation on Ser1524
BRCA1 is phosphorylated on a number of different sites following DNA damage [31,32]. To address the question of which sites on BRCA1 were phosphorylated after AA, we evaluated the phosphorylation status of three serine resides on BRCA1 that are known sites for DNA damage induced phosphorylation (Ser1387, 1457, or 1524) on BRCA1 using antibodies specific for each phosphorylated reside. We did not obtain consistent evidence of phosphorylation at Ser1387 or 1457 following AA exposure (data not shown). However, we did find that AA treatment resulted in an approximately 2.5 fold increase in BRCA1 phosphorylation at Ser1524 (Figure 3D).
3.3 AA Increases Phosphorylation of H2AX Independent of NER
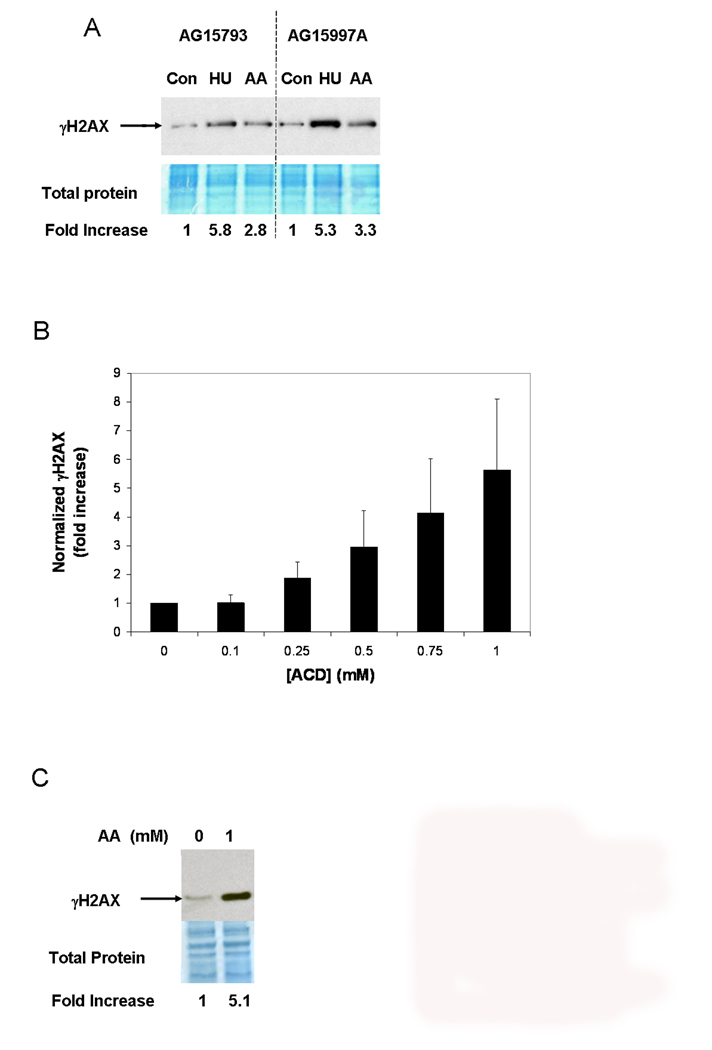
Recent studies have identified histone H2AX as a component the FANC-BRCA1 protein network [33]. Having observed increased FANCD2 monoubiquitination and BRCA1 phosphorylation by AA, we therefore asked whether the same treatment also increased phosphorylation of H2AX at Ser 139 (γH2AX). As shown in Figure 4A, Western blotting analysis indicated a clear increase in total cellular γH2AX levels after 1 mM AA. A dose response study (Figure 4B) indicates that increased levels of γH2AX are first observed at 0.25 mM AA.
Figure 4.
AA treatment increases γH2AX phosphorylation, and NER is not required. A. Western blotting of extracts from normal lymphoblastoid cell lines exposed to 1 mM AA or 2 mM HU using the sealed flask protocol for 24 hours and probed for γH2AX. A section of the blot that was stained for total protein using Amido black is shown below the corresponding film autoradiograph. For quantitation, the γH2AX signal was normalized to a scan of the total protein stain. B. γH2AX formation in AG15793 cells exposed to different AA concentrations (sealed flask protocol). C. 1 mM AA increases γH2AX levels in cells from an XP-A patient (filled flask protocol).
