Abstract
Aminoacyl-tRNA synthetases often rely on a proofreading mechanism to clear mischarging errors before they can be incorporated into newly synthesized proteins. Leucyl-tRNA synthetase houses a hydrolytic editing pocket in a domain that is distinct from its aminoacylation domain. Mischarged amino acids are transiently translocated ∼30 Å between active sites for editing by an unknown tRNA-dependent mechanism. A glycine within a flexible β-strand that links the aminoacylation and editing domains of LeuRS was determined to be important to tRNA translocation. The translocation-defective mutation also demonstrated that the editing site screens both correctly and incorrectly charged tRNAs prior to product release.
Keywords: fidelity, protein synthesis, amino acid editing, translocation
1. Introduction
The aminoacyl-tRNA synthetase (AARSs) family is responsible for the first step of translation of the genetic code by covalently linking tRNA with the correct amino acid. However, about half of the AARSs require editing to enhance fidelity because they cannot fully distinguish between structurally related amino acids [1]. Failure of AARS editing, even at modest levels, can result in cell death and mammalian disease [2-4].
Leucyl-tRNA synthetase (LeuRS) is challenged by standard and non-standard amino acids [2,5-7]. Its “double sieve” proofreading process [8] has separate active sites for aminoacylation and editing that are ∼30 Å from each other [9]. The editing active site resides in a discrete domain called CP1. Recently, a short CP1-based peptide in Escherichia coli LeuRS was implicated in the tRNA translocation between the two active sites [10], although this transient mechanism remains poorly defined. It has been proposed that the uncharged end of the tRNA binds near the CP1 domain initially and passes through the editing active site before binding at the canonical core for aminoacylation [11]. Once charged, the tRNA is translocated back to the editing active site for proofreading. In LeuRS, a T252A mutation in the editing active site cleared correctly charged Leu-tRNALeu, suggesting that all charged and mischarged tRNAs are translocated from the aminoacylation to the editing active site prior to product release [12].
The discrete CP1 domains of LeuRS, as well as the homologous isoleucyl- (IleRS) and valyl-tRNA synthetases (ValRS) are tethered to the ancient canonical aminoacylation core via two β-strands (Fig. 1) [13-16]. A dozen crystal structures for LeuRS show that the CP1 domain and the main body of these AARSs re-orient themselves as the enzyme undergoes its multi-step reaction cycle [1,14,16,17]. The β-strand tethers adopt different conformations to accommodate these dynamic movements of the CP1 domain.
Fig. 1.

Tertiary and primary structures of LeuRS. (a) Crystal structure of T. thermophilus LeuRS (PDB code 2BTE) [16]. The enzyme is displayed as a grey ribbon, with the CP1 domain inserted into the canonical core by two β-strands (black). (b, c) Sequence alignment of the N- and C-terminal β-strand linkers. Black and grey shading highlight highly conserved and homologous residues, respectively. The N- and C-terminal β-strands are marked by horizontal arrows and the glycine residues that were mutated are indicated by vertical arrows.
We hypothesized that flexible sites within the dynamic β-strand linkers facilitate movement and positioning of the CP1 domain during editing, aminoacylation as well as the transient tRNA-dependent translocation mechanism. Specific mutations within E. coli LeuRS β-strands can alternatively affect aminoacylation or editing activity [18]. We wondered if the β-strands also influenced the transient tRNA translocation event that moves the amino acid between active sites. We mutationally analyzed conserved glycine residues within each β-strand that might provide flexibility for tRNA translocation. Disruption of one of these glycine sites hindered charged and mischarged tRNA translocation, while retaining aminoacylation and tRNA deacylation activity.
2. Materials and Methods
Plasmid p15ec3-1 (encodes E. coli LeuRS gene) was mutated to G225P, G229P, G407P and G409P using PCR-based mutagenesis [18]. Likewise, a second mutation, T252A, in the editing active site was introduced into each mutant LeuRS gene-containing plasmids. Protein expression was carried out in E. coli strain BL21 (Novagen) and purified by affinity chromatography via an N-terminal fused six-histidine tag [18].
