Abstract
Machine learning techniques such as classification and regression trees (CART) have been suggested as promising alternatives to logistic regression for the estimation of propensity scores. The authors examined the performance of various CART-based propensity score models using simulated data. Hypothetical studies of varying sample sizes (n=500, 1000, 2000) with a binary exposure, continuous outcome, and ten covariates were simulated under seven scenarios differing by degree of non-linear and non-additive associations between covariates and the exposure. Propensity score weights were estimated using logistic regression (all main effects), CART, pruned CART, and the ensemble methods of bagged CART, random forests, and boosted CART. Performance metrics included covariate balance, standard error, percent absolute bias, and 95% confidence interval coverage. All methods displayed generally acceptable performance under conditions of either non-linearity or non-additivity alone. However, under conditions of both moderate non-additivity and moderate non-linearity, logistic regression had subpar performance, while ensemble methods provided substantially better bias reduction and more consistent 95% CI coverage. The results suggest that ensemble methods, especially boosted CART, may be useful for propensity score weighting.
Keywords: propensity score, weighting, CART, boosting, machine learning, ensemble methods, simulation, data mining
INTRODUCTION
The propensity score is the probability of receiving a treatment conditional on a set of observed covariates [1]. At each value of the propensity score, the distribution of observed covariates is the same across treatment groups. Thus, by conditioning on the propensity score, one can estimate treatment effects free from confounding due to the covariates that determined the propensity score. Conditioning on the propensity score typically is done by matching on the propensity score, subclassification into strata within which propensity scores are similar, regression adjustment on the propensity score, or weighting by the propensity score [2, 3]. Matching and subclassification approaches rely only on selecting subjects with similar propensity score values, relying less on the precise numerical propensity score values. In contrast, regression adjustment and weighting are especially sensitive to misspecification of the propensity score model due to the incorporation of the actual propensity scores or functions in the outcome model [4–6].
The literature has few guidelines for estimating propensity scores for any of these propensity score techniques. Propensity scores are generally estimated using logistic regression. However, parametric models require assumptions regarding variable selection, the functional form and distributions of variables, and specification of interactions. If any of these assumptions are incorrect, covariate balance may not be achieved by conditioning on the propensity score, which may result in a biased effect estimate [7]. In this paper we examine the use of machine learning methods as one alternative to logistic regression.
Machine learning is a general term for a diverse number of classification and prediction algorithms and has applications ranging from detection of credit card fraud to computerized facial recognition [8, 9]. Contrary to statistical approaches to modeling that assume a data model with parameters estimated from the data, machine learning tries to extract the relationship between an outcome and predictor through a learning algorithm without an a priori data model [10]. The suggestion to use such algorithms for propensity score model construction is not new [2, 11–16]. However, these methods have not been widely applied in the propensity score literature, perhaps because of the “black box” nature of some of the algorithms and difficulty in etiologic interpretations of results [17]. Because decision trees are common in medical research for diagnostic and prognostic purposes [18] and are intuitive to visualize and understand, they are a natural starting point for a discussion of machine learning algorithms.
Decision trees partition a dataset into regions such that within each region, observations are as homogeneous as possible [19]. Decision trees are referred to as classification trees if the predicted outcome is a class or regression trees if the outcome is numerical; we refer to these methods collectively as Classification and Regression Trees (CART). Within each node of the tree, observations will have similar probabilities for class membership. CART has advantageous properties for estimating propensity scores, including the ability to handle categorical, ordinal, continuous, and missing data. It is insensitive to outliers and monotonic transformations of variables. Additionally, interactions and non-linearities are modeled naturally as a result of the splits. However, CART can have difficulty in modeling smooth functions and main effects, and is sensitive to overfitting [20].
Several approaches have been proposed to remedy these limitations. To address overfitting, cost-complexity pruning can be implemented where the number of tree splits is reduced or “pruned” with the idea that a simpler tree will be less sensitive to noise and generalize better to new data [19]. While the single tree implementation of CART and pruned CART can perform poorly as a classifier, the predictive capabilities of a weak classifier can be strengthened when working together with other weak classifiers. Ensemble methods, which are somewhat related to iterative and bootstrap procedures, utilize multiple samplings and passes through data (i.e., multiple trees) to enhance the performance of prediction algorithms and reduce overfitting [8]. Because these methods are complex, we provide only a brief description of ensemble methods with application to CART and provide references for the interested reader. Bootstrap aggregated (bagged) CART involves fitting a CART to a bootstrap sample with replacement and of the original sample size, repeated many times. For each observation, the number of times it is classified into a category by the set of trees is counted, with the final assignment of class membership, or probability thereof, based on an average or majority vote over all the trees [21]. Random forests are similar to bagging but utilize a random subsample of predictors in the construction of each CART [22]. Like bagged CART and random forests, boosted CART goes through multiple iterations of tree fitting on random subsets of the data. However, with each iteration, a new tree gives greater priority to the data points that were incorrectly classified with the previous tree [15, 23].
Because in the real world the true treatment effects within any observational dataset are unknown, simulation-based research is needed to evaluate the performance of machine learning propensity score methods. Using simulated data, Setoguchi et al. compared neural networks, CART, pruned CART, and logistic regression in the context of propensity score matching and found that neural networks produced the least biased estimates in many scenarios [16]. However, they do not consider ensemble methods which perform extremely well in classification and prediction tasks while having desirable statistical properties [8]. Furthermore, it is important to determine whether the performance of machine learning methods in propensity score estimation varies based on how those propensity scores are applied. Finally, Setoguchi et al. do not assess covariate balance. In the present analysis, we evaluate the performance of several decision tree-based algorithms, including ensemble methods, in the context of propensity score weighting.