Following exposure to UV radiation, γH2AX can result from strand breaks introduced during attempted NER [34]. Because some PdG adducts are substrates for NER [35,36], we wondered whether the γH2AX formation we observed following AA exposure was also the result of attempted NER. Therefore, we repeated these experiments using lymphoblastoid cells from an XP-A patient, which are deficient in NER. As shown in Figure 4C, AA also stimulated γH2AX formation in these NER deficient cells, demonstrating that NER is not required for AA-stimulated γH2AX formation.
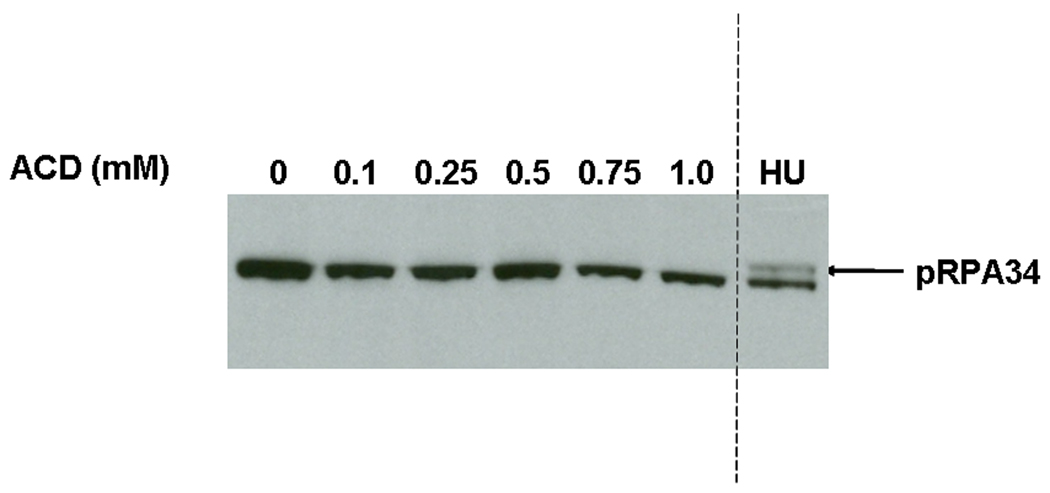
3.4 AA Does not Increase RPA34 Phosphorylation
FANCD2 monoubiquitination is produced by various genotoxic stressors, including UV and ionizing radiation, mitomycin C, and HU. However, the effects of these agents can be differentiated in terms of their ability to trigger phosphorylation of RPA, a response that results from production of single-stranded DNA during replication. Specifically, HU, which causes replication fork stalling by depleting cellular dNTP pools, is a powerful inducer of RPA34 phosphorylation, whereas MMC, which generates DNA ICLs, causes little or no RPA34 phosphorylation [37]. Therefore, we assessed whether exposure of cells to AA could result in hyperphosphorylation of RPA34, as monitored by a reduced mobility of RPA34 during gel electrophoresis. As shown in Figure 5, we could not detect RPA34 hyperphosphorylation at any concentration of AA tested. As a positive control, we showed that exposure of cells to HU stimulated RPA34 hyperphosphorylation as expected. Thus while AA exposure triggers FANCD2 monoubiquitination, BRCA1 phosphorylation, and γH2AX formation, it does not produce a detectable increase in RPA34 hyperphosphorylation.
Figure 5.
AA does not increase RPA34 hyperphosphorylation. AG15793 cells were exposed to increasing doses of AA using the sealed flask protocol, then analyzed for RPA34 hyperphosphorylation by Western blotting [37]. HU (2mM) was used as a positive control. The arrow points to the hyperphosphorylated RPA34 band.
4. Discussion
In this work we report that AA, at concentrations within the range produced in target tissues for alcohol-related carcinogenesis in humans, activates the FANC-BRCA network. Specifically, AA exposure dose-dependently stimulates monoubiquitination of the FANCD2 protein, phosphorylation of the BRCA1 at Ser1524, and γH2AX formation, independent of NER. However, under the conditions used, AA does not produce a detectable increase in RPA34 hyperphosphorylation. Each of these main findings was observed in multiple human lymphoblastoid cell lines and under two different protocols for AA exposure (Figure 2 and Figure 4 and data not shown).
