Abstract
In this work we present a new support vector machine (SVM)-based method for fMRI data analysis. SVM has been shown to be a powerful, efficient data-driven tool in pattern recognition, and has been applied to the supervised classification of brain cognitive states in fMRI experiments. We examine the unsupervised mapping of activated brain regions using SVM. Specifically, the mapping process is formulated as an outlier detection problem of one-class SVM (OCSVM) that provides initial mapping results. These results are further refined by applying prototype selection and SVM reclassification. Multiple spatial and temporal features are extracted and selected to facilitate SVM learning. The proposed method was compared with correlation analysis (CA), t-test (TT), and spatial independent component analysis (SICA) methods using synthetic and experimental data. Our results show that the proposed method can provide more accurate and robust activation mapping than CA, TT and SICA, and is computationally more efficient than SICA. Besides its applicability to typical fMRI experiments, the proposed method is also a powerful tool in fMRI studies where a reliable quantification of activated brain regions is required.
Introduction
Blood oxygenation level dependent (BOLD) contrast functional magnetic resonance imaging (fMRI) has made possible the noninvasive study of brain function during various cognitive activities. However, accurately mapping activated brain regions remains a challenging problem because of relatively weak BOLD effects corrupted by noise and interferences.
A number of different techniques have been developed for fMRI data analysis, including parametric model-driven methods, such as statistical parametric mapping (SPM) based on the general linear model (GLM), statistical tests, and correlations (Friston et al., 1995). Due to spatial coherence and temporal autocorrelation between voxels (Zarahn at al., 1997), parametric linear models are not sufficient to characterize BOLD effects (Birn et al., 2001, Friston et al., 1998, Miller et al., 2001). Wavelet-based methods that consider spatial correlations have been suggested (Ruttimann at al., 1998, Ville et al., 2006). Methods using more complex parametric models, such as Markov random field models (Descombes et al., 1998) and hidden Markov models (Faisan et al., 2005), have also been proposed for fMRI data analysis. These methods require prior knowledge about activation patterns to model the hemodynamic response, and are most efficient for analyzing data from simple tasks. However, since they impose limitations on the shape and timing of the BOLD signal, the underlying neuronal mechanisms of which are not yet thoroughly understood, they are less effective for detecting complex activation patterns with unknown shapes, or change over time or across subjects.
Nonparametric data-driven methods, such as clustering (Baumgartner et al. 2000, Chuang et al., 1999, Goutte et al., 1999), principal component analysis (PCA) (Backfriender et al., 1996, Hansen et al., 1999), independent component analysis (ICA) (McKeown et al., 1998, Calhoun et al., 2001), and self-organizing mapping (Chuang et al., 1999, Ngan et al., 1999), do not require prior knowledge of BOLD response, and can deal flexibly with complex or unknown activation patterns. However, each method has its own limitation. For example, K-means clustering was used (Goutte et al., 1999, Goutte et al., 2001) with an implicit assumption that clusters are spherically symmetric and separable in the feature space which is not always applicable. Additionally, K-means clustering may suffer from the curse of dimensionality if data size does not significantly outnumber feature dimension. PCA only considers the second order statistics that are not sufficient to characterize fMRI data structure (Friston et al., 2000). ICA considers high-order dependencies among multiple samples and performs better than PCA in some comparison studies (McKeown et al., 1998, Laudadio et al., 2004). However, the assumptions of linear combinations of spatial (or temporal) independent sources in ICA could result in biased decompositions (McKeown et al., 1998). In addition, it is difficult to obtain optimal solutions of ICA by finding a proper initialization of the un-mixing matrix as the often used random initialization generates different suboptimal solutions in different runs. The nonlinear extension of discriminant analysis techniques, such as kernel PCA (Thirion et al., 2003) and kernel canonical correlation analysis (Hardoon et al., 2007), have been used to overcome the limitations of the linear discriminant analysis. A nonparametric machine learning method, support vector machine (SVM), has received increased attention for fMRI data analysis because it can overcome all of the aforementioned limitations by optimizing margin-based criteria and using the kernel trick (Vapnik, 1998). In addition, the SVM learning does not require any thresholds that could be affected by experimental conditions, making it applicable to quantitative fMRI studies where accurate measurements of activated brain area are required. Previously, SVM has been applied to the supervised classification of cognitive states corresponding to different categories of stimuli (Cox et al., 2003, Davatzikos et al., 2005, Hardoon et al., 2006, Ji et al., 2004, LaConte et al., 2005, Mitchell et al., 2004, Onut et al., 2004, Wang et al., 2003, Zhang et al., 2005). However, this technique has not been used for general mapping of brain activation likely due to the inter- and intra- subject variabilities of the BOLD response (Aquirre et al., 1998), which make training prototypes from a single subject, or multiple subjects not sufficiently representative. This issue has been addressed for the supervised decoding of brain states using SVM (Fan et al., 2006). Selecting reliable training prototypes becomes considerably more difficult for general brain activation detection using supervised SVM where multiple functional states that differ among subjects may be involved.
In this work, we propose a new SVM-based method that maps brain activation in an unsupervised way. Specifically, the mapping of activated brain voxels is formulated as an outlier (minority class) detection process of one-class SVM (OCSVM) (Schőlkopf et al., 2001), where non-activated voxels (majority class) outnumber the activated ones. The OCSVM is used to obtain the initial activation map, where a parameter ν controls the outlier ratio (OR) that is defined as the ratio of activated voxels to all brain voxels and is typically unknown. The initial activation map is further refined by using prototype selection (PS) and two-class SVM (TCSVM) reclassification. The proposed method has several advantages. First, it is data-driven requiring no prior knowledge about hemodynamic response. Second, it does not require an accurate setting of ν. Third, it is computationally efficient. Fourth, it does not need thresholds that could be affected by experimental conditions. We evaluated the proposed method using synthetic and experimental fMRI data and our results show that the proposed method can provide robust and accurate mapping of functional activation.
Methods
Theory of Support Vector Machine
We briefly review supervised and unsupervised SVM learning that provide the basis for the development of our method.
