Abstract
Mental stress modifies both cholinergic neurotransmission and alternative splicing in the brain, via incompletely understood mechanisms. Here, we report that stress changes brain microRNA (miR) expression and that some of these stress-regulated miRs regulate alternative splicing. Acute and chronic immobilization stress differentially altered the expression of numerous miRs in two stress-responsive regions of the rat brain, the hippocampal CA1 region and the central nucleus of the amygdala. miR-134 and miR-183 levels both increased in the amygdala following acute stress, compared to unstressed controls. Chronic stress decreased miR-134 levels, whereas miR-183 remained unchanged in both the amygdala and CA1. Importantly, miR-134 and miR-183 share a common predicted mRNA target, encoding the splicing factor SC35. Stress was previously shown to upregulate SC35, which promotes the alternative splicing of acetylcholinesterase (AChE) from the synapse-associated isoform AChE-S to the, normally rare, soluble AChE-R protein. Knockdown of miR-183 expression increased SC35 protein levels in vitro, whereas overexpression of miR-183 reduced SC35 protein levels, suggesting a physiological role for miR-183 regulation under stress. We show stress-induced changes in miR-183 and miR-134 and suggest that, by regulating splicing factors and their targets, these changes modify both alternative splicing and cholinergic neurotransmission in the stressed brain.
Keywords: Stress, microRNA, miR-183, miR-134, SC35, Cholinergic
Introduction
Mammalian psychological stress is known to induce prominent changes in neuronal activity and gene regulation across multiple brain regions (McEwen 2007). Acute and chronic stress both lead to the remodeling of dendrites in the hippocampus (McEwen 1999), which controls learning functions via establishing spatial, episodic, and contextual memory formation (Kenney and Gould 2008; Otto and Eichenbaum 1992). Specifically, in the CA1 region of the hippocampus, both neurons and glia are affected by mental stress (Espinosa-Oliva et al. 2009; Hirata et al. 2009). In the amygdala, stress reactions impact emotion, addiction, fear, and anxiety (Vyas et al. 2002; Walker and Davis 2008). Chronic stress also increases aggression, likely to reflect hyperactivity of the amygdala (Wood et al. 2003). At the physiological level, cholinergic neurotransmission is altered under stress (Kaufer et al. 1998; Soreq and Seidman 2001). These stress-induced changes are mostly attributed to the combinatorial regulation of many genes’ altered transcription levels (Anguelova et al. 2000). However, the contribution of posttranscriptional regulation mechanisms to these stress-associated responses is increasingly acknowledged (Battaglia and Ogliari 2005; Gattoni et al. 1996; Meshorer et al. 2005). One such mechanism is the production of multiple proteins with divergent or even opposite functions from a single transcript via alternative splicing, which accounts for much of the proteome's flexibility (Stamm et al. 2005). A case in point is the stress-induced alternative splicing of the primary ACh-hydrolyzing enzyme acetylcholinesterase (AChE), which modifies cholinergic neurotransmission (Meshorer and Soreq 2006) and affects neuronal processes in a pathway involving the splicing factors SC35 and ASF/SF2 (Meshorer et al. 2005). However, the molecular mechanism(s) underlying the apparent relationship between stress, alternative splicing, and cholinergic neurotransmission remain incompletely understood.
An important posttranscriptional mechanism for gene regulation involves microRNAs (miRs). miRs are 20–28-nucleotide noncoding RNAs encoded in the genomes of plants and animals, exerting translational repression and/or degradation of target mRNAs via complementary binding to the 3′ untranslated region (UTR; Maniataki and Mourelatos 2005). miR levels vary across cell type, tissue, and developmental stages (Baek et al. 2008; Liang et al. 2007; Plasterk 2006; Sood et al. 2006). Each miR targets multiple mRNAs (Krek et al. 2005; Lewis et al. 2005), which in some cases code for proteins participating in the same signaling pathway (Li et al. 2007). Thus, both the overall profile of miR expression and the expression levels of particular key miRs impact diverse biological processes (Chen et al. 2004; He et al. 2005; Kluiver et al. 2007; Laneve et al. 2007; Li et al. 2007; Thai et al. 2007).
miR-18 and miR-124 were recently reported to regulate glucocorticoid receptors, suggesting involvement in a variety of systemic stress responses (Vreugdenhil et al. 2009). However, the mechanistic involvement of miRs at large in psychological stress remains unknown. We predicted that miR-mediated regulation contributes to the yet incompletely understood link(s) between the molecular and the physiological reactions of particular brain regions to psychological stress and more specifically to the regulation of alternative splicing. To test for miR involvement in governing the region-specific stress-induced changes in alternative splicing, we studied the expression profiles of miRs in the hippocampal CA1 and the central amygdala brain regions in stressed and control rats and challenged the relevance of observed changes by manipulating miR-183 levels in cultured cells.
Materials and Methods
Immobilization Stress
Adult male rats (Charles River; 200–225 g) were subjected to a single 4-h session of immobilization stress (acute stress group), 4 h of complete immobilization stress per day for 14 days (chronic stress group), or brief daily handling (no-stress group). Twenty-four hours after the last stress or handling session, the animals were sacrificed, and tissues were carefully dissected, flash-frozen, and stored at −80°C. RNA was then purified using the mirVana kit (Ambion), which preserves short RNAs, from the dissected central amygdala and hippocampus CA1 region. For each brain region in each of the three groups, pooled samples were generated from the RNA of three to four rats.