METHODS
Simulation setup
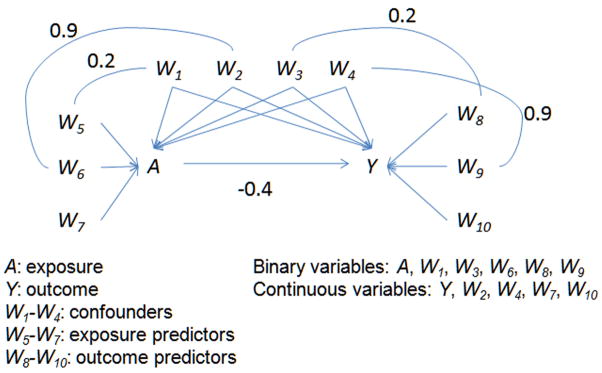
We followed the simulation structure described by Setoguchi and colleagues with slight modifications [16]. For each simulated dataset, ten covariates (four confounders associated with both exposure and outcome, three exposure predictors, and three outcome predictors) Wi were generated as standard normal random variables with zero mean and unit variance. Correlations were induced between several of the variables (Figure 1). The binary exposure A has Pr(A=1|Wi) = 1/(1+exp(−β · f(Wi))). The average exposure probability (in other words, the exposure probability at the average of covariates) was ≈ 0.5 and was modeled from Wi according to the scenarios below, using the formulae provided by Setoguchi et al. The continuous outcome Y was generated from a linear combination of A and Wi such that Y = αiWi + γA where the effect of exposure, γ, = −0.4.
Figure 1.
Variable Relationships and Form in Simulation Data Structure
We evaluated the performance of CART-based methods in seven scenarios that differed in degrees of linearity and additivity in the true propensity score model, specified with quadratic terms and interactions. The scenarios were designed such that the true propensity score model had the following properties [16]:
A: additivity and linearity (main effects only);
B: mild non-linearity (one quadratic term);
C: moderate non-linearity (three quadratic terms);
D: mild non-additivity (three two-way interaction terms);
E: mild non-additivity and non-linearity (three two-way interaction terms and one quadratic term);
F: moderate non-additivity (ten two-way interaction terms);
G: moderate non-additivity and non-linearity (ten two-way interaction terms and three quadratic terms).
To assess the performance of machine learning methods in small, medium, and large-sized datasets, data were simulated for cohort studies of size n=500, n=1000, and n=2000. One thousand datasets of each study size were generated for each of the seven scenarios.
Propensity score estimation methods
We used R version 2.6.1 [24] to estimate propensity scores using the following methods:
Logistic regression: standard logistic regression with a main effect for each covariate.
CART: recursive partitioning using the rpart package with default parameters [25].
Pruned CART: recursive partitioning as described above but with a cost-complexity parameter that controls tree growth. The cost-complexity parameter is automatically chosen to minimize the cross-validated error estimated from a complexity parameter table generated by the plotcp function.
Bagged CART: bootstrap aggregated CART is implemented using the ipred package [26]. We used 100 bootstrap replicates based on empirical evidence suggesting that with more replicates, misclassification rates improve and test errors are more stable [20].
Random forests: random forests are implemented using the randomForest package with the default parameters [27].
Boosted CART: boosted regression trees are implemented using the twang package [28]. We used the parameters recommended by McCaffrey et al., with 20,000 iterations and a shrinkage parameter of 0.0005 [15], with an iteration stopping point that minimizes the mean of the Kolmogorov-Smirnov test statistics.
Estimation of the treatment effect using propensity score weighting
Propensity score weighting is similar to the use of sampling weights in survey data analysis to account for unequal probabilities of inclusion in a study sample. A number of propensity score weighting schemes have been applied in the literature [3, 13, 29, 30]. For example, with inverse probability of treatment weighting, treated persons receive a weight of 1/pi and untreated persons receive a weight of 1/(1−pi), where pi is individual i’s estimated propensity score. In essence, this weights both the treated and untreated groups to look like the combined sample in order to estimate the average treatment effect in the combined sample.
An alternative estimand of interest is the average treatment effect on the treated -- the average treatment effect in a population with a distribution of risk factors similar to that of the treated group. Because this estimand is often of interest in observational studies, we elected to use a weighting scheme with this estimand in mind. We assigned treated persons a weight of 1 while untreated persons are assigned a weight of pi/(1−pi) [12, 13, 15, 29]. Thus, persons in the comparison group who are more similar to those in the treatment group are given greater weight and those more dissimilar are downweighted. If the propensity scores are properly estimated, then the weighted covariate distributions between treatment groups should be similar and the average treatment effect can be estimated as the difference of weighted means. Although it is a good idea in practice to perform “doubly robust” linear regression adjustment for covariates after weighting is applied [4, 31], we did not do so in order to better isolate and compare the performance of the various methods with regards to propensity score weighting.
Performance metrics
We evaluated the performance of the various propensity score fitting methods through several measures.
ASAM: average standardized absolute mean difference, a measure of covariate balance. After weights were applied, the absolute value of the standardized difference of means (standardized by the standard deviation of the particular covariate in the treatment group) between treatment and comparison groups was calculated for each covariate and the average taken across all the covariates. A lower ASAM indicates that the treatment and comparison groups are more similar with respect to the given covariates. We refer to the average value of the 1000 ASAMs in a simulation scenario as the mean ASAM.
