Abstract
Estrogen receptor alpha (ERα)-positive breast cancers that co-express trans cription factors GATA-3 and FOXA1 have a favorable prognosis. These transcription factors form an autoregulatory hormonal network that influences estrogen responsiveness and sensitivity to hormonal therapy. Disruption of this network may be a mechanism whereby ERα positive breast cancers become resistant to therapy. The transcription factor T-bet is a negative regulator of GATA-3 in the immune system. In this study, we report that insulin increases the expression of T-bet in breast cancer cells, which correlates with reduced expression of GATA-3, FOXA1 and the ERα:FOXA1:GATA-3 target gene GREB-1. The effects of insulin on GATA-3 and FOXA1 could be recapitulated through overexpression of T-bet in MCF-7 cells (MCF-7-T-bet). Chromatin immunoprecipitation assays revealed reduced ERα binding to GREB-1 enhancer regions in MCF-7-T-bet cells and in insulin treated MCF-7 cells. MCF-7-T-bet cells were resistant to tamoxifen in the presence of insulin and displayed prolonged ERK and AKT activation in response to epidermal growth factor treatment. ERα-positive cells with intrinsic tamoxifen resistance as well as MCF-7 cells with acquired tamoxifen and fulvestrant resistance expressed elevated levels of T-bet and/or reduced levels of FOXA1 and GATA-3. Analysis of publicly available databases revealed ERα-positive/T-bet-positive breast cancers expressing lower levels of FOXA1 (p=0.0137) and GATA-3 (p=0.0063) compared to ERα-positive/T-bet-negative breast cancers. Thus, T-bet expression in primary tumors and circulating insulin levels may serve as surrogate biomarkers to identify ERα-positive breast cancers with a dysfunctional hormonal network, enhanced growth factor signaling, and resistance to hormonal therapy.
Keywords: estrogen receptor, GATA-3, FOXA1, T-bet, breast cancer
Introduction
Gene expression studies have enabled classification of breast cancers into different prognostic subgroups; intrinsic subtype is one among them (1, 2). There are five intrinsic subtypes: luminal type A, luminal type B, HER2/Neu-positive, Basal-like, and Normal-like (3). Luminal type A cancers, which express estrogen receptor alpha (ERα), have one of the best prognosis with a 90% 5-year survival rate (3). This is partly attributed to their sensitivity to anti-estrogen therapy. However, resistance commonly develops to these hormonal therapies over time. Luminal type B breast cancers, which express either ERα or Progesterone Receptor (PR) and ki67high with few cases being HER2-positive, is associated with worse prognosis than luminal A cancers (4, 5).
Luminal type A tumors are characterized by elevated expression of three transcription factors: ERα, FOXA1, and GATA-3 (3, 6). The coexpression of these three factors is associated with a better prognosis (3, 7, 8). In vitro as well as gene knockout studies have revealed a mutual interdependence of these factors for expression. For example, FOXA1 is an estrogen-regulated gene, whereas GATA-3 and ERα regulate each other’s expression (6, 9, 10). GATA-3 is also essential for FOXA1 expression during mammary development (11, 12). FOXA1 is recruited to distal enhancer elements depending on the distribution of histone H3 lysine 4 mono and dimethylation; this facilitates ERα binding to regions that bind to both FOXA1 and ERα (13). Hence, FOXA1 is thought to be required for the expression of ~50% of estrogen’s target genes (9, 14). Similarly, GATA-3 binding sites are enriched in genomic regions that also bind to ERα (15). Based on their interdependence, these three transcription factors are suggested to constitute a cell-lineage specific hormonal transcription factor network (6).
Most studies on anti-estrogen resistance have focused on the role of ERα:estrogen axis, transcription co-regulatory molecules and the kinases that phosphorylate ERα and/or co-regulatory molecules (16–18). However, signaling pathways that may disrupt the ERα:GATA-3:FOXA1 hormonal network have received very little attention. GATA-3 was originally characterized as a signaling molecule involved in T-cell differentiation (19) and subsequently found to have a role in the differentiation of breast luminal progenitor cells (11). In T cells, the transcription factor T-bet, also known as Tbx21, is a negative regulator of GATA-3 activity (19). While T-bet is essential for differentiation of T helper progenitors to Th1 cells, GATA-3 performs an equivalent function in Th2 cells. T-bet prevents Th2 lineage commitment by inhibiting GATA-3 DNA binding (20). T-bet is expressed in epithelial cells of the female reproductive tract where, along with GATA-3, is expressed cyclically suggesting a hormonally regulated expression (21). From these studies, we considered the possibility of T-bet regulating GATA-3 activity in breast cancer cells and disrupting the ERα:GATA-3:FOXA1 signaling network. Furthermore, since previous studies showing a role for T-bet in insulin-dependent diabetes, we evaluated the role of insulin in disrupting hormonal network (22–24). Serum insulin level is an independent prognostic factor in breast cancer (25–28). We observed insulin-dependent overexpression of T-bet with subsequent reduction in GATA-3 expression in breast cancer cells. T-bet impaired estradiol (E2) and tamoxifen response in ERα-positive breast cancer cells implicating its role in the progression of luminal A breast cancers.
Materials and Methods
Cell types and cell culture: Cell culture
MCF-7 and T47D cells were maintained in minimum essential media (MEM) (Mediatech, Manassas, VA) plus 10% fetal bovine serum (FBS). ZR75-30 and BT474 cells were maintained in RPMI media with 10% FBS. MD-361 cells were grown in DMEM/F12 with 10% FBS, 100 nM E2, Sodium Pyruvate and non-essential amino acids. MCF-7 derivatives MCF-7p, MCF-7-T, and MCF-7-F cells corresponding to parental, tamoxifen-resistant, and fulvestrant-resistant cells, respectively, have been described (29). Prior to treatment, cells were placed in phenol red free MEM or RPMI supplemented with 5% charcoal-dextran treated serum (CCS) and L-glutamine for four days. Amphophenix cells were maintained in DMEM media supplemented with 10% FBS and Penicillin/Streptomycin.
Plasmids and retrovirus preparation
T-Bet cDNA (from ATCC, Manassas, VA) was cloned into the bicistronic retrovirus pcQXIN from Clontech (Mountain View, CA). Retrovirus packaging and transduction of MCF-7 cells have been described previously (17).
siRNA transfection
Cells were seeded with phenol red free MEM plus 5% CCS for 48 h and then transfected with 25 nM of siRNA (Dharmacon, Lafayette, CO) using TransIT-TKO Transfection Reagent (Mirus, Madison, WI) according to the manufacturer’s protocol.