A. Supervised SVM Learning
TCSVM was first developed for supervised learning and classification (Vapnik, 1998). Given N training data from two classes {(xi, yi), i = 1, · · ·, N}, xi ∈ Rm, yi {1, −1}, where xi indicates an m dimensional feature vector and y is its class label, the TCSVM learning aims to find a linear classification hyperplane that maximizes separation of the two classes. Mathematically, it is equivalent to maximizing the margin magnitude subject to yi ((xi · w) + b) ≥ 1, where w and b determine the hyperplane (w · x) + b = 0. In order to improve generalization performance or classify nonseparable samples, training errors are permitted by introducing slack variables ξi ≥ 0. The hyperplane is then obtained by minimizing:
| (1) |
where C controls the tradeoff between the hyperplane complexity and training error, with a small C value corresponding to a large margin size. By solving the Lagrangian dual of (1), the final decision function can be written as:
| (2) |
where αi are Lagrange multipliers, (·) is the inner product, and . The linear TCSVM can be extended to the nonlinear case by using kernel methods (Vapnik, 1998). Kernel methods project the original data into a higher dimensional feature space x → φ(x), and a linear TCSVM classification in the latter is equivalent to a nonlinear TCSVM classification in the former, which is represented as:
| (3) |
where k(x, xi) = (φ (x) · φ (xi) is a kernel function satisfying Mercer’s condition (Vapnik, 1998). In our study, we used the Radial Basis Function (RBF) kernel that can project the input into unlimited dimensional feature space and distinguish activated brain voxels from non-activated ones. The RBF kernel is defined as k(x, xi) = e− γ||x − xi||2, where γ is the width parameter, and a large γ value corresponds to a small kernel width.
B. Unsupervised SVM Learning
The one-class classification estimates an approximation function to categorize a majority of the learning data in the feature space, and is applicable to unsupervised learning where class labels of training prototypes are not provided. There are two typical SVM-based one-class classifiers. One is the Support Vector Data Description (SVDD) that constructs a hypersphere containing most data in the feature space with a minimum volume (Tax, 2001). The other is the one-class SVM (OCSVM) that computes a hyperplane in the feature space to separate a pre-specified fraction (1− ν) of data with the maximum distance to the origin with a margin size of (Schőlkopf et al., 2001). ν ∈ (0,1] is an upper bound for the fraction of margin error, and a lower bound for the ratio of support vectors. The OCSVM hyperplane is constructed by minimizing:
| (4) |
where i = 1, · · ·, N. When the RBF kernel is used, SVDD and OCSVM are equivalent, operate comparably in practice and perform best (Schőlkopf et al., 2001, Tax, 2001).
The value of ν is unknown in most applications and currently there is no universal method for ν estimation. In (Ratsch, et al., 2002), the separation distance between the majority and outlier is calculated with respect to different ν values using training data, and the ν resulting in the largest distance is selected. This method works well only when the majority and outlier are clearly separable in the feature space, which is not always the case. In this paper, we describe the use of OCSVM and TCSVM for unsupervised fMRI data analysis.
Problem Formulation
Activated brain voxels differ from non-activated voxels spatiotemporally. Since non-activated voxels usually significantly outnumber the activated ones, mapping brain activation is an unbalanced problem. If all voxels are characterized by one cluster in a feature space, activated ones can be considered as outliers. Therefore, mapping brain activation can be formulated as an outlier detection problem using OCSVM. This formulation also provides the basic assumption underlying the proposed approach that activated voxels constitute less than 50% of all brain voxels. For convenience, we assign the class label “1” to activated voxels, and “−1” to the non-activated.
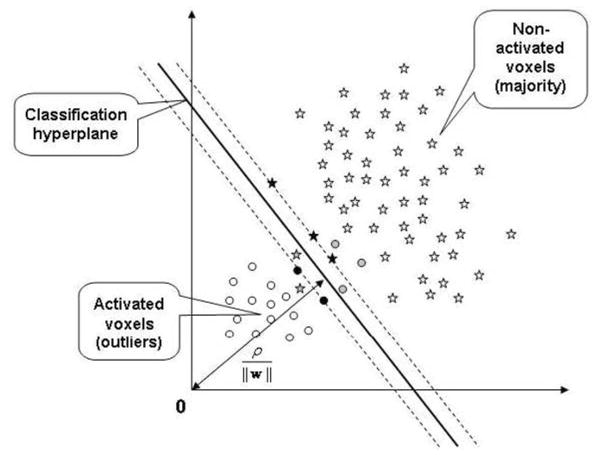
Fig. 1 illustrates the feature space distribution of activated (circle) and non-activated voxels (star) when OCSVM is used to provide the initial mapping. Stars and circles with dark fill are support vectors, and those with gray fill are mis-classifications. The outlier ratio (OR) obtained from OCSVM is upper bounded by ν, which is usually unknown but should be no larger than 0.5 according to our basic assumption. We aimed to develop a method that is not sensitive to ν set below 0.5 because finding a specific and precise ν is unrealistic due to inter- and intra-subject variation of BOLD signals. In a case where ORtrue the true ratio of activated voxels to all voxels, is known, it still cannot guarantee a clear separation by OCSVM as activated and non-activated voxels may overlap in the kernel projected feature spaces. Therefore, a ν greater than ORtrue is necessary to detect all activated voxels.
Fig. 1.
Feature space distribution of activated and non-activated voxels in terms of OCSVM learning, which builds a classification hyperplane in the feature space that maximally separates a majority of feature points from the origin. The feature points that are not classified as the majority are treated as outliers. Circles represent activated voxels (outliers) in the feature space, and stars indicate non-activated voxels (majority). Circles and stars with dark fill are support vectors, and those with gray fill are mis-classifications.
Proposed Approach
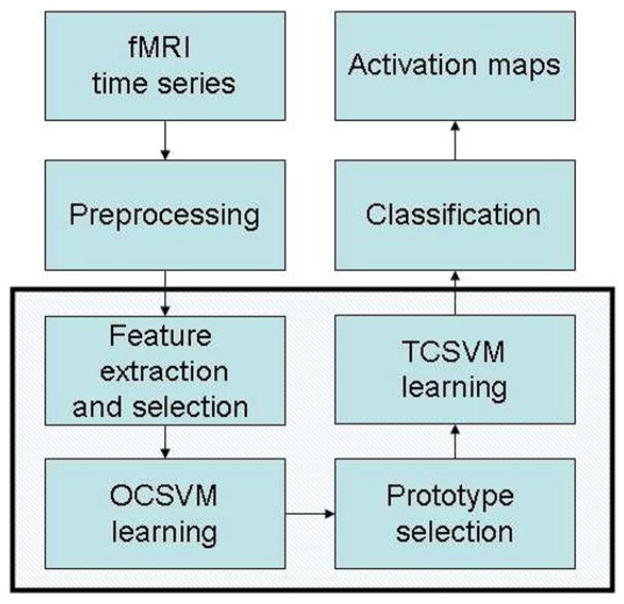
Fig. 2 illustrates the block diagram of the proposed method that follows a 4-step process after the preprocessing: 1) Features are extracted and selected. 2) OCSVM is used to generate an initial activation map, which provides prototypes for both activated and non-activated voxels. 3) Prototype selection (PS) is applied next to remove false prototypes (OCSVM mis-detections). 4) A TCSVM is trained using the prototypes selected in step 3 to reclassify the fMRI data. Steps 3 and 4 can be repeated to further refine the results.
