SUMMARY
The CASTOR (CST) transcription factor was initially identified for its role in maintaining stem cell competence in the Drosophila dorsal midline. Here we report that Xenopus CST affects cardiogenesis. In CST-depleted embryos, cardiomyocytes at the ventral midline arrest and are maintained as cardiac progenitors, while cells in more dorsal regions of the heart undergo their normal program of differentiation. Cardia bifida results from failed midline differentiation, even though cardiac cell migration and initial cell fate specification occur normally. Our fate mapping studies reveal that this ventral midline population of cardiomyocytes ultimately gives rise to the outer curvature of the heart; however, CST-depleted midline cells overproliferate and remain a coherent population of nonintegrated cells positioned on the outer wall of the ventricle. These midline-specific requirements for CST suggest the regulation of cardiomyocyte differentiation is regionalized along a dorsal-ventral axis and that this patterning occurs prior to heart tube formation.
INTRODUCTION
Terminal differentiation of progenitor populations is initiated in response to alterations in growth factor signaling, which leads to an arrest in cell cycle progression and the transcription of tissue-specific genes. In contrast to other cell types such as neural tissue and skeletal muscle, relatively little is known about the molecular pathways that trigger the onset of cardiomyocyte differentiation during heart development. Studies in tissue culture focusing on the ability of growth factors to drive the formation of cardiomyocytes from stem cells have identified numerous pathways that may be involved in cardiomyocyte differentiation (Guan and Hasenfuss, 2007; Liu et al., 2007); however, the transcriptional targets of these pathways or the endogenous role for their components during in vivo embryonic cardiomyocyte differentiation remains unclear. Furthermore, it is not known whether cardiomyocytes differentiate in a uniform manner in response to a single cue or if specific subsets of cardiomyocytes differentiate in response to different pathways.
To address these issues and to begin to identify the molecular pathways that function in vivo to regulate the onset of cardiomyocyte differentiation, we have characterized the Xenopus ortholog of Castor (Cst), a protein that regulates stem cell competence in Drosophila (Cui and Doe, 1992; Mellerick et al., 1992). In this report, we show that vertebrate Cst is expressed in the myocardial layer of the heart in a dorsal-to-ventral gradient and demonstrate that CST is required within a subset of cardiac progenitor cells for the initiation of cardiomyocyte differentiation at the ventral midline. Fate mapping of cardiac tissue indicates that cardiac progenitors at the ventral midline have a specific cell fate, giving rise to a population of cells in the outer curvature of the ventricle. In contrast, ventral midline cells depleted of CST overproliferate and fail to integrate into cardiac muscle. Collectively, these studies demonstrate that CST is required for the proper timing of differentiation within a subset of cardiac progenitors that are fated to give rise to the outer curvature of the ventricle, and suggest that regulation of cardiomyocyte differentiation is regionalized along a dorsal-ventral axis prior to heart tube formation.
RESULTS
Cst Is Expressed in the Myocardium Prior to the Onset of Cardiomyocyte Differentiation
To begin to identify the molecular pathways that are involved in the decisions of cardiac progenitor cells to proliferate or differentiate, we have focused on the vertebrate orthologs of Castor (Cst), a known regulator of stem cell competence in Drosophila. We have identified two alternatively 5-prime-spliced variants of vertebrate Castor (Cstα and Cstβ) from Xenopus laevis (X. laevis) as well as Xenopus tropicalis (X. tropicalis) (Figure 1A; see Figure S1A in the Supplemental Data available with this article online). Synteny and sequence analysis confirmed that these are the true orthologs of jawed vertebrate and Drosophila Castor (Figures S1B–S1D). In vitro translation of CSTα and CSTβ give proteins of the expected size (Figure S1E) and injection of mRNAs into Xenopus show CSTα and CSTβ both localize to the nucleus in Xenopus tissue (Figures S1F–S1G). RT-PCR analysis with Cstα- and Cstβ-specific primers (Figure S2A) indicate Cstα is expressed at the onset of neurulation (Stage 13), whereas Cstβ is expressed slightly later (Stage 15), with both transcripts continuing to be expressed throughout embryonic development. Using a probe common to Cstα and Cstβ shows a nearly identical spatial pattern of expression in X. laevis and X. tropicalis (Figure 1B; Figures S2B–S2K and S3), with Cst expression first observed at the dorsal midline in the developing hindbrain region of the embryo (Figure S2B) (Stage 13) and by early tailbud stage in the developing somites (Figure 1B). By early tailbud stage (Stage 27) we also observe expression of Cst in the heart primordium (Figure 1B) during the period when the bilateral heart fields begin to fuse across the ventral midline, and at Stage 29 we find Cst coexpressed with the cardiac maker Nkx2.5 throughout the heart tube (Figures 1B–1C; Figure S4). On more detailed analysis, we observe Cst expression within the myocardial layer of the heart in a dorsal-to-ventral gradient (Figure 1D). We further note that onset and maintenance of Cst expression is not disrupted in TBX5-, TBX20-, or HSP27-depleted embryos (Brown et al., 2005, 2007), suggesting that CST is not a downstream component of these pathways (data not shown).