Aminoacylation reactions containing 60 mM Tris, pH 7.5, 10 mM MgCl2, 150 mM KCl, 1 mM dithiothreitol, 22 μM [3H]-leucine (167 μCi/ml), 4 μM in vitro transcribed E. coli tRNALeu [18] and catalytic amounts of enzyme were initiated with 4 mM ATP. Aliquots of 10 μl were quenched and processed as described [18]. The measured charged tRNA exceeded the actual amount of tRNA for some mutant LeuRSs. This could be due to co-precipitation of mutant enzyme that is tightly bound to amino acid or covalently self-labelled with amino acid [19]. Misaminoacylation assays were carried out similarly in reactions containing 22 μM [3H]-isoleucine (93 μCi/ml) and 1 μM enzyme. Error analyses were based on triplicated reactions.
Production of [3H]-Leu-tRNALeu or [3H]-Ile-tRNALeu was prepared as previously described [18]. Approximately 4 μM charged or mischarged tRNALeu were incorporated into deacylation reactions containing 60 mM Tris, pH 7.5, 10 mM MgCl2 that were initiated by 100 nM enzyme. Aliquots of 5 μL were quenched at specific time points and processed [18].
3. Results
The CP1 domain of LeuRS is attached to the canonical core of the enzyme by two flexible β-strands (Fig. 1a). These β-strand linkers would be expected to play a central role in positioning the CP1 domain to accommodate and stabilize different tRNA-bound complexes at various stages of enzyme activity including aminoacylation and editing [18]. We hypothesized that dynamic changes of the protein and protein-tRNA complex could be facilitated by one or more glycine residues that might confer flexibility to these linker regions.
In E. coli LeuRS, Gly229, Gly407, and Gly409 are located within the β-strand linkers and conserved (Fig. 1b, c). In addition, Gly225 is just upstream of the N-terminal β-strand tether and also conserved. We substituted proline at each of these residues to introduce rigidity that might restrict movement at these specific sites within the β-strand linkers. The mutant LeuRS enzymes were stably expressed and purified. We then characterized the proline substitutions to determine the effect of decreased flexibility at each individual site on LeuRS enzyme activity.
The four LeuRS glycine to proline variants exhibited varying tRNALeu aminoacylation efficiencies (Fig. 2a). The G407P variant aminoacylated tRNALeu as well as wild-type LeuRS (not shown). Deacylation of Ile-tRNALeu was also robust for G407P (not shown) and thus, this variant will not be discussed further. The G225P LeuRS mutant in the N-terminal β-strand abolished tRNA leucylation (Fig. 2a), but this was due to a defect in leucine activation (not shown). The G229P and G409P mutant LeuRSs consistently yielded higher aminoacylation activity compared to the wild-type enzyme.
Fig. 2.

Aminoacylation and deacylation activities. (a) Leucylation activities were obtained using 50 nM enzyme and 4 μM tRNALeu. (b) Deacylation reactions contained 100 nM enzyme and approximately 4 μM [3H]-Ile-tRNALeu.
We also tested the β-strand glycine mutations to determine if the LeuRS proline substitutions disrupted editing. The G229P and G409P mutant LeuRSs deacylated Ile-tRNALeu similar to wild type LeuRS (Fig. 2b). The deacylation activity of the G225P LeuRS mutant was decreased. We hypothesized that the β-strand was kinked by the proline mutation into an orientation that prohibited proper binding of substrate to form a competent editing complex.
The LeuRS proline substitutions were tested for misaminoacylation activity. As would be expected, the G225P mutant LeuRS did not generate mischarged product because its amino acid activation activity was abolished (Fig. 3). However, the N-terminal β-strand G229P and the C-terminal β-strand G409P mutations in LeuRS resulted in a weak mischarging activity to yield Ile-tRNALeu (relative to strong mischarging mutants that are defined by a plateau that reaches 20 pmol of product [10]). Because both the G229P and G409P mutant LeuRSs had significant deacylation activity that cleared Ile-tRNALeu (Fig. 2b), we hypothesized that this mischarging activity was due to a disruption of the transient tRNA translocation mechanism that moves the 3′ end of the tRNA from the aminoacylation to the editing active site.
Fig. 3.

Misaminoacylation activity. Reactions contained 1 μM enzyme and 4 μM tRNALeu.
Aminoacylated tRNA is translocated from the aminoacylation to the editing active site and proofread in the latter before it is released from the enzyme (Fig. 4a). Previously, we isolated a T252A mutation in the LeuRS editing active site that conferred hydrolysis of correctly charged Leu-tRNALeu [12]. Thus, aminoacylation activity of this mutant appears to be abolished, because the correctly charged tRNA is cleared in the editing active site prior to product release (Fig. 4b).
