Abstract
Six family transcription factor genes play multiple and crucial roles in the development of the vertebrate sensory system including the eye, olfactory epithelium and otic vesicle, and these genes are highly expressed in the neural crest-derived cranial mesenchymal cells in the mouse embryo. However, expression patterns have yet to be determined for the Six family genes in the developing tooth germ. In this study, we examined expression of six members of the Six family genes in the dental mesenchyme and the dental epithelium of the developing tooth germs in mice by in situ hybridization. We found dynamic expression patterns for Six1, Six2, Six4 and Six5 in the oral epithelium and mesenchymal cells with distinct expression patterns at the early stage before invagination of the dental epithelium. In addition, expression of Six1 and Six4 was observed in the inner enamel epithelium of the incisor and molar tooth germs at the cap stage. Expression of Six5 was maintained in the bell stage tooth germs, and intense expression of Six1 and Six4 was detected not only in the mesenchyme-derived dental follicle but also in the proliferating inner enamel epithelium of the labial cervical loop of the incisor tooth germ. Taken together, our results suggest that dynamic expression of Six family genes represents specific stages of the developing tooth germ. This dynamic expression is embodied in changes in both space and over time, and these changes in expression suggest that Six family genes may participate in tooth germ morphogenesis and the proliferation and/or differentiation of the incisor ameloblast stem/progenitor cells.
Keywords: ameloblast stem/progenitor cells, incisor, inner enamel epithelium, mesenchyme, molar, Six family genes
Introduction
Six family transcription factor genes have been found to be important in the development of the vertebrate sensory system, including the eye, olfactory epithelium and otic vesicle. Vertebrate Six family transcription factor genes are homologues of the Drosophila sine oculis gene, which is known to play an essential role in eye formation (Cheyette et al. 1994; Serikaku & O’Tousa, 1994), and six members (Six1–6) of this gene family have been identified in mammals (Oliver et al. 1995a,b;, Kawakami et al. 1996, 2000). The Six proteins are characterized by the Six domain and Six-type homeodomain, which are necessary for DNA binding and interaction with Eya proteins (Ikeda et al. 2002; Li et al. 2003). Six3 and Six6 (optx2) are necessary for the development of the eye and forebrain (Oliver et al. 1995a; Kobayashi et al. 1998; Toy et al. 1998; Jean et al. 1999; Lagutin et al. 2003). The lack of the Six1 gene is associated with decreased cell proliferation in the otic placode (Zheng et al. 2003; Ozaki et al. 2004), and loss of pioneer olfactory neurons (Ikeda et al. 2007; Chen et al. 2008). Six1 and Six4 are both required for the promotion of cell survival of the trigeminal sensory neurons (Konishi et al. 2006). Furthermore, Six1, Six2, Six4, and Six5 have been found to be highly expressed in the neural crest-derived mesenchymal cells (Oliver et al. 1995a; Ohto et al. 1998; Gray et al. 2004). However, the expression patterns of Six family genes have yet to be characterized in the developing tooth germ.
We selected the mouse to examine the developing tooth germ because rodent incisors grow continuously throughout the life span of the animal. A comparison can thus be made between murine incisors and molars, which may provide unique insights into development patterns, gene expression and cell differentiation. The incisor and molar tooth germs are formed at the proximal and distal positions of the maxilla and mandible through a series of epithelial–mesenchymal interactions mediated by the signaling pathway of the signaling molecules and transcription factors (Grobstein, 1967; Thesleff et al. 1995; Tucker et al. 1998; Tucker & Sharpe, 2004). In the early stage of murine tooth development, the signaling molecules FGF8 and BMP4 are secreted from the proximal and distal oral epithelium of the maxilla and mandible, respectively. These signaling molecules then induce expression of transcription factors and affect the fate of the underlying oral mesenchymal cells (Tucker et al. 1998). The discrete regions of the oral epithelium subsequently thicken and specialize into the dental placode. The dental placode invaginates into the underlying dental mesenchyme, which consists of mesodermal cells and neural crest-derived cells (NCCs) that have migrated from the anterior midbrain to the presumptive tooth-forming regions (Imai et al. 1996; Miletich & Sharpe, 2004; Yoshida et al. 2008). The condensed NCCs form the dental papilla and the dental follicle cells surrounding the enamel epithelium (Lumsden, 1988; Imai et al. 1996; Chai & Maxson, 2006). At the later stage, the surface cells of the invaginating dental placode form the enamel epithelium, including proliferating epithelial cells. These epithelial cells differentiate into enamel-secreting ameloblasts. At the same time, the mesenchymal cells within the dental papilla differentiate into both the dentin-producing odontoblasts and the dental follicle cells (Thesleff & Sharpe, 1997; Chai et al. 2000; Chai & Maxson, 2006; Fleischmannova et al. 2008).
Regeneration of teeth is limited in most mammals. However, some rodent species have continuously growing teeth (Thesleff et al. 2007). The continuous growth of murine incisors is caused by an increase in enamel-forming ameloblasts originating from a type of epithelial progenitor cells known as transit-amplifying cells (TA), which surface throughout the life of the animal. TAs are derived from ameloblast stem cells (ASCs), which are asymmetrically located in the stellate reticulum compartment of the labial cervical loop (Smith & Warshawsky, 1975; Harada et al. 1999; Thesleff et al. 2007). In contrast, the proliferation of epithelial cells in the lingual cervical loop is less than that of the labial epithelial cells, and these cells in the lingual cervical loop do not produce ameloblasts. The increase in the ameloblasts in the murine incisors but not in the molars suggests that distinct genetic pathways are involved in the control of cell proliferation and differentiation of the inner enamel epithelial cells of the incisor and molar tooth germs.
In this study, we have found expression of four members of the Six family in the dental mesenchymal and dental epithelial cells of the developing tooth germs by in situ hybridization. These four members, Six1, Six2, Six4 and Six5, exhibited dynamic expression patterns in the inner enamel epithelium, dental papilla and dental follicle of the mesenchyme at the cap and bell stages. Furthermore, Six1 and Six4 were asymmetrically expressed in the inner enamel epithelium of the incisor tooth germs in the posterior side of the labial cervical loop, which corresponds to the proliferation zone. These results suggest that changes in the combined expression levels of Six family genes represent specific stages of tooth germ development and that the Six family genes may regulate tooth germ morphogenesis and the proliferation and/or differentiation of ameloblast stem/progenitor cells in the developing incisors.
Materials and methods
Animals
Animal experiments were carried out in accordance with the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health. The Committee for Animal Experimentation of the Tohoku University Graduate School of Medicine approved the experimental procedures described herein. The midday of the vaginal plug was designated as embryonic day 0.5 (E0.5). Pregnant ICR mice were purchased from Charles River Japan (Yokohama, Japan).