Spotted Array Methods
Spotted array methods were adapted from Ben-Ari et al. (2006). The mirVana oligo set (Ambion, Austin, TX, USA; Cat. Num. 1564V1) was used to construct an in-house array with >200 spotted probes complementary to known human and mouse miRs. Dye-swapping tests served to exclude dye-specific labeling differences (Dombkowski et al. 2004). Labeling used the CyDye reactive dye pack (Amersham, NSW, Australia), as instructed 2 dye-swapped arrays were used for every comparison, each with at least 6 replicate spots per miRNA probe. Hybridization was performed in chambers (Corning, NY, USA) for 15 h at 64°C. Scanning used an Affymetrix 428 Array Scanner at 532 and 650 nm, controlled by the “Jaguar” software (Affymetrix, CA, USA), and results were exported to the “Imagene” program (BioDiscovery Inc., CA, USA) for quantification. Data normalization, exclusion of unreliable spots, and combination of the information from all slides were performed using the Normalize Suite (Beheshti et al. 2003). Significantly altered transcripts (with a p value of the sign-test smaller than 0.05), which were not disqualified due to any quality parameter, were identified by setting an arbitrary threshold.
Cell Culture
Chinese hamster ovary (CHO) cells were grown in 5-ml flasks in Dulbecco’s modified Eagle’s medium (Biological Industries, Beit Haemek, Israel) supplemented with 10% fetal bovine serum and 2 mM l-glutamine (Biological Industries). Transfection of pre-miR-183 and anti-miR-183 oligonucleotides (Ambion) was performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) at 80–90% confluence. For each transfection sample, 3 μg of oligonucleotide or plasmid and the lipofectamine reagent were both diluted in 0.25–1 ml of unsupplemented growth medium. After 5 min, the diluted oligonucleotide and lipofectamine were combined, incubated for 20 min, and added to the cells. Medium was replaced at 6 h, and cells were harvested 24 h posttransfection. A scrambled sequence oligonucleotide and lipofectamine treatment alone served as negative controls; a green fluorescent protein (GFP) expression vector served to assay transfection efficiency.
Quantitative RT-PCR
Total RNA including transcripts that are 200 bases and smaller was extracted using the mirVana (Ambion, Austin, TX, USA) and the miRNeasy (QIAGEN) isolation kits from cultured cell samples. Contaminating DNA was removed with DNA-free (Ambion). RNA concentration was determined using the NanoDrop ND-1000 instrument (NanoDrop Technologies, Wilmington, DE, USA). Reverse transcription (RT) of miRs was performed using SuperScript III First-Strand Synthesis Systems kit reagents for RT-polymerase chain reaction (PCR; Invitrogen) as detailed elsewhere (Raymond et al. 2005). Primer extension quantitative PCR (QPCR) was conducted as previously described (Raymond et al. 2005) using Power SYBR Green PCR Master Mix (Applied Biosystems), LNA-modified forward primer, and a universal reverse primer and amplified using the ABI-7900HT instrument (Applied Biosystems) equipped with dedicated software (ver. 2). Triplicate values of each treatment were normalized to β-actin mRNA, 5S rRNA, or the U6 small RNA. Absolute quantification of miR levels was performed using standards containing known dilutions of a commercial pre-miR-183 (Ambion). mRNA RT and QPCR were performed as previously described (Gilboa-Geffen et al. 2007). Triplicate values of each sample were normalized to β-actin or GAPDH mRNA.
Immunoblots
These were performed as previously described (Berson et al. 2008) using mouse monoclonal antibodies (Zymed 339400) against arginine–serine-rich (SR) proteins and mouse monoclonal antibodies (SC-32293) targeted to α-tubulin.
Statistics
Student's T test, the Kolmogorov–Smirnov two-sample test, and the nonparametric Wilcoxon test were employed for defining the significance of differences between analyzed groups. p values <0.05 were considered significant. Hierarchical clustering of array data was performed using MeV rev.4 software (Saeed et al. 2003) for Mac OS X, with Euclidian metrics and default parameters.