Bias: the percentage difference from the true treatment effect of −0.4. Both absolute bias and bias (either positive or negative) are considered.
SE: the standard error of the effect estimate. To calculate the standard error of estimates using the weights we used the survey sampling analysis methods implemented by the survey package [32].
95% CI coverage: the percentage of the 1000 datasets in which the estimated 95% confidence interval included the true treatment effect.
Weights: The performance of weighting methods can be adversely affected if weights are extreme, as a result of estimated propensity scores that are close to 0 or to 1. We therefore examined the distribution of weights for the untreated observations.
RESULTS
Simulations of N=1000
Covariate balance
One rule of thumb for assessing the covariate balance between treatment groups is that an absolute standardized difference in means of 0.2 or greater may be of concern [15, 33]. The average covariate balancing performance of logistic regression propensity score models was acceptable, with low mean ASAMs in all scenarios (range: 0.041, 0.094) (Table 1). However, although the mean ASAMs for logistic regression were relatively and absolutely low, if not the lowest, for each scenario, the ASAMs were skewed with a number of high outliers (Figure 2).
Table 1.
Performance Metrics of Propensity Score Estimation Methods in 1000 Simulated Datasets of N=1000.
| Metric | Method1 | Scenario4 | ||||||
|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | ||
| ASAM2 | LGR | 0.041 | 0.042 | 0.058 | 0.056 | 0.061 | 0.068 | 0.094 |
| CART | 0.159 | 0.148 | 0.143 | 0.171 | 0.162 | 0.15 | 0.143 | |
| PRUNE | 0.175 | 0.164 | 0.148 | 0.182 | 0.173 | 0.161 | 0.151 | |
| BAG | 0.132 | 0.127 | 0.121 | 0.144 | 0.141 | 0.119 | 0.112 | |
| RFRST | 0.08 | 0.076 | 0.076 | 0.089 | 0.086 | 0.077 | 0.075 | |
| BOOST | 0.068 | 0.065 | 0.067 | 0.073 | 0.071 | 0.065 | 0.067 | |
| Absolute bias | LGR | 7.6% | 8.3% | 13.9% | 12.1% | 16.0% | 16.8% | 29.6% |
| CART | 20.5% | 15.2% | 18.2% | 20.1% | 16.9% | 22.5% | 19.0% | |
| PRUNE | 26.3% | 19.5% | 19.1% | 22.9% | 19.1% | 23.9% | 19.9% | |
| BAG | 12.3% | 9.1% | 11.0% | 11.2% | 9.0% | 10.2% | 9.4% | |
| RFRST | 7.4% | 6.2% | 8.9% | 7.6% | 7.3% | 7.5% | 9.0% | |
| BOOST | 8.1% | 6.9% | 6.8% | 7.0% | 6.4% | 6.1% | 6.2% | |
| Standard error | LGR | 0.066 | 0.066 | 0.062 | 0.075 | 0.076 | 0.075 | 0.071 |
| CART | 0.059 | 0.059 | 0.068 | 0.06 | 0.06 | 0.061 | 0.066 | |
| PRUNE | 0.057 | 0.057 | 0.066 | 0.059 | 0.059 | 0.059 | 0.065 | |
| BAG | 0.055 | 0.055 | 0.06 | 0.056 | 0.055 | 0.056 | 0.058 | |
| RFRST | 0.062 | 0.06 | 0.064 | 0.064 | 0.061 | 0.063 | 0.062 | |
| BOOST | 0.059 | 0.058 | 0.059 | 0.061 | 0.059 | 0.06 | 0.059 | |
| 95% CI3 coverage | LGR | 97.9% | 96.8% | 89.7% | 88.5% | 80.0% | 76.0% | 32.5% |
| CART | 63.3% | 78.2% | 69.5% | 63.7% | 74.7% | 58.3% | 67.2% | |
| PRUNE | 49.1% | 66.0% | 68.0% | 56.6% | 68.4% | 54.9% | 65.7% | |
| BAG | 90.9% | 96.8% | 90.4% | 91.8% | 95.7% | 93.6% | 94.7% | |
| RFRST | 98.3% | 99.6% | 95.3% | 98.2% | 98.6% | 98.4% | 95.1% | |
| BOOST | 98.6% | 99.8% | 99.5% | 99.9% | 99.9% | 100.0% | 99.8% | |
LGR – logistic regression, CART – classification and regression tree, PRUNE – pruned CART, BAG – bagged CART, RFRST – random forests, BOOST – boosted CART;
ASAM – average standardized absolute mean distance;
CI – confidence interval
A: additivity and linearity; B: mild non-linearity; C: moderate non-linearity; D: mild non-additivity; E: mild non-additivity and non-linearity; F: moderate non-additivity; G: moderate non-additivity and non-linearity.
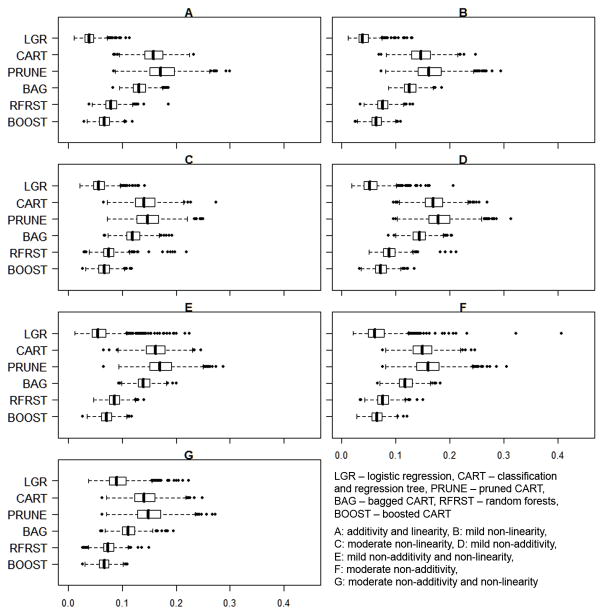
Figure 2.