Cloning of murine Six family cDNAs
Murine Six family cDNAs were amplified by RT-PCR using mRNA prepared from the E14.5 ICR mouse embryos (Six1–5′, Six1–3′, Six2–5′, Six2–3′, Six4–5′, Six4–3′, Six5–3′, and Six6–5′) or genomic PCR using adult liver DNA of ICR mice (Six5–5′). Primer sets were designed based on the mouse sequences in the DDBJ/EMBL/GenBank database (accession numbers: Six1, AK031122; Six2, BC068021; Six4, D50416; Six5, D83146, and Six6, AK029309). The oligonucleotide primers which were used are as follows: Six1–5′ (44–336 bp, 3′-GTCGATGCTGCCGTCGTTTG-5′ and 5′-GAAGTTTCTCGGCCTCCACGTAG-3′); Six1–3′ (558–571 bp, 5′-AAGAACCGGAGGCAAAGAGCA-3′ and 5′-TACCCTAACCGCCTCCCATAG-3′); Six2–5′ (302–821 bp, 5′-CATGTCCATGCTGCCCACCTTC-3′ and 5′-CTTGAACCAGTTGCTGACTTGCGT-3′); Six2–3′ (875–1426 bp, 5′-GAACTCCAATTCCAGCAGCCACAAC-3′ and TGCCTAGTTCAAGACTCGGTCTCTG-3′); Six4–5′ (887–1890 bp, 5′-GAGACCCAGTCCAAAAGCGAA-3′ and 5′-GCGGACACATTAACCAACGAC-3′); Six4–3′ (2067–2454 bp, TATTGTCCGGCCCGATGACC-3′ and 5′-GCTTCGCAAAGAAAAGGATTTGCTC-3′); Six5–5′ (627–1413 bp, 5′-CGAGTCTGATGGGAACCCCACTAC-3′ and 5′-TGGAGCAGTGGACGAGGGAA-3′); Six5–3′ (1637–2049 bp, 5′-GTCACACTCTGGGGCCAATC-3′ and 5′-AGCACCTTGACACCATTGTCAG-3′); and Six6–5′ (150–1111 bp, 5′-AAGTAGCCGGGGTATGTGAGAC-3′ and 5′-TTCTGTTCCAAAGGAGTCTTTGCAG-3′). Amplification was performed for 35 cycles under the following conditions: denaturation, 95 °C, 5 min; annealing, 60.8 °C (Six1–5′, Six4–1′), 63.5 °C (Six6–5′), 66.0 °C (Six1–3′, Six2–5′, Six2–3′, Six4–3′, Six5–5′, and Six5–3′), 1 min; and extension, 72 °C, 1 min. The amplified products were cloned by blunting these fragments using T4 DNA polymerase (Invitrogen, Carlsbad, CA) and inserting the fragments into the EcoRV site of pBluescriptII SK (−) (Stratagene, La Jolla, CA).
In situ hybridization
In situ hybridization on frozen sections for embryonic tissues was performed as described previously with minor modifications (Ishii et al. 1998). E10.5 and E13.5 ICR mouse heads were fixed in 4% paraformaldehyde (PFA)/phosphate-buffered saline (PBS) overnight at 4 °C. E16.5 and E18.5 mouse fetuses were fixed by perfusion with 4% PFA/PBS and additionally fixed in 4% PFA/PBS overnight at 4 °C. Embryos and fetuses were embedded in O.T.C. compound (Sakura, Tokyo), and cut into 12-μm sections with a cryostat (CM-3050, Leica, Nussloch, Germany). Digoxigenin (DIG)-labeled riboprobes were synthesized with T3 or T7 RNA polymerase (Promega, Madison, WI). Distinct riboprobes were generated from different cDNA fragments, and these riboprobes were used as a mixture to enhance the signals of Six1, Six2, Six4 and Six5. Alkaline phosphatase (AP)-conjugated anti-DIG antibody (1 : 5000, Roche) and NBT/BCIP (Wako Pure Chemical Industries, Osaka, Japan) were used to promote colour reactions. Images were recorded using a colour CCD camera (HC-25000 3CCD, Fuji, Japan).
BrdU labeling
For short-pulse labeling of S-phase cells using bromodeoxyuridine (BrdU, Sigma Chemical Co., St. Louis, MO), 50 mg mL−1 BrdU/PBS solution was directly injected into the abdominal cavity of pregnant mice. Two hours later, the fetuses were fixed by perfusion with 4% PFA/PBS and fixed in the same solution overnight at 4 °C. To detect BrdU-incorporated cells, sections were treated with a 2 N HCl solution for 30 min at 37 °C and neutralized in Tris-buffered saline Tween-20 (TBST). The sections were then blocked with 2% goat serum/TBST and subsequently incubated overnight at 4 °C with anti-BrdU mouse monoclonal antibody (1 : 50, Becton Dickinson, Mountain View, CA) which was diluted with 2% goat serum/TBST. A biotin-conjugated affinity purified anti-mouse IgG donkey antibody (1 : 400, Chemicon International, Inc., Temecula, CA) was used as a secondary antibody. The signal was enhanced using both an ABC kit (Vector Laboratories, Burlingame, CA) and enhanced DAB kit (Pierce, Rockford, IL). Images were recorded using an Axioplan fluorescent microscope equipped with a colour CCD camera (HC-25000 3 CCD, Fuji, Tokyo, Japan).
Results
Expression of Six family genes in the tooth-forming areas of the maxilla and mandible
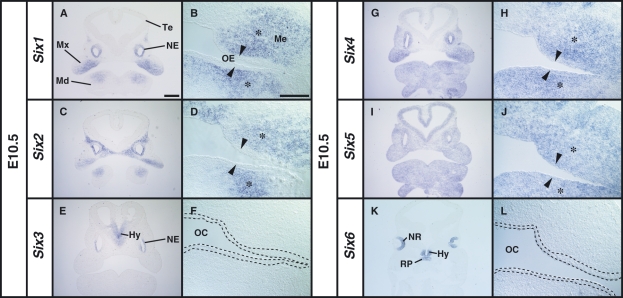
In situ hybridization on serial frontal sections revealed that four members of the Six family genes (Six1, Six2, Six4 and Six5) were expressed in the oral epithelium and the underlying mesenchyme with distinct spatial expression patterns at E10.5 (Fig. 1A–L). In contrast, Six3 and Six6 expression was not observed in the developing tooth-forming regions (Fig. 1E,F,K,L). Six1 was highly expressed in the proximal mesenchyme of the maxilla and mandible (Fig. 1A,B), and Six2 was expressed in the maxillary mesenchymal cells and in the proximal mandibular mesenchymal cells (Fig. 1C,D). The Six4 signal was detected proximally in the oral epithelium and mesenchyme of the maxilla, and its expression in the mandible was uniform in both the oral epithelium and the underlying mesenchyme (Fig. 1G,H). The Six4 expression domain partially overlapped with Six1 and Six2 domains (Fig. 1A,C,G). Six5 was uniformly expressed in the epithelium and mesenchyme of the maxilla and mandible (Fig. 1I,J). The sense RNA probes showed no hybridization signal for any Six family gene (data not shown). Our results revealed that Six1, Six2, Six4 and Six5 of the Six family genes are expressed in the tooth-forming regions, including the presumptive dental epithelium and the mesenchyme in the early stage of murine tooth development.