Results
Amygdala and Hippocampus Display Distinct Stress-Induced Changes in miR Profiles Following Acute or Chronic Stress
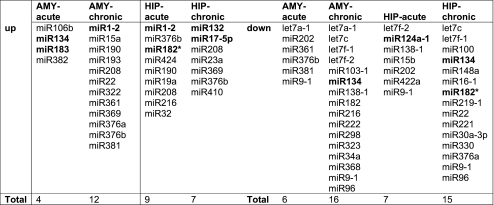
To test if stress-responsive brain regions display altered miR profiles following psychological stress, miR-specific spotted arrays were hybridized with short RNAs from the central amygdala or the CA1 region of the hippocampus of rats subjected to acute or chronic stress or from the corresponding regions of nonstressed controls. Several miRs appeared to be differentially regulated in the stress-responsive brain regions by acute and chronic stress (Table 1). In both regions, we observed, more decreases than increases in miR levels under chronic stress (Table 1), possibly indicating a tendency of upregulated expression of the target mRNAs of these miRs under stress. Additionally, chronic stress resulted in numerically larger changes than acute stress in both brain regions. In the central amygdala, ten different miRs were modified (upregulated or downregulated) under acute stress, as compared to 28 following chronic stress, whereas the hippocampal CA1 region showed 16 and 22 modified miRs after acute and chronic stress, respectively. Moreover, the expression profiles of stress-responsive miRs were different in the analyzed regions, so that there was very limited overlap between miRs modified in each of these regions under acute and chronic stress. For example, in the central amygdala, miR Let-7a-1 was the only miR affected by both acute and chronic stress, whereas, in the hippocampal CA1 region, miR-376b and miR-208 both increased under either acute or chronic stress whereas miR-9-1 decreased in both these conditions. All of the other changes were unique to either acute or chronic stress and particular to the analyzed brain regions.
Table 1.
miRs with a mean LR change >0.25 in absolute value (p < 0.05) 24 h after acute or chronic immobilization stress in rats, sorted by brain region and stress regimen (left to right)
miRs with established brain or stress-related functions are shown in italics. miR-1 targets the nAChR (Simon et al. 2008); miR-134 regulates dendritic spine development (Schratt et al. 2006); miR-132 and miR-182* regulate AChE levels (Shaked et al., submitted). miR-17-5p (Hebert et al. 2009) and miR-124 (Makeyev et al. 2007) control neuronal development and differentiation
Clustering analysis of rat central amygdala and hippocampus CA1 array data (Fig. 1a) confirmed that the majority of miRs represented on the array are differentially regulated by acute and chronic stress. The stress-induced changes in miR profiles in the two brain regions were also dissimilar in the clustered profiles. Thus, the changes observed varied both for the different brain regions and for the different stress paradigms employed. Furthermore, several of the miRs showed distinct patterns of regulation, suggesting functional relevance. Thus, miR-17-5p is upregulated in the hippocampus CA1 region under chronic stress and was recently shown to suppress the amyloid precursor protein (APP) and to undergo downregulation in the course of neuronal development and differentiation (Hebert et al. 2009). Additionally, several let-7 family members were downregulated in both the central amygdala and the hippocampal CA1 under both acute and chronic stress and were recently reported to similarly suppress APP in the course of development in Caenorhabditis elegans (Niwa et al. 2008).
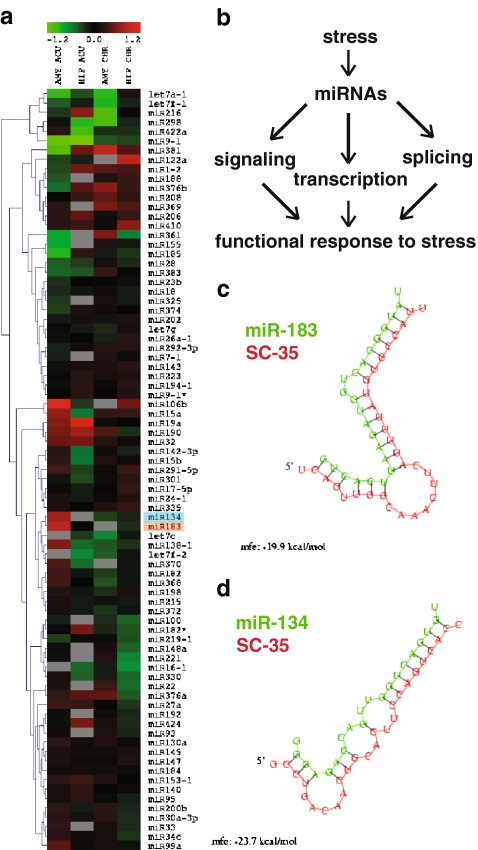
Figure 1.
Stress-responsive miR-134 and miR-183 are predicted to target the splicing factor SC35. a Hierarchical clustering of signal ratios of miRs that passed the quality control in at least three out of four comparisons, from left to right: amygdala (central nucleus) acute/control, hippocampus (CA1) acute/control, amygdala chronic/control, CA1 chronic/control. SC35-targeting miR-134 and miR-183 are emphasized in blue and orange, respectively. Gray fields denote signals that did not pass the more stringent quality control due to low expression levels or high variability between spots. b Predicted targets of stress-regulated miRs seem to affect all the major stages of gene regulation. c, d PicTar-predicted (pictar.bio.nyu.edu) duplexes for miR-183 (b) and miR-134 (c; in green) with 3′ UTR of SC35 mRNA (in red)
Several of the stress-regulated miRs have validated target mRNA transcripts and functional significance. Importantly, the functions of several of these targets are largely relevant to brain stress responses in general and to cholinergic stress reactions in particular. For example, miR-1 targets and regulates two different subunits of the nicotinic ACh receptor (nAChR) and alters presynaptic ACh secretion (Simon et al. 2008). We have recently shown that miR-132 and miR-182* regulate AChE levels downstream of the transcriptional regulator nuclear factor kappa B and AP1 signaling (Shaked et al., submitted). Other miRs appear to control neuronal development and differentiation, both of which are modified under stress. These include miR-17-5p (Hebert et al. 2009) and miR-124, which promotes neuronal differentiation by targeting PTBP1, a repressor of alternative splicing in nonneuronal cells (Makeyev et al. 2007).
