Abstract
Molecular markers linked to phenotypically important traits are of great interest especially when traits are difficult and/or costly to be observed. In tomato where a strong focus on resistance breeding has led to the introgression of several resistance genes, resistance traits have become important characteristics in distinctness, uniformity and stability (DUS) testing for Plant Breeders Rights (PBR) applications. Evaluation of disease traits in biological assays is not always straightforward because assays are often influenced by environmental factors, and difficulties in scoring exist. In this study, we describe the development and/or evaluation of molecular marker assays for the Verticillium genes Ve1 and Ve2, the tomato mosaic virus Tm1 (linked marker), the tomato mosaic virus Tm2 and Tm2 2 genes, the Meloidogyne incognita Mi1-2 gene, the Fusarium I (linked marker) and I2 loci, which are obligatory traits in PBR testing. The marker assays were evaluated for their robustness in a ring test and then evaluated in a set of varieties. Although in general, results between biological assays and marker assays gave highly correlated results, marker assays showed an advantage over biological tests in that the results were clearer, i.e., homozygote/heterozygote presence of the resistance gene can be detected and heterogeneity in seed lots can be identified readily. Within the UPOV framework for granting of PBR, the markers have the potential to fulfil the requirements needed for implementation in DUS testing of candidate varieties and could complement or may be an alternative to the pathogenesis tests that are carried out at present.
Electronic supplementary material
The online version of this article (doi:10.1007/s00122-009-1183-2) contains supplementary material, which is available to authorized users.
Introduction
For the past 30–40 years, breeding for disease resistance has been an important objective in tomato improvement. Numerous resistance genes, most of them originating from wild relatives, have been identified and introgressed into cultivated tomato (for a recent review see Labate et al. 2007). Currently, resistance genes for over 35 different pathogens have been mapped amongst which 17 have been cloned (van Ooijen et al. 2007), making tomato a model system for studying plant–pathogen interactions.
The available sets of linked markers and the sequence data of cloned resistance genes open up the possibility for multiple resistance screening, i.e. the screening of varieties and populations for many resistance genes at the same time. Most preferable are markers detecting DNA polymorphisms directly within the target gene so that there is no risk of losing linkage due to recombination. Such markers are allele specific and remain informative whatever the genetic background. As the number of cloned genes is steadily increasing, the latter approach is becoming more and more feasible. However, to be able to design a resistance-specific assay for many of the cloned resistance genes, the corresponding susceptibility allele still has to be identified. When identifying the susceptibility allele, one can expect two situations: either there is an almost identical gene that has lost its function because of a number of SNPs or indel mutations, or the corresponding gene is lacking. Whereas in the first situation the development of a co-dominant marker based on SNPs can be considered, in the latter case it is likely that a dominant marker will be developed. In both scenarios, the presence of very similar paralogues has to be taken into account, as many resistance genes remain in (large) clusters, containing highly similar paralogues (Simons et al. 1998).
As a consequence of the strong focus on disease resistance in tomato breeding, disease resistances have become an important discriminating characteristic in Plant Breeders Rights (PBR) research. The PBR system sanctioned by the International Union for the Protection of new varieties of Plants (UPOV) involves the evaluation of distinctness, uniformity and stability (DUS) of the candidate variety using morphological and physiological characteristics. A candidate variety is granted PBR when it is shown to be distinct from all existing varieties and is uniform and stable. Molecular markers are currently not accepted for indicating DUS. However, UPOV has established a working group on the use of biochemical and molecular techniques (BMT group), in which discussions with respect to the use of molecular marker for granting PBR are ongoing. This working group has developed three options for implementing molecular markers in DUS testing. The first one is relevant to the work presented in this study and deals with the use of molecular characteristics as a predictor of traditional characteristics. It was split up into two sub options: option 1(a), the use of molecular characteristics, which are directly linked to phenotypic characteristics (gene-specific markers); and option 1(b), the use of molecular characteristics that can be used reliably to estimate traditional characteristics, e.g. quantitative trait loci (QTLs). Following the discussions within UPOV it can be concluded that an option 1(a) approach is acceptable within the terms of the UPOV convention and would not necessarily undermine the effectiveness of protection offered under the UPOV system (Button 2006).
Within DUS testing of tomato, a number of disease tests have become obligatory, i.e. resistance or susceptibility of a variety has to be assessed. Unfortunately, resistance testing is not always straightforward: several factors might influence the expression of the characteristic (environmental conditions, level of heterozygosity for monogenic dominant genes, developmental stage of the plant, etc.) and there are problems regarding the multiplication and stability of the inocula, which require continuous attention.
In the current study, we have investigated the possibilities for multiple resistances screening using molecular markers within the framework of an option 1a approach and provide data on robustness, i.e., evaluation of the marker assays in several laboratories on a common set of varieties/genotypes. The study was limited to the diseases that were obligatory in the DUS testing of candidate varieties that were submitted for granting of PBR within the EU (see http://www.cpvo.europa.eu) i.e., Meloidogyne incognita resistance, controlled by the Mi1 gene, tomato mosaic virus (ToMV) resistances controlled by the Tm1, Tm2 and Tm2 2 genes, Verticillium dahliae resistance controlled by the Ve1 and Ve2 gene, and Fusarium oxysporum f. sp. lycopersici resistance controlled by the I and I2 genes.
For these resistances, either a comparison of existing PCR assays was made (Mi1, Tm1) or a molecular assay was developed (Tm2 and Tm2 2 alleles, Ve1, Ve2, I and I2). These assays have subsequently been tested in a ring test to assess their versatility and evaluated on a set of varieties recently submitted for DUS testing.