Fig. 2.
Implementation diagram of the proposed method: OCSVM provides an initial activation map, based on which training data are selected via prototype selection, and a TCSVM is trained to re-classify the data.
1) Feature Extraction and Selection
Most SVM-based methods for supervised classification of fMRI data have used voxel intensity in spatial and/or temporal dimension as the input feature. This approach decreases the efficiency of SVM learning if the data size is very large. To increase the SVM learning efficiency, we extract intensity and correlation-based features that contain spatial, temporal, or spatiotemporal information related to brain activity from a voxel’s time course (TC) in the fMRI time series. Since the increase of temporal dimension will not increase the feature vector size, it is computationally more efficient than using intensity values as the input feature.
The extracted features are categorized as model-independent and model-dependent features. The following model-independent features are derived: the maximum magnitude of the TC, the average, maximum and minimum correlation coefficient (cc) values between the TC and other TCs within its 2nd-order neighborhood, as well as the average signed extreme value and delay of the cross correlation functions (ccf) between them. SVM learning assumes that input samples are independently drawn from an unknown distribution. However, fMRI data show strong local spatial correlations and ignoring such correlation may affect the detection performance. There are two general approaches to incorporate spatial correlation of fMRI data into machine learning. One approach is to integrate the spatial correlation into the objective function of machine learning (Liang, L., et al., 2006). The second approach is to use features containing information on fMRI spatial correlation for machine learning (Goutte et al., 2001), such as the above cc value and ccf based features. In order to facilitate event-related fMRI analysis with multiple trials, a temporal self-correlation measure was also used that is computed by averaging cc values between all pairs of trials’ TCs of the voxel (Ngan et al., 2001). When the hemodynamic model is available, model-dependent features can also be computed, including: the cc between the TC and the model, the signed extreme value and its delay in the cross correlation function between the TC and the model (Goutte et al., 2001). All features are normalized to a range between 0 and 1. Additional features that are helpful to the analysis can be added as well.
A heuristic method is applied next to select most salient features to facilitate the SVM learning. This method measures the contribution of each feature to SVM learning by evaluating its effect on the hyperplane construction (Evgeniou et al., 2003). The contribution of the dth feature Id is quantified by the integration of the first derivative of the decision function fc with respect to this feature around the hyperplane, and is approximated by (Evgeniou et al., 2003):
| (5) |
where is the dth feature of the ith support vector, NSV is the number of support vectors, αj are defined in equation (2), and Kd(xj, xi) is the derivative of the kernel regarding . Features with relative large Id values will be selected for SVM learning, and those with small values will be discarded. After feature selection, each brain voxel is represented by a feature vector composed of selected features, and used as input to the following step.
2) Initial Mapping Using OCSVM
Since brain activation mapping is formulated as an outlier detection problem, OCSVM is used to provide an initial classification of activated and non-activated voxels. For this step, ν is randomly set within a range Rν, which is either unknown but upper bounded by 0.5, or known a priori from previous experiments. We could run a risk of under- or over-detecting the activation if ν is too large or too small. A too-large ν might generate significant mis-detections that cannot be completely removed by prototype selection, whereas a too-small ν could under-detect. In the event of under-detection, if omitted activated voxels are spatiotemporally similar to those already detected they can be found by steps 3 and 4. However, if they have distinct spatiotemporal patterns from the detected voxels they cannot be completely recovered, and we need to increase ν to improve the sensitivity. For a known Rν, ν can be set randomly within this range.
3) Prototype Selection
OCSVM results are used to select training prototypes as the input for TCSVM learning that is applied to refine the initial activation map. A prototype consists of a data sample (a feature vector representing a voxel) and its class label (activated and non-activated). Since a ν that is greater than ORtrue could result in over detection, which could mislead the TCSVM learning, special considerations must be made to remove false detections. Activated brain regions could be in different anatomic locations, and in each location activated voxels are clustered together spatially. We use an editing method that takes advantage of this spatial connectivity to remove OCSVM mis-detections, which are less likely to cluster together spatially, so that the subsequent TCSVM training can be improved. Editing is a PS method that aims to remove erroneously labeled training data to improve classification accuracy (Dasarathy et al., 2000). The neighborhood connectivity of brain voxels can be described by one type of proximity graphs (PG), i.e., Gabriel Graph (GG) (Jaromczyk et al., 1992). Given a set of n points Z = {z1, · · ·, zn} in a d-dimensional feature space Rd, a PG G(V,E) is a graph with a set of vertices V=Z and a set of edges E, such that (zi, zj) ∈ E if and only if zi and zj satisfy certain neighborhood property. A GG is a PG with the set of edges:
| (6) |
where zk ∈ Z, and Dis(·, ·) is the Euclidean distance in Rd. When Z is the spatial coordinate of all brain voxels, d=2. Given the 2nd-order neighborhood system of zi, the corresponding PG G(V,E) is a GG. By applying the GG’s 1st-order graph neighborhood editing technique with voting strategy (Dasarathy et al., 2000), any voxel which label is not dominant in its 2nd-order neighborhood is removed from the training data.
4) TCSVM Reclassification
The selected prototypes for activated and non-activated voxels in the previous step are used to train a TCSVM. The trained TCSVM is then applied to reclassify the OCSVM input to obtain a refined activation map. In order to recover under-detected activations in step 2, possibly removed activations in step 3, or reduce the effects from possible remaining erroneous prototypes after step 3, the TCSVM capacity is carefully controlled to favor high generalization performance that is achieved by using large RBF kernel width and small C values.
Method Evaluation
The proposed method was evaluated by using both synthetic and experimental fMRI time series, and compared with two model-driven methods, correlation analysis (CA) between the task paradigm and voxels’ time courses, and t-test (TT), and a data-driven method Spatial ICA (SICA). A software package LIBSVM was used to implement OCSVM and TCSVM (Chang et al., 2001). FastICA with nonlinear contrast function Tanh and symmetric de-correlation (Hyvärinen et al., 1999) was applied to implement SICA, where the data were first pre-whitened and reduced in dimension using PCA. Currently there is no method that can properly initialize FastICA to achieve a global optimization, and random initialization is often used that leads to different convergence rates and suboptimal decomposition results. Multiple runs were therefore compared to evaluate the consistency of results. All computations were implemented in Matlab on a HP6200 Linux workstation with an Intel 3.4G Hz CPU and 2G Bytes RAM.