Fig. 1.
Localization of Six1, Six2, Six4 and Six5 mRNAs in the craniofacial region of the E10.5 mouse embryo before thickening of the dental epithelium. (B,D,F,H,J,L) High magnification images of the oral region of serial sections shown in A, C, E, G, I and K, respectively. (A,B) Expression of Six1 in the mesenchyme (Me) in the proximal region of maxillary (Mx) and mandibular (Md) arches (asterisk). No expression of Six1 in the oral epithelium (arrowhead). (C,D) Expression of Six2 in the Me of the maxillary and mandibular arches. (E,F) The expression of Six3 in the hypothalamus (Hy) and nasal epithelium (NE). The area within the broken line in F corresponds to the oral epithelium. (G,H) Expression of Six4 in the proximal mesenchymal cells (asterisk) and the oral epithelium (arrowhead) in the Mx and Md. (I,J) Uniform expression of Six5 in the craniofacial regions including the oral epithelium (arrow) and the mesenchyme (asterisk). (K,L) Expression of Six6 in the neural retina (NR), Hy and Rathke’s pouche (RP). Te, telencephalon; OE, oral epithelium; OC, oral cavity. Scale bars = 400 μm (A,C,E,G,I,K), 200 μm (B,D,F,H,J,L).
Six1 and Six2 are expressed in the dental mesenchyme, and Six4 and Six5 are expressed both in the dental epithelium and mesenchyme at the bud stage
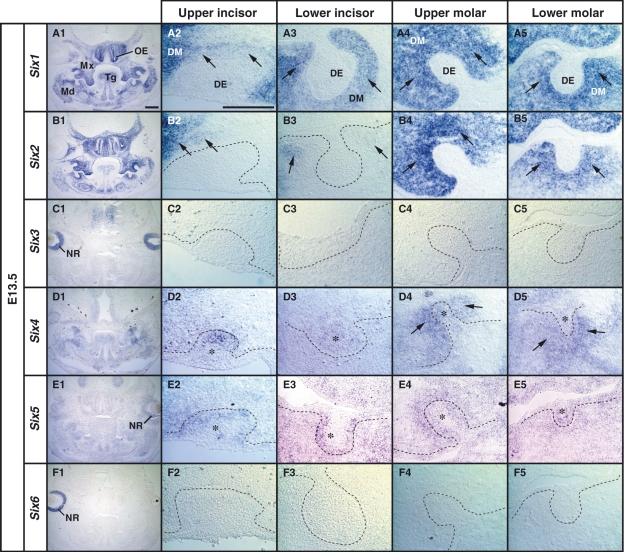
We next examined the expression patterns of Six family genes in the tooth germ at the bud stage (E13.5). Six1 expression was not detected in the dental epithelium of the upper and lower tooth germs (Fig. 2A2–A5). In the molar tooth germs, Six1 expression was detected in the dental mesenchyme facing the dental epithelium (Fig. 2A4,A5), and intense Six2 expression was detected in the dental mesenchymal cells with a pattern similar to Six1 (Fig. 2A4,A5,B4,B5). However, in the incisor-forming region, the Six2 signal was detected only in a population of mesenchymal cells surrounding the dental epithelium at a low expression level (Fig. 2B2,B3), and Six2 expression was not detected in the dental epithelium (Fig. 2B4,B5). Six4 and Six5 expression was observed in the dental epithelium and dental mesenchyme of the incisor and the molar tooth germs (Fig. 2D1–D5 and E1–E5). Six3 and Six6 expression was detected in the neural retina (Fig. 2C1,F1), but not in the tooth-forming regions (Fig. 2C2–C5 and F2–F5). These results suggest that Six1 and Six2 are the dominant Six genes expressed in the dental mesenchyme, and Six4 and Six5 are similarly expressed both in the dental epithelium and the mesenchyme at the bud stage.
Fig. 2.
Localization of Six1, Six2, Six4 and Six5 mRNAs in the tooth germ on serial frontal sections of E13.5 mouse embryo. (A1,B1,C1,D1,E1,F1) Low magnification images of expression patterns of Six family genes at the anterior level including first molars. (A2–A5) Expression of Six1 in condensed dental mesenchyme (DM) (arrow), and the exclusion from the dental epithelium (DE) in the upper and the lower tooth germs at the bud stage. (B2,B5) Partial expression of Six2 in the mesenchyme surrounding DE of the incisor tooth germs (arrows in B2,B3) and the intense expression in the DM of the upper and lower molar tooth germs in the posterior mandible (Md) (arrows in B4,B5). (C2–C5) Expression of Six3 in the neural retina (NR) as shown in C1. (D2–D5) Weak expression of Six4 in the DE of the upper and lower incisor tooth germs (asterisk in D2,D3), and in the DE (asterisk in D4,D5) and the DM (arrow in D4,D5) at the position of molars. (E2,E5) Expression of Six5 in the DE (asterisk) and mesenchyme surrounding the DE. (F2–F5) Expression of Six6 in the NR as shown in F1. OE, olfactory epithelium; Mx, maxilla; Tg, tongue. Scale bars = 400 μm (A1,B1,C1,D1,E1,F1), 200 μm (A2–A5,B2–B5,C2–C5,E2–E5,F2–F5).