Fig. 4.
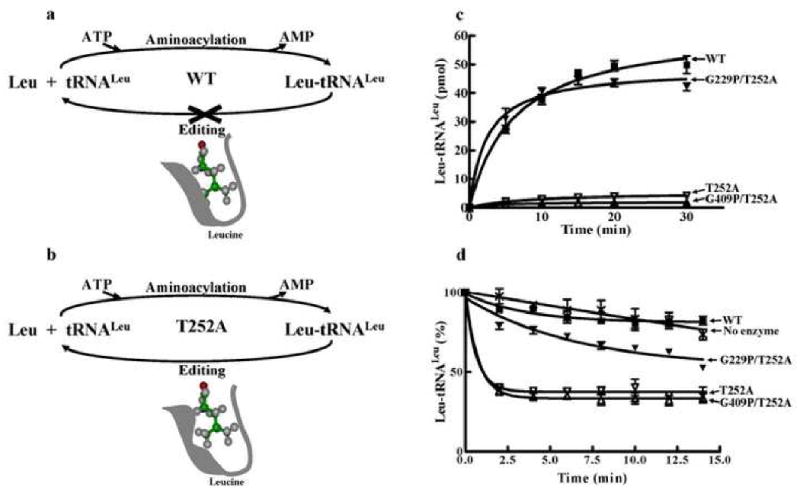
Rescue of leucylation activity by T252A double mutant LeuRSs. (a) Aminoacylation reaction catalyzed by wild type LeuRS showing release of Leu-tRNALeu. (b) Aminoacylation reaction catalyzed by T252A LeuRS showing editing of correctly charged Leu-tRNALeu. [12] (c) Leucylation activities were obtained using 50 nM enzyme and 4 μM tRNALeu. (d) Deacylation activities were obtained using 100 nM enzyme and approximately 4 μM [3H]-Leu-tRNALeu.
We combined the T252A mutation with the glycine substitutions G409P and G229P, which we hypothesized were defective in translocation. The G229P/T252A double mutant LeuRSs rescued leucylation activity to levels that were comparable to wild type (Fig. 4c). This double mutant retained deacylation activity of Leu-tRNALeu that is characteristic of the T252A mutation (data not shown) and supports that it binds effectively to the editing complex for tRNA deacylation (Fig. 4d). Significantly, the tRNA deacylation assay does not require a translocation step from the aminoacylation active site. Thus, we hypothesize that the double mutation restores aminoacylated product formation in the presence of the T252A mutation, because disruption of the transient translocation step is disrupted and the charged tRNA end bypasses the editing active site.
The G409P/T252A double mutant failed to rescue the T252A mutation in LeuRS. It is possible that re-binding of charged and mischarged tRNA products to form a competent hydrolytic complex similar to the wild type in this assay masked translocation-specific effects. We propose that hindering the flexibility of either the N- or C-terminal β-strands at specific sites, in particular the Gly229 position disrupts transient translocation of the charged tRNA from the aminoacylation to the editing active site. These translocation-defective mutations bypass the editing active site resulting in premature product release (Fig. 5).
Fig. 5.

Proposed LeuRS reaction pathway and translocation determinant. The Gly229 residue is located within the N-terminal β-strand linker. The T252A mutation uncouples specificity in the editing active site [12].
4. Discussion
Translocation of mischarged tRNA from the aminoacylation to the editing active site is necessary for the “double sieve” proofreading mechanism that relies on two distinct active sites. The tRNA-dependent translocation event is transient and thus has proven difficult to capture. Recently published crystal structures of LeuRS complexed with tRNALeu provided snapshots of different orientations of the enzyme [11,14,16]. The enzyme cycles through aminoacylation, editing and exit complexes in coordinated movements with the tRNA. An “intermediate” complex where the 3′ end of the tRNA lies nears a conserved motif on the canonical core that neighbors the C-terminus of the C-terminal β-strand has been suggested to represent a state of the tRNA-bound LeuRS complex which is between the aminoacylation and editing complexes (PDB code 1WZ2) [14]. In addition, a peptide within the CP1 domain of E. coli LeuRS has been identified, which facilitates transient tRNA translocation between the aminoacylation and editing active sites, but its mechanism remains obscure [10].