RNA preparation and quantitative reverse transcription polymerase chain reaction (qRT-PCR)
RNA was isolated using the RNAeasy kit (Qiagen, Valencia, CA). For qRT-PCR, RNA was reverse transcribed using single stranded cDNA synthesis kit (Invitrogen, Carlsbad, CA) and subjected to Q-PCR using Syber Green or TaqMan (for T-bet) (Applied Biosciences, Foster City, CA). Primers for GREB-1 were as follows: forward 5′-TGC CAG ATG ACA ATG GCC ACA ATG -3′; reverse 5′-TCT GCT TCT TGG GTT GAG TGG TCA -3′. Primers for XBP-1 were: forward 5′-AGT GAG CTG GAA CAG CAA GTG GTA -3′; reverse 5′-AGC GCT GTC TTA ACT CCT GGT TCT -3′. Primers for GATA-3 were: forward 5′-ACA CTC TGG AGG AGG AAT GCC AAT-3′; reverse 5′-TTC GGT TTC TGG TCT GGA TGC CTT-3′. Primers for β-Actin as a control were as follows: forward 5′-AAT GAG GCC GAG GAC TTT GAT TGC-3′; reverse 5′-AGG ATG GGA AGG GAG TTC GTG TAA-3′. TaqMan probes for T-bet was purchased from Applied Biosciences.
Chromatin immunoprecipitation (ChIP)
ChIP assays with antibodies directed towards ERα for XBP-1 (enhancers 1, 2, and 3), GREB-1 enhancer 3, and SMRT enhancer ERα binding regions followed by quantitative PCR with specific primers were conducted as described previously (17).
Western blot analysis
Cell lysates prepared in radioimmunoassay buffer were subjected to western blot. Antibodies against T-bet, FOXA1, GATA-3, ERα, EGFR, ErBB2, and ErBB3 were purchased from Santa Cruz Biotechnologies (Santa Cruz, CA). Antibodies against pAKT, AKT, pERK, and ERK were from Cell Signaling/Millipore (Billerica, MA). Antibody against β-Actin was from Sigma (St. Louis, MO). Densitometry was performed to determine band intensity. Representative autoradiography and numerical values normalized for β-Actin from 3–5 experiments are presented.
Cell Proliferation Assay
Cells were placed in phenol red free MEM or RPMI (for BT474) supplemented with 5% CCS, and L-glutamine for 48 hours prior to plating. 1000 cells/well were plated in a 96 well plate and treatment was initiated after two days with subsequent media change on fifth day. Cell proliferation was measured on seventh day using Bromodeoxyuridine incorporation-ELISA assay (Calbiochem, San Diego, CA).
Statistical Analysis
All statistical analysis was performed using the GraphPad software (graphpad.com). Analysis of variance was employed to determine the p-values between mean measurements. A p-value of <0.05 was deemed significant.
Results
T-bet is overexpressed in a subset of ERα-positive breast cancers
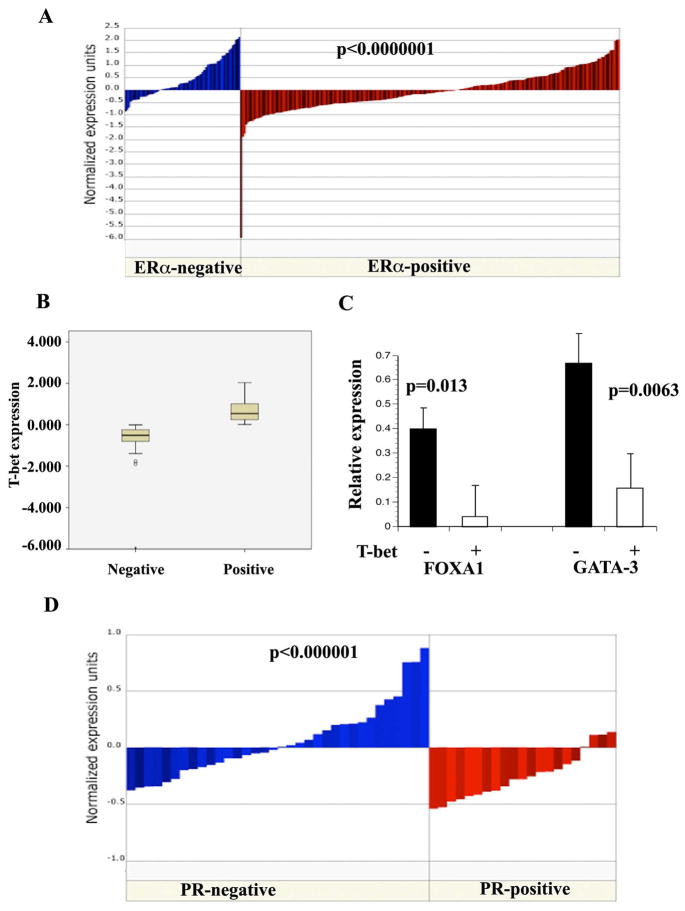
To consider the possibility of T-bet, a negative regulator of GATA-3 activity in T cells (19), controlling the function of hormonal network involving GATA-3 in breast cancer, we first examined the expression pattern of T-bet in the publicly available microarray databases. Although T-bet is expressed at higher levels in ERα-negative breast cancers compared to ERα-positive breast cancers, a subset of ERα-positive breast cancers expressed higher levels of T-bet (Figure 1A) (30). Differences in the expression between T-bet-positive (N=94) and T-bet negative (N=131) subgroups within ERα-positive breast cancer are statistically significant (p=0.0001) (Figure 1B). Using the same dataset, we then analyzed the expression levels of GATA-3 and FOXA1 in T-bet-positive and T-bet-negative subgroups. T-bet expression negatively correlated with FOXA1 (p=0.0137) and GATA-3 (p=0.0063) (Figure 1C). T-bet expression was also associated with progesterone receptor (PR) negativity (p<0.00005) (31), a subgroup that is known to be associated with resistance to endocrine therapy (32) (Figure 1D). Analysis of T-bet expression among ERα/PR-positive breast cancer patients who received tamoxifen treatment in a different dataset revealed a trend of elevated T-bet expression in tumors of patients with recurrence (n=28) compared to patients who were disease-free (n=32) after five years of treatment, although this did not reach statistical significance (p=0.066) (33). Since microarray analysis was performed on laser capture micro-dissected tumor samples, this study demonstrates that T-bet is present in cancer cells.
Figure 1. T-bet, GATA-3, and FOXA1 expression in primary breast cancer.
A) Expression pattern of T-bet in ERα-positive and ERα-negative breast cancer. Gene expression levels from a published study (30) were extracted from Oncomine (www.oncomine.org). Difference in expression between two groups is statistically significant. B). ERα-positive breast cancers from the above study were classified into T-bet positive and T-bet negative subgroups based on significant differences in the expression levels. C). Expression levels of FOXA1 and GATA-3 in ERα+/T-bet- and ERα+/T-bet+ subgroups. D). T-bet expression negatively correlates with PR negativity. As in A, data was extracted from a published study (31).