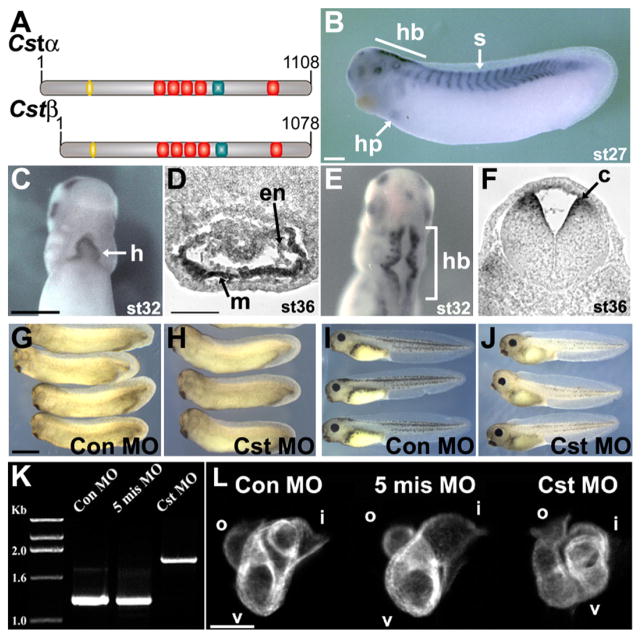
Figure 1. CST Is Required for Vertebrate Heart Development.
(A) Predicted schematic representation of CSTα and CSTβ proteins; nuclear localization signal (yellow), zinc finger repeats (red), serine-rich region (red).
(B–F) Whole-mount in situ analysis of Stage 27 (early tailbud), Stage 32 (tailbud), and Stage 36 (early tadpole) embryos using a Cst-specific probe common to Cstα and Cstβ. ([B], lateral view with anterior to the left; [C and D] ventral and dorsal views, respectively, with anterior to the top). (D and F) Transverse sections of whole-mount in situ Stage 36 embryos through (D) the heart and (F) the hindbrain: hindbrain (hb), somites (s), heart primordium (hp), heart (h), myocardium (m), endocardium (en), commissural neurons (c).
(G–J) Representative (G and I) control MO and (H and J) CstMO embryos. (G) Stage 32 control MO and (H) CstMO embryos are indistinguishable. (I) Stage 41 control MO and (J) CstMO embryos. CST-depleted embryos present with dorsal fin edema and no gross ventral region abnormalities.
(K) RT-PCR analysis of Stage 42 tadpoles injected at the one-cell stage with the CstMO demonstrating inhibition of proper slicing of Cst pre-mRNA. Control MO (Con MO) and 5-mismatch MO (5-mis MO) are negative controls.
(L) Whole-mount MHC antibody staining of tadpole Stage 37 CST-depleted embryos (lateral views with anterior to the left); inflow tract (i), ventricle (v), outflow tract (o). Scale bars: (B–C) = 0.5 mm, (G) = 1 mm, (D and L) = 100 μm.
CST Is Required for Heart Development
To determine the requirement for CST in development, both CSTα and CSTβ were depleted using morpholinos that block splicing of a common conserved region of the transcripts located between exon 8 (ex8D MO) and exon 9 (ex9A MO) (Figure S5A). These splice junction morpholinos, referred to collectively as CstMO, abrogate proper splicing of both pre-mRNAs and introduce a stop codon after the second amino acid following exon 8. CstMO, but not a five-nucleotide mismatched CstMO, properly targets Cst until at least tadpole stage (Stage 42), as shown by RT-PCR with the CstMO giving an amplified product that is larger than that in control-MO-injected embryos by the size of the corresponding intron (679 bp) (Figure 1K). Cloning and sequencing of respective PCR bands confirmed that they were the expected products derived from the Cst locus. Together, these data demonstrate that CstMO depletes the embryo of CSTα and CSTβ until at least Stage 42.
Examination of CST-depleted embryos indicate that they are indistinguishable from control MO-injected embryos until Stage 41 (Figures 1G–1J) when fluid begins to accumulate over the dorsal fin and they have small, compact hearts (Figure 1J). Shortly thereafter (Stage 42), CST-depleted embryos develop ventral edema (Figure S6) and die when sibling embryos develop to Stage 46. Despite the normal appearance of earlier CST-depleted tadpole-stage embryos (Stage 37) (Figures 1G–1H), upon closer examination, CST-depleted hearts had improperly looped, giving the overall appearance of a twisted, kinked tube (Figure 1L) when examined by MHC staining. Cst-specific morpholinos against the 5′ regions of Cstα and Cstβ (Figure S5B) gave identical results, confirming that the phenotype is due to depletion of CSTα and CSTβ. This strongly implies that any protein associated with the Cst splice junction morpholinos is nonfunctional rather than acting as a dominant-negative protein. These data demonstrate that CST is required for early cardiac development in Xenopus.
CST Is Required for the Onset of Cardiomyocyte Differentiation at the Ventral Midline
Since Cst is first expressed in heart primordium at Stage 27, the abnormal heart morphology at tadpole Stage 37 is likely a consequence of an earlier requirement for CST in cardiac development. During the process of early cardiogenesis, specified cardiac progenitor cells migrate toward the ventral midline, where they meet (Stage 26) and fuse at the ventral midline (Stage 29). Cardiomyocytes then undergo epithelialization, initiate the expression of cardiac structural genes, and begin myofibrillogenesis. To determine when CST is required in heart development, in situ analysis was performed on control and CST-depleted embryos with a panel of early cardiac markers that include Nkx2.5, Tbx5, Tbx20, Gata4, Gata5, and Gata6. No alteration in temporal or spatial expression of any of these markers was observed between control and CST-depleted embryos up to Stage 29 (Figures 2A–2L; Figure S7), suggesting that CST is not required for the determination, migration, or, most critically, fusion of cardiac precursor cells.