Stress-Regulated miR-134 and miR-183 Are Both Predicted to Target Stress-Responsive Transcripts
miR-134 and miR-183 were both upregulated by acute stress in the central amygdala; mir-134 additionally showed a trend of downregulation following chronic stress in both the central amygdala and the hippocampus CA1 region (Table 1, Fig. 1a). These two miRs share a number of putative targets as predicted by the PicTar algorithm (Krek et al. 2005; Table 2), suggesting common regulatory roles. Specifically, these predicted targets included two different transcription factors (ZPFM2, CBFA2T1), one of which was reported to be causally involved with controlling oxidative stress and inflammation (Martinez et al. 2004), and several signaling molecules (integrin-β, casein kinase 1G3, OGT), one of which (integrin-β) is involved in regulating sheer stress (Chen et al. 1999) and another (OGT) functioning as a chaperone to control heat shock reactions (Sohn et al. 2004). Finally, three of these predicted targets are known regulators of alternative splicing (MBNL1, CUGBP2, SFRS2), with CUGBP2 also prominently involved in RNA editing (Anant et al. 2001). Together, this agrees with a central functional role of the stress-regulated miRs, impacting all the major tiers of gene regulation (scheme, Fig.1b). Notably, miR-134 and miR-183 were both predicted by PicTar to bind the 3′ UTR of SFRS2 (SC35) mRNA (Fig. 1c, d). The SC35 protein accumulates in the prefrontal cortex (PFC) following acute stress and regulates the stress-induced alternative splicing of AChE mRNA in brain neurons (Meshorer et al. 2005). However, long-term excess of AChE induces a reduction in SC35 (Ben-Ari et al. 2006). Therefore, we selected the miR-183/SC35 regulatory module for a functional validation study.
Table 2.
PicTar-predicted targets common to miR-134 and miR-183
| Human RefSeq | miR-134 score | miR-183 score | Annotation |
|---|---|---|---|
| NM_012082 | 6.49 | 3.37 | Zinc finger protein, multitype 2 (ZFPM2) |
| NM_002211 | 5.61 | 5.93 | Integrin, beta 1 (ITGB1), transcript variant 1A |
| NM_019063 | 4.56 | 2.15 | Echinoderm microtubule-associated protein-like 4 (EML4) |
| NM_016279 | 4.12 | 3.51 | ZFPM2 |
| NM_004349 | 3.97 | 2.27 | Core-binding factor, runt domain, alpha subunit 2 (CBFA2T1), transcript variant 1 |
| NM_175634 | 3.97 | 2.27 | CBFA2T1 transcript variant 2 |
| NM_175635 | 3.97 | 2.27 | CBFA2T1 transcript variant 3 |
| NM_175636 | 3.97 | 2.27 | CBFA2T1 transcript variant 4 |
| NM_003016 | 3.9 | 2.22 | Splicing factor, arginine/serine-rich 2 (SFRS2, SC35) |
| NM_004384 | 3.21 | 2.91 | Casein kinase 1, gamma 3 (CSNK1G3) |
| NM_173683 | 2.99 | 2.48 | Chromosome 8 open reading frame 21 (C8orf21) |
| NM_182485 | 2.38 | 1.46 | Cytoplasmic polyadenylation element binding protein 2 (CPEB2), transcript variant B |
| NM_182646 | 2.38 | 1.46 | CPEB2 transcript variant A |
| NM_003605 | 2.36 | 2.97 | O-linked N-acetylglucosamine (GlcNAc) transferase (OGT), transcript variant 3 |
| NM_006561 | 2.32 | 1.34 | CUG triplet repeat, RNA binding protein 2 (CUGBP2) |
| NM_021038 | 2.22 | 1.55 | Muscle blind-like (Drosophila; MBNL1), transcript variant 1 |
| NM_207292 | 2.22 | 1.55 | MBNL1 transcript variant 2 |
| NM_207293 | 2.22 | 1.55 | MBNL1 transcript variant 3 |
| NM_207294 | 2.22 | 1.55 | MBNL1 transcript variant 4 |
| NM_207295 | 2.22 | 1.55 | MBNL1 transcript variant 5 |
| NM_207296 | 2.22 | 1.55 | MBNL1 transcript variant 6 |