4.1 Mechanistic Considerations
The characteristics of the AA DNA damage response we report here are qualitatively similar to the cellular response to MMC, which generates ICLs as well as monoadducts. Consistent with this similarity, AA can produce ICLs via the formation of CrPdG adducts, which in some sequence contexts can form ICLs (Figure 1). Our current results are also consistent with the observations of Mechilli et al [38] who found that cells lacking either the homologous recombination repair or FANC proteins were most sensitive to chromosomal aberrations from AA. Based on their results, they concluded that DNA ICLs are the biologically relevant AA-induced DNA lesions for producing chromosomal aberrations.
Despite these considerations, it is unlikely that the responses we detect here are entirely related to AA generated ICLs. The CrPdG adducts can only form ICLs in a specific sequence context (5′GpC3′) [13,14]. The aldehyde moiety present in the open form of the CrPdG adduct can also form DNA-protein crosslinks, most likely with histones [16], which would be expected to be sequence independent. Also, it was recently shown that AA generates crosslinks that can be detected via the comet assay, but most of these were DNA-protein crosslinks (DPCLs) [17]. Thus, the extent to which activation of the FANC-BRCA network we have observed is due to ICL versus DPCLs remains to be determined. Furthermore, the FANC-BRCA network not only promotes homologous recombination repair of broken replication forks, it has a role during DNA replication in coordinating translesion synthesis (TLS), thereby preventing replication fork breakage [39]. CrPdGs lesions have substantial miscoding properties [40], and multiple TLS polymerases have been proposed to be involved in efficient and error-free bypass of structurally related PdG adducts (e.g. [41]). Interestingly, a recent study provided evidence that the nucleolytic processing of an ICL can yield a modified DNA template that can be bypassed by pol κ [42].
In apparent contrast to our findings, a recent publication [43] showed that ethanol increased the expression of FANCD2 mRNA in the rat brain, and both ethanol and AA increased the amount of FANCD2 protein but did not stimulate FANCD2 monoubiquitination in brain cells. This is an interesting finding but is not directly relevant to our results with replicating cells, in contrast to neuronal cells, which are terminally differentiated. Furthermore, the concentration of AA used in that study (5 mM) was higher than the concentrations used in our work (0.1–1 mM). Thus the two studies address fundamentally different questions, and the findings are not mutually exclusive.
4.2 Clinical Implications
In this work we used AA concentrations between 0.1–1 mM, because cells in the human and animal gastrointestinal tract are exposed to levels of AA within this range due to the local metabolism of ethanol by microbes [23,27]. Therefore, our observations are likely to be of relevance to the human gastrointestinal tract following ethanol consumption.
Our observation that increased FANCD2 monoubiquitination is detected at 0.1 mM AA and is maximal at 0.5–0.75 mM AA (Figure 2C) implies that the FANC proteins play a protective role against AA at these concentrations. Consistent with this idea, an early study showed that lymphocytes from a FA patient were hypersensitive to chromosomal aberrations from AA at a concentration of ≈ 0.2 mM [30], an effect we confirmed in FA-A fibroblasts (Table 1). Likewise, Mechilli et al [38] observed a protective effect of the CHO Fancg gene against AA-generated chromosomal aberrations in an overlapping range of AA concentrations (0.3 – 1.8 mM). Interestingly, in WT CHO cells these authors observed increased chromosomal aberrations up to 0.6 mM, but no additional increase at concentrations up to 1.8 mM, suggestive of a plateau effect that is also consistent with our observations (Figure 2C).
FA patients have a high risk of several different types of epithelial cancers, including esophageal cancer [44], and our results, in addition to those of others [30,38,45], indicate that ethanol consumption would significantly elevate that risk. FA patients are already cautioned against drinking alcohol [46], and our results provide further experimental support for this advice.
While the role of BRCA1 in protecting against breast cancer is well known [47], studies in both humans [48] and mice [49] suggest a protective role for BRCA1 against esophageal cancer as well. Of potentially broader significance to the human population, it will be important to determine whether functional SNPs in FANC or BRCA genes can affect susceptibility to acetaldehyde-induced genotoxicity.