Insulin induces T-bet and/or reduces GATA-3 and FOXA1 expression
To determine whether T-bet is expressed in breast cancer cell lines and the expression is regulated by extracellular signals, we investigated the effects of growth hormone, insulin, insulin-like growth factor I and II (IGF I and IGF II), epidermal growth factor (EGF), inflammatory cytokines, and estrogen on T-bet expression in MCF-7 cells. Only insulin and IGF I induced T-bet expression (Figure 2A and Figure S1 in the supplementary file, data not shown). Insulin increased T-bet mRNA levels and this increase in mRNA correlated with insulin-dependent increase in STAT-1 phosphorylation, a transcription factor involved T-bet expression (34) (Figure S2A and B).
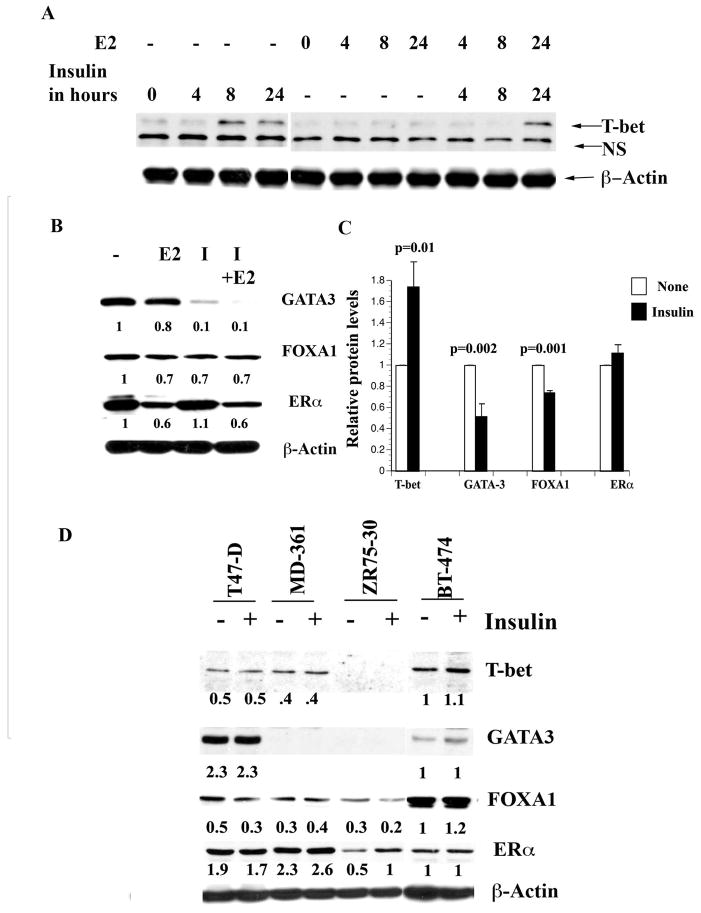
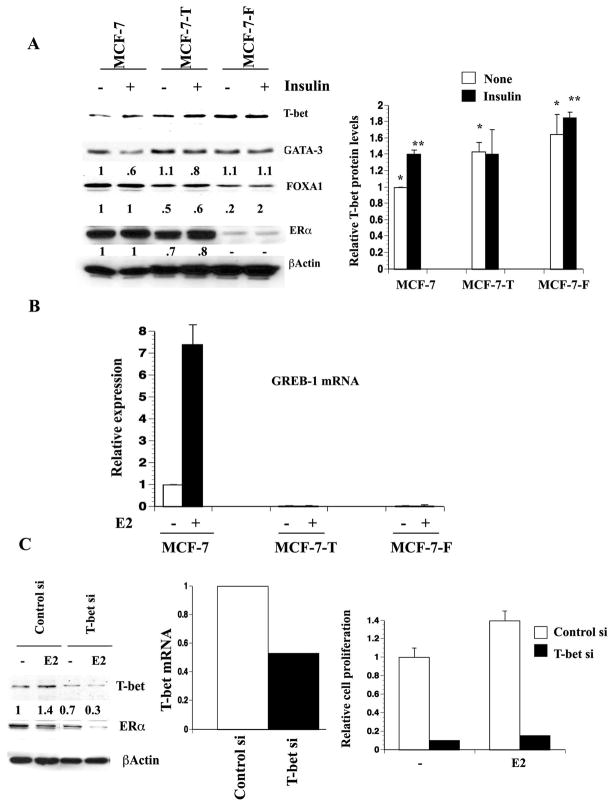
Figure 2. Insulin alters T-bet, GATA-3, and FOXA1 expression in breast cancer cells.
A) Insulin increases T-bet expression in MCF-7 cells. Cells were treated with insulin (50 nM) and/or E2 (0.1 nM) for indicated time and T-bet expression was measured by western blotting. B) The effect of insulin on GATA-3, FOXA1, and ERα expression. Cells were treated with insulin and/or E2 for 24 hours and the expression levels of different proteins were measured by western blotting. C) Densitometric scanning data of 3 or more experiments showing insulin-mediated significant increase in T-bet expression and reduction of GATA-3 and FOXA1 expression in MCF-7 cells. Mean and standard error of the mean are presented. D). Variable expression of T-bet, GATA-3, FOXA1, and ERα in luminal B cell lines. All luminal B cell lines (BT-474, MD-361, and ZR75-30) showed significantly lower levels of GATA-3 compared to the luminal A cell line T47-D. Note that insulin reduced FOXA1 expression in T47-D and ZR75-30 cell lines.
We next examined the effects of insulin on GATA-3 and FOXA1 expression. If crosstalk between T-bet and GATA-3 is similar in both T cells and breast epithelial cells, insulin is expected to reduce the expression and/or activity of GATA-3. As expected, insulin reduced the expression of GATA-3 both at protein (Figure 2B) and transcript level (Figure S3A). Results of multiple experiments on the effects of insulin on T-bet, GATA-3, FOXA1, and ERα proteins are shown in Figure 2C. Insulin-mediated reduction in GATA-3 correlated with 30% reduction in FOXA1 expression. However, it was noted that not all MCF-7 variants or breast cancer cell lines showed insulin-dependent reduction of FOXA1 (see below).
To determine cell type specificity of insulin action, we examined additional ERα-positive luminal A (T47-D) and luminal B (BT-474, MD-361, and ZR75-30) cell lines for T-bet, GATA- 3, FOXA1, and ERα expression (35). Luminal B phenotype appears to be associated with reduced or loss of GATA-3 expression (Figure 2D). T-bet expression was markedly higher in BT-474 cells compared to other cell lines (Figure 2D). Insulin reduced FOXA1 expression in T47-D and ZR75-30 cells by ~30%. GATA-3 dependency of FOXA1 expression is cell type specific because all luminal B cell lines expressed significant FOXA1. Taken together, these results reveal a cell type specific association between T-bet, GATA-3, and FOXA1 expression in breast cancer cells and the effects of insulin on their expression.