Materials and methods
Plant materials used
The following tomato varieties were used for the development of the molecular marker-based resistance assays: the old and introgression-free varieties, Marmande and Moneymaker, which are susceptible to all the diseases considered here and the modern varieties, Campeon and Persica, which are resistant to all the diseases described as the obligatory disease tests within the technical protocol for tomato DUS testing CPVO-TP/44/2 (http://www.cpvo.eu.int). Additional varieties and isogenic lines were included for the specific tests: Master no 2 and NAK 83 (Tm-1); Moperou (Tm2), Monalbo x Momor (Tm2 2) and Draco (tm2); Kamonium (ve1ve2) and Chablis (Ve1Ve2); Marmande verte and Marporum (I); Ideucenzi (I2) and Motelle (I2 +); Anahu and Casaque Rouge (Mi-1). A total of 71 anonymous genotypes for the evaluation of markers were derived from DUS testing of 2006.
Development of molecular tests
The starting point for the development of molecular assays was the requirement that the assays needed to be performed with PCR and analysis on agarose gels. For each of the resistance genes, different starting situations existed and research efforts were needed depending on the already available data, i.e. assays already described in the literature and availability of the sequence of the resistance and susceptibility alleles. In case of dominant markers, controls for PCR amplification, i.e. LAT52 (Yang and Francis 2005; GenBank no. X15855) and Rubisco (Genbank no. CAJ32401) primers were used. Primers used in this study are described in Table 1. In the following, the starting situation for each of the targeted resistance genes has been described.
Table 1.
Primers used, amplification products and PCR conditions
| Pathogen and gene involved | Primer name | Sequence (5′–3′)a | Assay type | Amplification products (bp) | Origin of test | ||
|---|---|---|---|---|---|---|---|
| R-band | S-band | Other | |||||
| M. incognita Mi1-2 | PMiF3 | GGTATGAGCATGCTTAATCAGAGCTCTC | SCAR co-dominant | 550 | 350 | Literature El Mehrach et al. (2005) | |
| PMiR3 | CCTACAAGAAATTATTGTGCGTGTGAATG | ||||||
| ToMV Tm-1 | SCN20F | GGTGCTCCGTCGATGCAAAGTGCA | SCAR dominant + LAT primersb | 1400 | – | 92 (Lat product) | Literature Ohmori et al. (1996) |
| SCN20R | GGTGCTCCGTAGACATAAAATCTA | ||||||
| ToMV Tm-2/Tm2 c Assay 1 SNP2493/2494 | Outer primer TMV-2262F | GGGTATACTGGGAGTGTCCAATTC | Tetra primer ARMS co-dominant | 214 (Tm22) 255 (Tm2) | 417 | Primer design based on Lanfermeijer et al. (2005) | |
| Outer primer TMV-2678R | CCGTGCACGTTACTTCAGACAA | ||||||
| Tm22 SNP 2494F | CTCATCAAGCTTACTCTAGCCTACTTTAGT | ||||||
| Tm2 SNP2493 R | CTGCCAGTATATAACGGTCTACCG | ||||||
| ToMV Tm-2/Tm2 c Assay 2 SNP901 | Outer primer TM2-748F | CGGTCTGGGGAAAACAACTCT | Tetra primer ARMS co-dominant | 179 (Tm2 or Tm22) | 382 (tm2) | 509 | Primer design based on Lanfermeijer et al. (2005) |
| Outer primer TM2-1256R | CTAGCGGTATACCTCCACATCTCC | ||||||
| TM2- SNP901misR | GCAGGTTGTCCTCCAAATTTTCCATC | ||||||
| TM2- SNP901misF | CAAATTGGACTGACGGAACAGAAAGTT | ||||||
| V. dahliae Ve1 Assay SNP2199 | Outer primer Ve1_2072F | CCTTGATGGGGTTGATCTTTCGT | Tetra primer ARMS co-dominant | 476 | 158 | 580 | Sequence retrieval (based on Kawchuk et al. 2001) and development of assay this study |
| Outer primer Ve1_2651R | GTAGGTGAGTTTCTTGGACAGTCGA | ||||||
| Ve1_SNP2199Ft | CAGGCCCTTTGGATGAATCACATT | ||||||
| Ve1_SNP2199Ra | GTTGGACAAAAGAGAGAAAGTGAAGCTAAGT | ||||||
| Ve1_SNP2199F1tc | CAGGCCCTTTGGATGAATCACTA T | ||||||
| Ve1_SNP2199R1ac | GTTGGACAAAAGAGAGAAAGTGAAGCTTAC T | ||||||
| V. dahliae Ve2 Assay SNP 2827 | Outer primer Ve2_2720F | GGATCTTAGCTCACTTTATGTTTTGAAC | Tetra primer ARMS co-dominant | 242 | 131 | 321 | Sequence retrieval (based on Kawchuk et al. 2001) and development of assay this study |
| Outer primer Ve2_3040R | GGTGCTGGTTTCAACTCTGAAGT | ||||||
| Ve2_SNP2827F | CAAATGCTTGAATCACTAGACCTGTGAAC | ||||||
| Ve2_SNP2827R | GGATCTCCCCGGACAGGTGGATTC | ||||||
| F. oxysporum race 0, I | At2-F3 | CGAATCTGTATATTACATCCGTCGT | SCAR dominant + LAT primersb | 130 | 92 (Lat product) | Literature on map position (Ori et al. 1997; Scott et al. 2004) sequence retrieval of markers and development of assay | |
| At2-R3 | GGTGAATACCGATCATAGTCGAG | ||||||
| F. oxysporum race 1, I2 | Z1063F | ATTTGAAAGCGTGGTATTGC | SCAR dominant + Rubisco primersd | 940 | 1380 (Rub product) | Primer design based on sequences Simons et al. (1998) | |
| Z1063R | CTTAAACTCACCATTAAATC | ||||||
aIn bold SNP-specific base, underlined base deliberately changed to create mismatch with template
bLat control primers: Lat1-F AGACCACGAGAACGATATTTGC, Lat2-R TTCTTGCCTTTTCATATCCAGACA
cSecond set of primers with deliberate mismatch closer to 3’-SNP end of primer gives better results at some laboratories
dRubisco control primers: Rub-F ATGTCACCACAAACAGAGAC, Rub-R CTCACAAGCAGCAGCTAG
Mi1-2
The Mi region (1 Mb), originating from Solanum peruvianum, is localised on the short arm of chromosome 6. The locus contains 3 ORFs of which two encode intact genes named Mi1-1 and Mi1-2, and the third is a pseudo gene (Milligan et al. 1998). Mi1-2, but not Mi1-1, is sufficient to confer resistance to Meloidogyne incognita, M. javanica and M. arenaria. Four PCR markers were described for the specific detection of Mi1-2 (Goggin et al. 2004; Devran and Elekçioglu 2004; Bendezu 2004; El Mehrach et al. 2005). These markers were tested on a set of ten discriminative varieties to evaluate their reproducibility and possibilities for unequivocal scoring.