Subsequent to aminoacylation, movement of the tRNA 3′ end to the editing active site is accompanied by CP1 domain rotation and other conformational changes [16,20]. The rapid rate of translocation has likely precluded isolation of its associated conformational states. In E. coli IleRS, mutations at Lys183 and Trp421 identified a “hinge” region near the N- and C-terminus of the CP1 domain insert, which resulted in translocation deficiencies [21]. The Lys183 correlates to a critical Lys186 in E. coli LeuRS [22]. Herein, we have identified a translocation specific site within a β-strand tether that links the CP1 domain to the aminoacylation core. We hypothesize that a conserved glycine residue contributes to a translocation-specific complex that moves the tRNA end from the aminoacylation to the editing site.
Conserved glycines on the β-strands that tether the CP1 domain to the aminoacylation core would be expected to lend flexibility to allow rigid-body rotation and movement of the editing domain. The CP1 domain rotates ∼20° away from the main body to avoid a clash with the 5′ end of tRNA during aminoacylation [14] and swings ∼35° closer to the core for editing to occur [16]. In spite of efficient deacylation activity of mischarged tRNALeu, G229P and G409P mutant LeuRSs produced mischarged tRNA, suggesting that the tRNA translocation pathway had been disrupted. The G229P translocation defect also rescued the T252A phenotype [12] to produce correctly charged tRNA. We hypothesize that the G229P translocation defect interrupts the enzyme reaction cycle to bypass the editing active site and proceeds to premature product release (Fig. 5).
A proline substitution at a critical Gly229 position likely locks just a portion of the β-strand tether into an orientation that impedes movement between the aminoacylation and editing active sites. Significantly though, enough flexibility is retained throughout the two β-strand tethers to allow independent formation of competent aminoacylation and editing complexes to confer their respective activities. We propose then that the conserved Gly229 position is an essential element of the transient molecular mechanism for tRNA translocation between the aminoacylation and editing active sites.
Acknowledgments
We are grateful to Dr. Stephen Cusack for valuable advice and Drs. Michael Vu and Michal Boniecki for technical and insightful suggestions. This work was supported by the NIH (GM063789).
Abbreviations
- LeuRS
leucyl-tRNA synthetase
- CP1
connective polypeptide 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mascarenhas AP, An S, Rosen AE, Martinis SA, Musier-Forsyth K. Fidelity Mechanisms of the Aminoacyl-tRNA Synthetases. In: RajBhandary UL, Köhrer C, editors. Protein Engineering. Springer Verlag; 2007. pp. 153–200. [Google Scholar]
- 2.Karkhanis VA, Mascarenhas AP, Martinis SA. Amino Acid Toxicities of Escherichia coli that are prevented by leucyl-tRNA synthetase amino acid editing. J Bacteriol. 2007;189:8765–8768. doi: 10.1128/JB.01215-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JW, Beebe K, Nangle LA, Jang J, Longo-Guess CM, Cook SA, Davisson MT, Sundberg JP, Schimmel P, Ackerman SL. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–5. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- 4.Nangle LA, Motta CM, Schimmel P. Global effects of mistranslation from an editing defect in mammalian cells. Chem Biol. 2006;13:1091–100. doi: 10.1016/j.chembiol.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 5.Englisch S, Englisch U, von der Haar F, Cramer F. The proofreading of hydroxy analogues of leucine and isoleucine by leucyl-tRNA synthetases from E. coli and yeast. Nucleic Acids Res. 1986;14:7529–39. doi: 10.1093/nar/14.19.7529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karkhanis VA, Boniecki MT, Poruri K, Martinis SA. A viable amino acid editing activity in the leucyl-tRNA synthetase CP1-splicing domain is not required in the yeast mitochondria. J Biol Chem. 2006;281:33217–25. doi: 10.1074/jbc.M607406200. [DOI] [PubMed] [Google Scholar]