Finally, in view of the recent evidence that alcohol drinking increased breast cancer risk in women [2], our observation that AA exposure stimulated BRCA1 phosphorylation is of potential clinical relevance. However, it is important to note that in contrast to its role in alcohol related UADT cancer risk, the role of AA in breast cancer risk from alcohol drinking is less clear [3]. Blood AA levels are very low or undetectable after ethanol consumption in humans [50]. Therefore, cells in the human breast would not be exposed to exogenous AA at the concentrations used in this work. On the other hand, human breast cells express alcohol dehydrogenases, which are capable of oxidizing ethanol into AA at ethanol concentrations attainable after social drinking [51]. Therefore, the intracellular generation of AA in the human breast cells after ethanol consumption is of potential significance for alcohol-related breast cancer. While alcohol dehydrogenase gene polymorphisms have been associated with breast cancer risk from ethanol consumption in some studies, other factors are likely to be important as well (for discussion see [3]). Further studies will be necessary to determine whether AA generated in cells from ethanol metabolism can activate the FANC-BRCA network in the human body.
Supplementary Material
Acknowledgments
We thank Sandra Burkett, NCI-Frederick, for analysis of chromosomal aberrations, Sharon Cantor for reagents used in preliminary studies and helpful discussions, and Jessy Abraham for helpful comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.P.H.S. U.S. Department of Health and Human Services. Report on Carcinogens. Eleventh Edition 2005. National Toxicology Program Alcoholic Beverage Consumption. [Google Scholar]
- 2.Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, Bouvard V, Altieri A, Cogliano V. Carcinogenicity of alcoholic beverages. Lancet Oncol. 2007;8:292–293. doi: 10.1016/s1470-2045(07)70099-2. [DOI] [PubMed] [Google Scholar]
- 3.Seitz HK, Stickel F. Molecular mechanisms of alcohol-mediated carcinogenesis. Nat Rev Cancer. 2007;7:599–612. doi: 10.1038/nrc2191. [DOI] [PubMed] [Google Scholar]
- 4.Bosron WF, Li TK. Genetic polymorphism of human liver alcohol and aldehyde dehydrogenases, and their relationship to alcohol metabolism and alcoholism. Hepatology. 1986;6:502–510. doi: 10.1002/hep.1840060330. [DOI] [PubMed] [Google Scholar]
- 5.Crabb DW, Edenberg HJ, Bosron WF, Li TK. Genotypes for aldehyde dehydrogenase deficiency and alcohol sensitivity. The inactive ALDH2(2) allele is dominant. J Clin Invest. 1989;83:314–316. doi: 10.1172/JCI113875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yokoyama A, Muramatsu T, Ohmori T, Yokoyama T, Okuyama K, Takahashi H, Hasegawa Y, Higuchi S, Maruyama K, Shirakura K, Ishii H. Alcohol-related cancers and aldehyde dehydrogenase-2 in Japanese alcoholics. Carcinogenesis. 1998;19:1383–1387. doi: 10.1093/carcin/19.8.1383. [DOI] [PubMed] [Google Scholar]
- 7.Lewis SJ, Smith GD. Alcohol, ALDH2, and esophageal cancer: a meta-analysis which illustrates the potentials and limitations of a Mendelian randomization approach. Cancer Epidemiol Biomarkers Prev. 2005;14:1967–1971. doi: 10.1158/1055-9965.EPI-05-0196. [DOI] [PubMed] [Google Scholar]
- 8.Fang JL, Vaca CE. Detection of DNA adducts of acetaldehyde in peripheral white blood cells of alcohol abusers. Carcinogenesis. 1997;18:627–632. doi: 10.1093/carcin/18.4.627. [DOI] [PubMed] [Google Scholar]
- 9.Wang M, McIntee EJ, Cheng G, Shi Y, Villalta PW, Hecht SS. Identification of DNA adducts of acetaldehyde. Chem Res Toxicol. 2000;13:1149–1157. doi: 10.1021/tx000118t. [DOI] [PubMed] [Google Scholar]