T-bet overexpression in MCF-7 cells leads to altered E2 and tamoxifen response
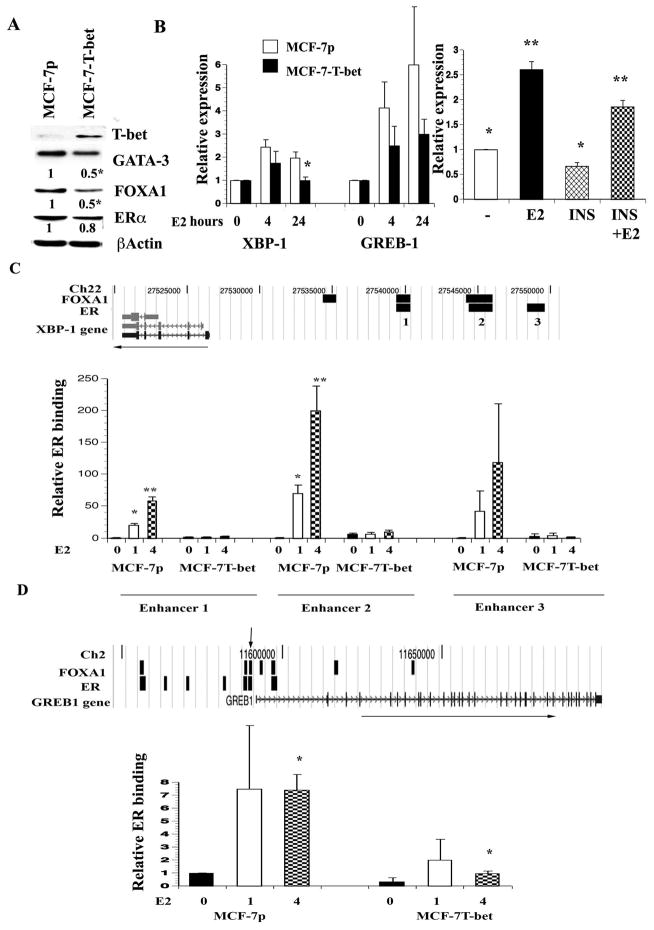
To determine whether T-bet negatively regulates E2-inducible expression of specific ERα, GATA-3, and FOXA1 target genes, we generated MCF-7 cells overexpressing T-bet (MCF-7-T-bet, Figure 3A). The expression of GATA-3, FOXA1, and ERα was lower in MCF-7-T-bet cells compared to parental (MCF-7p) cells, which is consistent with the effect of insulin on the expression of GATA-3 and FOXA1. As with insulin, T-bet overexpression resulted in lower levels of reduced GATA-3 transcripts (Figure S3B).
Figure 3. T-bet inhibits chromatin binding of ERα and E2-regulated gene expression.
A) T-bet overexpression results in general reduction in GATA-3, FOXA1, and ERα expression. Expression levels of indicated transcription factors were measured in parental cells transduced with empty retrovirus (MCF-7p) or T-bet expressing virus (MCF-7-T-bet). Densitometric scanning data from two or more experiments normalized to the control β-Actin are presented. *p<0.001, MCF-7p vs MCF-7-T-bet. B) E2-inducible expression of XBP-1 and GREB-1 in MCF-7p and MCF-7-T-bet cells (left panel). Results of three or more experiments are presented (mean ±SEM). *p=0.01. The effect of insulin (INS) on basal and E2-inducible expression of GREB-1 is shown in the right panel. Cells were pre-treated with insulin overnight and then exposed to ethanol or E2 for four hours. *p=0.01 control versus insulin treatment; **p=0.02, E2 vs E2+ insulin (n=5). C) ERα binding to enhancer regions of XBP-1 was markedly lower in MCF-7-T-bet cells compared to MCF-7p cells. ERα and FOXA1 binding sites associated with XBP-1 from previous ChIP-on-Chip (13, 17) are shown on the top (black bars) along chromosomal location and direction of the gene (horizontal arrow). ERα binding in untreated and E2 treated MCF-7p and MCF-7-T-bet cells was determined by ChIP analysis followed by q-PCR. Asterisks denote statistically significant differences in ERα binding under identical treatment conditions between two cell types. D) ChIP assay was used to measure ERα binding to one of the ERα binding regions (black bars) associated with GREB-1 gene (indicated by inverted arrow on the top).
XBP-1 is a potential downstream target of ERα, FOXA1, and GATA-3 network based on a meta-analysis and contains binding sites for all three transcription factors (7, 14). While E2 readily induced XBP-1 expression in MCF-7p cells, it was markedly lower in T-bet overexpressing cells (Figure 3B, left panel). E2-inducible expression of GREB-1, which also contains both ERα and FOXA1 binding sites, was lower in MCF-7-T-bet cells compared to MCF-7p cells, although the magnitude of this effect was not as dramatic (Figure 3B, left panel). T-bet did not have an effect on E2-inducible expression of EBP50, which suggests gene specific effects of T-bet on E2-regulated gene expression (data not shown).
We next investigated whether reduced E2-inducible expression of XBP-1 in MCF-7-T- bet cells correlates with lower ERα binding to regulatory regions by performing a ChIP assay. XBP-1 has three distinct enhancer elements with ERα binding sites; enhancers 1 and 2 also contain FOXA1 binding sites (Figure 3C). E2-induced ERα binding to all three ERα binding sites of XBP-1 was substantially lower in MCF-7-T-bet cells compared to MCF-7p cells (Figure 3C). Like XBP-1, GREB-1 is associated with multiple ERα binding sites; a few of these sites are enriched for FOXA1 binding (Figure 3D). ERα binding to one of these binding sites that we examined was lower in T-bet overexpressing cells compared to parental cells. Taken together, these results suggest a negative effect of T-bet on ERα binding to the genome.
To determine whether insulin can mimic T-bet overexpression on E2-inducible expression of the above genes, we pretreated MCF-7 cells with insulin overnight and then exposed cells to ethanol or E2 for four hours. Insulin significantly reduced basal and E2-inducible expression of GREB-1 (Figure 3B, right panel). We performed ChIP assay to determine whether insulin alters ERα binding to GREB-1 enhancer. Indeed, insulin reduced basal and E2-inducible ERα binding to GREB-1 enhancer (Figure S4). Insulin similarly reduced ERα binding to XBP-1 enhancer 1 (Figure S4). Interestingly, insulin did not alter ERα binding to enhancer region of another E2-regulated gene SMRT indicating gene specific effects of insulin on ERα binding (Figure S4).