Intense Six1 and Six4 signals appear in the inner enamel epithelium of the incisor and molar tooth germs at the late cap stage
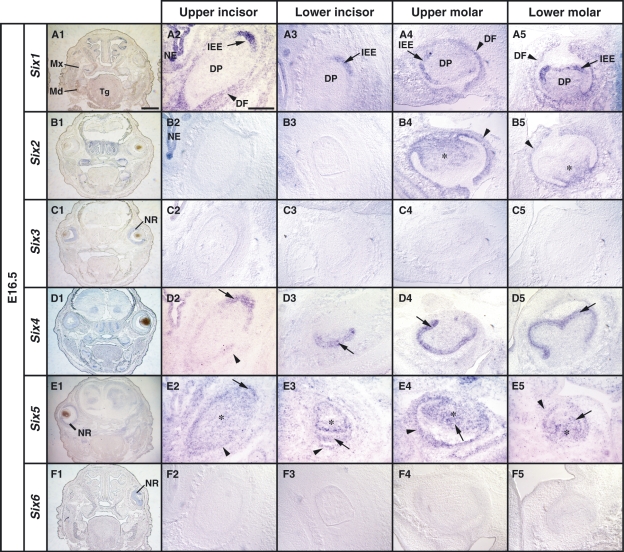
We next looked at the expression patterns of the Six family genes in the developing tooth germs at the late cap stage (E16.5). Interestingly, an intense Six1 signal emerged in the inner enamel epithelium of the upper and lower molar tooth germs. Moreover, Six1 was also expressed in the dental follicle (Fig. 3A4,A5). However, Six1 expression was not detected in the primordia of these epithelia at earlier stages (Fig. 1A,B). In the incisor tooth germs, Six1 expression was also detected in the epithelial cells forming the inner enamel epithelium (Fig. 3A2,A3) and was maintained in the dental mesenchyme forming the dental follicle of the molar and incisor tooth germs at lower levels (Fig. 3A2–A5). Six2 expression was maintained in the dental mesenchyme forming the dental papilla and the dental follicle in the molar tooth germs (Fig. 3B4,B5). Interestingly, we also observed pronounced expression of Six4 in the inner enamel epithelium of the molar and incisor tooth germs (Fig. 3D2–D5). We also detected Six4 expression in the dental follicle of the upper incisor tooth germ (Fig. 3D2). Six5 expression was detected in the inner enamel epithelium, outer enamel epithelium, stellate reticulum and dental mesenchyme (Fig. 3E1–E5). No expression of Six3 and Six6 was observed in the tooth germs (Fig. 3C2–C5 and F2–F5). These results suggest that Six1 and Six4 are highly expressed in the enamel epithelial tissue at the later cap stage.
Fig. 3.
Localization of Six1, Six2, Six4 and Six5 mRNAs in the tooth germs at the late cap stage (E16.5). (A1,B1,C1,D1,E1,F1) Low magnification images of expression patterns of Six family genes at the anterior level including first molars on serial frontal sections. (A2–A5) Six1 expression in the inner enamel epithelium (IEE) of the upper incisor tooth germ (arrow in A2,A3), and in the IEE and dental follicle (DF) in the upper and lower molar tooth germs (arrow and arrowhead in A4,A5). (B2–B5) Restricted expression of Six2 in the dental papilla (DP) and DF (asterisk and arrowhead in B4,B5, respectively). (C2–C5) No expression of Six3 mRNA in any tooth germs. (D2–D5) Expression of Six4 in the IEE and DF in the upper incisor tooth germ (arrow and arrowhead in D2,D3) and the intense expression in the IEE of the molar tooth germs (arrow in D4,D5). (E2–E5) Six5 is expressed in the neural retina (NR) shown as in E1. Expression of Six5 in the IEE (arrow in E2,E3) and DF (arrowhead in E2–E5) in the tooth germs, and the expression in the DP (asterisk in E2–E5). (F2–F5) Expression of Six6 in the NR shown as in F1. NE, nasal epithelium; Md, mandible; Mx, maxilla; Tg, tongue. Scale bars = 400 μm (A1,B1,C1,D1,E1,F1), 200 μm (A2–A5,B2–B5,C2–C5,E2–E5,F2–F5).
Expression of Six1 and Six4 is maintained in the inner enamel epithelium of the incisor and molar tooth germs at the bell stage
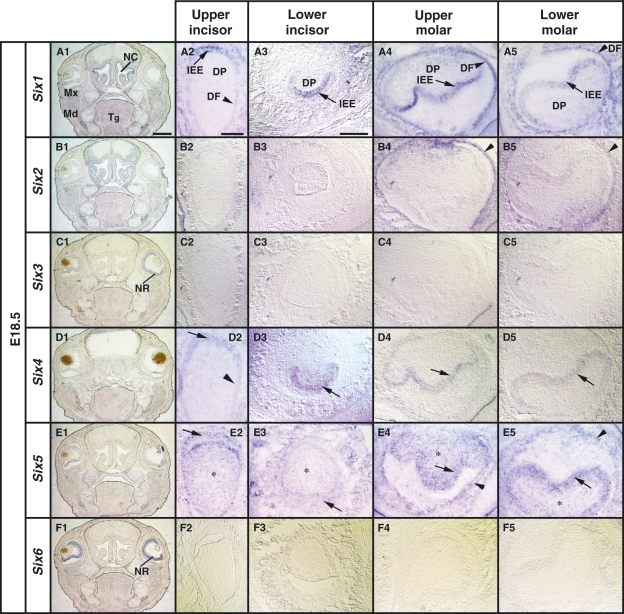
The morphological structure of the tooth germs changes into the bell shape with cell proliferation and differentiation in the later stages. At E18.5, the Six1 and Six4 expression in the inner enamel epithelium was maintained both in the incisor and the molar tooth germs (Fig. 4A2–A5 and D2–D5). Six1 exhibited different expression levels in individual epithelial cells in the molar tooth germs (Fig. 4A4,A5). In contrast to the Six1 expression, Six4 was uniformly expressed in the inner enamel epithelium in the molar tooth germs (Fig. 4D4,D5). Six1 expression was also detected in the dental follicle of the molar tooth germs (Fig. 4A4,A5). Six2 expression was observed in the dental papilla and the dental follicle of the molar tooth germs (Fig. 4B4,B5). The Six5 signal was detected in the inner enamel epithelium, outer enamel epithelium, and stellate reticulum and in the peripheral region of the dental papilla (Fig. 4E1–E5). These results demonstrate that Six family gene expression overlaps in the inner epithelium and the dental follicle mesenchyme of the developing tooth germs at the bell stage, and Six1 and Six4 expression is maintained in the inner epithelium from the late cap to the bell stage.
Fig. 4.
Localization of Six1, Six2, Six4 and Six5 mRNA on frontal sections at the bell stage (E18.5). (A1,B1,C1,D1,E1,F1) Low magnification images of expression patterns of the Six family genes at the anterior level including first molar tooth germs. (A1–A5) Expression of Six1 in the inner enamel epithelium (IEE) of the upper incisor tooth germ (arrow in A2,A3) and the IEE in the molar tooth germs (arrow in A4,A5). The expression of Six1 is detected in the dental follicle (DF) of the incisor and the molar tooth germs (arrowhead in A2,A4,A5). (B2–B5) Expression of Six2 in the DF (arrowhead in B4,B5) in the molar but not in the incisor tooth germs (B2,B3). (C2–C5) No expression of Six3 in the tooth germs. (D2–D5) Expression of Six4 in the IEE of the upper incisor and the molar tooth germs (arrow in D2–D5). (E2–E5) Expression of Six5 in the IEE of the incisor tooth germs (arrow in E2,E3) and the IEE and DF of the molar tooth germs (arrow and arrowhead in E4,E5). Asterisk indicates the expression of Six5 in the DF. (F2–F5) No expression of Six6 in the tooth germs. Md, mandible; Mx, maxilla; Tg, tongue; NC, nasal cavity; NR, neural retina. Scale bars = 400 μm (A1,B1,C1,D1,E1,F1), 200 μm (A2–A5,B2–B5,C2–C5,E2–E5,F2–F5).