Distribution of the Average Standardized Absolute Mean Difference by Propensity Score Estimation Method for 1000 Datasets in each of 7 Scenarios (N=1000)
CART and pruned CART propensity score models produced higher mean ASAMs than other methods, with respective ranges of 0.143 to 0.171 and 0.148 to 0.182 across the 7 scenarios. However, CART and pruned CART propensity score models did not provide consistent covariate balance within all datasets, as indicated by the high dispersion of mean ASAMs as well as the large number of high outliers (Figure 2). In contrast, the ensemble methods of bagged CART, random forests, and boosted CARTs produced low mean ASAMs in all scenarios (ranges: bagged CART: 0.112, 0.144; random forests: 0.075, 0.089; boosted CART: 0.065, 0.073) and the ASAMs were much less dispersed than those from the other methods. For example, boosted CART produced no ASAMs > 0.2 in any of the scenarios.
Estimate of effect and standard error
The performance of logistic regression was generally acceptable in the scenario of additivity and linearity (scenario A) with a mean absolute bias of 7.6% and 95% CI coverage of 98.0% (Table 1). However, with increasing non-additivity and non-linearity, logistic regression performed poorly; with moderate non-additivity and nonlinearity (scenario G), logistic regression had a mean absolute bias of 29.6% and 95% CI coverage of 32.5%. Logistic regression propensity score models overestimated the true effect with increasing frequency as non-additivity and non-linearity increased (Figure 3). Both CART and pruned CART had high absolute biases with respective averages of 18.9% and 21.5% across all scenarios as well as low 95% CI coverage (respective averages of 67.8% and 61.2%). In contrast, the ensemble methods displayed low absolute biases as well as high 95% CI coverage. Across all scenarios, bagged CART, random forests, and boosted CART averaged mean absolute biases of 10.3%, 7.7%, and 6.8%, respectively. Boosted CART displayed the best 95% CI coverage with ≥ 98.6% coverage in all scenarios.
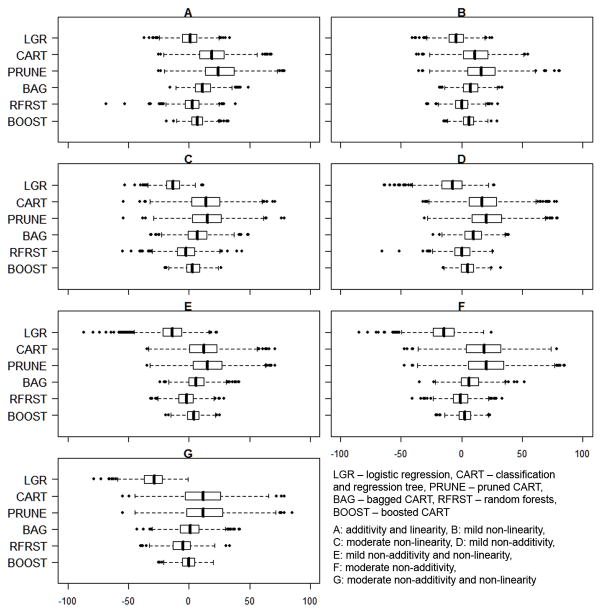
Figure 3.
Distribution of the Percent Bias by Propensity Score Estimation Method for 1000 Datasets in each of 7 Scenarios (N=1000)
The superior CI coverage of boosted CART did not come at the expense of relatively large standard errors. The different methods did not yield substantially different SE estimates although logistic regression tended to produce the largest standard errors compared with the other methods (Table 1). For example, logistic regression produced standard errors that ranged on average from 1.06 times (scenario C) to 1.29 times (scenario E) larger than the errors produced by boosted CART, even though both methods had similar performance in terms of coverage rates.
Weights
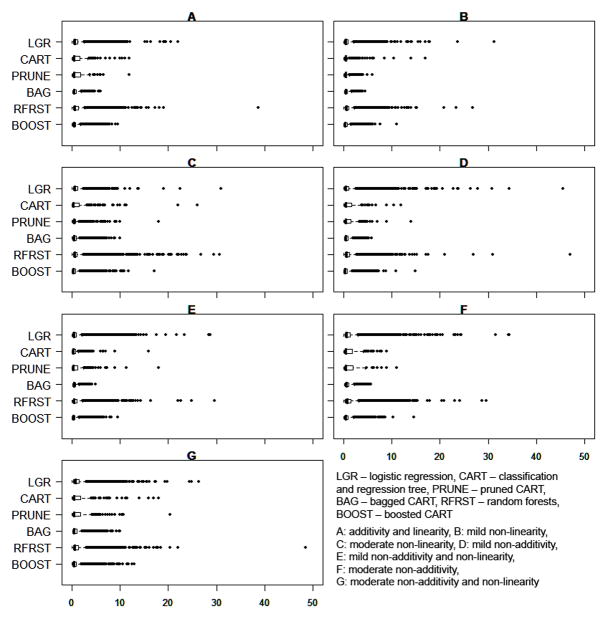
The average weight did not differ greatly by estimation method (Figure 4). For example, the mean weight assigned to a comparison group observation in Scenario E (mild non-additivity and non-linearity) by method was: logistic regression: 0.90, CART: 0.88, pruned CART: 0.88, bagged CART: 0.67, random forests: 0.92, boosted CART: 0.60. However, in all scenarios, logistic regression and random forests tended to produce a relatively large number of extreme high weights while bagged CART had the fewest number of extreme high weights. For example, in Scenario E, the proportion of comparison group weights greater than ten by method was: logistic regression: 3.5%, CART: 0.9%, pruned CART: 0.7%, bagged CART: 0.0%, random forests: 3.1%, boosted CART 0.7%. This may partially explain the relatively large standard errors produced by the logistic regression approach.