T-bet overexpressing cells are less sensitive to tamoxifen in the presence of insulin
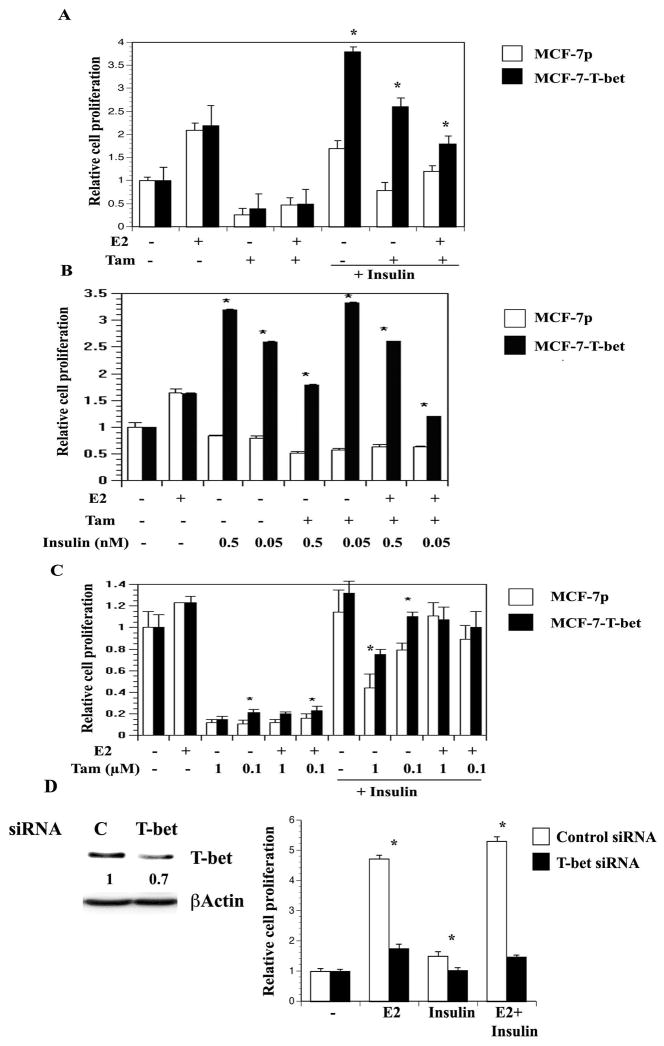
To further evaluate the effects of T-bet mediated changes in E2 signaling, we examined tamoxifen sensitivity of MCF-7p versus MCF-7-T-bet cells. Both cell types demonstrated similar sensitivity to 1 μM 4-hydroxy tamoxifen in the absence of insulin (Figure 4A). Insulin increased the proliferation of MCF-7p as well as MCF-7-T-bet cells; the magnitude of stimulation was significantly higher with MCF-7-T-bet cells. Although tamoxifen treatment reduced insulin-stimulated growth in both cell types, overall level of proliferation was significantly higher in MCF-7-T-bet cells compared to parental cells (under insulin plus tamoxifen or a combination of insulin, tamoxifen and E2). Similar results were obtained at variable insulin and tamoxifen concentrations (Figure 4B and C). Concentrations of insulin used in these experiments are similar to the levels seen in breast cancer patients with hyperinsulinemia (36). Note that at 0.1 μM tamoxifen, MCF-7-T-bet cells showed modest yet significant resistance to tamoxifen compared to MCF-7p cells and this resistance was further enhanced in the presence of insulin (Figure 4C).
Figure 4. Insulin confers resistance to tamoxifen.
A) The effect of insulin on proliferation and tamoxifen sensitivity of MCF-7p and MCF-7-T-bet cells. Cells were plated on 96 well plates and treated with E2 (0.1 nM), 4-hydroxy-tamoxifen (1 μM) and/or insulin (50 nM) as described in materials and methods. *p= 0.0001, MCF-7p vs MCF-7-T-bet cells under identical treatment condition. B) The effect of different concentrations of insulin on proliferation and tamoxifen sensitivity (1 μM) of MCF-7p and MCF-7-T-bet cells. *p<0.01 between MCF-7p and MCF-7-T-bet cells under identical treatment condition. C) The effect of insulin on proliferation under variable concentration of tamoxifen. Experiments are done as in B. D) T-bet is required for E2- and insulin-dependent proliferation of T47-D cells. T47-D cells were treated with siRNA against T-bet or control non-specific siRNA targeting luciferase gene for four days. T-bet siRNA reduced T-bet protein levels by 30% (left panel). Cells were treated with E2, insulin or both for six days. *p<0.01 control vs T-bet siRNA.
To examine the role of endogenous T-bet on cell proliferation, we treated MCF-7 and T47-D cells with siRNA against T-bet. Due to low basal levels of T-bet and the failure of T-bet siRNA treated cells to proliferate, interpretable results could not be obtained in MCF-7 cells (data not shown). Even with only a 30% reduction, T-bet siRNA treated T47-D cells showed reduced proliferation upon insulin or E2 stimulation (Figure 4D). Similar results were obtained with BT-474 cells (data not shown).
Anti-estrogen resistant cells express higher levels of T-bet
We used clonal variants of MCF-7 that had acquired tamoxifen (MCF-7-T) or fulvestrant resistance (MCF-7-F) (29) to determine whether there is a correlation between anti-estrogen resistance and T-bet expression. Both MCF-7-T and MCF-7-F cells expressed higher levels of T-bet (Figure 5A). FOXA1 expression was significantly reduced in these resistant cells compared to parental cells. Basal GATA-3 expression was unchanged in all three-cell types. Insulin reproducibly reduced GATA-3 expression in MCF-7 and MCF-7-T cells. Reduced FOXA1 expression and T-bet overexpression in MCF-7-T and MCF-7-F cells correlated with absence of E2-inducible GREB-1 expression (Figure 5B). Thus, T-bet overexpression and reduced expression of either FOXA1 or GATA-3 are consistent features associated with acquired (MCF-7 derivatives) or intrinsic (BT-474, ZR75-30, and MD-361) anti-estrogen resistance of breast cancer cell lines.
Figure 5. Changes in ERα:FOXA1:GATA-3 axis in MCF-7 cells that acquired resistance to tamoxifen (MCF-7-T) or fulvestrant (MCF-7-F).
A). Basal and insulin-regulated expression pattern of T-bet, ERα, FOXA1, and GATA-3 in MCF-7, MCF-7-T and MCF-7-F cells. Right panel displays densitometric scanning results of T-bet from three or more experiments. The difference in T-bet expression between different cell types is significant (*p=0.01, **p=0.03). Similarly, reduction in FOXA1 expression in MCF-7-T and MCF-7-F cells compared to parental cells is significant (p<0.05). B) E2 fails to induce ERα:FOXA1:GATA-3 target gene GREB-1 in MCF-7-T and MCF-7-F cells. GREB-1 expression was measured by qRT-PCR (n=3). C) T-bet siRNA inhibits growth of MCF-7-T cells. Cells were treated with siRNA as in Figure 4D and cell proliferation was measured by BrDU-ELISA. As in T47-D cells, T-bet siRNA reduced T-bet protein levels by 30% (left panel) and transcript levels by 50% (middle panel).