- 10.Brooks PJ, Theruvathu JA. DNA adducts from acetaldehyde: implications for alcohol-related carcinogenesis. Alcohol. 2005;35:187–193. doi: 10.1016/j.alcohol.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 11.Theruvathu JA, Jaruga P, Nath RG, Dizdaroglu M, Brooks PJ. Polyamines stimulate the formation of mutagenic 1,N2-propanodeoxyguanosine adducts from acetaldehyde. Nucleic Acids Res. 2005;33:3513–3520. doi: 10.1093/nar/gki661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung FL, Young R, Hecht SS. Hecht Formation of cyclic 1,N2-propanodeoxyguanosine adducts in DNA upon reaction with acrolein or crotonaldehyde. Cancer Res. 1984;44:990–995. [PubMed] [Google Scholar]
- 13.Cho YJ, Wang H, Kozekov ID, Kurtz AJ, Jacob J, Voehler M, Smith J, Harris TM, Lloyd RS, Rizzo CJ, Stone MP. Stereospecific formation of interstrand carbinolamine DNA cross-links by crotonaldehyde- and acetaldehyde-derived alpha-CH3-gamma-OH-1,N2-propano-2'-deoxyguanosine adducts in the 5'-CpG-3' sequence. Chem Res Toxicol. 2006;19:195–208. doi: 10.1021/tx050239z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stone MP, Cho YJ, Huang H, Kim HY, Kozekov ID, Kozekova A, Wang H, Minko IG, Lloyd RS, Harris TM, Rizzo CJ. Interstrand DNA cross-links induced by alpha,beta-unsaturated aldehydes derived from lipid peroxidation and environmental sources. Acc Chem Res. 2008;41:793–804. doi: 10.1021/ar700246x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurtz AJ, Lloyd RS. 1,N2-deoxyguanosine adducts of acrolein, crotonaldehyde, and trans-4-hydroxynonenal cross-link to peptides via Schiff base linkage. J Biol Chem. 2003;278:5970–5976. doi: 10.1074/jbc.M212012200. [DOI] [PubMed] [Google Scholar]
- 16.Kuykendall JR, Bogdanffy MS. Reaction kinetics of DNA-histone crosslinking by vinyl acetate and acetaldehyde. Carcinogenesis. 1992;13:2095–2100. doi: 10.1093/carcin/13.11.2095. [DOI] [PubMed] [Google Scholar]
- 17.Lorenti Garcia C, Mechilli M, Proietti De Santis L, Schinoppi A, Kobos K, Palitti F. Relationship between DNA lesions, DNA repair and chromosomal damage induced by acetaldehyde. Mutation Research. 2008;662:3–9. doi: 10.1016/j.mrfmmm.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 18.Matsuda T, Yabushita H, Kanaly RA, Shibutani S, Yokoyama A. Increased DNA damage in ALDH2-deficient alcoholics. Chem Res Toxicol. 2006;19:1374–1378. doi: 10.1021/tx060113h. [DOI] [PubMed] [Google Scholar]
- 19.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 20.Thompson LH, Hinz JM, Yamada NA, Jones NJ. How Fanconi anemia proteins promote the four Rs: replication, recombination, repair, and recovery. Environ Mol Mutagen. 2005;45:128–142. doi: 10.1002/em.20109. [DOI] [PubMed] [Google Scholar]
- 21.Eriksson CJ, Fukunaga T, Sarkola T, Lindholm H, Ahola L. Estrogen-related acetaldehyde elevation in women during alcohol intoxication. Alcohol Clin Exp Res. 1996;20:1192–1195. doi: 10.1111/j.1530-0277.1996.tb01110.x. [DOI] [PubMed] [Google Scholar]
- 22.Salaspuro MP. Alcohol consumption and cancer of the gastrointestinal tract. Best Pract Res Clin Gastroenterol. 2003;17:679–694. doi: 10.1016/s1521-6918(03)00035-0. [DOI] [PubMed] [Google Scholar]
- 23.Homann N, Jousimies-Somer H, Jokelainen K, Heine R, Salaspuro M. High acetaldehyde levels in saliva after ethanol consumption: methodological aspects and pathogenetic implications. Carcinogenesis. 1997;18:1739–1743. doi: 10.1093/carcin/18.9.1739. [DOI] [PubMed] [Google Scholar]