Tm-1
The Tm1 gene, originating from S. habrochaites, confers resistance to ToMV strains 0 and 2. The gene has been mapped to the short arm of chromosome 2, but its sequence remains to be determined. Ohmori et al. (1996) described six closely linked SCAR markers; the distance between these markers and Tm1 is not precisely known, but no recombinants could be detected in 125 BC1 plants for any of these markers. This set of six markers has been tested for their applicability on a selection of tomato varieties in conjunction with primers Lat1F/Lat2R.
Tm2 and Tm22
Two alleles, both originating from S. peruvianum, were identified at the Tm2 locus. Allele Tm2 confers resistance to ToMV strains 0 and 1, whereas allele Tm2 2 confers resistance to ToMV strains 0, 1 and 2. The Tm2 locus was mapped close to the centromere of chromosome 9 and sequences of the Tm2 and Tm2 2 resistance alleles as well as the susceptible allele tm2 (S. lycopersicum) were published by Lanfermeijer et al. (2003, 2005). Sequences can be found under Genbank numbers AF536199–AF536201. Based on the differences between the three alleles at this locus, Lanfermeijer et al. (2005) designed two CAPS markers with which the genotypes could be discriminated co-dominantly. These two CAPS markers reside in close proximity from each other and can be assessed using the same PCR fragment, but using different restriction enzymes. In this study, a co-dominant assay based on PCR discrimination of the three alleles has been developed using tetra primer ARMS-PCR SNP detection (Ye et al. 2001). This assay has been compared to the CAPS assay and further tested for its robustness in a number of varieties.
Ve1 and Ve2
Resistance against Verticillium dahliae race 0 was introgressed from S. lycopersicum line Peru Wild (Schaible et al. 1951) and was mapped on chromosome 9. Characterisation of the Ve locus showed two closely linked inverted genes, both of which conferred resistance to Verticillium albo-atrum in susceptible potato plants (Kawchuk et al. 2001). The two genes were named Ve1 (AF272367 genomic DNA; AF272366 cDNA) and Ve2 (AF365929 genomic DNA; AF365930 cDNA). The genes have no introns and share 84% of amino acid identity. To develop molecular assays for these two genes, sequence information of susceptible varieties has to be obtained, as well as additional sequences from different resistant varieties to look for possible allelic variation in both genes. Based on an alignment of the sequences deposited by Kawchuk et al. (2001), sequence-specific primers have been developed to amplify either Ve1 or Ve2-specific fragments (please note that a number of inconsistencies have slipped into the paper of Kawchuk et al. (2001), which include annotation and specificity of primers as well as SNPs between the genomic and cDNA sequences of the same gene). Ve1 and Ve2-specific primers have been developed and used to amplify fragments in both susceptible and resistant varieties. These fragments have been sequenced to look for SNPs. In total, one fragment of 721 bp of the Ve2 gene was sequenced and four overlapping segments of Ve1 gene between position 1,466 in the 5′UTR to position 4,481 (positions on AF272367) spanning 2,988 bp of the gene (3,016 bp in total) were sequenced. The SNPs found between the sequences of V. dahliae susceptible and resistant varieties were used for development of tetra-primer ARMS-PCR assays.
I
Resistance to Fusarium oxysporum f. sp. lycopersici race 0 (Fol:0, ex race 1) was introgressed from S. pimpinellifolium. The I locus was mapped on the short arm of chromosome 11 (Ori et al. 1997; Sela-Buurlage et al. 2001; Scott et al. 2004) positioned between the RFLP markers TG523 and TG7 (approximate interval of 3.8 cM). Sequence data, from both ends of these RFLP fragments, were available and these were used to design primers to amplify fragments in resistant and susceptible varieties that can be screened for polymorphisms associated with the resistance. Similarly, primers from two COS markers C2_At5g16710 and C2_At2g22570 closely linked to TG523 were available from the Sol Genomics Network (Mueller et al. 2005; http://sgn.cornell.edu). Amplified fragments from all varieties were sequenced to find polymorphisms between the resistant and susceptible varieties. New primers were designed to amplify fragments in resistant varieties only.
I2
The I2 locus was introduced into tomato from S. pimpinellifolium and confers resistance to race 1 of the soil-borne fungus F. oxysporum f. sp. lycopersici (Sela-Buurlage et al. 2001). I2 was mapped on the long arm of chromosome 11 and cloned (Ori et al. 1997). It encodes for a protein of 1,266 amino acids and belongs to the nucleotide-binding sites (NBS), leucine-rich repeat (LRR) superfamily of plant resistance genes (GenBank no. AF118127). I2 is a member of a complex resistance locus with six members (I2, I2C1, I2C2, I2C3, I2C4 and I2C5) within 90 kb named the I2C gene family (Simons et al. 1998). In addition to I2, the complete sequences of I2C1, I2C2 and I2C5 were available (GenBank no. AF004878, AF004879 and AF408705), whereas for I2C3 and I2C4 only the sequences encompassing the LRR coding sequence were available (GenBank no. AF004880 and AF004881). Some of these members share strong sequence similarities with I2, but cannot confer complete resistance to Fol. Three sets of primers were tested for their ability to amplify an I2-specific fragment based on differences between I2 and its I2C homologues.