We used short interfering RNA (siRNA) against T-bet to determine whether tamoxifen resistance of MCF-7-T cells can be partially reversed by reducing the levels of T-bet (Figure 5C). T-bet siRNA treated cells failed to grow and, as in T47-D cells, E2 treatment did not result in cell proliferation. These results suggest that T-bet is required for redirecting ERα to genes that may be essential for E2-stimulated proliferation of cells.
T-bet overexpressing cells display elevated EGF stimulated MAP kinase activation
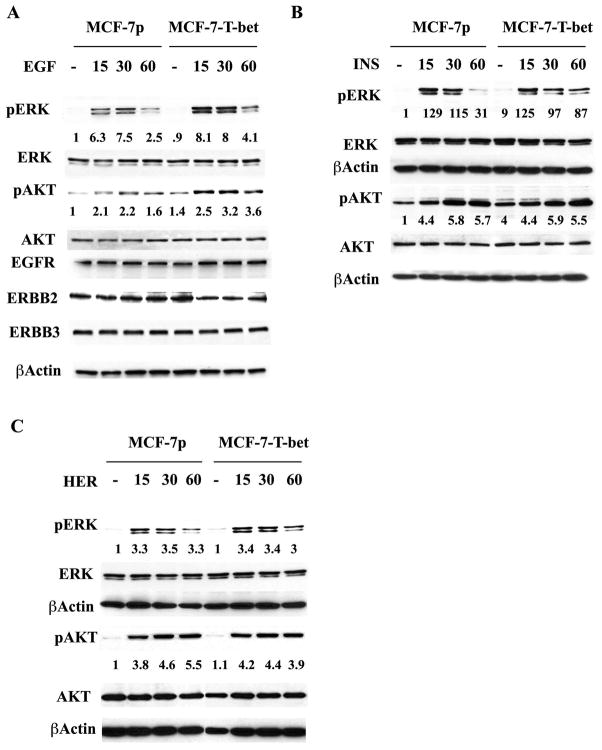
A functional ERα transcriptional network has previously been shown to suppress growth factor-activated signaling and this network is thought to be essential for ERα-positive breast cancers to respond to anti-estrogen treatment (37). Conversly, elevated growth factor-dependent MAPK and/or AKT activation is associated with anti-estrogen resistance in breast cancers (38–40). To determine whether T-bet-mediated disruption of the ERα FOXA1:GATA-3 transcriptional network leads to altered growth factor-dependent MAPK and AKT activation, we examined the levels of phospho-ERK and phospho-AKT in MCF-7p and MCF-7-T-bet cells upon treatment with EGF, heregulin, or insulin. pERK levels were higher and prolonged in EGF treated MCF-7-T-bet cells compared to MCF-7p cells (Figure 6A). Basal pAKT levels were consistently higher in MCF-7-T-bet cells (1.44 fold, p=0.006, n=6) compared to MCF-7p cells. Consequently, overall EGF stimulated pAKT level was also elevated in MCF-7-T-bet cells compared to MCF-7p cells. Similar to EGF, insulin stimulated ERK activation was prolonged in MCF-7-T-bet cells compared to MCF-7p cells (Figure 6B). Interestingly, heregulin-mediated ERK and AKT activation, which relies mostly on ERBB2:ERBB3 heterodimers, was similar in both cell types (Figure 6C). The effects of T-bet on EGF and insulin-mediated ERK and AKT is independent of growth factor receptor levels as the levels of EGFR, ERBB2/HER2, and ERBB3 were similar in both cell types.
Figure 6. MCF-7p and MCF-7-T-bet cells show differential ERK and AKT activation in response to growth factor signaling.
A) EGF-inducible ERK activation in MCF-7p and MCF-7-T-bet cells. Cells were treated with 20 ng/ml EGF for indicated time and ERK and AKT activation was measured using phospho-specific antibodies. Ratio between phosphorylated ERK and the loading control β-Actin is presented. B) Insulin-inducible (50 ng/ml) ERK and AKT activation in MCF-7p and MCF-7-T-bet cells. C) Heregulin-inducible (50 ng/ml) ERK and AKT activation in MCF-7p and MCF-7-T-bet cells.
Discussion
The intrinsic subtype classification scheme and several other studies have identified the ERα:FOXA1:GATA-3 transcription factor network in luminal type A breast cancers as well as in ERα-positive breast cancer cell lines (3, 6, 35). FOXA1 is a positive regulator of ERα binding to genome (14, 41). GATA-3, similar to FOXA1, binds to regions on chromatin that are enriched for ERα binding and may function as a co-factor for ERα binding (15). In contrast to FOXA1 and GATA-3, the transcription factors Nkx3-1 and LEF-1 inhibit ERα binding to genome (42). In this study, we have identified T-bet as an additional transcription factor that can disrupt the activity of ERα. T-bet significantly inhibited the ability of ERα to bind chromatinized DNA despite having a modest effect on ERα protein levels. T-bet may inhibit ERα binding by reducing the levels of GATA-3. T-bet additionally may inhibit GATA-3 DNA binding through protein-protein interaction as in T cells (20).
The major effects of T-bet and insulin on ERα, FOXA1, and GATA-3 positive cells are on E2-dependent proliferation, response to tamoxifen, and growth factor receptor activation. In both MCF-7-T and T47-D cells, even a modest reduction (30%) of T-bet levels caused marked decrease in E2-dependent proliferation. These results are somewhat incompatible with results of ERα binding studies, which showed T-bet-mediated reduction in ERα binding to XBP-1 and GREB-1. We noted that T-bet overexpression did not significantly affect ERα binding to another E2-regulated gene SMRT (Figure S4). This suggests that the effects of T-bet on ERα DNA binding are gene specific. It is possible that T-bet redirects ERα to proliferation-associated genes. In this context, studies in mammary and prostate epithelial cells suggest a key role for GATA-3 and FOXA1 in the expression of differentiation-associated genes (11, 12, 43). Therefore, ERα:FOXA1:GATA-3 may be involved in the expression of two independent gene expression pathways; one involved in differentiation and the other proliferation. T-bet may abrogate the differentiation-associated gene expression program while maintaining or enhancing the proliferation-associated gene expression program.
T-bet, ERα, FOXA1, and GATA-3 are rate limiting cell lineage-specific transcription factors compared to housekeeping transcription factors such as SP-1; therefore, a subtle change in their expression and/or activity levels is sufficient to have a biological effect. In this context, an insulin-dependent increase in T-bet expression (2-fold) correlated with a 50% reduction in GATA-3 expression (Figure 2C) and 70% increase in proliferation rate (Figure 4A). As a consequence, tamoxifen plus insulin treated MCF-7 cells maintained a proliferation rate similar to untreated cells (Figure 4A). Further elevation of T-bet levels through overexpression augmented insulin effects both in terms of overall proliferation and response to tamoxifen.