Six1 and Six4 are expressed in the ameloblast stem/progenitor cells at the labial cervical loop of the incisor tooth germs
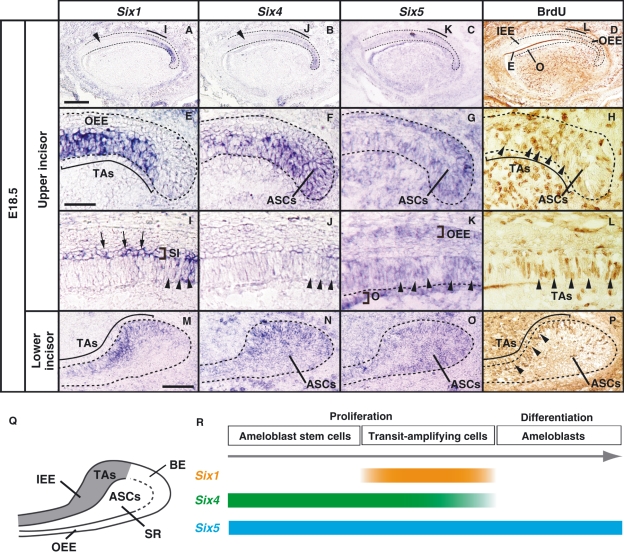
The expression of amelogenin (Amelx), a gene crucial for enamel formation, has been detected at the anterior (apical) region of the labial inner enamel epithelium at E16.5 in the murine incisor tooth germs (Klein et al. 2008). The presence of Amelx suggests that the anterior enamel epithelial cells differentiate into ameloblasts and that the posterior (proximal) enamel epithelial cells remain undifferentiated as the ameloblast stem/progenitor cells. To determine whether the expression of Six1 and Six4 observed in the incisor enamel epithelium could be correlated with cell proliferation, we compared the expression domains of Six1 and Six4 with proliferation regions along the antero–posterior axis by the BrdU-pulse labeling method at E18.5 (Fig. 5). We found that Six1 expression was higher in the posterior region of the upper and the lower incisor germs, and this expression pattern overlapped with the BrdU-incorporated proliferation region in which TAs reside (Fig. 5A,D,E,H,I,L,M,P). Furthermore, Six1 expression was also detected in the stratum intermedium, which is located between the inner enamel epithelium and newly forming cells of the stellate reticulum (Fig. 5I). We also found that Six4 expression was detected in the posterior inner enamel epithelium, which included TAs, but not in the anterior inner enamel epithelium of the incisor tooth germs at E18.5 (Fig. 5B,D,F,H,J,L,N,P). Six1 and Six4 expression decreased in anterior TAs, which were labeled with BrdU (Fig. 5I,J,L). Six1 and Six5 were expressed in the stellate reticulum including ASCs (Fig. 5F,G,H,N,O,P). Six5 was also detected in the anterior enamel epithelium including differentiated ameloblasts, and in the dental follicle and odontoblast (Fig. 5C,G,K,O). Six1 and Six4 expression was only detected in the labial side of the upper and lower incisor tooth germs (Fig. 5A,B and Table 1). Therefore, these findings indicate that epithelial cells expressing Six1 and Six4 would likely include proliferating ameloblast stem/progenitor cells.
Fig. 5.
Comparative analysis of Six1/Six4/Six5 expression and the distribution of BrdU-positive cells in the incisor tooth germs along the anterior–posterior axis at E18.5 using serial sagittal sections. (A–D) In the upper incisor tooth germ, the expression of Six1 and Six4 is restricted to the posterior inner enamel epithelium including BrdU-incorporated cells (D). Six1 and Six4 signals are not detected in the BrdU-negative anterior inner enamel epithelium (arrowhead in A,B). Six5 is detected in the enamel epithelium and mesenchymal cells. (E–H) High magnification images of the cervical loop of the upper incisor tooth germ shown as in A–D. Six1 expression is detected in the posterior enamel epithelium (E) corresponding to the area in which BrdU-labeled transit-amplifying cells (TAs) reside (arrowheads in H). Six4 is expressed in the posterior region including both TAs and ameloblast stem cells (ASCs) (F,H). (I–L) Six1 and Six4 were expressed in the epithelium (arrowheads in I,J) including BrdU-labeled TAs (arrowheads in L) and become decreased in the anterior region. Six1 expression is also detected in the stratum intermedium (SI) (arrows in I). Six5 is detected in the outer enamel epithelium (OEE), inner enamel epithelium (IEE) (arrowheads in K), and the odontoblast (K). (M–P) In the cervical loop of the lower incisor tooth germ, Six1 expression is restricted in TAs (M,P). Six4 and Six5 are detected in TAs and the stellate reticulum in which ASCs reside (N,O,P). (Q) Localization of TAs and ASCs in the labial cervical loop of the lower incisor. (R) Time course of Six1/Six4/Six5 in the incisor tooth germs. E, enamel; O, odontoblasts; BE, basal epithelium. Scale bars = 100 μm (A–I), 200 μm (E–L), 100 μm (M–P).
Table 1.
Expression of Six1, Six2, Six4 and Six5 in murine incisors and molars.