| NM_207297 | 2.21 | 1.54 | MBNL1 transcript variant 7 |
ZPFM2 and CBFA2T1 are transcription factors (CBFA2T1 is involved with controlling oxidative stress and inflammation (Martinez et al. 2004)); integrin-β, casein kinase 1G3, and OGT are signaling molecules (integrin-β is involved in regulating sheer stress (Chen et al. 1999)); OGT controls heat shock reactions (Sohn et al. 2004). MBNL1, CUGBP2, and SFRS2 are known regulators of alternative splicing (CUGBP2 is involved in RNA editing (Anant et al. 2001); SC35 accumulates in the PFC following acute stress and regulates the stress-induced alternative splicing of AChE mRNA in brain neurons (Meshorer et al. 2005))
Manipulating miR-183 Levels Affects SC35 Levels in Cultured Cells
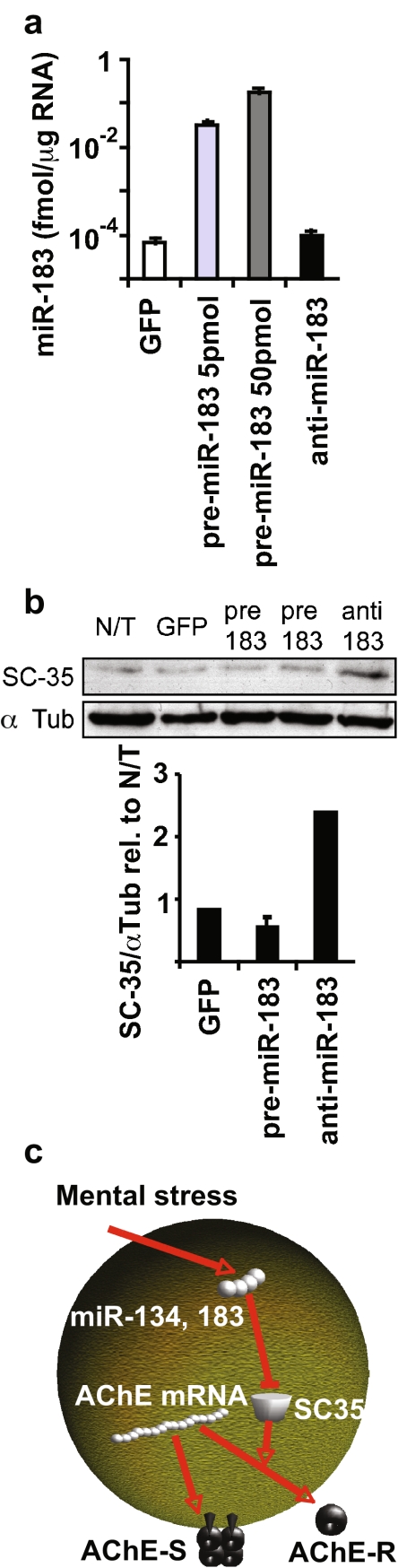
To test the direct effects of miR-183 on SC35 levels, CHO cells were transfected with a pre-miR-183 precursor stem loop, an anti-miR-183 oligo, or a GFP expression vector as negative control. Transfection with pre-miR-183 elevated miR-183 levels by up to four orders of magnitude; the QRT-PCR analysis did not differentiate between the mature and precursor forms (as tested in QRT-PCR calibration; data not shown). Anti-miR-183 did not alter the amount of endogenous miR-183 (Fig. 2a), consistent with a functional interference effect rather than degradation of the endogenous miR. Anti-miR-183 elevated SC35 protein levels by more than twofold (quantification of immunoblot, Fig. 2b). Conversely, pre-miR-183 suppressed SC35 protein levels, although not dramatically; this can be attributed to only partial processing of the exogenous precursor. Thus, tests in cultured cells validated miR-183-mediated suppression of SC35 protein levels.
Figure 2.
miR-183 regulates the splicing factor SC35. a QRT-PCR for miR-183 in CHO cells transfected with pre-miR-183 or anti-miR-183 (Ambion) and GFP-transfected controls. Bars: SD; t test p < 10−5 for both concentrations of pre-miR-183 compared to GFP control. b Scan and quantification of immunoblot for SC35 in cells in a with α-tubulin used for normalization. c Suggested interplay of alternative splicing and miRs (exemplified by miR-134, miR-183, and their target, the splicing factor SC35) contributes to the complexity of stress responses in the brain, including regulation of the alternative splicing choice between the major “synaptic” AChE transcript AChE-S and the normally minor stress-induced variant AChE-R
Discussion
The molecular mechanisms underlying the brain’s response to psychological stress are still incompletely understood. Posttranscriptional regulation by miRs can add flexibility and robustness to the control of alternative splicing, in line with the observed complexity of the mammalian stress response at the physiological level (McEwen 2007). In our current study, changes in miR expression profiles were characterized within the stress-responsive amygdala and the CA1 region of the hippocampus of rats subjected to immobilization stress. Microarray analysis and cell culture validation experiments identified specific miRs (e.g., miR-183, miR-134) as regulators of key splicing factors (e.g., SC35 which controls the stress-induced alternative splicing of AChE mRNA; scheme, Fig. 2c). Together, these findings support the notion that stress responses in the mammalian brain involve miR-mediated control of other posttranscriptional regulators of gene expression, including (but not limited to) proteins regulating alternative splicing.