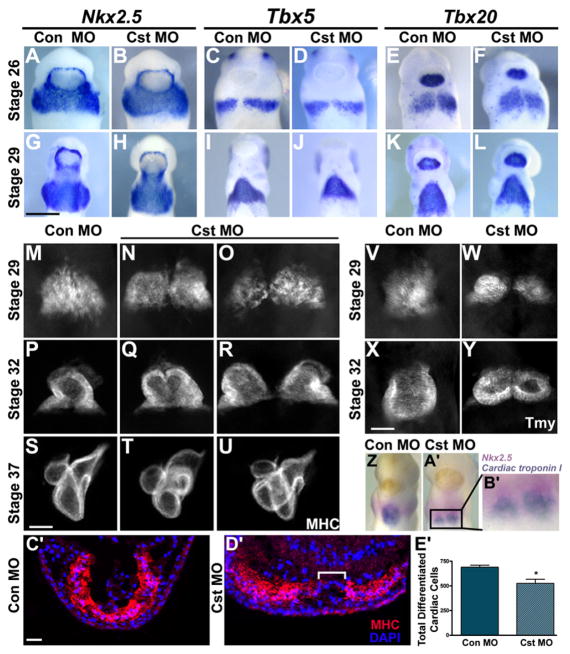
Figure 2. CST Is Required for Cardiomyocyte Differentiation at the Ventral Midline.
(A–L) Whole-mount in situ analysis with early cardiac markers Nkx2.5 (A, B, G, and H), Tbx5 (C, D, I, and J), and Tbx20 (E, F, K, and L) of tailbud Stage 26 and 29 control and CST-depleted embryos (ventral view with anterior to the top). Cardiac progenitors have properly migrated and completely fused across the ventral midline.
(M–U) Whole-mount MHC antibody staining at Stage 29 (onset of cardiac differentiation), Stage 32 (completion of linear heart tube formation), and Stage 37 (chamber formation) (ventral view with anterior to the top). (M) Stage 29 control MO embryos and (N and O) CST-depleted embryos. (P) Stage 32 control MO embryos and (Q and R) CST-depleted embryos display varying degrees of cardia bifida of the linear heart tube upon CST depletion. (S) Stage 37 control MO embryos and (T and U) CST-depleted embryos display morphological consequences of CST depletion on chamber formation.
(V–Y) Whole-mount Tmy antibody staining of Stage 29 and 32 (V and X) control MO and (W and Y) CST-depleted embryos demonstrates that lack of differentiation is not specific to MHC. (Z–B′) Simultaneous detection of cardiac progenitor cells and differentiated cardiac cells in a Stage 29 CST-depleted embryo.
(Z and A′) Whole-mount double in situ analysis using a Nkx2.5-specific probe (pink) to mark cardiac progenitor cells and Cardiac troponin I-specific probe (blue) to mark differentiated cardiac cells in (Z) control MO and (A′) CST-depleted embryos.
(B′) Magnified image of the cardiac region in the CST-depleted embryo in (A′).
(C′ and D′) Transverse sections of Stage 29 (C′) control MO and (D′) CST-depleted embryos stained with MHC antibody and DAPI. Brackets highlight the lack of differentiation at the ventral midline.
(E′) Quantification of differentiated cardiomyocytes determined by counting the total MHC-positive cells derived from serial sectioned embryos. Bars represent the average of at least six embryos per condition ± SEM; *p < 0.01. Representative images are derived from a single experiment, and all experiments were repeated at least twice with independent batches of embryos. Scale bars: (G) = 0.5 mm, (S and X) = 100 μm, (C′) = 200 μm.
To determine if CST is required for cardiomyocyte differentiation, whole-mount antibody staining was conducted with antibodies against MHC (Figures 2M–2U) and Tropomyosin (Tmy; Figures 2V–2Y). Although CST-depleted embryos have a single normal-sized continuous cardiac field that uniformly expresses Nkx2.5, Tbx5, Tbx20, Gata4, Gata5, and Gata6 (Figures 2A–2L; Figure S7), we found that CST-depleted embryos fail to initiate cardiac differentiation at the ventral midline (Figures 2M–2O and 2V–2W). We confirmed that the cells at the ventral midline are Nkx2.5 positive by double whole-mount in situ analysis with Nkx2.5- and cardiac troponin I-specific probes (Figures 2Z–2B′). Additionally, we find CST-depleted hearts have architectural defects including distention along the left-right axis in the ventral region of the embryo (Figures 2N–2O and D′). These results collectively suggest that CST is required within a 6 hr period of heart development between Stage 27 and Stage 29 for proper differentiation of a subset of cardiomyocyte progenitors at the ventral midline.
To verify that alteration in heart morphology in CST-depleted embryos is associated with a failure of ventral cardiomyocytes to undergo differentiation, we determined the total number of differentiated cardiomyocytes in control and CST-depleted heart tissue staining with DAPI, to mark cell nuclei, and anti-MHC, to mark differentiated cardiomyocytes (Figures 2C′–2E′). Consistent with a failure of the ventral midline cells to express MHC, there is a significant reduction in the number of terminally differentiated cardiomyocytes in CST-depleted hearts relative to control hearts, 525.90 ± 42.16 versus 690.5 ± 19.76, respectively (Figure 2E′). We further note that this decrease is not a reflection of a role for CST in cardiac cell survival since we observed no increase in the total number of cardiac cells that were positive for TUNEL or the apoptotic marker Capase-3 during these stages (data not shown). Taken together, these data demonstrate a role for CST in cardiac differentiation at the ventral midline and suggest that regulation of cardiomyocyte differentiation is regionalized along the dorsal-ventral axis.