Robustness test
For each of the resistance genes, one assay (either developed or from literature and tested) was selected by the laboratory that evaluated the markers for that gene. Primers and conditions used for amplification are presented in Table 1. A protocol for each of the selected assays was sent to the five laboratories involved in the robustness test. Robustness tests were performed on a common set of varieties consisting of two resistant varieties (with the targeted resistance gene, except for the Tm-1 gene) and two susceptible varieties (without the targeted gene) supplemented with some specific varieties that have been used in the testing of the assays (see “Plant materials used”).
Validation of markers
Evaluation of marker assays was carried out on sets of 49–51 varieties, depending on the gene that had been tested for DUS characteristics, including the biological resistance assays. For each variety, at least two plants were independently analysed with molecular markers. In varieties where results of the biological assays showed heterogeneous or inconclusive results, five plants per variety were analysed.
Results
Selection amongst existing molecular assays: Mi1 and Tm1
The four PCR markers described as Mi1-2 specific (Goggin et al. 2004; Devran and Elekçioglu 2004; Bendezu 2004; El Mehrach et al. 2005) were able to discriminate resistant and susceptible varieties in our assays. From these markers, the co-dominant marker PMiF3-R3 (El Mehrach et al. 2005, Table 1) fits best to the requirement of a routine application, i.e. no a-specific bands and a clear separation of resistant (R) and susceptibility (S)-specific bands on agarose gels. The primers amplify a DNA fragment of 1,000 bp in the 3′-UTR outside the Mi1-2 coding sequence.
Concerning Tm1, the six markers described by Ohmori et al. (1996) were tested. The dominant SCARs markers, SCA15 and SCN20, amplified a band in the resistant varieties and were further tested in conjunction with primers Lat1F/Lat2R (Table 1) as PCR controls. Other markers (SCB10, SCL10 and SCN9) either did not amplify a specific band in our material or gave a weak amplification in susceptible plants of identical length (SCG12). Sequencing of bands from SCG12 that show differences in amplification yields between resistant and susceptible varieties did not reveal differences in the internal sequence. SCN20 was chosen as the final marker.
Development of new molecular assays: Tm2/Tm22, Ve1/Ve2, I and I2
Tm2 locus
Between Tm2 and Tm2 2 seven SNPs can be detected, amongst which five result in four different amino acids (Lanfermeijer et al. 2005; GenBank no. AF536199–536201). Similarly, SNPs can be detected between Tm2 and Tm2 2 compared to the recessive tm2 allele from S. lycopersicum. At two positions (901 and 2493/2494), the non-synonymous SNPs were used to develop co-dominant assays based on tetra ARMS-PCR (see Table 1 for primer sequences) as an alternative to the CAPS markers of Lanfermeijer et al. (2005). Tetra-primer ARMS-PCR assays use two primer pairs, which amplify the two different alleles of an SNP in a single PCR reaction (Ye et al. 2001). With the assay on SNP position 901, the presence of the allele tm2 (382 bp fragment) can be assessed as well as the presence of either Tm2 or Tm2 2 (179 bp fragment). With the assay on SNP positions 2,493/2,494, the presence of the alleles Tm2 (band of 255 bp) and Tm2 2 (band of 214 bp) can be assessed.
Ve1 and Ve2 genes
Using the primers that amplify Ve1 and Ve2-specific fragments in both susceptible and resistant plants (heterozygous and homozygous resistance), the sequence for the resistant and susceptible alleles was determined. By comparison of the sequence data from the resistant and susceptible alleles of the Ve1-specific fragments, seven SNPs were identified. Four SNPs were in the gene coding sequence and three in the 5′UTR. For the Ve2 gene, two SNPs were found (positions 2,827 and 2,949) between the resistant and susceptible alleles. Comparison of the sequence information obtained in our study with the study of Kawchuk et al. (2001), in which the cloning of the two genes was described, showed some inconsistencies. For the Ve1 gene, Kawchuk et al. (2001) published an mRNA sequence (AF272366), which was not identical to their published genomic sequence (AF272367) of the same resistance gene. In our study, the SNP haplotype found in susceptible varieties was identical to the mRNA sequence published by Kawchuk et al. (2001), whereas the genomic sequence was identical to the sequence only found in resistant plants. Therefore, we concluded that the mRNA sequence accession AF272366 represents the susceptible allele and the genomic sequence accession AF272367 represents the resistant allele.
For Ve2, both the mRNA sequence (AF365930) and the genomic sequence (AF365929) of Kawchuk et al. (2001) were identical to the sequences found in susceptible varieties in our study, indicating that these two sequence accessions represent the susceptible allele (EMBL accession no. FN548097, FN548098 and FN548099 represent sequences found in, respectively, a susceptible, resistant and heterozygous resistant variety). As a control for this conclusion, the linked markers described by Kawchuk et al. (1998) were tested on the same set of varieties and the results with these markers fitted with our results. Therefore, it is likely that a mistake has been made somewhere between the cloning of the Ve1 and Ve2 genes and the donation of the sequences to the database by Kawchuk et al. (2001), as the sequences donated were those from the susceptible alleles except for the genomic Ve1 sequence (AF272367). Identical conclusions can be drawn from the sequence data provided by Acciarri et al. (2007) and Fradin et al. (2009).
Amongst the SNPs of the resistant and susceptible alleles, two non-synonymous SNPs were selected for marker development. Based on Ve1 SNP pos. 2199 (base positions related to AF272367) and Ve2 SNP pos. 2827 (base positions related to AF365929) two tetra primer ARMS-PCR assays have been developed (Fig. 1).