Figure 4.
Distribution of Propensity Score Weights for the Comparison Group for Ten Random Datasets of (N=1000)
Simulations of N=500, N=2000 sample sizes
The comparative results of the differently sized studies did not qualitatively differ from the N=1000 studies (Tables 2 and 3). For all methods, covariate balance increased as the sample size increased. This resulted in less biased effect estimates for all methods. However, because larger sample sizes produced smaller errors and thus attributed greater precision to estimates, the methods that had higher bias (logistic regression, CART, and pruned CART) had notably poor 95% CI coverage in the N=2000 studies: for example, with moderate non-additivity and non-linearity (Scenario G), the 95% CI produced by logistic regression included the true effect size only 2.9% of the time, in contrast with coverage of 99.1% for boosted CART (Table 3).
Table 2.
Performance Metrics of Propensity Score Estimation Methods in 1000 Simulated Datasets of N=500.
| Metric | Method1 | Scenario4 | ||||||
|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | ||
| ASAM2 | LGR | 0.059 | 0.057 | 0.065 | 0.077 | 0.078 | 0.081 | 0.103 |
| CART | 0.143 | 0.138 | 0.152 | 0.155 | 0.151 | 0.142 | 0.149 | |
| PRUNE | 0.179 | 0.17 | 0.165 | 0.184 | 0.181 | 0.166 | 0.165 | |
| BAG | 0.129 | 0.126 | 0.125 | 0.143 | 0.141 | 0.126 | 0.121 | |
| RFRST | 0.099 | 0.095 | 0.093 | 0.108 | 0.107 | 0.101 | 0.095 | |
| BOOST | 0.089 | 0.085 | 0.084 | 0.096 | 0.094 | 0.088 | 0.086 | |
| Absolute bias | LGR | 11.2% | 11.2% | 13.8% | 15.3% | 17.7% | 18.1% | 30.3% |
| CART | 19.0% | 16.6% | 21.8% | 19.3% | 19.0% | 20.1% | 21.6% | |
| PRUNE | 27.6% | 22.6% | 24.5% | 24.8% | 22.6% | 22.9% | 23.1% | |
| BAG | 12.6% | 10.4% | 13.5% | 11.6% | 10.3% | 10.8% | 11.4% | |
| RFRST | 10.7% | 9.2% | 11.4% | 10.5% | 9.5% | 10.5% | 11.6% | |
| BOOST | 11.0% | 9.6% | 9.2% | 10.3% | 9.3% | 9.0% | 8.6% | |
| Standard error | LGR | 0.094 | 0.094 | 0.087 | 0.105 | 0.105 | 0.103 | 0.102 |
| CART | 0.095 | 0.096 | 0.105 | 0.097 | 0.098 | 0.096 | 0.104 | |
| PRUNE | 0.085 | 0.085 | 0.097 | 0.089 | 0.089 | 0.088 | 0.097 | |
| BAG | 0.08 | 0.079 | 0.085 | 0.081 | 0.079 | 0.081 | 0.082 | |
| RFRST | 0.086 | 0.083 | 0.085 | 0.088 | 0.084 | 0.087 | 0.085 | |
| BOOST | 0.084 | 0.082 | 0.081 | 0.086 | 0.083 | 0.085 | 0.083 | |
| 95% CI3 coverage | LGR | 97.0% | 97.3% | 96.2% | 91.1% | 87.5% | 86.5% | 64.3% |
| CART | 84.5% | 88.3% | 76.9% | 82.4% | 84.0% | 81.6% | 75.7% | |
| PRUNE | 65.3% | 75.3% | 71.5% | 71.2% | 75.8% | 76.6% | 73.8% | |
| BAG | 97.5% | 98.2% | 95.4% | 98.7% | 99.1% | 99.0% | 98.1% | |
| RFRST | 98.5% | 99.7% | 97.9% | 98.5% | 98.7% | 99.0% | 97.0% | |
| BOOST | 99.0% | 99.2% | 99.5% | 99.1% | 99.6% | 99.9% | 99.8% | |
LGR – logistic regression, CART – classification and regression tree, PRUNE – pruned CART, BAG – bagged CART, RFRST – random forests, BOOST – boosted CART;
ASAM – average standardized absolute mean distance;
CI – confidence interval
A: additivity and linearity; B: mild non-linearity; C: moderate non-linearity; D: mild non-additivity; E: mild non-additivity and non-linearity; F: moderate non-additivity; G: moderate non-additivity and non-linearity.
Table 3.