| Age | Region | Six1 | Six2 | Six4 | Six5 | ||
|---|---|---|---|---|---|---|---|
| E10.5 | Medial | Upper | Oral epithelium | − | − | − | + |
| Oral mesenchyme | − | + | − | + | |||
| Lower | Oral epithelium | − | − | + | + | ||
| Oral mesenchyme | − | − | + | + | |||
| Proximal | Upper | Oral epithelium | − | − | + | + | |
| Oral mesenchyme | + | + | + | + | |||
| Lower | Oral epithelium | − | − | + | + | ||
| Oral mesenchyme | + | + | + | + | |||
| E13.5 (bud stage) | Incisors | Upper | Dental epithelium | − | − | + | + |
| Dental mesenchyme | + | +* | − | + | |||
| Lower | Dental epithelium | − | − | + | + | ||
| Dental mesenchyme | + | +* | − | + | |||
| Molars | Upper | Dental epithelium | − | − | + | + | |
| Dental mesenchyme | + | + | + | + | |||
| Lower | Dental epithelium | − | − | + | + | ||
| Dental mesenchyme | + | + | + | + | |||
| E16.5/E18.5 (late cup stage/bell stage) | Incisors | Upper | Inner enamel epithelium | +** | − | +** | + |
| Outer enamel epithelium | − | − | − | + | |||
| Basal epithelium | − | − | + | + | |||
| Stellate reticulum | − | − | +*** | + | |||
| Dental follicle | + | − | + | + | |||
| Dental papilla | − | − | − | + | |||
| Lower | Inner enamel epithelium | +** | − | +** | + | ||
| Outer enamel epithelium | − | − | − | + | |||
| Basal epithelium | +* | − | + | + | |||
| Stellate reticulum | − | − | +*** | + | |||
| Dental follicle | − | − | − | + | |||
| Dental papilla | − | − | − | + | |||
| Molars | Upper | Inner enamel epithelium | + | − | + | + | |
| Outer enamel epithelium | − | − | − | + | |||
| Stellate reticulum | − | − | − | + | |||
| Dental follicle | + | + | − | + | |||
| Dental papilla | +* | +* | − | + | |||
| Lower | Inner enamel epithelium | + | − | + | + | ||
| Outer enamel epithelium | − | − | − | + | |||
| Stellate reticulum | − | − | − | + | |||
| Dental follicle | + | + | − | + | |||
| Dental papilla | +* | +* | − | + |
*Partial expression.
**Expression in the posterior epithelial cells on the labial side.
***Expression on the labial side.
Discussion
Expression of Six family genes during murine tooth development
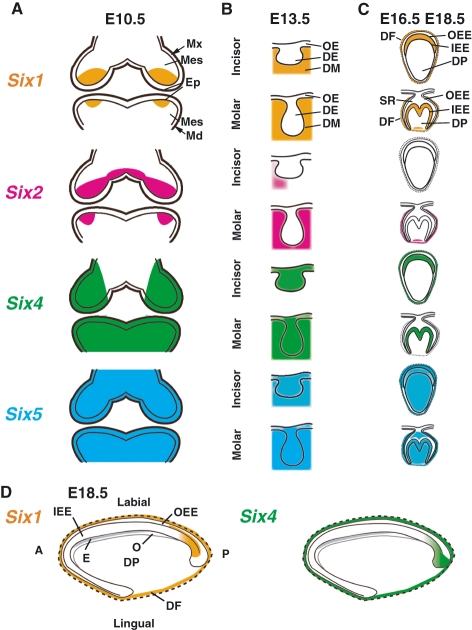
Previous reports suggest that maxillary and mandibular mesenchymal cells highly express Six1, Six2, and Six4 in the mouse embryo, and these genes are simultaneously in the same region in the early developmental stages (Oliver et al. 1995b; Ohto et al. 1998; Kawakami et al. 2000). We found that Six1, Six2 and Six4 overlapped in the proximal mesenchyme, whereas Six1, Six2 and Six4 were differentially expressed in the distal mesenchyme in the E10.5 maxillary and mandibular arches (summarized in Table 1 and Fig. 6A). Furthermore, while Six4 was also found in the oral epithelium including the presumptive dental epithelium, Six1 expression was restricted to the oral mesenchyme. The expression of Six5 in E10.5 mouse pharyngeal region has been suggested by whole-mount staining (Gray et al. 2004), and we also clearly demonstrated that Six5 is expressed not only in the cranial mesenchymal cells but also in the oral epithelial cells. Taken together, the expression patterns of Six1, Six2, Six4, and Six5 suggest that Six family genes may have a role in regionalization of the cranial mesenchyme, possibly in a redundant manner, at the early stage of tooth development.
Fig. 6.
Schematic illustration showing expression patterns of Six family genes during murine tooth development. (A) Expression patterns at E10.5. (B) Expression patterns in the lower incisor and molar tooth germs at the bud stage (E13.5). (C) Expression patterns in the upper incisor and lower molar tooth germs at the late cap (E16.5) and the bell (E18.5) stages. (D) Expression patterns in the upper incisor germs at E18.5 along the anterior–posterior (apical–proximal) axis. Six1 expression, orange; Six2 expression, pink; Six4 expression, green; Six5 expression, blue; Md, mandible; Mx, maxilla; Mes, mesenchyme; OE, oral epithelium; DE, dental epithelium; DM, dental mesenchyme; DF, dental follicle; IEE, inner enamel epithelium; OEE, outer enamel epithelium; O, odontoblast; SR, stellate reticulum; DP, dental papilla; A, anterior; P, posterior.
The shape of the tooth germ changes through a series of epithelial–mesenchymal interactions. At the bud stage (E13.5), Six1 and Six2 transcripts were widely found in the maxillary and mandibular mesenchymal tissues. However, Six2 expression was not detected in most of the dental mesenchymal cells surrounding the dental epithelium of the upper and lower incisor tooth germs, in contrast to Six1 and Six4 expression. Therefore, development of the incisor and molar tooth germs seems to be characterized by distinct combinations of Six family gene expression.
At the late cap stage (E16.5), Six4 expression diminishes in the dental follicle cells of the molar tooth germ, while Six1 is expressed in the inner enamel epithelial cells in the molar tooth germ. Our results indicate that the expression levels of Six1 are unequal in the E16.5 and E18.5 molar inner enamel epithelial cells, whereas Six4 expression is detected in these cells at equal expression levels. This observation raises the possibility that Six4 expression initially starts in the undifferentiated epithelial cells, and that Six1 expression may be detected in differentiated cells such as the preameloblasts in ameloblast development.
Comparative analysis of Six family gene expression also revealed that individual members of the Six family are expressed in specific regions and stages of the molar and incisor tooth germs. An intense Six1 signal was detected in the stratum intermedium in incisor tooth germs at the bell stage. Six5 expression was pronounced in the mesenchyme-derived dental papilla and odontoblasts in incisor and molar tooth germs at the cup and bell stages, whereas Six1, Six2, and Six4 were downregulated in the same regions and stages. Thus, Six1 and Six5 may possess specific and discrete functions that operate in the later development of dental epithelial and mesenchymal cells.