- 7.Xu MG, Li J, Du X, Wang ED. Groups on the side chain of T252 in Escherichia coli leucyl-tRNA synthetase are important for discrimination of amino acids and cell viability. Biochem Biophys Res Commun. 2004;318:11–6. doi: 10.1016/j.bbrc.2004.03.180. [DOI] [PubMed] [Google Scholar]
- 8.Fersht AR. Sieves in sequence. Science. 1998;280:541. doi: 10.1126/science.280.5363.541. [DOI] [PubMed] [Google Scholar]
- 9.Lincecum TL, Jr, et al. Structural and mechanistic basis of pre- and posttransfer editing by leucyl-tRNA synthetase. Mol Cell. 2003;11:951–63. doi: 10.1016/s1097-2765(03)00098-4. [DOI] [PubMed] [Google Scholar]
- 10.Hellmann RA, Martinis SA. Defects in transient tRNA translocation bypass tRNA synthetase quality control mechanisms. J Biol Chem. 2009;284:11478–84. doi: 10.1074/jbc.M807395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rock FL, et al. An antifungal agent inhibits an aminoacyl-tRNA synthetase by trapping tRNA in the editing site. Science. 2007;316:1759–61. doi: 10.1126/science.1142189. [DOI] [PubMed] [Google Scholar]
- 12.Mursinna RS, Lincecum TL, Jr, Martinis SA. A conserved threonine within Escherichia coli leucyl-tRNA synthetase prevents hydrolytic editing of leucyl-tRNALeu. Biochemistry. 2001;40:5376–81. doi: 10.1021/bi002915w. [DOI] [PubMed] [Google Scholar]
- 13.Fukai S, Nureki O, Sekine S, Shimada A, Tao J, Vassylyev DG, Yokoyama S. Structural basis for double-sieve discrimination of L-valine from L-isoleucine and L-threonine by the complex of tRNA(Val) and valyl-tRNA synthetase. Cell. 2000;103:793–803. doi: 10.1016/s0092-8674(00)00182-3. [DOI] [PubMed] [Google Scholar]
- 14.Fukunaga R, Yokoyama S. Aminoacylation complex structures of leucyl-tRNA synthetase and tRNALeu reveal two modes of discriminator-base recognition. Nat Struct Mol Biol. 2005;12:915–22. doi: 10.1038/nsmb985. [DOI] [PubMed] [Google Scholar]
- 15.Nureki O, Vassylyev DG, Tateno M, Shimada A, Nakama T, Fukai S, Konno M, Hendrickson TL, Schimmel P, Yokoyama S. Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science. 1998;280:578–82. doi: 10.1126/science.280.5363.578. [DOI] [PubMed] [Google Scholar]
- 16.Tukalo M, Yaremchuk A, Fukunaga R, Yokoyama S, Cusack S. The crystal structure of leucyl-tRNA synthetase complexed with tRNALeu in the post-transfer-editing conformation. Nat Struct Mol Biol. 2005;12:923–30. doi: 10.1038/nsmb986. [DOI] [PubMed] [Google Scholar]
- 17.Cusack S, Yaremchuk A, Tukalo M. The 2 Å crystal structure of leucyl-tRNA synthetase and its complex with a leucyl-adenylate analogue. EMBO J. 2000;19:2351–61. doi: 10.1093/emboj/19.10.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mascarenhas AP, Martinis SA. Functional segregation of a predicted “hinge” site within the β-strand linkers of Escherichia coli leucyl-tRNA synthetase. Biochemistry. 2008;47:4808–16. doi: 10.1021/bi702494q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gillet S, Hountondji C, Smitter JM, Blanquet S. Covalent methionylation of Escherichia coli methiony-tRNA synthetase: Identification of the labeled amino acid residues by matrix-assisted laser desorption-ionization mass spectrometry. Protein Sci. 1997;6:2426–35. doi: 10.1002/pro.5560061116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silvian LF, Wang J, Steitz TA. Insights into editing from an Ile-tRNA synthetase structure with tRNAIle and mupirocin. Science. 1999;285:1074–7. [PubMed] [Google Scholar]
- 21.Bishop AC, Beebe K, Schimmel PR. Interstice mutations that block site-to-site translocation of a misactivated amino acid bound to a class I tRNA synthetase. Proc Natl Acad Sci U S A. 2003;100:490–4. doi: 10.1073/pnas.0237335100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams AM, Martinis SA. Mutational unmasking of a tRNA-dependent pathway for preventing genetic code ambiguity. Proc Natl Acad Sci U S A. 2006;103:3586–91. doi: 10.1073/pnas.0507362103. [DOI] [PMC free article] [PubMed] [Google Scholar]