A. Synthetic Data
A synthetic fMRI time series of 30 images was generated with a boxcar paradigm of 10 images off, 10 on, and 10 off. All images in the time series were generated from a single EPI image by adding artificial activations and noise. Fig. 3(a) shows the image with two artificially activated regions that differ in area and signal magnitude. The activated region on the left represents 1.6% of the image area and 4% signal increase. The region on the right represents 2.5% of the image area and 7% signal increase. The noise in fMRI images is assumed to follow a Rician distribution, and was generated using the method in (Wink et al., 2004). Given a clean image I and two images I1 and I2 that only contain independently and identically distributed Gaussian noise with zero mean, we use to get the noisy image In, and the corresponding Rician noise is R=In−I. The baseline average calculated using the first 10 images was subtracted from individual images to generate the difference image time series with a SNR of −21.66dB.
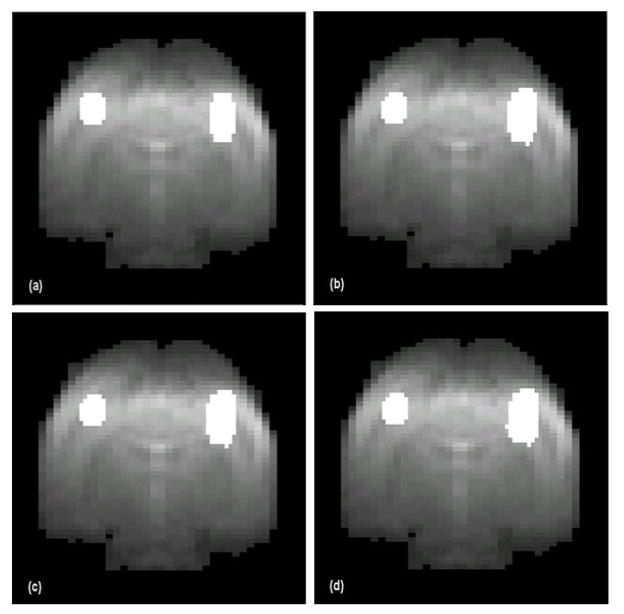
Fig. 3.
Evaluation of the proposed method with synthetic data. (a) Synthetic image with two activated regions comprising 4.1% of the image area. (b)–(d) are activation maps generated by the proposed method from the synthetic data using ν values of 0.18 (b), 0.22 (c), and 0.25 (d). These maps indicate that the proposed method can provide essentially uniform results over a range of ν values within Rν = [0.1, 0.3].
For the evaluation of proposed method, γ was set to 0.1 and 0.01 for the RBF kernel used with OCSVM and TCSVM, respectively. C of the TCSVM was set to 1, indicating the mis-classification of 1 and −1 will be equally treated during the learning. This setting favors high recall rate (sensitivity) of detection as opposed to high precision. For CA and TT, the tradeoff between the recall and precision rates can be changed by varying the significance level. For FastICA, the data were first reduced in dimension to 28 components (retaining 98.9% original variation) by PCA, then 28 ICA components were estimated. The values in each ICA component were scaled to z-scores with zero mean and standard deviation. Voxels whose absolute z-scores are above a pre-specified threshold were considered to be active for that component (McKeown et al., 1998).
B. Experimental Data
Experimental data were acquired for two different paradigms, a simple block-design sensory stimulation paradigm and an event-related learning paradigm, to evaluate the performance of proposed method.
The sensory paradigm was designed to characterize the BOLD response to a whisker stimulus (65Hz sinusoidal vibration) and consisted of 22 images off, 20 on, and 20 off. BOLD contrast fMRI data were acquired from the somatosensory cortex of a Dutch-belted rabbit using a 4.7T Bruker Avance imaging spectrometer. Four contiguous 1mm-thick slices were acquired using a single-shot gradient echo EPI with a 48mm × 48mm FOV and a 128 × 64 matrix size, corresponding to a voxel size of 375μm × 750μm, a 2s repetition time (TR) and a 20ms echo time (TE). The initial three images were deleted to allow for longitudinal relaxation equilibration. Ten trials were averaged to improve SNR.
The fMRI data were first preprocessed by affine transformation-based registration to remove motion artifacts, followed by denoising using a newly developed method (Song et al., 2006). In this method, a statistical t-test is first applied to each voxel’s time course in the wavelet domain. The test result is used as an initial significance estimation of wavelet coefficients, and applied to the Gaussian mixture model (GMM)-based Bayesian wavelet shrinkage in the spatial domain. OCSVM settings were the same as those selected for the synthetic data. In implementing TCSVM, γ was set to 0.05 for the RBF kernel and C was set to 10 after experimentally comparing different values. For FastICA, the first 50 components (retaining 98.9% of original variance) extracted by PCA were used to estimate 50 ICA components.
The event-related learning paradigm consisted of a 250ms visual conditioned stimulus (CS), followed by 500ms interval and a 100ms airpuff as unconditioned stimulus (US). The stimuli were delivered as described previously (Wyrwicz et al., 2000). It was designed to identify regions of functional activation related to learning from a Dutch-belted rabbit trained with a trace eyeblink conditioning paradigm. The subject received two pseudo-conditioning sessions (40 trials/session), followed by training sessions (40 trials/session) until it reached a learning criterion. Functional images were acquired on the 4.7T Bruker Avance from eight contiguous 1mm thick coronal slices perpendicular to the plane of a 60mm flat, circular surface coil using a single-shot gradient echo EPI with the same FOV, matrix size, TR, and TE as the block-design experiment, and 35 images collected per trial. The initial three images were deleted from each trial and the trials were averaged.
For this learning paradigm, the hemodynamic response is not known and only the model-independent features were computed and used in the OCSVM learning. For FastICA, the first 26 components (retaining 96% original variance) extracted by PCA were used to estimate 26 ICA components. For TT analysis, the first two images acquired after the presentation of stimuli were used to extract activation statistics.
Results and Discussion
Synthetic Data
Fig. 3(b)–(d) shows the activation maps generated for three ν values, ν=0.18, 0.22, and 0.25 with one iteration of steps 3 and 4 described in the proposed method section. Although these ν values and their relative difference (0.25−0.18=0.07) are greater than ORtrue, the mapping results are essentially the same for the three, suggesting little dependence on the initial v setting.