Possible roles of Six1 and Six4 in the ameloblast stem/progenitor cells in the incisor tooth germ
In the rodent incisor tooth germ, ASCs in the stellate reticulum compartment of the labial cervical loop produce TAs, and these cells anteriorly translocate and differentiate into enamel-forming ameloblasts. Recent studies have shown that the FGF3 and FGF10 signaling molecules from the dental papilla mesenchyme promote the proliferation of ASCs and TAs in the labial cervical loop, while follistatin and Sprouty inhibit cell proliferation of epithelial cells in the lingual cervical loop through inhibition of the BMP and FGF signaling pathways (Harada et al. 2002; Wang et al. 2004, 2007; Klein et al. 2008). In the mouse, transcripts of Amelx have been detected in the anterior inner enamel epithelium at E16.5 (Klein et al. 2008). This observation suggests that the anterior epithelial cells start to differentiate into ameloblasts that secrete enamel matrix, which is independent of ameloblast formation from epithelial stem cells in the cervical loop (Lesot et al. 2002). The intense expression of Six1 is notably only detected in the labial-posterior side of the incisor tooth germ, which includes BrdU-positive TAs (Harada et al. 1999, 2002; Wang et al. 2007). Furthermore, Six4 expression also becomes restricted to the labial-posterior epithelium and the stellate reticulum in which BrdU-positive TAs/ASCs reside. In contrast to Six1 and Six4 expression, Six5 expression was detected in the inner enamel epithelium and stellate reticulum on both the labial and lingual sides. These results suggest that Six1 and Six4 rather than Six5 contribute to ameloblast proliferation and differentiation after E16.5.
It has been reported that Six1 and Six4 are involved in the regulation of proliferation and differentiation of cells derived from the sensory placodes and the neural crest (Zheng et al. 2003; Ozaki et al. 2004; Ikeda et al. 2007; Chen et al. 2008). For example, apoptosis increased in the otic vesicle epithelium in the Six1 homozygous mutant embryo (Ozaki et al. 2004), and Hes1 and Hes5, which inhibit neuronal differentiation, were upregulated in the olfactory epithelium (Ikeda et al. 2007), suggesting that Six1 has bimodal functions in the morphogenesis of various tissues. Six1 is also necessary for cyclinA1 expression in the embryonic mammary gland, and Six1-overexpression facilitates cell proliferation in mouse embryonic fibroblasts (Coletta et al. 2004). Therefore, it is possible that Six4 maintains stemness of ASCs in the labial cervical loop, and that Six1 and Six4 cooperatively regulate the proliferation and/or differentiation of TAs.
Interestingly, the region expressing Six1 faces the FGF3-producing dental mesenchyme in the incisor tooth germs while Six4 expression is extended into the basal epithelium neighboring FGF10-producing mesenchymal cells (Harada et al. 2002). FGF10 is known to inhibit apoptosis of ASCs and to maintain a stem cell pool of self-renewing ASCs (Harada et al. 2002). However, transcription factors regulated by FGF signaling pathway and markers of TAs/ASCs have not yet been identified in the incisor tooth germs. In the wild-type mice, Fgf3 is asymmetrically expressed only in the labial side by E16.5 (Harada et al. 1999). In contrast, the Fgf3 expression was ectopically observed in the lingual mesenchyme of the Sprouty4 mutant mice at E16.5, suggesting that epithelial–mesenchymal FGF signaling loop in the labial incisor establishes ASCs in the cervical loop by E16.5 (Klein et al. 2008). The Six1 and Six4 genes are most likely regulated by FGF3 and FGF10 on the labial side in light of the onset of expression of these two genes in the incisor epithelium, and Six1 is characterized as a first TAs marker, although the role of Six1 in ameloblast formation remains to be studied.
Concluding remarks
We found that dynamic changes in the combinations of Six1, Six2, Six4 and Six5 expression represent specific stages of murine tooth germ development, and the expression of Six1 and Six4 was also detected in the inner enamel epithelium of the molar and incisor tooth germs in the later stages. Furthermore, the expression of Six1 and Six4 became restricted to the proliferation zone within the labial incisor cervical loop in which the epithelial ameloblast stem/progenitor cells reside.
Acknowledgments
We thank Drs Ahmed Mansouri and Peter Gruss for a gift of mouse Six3 plasmid. We also thank Drs Keiko Ikeda and Satoshi Fukumoto for their critical reading of the manuscript and valuable comments. We also thank Dr Hiroshi Yajima and all members of the Osumi Laboratory for valuable comments and discussions. K.N. was supported by the Global COE Program ‘Basic and Translational Research Center for Global Brain Science’ of MEXT as a research associate.
Author contributions
K.N., Y.W. and M.T. designed experiments. K.N. and M.T. carried out all experiments. K.N., N.O. and M.T. wrote the manuscript. K.N., M.T., Y.W., N.O. and T.T-Y. discussed the results. All authors read and approved the final manuscript.
References
- Chai Y, Maxson RE., Jr Recent advances in craniofacial morphogenesis. Dev Dyn. 2006;235:2353–2375. doi: 10.1002/dvdy.20833. [DOI] [PubMed] [Google Scholar]
- Chai Y, Jiang X, Ito Y, et al. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- Chen B, Kim EH, Xu PX. Initiation of olfactory placode development and neurogenesis is blocked in mice lacking both Six1 and Six4. Dev Biol. 2008;326:75–85. doi: 10.1016/j.ydbio.2008.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheyette BN, Green PJ, Martin K, et al. The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron. 1994;12:977–996. doi: 10.1016/0896-6273(94)90308-5. [DOI] [PubMed] [Google Scholar]
- Coletta RD, Christensen K, Reichenberger KJ, et al. The Six1 homeoprotein stimulates tumorigenesis by reactivation of cyclin A1. Proc Natl Acad Sci U S A. 2004;101:6478–6483. doi: 10.1073/pnas.0401139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmannova J, Matalova E, Tucker AS, et al. Mouse models of tooth abnormalities. Eur J Oral Sci. 2008;116:1–10. doi: 10.1111/j.1600-0722.2007.00504.x. [DOI] [PubMed] [Google Scholar]
- Gray PA, Fu H, Luo P, et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–2257. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- Grobstein C. Mechanisms of organogenetic tissue interaction. Natl Cancer Inst Monogr. 1967;26:279–299. [PubMed] [Google Scholar]
- Harada H, Kettunen P, Jung HS, et al. Localization of putative stem cells in dental epithelium and their association with Notch and FGF signaling. J Cell Biol. 1999;147:105–120. doi: 10.1083/jcb.147.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Toyono T, Toyoshima K, et al. FGF10 maintains stem cell compartment in developing mouse incisors. Development. 2002;129:1533–1541. doi: 10.1242/dev.129.6.1533. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Watanabe Y, Ohto H, et al. Molecular interaction and synergistic activation of a promoter by Six, Eya, and Dach proteins mediated through CREB binding protein. Mol Cell Biol. 2002;22:6759–6766. doi: 10.1128/MCB.22.19.6759-6766.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Ookawara S, Sato S, et al. Six1 is essential for early neurogenesis in the development of olfactory epithelium. Dev Biol. 2007;311:53–68. doi: 10.1016/j.ydbio.2007.08.020. [DOI] [PubMed] [Google Scholar]