It is recently suggested that while resistance to anti-estrogen aromatase inhibitors is due to ligand-independent activation of ERα, resistance to tamoxifen is due to activation of growth factor signaling pathways, (40, 44). Consistent with this possibility, elevated activity of growth factor signaling pathway molecules Raf and ERK is associated with tamoxifen resistance (38). These results raise the possibility that E2 and tamoxifen actively repress growth factor signaling pathways through ERα in tamoxifen sensitive cells and the loss of this repressive mechanism leads to tamoxifen resistance. A recent study supports this possibility: E2:ERα and tamoxifen:ERα repress ERBB2 expression through Pax2-dependent recruitment of ERα to the ERBB2 enhancer and loss of this association results in tamoxifen resistance (37). Although we did not observe an effect of T-bet on any growth factor receptor expression, we did observe a prolonged EGF and insulin-mediated induction of ERK and an increased basal level of activated AKT. T-bet-mediated changes in ERα activity could influence the expression of other positive or negative regulators of growth factor signaling.
Insulin mediated upregulation of T-bet accompanied with repression of GATA-3 is the major finding of this study. The loss of GATA-3 is an important step in the transformation process as it marks the progression from adenoma to early carcinoma followed by metastasis in animal models of breast cancer (45). Hyperinsulinemia is an independent risk factor for breast cancer and the presence of diabetes in breast cancer patients is linked to a 40% increase in mortality within the first five years after diagnosis (25, 46). Elevated fasting insulin and estrogen levels but not IGF1 is associated with increased risk of breast cancer (47). A recent systematic review and meta-analysis revealed increased breast cancer among women with diabetes and it is suggested that diabetes contributes to cancer progression and mortality (28). Consistent with these observations, use of Metformin, a diabetic therapy, is associated with reduced cancer incidence, lower cancer-related mortality, and higher response to neoadjuvant therapy (48, 49). Our in vitro studies show the effect of elevated insulin on ERα-positive cells with respect to growth, response to anti-estrogen treatment, and growth factor signaling. Further epidemiological studies focusing on outcomes of anti-estrogen therapy on ERα-positive patients with or without diabetes are needed.
Supplementary Material
Acknowledgments
We thank Dr. Mark Kaplan for advise and reagents and Dr. Jay Patel for critical reading of the manuscript. This work is supported by the Department of Defense Concept Award BC0632283 (to HN). K.M. is supported by NCI T32 training grant CA111198. HN is Marian J. Morrison Professor of Breast Cancer Research.
References
- 1.Desmedt C, Ruiz-Garcia E, Andre F. Gene expression predictors in breast cancer: current status, limitations and perspectives. Eur J Cancer. 2008;44:2714–20. doi: 10.1016/j.ejca.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 2.Badve S, Nakshatri H. Oestrogen-receptor-positive breast cancer: towards bridging histopathological and molecular classifications. J Clin Pathol. 2009;62:6–12. doi: 10.1136/jcp.2008.059899. [DOI] [PubMed] [Google Scholar]
- 3.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hugh J, Hanson J, Cheang MC, et al. Breast Cancer Subtypes and Response to Docetaxel in Node-Positive Breast Cancer: Use of an Immunohistochemical Definition in the BCIRG 001 Trial. J Clin Oncol. 2009 doi: 10.1200/JCO.2008.18.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sotiriou C, Wirapati P, Loi S, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–72. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 6.Eeckhoute J, Keeton EK, Lupien M, Krum SA, Carroll JS, Brown M. Positive cross-regulatory loop ties GATA-3 to estrogen receptor alpha expression in breast cancer. Cancer Res. 2007;67:6477–83. doi: 10.1158/0008-5472.CAN-07-0746. [DOI] [PubMed] [Google Scholar]
- 7.Wilson BJ, Giguere V. Meta-analysis of human cancer microarrays reveals GATA3 is integral to the estrogen receptor alpha pathway. Mol Cancer. 2008;7:49. doi: 10.1186/1476-4598-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Badve S, Turbin D, Thorat MA, et al. FOXA1 Expression in Breast Cancer Correlation with Luminal Subtype A and Survival. Clin Cancer Res. 2007;13:4415–21. doi: 10.1158/1078-0432.CCR-07-0122. [DOI] [PubMed] [Google Scholar]
- 9.Laganiere J, Deblois G, Lefebvre C, Bataille AR, Robert F, Giguere V. From the Cover: Location analysis of estrogen receptor alpha target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc Natl Acad Sci U S A. 2005;102:11651–6. doi: 10.1073/pnas.0505575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolf I, Bose S, Williamson EA, Miller CW, Karlan BY, Koeffler HP. FOXA1: Growth inhibitor and a favorable prognostic factor in human breast cancer. Int J Cancer. 2007;120:1013–22. doi: 10.1002/ijc.22389. [DOI] [PubMed] [Google Scholar]
- 11.Asselin-Labat ML, Sutherland KD, Barker H, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2007;9:201–9. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- 12.Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 2006;127:1041–55. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lupien M, Eeckhoute J, Meyer CA, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–70. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carroll JS, Liu XS, Brodsky AS, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 15.Hua S, Kittler R, White KP. Genomic Antagonism between Retinoic Acid and Estrogen Signaling in Breast Cancer. Cell. 2009;137:1259–71. doi: 10.1016/j.cell.2009.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jordan VC, O’Malley BW. Selective estrogen-receptor modulators and antihormonal resistance in breast cancer. J Clin Oncol. 2007;25:5815–24. doi: 10.1200/JCO.2007.11.3886. [DOI] [PubMed] [Google Scholar]
- 17.Bhat-Nakshatri P, Wang G, Appaiah H, et al. AKT Alters genome-wide estrogen receptor alpha binding and impacts estrogen signaling in breast cancer. Mol Cell Biol. 2008;28:7487–503. doi: 10.1128/MCB.00799-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ali S, Coombes RC. Endocrine-responsive breast cancer and strategies for combating resistance. Nat Rev Cancer. 2002;2:101–12. doi: 10.1038/nrc721. [DOI] [PubMed] [Google Scholar]
- 19.Usui T, Preiss JC, Kanno Y, et al. T-bet regulates Th1 responses through essential effects on GATA-3 function rather than on IFNG gene acetylation and transcription. J Exp Med. 2006;203:755–66. doi: 10.1084/jem.20052165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang ES, Szabo SJ, Schwartzberg PL, Glimcher LH. T helper cell fate specified by kinase-mediated interaction of T-bet with GATA-3. Science. 2005;307:430–3. doi: 10.1126/science.1103336. [DOI] [PubMed] [Google Scholar]