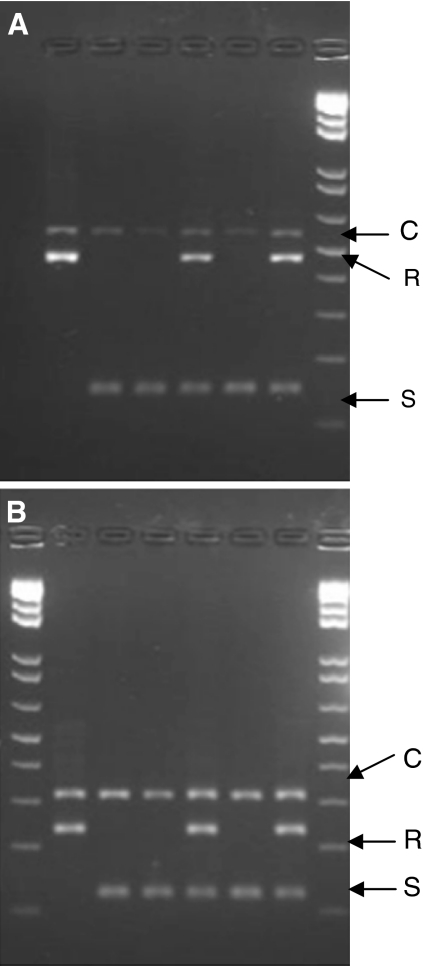
Fig. 1.
Example of molecular assay: results of Verticillium assays on test set for robustness assessment (2% agarose). Varieties are Campeon, Marmande, Moneymaker, Persica, Kamonium and Chablis. a Ve1 assay: C control band (580 bp), R resistant allele (476 bp), S susceptible allele (158 bp) b Ve2 assay: C control band (321 bp), R resistant allele (242 bp), S susceptible allele (131 bp)
I locus
Amongst the 16 primer pairs tested, only the primers based on the COS marker from the nicotinamidase 1 gene from Arabidopsis thaliana At2-F/At2-R showed a difference between the resistant variety Marporum and the susceptible Marmande verte, i.e. the presence of a 7-bp deletion in Marmande verte. Because amplification in other varieties failed, new primers (At2-F1/R1) were designed to border the gap region and facilitate the sequencing of this region. The PCR product from five additional varieties were then sequenced and clearly showed again the presence of a 7-nucleotide gap in all susceptible varieties. This deletion was used to develop a dominant PCR-based marker (At2-F3/At2-R3) with the reverse primer matching the additional 7-bp present only in resistant varieties and the forward primer matching a region common to resistant and susceptible varieties (Table 1). The primer pair amplifies a product of 130 bp. As a positive control for amplification, use is made of the Lat1F/Lat2R primers.
I2 gene
Of the three primer pairs developed for this locus, the primers Z1063F/R (Table 1) amplified a single product of 940 bp only in genotypes carrying the I2 gene from S. pimpinellifolium. Sequencing of this PCR fragment confirmed that it corresponded to I2. Interestingly, these primers failed to amplify the I2 orthologue from S. pennellii, which controls only partial resistance to race 1 of Fol (Sela-Buurlage et al. 2001). Because of the dominant nature of this marker, the Rubisco control for amplification was included in the PCR reaction. Optimisation of multiplex PCR conditions gave specific and reproducible results by using three times more I2 primers in comparison with Rubisco primers.
Results of robustness test
An inter-laboratory ring test was conducted to test the methodology and interpretation of the results for each of the assays on a standardised set of four varieties included in all assays supplemented with two or three varieties that were specific for the disease and used in the assay development. Without optimisation, most of the assays could be reproduced and results obtained were according to expectations in at least three of the five laboratories for each assay. In all assays (except for the Mi1 assay), one or two laboratories encountered results that were not optimal (mainly weak amplification). In these cases, the assays were further optimised (adapting cycling conditions and in one case changing the Taq polymerase used) towards the specific conditions for the laboratory in question. In all cases, optimisations were successful.
Validation of markers
The set of varieties chosen for the validation of the marker assays could be divided into two categories, one a group of varieties with a clear biological assay phenotype and the other a group of varieties with inconclusive results in the biological assays.
The results presented in Table 2 show that not all biological assays give clear results. This is the case for all tests performed and in total around 8% of the samples could not be classified conclusively. In the group of varieties with a clear phenotype in the biological assays, the results obtained with the molecular assays supported the results from the biological assays (Table 2), with a few exceptions. For Ve, one variety was found resistant in the biological assay, whereas in the molecular assay it was scored susceptible for both Ve1 and Ve2. Another variety that was susceptible in the biological assay was scored resistant based on the molecular assay for Ve1. For ToMV, biological and marker assays gave identical results. Similarly, for the nematode resistance gene Mi1-2 biological and marker assays gave identical results. In one of the varieties, an additional band around 500 bp was observed, which may be linked to a variant of the Mi1 resistance gene from S. chilense (see below). For Fusarium, the test for the I gene showed two varieties for which the biological assay indicated susceptibility, whereas in the marker assay they were scored resistant. Also, the test for the I2 gene showed a discrepancy between the biological test (susceptible) and the molecular assay (resistant) for one variety.
Table 2.
Overview of results in biological and molecular assays
| Test | Bioassay uncleara | Bio assay resultb | Molecular assay resultb | Overall result | ||
|---|---|---|---|---|---|---|
| R | S | R | S | Agreement/total | ||
| Verticillium | 4 | 36 | 9 | 36 | 9 | 43/45c |
| TMV | 2 | 39 | 10 | 39 | 10 | 49/49 |
| Mi | 4 | 26 | 21 | 26 | 21 | 47/47 |
| Fusarium I | 7 | 37 | 6 | 37 | 6 | 41/43c |
| Fusarium I2 | 5 | 36 | 9 | 37 | 8 | 44/45 |
aVarieties for which biological assay was unclear or inconsistent were analysed separately, see text
bOnly the varieties with conclusive biological test results
cOne variety was tested to be resistant using the bio assay, but was tested susceptible in the molecular assay; for one other variety, it was the other way about
Varieties that showed inconclusive results in the biological assay were also selected for analyses with the molecular assays to test the potential of the molecular assays to clarify these inconclusive results. The inconclusive results in the biological assays were mainly due to heterogeneity in scoring between plants and also to unclear symptoms in subsets of individuals from a variety. For these varieties, the marker assay gave clear homogeneous results for most varieties and only in two cases (one with Verticillium and one with tomato mosaic virus) confirmed resistance heterogeneity.