Performance Metrics of Propensity Score Estimation Methods in 1000 Simulated Datasets of N=2000.
| Metric | Method1 | Scenario4 | ||||||
|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | ||
| ASAM2 | LGR | 0.029 | 0.031 | 0.052 | 0.047 | 0.05 | 0.057 | 0.09 |
| CART | 0.177 | 0.165 | 0.142 | 0.186 | 0.178 | 0.165 | 0.153 | |
| PRUNE | 0.181 | 0.171 | 0.143 | 0.189 | 0.182 | 0.168 | 0.156 | |
| BAG | 0.155 | 0.146 | 0.127 | 0.164 | 0.158 | 0.137 | 0.121 | |
| RFRST | 0.062 | 0.061 | 0.059 | 0.068 | 0.067 | 0.058 | 0.057 | |
| BOOST | 0.052 | 0.049 | 0.054 | 0.054 | 0.053 | 0.048 | 0.053 | |
| Absolute bias | LGR | 5.5% | 6.8% | 13.5% | 10.9% | 14.7% | 16.1% | 30.2% |
| CART | 27.6% | 18.8% | 16.5% | 25.1% | 18.9% | 32.2% | 21.1% | |
| PRUNE | 29.2% | 20.9% | 16.7% | 25.8% | 19.9% | 32.7% | 21.7% | |
| BAG | 19.9% | 13.0% | 11.3% | 16.7% | 11.6% | 16.3% | 9.1% | |
| RFRST | 5.0% | 4.5% | 7.5% | 5.2% | 5.2% | 5.5% | 8.0% | |
| BOOST | 5.6% | 4.7% | 4.8% | 4.7% | 4.1% | 4.1% | 4.7% | |
| Standard error | LGR | 0.047 | 0.047 | 0.043 | 0.054 | 0.053 | 0.054 | 0.051 |
| CART | 0.039 | 0.039 | 0.045 | 0.04 | 0.04 | 0.041 | 0.044 | |
| PRUNE | 0.039 | 0.039 | 0.045 | 0.04 | 0.04 | 0.041 | 0.044 | |
| BAG | 0.038 | 0.038 | 0.042 | 0.038 | 0.038 | 0.039 | 0.04 | |
| RFRST | 0.045 | 0.043 | 0.048 | 0.046 | 0.044 | 0.046 | 0.047 | |
| BOOST | 0.042 | 0.041 | 0.042 | 0.043 | 0.042 | 0.043 | 0.042 | |
| 95% CI3 coverage | LGR | 98.1% | 95.9% | 77.6% | 78.8% | 63.4% | 57.6% | 2.9% |
| CART | 18.1% | 45.0% | 55.6% | 27.3% | 47.5% | 20.4% | 41.6% | |
| PRUNE | 15.5% | 39.0% | 54.4% | 26.0% | 43.9% | 20.0% | 40.1% | |
| BAG | 31.6% | 69.8% | 76.1% | 51.9% | 78.5% | 52.9% | 85.8% | |
| RFRST | 99.2% | 99.3% | 90.8% | 98.1% | 98.3% | 98.2% | 89.5% | |
| BOOST | 99.0% | 99.9% | 99.9% | 99.8% | 99.9% | 99.9% | 99.1% | |
LGR – logistic regression, CART – classification and regression tree, PRUNE – pruned CART, BAG – bagged CART, RFRST – random forests, BOOST – boosted CART;
ASAM – average standardized absolute mean distance;
CI – confidence interval
A: additivity and linearity; B: mild non-linearity; C: moderate non-linearity; D: mild non-additivity; E: mild non-additivity and non-linearity; F: moderate non-additivity; G: moderate non-additivity and non-linearity.
DISCUSSION
The primary objective of propensity score adjustment is to achieve covariate balance between comparison groups so that valid estimates of the treatment effect can be obtained. Logistic regression propensity score models with only main effect terms generally provided adequate covariate balance. However, the bias reducing capabilities of logistic regression propensity score models substantially degraded when the models did not account for interactions and non-linearities. In contrast, regardless of sample size or the extent of non-additivity or nonlinearity, the ensemble methods of bagged CART, random forests, and boosted CART propensity score models provided excellent performance in terms of covariate balance and effect estimation. The consistently superior performance of boosted CART and random forests leads us to recommend these two machine learning techniques for future consideration in propensity score estimation.
In this study we used only the basic, off-the-shelf versions of each of the methods, since that is likely what most applied researchers would do. It is likely that any method may perform better when implemented by a highly skilled user. For example, logistic regressions with carefully chosen interactions may perform better than the simple main effects-only model used here. Similarly, while random forests sometimes produced large propensity score weights, the estimation algorithm could be calibrated to reduce the likelihood of extreme weights.
In our simulations, the outcome Y is fully determined by the observed covariates Wi and the treatment A, which is not what we normally expect to see in practice but is a common strategy for assessing bias in propensity score settings [34, 35]. However, this fact should have no effect on the performance of the propensity score estimation techniques. To check this supposition we performed a sensitivity analysis on a subset of the data where we added random error to Y such that Y = αiWi + γA + ε, ε~N(0,σ). We performed analyses for σ = 0.1 and σ = 0.2, equivalent to 25% and 50%, respectively, of the effect of exposure (γ = −0.4). As expected, this sensitivity analysis showed no changes in the relative performance of the methods considered (results not shown).