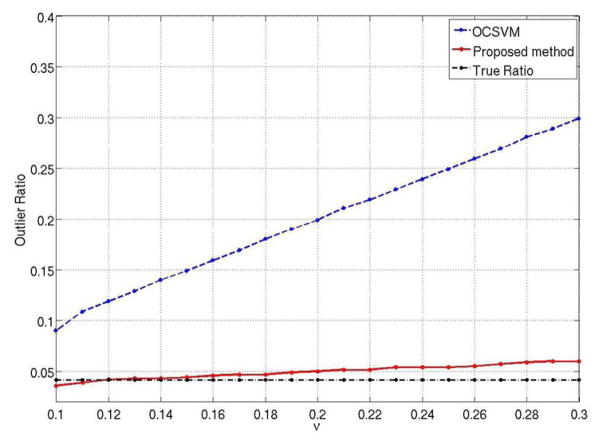
The dependency of the proposed method on initial ν setting was evaluated further and compared to the OCSVM. A set of ν values greater than ORtrue=0.04 were selected from Rν=[0.1, 0.3], beginning with 0.1 and incremented by 0.01 to generate activation maps. The ORs calculated from the activation maps generated by the two methods as a function of ν are compared with ORtrue in Fig. 4. The ORs from the proposed method compare well with the ORtrue and show 8.7 times less dependence on ν (measured by the slope) than those from OCSVM.
Fig. 4.
Outlier ratio (OR) calculated as a function of ν for the synthetic data using OCSVM (dashed blue line), and the proposed method (solid red line). The later provides OR close to its true value (0.04, dash-dot dark line) with negligible dependence on ν over this range, as compared to OCSVM.
The results in Figs. 3 and 4 indicate that (a) the method can provide nearly uniform results over Rν; and (b) an increase in the initial ν value slightly increases the sensitivity of activation detection without significant mis-detections. The results also verified that an initial ν greater than ORtrue is appropriate for the proposed method.
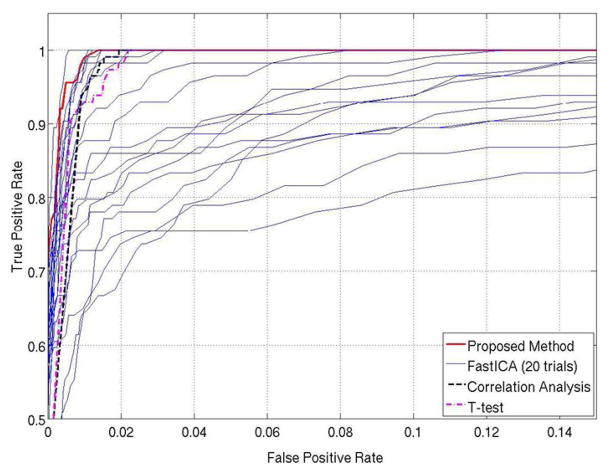
Fig. 5 shows a comparison of the proposed method performance with CA, TT, and FastICA using receiver operating characteristic (ROC) curves. 20 runs of FastICA were performed and resulted in 20 ROC curves that vary significantly due to the random initialization. In terms of ROC performance, the proposed method outperforms CA and TT. In 20 runs of FastICA, only one run achieves somewhat better ROC than the proposed method. However, obtaining a proper initialization is difficult for FastICA, and selecting the best from multiple runs might not be a practical approach due to increased computational time. The computational time of each of 20 trials of FastICA is between 30 and 200 seconds, in comparison to 3–6 seconds for the proposed method.
Fig. 5.
A comparison of the receiver operating characteristic (ROC) performance between the proposed method (red thick solid line), correlation analysis (dark thick dashed line), t-test (pink thick dash-dot line), and FastICA (blue thin solid lines) using the synthetic data. The best ROC performances were obtained by the proposed method and only one of twenty FastICA runs.
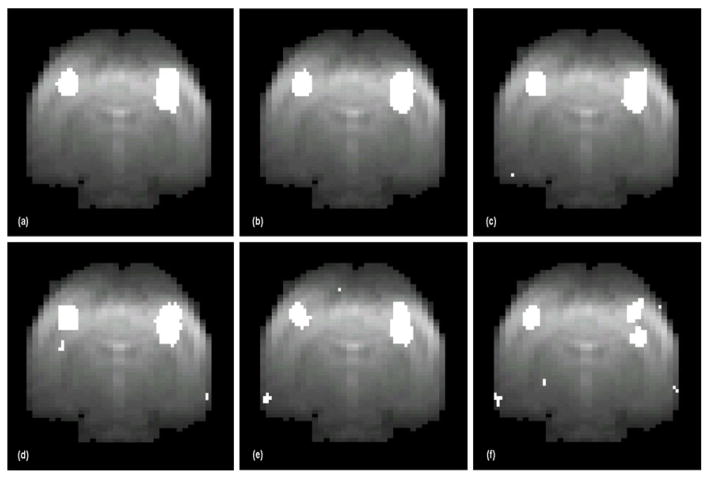
The four methods were also compared in terms of detection sensitivity by specifying the same false positive rate (FPR) at 0.01, and the results are shown in Fig. 6. The corresponding detection sensitivities are 99.12% for the proposed method, 94.74% for CA, 92.98% for TT. The highest, median, and lowest sensitivities of FastICA results are 100%, 83%, and 65%, respectively. With the same FPR, FastICA can achieve 100% detection sensitivity in one of its 20 runs, but it tends to generate relatively sparse and discrete activation maps by splitting continuous activated areas into multiple ones, as mentioned in (McKeown et al., 1998). This effect is most obvious in the run with the 65% sensitivity as shown in Fig. 6(f). In contrast, the sensitivity of proposed method is comparable to the best case of FastICA with continuous activated areas, and is higher than that of CA and TT. The upper bound of the FPR of the proposed method can be determined by ν specified at the level of the OCSVM step. Typically, the OCSVM results fall well below the FPR upper bound, and the prototype selection step can reduce it further. However, since prototype selection is a nonlinear process it is difficult to know exactly how much lower FPR is and its relationship to ν. To target a specific FPR, the ν value that generates the desired FPR in the ROC curve may be used (Sorenson et al., 1996).
Fig. 6.
Comparison of detection sensitivity among the four methods at 1% false positive rate (FPR). The activation maps were generated from the synthetic data by (a) the proposed method (sensitivity 99.12%, (b) correlation analysis (sensitivity 94.47%), (c) t-test (sensitivity 92.98%), (d)–(f) FastICA (sensitivity 100%, 83%, and 65%, respectively) from three different runs. The proposed method provides higher detection sensitivity than correlation analysis and t-test. Only one FastICA run achieves a high detection sensitivity, and the map in (f) indicates that ICA tends to split a continuous activated region into discrete ones (McKeown et al., 1998).
The evaluation with the synthetic data numerically validates the efficiency and robustness of the proposed method measured by the ν-dependency, ROC, and computational time. We further evaluated this method using block-design and event-related experimental data.