For Ve, four varieties showed non-conclusive results. Two varieties showed low percentages of plants with weak infection symptoms, but of which none died. All the tested plants from these two varieties showed the presence of both resistance alleles, although in one variety heterogeneity in the zygotic state could be detected for the Ve2 gene (2 individuals: Ve1Ve1/Ve2ve2 and 3 individuals: Ve1Ve1/Ve2Ve2). In the third variety, a 4:1 segregation of symptoms:no symptoms was found in the biological assay, whereas the molecular assay indicated a 4:1 presence of the haplotypes ve1ve1/Ve2ve2: Ve1ve1/Ve2ve2 that both should be considered resistant. The fourth variety showed susceptible plants with some escapes in the biological assay which was confirmed in the molecular assay by the presence of a 4:1 segregation of the haplotype’s ve1ve1/ve2ve2:Ve1ve1/ve2ve2.
Within the ToMV biological assays, one variety shows unclear results (resistant-like, but with necrosis), whereas the molecular analysis shows the homozygous presence of the Tm2 2 allele. For another variety, the heterogeneous results from the biological assay were confirmed with the molecular assay in which tm2tm2 (susceptible) as well as Tm2 2 tm2 and Tm2 2 Tm2 2 (resistant) individuals were detected.
In the Mi biological assay, four varieties (including the variety Madyta that is used as an intermediate resistant control variety) showed heterogeneous results with both susceptible and resistant plants. The molecular assays for these varieties showed two bands, one at 350 bp, which is the expected mi1-2 susceptibility allele, the other band at around 510 bp (instead of the expected 550-bp resistance Mi1-2 allele) and probably representing the so-called Mi-J allele, which is a variant of the Mi1 resistance gene. This variant gives intermediate resistance to M. incognita and is linked to the Ty-1 gene TYLCV resistance introgressed from S. chilense (Hoogstraten and Braun 2005).
In the I biological assay, seven varieties showed heterogeneous results (including the control variety, Marporum), whereas the molecular assay for the two plants tested indicated resistance for five varieties and susceptibility for two varieties. Finally, the I2 biological assay showed five varieties for which the results were inconclusive; all of them were scored as susceptible in the molecular marker assay.
Discussion
Development and evaluation of robustness of molecular marker assays for disease resistance testing
In this study, we present the development and evaluation of molecular assays for seven disease resistance loci in tomato. The assays are based on either tetra ARMS-PCR, CAPS or SCAR markers. When possible, the cloned resistance gene was taken as the starting point for marker development. Only for the ToMV resistance gene Tm1 and the Fusarium I gene, linked markers had to be used. Because the purpose of this study was to assess the potential of the markers for usage in DUS testing by different testing stations, the robustness of the markers were tested in a ring test at the laboratories of all five parties involved.
Transfer of molecular markers from one laboratories to the next is not always easy (Jones et al. 1997). It often requires optimisation, because between most laboratories differences exist in the equipment (notably PCR machines) and in the reaction components used (mainly Taq polymerases). In this study, for each assay, results could be reproduced without prior optimisation in at least two other laboratories. From this, it can be concluded that the assays are robust and it should not be a major problem to implement them in other laboratories as well. Only in some laboratories, optimisation was needed to get all assays at the required level of scorability. Optimisation mainly involved adjustment of annealing temperature in PCR reactions, use of a touch-down PCR protocol and in one case use of another Taq polymerase. Because the markers could be reproduced fairly easily and gave consistent results, they were further used to assess their potential in DUS testing for disease resistance.
Comparing biological and marker assays
From our results (Table 2), it is clear that in 98% of the cases the molecular assay gave results identical to the biological assay. In a number of varieties, the inconsistent results from the biological assays could be confirmed by the molecular assay that showed that individuals were heterogeneous for the resistance, like for example with the variety showing “susceptible plants with escapes”, the molecular results indicated that genotypes ve1ve1/ve2ve2 and Ve1ve1/ve2ve2 were found in a 4:1 ratio. Nevertheless, in the majority of cases with unclear biological assay results, the molecular assays gave clear results on the presence or absence of the resistance gene without indication of heterogeneity at disease resistance loci. In such cases, the markers may be used as supporting evidence to resolve the unclear biological assay results. However, care should be taken, as the genetic background in which the resistance gene is present may also play an important role and influence the expression of the resistance.
In conclusion, for ToMV and Mi resistance, there was a 100% agreement between both assay types. For the fungal diseases (Ve, I and I2), there were a few disagreements between the biological and molecular marker assays. Although the biological and molecular assays were not performed simultaneously and on the same individual plants, some conclusions can be drawn. The inconclusive results of the biological assays for a number of varieties suggest that environmental factors can be very important for the results from these assays, but such factors do not obscure the molecular marker assays. However, the molecular assay used for the Fusarium I is based on markers linked to the resistance gene (within a 3.8 cM area). This linkage may have been lost in the two varieties showing the deviating results and can thus be the explanation for the conflicting results as well.
Molecular marker assays within the UPOV framework
Option 1(a) describes the possible use of molecular markers for DUS testing as the use of molecular characteristics, which are directly linked to phenotypic characteristics (gene-specific markers). Regarding this, markers that are associated with phenotypic traits are most valuable when the linkage between the trait and the marker is high, as this will lower the risk of losing the association. In this respect, markers that target the actual resistance gene itself are preferred as this will prevent erroneous results due to recombination between the trait and the marker. The use of markers within the untranslated region or the ORF of a gene was achieved for six out of the eight genes involved in this study. For the Tm1 and I gene, the marker used was only linked to the gene of interest. Because the Tm1 locus does not play an important role due to the broader protection given by the Tm22 allele, it is not a breeding objective and of little importance. For the Fusarium I this is different as the locus is taken into consideration in breeding. The fact that for the Fusarium I, two varieties tested susceptible in the biological test, but resistant in the molecular test, may have been caused by loss of linkage between the molecular marker and the resistance gene raises question whether or not linked markers should be used at all. In any case, results with linked markers should be treated with great caution.