Failure of the cardiac cells in CST-depleted embryos to uniformly differentiate across the ventral midline leads to a bifurcation of the developing linear heart tube by slightly later stages (Stage 32) (Figures 2Q–2R and 2Y). Similar to genetic mutants in zebrafish which lead to cardia bifida, e.g., natter, miles apart, mtx, and bon (Chen et al., 1996; Sakaguchi et al., 2006; Stainier et al., 1996), severity of the bifida ranges from extreme phenotypes (30% penetrance) (Figures 2R and 2Y), in which CST depletion results in two separate linear heart tubes, to moderate phenotypes (60% penetrance) (Figure 2Q) that manifest as two heart fields joined along the ventral mid-line either posteriorly or along the length of the heart field to form an irregular heart tube. Variation in linear heart tube formation ultimately disrupts the morphogenic movements of the heart tube, resulting in a secondary cardiac looping defect seen in the chambered heart (Stage 37) (Figures 2T–2U). An identical cardiac phenotype is observed when a combination of morpholinos targeting the 5′ UTR of Cstα and Cstβ is injected (Figure S5C). Taken together, these results imply that, in contrast to other cardia bifida phenotypes, CST depletion causes a bifurcation of the heart due to lack of uniform differentiation of cardiomyocytes across the midline.
CST Depletion Does Not Affect Endoderm Formation or Patterning, Components of the Extracellular Matrix, or Cardiac Polarity
Signals from the endoderm are required for proper cardiac midline development (Chen et al., 1996; Stainier et al., 1996); however, we were unable to detect Cst expression within the endoderm or endodermal derivatives over the periods in which we observed the phenotype in CST-depleted embryos (Figure 1B; Figure S2). To further exclude the possibility that the inability of cardiomyocytes to differentiate at the ventral midline in CST-depleted embryos is a secondary consequence of alterations in the formation, maintenance, or patterning of endodermal tissue, endodermal markers Endodermin, Sox2, Endocut, and Vito were assayed for spatial expression by in situ analysis in both whole-mount and sectioned embryos, and for quantitative expression by SYBR Green quantitative PCR (Figure 3). No alterations in spatial or quantitative expression of any endodermal markers were observed in CST-depleted embryos, further suggesting that CST is not required for the proper formation, maintenance, or patterning of endodermal tissue.
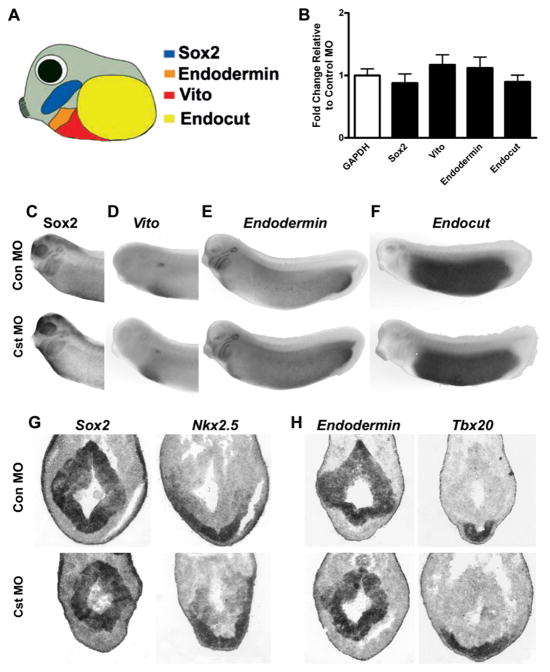
Figure 3. CST Is Not Required for Formation or Patterning of Endodermal Tissue.
(A) Schematic representation of endodermal tissue markers that demarcate pharyngeal endoderm (Sox2 and Endodermin), ventral midgut (Vito and Endodermin), and posterior endoderm (Endocut).
(B) Relative expression levels of endodermal markers Sox2, Vito, Endodermin, and Endocut in Stage 29 CST-depleted embryos (n = 5) relative to control MO embryos (n = 5) using GAPDH as the housekeeping gene. Bars represent the relative expression levels ± SEM.
(C–F) Whole-mount in situ analysis of endodermal markers (C) Sox2, (D) Vito, (E) Endodermin, and (F) Endocut in Stage 29 (top) control MO and (bottom) CST-depleted embryos (lateral views with anterior to the left).
(G–H) In situ analysis of endodermal and cardiac markers on adjacent transverse sections through the cardiac region of (top) control MO and (bottom) CST-depleted Stage 29 embryos. (G) Sox2 and Nkx2.5 expression on adjacent sections demonstrating proper expression within pharyngeal tissue of CST-depleted embryos. (H) Endodermin and Tbx20 expression on adjacent sections demonstrating proper relative spatial expression within the cardiac tissue and endoderm of the embryo.
To determine if CST functions in a tissue-autonomous fashion, we assayed for alteration in components of the extracellular matrix (Fibronectin and Fibrillin), cardiomyocyte polarity (β-Catenin), and endocardium/BMP signaling (SMAD1/5/8). No alteration in the staining pattern of any of these markers was observed between control MO and CstMO-derived hearts (data not shown). Collectively, these data demonstrate that the midline defect as a consequence of CST depletion is due to a block or delay in cardiomyocyte differentiation and is not a secondary consequence of alterations in cardiac migration or polarity. Moreover, these results suggest that the phenotype of CST depletion is not associated with inappropriate activation of the canonical Wnt pathway or inactivation of the BMP2/4/7 pathway.