Experimental Data
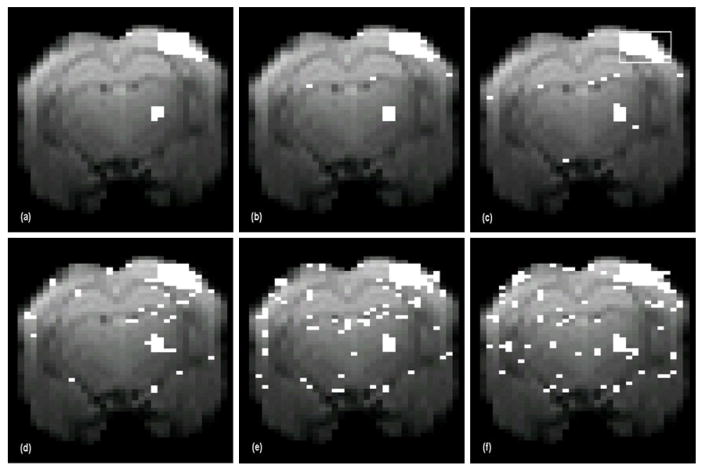
Fig. 7(a)–(c) shows activation maps in a slice containing the somatosensory cortex generated for the whisker stimulation paradigm with the proposed method for ν=0.22, 0.25, and 0.28. All three values are greater than ORtrue ∈ [0.07, 0.1], estimated from previous experiments. Two activated regions were detected: a larger area in the whisker region of the somatosensory cortex (upper right) and a smaller area in the somatosensory thalamic nuclei (medial and lateral ventral posterior thalamic nuclei). The results of all three ν values are similar and are consistent with the anatomically defined cortical and thalamic representation of the stimulated whiskers. The maximum increase in BOLD signal was 1.7% for the somatosensory cortex and 2.0% for the somatosensory thalamic nuclei. The relative performance of the four methods was evaluated in terms of detection sensitivity. For uniform comparison, the threshold settings for CA, TT and FastICA were set to generate an equal number of activated voxels (contiguously clustered) in the somatosensory cortex to those detected with our proposed method for ν =0.28, indicated by the box in Fig. 7(c). The resulting activation maps are shown in Fig. 7(d)–(f). At comparable detection sensitivity in the somatosensory cortex, CA, TT, and FastICA methods generate a much greater number of mis-detections than the proposed method.
Fig. 7.
Activation maps (overlaid on an EPI image) generated for the whisker stimulation paradigm by the proposed method with (a) ν = 0.22, (b) ν = 0.25, (c) ν = 0.28; by (d) correlation analysis, (e) t-test, and (f) FastICA under a comparable activation detection sensitivity to the proposed method using ν = 0.28. The box in (c) indicates an 8 voxels × 8 voxels region in the somatosensory cortex used for detection sensitivity comparison between the different methods. All three maps in (a)–(c) are similar and consistent with the anatomically defined cortical and thalamic representation of the stimulated whiskers. Note the visibly greater number of mis-detections in (d)–(f), in contrast to the proposed method.
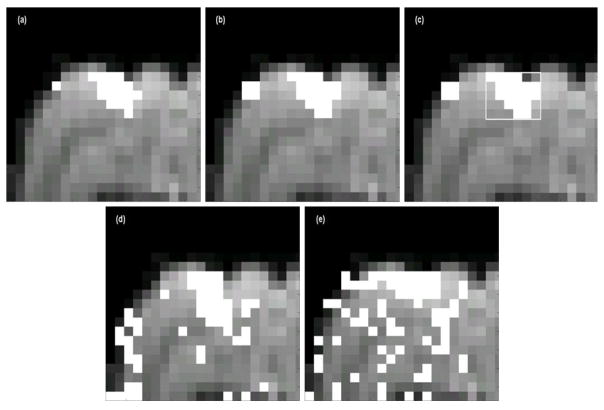
fMRI data acquired using the learning paradigm with the visual stimulus were preprocessed as described previously and analyzed using the proposed method, TT, and FastICA. Fig. 8 shows the activation maps for a representative visual cortex slice generated by the proposed method using three randomly selected ν values: 0.3, 0.33, and 0.35. The three maps are very close in terms of the activated area (17, 19, 19 activated voxels, respectively) in the visual cortex although there is a 5% increase in ν.
Fig. 8.
Activation maps (overlaid on an EPI image) generated for the trace conditioning data by the proposed method with (a)ν = 0.3, (b) ν = 0.33, (c) ν = 0.35; by (d). t-test and (e) FastICA under a comparable activation detection sensitivity to the proposed method using ν = 0.35. Only the expanded section of the image containing the primary visual cortex region is shown for better comparison. The three maps in (a)–(c) are similar in terms of the activated area (17, 19, 19 activated voxels, respectively). Note the visibly greater number of mis-detections produced by these methods, in contrast to the proposed method.
The relative performance of the methods was evaluated in terms of detection sensitivity by the same approach used for the block-design experiment. The threshold settings for TT and FastICA were set to generate a contiguous cluster of activated voxels in the visual cortex equal in number to that detected with the proposed method when ν=0.35, indicated by the box in Fig. 8(c). Both TT and FastICA methods generate a large number of mis-detections as compared to the proposed method as shown in Fig. 8(d) and (e). In addition, TT results are less reliable as only two images after the stimulus presentation were used in the analysis. However, when the number of images in the analysis is increased, the transient activation in the visual cortex is not detectable with the TT method.
In addition to identifying regions of functional activation related to learning, these type of experiments also need to measure and compare the size of the activated areas during learning acquisition (Miller et al., 2008). However, both SICA and TT use thresholds and thus cannot provide a reliable measurement of the activated area over time as it cannot be certain that the change in the area is due to learning or to change in the thresholds. Using the same threshold for different stages of learning is not reliable either because the fMRI data recorded at different times may have very distinct intensity level and noise conditions. The proposed method does not require such thresholds during the entire process and therefore is uniquely suited for the quantitative analysis in these types of experiments. A comparison study of brain activation at different phases of the learning acquisition using the proposed method is presented in detail in (Miller et al., 2008).
Conclusion
We proposed an unsupervised SVM-based method for fMRI data analysis and compared it with existing model-dependent and model-independent methods using synthetic and experimental data. The approach is novel as it treats the activated voxels as outliers when generating the initial activation map using OCSVM. Further refinement of the activation map is achieved with efficient prototype selection and TCSVM reclassification that improves accuracy and sensitivity of the activation detection. The proposed method is also computationally efficient as a result of using spatial and temporal features instead of the intensity values as input to facilitate the SVM learning. Although this method does not require prior knowledge of the fMRI response to the paradigm it can, however, incorporate prior information into the analysis.