Molecular markers, as developed and evaluated in this project, could potentially fulfil the requirements needed for implementation in daily DUS testing as an alternative to the pathogenesis tests that are carried out at present. In the large majority of the varieties tested, the results of the biological and molecular assays were in agreement with each other, which enable the possible introduction of markers as tools in DUS testing. This is especially true for ToMV and Meloidogyne where the biological and molecular assays gave identical results. For fungal diseases, this situation may be slightly different. Although the assays for the fungal diseases Verticillium and Fusarium also predicted the right phenotype in almost all cases, there were a few varieties in which there were some disagreements between marker and biological assays, which may cause some hesitation in introducing the marker assays.
One aspect that will need attention is the issue of variety heterogeneity. Previous studies have shown that marker assays can be very effectively used to spot heterogeneity or impurities in a seed sample (Bredemeijer et al. 2002; Röder et al. 2002; Cooke et al. 2003). As within the current study also varieties were found with different genotype constitutions, agreement has to be reached in which diseases (additive effects or not) and to what extent this should be considered within the limits for PBR approval. Similar to some of the biological disease tests where resistance phenotype can be pathogen race dependent, the usability of the molecular marker assays is directly linked to a particular resistance gene. Therefore, if new sources of resistance are introduced involving different genes, marker assays have to be developed for these as well.
The development of the assays may be expanded to other disease resistance genes, which can be claimed as discriminating traits, as well. Especially, the development of tests for quarantine organisms is interesting, as handling of the pathogen can be avoided. Genes to be included could be those for resistance against, e.g. Cladosporium, Stemphylium, Oidium (two species), Phytophthora infestans, Pyrenochaeta, tomato spotted wilt virus, tomato yellow leaf curl virus, Pseudomonas tomato, Ralstonia solanacearum and Clavibacter michiganense (for reviews see Barone and Frusciante 2007; Van Berloo and Lindhout 2001). The current study may form the starting point for the discussions around the actual implementation of marker data in DUS testing.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This project was co-funded by the Community Plant Variety Office (CPVO), the Ministry of Agriculture, Nature and Food Quality of the Netherlands, GEVES, INRA, the French Ministry of Agriculture and INIA. The authors wish to thank Sergio Semon of the CPVO for fruitful discussions.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Acciarri N, Rotino GL, Tamietti G, Valentino D, Voltattorni S, Sabatini E. Molecular markers for Ve1 and Ve2Verticillium resistance genes from Italian germplasm. Plant Breed. 2007;126:617–621. doi: 10.1111/j.1439-0523.2007.01398.x. [DOI] [Google Scholar]
- Barone A, Frusciante L (2007) Molecular marker-assisted selection for resistance to pathogens in tomato. In: Guimaraes E, Ruane J, Scherf BD, Sonnino A, Dargie JD (eds) Marker-assisted selection: current status and future perspectives in crops, livestock, forestry and fish/. FAO, Rome (Italy). Agriculture and Consumer Protection Dept., 978-92-5-105717-9, A1120 (ftp://ftp.fao.org/docrep/fao/010/a1120e/a1120e02.pdf), pp 151–164
- Bendezu IF. Detection of the Mi 1.2 tomato gene by PCR using non-organic DNA purification. Nematropica. 2004;34:23–30. [Google Scholar]
- Bredemeijer GMM, Cooke RJ, Ganal MW, Peeters R, Isaac P, Noordijk Y, Rendell S, Jackson J, Röder MS, Wendehake K, Dijcks M, Amelaine M, Wickaert R, Bertrand L, Vosman B. Construction and testing of a microsatellite database containing more than 500 tomato varieties. Theor Appl Genet. 2002;105:1019–1026. doi: 10.1007/s00122-002-1038-6. [DOI] [PubMed] [Google Scholar]
- Button P. New Developments in the International Union for the Protection of New varieties of plants. Acta Hortic. 2006;714:195–210. [Google Scholar]
- Cooke RJ, Bredemeijer GMM, Ganal MW, Peeters R, Isaac P, Rendell S, Jackson J, Röder MS, Korzun V, Wendehake K, Areshchenkova T, Dijcks M, Laborie D, Bertrand L, Vosman B. Assessment of the uniformity of wheat and tomato varieties at dna microsatellite loci. Euphytica. 2003;132:331–341. doi: 10.1023/A:1025046919570. [DOI] [Google Scholar]
- Devran Z, Elekçioğlu IH. The screening of F2 plants for the Root-knot nematode resistance gene, Mi by PCR in tomato. Turk J Agric For. 2004;28:253–257. [Google Scholar]
- El Mehrach K, Mejía L, Gharsallah-Couchane S, Salus MS, Martin CT, Hatimi A, Vidavski F, Williamson V, Maxwell DP. PCR-based methods for tagging the Mi-1 locus for resistance to root-knot nematode in begomovirus-resistant tomato germplasm. Acta Hortic. 2005;695:263–270. [Google Scholar]
- Fradin EF, Zhang Z, Juarez Ayala JC, Castroverde CDM, Nazar RN, Robb J, Liu CM, Thomma BPHJ. Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol. 2009;150:320–332. doi: 10.1104/pp.109.136762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goggin FL, Shah G, Williamson VM, Ullman DE. Instability of Mi-mediated nematode resistance in transgenic tomato plants. Mol Breed. 2004;13:391–394. doi: 10.1023/B:MOLB.0000034090.73857.b1. [DOI] [Google Scholar]