- 24.Visapaa JP, Gotte K, Benesova M, Li J, Homann N, Conradt C, Inoue H, Tisch M, Horrmann K, Vakevainen S, Salaspuro M, Seitz HK. Increased cancer risk in heavy drinkers with the alcohol dehydrogenase 1C*1 allele, possibly due to salivary acetaldehyde. Gut. 2004;53:871–876. doi: 10.1136/gut.2003.018994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Homann N, Tillonen J, Rintamaki H, Salaspuro M, Lindqvist C, Meurman JH. Poor dental status increases acetaldehyde production from ethanol in saliva: a possible link to increased oral cancer risk among heavy drinkers. Oral Oncol. 2001;37:153–158. doi: 10.1016/s1368-8375(00)00076-2. [DOI] [PubMed] [Google Scholar]
- 26.Salaspuro V, Salaspuro M. Synergistic effect of alcohol drinking and smoking on in vivo acetaldehyde concentration in saliva. Int J Cancer. 2004;111:480–483. doi: 10.1002/ijc.20293. [DOI] [PubMed] [Google Scholar]
- 27.Visapaa JP, Jokelainen K, Nosova T, Salaspuro M. Inhibition of intracolonic acetaldehyde production and alcoholic fermentation in rats by ciprofloxacin. Alcohol Clin Exp Res. 1998;22:1161–1164. [PubMed] [Google Scholar]
- 28.Nagayoshi H, Matsumoto A, Nishi R, Kawamoto T, Ichiba M, Matsuda T. Increased formation of gastric N(2)-ethylidene-2'-deoxyguanosine DNA adducts in aldehyde dehydrogenase-2 knockout mice treated with ethanol. Mutat Res. 2008 doi: 10.1016/j.mrgentox.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 29.Peng M, Litman R, Jin Z, Fong G, Cantor SB. BACH1 is a DNA repair protein supporting BRCA1 damage response. Oncogene. 2006;25:2245–2253. doi: 10.1038/sj.onc.1209257. [DOI] [PubMed] [Google Scholar]
- 30.Obe G, Natarajan AT, Meyers M, Hertog AD. Induction of chromosomal aberrations in peripheral lymphocytes of human blood in vitro, and of SCEs in bone-marrow cells of mice in vivo by ethanol and its metabolite acetaldehyde. Mutat Res. 1979;68:291–294. doi: 10.1016/0165-1218(79)90160-5. [DOI] [PubMed] [Google Scholar]
- 31.Gatei M, Zhou BB, Hobson K, Scott S, Young D, Khanna KK. Ataxia telangiectasia mutated (ATM) kinase and ATM and Rad3 related kinase mediate phosphorylation of Brca1 at distinct and overlapping sites. In vivo assessment using phospho-specific antibodies. J Biol Chem. 2001;276:17276–17280. doi: 10.1074/jbc.M011681200. [DOI] [PubMed] [Google Scholar]
- 32.Ouchi T. BRCA1 phosphorylation: biological consequences. Cancer Biol Ther. 2006;5:470–475. doi: 10.4161/cbt.5.5.2845. [DOI] [PubMed] [Google Scholar]
- 33.Bogliolo M, Lyakhovich A, Callen E, Castella M, Cappelli E, Ramirez MJ, Creus A, Marcos R, Kalb R, Neveling K, Schindler D, Surralles J. Histone H2AX and Fanconi anemia FANCD2 function in the same pathway to maintain chromosome stability. Embo J. 2007;26:1340–1351. doi: 10.1038/sj.emboj.7601574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanasoge S, Ljungman M. H2AX phosphorylation after UV irradiation is triggered by DNA repair intermediates and is mediated by the ATR kinase. Carcinogenesis. 2007;28:2298–2304. doi: 10.1093/carcin/bgm157. [DOI] [PubMed] [Google Scholar]
- 35.Johnson KA, Fink SP, Marnett LJ. Repair of propanodeoxyguanosine by nucleotide excision repair in vivo and in vitro. J Biol Chem. 1997;272:11434–11438. doi: 10.1074/jbc.272.17.11434. [DOI] [PubMed] [Google Scholar]
- 36.Choudhury S, Pan J, Amin S, Chung FL, Roy R. Repair kinetics of trans-4-hydroxynonenal-induced cyclic 1,N2-propanodeoxyguanine DNA adducts by human cell nuclear extracts. Biochemistry. 2004;43:7514–7521. doi: 10.1021/bi049877r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robison JG, Lu L, Dixon K, Bissler JJ. DNA lesion-specific co-localization of the Mre11/Rad50/Nbs1 (MRN) complex and replication protein A (RPA) to repair foci. J Biol Chem. 2005;280:12927–12934. doi: 10.1074/jbc.M414391200. [DOI] [PubMed] [Google Scholar]