miR-134, a previously reported regulator of dendritic spine development in the brain (Schratt et al. 2006), is elevated in both the hippocampus CA1 and the central amygdala under acute stress and downregulated in both these brain regions under chronic stress, while miR-183, also upregulated in both regions under acute stress, maintains unchanged levels in the chronically stressed hippocampus CA1 and central amygdala. This tentatively suggests the relevance of the observed miR-183 and miR-134 changes to both the acute and chronic stress-induced alterations in SC35. Apart from targeting the splicing factors SC35, MBNL1, and CUGBP2, both miR-134 and miR-183 are predicted to target other splicing factors (e.g., SRP46 and SFRS11, respectively), potentially expanding the impact of these miRs on the pre-mRNA splicing machinery. Both SRP46 and SFRS11 are SR proteins that participate in composing the multicomponent splicing complex and play pivotal roles in both regular and alternative splicing. SRP46 is an SC35 homolog (Soret et al. 1998), while SFRS11 regulates the alternative splicing of Tau, the missplicing of which causes frontotemporal dementia with Parkinsonism linked to chromosome 17, an autosomal dominant neurodegenerative disorder (Wu et al. 2006). Additionally, miR-183 is predicted to bind two sites on the 3′ UTR of profilin 2 (PFN2) mRNA. PFN2 regulates actin polymerization, determining dendritic spine morphology in neurons; its knockout severely affects neurotransmitter homeostasis and behavior in mice (Witke 2004). Elevation of PFN2 mRNA was observed in nucleated blood cells from bipolar-disorder-diagnosed twins in five monozygotic twin pairs, compared with their healthy twins (Matigian et al. 2007). Thus, miR-134 and miR-183 both appear to regulate known mediators of neuronal stress reactions, compatible with our current findings and relevant to stress-associated diseases.
There is compelling evidence that bipolar disorder, as well as schizophrenia and some neurosensory pathologies, involves hypercholinergic neurotransmission, and nAChRs were suggested as targets for treatment (McEvoy and Allen 2002; Shytle et al. 2002). The local and temporal regulations of cholinergic neurotransmission via the alternative splicing of AChE are therefore of high relevance to the understanding and treatment of mental stress and disease. We previously demonstrated a shift in the splicing pattern of AChE following acute stress (Kaufer et al. 1998). Furthermore, alternative splicing of the AChE mRNA transcript modifies the effect of the resultant protein product with regards to fibril formation from the APP C-terminal peptides, from an upregulation by the primary “synaptic” AChE-S protein (Inestrosa et al. 1996) to suppression by the stress-induced AChE-R variant (Berson et al. 2008). Our current findings suggest that SC35 mediated alternative splicing of AChE, and, downstream to it, the amyloid fibril formation outcome in neurodegeneration processes is subject to regulation by stress-responding miRs.
In conclusion, our data point at miR functions in regulating alternative splicing under mental stress. The micromanagement of alternative splicing thus merits special attention.
Acknowledgments
This research was supported by the Israel Science Fund, the European Union (LSH-2004-1.1.5-3) and STREP (LSHG-CT-2006-037277), The German Ministry of Science and German-Israel Project, DIP-G 3.2, and The Hebrew University's Interdisciplinary Center for Neuronal Computation (ICNC).The authors are grateful to Shani Ben-Ari and Lilach Soreq for their contributions.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Proceedings of the XIII International Symposium on Cholinergic Mechanisms
Contributor Information
Hermona Soreq, Phone: +972-2-6585109, FAX: +972-2-6520258, Email: soreq@cc.huji.ac.il.
Daniela Kaufer, Phone: +1-650-8144816, FAX: +1-510-6436264, Email: danielak@berkeley.edu.
References
- Anant S, Henderson JO, Mukhopadhyay D, Navaratnam N, Kennedy S, Min J, et al. Novel role for RNA-binding protein CUGBP2 in mammalian RNA editing. CUGBP2 modulates C to U editing of apolipoprotein B mRNA by interacting with apobec-1 and ACF, the apobec-1 complementation factor. Journal of Biological Chemistry. 2001;276:47338–47351. doi: 10.1074/jbc.M104911200. [DOI] [PubMed] [Google Scholar]
- Anguelova E, Boularand S, Nowicki JP, Benavides J, Smirnova T. Up-regulation of genes involved in cellular stress and apoptosis in a rat model of hippocampal degeneration. Journal of Neuroscience Research. 2000;59:209–217. doi: 10.1002/(SICI)1097-4547(20000115)59:2<209::AID-JNR7>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia M, Ogliari A. Anxiety and panic: from human studies to animal research and back. Neuroscience and Biobehavioral Reviews. 2005;29:169–179. doi: 10.1016/j.neubiorev.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Beheshti B, Braude I, Marrano P, Thorner P, Zielenska M, Squire JA. Chromosomal localization of DNA amplifications in neuroblastoma tumors using cDNA microarray comparative genomic hybridization. Neoplasia. 2003;5:53–62. doi: 10.1016/s1476-5586(03)80017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ari S, Toiber D, Sas AS, Soreq H, Ben-Shaul Y. Modulated splicing-associated gene expression in P19 cells expressing distinct acetylcholinesterase splice variants. Journal of Neurochemistry. 2006;97(Suppl 1):24–34. doi: 10.1111/j.1471-4159.2006.03725.x. [DOI] [PubMed] [Google Scholar]