The proposed method is robust as indicated by insensitivity to the initial ν value, and provides more accurate results than the conventional correlation analysis, statistical t-test or SICA as illustrated by the ROC analysis results. It can be applied to block design or event-related paradigms. In addition, it allows for accurate measurements of activated brain area from fMRI data acquired over multiple scanning sessions over time that is not feasible with other existing methods. This capability is important for functional studies of learning and memory where an increase in size of the activated area may be linked to recruitment of activated neurons during processing and/or storage of the acquired information.
Acknowledgments
The authors thank Dr. Michael Miller for providing the trace eyeblink conditioning data. This research is supported by National Institute of Health (NIH) RO1 NS44617 and S10 RR15685 grants.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aquirre GK, Zarahn E, D’Esposito M. The variability of human, BOLD hemodynamic responses. NeuroImage. 1998;8 (4):360–369. doi: 10.1006/nimg.1998.0369. [DOI] [PubMed] [Google Scholar]
- Backfriender W, Baumgartner R, Samal M, Moser E, Bergmann Quantification of intensity variations in functional MR images using rotated principal components. Physics in Medicine and Biology. 1996;41 (8):1425–1438. doi: 10.1088/0031-9155/41/8/011. [DOI] [PubMed] [Google Scholar]
- Baumgartner R, Ryner L, Richter W, Summers R, Jarmasz M, Somorjai R. Comparison of two exploratory data analysis methods for fMRI: Fuzzy clustering vs. principal component analysis. Magnetic Resonance in Medicine. 2000;18:89–94. doi: 10.1016/s0730-725x(99)00102-2. [DOI] [PubMed] [Google Scholar]
- Birn RM, Saad ZS, Bandettini PA. Spatial heterogeneity of the nonlinear dynamics in the fMRI BOLD response. NeuroImage. 2001;14:817–826. doi: 10.1006/nimg.2001.0873. [DOI] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pearlson GD, Pekar JJ. Spatial and temporal independent component analysis of functional MRI data containing a pair of block-design waveforms. Human Brain Mapping. 2001;13:43–53. doi: 10.1002/hbm.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C-C, Lin C-J. LIBSVM: a library for support vector machines 2001 [Google Scholar]
- Chuang KH, Chiu MJ, Lin CC, Chen JH. Model-Free functional MRI analysis using Kohonen clustering neural network and fuzzy C means. IEEE Trans on Medical Imaging. 1999;18 (2):1117–1128. doi: 10.1109/42.819322. [DOI] [PubMed] [Google Scholar]
- Cox DD, Savoy RL. Functional magnetic resonance imaging (fMRI) “brain reading”: detecting and classifying distributed patterns of fMRI activity in human visual cortex. NeuroImage. 2003;19:261–270. doi: 10.1016/s1053-8119(03)00049-1. [DOI] [PubMed] [Google Scholar]
- Dasarathy BV, Sanchez JS, Townsend S. Nearest neighbour editing and condensing tools-synergy exploitation. Pattern Analysis & Applications. 2000;3 (1):19–30. [Google Scholar]
- Davatzikos C, Ruparel K, Fan Y, Shen D, Acharyya M, Loughead J, Gur R, Langlebenb D. Classifying spatial patterns of brain activity with machine learning methods: Application to lie detection. NeuroImage. 2005;28:663–668. doi: 10.1016/j.neuroimage.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Descombes X, Kruggel F, Cramon DYV. Spatio-temporal fMRI analysis using Markov random fields. IEEE Trans on Medical Imaging. 1998;17:1028–1039. doi: 10.1109/42.746636. [DOI] [PubMed] [Google Scholar]
- Evgeniou T, Pontil M, Papageorgiou C, Poggio T. Image representations and feature selection for multimedia database search. IEEE Trans on Pattern Analysis and Machine Intelligence. 2003;15 (4):911–920. [Google Scholar]
- Faisan S, Throava L, Armspach JP, Metz-Lutz MN, Heitz F. Unsupervised learning and mapping of active brain functional MRI signals based on hidden semi-Markov event sequence models. IEEE Trans on Medical Imaging. 2005;24 (2):263–276. doi: 10.1109/tmi.2004.841225. [DOI] [PubMed] [Google Scholar]
- Fan Y, Sheng D, Davatzikos C. Detecting cognitive states from fMRI images by machine learning and multivariate classification. Proc. Conf. on Computer Vision and Pattern Recognition Workshop; 2006. pp. 89–96. [Google Scholar]
- Friston K, Holmes A, Worsley, Poline KJ, Frith C, Frackowiak R. Statistical parametric maps in functional imaging: A general linear approach. Human Brain Mapping. 1995;2:189–210. [Google Scholar]
- Friston K, Josephs O, Rees G, Tuner R. Nonlinear event-related responses in fMRI. Magn Reson Med. 1998;39:41–52. doi: 10.1002/mrm.1910390109. [DOI] [PubMed] [Google Scholar]
- Friston K, Phillips J, Chawla D, Buchel C. Nonlinear PCA: Characterizing interactions between modes of brain activity. Philos Trans R Soc Lond B Biol Sci. 2000;355 (1393):135–146. doi: 10.1098/rstb.2000.0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goutte C, Toft P, Rostrup F, Nielsen FA, Hansen LK. On clustering fMRI time series. Neuroimage. 1999;9:298–310. doi: 10.1006/nimg.1998.0391. [DOI] [PubMed] [Google Scholar]
- Goutte C, Hansen LK, Liptrot MG, Rostrup E. Feature space clustering for fMRI meta-analysis. Human Brain Mapping. 2001;13:165–183. doi: 10.1002/hbm.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen LK, Larsen FA, Nielsen SC, Strother E, Rostrup E, Savoy R, Svarer C, Paulson OB. Generalizeable patterns in neuroimaging: How many principal components. Neuroimage. 1999;9:534–544. doi: 10.1006/nimg.1998.0425. [DOI] [PubMed] [Google Scholar]
- Hardoon DR, Mourão-Miranda J, Brammer M, Shawe-Taylor J. Unsupervised analysis of fMRI data using kernel canonical correlation. Neuroimage. 2007;37:1250–1259. doi: 10.1016/j.neuroimage.2007.06.017. [DOI] [PubMed] [Google Scholar]