Fate Mapping of Cardiac Ventral Midline Cells
Collectively, our results imply cardiomyocytes at the ventral mid-line have a fate different from that of neighboring cardiomyocyte progenitors. To determine the ultimate fate of these cardiomyocytes at the ventral midline, we conducted fate mapping studies at the time when we first detect a phenotype in CST-depleted embryos; i.e., when cardiac progenitors have fused at the anterior ventral midline (Stage 29). Using anatomical landmarks and a cardiac actin-GFP (CA-GFP) transgenic reporter host strain of Xenopus to unambiguously identify cardiac tissue, we injected MitoTracker dye into the ventral midline of control MO-injected CA-GFP-positive embryos (Figures 4A and 4B), identified by GFP expression in the developing somites. We then scored embryos that incorporated the dye into the underlying mesoderm. Broadly consistent with studies in chick (Abu-Issa and Kirby, 2007; De La Cruz et al., 1989; Moreno-Rodriguez et al., 2006), we find that by the stage at which the heart initiates chamber morphogenesis, cardiomyocytes derived from the ventral mid-line give rise to cells that occupy positions in the medial outer curvature of the heart (Stage 35) (Figures 4C and 4E–4I). At later stages, during the period after completion of chamber and valve formation (Stage 45), cells preferentially colonize the outer curvature of the mature ventricle and, to a lesser extent, the atrium (Figures 4D and 4V–4Z).
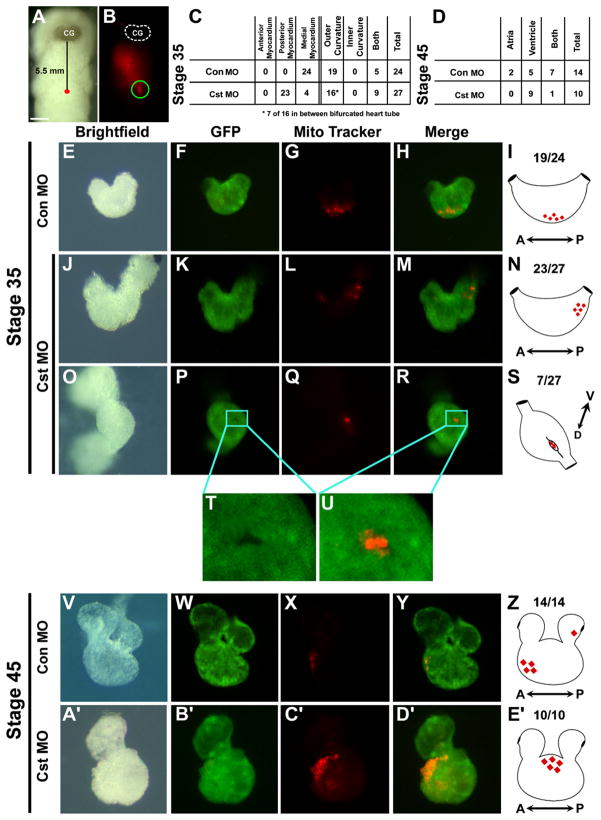
Figure 4. Fate Mapping Cardiac Ventral Midline Cells.
(A) Bright-field image of a living cardiac actin-GFP transgenic embryo injected with MitoTracker at Stage 29 along the ventral midline 5.5 mm posterior to the cement gland (CG). (B) Fluorescent image of the same embryo demonstrating the location of incorporated MitoTracker into cells at the ventral midline (ventral views with anterior to the top). Fluorescence anterior to site of injection is reflection off the surface of the live embryo. (C and D) Tabulation of the location of MitoTracker-labeled cardiac cells of control MO and CST-depleted embryo at (C) midtailbud Stage 35 and (D) tadpole Stage 45. Images of CA-GFP transgenic control MO-injected and CST-depleted (E–H, J–M, and O–U) Stage 35 and (V–Y and A′–D′) Stage 45 dissected hearts. (F, K, P, T, U, and Z) Corresponding images of GFP expression. (G, L, Q, X, and C′) Corresponding images of fated MitoTracker-labeled cardiac ventral midline cells. (H, M, R, U, Y, and D′) Merged images of GFP and fated MitoTracker-labeled ventral midline cells. (T and U) Note the fated ventral midline cells in a pocket of undifferentiated (GFP-negative) cardiomyocytes. (I, N, S, Z, and E′) Schematics representing fate of the cardiac ventral midline cells to the outer curvature of the ventricle in (I) Stage 35 and (Z) Stage 45 control MO-injected hearts. CST-depleted fated ventral midline cells located in the (N) posterior midline or (S) in an undifferentiated cleft in the outer ventricular myocardium in Stage 35 CST-depleted hearts and (E′) in a condensed mass of cells on the outer ventricle in Stage 45 CST-depleted hearts.
CST-Depleted Cardiac Ventral Midline Cells Fail to Integrate into the Mature Heart
To further establish the role of CST in cardiac midline development, and to confirm that cells that occupy a position in cardiac ventral midline in CST-depleted embryos are cardiomyocyte progenitors, we fate mapped ventral midline cells in CA-GFP embryos in which we depleted CST. In stark contrast to results from wild-type embryos, ventral midline cells in CST-depleted embryos preferentially give rise to cells that occupy positions in the posterior midline of the developing heart (Figures 4D and 4J–4N). Interestingly, we also found a portion of fated CST-depleted ventral midline cells located in a posterior undifferentiated ventricular cleft (Figures 4O–4U). Although we often observe that CST-depleted ventral midline cells are delayed but not blocked in cardiomyocyte differentiation, in all case cells fail to integrate into the heart and remain as a condensed population of cells attached to the outer ventricular wall (Figures 4D and 4V–4Z).