- Berson A, Knobloch M, Hanan M, Diamant S, Sharoni M, Schuppli D, et al. Changes in readthrough acetylcholinesterase expression modulate amyloid-beta pathology. Brain. 2008;131:109–119. doi: 10.1093/brain/awm276. [DOI] [PubMed] [Google Scholar]
- Chen KD, Li YS, Kim M, Li S, Yuan S, Chien S, et al. Mechanotransduction in response to shear stress. Roles of receptor tyrosine kinases, integrins, and Shc. Journal of Biological Chemistry. 1999;274:18393–18400. doi: 10.1074/jbc.274.26.18393. [DOI] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Dombkowski AA, Thibodeau BJ, Starcevic SL, Novak RF. Gene-specific dye bias in microarray reference designs. FEBS Letters. 2004;560:120–124. doi: 10.1016/S0014-5793(04)00083-3. [DOI] [PubMed] [Google Scholar]
- Espinosa-Oliva, A. M., de Pablos, R. M., Villaran, R. F., Arguelles, S., Venero, J. L., Machado, A., et al. (2009) Stress is critical for LPS-induced activation of microglia and damage in the rat hippocampus. Neurobiology of Aging, in press. [DOI] [PubMed]
- Gattoni R, Mahe D, Mahl P, Fischer N, Mattei MG, Stevenin J, et al. The human hnRNP-M proteins: structure and relation with early heat shock-induced splicing arrest and chromosome mapping. Nucleic Acids Research. 1996;24:2535–2542. doi: 10.1093/nar/24.13.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa-Geffen A, Lacoste PP, Soreq L, Cizeron-Clairac G, Le Panse R, Truffault F, et al. The thymic theme of acetylcholinesterase splice variants in myasthenia gravis. Blood. 2007;109:4383–4391. doi: 10.1182/blood-2006-07-033373. [DOI] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert SS, Horre K, Nicolai L, Bergmans B, Papadopoulou AS, Delacourte A, et al. MicroRNA regulation of Alzheimer's amyloid precursor protein expression. Neurobiology of Disease. 2009;33:422–428. doi: 10.1016/j.nbd.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Hirata R, Matsumoto M, Judo C, Yamaguchi T, Izumi T, Yoshioka M, et al. Possible relationship between the stress-induced synaptic response and metaplasticity in the hippocampal CA1 field of freely moving rats. Synapse. 2009;63:549–556. doi: 10.1002/syn.20631. [DOI] [PubMed] [Google Scholar]
- Inestrosa NC, Alvarez A, Perez CA, Moreno RD, Vicente M, Linker C, et al. Acetylcholinesterase accelerates assembly of amyloid-beta-peptides into Alzheimer's fibrils: possible role of the peripheral site of the enzyme. Neuron. 1996;16:881–891. doi: 10.1016/S0896-6273(00)80108-7. [DOI] [PubMed] [Google Scholar]
- Kaufer D, Friedman A, Seidman S, Soreq H. Acute stress facilitates long-lasting changes in cholinergic gene expression. Nature. 1998;393:373–377. doi: 10.1038/30741. [DOI] [PubMed] [Google Scholar]
- Kenney JW, Gould TJ. Modulation of hippocampus-dependent learning and synaptic plasticity by nicotine. Molecular Neurobiology. 2008;38:101–121. doi: 10.1007/s12035-008-8037-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluiver J, van den Berg A, de Jong D, Blokzijl T, Harms G, Bouwman E, et al. Regulation of pri-microRNA BIC transcription and processing in Burkitt lymphoma. Oncogene. 2007;26:3769–3776. doi: 10.1038/sj.onc.1210147. [DOI] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial microRNA target predictions. Nature Genetics. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Laneve P, Di Marcotullio L, Gioia U, Fiori ME, Ferretti E, Gulino A, et al. The interplay between microRNAs and the neurotrophin receptor tropomyosin-related kinase C controls proliferation of human neuroblastoma cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:7957–7962. doi: 10.1073/pnas.0700071104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The microRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Molecular Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes and Development. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez N, Drescher B, Riehle H, Cullmann C, Vornlocher HP, Ganser A, et al. The oncogenic fusion protein RUNX1–CBFA2T1 supports proliferation and inhibits senescence in t(8;21)-positive leukaemic cells. BMC Cancer. 2004;4:44. doi: 10.1186/1471-2407-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matigian N, Windus L, Smith H, Filippich C, Pantelis C, McGrath J, et al. Expression profiling in monozygotic twins discordant for bipolar disorder reveals dysregulation of the WNT signalling pathway. Molecular Psychiatry. 2007;12:815–825. doi: 10.1038/sj.mp.4001998. [DOI] [PubMed] [Google Scholar]
- McEvoy JP, Allen TB. The importance of nicotinic acetylcholine receptors in schizophrenia, bipolar disorder and Tourette's syndrome. Current Drug Targets—CNS & Neurological Disorders. 2002;1:433–442. doi: 10.2174/1568007023339210. [DOI] [PubMed] [Google Scholar]