Our results suggest that approximately unbiased estimates of the population average treatment effect for the treated can be obtained from machine learning propensity score methods in a variety of scenarios differing by additivity and linearity. These results support the findings presented by Setoguchi et al. in a comparison of logistic regression, neural nets, CART, and pruned CART in propensity score model building [16]. Setoguchi et al. used the propensity score to select matched samples while we use the propensity score to weight the comparison subjects to appear similar to the treated subjects. For both uses of the propensity scores the machine learning methods performed well in a variety of scenarios, indicating the broad applicability of these results. Furthermore, our results indicate that even while machine learning methods are traditionally applied to larger datasets, machine learning methods can also be applied to smaller datasets as well (e.g., N=500).
One interesting observation is that logistic regression often yielded the lowest ASAM but also produced large biases in the estimated treatment effect; conversely, boosted CART often did not have the lowest ASAM but frequently produced better bias reduction. This may have important implications for diagnostic methods to assess propensity score methods: is good covariate balance not enough for ensuring low bias, or is it perhaps that the ASAM is not an adequate measure of balance? For example, in the studies of N=1000, the correlations of ASAM with absolute bias ranged from 0.38 to 0.66 across scenarios. In contrast, the average standardized mean distance calculated using all possible two-way interaction terms (10 choose 2 = 45), not just the covariates themselves, was correlated more strongly with absolute bias in all scenarios, with a range of correlations from 0.56 to 0.72. While further investigation of this issue is needed, these results suggest that covariate balance in interactions may be important to account for in propensity score models and balance checks, particularly when the true outcome models themselves have interaction terms. Although some researchers have recommended checking balance on interactions [36, 37], it is unfortunately rarely done in practice. This may also indicate why logistic regression, with no interaction terms, did not perform well in these simulations.
Boosted CART provided consistently excellent performance in propensity score model building. The efficiency of boosting as a general algorithm to improve estimates is well-known in the machine learning world [20]. What has been less known was whether those benefits would carry over to the world of propensity score estimation and use. As discussed by McCaffrey and colleagues [5, 15], the boosting algorithm we used has a number of features that improves propensity score estimation performance. Boosted CART estimates the propensity score using a piecewise linear combination of multiple CARTs. To reduce prediction error, each successive CART is estimated from a random subsample of the data. In addition, the application of a shrinkage coefficient to downweight each additional CART helps to prevent overfitting. Finally, the use of the piecewise constants has the effect of flattening the estimated propensity scores at the extreme values of the predictors. This minimizes the chance of obtaining predicted probabilities near 0 or 1, thus preventing the high variability in weights that can be problematic for propensity score weighting.
One criticism of machine learning is that the “black box” nature of the algorithms obscures the relationships between predictors and outcome. However, etiologic inference is not a necessary component of propensity score estimation [2]. Therefore, machine learning techniques may be well-suited to the task of creating propensity scores from high-dimensional data where improper parametric specification of relationships may lead to biased estimates. We also note that the available software can offer insight into the relationships among variables. For example, the boosting package twang can analyze the relative contributions of variables to improvements in the model log-likelihood [28] and Elith et al. describe and provide code for visualizing partial dependences and interactions in a boosted regression tree model [23].
In conclusion, our simulation results complement previous work by Setoguchi et al. to show that using machine learning techniques to estimate propensity scores can greatly reduce bias across a range of sample sizes, scenarios, and propensity score application methods. These techniques offer a number of advantages over logistic regression in propensity score estimation and may be implemented using freely available software packages.
Acknowledgments
Grant support: This work was supported by Award Number K25MH083846 from the National Institute of Mental Health (PI: Stuart). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Mental Health or the National Institutes of Health.
Abbreviations
- ASAM
average standardized absolute mean distance
- CART
classification and regression trees
- CI
confidence interval
- SE
standard error
WORKS CITED
- 1.Rosenbaum PR, Rubin DB. The central role of the propensity score in observational studies for causal effects. Biometrika. 1983;70:41–55. [Google Scholar]
- 2.D’Agostino RB., Jr Propensity score methods for bias reduction in the comparison of a treatment to a non-randomized control group. Stat Med. 1998;17(19):2265–81. doi: 10.1002/(sici)1097-0258(19981015)17:19<2265::aid-sim918>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 3.Hirano K, Imbens G. Estimation of causal effects using propensity score weighting: an application to data on right heart catheterization. Health Services and Outcomes Research Methodology. 2001;2:259–278. [Google Scholar]