CST Is Required for the Regulation of Cardiomyocyte Proliferation
We observe from our fate mapping studies at later stages that CST-depleted, labeled ventral midline cells consistently give rise to a much larger population of cells relative to controls (compare Figure 4Y with 4D′). Consistent with this observation, we find cardiomyocytes undergoing differentiation in the more dorsal regions of the CST-depleted hearts have a significantly increased mitotic index (Figure S8). Collectively, these data imply that CST plays a critical role in regulating the proliferation and onset of differentiation of cardiac progenitors.
DISCUSSION
Our findings collectively imply that cardiomyocytes do not differentiate in a uniform manner and strongly suggest that cells in the ventral region of the heart field differentiate in response to localized signals. Our results further imply that cardiomyocytes differentiate along a dorsal-ventral axis before the onset of terminal cardiomyocyte differentiation; thus, cardiac progenitor fates appear to be determined prior to the formation of the linear heart tube.
We observe that the requirement for CST in cardiomyocyte differentiation is within a subset of cardiac progenitor cells in which Cst is expressed. What then limits CST function to the ventral midline? One possibility is that CST functions redundantly with other transcription factors in regulating cardiomyocyte differentiation in more dorsal regions of the heart; however, CST binds DNA through a unique para-zinc finger domain (Mellerick et al., 1992). Similar to studies in Drosophila, we are unable to identify any protein in the genomes of Xenopus, chick, or mammals that contains a similar domain, suggesting that CST acts on an exclusive set of transcriptional targets. Based on these observations, our phenotypic analysis of cardiac tissue depleted of CST, and by inference with Drosophila (Kambadur et al., 1998), we hypothesize that Cst activity is regulated by as yet unidentified factors. Consistent with this proposal, we have shown that overexpression of CST throughout the embryo leads to cellular defects only in tissue types that normally express CST (data not shown).
We note that our fate mapping studies of early Xenopus hearts are consistent with those performed in chick showing that cardiac progenitors at the ventral midline preferentially give rise to terminally differentiated cardiomyocytes in the outer ventricular wall (De La Cruz et al., 1989; Moreno-Rodriguez et al., 2006). Studies in chick further demonstrated that cardiomyocyte progenitors at the ventral midline are derived from the cephalic mesoderm; i.e., the anterior heart field. Therefore, it is intriguing to speculate that the regulation of CST activity restricts its role to cardiac cells derived from the anterior heart field.
In light of the role for CST in cardiac differentiation and its high degree of sequence conservation and expression between vertebrates (Liu et al., 2006; Vacalla and Theil, 2002), it will be interesting in the future to examine the relationship between CST and congenital heart disease and cardiac hypertrophy.
EXPERIMENTAL PROCEDURES
Embryo Collection, Cst Cloning, and Morpholinos
Preparation and collection of X. tropicalis and X. laevis were performed as described (Showell et al., 2006) and staged according to Nieuwkoop and Faber (1967). Cst-specific morpholinos were obtained from Gene Tools, LLC. Sequences of all morpholinos are available upon request. To demonstrate morpholino efficacy, RNA was extracted from homogenized Stage 42 tadpoles in lysis buffer (Goetz et al., 2006) followed by RT-PCR as described in Supplemental Experimental Procedures.
Developmental Whole-Mount In Situ Hybridization
Whole-mount in situ analysis and double detection whole-mount in situ analysis was carried out as described (Harland, 1991) using α-sense RNA probes of Nkx2.5 (Tonissen et al., 1994), Tbx5 (Horb and Thomsen, 1999), Tbx20 (Brown et al., 2003), cardiac troponin I (Logan and Mohun, 1993), Sox2 (Kishi et al., 2000), Endodermin (Langdon et al., 2007), Endocut (Costa et al., 2003), and Vito (Xenopus EST clone XL043k17 [Costa et al., 2003]). For double in situs the protocol was modified so embryos were simultaneously analyzed with fluorosceine-labeled Nkx2.5 probe and either Cst or cardiac troponin I DIG-labeled probe. Following incubation with α-fluorosceine alkaline phosphatase antibody, Nkx2.5 expression was detected using 175 μg/ml of magenta phosphate and 225 μg/ml tetrazolium red in AP buffer at 4°C for 3–5 days. Embryos were washed three times in PBS incubated in 0.1 M glycine-HCl (pH 2.2), 0.1% Tween-20 for 10 min at RT. DIG-labeled probes were detected as described. Embryos were bleached then photographed on a Leica MZF III fluorescent dissecting microscope. Stained embryos were prepared for 20 μm frozen sectioning in OTC freezing media. Sections were rinsed with PBS, coverslipped, and imaged on a Zeiss Axiovert 35 microscope.
Immunohistochemistry
Immunohistochemistry was carried out with mouse anti-Myosin Heavy Chain, mouse anti-Tropomyosin (Developmental Studies Hybridoma Bank), and rabbit anti-phospho-histone H3 (Cell Signaling) using Cy3 anti-rabbit, Cy3 anti-mouse, and Alexa 488 anti-mouse as described (Goetz et al., 2006). Embryos were cleared in 2:1 benzyl benzoate:benzyl alcohol and viewed on a Leica MZFIII fluorescence dissecting microscope. Sections were imaged on a Nikon Eclipse E800 fluorescent microscope.