- McEwen BS. Stress and hippocampal plasticity. Annual Review of Neuroscience. 1999;22:105–122. doi: 10.1146/annurev.neuro.22.1.105. [DOI] [PubMed] [Google Scholar]
- McEwen BS. Physiology and neurobiology of stress and adaptation: Central role of the brain. Physiological Reviews. 2007;87:873–904. doi: 10.1152/physrev.00041.2006. [DOI] [PubMed] [Google Scholar]
- Meshorer E, Soreq H. Virtues and woes of AChE alternative splicing in stress-related neuropathologies. Trends in Neurosciences. 2006;29:216–224. doi: 10.1016/j.tins.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Meshorer E, Bryk B, Toiber D, Cohen J, Podoly E, Dori A, et al. SC35 promotes sustainable stress-induced alternative splicing of neuronal acetylcholinesterase mRNA. Molecular Psychiatry. 2005;10:985–997. doi: 10.1038/sj.mp.4001735. [DOI] [PubMed] [Google Scholar]
- Niwa R, Zhou F, Li C, Slack FJ. The expression of the Alzheimer's amyloid precursor protein-like gene is regulated by developmental timing microRNAs and their targets in Caenorhabditis elegans. Developmental Biology. 2008;315:418–425. doi: 10.1016/j.ydbio.2007.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto T, Eichenbaum H. Neuronal activity in the hippocampus during delayed non-match to sample performance in rats: Evidence for hippocampal processing in recognition memory. Hippocampus. 1992;2:323–334. doi: 10.1002/hipo.450020310. [DOI] [PubMed] [Google Scholar]
- Plasterk RH. Micro RNAs in animal development. Cell. 2006;124:877–881. doi: 10.1016/j.cell.2006.02.030. [DOI] [PubMed] [Google Scholar]
- Raymond CK, Roberts BS, Garrett-Engele P, Lim LP, Johnson JM. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA New York, NY. 2005;11:1737–1744. doi: 10.1261/rna.2148705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, et al. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Shytle RD, Silver AA, Sheehan KH, Sheehan DV, Sanberg PR. Neuronal nicotinic receptor inhibition for treating mood disorders: Preliminary controlled evidence with mecamylamine. Depression and Anxiety. 2002;16:89–92. doi: 10.1002/da.10035. [DOI] [PubMed] [Google Scholar]
- Simon DJ, Madison JM, Conery AL, Thompson-Peer KL, Soskis M, Ruvkun GB, et al. The microRNA miR-1 regulates a MEF-2-dependent retrograde signal at neuromuscular junctions. Cell. 2008;133:903–915. doi: 10.1016/j.cell.2008.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn KC, Lee KY, Park JE, Do SI. OGT functions as a catalytic chaperone under heat stress response: A unique defense role of OGT in hyperthermia. Biochemical and Biophysical Research Communications. 2004;322:1045–1051. doi: 10.1016/j.bbrc.2004.08.023. [DOI] [PubMed] [Google Scholar]
- Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soreq H, Seidman S. Acetylcholinesterase—New roles for an old actor. Nature Reviews Neuroscience. 2001;2:294–302. doi: 10.1038/35067589. [DOI] [PubMed] [Google Scholar]
- Soret J, Gattoni R, Guyon C, Sureau A, Popielarz M, Le Rouzic E, et al. Characterization of SRp46, a novel human SR splicing factor encoded by a PR264/SC35 retropseudogene. Molecular and Cellular Biology. 1998;18:4924–4934. doi: 10.1128/mcb.18.8.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, et al. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- Thai T-H, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, et al. Regulation of the germinal center response by MicroRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- Vreugdenhil E, Verissimo CS, Mariman R, Kamphorst JT, Barbosa JS, Zweers T, et al. MicroRNAs miR-18 and miR-124a downregulate the glucocorticoid receptor: Implications for glucocorticoid responsiveness in the brain. Endocrinology. 2009;150:2220–2228. doi: 10.1210/en.2008-1335. [DOI] [PubMed] [Google Scholar]
- Vyas A, Mitra R, Shankaranarayana Rao BS, Chattarji S. Chronic stress induces contrasting patterns of dendritic remodeling in hippocampal and amygdaloid neurons. Journal of Neuroscience. 2002;22:6810–6818. doi: 10.1523/JNEUROSCI.22-15-06810.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DL, Davis M. Role of the extended amygdala in short-duration versus sustained fear: A tribute to Dr. Lennart Heimer. Brain Structure and Function. 2008;213:29–42. doi: 10.1007/s00429-008-0183-3. [DOI] [PubMed] [Google Scholar]
- Witke W. The role of profilin complexes in cell motility and other cellular processes. Trends in Cell Biology. 2004;14:461–469. doi: 10.1016/j.tcb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Wood GE, Young LT, Reagan LP, McEwen BS. Acute and chronic restraint stress alter the incidence of social conflict in male rats. Hormones and Behavior. 2003;43:205–213. doi: 10.1016/S0018-506X(02)00026-0. [DOI] [PubMed] [Google Scholar]
- Wu JY, Kar A, Kuo D, Yu B, Havlioglu N. SRp54 (SFRS11), a regulator for tau exon 10 alternative splicing identified by an expression cloning strategy. Molecular and Cellular Biology. 2006;26:6739–6747. doi: 10.1128/MCB.00739-06. [DOI] [PMC free article] [PubMed] [Google Scholar]