- 4.Kang J, Schafer J. Demystifying double robustness: a comparison of alternative strategies for estimating a population mean from incomplete data. Statistical Science. 2007;22:523–580. doi: 10.1214/07-STS227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ridgeway G, McCaffrey DF. Comment: Demystifying double robustness: a comparison of alternative strategies for estimating a population mean from incomplete data. Statistical Science. 2007;22(4):540–543. doi: 10.1214/07-STS227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rubin DB. On principles for modeling propensity scores in medical research. Pharmacoepidemiol Drug Saf. 2004;13(12):855–7. doi: 10.1002/pds.968. [DOI] [PubMed] [Google Scholar]
- 7.Drake C. Effects of misspecification of the propensity score on estimators of treatment effect. Biometrics. 1993;49:1231–1236. [Google Scholar]
- 8.Berk R. An Introduction to Ensemble Methods for Data Analysis. Department of Statistics Papers; Paper 2005032701 2005 [cited; Available from: http://repositories.cdlib.org/uclastat/papers/2005032701. [Google Scholar]
- 9.Mitchell TM. Machine Learning. 1. McGraw-Hill Science/Engineering/Math; 1997. [Google Scholar]
- 10.Breiman L. Statistical modeling: the two cultures. Statistical Science. 2001;16:199–215. [Google Scholar]
- 11.Glynn RJ, Schneeweiss S, Sturmer T. Indications for propensity scores and review of their use in pharmacoepidemiology. Basic Clin Pharmacol Toxicol. 2006;98(3):253–9. doi: 10.1111/j.1742-7843.2006.pto_293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harder VS, Morral AR, Arkes J. Marijuana use and depression among adults: Testing for causal associations. Addiction. 2006;101(10):1463–72. doi: 10.1111/j.1360-0443.2006.01545.x. [DOI] [PubMed] [Google Scholar]
- 13.Harder VS, Stuart EA, Anthony JC. Adolescent cannabis problems and young adult depression: male-female stratified propensity score analyses. Am J Epidemiol. 2008;168(6):592–601. doi: 10.1093/aje/kwn184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luellen JK, Shadish WR, Clark MH. Propensity scores: an introduction and experimental test. Eval Rev. 2005;29(6):530–58. doi: 10.1177/0193841X05275596. [DOI] [PubMed] [Google Scholar]
- 15.McCaffrey DF, Ridgeway G, Morral AR. Propensity score estimation with boosted regression for evaluating causal effects in observational studies. Psychol Methods. 2004;9(4):403–25. doi: 10.1037/1082-989X.9.4.403. [DOI] [PubMed] [Google Scholar]
- 16.Setoguchi S, Schneeweiss S, Brookhart MA, Glynn RJ, Cook EF. Evaluating uses of data mining techniques in propensity score estimation: a simulation study. Pharmacoepidemiology and drug safety. 2008 Jun;17(6):546–55. doi: 10.1002/pds.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Westreich D, Lessler J, Jonsson Funk M. Journal of Clinical Epidemiology. 2009 doi: 10.1016/j.jclinepi.2009.11.020. Provisionally accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marshall RJ. The use of classification and regression trees in clinical epidemiology. J Clin Epidemiol. 2001;54(6):603–9. doi: 10.1016/s0895-4356(00)00344-9. [DOI] [PubMed] [Google Scholar]
- 19.Breiman L, Friedman JH, Olshen RA, Stone CJ. Classification and Regression Trees. Chapman & Hall; 1984. [Google Scholar]
- 20.Hastie T, Tibshirani R, Friedman J. The elements of statistical learning: data mining, inference, and prediction. New York: Springer-Verlag; 2001. [Google Scholar]
- 21.Breiman L. Bagging predictors. Machine Learning. 1996;24:123–40. [Google Scholar]
- 22.Breiman L. Random Forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 23.Elith J, Leathwick JR, Hastie T. A working guide to boosted regression trees. J Anim Ecol. 2008 doi: 10.1111/j.1365-2656.2008.01390.x. [DOI] [PubMed] [Google Scholar]
- 24.R_Development_Core_Team. R: A language and environment for statistical computing. 2008 [Google Scholar]
- 25.Therneau TM, Atkinson B. rpart: Recursive Partitioning. R port by Brian Ripley. R package version 3.1–41. 2008 [Google Scholar]
- 26.Peters A, Hothorn T. ipred: Improved Predictors. R package version 0.8–6. 2008 [Google Scholar]
- 27.Liaw A, Wiener M. Classification and regression by randomForest. R News. 2002;2(3):18–22. [Google Scholar]
- 28.Ridgeway G, McCaffrey DF, Morral AR. twang: Toolkit for Weighting and Analysis of Nonequivalent Groups. R package veresion 1.0–1. 2006 [Google Scholar]
- 29.Kurth T, Walker AM, Glynn RJ, Chan KA, Gaziano JM, Berger K, Robins JM. Results of multivariable logistic regression, propensity matching, propensity adjustment, and propensity-based weighting under conditions of nonuniform effect. Am J Epidemiol. 2006;163(3):262–70. doi: 10.1093/aje/kwj047. [DOI] [PubMed] [Google Scholar]
- 30.Lunceford JK, Davidian M. Stratification and weighting via the propensity score in estimation of causal treatment effects: a comparative study. Stat Med. 2004;23(19):2937–60. doi: 10.1002/sim.1903. [DOI] [PubMed] [Google Scholar]
- 31.Bang H, Robins JM. Doubly robust estimation in missing data and causal inference models. Biometrics. 2005;61:962–973. doi: 10.1111/j.1541-0420.2005.00377.x. [DOI] [PubMed] [Google Scholar]
- 32.Lumley T. survey: analysis of complex survey samples. R package version 3.9–1. 2008 [Google Scholar]
- 33.Stuart EA, Rubin DB. Best Practices in Quantitative Methods. Sage Publications; New York: 2007. Best practices in quasi-experimental designs: matching methods for causal inference; pp. 155–176. [Google Scholar]
- 34.Rubin DB, Thomas N. Combining propensity score matching with additional adjustments for prognostic covariates. Journal of the American Statistical Association. 2000;95(450):573–585. [Google Scholar]
- 35.Stuart EA, Rubin DB. Matching with multiple control groups and adjusting for group differences. Journal of Educational and Behavioral Statistics. 2008;33(3):279–306. [Google Scholar]
- 36.Ho DE, Imai K, King G, Stuart EA. Matching as nonparametric preprocessing for reducing model dependence in parametric causal inference. Political Analysis. 2007;15:199–236. [Google Scholar]
- 37.Rubin DB. Using propensity scores to help design observational studies: application to the tobacco litigation. Health Services and Outcomes Research Methodology. 2001;2(3–4) [Google Scholar]