SYBR Green Quantitative PCR
RNA was isolated from Stage 29 embryos (ten embryos per condition) using Trizol (Invitrogen) and purified using an RNeasy column (QIAGEN) according to the manufacture’s protocol. cDNA synthesis with 1 μg of RNA was performed with random primers using SuperScript II reverse transcriptase (Invitrogen) according to the manufacture’s protocol. Expression levels were assessed by quantitative PCR using SYBR Green Master Mix (Sigma) on a Rotogene 3000. Primers (19–21 bp) were designed for each mRNA to amplify a 100–121 bp product. Prior to quantitative PCR analysis, primer sets were tested by standard PCR and analyzed by 2.5% agarose gel electrophoresis to determine amplification of a single PCR product. GAPDH was used as the housekeeping gene. Reactions with 5 μl of a 1:60 dilution of the cDNA were heated to 94° for 2 min, followed by 50 cycles of 94°C, 15 s; 55°C, 15 s; 72°C, 20 s. Following amplification, melting curves were generated to verify the presence of a single amplification product. Each sample was analyzed in triplicate with a corresponding minus-RT control. Five samples (n = 5) per condition were analyzed. Analysis generated Ct values based on thresholds determined by Rotogene 3000 software. Data were analyzed by the Pfaffl method (Pfaffl et al., 2002) and represented as relative fold change ± SEM.
Fate Mapping
Fertilized embryos were injected at the one-cell stage with either Cst MOs or control MOs as described (Goetz et al., 2006). Stage 29 GFP-positive embryos were embedded in 3% methyl-cellulose and injected with 0.3 nl of 1 mM Mito Tracker Red CMXRos (Molecular Probes) at the ventral midline 5.5 mm from the posterior boundary of the cement gland. Two hours postinjection, embryos were analyzed and scored for incorporation into GFP-positive cardiac tissue. Stage 35 and 45 hearts were dissected in 1 × Modified Barths Solution (MBS) and placed in 1 × MBS supplemented with 0.1 M KCl to arrest contraction. Stage 35 labeled clones were scored based on anterior-posterior axis and inner-outer curvature of the heart, and Stage 45 based on location of labeled clones in the atrium or ventricle. Transgenic animals expressing cardiac actin-GFP were a generous gift from Dr. Tim Mohun (Latinkic et al., 2002).
Statistical Methods
Morpholino injections were performed on >15 batches of embryos. Similar results were seen in each experiment. Statistical differences of cell counts between injected embryos were determined using one-tailed, unpaired t tests. The number of embryos used in each experiment is noted in the results.
Supplementary Material
Acknowledgments
This work is supported by grants to F.L.C. from the NIH/NHLBI (R01 HL075256 and R21 HL083965). K.S.C. is a trainee in the Integrative Vascular Biology program supported by T32HL69768 from the NIH. We are extremely grateful to Sukwon Jin, Dazhi Wang, Larysa Pevny, Cam Patterson, Mark Peifer, and Mark Majesky for their helpful discussions and critical reading of the manuscript.
Footnotes
Supplemental Data include Supplemental Experimental Procedures and eight figures containing CASTOR gene and protein characterization, Castor developmental in situ analysis, morpholino design, additional cardiac and endoderm marker expression, and cardiac mitotic index assay. They can be found with this article online at http://www.developmentalcell.com/cgi/content/full/14/4/616/DC1/.
References
- Abu-Issa R, Kirby ML. Heart field: from mesoderm to heart tube. Annu Rev Cell Dev Biol. 2007;23:45–68. doi: 10.1146/annurev.cellbio.23.090506.123331. [DOI] [PubMed] [Google Scholar]
- Brown DD, Binder O, Pagratis M, Parr BA, Conlon FL. Developmental expression of the Xenopus laevis Tbx20 orthologue. Dev Genes Evol. 2003;212:604–607. doi: 10.1007/s00427-002-0276-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DD, Martz SN, Binder O, Goetz SC, Price BMJ, Smith JC, Conlon FL. Tbx5 and Tbx20 act synergistically to control vertebrate heart morphogenesis. Development. 2005;132:553–563. doi: 10.1242/dev.01596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DD, Christine KS, Showell C, Conlon FL. Small heat shock protein Hsp27 is required for proper heart tube formation. Genesis. 2007;45:667–678. doi: 10.1002/dvg.20340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JN, Haffter P, Odenthal J, Vogelsang E, Brand M, van Eeden FJ, Furutani-Seiki M, Granato M, Hammerschmidt M, Heisenberg CP, et al. Mutations affecting the cardiovascular system and other internal organs in zebrafish. Development. 1996;123:293–302. doi: 10.1242/dev.123.1.293. [DOI] [PubMed] [Google Scholar]
- Costa RM, Mason J, Lee M, Amaya E, Zorn AM. Novel gene expression domains reveal early patterning of the Xenopus endoderm. Gene Expr Patterns. 2003;3:509–519. doi: 10.1016/s1567-133x(03)00086-3. [DOI] [PubMed] [Google Scholar]