- 21.Inman D, Kawana K, Schust D, Lininger R, Young S. Cyclic regulation of T-Bet and GATA-3 in human endometrium. Reprod Sci. 2008;15:83–90. doi: 10.1177/1933719107309690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu J, Wei M, Boyd Z, et al. Transcriptional control of human T-BET expression: the role of Sp1. Eur J Immunol. 2007;37:2549–61. doi: 10.1002/eji.200737088. [DOI] [PubMed] [Google Scholar]
- 23.Neurath MF, Weigmann B, Finotto S, et al. The transcription factor T-bet regulates mucosal T cell activation in experimental colitis and Crohn’s disease. J Exp Med. 2002;195:1129–43. doi: 10.1084/jem.20011956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juedes AE, Rodrigo E, Togher L, Glimcher LH, von Herrath MG. T-bet controls autoaggressive CD8 lymphocyte responses in type 1 diabetes. J Exp Med. 2004;199:1153–62. doi: 10.1084/jem.20031873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lipscombe LL, Goodwin PJ, Zinman B, McLaughlin JR, Hux JE. The impact of diabetes on survival following breast cancer. Breast Cancer Res Treat. 2008;109:389–95. doi: 10.1007/s10549-007-9654-0. [DOI] [PubMed] [Google Scholar]
- 26.Lipscombe LL, Goodwin PJ, Zinman B, McLaughlin JR, Hux JE. Diabetes mellitus and breast cancer: a retrospective population-based cohort study. Breast Cancer Res Treat. 2006;98:349–56. doi: 10.1007/s10549-006-9172-5. [DOI] [PubMed] [Google Scholar]
- 27.Jee SH, Ohrr H, Sull JW, Yun JE, Ji M, Samet JM. Fasting serum glucose level and cancer risk in Korean men and women. JAMA. 2005;293:194–202. doi: 10.1001/jama.293.2.194. [DOI] [PubMed] [Google Scholar]
- 28.Barone BB, Yeh HC, Snyder CF, et al. Long-term all-cause mortality in cancer patients with preexisting diabetes mellitus: a systematic review and meta-analysis. JAMA. 2008;300:2754–64. doi: 10.1001/jama.2008.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan M, Yan PS, Hartman-Frey C, et al. Diverse gene expression and DNA methylation profiles correlate with differential adaptation of breast cancer cells to the antiestrogens tamoxifen and fulvestrant. Cancer Res. 2006;66:11954–66. doi: 10.1158/0008-5472.CAN-06-1666. [DOI] [PubMed] [Google Scholar]
- 30.van de Vijver MJ, He YD, van’t Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 31.Ginestier C, Cervera N, Finetti P, et al. Prognosis and gene expression profiling of 20q13-amplified breast cancers. Clin Cancer Res. 2006;12:4533–44. doi: 10.1158/1078-0432.CCR-05-2339. [DOI] [PubMed] [Google Scholar]
- 32.Cui X, Schiff R, Arpino G, Osborne CK, Lee AV. Biology of progesterone receptor loss in breast cancer and its implications for endocrine therapy. J Clin Oncol. 2005;23:7721–35. doi: 10.1200/JCO.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 33.Ma XJ, Wang Z, Ryan PD, et al. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5:607–16. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 34.Afkarian M, Sedy JR, Yang J, et al. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat Immunol. 2002;3:549–57. doi: 10.1038/ni794. [DOI] [PubMed] [Google Scholar]
- 35.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goodwin PJ, Ennis M, Pritchard KI, et al. Fasting insulin and outcome in early-stage breast cancer: results of a prospective cohort study. J Clin Oncol. 2002;20:42–51. doi: 10.1200/JCO.2002.20.1.42. [DOI] [PubMed] [Google Scholar]
- 37.Hurtado A, Holmes KA, Geistlinger TR, et al. Regulation of ERBB2 by oestrogen receptor-PAX2 determines response to tamoxifen. Nature. 2008;456:663–6. doi: 10.1038/nature07483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGlynn LM, Kirkegaard T, Edwards J, et al. Ras/Raf-1/MAPK pathway mediates response to tamoxifen but not chemotherapy in breast cancer patients. Clin Cancer Res. 2009;15:1487–95. doi: 10.1158/1078-0432.CCR-07-4967. [DOI] [PubMed] [Google Scholar]
- 39.Kirkegaard T, Witton CJ, McGlynn LM, et al. AKT activation predicts outcome in breast cancer patients treated with tamoxifen. J Pathol. 2005;207:139–46. doi: 10.1002/path.1829. [DOI] [PubMed] [Google Scholar]
- 40.Massarweh S, Osborne CK, Creighton CJ, et al. Tamoxifen resistance in breast tumors is driven by growth factor receptor signaling with repression of classic estrogen receptor genomic function. Cancer Res. 2008;68:826–33. doi: 10.1158/0008-5472.CAN-07-2707. [DOI] [PubMed] [Google Scholar]
- 41.Lacroix M, Leclercq G. About GATA3, HNF3A, and XBP1, three genes co-expressed with the oestrogen receptor-alpha gene (ESR1) in breast cancer. Mol Cell Endocrinol. 2004;219:1–7. doi: 10.1016/j.mce.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 42.Holmes K, Song JS, Liu XS, Brown M, Carroll JS. Nkx3–1 and LEF-1 function as transcriptional inhibitors of estrogen receptor activity. Cancer Research. 2008;68:7380–5. doi: 10.1158/0008-5472.CAN-08-0133. [DOI] [PubMed] [Google Scholar]
- 43.Matusik RJ, Jin RJ, Sun Q, et al. Prostate epithelial cell fate. Differentiation. 2008;76:682–98. doi: 10.1111/j.1432-0436.2008.00276.x. [DOI] [PubMed] [Google Scholar]
- 44.Masri S, Phung S, Wang X, et al. Genome-wide analysis of aromatase inhibitor-resistant, tamoxifen-resistant, and long-term estrogen-deprived cells reveals a role for estrogen receptor. Cancer Res. 2008;68:4910–8. doi: 10.1158/0008-5472.CAN-08-0303. [DOI] [PubMed] [Google Scholar]
- 45.Kouros-Mehr H, Bechis SK, Slorach EM, et al. GATA-3 links tumor differentiation and dissemination in a luminal breast cancer model. Cancer Cell. 2008;13:141–52. doi: 10.1016/j.ccr.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pisani P. Hyper-insulinaemia and cancer, meta-analyses of epidemiological studies. Arch Physiol Biochem. 2008;114:63–70. doi: 10.1080/13813450801954451. [DOI] [PubMed] [Google Scholar]
- 47.Gunter MJ, Hoover DR, Yu H, et al. Insulin, insulin-like growth factor-I, and risk of breast cancer in postmenopausal women. J Natl Cancer Inst. 2009;101:48–60. doi: 10.1093/jnci/djn415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiralerspong S, Palla SL, Giordano SH, et al. Metformin and Pathologic Complete Responses to Neoadjuvant Chemotherapy in Diabetic Patients With Breast Cancer. J Clin Oncol. 2009;27:3297–302. doi: 10.1200/JCO.2009.19.6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bowker SL, Majumdar SR, Veugelers P, Johnson JA. Increased cancer-related mortality for patients with type 2 diabetes who use sulfonylureas or insulin. Diabetes Care. 2006;29:254–8. doi: 10.2337/diacare.29.02.06.dc05-1558. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.