- Hardoon DR, Manevitz LM. fMRI analysis via one-class machine learning techniques. Proc. Conf. on Data Mining, Systems Analysis and Optimization in Neuroscience.2006. [Google Scholar]
- Hyvärinen A. Fast and Robust fixed-point algorithms for independent component analysis. IEEE Trans on Neural Networks. 1999;10 (3):626–634. doi: 10.1109/72.761722. [DOI] [PubMed] [Google Scholar]
- Jaromczyk J, Toussaint G. Relative neighborhood graphs and their relatives. Proceedings of the IEEE; pp. 1502–1517. [Google Scholar]
- Ji Y, Liu H, Wang X, Tang Y. Cognitive states classification from fMRI data using support vector machines. Proc Int Conf on Machine Leaning and Cybernetics. 2004;5:2919–2923. [Google Scholar]
- LaConte S, Strother S, Cherkassky V, Anderson J, Hu X. Support vector machines for temporal classification of black design fMRI data. NeuroImage. 2005;26:317.329. doi: 10.1016/j.neuroimage.2005.01.048. [DOI] [PubMed] [Google Scholar]
- Laudadio T, Huffel SV. Extraction of independent components from fMRI data by principal component analysis, independent component analysis and canonical correlation analysis. ESAT-SISTA 2004 [Google Scholar]
- Liang L, Cherkassky V, Rottenberg DA. Spatial SVM for feature selection and fMRI activation detection. Proc. Int. Joint Conf. on Neural Networks; 2006. pp. 1463–1469. [Google Scholar]
- McKeown MJ, Makeig S, Brown GG, Jung TP, Kindermann SS, Bell AJ, Spjnowski TJ. Analysis of fMRI data by blind separation into independent spatial components. Human Brain Mapping. 1998;6:160–188. doi: 10.1002/(SICI)1097-0193(1998)6:3<160::AID-HBM5>3.0.CO;2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K, Luh W, Lie T, Martinez A, Obata T, Wong E, Frank L, Buxton R. Nonlinear temporal dynamics of the cerebral blood flow response. Human Brain Mapping. 2001;13:1–12. doi: 10.1002/hbm.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M, Weiss C, Song X, Iordanescu G, Disterhoft JF, Wyrwicz AM. fMRI of delay and trace eyeblink conditioning in the primary visual cortex of the rabbit. Journal of Neuroscience. 2008;28 (19):4974–4981. doi: 10.1523/JNEUROSCI.5622-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell T, Hutchinson R, Niculescu R, Pereira R, Wang X, Just M, Newman S. Learning to decode cognitive states from brain images. Machine Learning. 2004;57 (1–2):145–175. [Google Scholar]
- Ngan SC, Hu X. Analysis of functional magnetic resonance imaging data using self-organizing mapping with spatial connectivity. Magn Reson Med. 1999;41:936–946. doi: 10.1002/(sici)1522-2594(199905)41:5<939::aid-mrm13>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Ngan SC, Auffermann WF, Sarkar S, Hu X. Activation detection in event-related fMRI data based on spatio-temporal properties. Magnetic Resonance Imaging. 2001;19:1149–1158. doi: 10.1016/s0730-725x(01)00444-1. [DOI] [PubMed] [Google Scholar]
- Onut L, Ghorbani AA. Classifying cognitive states from fMRI data using neural networks. IEEE Int. Joint conf. on Neural Networks; 2004. pp. 2871–2875. [Google Scholar]
- Ratsch G, Mika S, Scholkopf B, Muller KR. Constructing boosting algorithms from SVMs: An application to one-class classification. IEEE Trans on Pattern Analysis and Machine Intelligence. 2002;24 (9):1184–1199. [Google Scholar]
- Ruttimann UE, Unser M, Rawlings R, Rio D, Ramsey NF, Mattay VS, Hommer DW, Frank JA, Weinberger DR. Statistical analysis of functional MRI data in the wavelet domain. IEEE Trans on Medical Imaging. 1998;17 (2):142–154. doi: 10.1109/42.700727. [DOI] [PubMed] [Google Scholar]
- Schőlkopf B, Platt JC, Shawe-Taylor J, Smola AJ, Williamson RC. Estimating the support of high-dimensional distribution. Neural Computation. 2001;13 (7):1443–1471. doi: 10.1162/089976601750264965. [DOI] [PubMed] [Google Scholar]
- Song X, Murphy M, Wyrwicz AM. Spatiotemporal denoising and clustering of fMRI date. Proc. IEEE Int. Conf. on Image Processing..2006. [Google Scholar]
- Sorenson JA, Wang X. ROC methods for evaluation of fMRI techniques. Magn Reson Med. 1996;36:737–744. doi: 10.1002/mrm.1910360512. [DOI] [PubMed] [Google Scholar]
- Tax DMJ. Ph.D. Dissertation. Technische Universiteit Delft; The Netherlands: 2001. One-class classification. [Google Scholar]
- Thirion B, Faugeras O. Dynamical component analysis of fMRI data through kernel PCA. NeuroImage. 2003;20:34–49. doi: 10.1016/s1053-8119(03)00316-1. [DOI] [PubMed] [Google Scholar]
- Vapnik VN. Statisical Learning Theory. 1998 doi: 10.1109/72.788640. [DOI] [PubMed] [Google Scholar]
- Ville DVD, Blu T, Unser M. Surfing the brain: An overview of wavelet-based techniques for fMRI data analysis. IEEE Engineering in Medicine and Biology Magazine. 2006;25 (2):64–78. doi: 10.1109/memb.2006.1607671. [DOI] [PubMed] [Google Scholar]
- Wang X, Hutchinson R, Mitchell TM. Training fMRI classifiers to detect cognitive states across multiple human subjects. Neural Information Processing Systems 2003 [Google Scholar]
- Wink AM, Roerdink Jos BTM. Denoising functional MR images: A comparison of wavelet denoising and Gaussian smoothing. IEEE Trans on Medical Imaging. 2004;23 (3):374–387. doi: 10.1109/TMI.2004.824234. [DOI] [PubMed] [Google Scholar]
- Wyrwicz AM, Chen N, Li L, Weiss C, Disterhoft JF. fMRI visual system activation in the conscious rabbit. Magnetic Resonance in Medicine. 2000;44:474–478. doi: 10.1002/1522-2594(200009)44:3<474::aid-mrm19>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Zarahn E, Aquirre GK, D’Esposito M. Empirical analysis of BOLD fMRI statistics. NeuroImage. 1997;5:179–197. doi: 10.1006/nimg.1997.0263. [DOI] [PubMed] [Google Scholar]
- Zhang L, Samaras D, Tomasi D, Goldstein N. Machine learning for clinical diagnosis from functional magnetic resonance imaging. Proc. IEEE conf. on Computer Vision and Pattern Recognition; 2005. pp. 1211–1217. [Google Scholar]