- Cui X, Doe CQ. ming is expressed in neuroblast sublineages and regulates gene expression in the Drosophila central nervous system. Development. 1992;116:943–952. doi: 10.1242/dev.116.4.943. [DOI] [PubMed] [Google Scholar]
- De La Cruz MV, Sanchez-Gomez C, Palomino MA. The primitive cardiac regions in the straight tube heart (Stage 9) and their anatomical expression in the mature heart: an experimental study in the chick embryo. J Anat. 1989;165:121–131. [PMC free article] [PubMed] [Google Scholar]
- Goetz SC, Brown DD, Conlon FL. TBX5 is required for embryonic cardiac cell cycle progression. Development. 2006;133:2575–2584. doi: 10.1242/dev.02420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan K, Hasenfuss G. Do stem cells in the heart truly differentiate into cardiomyocytes? J Mol Cell Cardiol. 2007;43:377–387. doi: 10.1016/j.yjmcc.2007.07.056. [DOI] [PubMed] [Google Scholar]
- Harland RM. In situ hybridization: an improved whole mount method for Xenopus embryos. Methods Cell Biol. 1991;36:675–685. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Horb ME, Thomsen GH. Tbx5 is essential for heart development. Development. 1999;126:1739–1751. doi: 10.1242/dev.126.8.1739. [DOI] [PubMed] [Google Scholar]
- Kambadur R, Koizumi K, Stivers C, Nagle J, Poole SJ, Odenwald WF. Regulation of POU genes by castor and hunchback establishes layered compartments in the Drosophila CNS. Genes Dev. 1998;12:246–260. doi: 10.1101/gad.12.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi M, Mizuseki K, Sasai N, Yamazaki H, Shiota K, Nakanishi S, Sasai Y. Requirement of Sox2-mediated signaling for differentiation of early Xenopus neuroectoderm. Development. 2000;127:791–800. doi: 10.1242/dev.127.4.791. [DOI] [PubMed] [Google Scholar]
- Langdon YG, Goetz SC, Berg AE, Swanik JT, Conlon FL. SHP-2 is required for the maintenance of cardiac progenitors. Development. 2007;134:4119–4130. doi: 10.1242/dev.009290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latinkic BV, Cooper B, Towers N, Sparrow D, Kotecha S, Mohun TJ. Distinct enhancers regulate skeletal and cardiac muscle-specific expression programs of the cardiac α-actin gene in Xenopus embryos. Dev Biol. 2002;245:57–70. doi: 10.1006/dbio.2002.0639. [DOI] [PubMed] [Google Scholar]
- Liu L, Zhang X, Qian B, Min X, Gao X, Li C, Cheng Y, Huang J. Over-expression of heat shock protein 27 attenuates doxorubicin-induced cardiac dysfunction in mice. Eur J Heart Fail. 2007;9:762–769. doi: 10.1016/j.ejheart.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Liu Z, Yang X, Tan F, Cullion K, Thiele CJ. Molecular cloning and characterization of human Castor, a novel human gene upregulated during cell differentiation. Biochem Biophys Res Commun. 2006;344:834–844. doi: 10.1016/j.bbrc.2006.03.207. [DOI] [PubMed] [Google Scholar]
- Logan M, Mohun T. Induction of cardiac muscle differentiation in isolated animal pole explants of Xenopus laevis embryos. Development. 1993;118:865–875. doi: 10.1242/dev.118.3.865. [DOI] [PubMed] [Google Scholar]
- Mellerick DM, Kassis JA, Zhang SD, Odenwald WF. castor encodes a novel zinc finger protein required for the development of a subset of CNS neurons in Drosophila. Neuron. 1992;9:789–803. doi: 10.1016/0896-6273(92)90234-5. [DOI] [PubMed] [Google Scholar]
- Moreno-Rodriguez RA, Krug EL, Reyes L, Villavicencio L, Mjaatvedt CH, Markwald RR. Bidirectional fusion of the heart-forming fields in the developing chick embryo. Dev Dyn. 2006;235:191–202. doi: 10.1002/dvdy.20601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal Table of Xenopus laevis (Daudin) Amsterdam: North Holland: 1967. [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi T, Kikuchi Y, Kuroiwa A, Takeda H, Stainier DY. The yolk syncytial layer regulates myocardial migration by influencing extracellular matrix assembly in zebrafish. Development. 2006;133:4063–4072. doi: 10.1242/dev.02581. [DOI] [PubMed] [Google Scholar]
- Showell C, Christine KS, Mandel EM, Conlon FL. Developmental expression patterns of Tb x 1, Tb x 2, Tb x 5, and Tb x 20 in Xenopus tropicalis. Dev Dyn. 2006;235:1623–1630. doi: 10.1002/dvdy.20714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stainier DY, Fouquet B, Chen JN, Warren KS, Weinstein BM, Meiler SE, Mohideen MA, Neuhauss SC, Solnica-Krezel L, Schier AF, et al. Mutations affecting the formation and function of the cardiovascular system in the zebrafish embryo. Development. 1996;123:285–292. doi: 10.1242/dev.123.1.285. [DOI] [PubMed] [Google Scholar]
- Tonissen KF, Drysdale TA, Lints TJ, Harvey RP, Krieg PA. XNkx-2.5, a Xenopus gene related to Nkx-2.5 and tinman: evidence for a conserved role in cardiac development. Dev Biol. 1994;162:325–328. doi: 10.1006/dbio.1994.1089. [DOI] [PubMed] [Google Scholar]
- Vacalla CM, Theil T. Cst, a novel mouse gene related to Drosophila Castor, exhibits dynamic expression patterns during neurogenesis and heart development. Mech Dev. 2002;118:265–268. doi: 10.1016/s0925-4773(02)00282-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.