- Imai H, Osumi-Yamashita N, Ninomiya Y, et al. Contribution of early-emigrating midbrain crest cells to the dental mesenchyme of mandibular molar teeth in rat embryos. Dev Biol. 1996;176:151–165. doi: 10.1006/dbio.1996.9985. [DOI] [PubMed] [Google Scholar]
- Ishii Y, Rex M, Scotting PJ, et al. Region-specific expression of chicken Sox2 in the developing gut and lung epithelium: regulation by epithelial–mesenchymal interactions. Dev Dyn. 1998;213:464–475. doi: 10.1002/(SICI)1097-0177(199812)213:4<464::AID-AJA11>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Jean D, Bernier G, Gruss P. Six6 (Optx2) is a novel murine Six3-related homeobox gene that demarcates the presumptive pituitary/hypothalamic axis and the ventral optic stalk. Mech Dev. 1999;84:31–40. doi: 10.1016/s0925-4773(99)00068-4. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Ohto H, Takizawa T, et al. Identification and expression of six family genes in mouse retina. FEBS Lett. 1996;393:259–263. doi: 10.1016/0014-5793(96)00899-x. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Sato S, Ozaki H, et al. Six family genes – structure and function as transcription factors and their roles in development. BioEssays. 2000;22:616–626. doi: 10.1002/1521-1878(200007)22:7<616::AID-BIES4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Klein OD, Lyons DB, Balooch G, et al. An FGF signaling loop sustains the generation of differentiated progeny from stem cells in mouse incisors. Development. 2008;135:377–385. doi: 10.1242/dev.015081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Toyama R, Takeda H, et al. Overexpression of the forebrain-specific homeobox gene six3 induces rostral forebrain enlargement in zebrafish. Development. 1998;125:2973–2982. doi: 10.1242/dev.125.15.2973. [DOI] [PubMed] [Google Scholar]
- Konishi Y, Ikeda K, Iwakura Y, et al. Six1 and Six4 promote survival of sensory neurons during early trigeminal gangliogenesis. Brain Res. 2006;1116:93–102. doi: 10.1016/j.brainres.2006.07.103. [DOI] [PubMed] [Google Scholar]
- Lagutin OV, Zhu CC, Kobayashi D, et al. Six3 repression of Wnt signaling in the anterior neuroectoderm is essential for vertebrate forebrain development. Genes Dev. 2003;17:368–379. doi: 10.1101/gad.1059403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesot H, Kieffer-Combeau S, Fausser JL, et al. Cell–cell and cell–matrix interactions during initial enamel organ histomorphogenesis in the mouse. Connect Tissue Res. 2002;43:191–200. doi: 10.1080/03008200290000529. [DOI] [PubMed] [Google Scholar]
- Li X, Oghi KA, Zhang J, et al. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003;426:247–254. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- Lumsden AG. Spatial organization of the epithelium and the role of neural crest cells in the initiation of the mammalian tooth germ. Development. 1988;103(Suppl):155–169. doi: 10.1242/dev.103.Supplement.155. [DOI] [PubMed] [Google Scholar]
- Miletich I, Sharpe PT. Neural crest contribution to mammalian tooth formation. Birth Defects Res C Embryo Today. 2004;72:200–212. doi: 10.1002/bdrc.20012. [DOI] [PubMed] [Google Scholar]
- Ohto H, Takizawa T, Saito T, et al. Tissue and developmental distribution of Six family gene products. Int J Dev Biol. 1998;42:141–148. [PubMed] [Google Scholar]
- Oliver G, Mailhos A, Wehr R, et al. Six3, a murine homologue of the sine oculis gene, demarcates the most anterior border of the developing neural plate and is expressed during eye development. Development. 1995a;121:4045–4055. doi: 10.1242/dev.121.12.4045. [DOI] [PubMed] [Google Scholar]
- Oliver G, Wehr R, Jenkins NA, et al. Homeobox genes and connective tissue patterning. Development. 1995b;121:693–705. doi: 10.1242/dev.121.3.693. [DOI] [PubMed] [Google Scholar]
- Ozaki H, Nakamura K, Funahashi J, et al. Six1 controls patterning of the mouse otic vesicle. Development. 2004;131:551–562. doi: 10.1242/dev.00943. [DOI] [PubMed] [Google Scholar]
- Serikaku MA, O’Tousa JE. sine oculis is a homeobox gene required for Drosophila visual system development. Genetics. 1994;138:1137–1150. doi: 10.1093/genetics/138.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE, Warshawsky H. Histological and three dimensional organization of the odontogenic organ in the lower incisor of 100 gram rats. Am J Anat. 1975;142:403–429. doi: 10.1002/aja.1001420402. [DOI] [PubMed] [Google Scholar]
- Thesleff I, Sharpe P. Signalling networks regulating dental development. Mech Dev. 1997;67:111–123. doi: 10.1016/s0925-4773(97)00115-9. [DOI] [PubMed] [Google Scholar]
- Thesleff I, Vaahtokari A, Kettunen P, et al. Epithelial-mesenchymal signaling during tooth development. Connect Tissue Res. 1995;32:9–15. doi: 10.3109/03008209509013700. [DOI] [PubMed] [Google Scholar]
- Thesleff I, Wang XP, Suomalainen M. Regulation of epithelial stem cells in tooth regeneration. C R Biol. 2007;330:561–564. doi: 10.1016/j.crvi.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Toy J, Yang JM, Leppert GS, et al. The optx2 homeobox gene is expressed in early precursors of the eye and activates retina-specific genes. Proc Natl Acad Sci U S A. 1998;95:10643–10648. doi: 10.1073/pnas.95.18.10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker A, Sharpe P. The cutting-edge of mammalian development; how the embryo makes teeth. Nat Rev Genet. 2004;5:499–508. doi: 10.1038/nrg1380. [DOI] [PubMed] [Google Scholar]
- Tucker AS, Matthews KL, Sharpe PT. Transformation of tooth type induced by inhibition of BMP signaling. Science. 1998;282:1136–1138. doi: 10.1126/science.282.5391.1136. [DOI] [PubMed] [Google Scholar]
- Wang XP, Suomalainen M, Jorgez CJ, et al. Follistatin regulates enamel patterning in mouse incisors by asymmetrically inhibiting BMP signaling and ameloblast differentiation. Dev Cell. 2004;7:719–730. doi: 10.1016/j.devcel.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Wang XP, Suomalainen M, Felszeghy S, et al. An integrated gene regulatory network controls stem cell proliferation in teeth. PLoS Biol. 2007;5:e159. doi: 10.1371/journal.pbio.0050159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Vivatbutsiri P, Morriss-Kay G, et al. Cell lineage in mammalian craniofacial mesenchyme. Mech Dev. 2008;125:797–808. doi: 10.1016/j.mod.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Zheng W, Huang L, Wei ZB, et al. The role of Six1 in mammalian auditory system development. Development. 2003;130:3989–4000. doi: 10.1242/dev.00628. [DOI] [PMC free article] [PubMed] [Google Scholar]