- Hoogstraten JGJ, Braun C (2005) Methods for coupling resistance alleles in tomato. Patent WO/2005/079342
- Jones CJ, Edwards KJ, Castiglione S, Winfield MO, Sala F, Van de Wiel C, Bredemeijer G, Vosman B, Matthes M, Daly A, Brettschneider R, Bettini P, Buiatti M, Maestri E, Malcevschi A, Marmiroli N, Aert R, Volckaert G, Rueda J, Linacero R, Vazquez A, Karp A. Reproducibility testing of RAPD, AFLP and SSR markers in plants by a network of European laboratories. Mol Breed. 1997;3:381–390. doi: 10.1023/A:1009612517139. [DOI] [Google Scholar]
- Kawchuk LM, Hachey J, Lynch DR. Development of sequence-charaterized DNA markers linked to a dominant Verticillium wilt resistance gene in tomato. Genome. 1998;41:91–95. doi: 10.1139/gen-41-1-91. [DOI] [PubMed] [Google Scholar]
- Kawchuk LM, Hachey J, Lynch DR, Kulcsar F, van Rooijen G, Waterer DR, Robertson A, Kokko E, Byers R, Howard RJ, Fischer R, Prüfer D. Tomato Ve disease resistance genes encode cell surface-like receptors. PNAS. 2001;98:6511–6515. doi: 10.1073/pnas.091114198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labate JA, Grandillo S, Fulton T, Muños S, Caicedo AL, Peralta I, Ji Y, Chetelat RT, Scott JW, Gonzalo MJ, Francis D, Yang W, van der Knaap E, Baldo AM, Smith-White B, Mueller LA, Prince JP, Blanchard NE, Storey DB, Stevens MR, Robbins MD, Fen Wang J, Liedl BE, O’Connell MA, Stommel JR, Aoki K, Iijima Y, Slade AJ, Hurst SR, Loeffler D, Steine MN, Vafeados D, McGuire C, Freeman C, Amen A, Goodstal J, Facciotti D, Van Eck J, Causse M. Tomato. In: Kole C, editor. Genome mapping and molecular breeding in plants, volume 5, vegetables. Berlin: Springer; 2007. pp. 11–135. [Google Scholar]
- Lanfermeijer FC, Dijkhuis J, Sturre MJG, de Haan P, Hille J. Cloning and characterization of the durable tomato mosaic virus resistance gene Tm-22 from Lycopersicon esculentum. Plant Mol Biol. 2003;52:1037–1049. doi: 10.1023/A:1025434519282. [DOI] [PubMed] [Google Scholar]
- Lanfermeijer FC, Warmink J, Hille J. The products of the broken Tm-2 and the durable Tm-22 resistance genes from tomato differ in four amino acids. J Exp Bot. 2005;56:2925–2933. doi: 10.1093/jxb/eri288. [DOI] [PubMed] [Google Scholar]
- Milligan SB, Bodeau J, Yaghoobi J, Kaloshian I, Zabel P, Williamson VM. The root-knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell. 1998;10:1307–1320. doi: 10.1105/tpc.10.8.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller LA, Solow TH, Taylor N, Skwarecki B, Buels R, Binns J, Lin C, Wright MH, Ahrens R, Wang Y, Herbst EV, Keyder ER, Menda N, Zamir D, Tanksley SD. The SOL Genomics Network: a comparative resource for Solanaceae biology and beyond. Plant Physiol. 2005;138(3):1310–1317. doi: 10.1104/pp.105.060707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori T, Murata M, Motoyoshi F. Molecular characterization of RAPD and SCAR markers linked to the Tm-1 locus in tomato. Theor Appl Genet. 1996;92:151–156. doi: 10.1007/BF00223369. [DOI] [PubMed] [Google Scholar]
- Ori N, Eshed Y, Paran I, Presting G, Aviv D, Tanksley S, Zamir D, Fluhr R. The I2C family from the wilt disease resistance locus I2 belongs to the nucleotide binding, leucine-rich repeat superfamily of plant resistance genes. Plant Cell. 1997;9(4):521–532. doi: 10.1105/tpc.9.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röder MS, Wendehake K, Korzun V, Bredemeijer G, Laborie D, Bertrand L, Isaac P, Rendell S, Jackson J, Cooke RJ, Vosman B, Ganal MW. Construction and analysis of a microsatellite-based database of European wheat varieties. Theor Appl Genet. 2002;106:67–73. doi: 10.1007/s00122-002-1061-7. [DOI] [PubMed] [Google Scholar]
- Schaible L, Cannon OS, Waddoups V. Inheritance of resistance to Verticillium wilt in a tomato cross. Phytopathology. 1951;41:986–990. [Google Scholar]
- Scott JW, Agrama HA, Jones JP. RFLP-based analysis of recombination among resistance genes to Fusarium wilt races 1, 2 and 3 in tomato. J Am Soc Hortic Sci. 2004;129(3):394–400. [Google Scholar]
- Sela-Buurlage MB, Budai-Hadrian O, Pan Q, Carmel-Goren L, Vunsch R, Zamir D, Fluhr R. Genome-wide dissection of Fusarium resistance in tomato reveals multiple complex loci. Mol Genet Genomics. 2001;265:1104–1111. doi: 10.1007/s004380100509. [DOI] [PubMed] [Google Scholar]
- Simons G, Groenendijk J, Wijbrandi J, Reijans M, Groenen J, Diergaarde P, Van der Lee T, Bleeker M, Onstenk J, de Both M, Haring M, Mes J, Cornelissen B, Zabeau M, Vos P. Dissection of the Fusarium I2 gene cluster in tomato reveals six homologs and one active gene copy. Plant Cell. 1998;10:1055–1068. doi: 10.1105/tpc.10.6.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Berloo R, Lindhout P. Mapping disease resistance genes in tomato. In: Zhu D, Hawtin G, Wang Y, editors. International symposium on biotechnology applications in horticultural crops. Beijing: China Agricultural Scientech Press; 2001. pp. 343–356. [Google Scholar]
- Van Ooijen G, van den Brug HA, Cornelissen BLC, Takken F. Structure and function of resistance proteins in solanaceous plants. Annu Rev Phytopathol. 2007;45:3.1–3.30. doi: 10.1146/annurev.phyto.45.062806.094430. [DOI] [PubMed] [Google Scholar]
- Yang W, Francis DM. Marker-assisted selection for combining resistance to bacterial spot and bacterial speck in tomato. J Am Soc Hortic Sci. 2005;130:716–721. [Google Scholar]
- Ye S, Dhillon S, Ke X, Collins AR, Day IN. An efficient procedure for genotyping single-nucleotide polymorphisms. Nucleic Acids Res. 2001;29:e88-8. doi: 10.1093/nar/29.17.e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.