- 38.Mechilli M, Schinoppi A, Kobos K, Natarajan AT, Palitti F. DNA repair deficiency and acetaldehyde-induced chromosomal alterations in CHO cells. Mutagenesis. 2008;23:51–56. doi: 10.1093/mutage/gem042. [DOI] [PubMed] [Google Scholar]
- 39.Hinz JM, Nham PB, Salazar EP, Thompson LH. The Fanconi anemia pathway limits the severity of mutagenesis. DNA Repair (Amst) 2006;5:875–884. doi: 10.1016/j.dnarep.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 40.Stein S, Lao Y, Yang IY, Hecht SS, Moriya M. Genotoxicity of acetaldehyde- and crotonaldehyde-induced 1,N2-propanodeoxyguanosine DNA adducts in human cells. Mutat Res. 2006;608:1–7. doi: 10.1016/j.mrgentox.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 41.Washington MT, Minko IG, Johnson RE, Wolfle WT, Harris TM, Lloyd RS, Prakash S, Prakash L. Efficient and error-free replication past a minor-groove DNA adduct by the sequential action of human DNA polymerases iota and kappa. Mol Cell Biol. 2004;24:5687–5693. doi: 10.1128/MCB.24.13.5687-5693.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Minko IG, Harbut MB, Kozekov ID, Kozekova A, Jakobs PM, Olson SB, Moses RE, Harris TM, Rizzo CJ, Lloyd RS. Role for DNA polymerase kappa in the processing of N2-N2-guanine interstrand cross-links. J Biol Chem. 2008;283:17075–17082. doi: 10.1074/jbc.M801238200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rulten SL, Hodder E, Ripley TL, Stephens DN, Mayne LV. Alcohol induces DNA damage and the Fanconi anemia D2 protein implicating FANCD2 in the DNA damage response pathways in brain. Alcohol Clin Exp Res. 2008;32:1186–1196. doi: 10.1111/j.1530-0277.2008.00673.x. [DOI] [PubMed] [Google Scholar]
- 44.Alter BP, Greene MH, Velazquez I, Rosenberg PS. Cancer in Fanconi anemia. Blood. 2003;101:2072. doi: 10.1182/blood-2002-11-3597. [DOI] [PubMed] [Google Scholar]
- 45.Ridpath JR, Nakamura A, Tano K, Luke AM, Sonoda E, Arakawa H, Buerstedde JM, Gillespie DA, Sale JE, Yamazoe M, Bishop DK, Takata M, Takeda S, Watanabe M, Swenberg JA, Nakamura J. Cells deficient in the FANC/BRCA pathway are hypersensitive to plasma levels of formaldehyde. Cancer Res. 2007;67:11117–11122. doi: 10.1158/0008-5472.CAN-07-3028. [DOI] [PubMed] [Google Scholar]
- 46.Frohnmayer L, Frohnmayer D. FANCONI ANEMIA: A Handbook for Families and Their Physicians. Eugene, Oregon: Fanconi Anemia Research Fund, Inc.; 2000. [Google Scholar]
- 47.Ford D, Easton DF, Bishop DT, Narod SA, Goldgar DE. Risks of cancer in BRCA1-mutation carriers. Breast Cancer Linkage Consortium. Lancet. 1994;343:692–695. doi: 10.1016/s0140-6736(94)91578-4. [DOI] [PubMed] [Google Scholar]
- 48.Mori T, Aoki T, Matsubara T, Iida F, Du X, Nishihira T, Mori S, Nakamura Y. Frequent loss of heterozygosity in the region including BRCA1 on chromosome 17q in squamous cell carcinomas of the esophagus. Cancer Res. 1994;54:1638–1640. [PubMed] [Google Scholar]
- 49.Cao L, Xu X, Cao LL, Wang RH, Coumoul X, Kim SS, Deng CX. Absence of full-length Brca1 sensitizes mice to oxidative stress and carcinogen-induced tumorigenesis in the esophagus and forestomach. Carcinogenesis. 2007;28:1401–1407. doi: 10.1093/carcin/bgm060. [DOI] [PubMed] [Google Scholar]
- 50.Eriksson CJ. The role of acetaldehyde in the actions of alcohol (update 2000) Alcohol Clin Exp Res. 2001;25:15S–32S. doi: 10.1097/00000374-200105051-00005. [DOI] [PubMed] [Google Scholar]
- 51.Triano EA, Slusher LB, Atkins TA, Beneski JT, Gestl SA, Zolfaghari R, Polavarapu R, Frauenhoffer E, Weisz J. Class I alcohol dehydrogenase is highly expressed in normal human mammary epithelium but not in invasive breast cancer: implications for breast carcinogenesis. Cancer Res. 2003;63:3092–3100. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.