Abstract
Recent transcriptome analysis indicates that >90% of human genes undergoes alternative splicing, underscoring the contribution of differential RNA processing to diverse proteomes in higher eukaryotic cells. The polypyrimidine tract binding protein PTB is a well-characterized splicing repressor, but PTB knockdown causes both exon inclusion and skipping. Genome-wide mapping of PTB-RNA interactions and construction of a functional RNA map now revealed that dominant PTB binding near a competing constitutive splice site generally induces exon inclusion whereas prevalent binding close to an alternative site often causes exon skipping. This positional effect was further demonstrated by disrupting or creating a PTB binding site on minigene constructs and testing their responses to PTB knockdown or overexpression. These findings suggest a mechanism for PTB to modulate splice site competition to produce opposite functional consequences, which may be generally applicable to RNA binding splicing factors to positively or negatively regulate alternative splicing in mammalian cells.
Introduction
Alternative splicing has been increasingly appreciated as a major mechanism to generate structural and functional diversity of gene products in higher eukaryotic cells (Black, 2003; Maniatis and Tasic, 2002). A recent transcriptome analysis indicated that more than 90% of human genes undergo alternative splicing and many mRNA isoforms appear to be regulated in a tissue-specific manner (Wang et al., 2008). Differential RNA splicing are controlled by many RNA binding proteins that recognize intronic and exonic cis-regulatory RNA elements, a second code of the genome for post-transcriptional regulation of gene expression (Black, 2003). Characterized cis-acting elements can be generally classified into intronic splicing enhancers (ISEs) or silencers (ISSs) and exonic splicing enhancers (ESEs) or silencers (ESSs), which act to positively or negatively influence the selection of alternative splice sites (Fu, 2004). However, splicing regulators can often affect alternative splicing in a position-dependent manner as recently emerged from genome-wide analysis of RNA binding splicing regulators (Licatalosi et al., 2008; Yeo et al., 2009).
The polypyrimidine tract binding protein PTB (also known as hnRNP I) is a well-characterized splicing suppressor on model minigene constructs (Spellman and Smith, 2006). PTB binds to CU-rich elements, often overlapping with the U2AF65 binding sites near the 3’ splice site. Therefore, one of the mechanisms for PTB-mediated splicing suppression is thought to compete with U2AF65 binding (Sauliere et al., 2006; Singh et al., 1995). PTB also binds to CU-rich sequences in many exonic and intronic regions to influence splice site selection by interfering with the process of exon definition (Izquierdo et al., 2005), obstructing intron definition (Chou et al., 2000; Sharma et al., 2005), or preventing the transition from exon to intron definition (Sharma et al., 2008).
To explain how PTB prevents spliceosome assembly events across exons or introns, it was initially proposed that PTB homodimers might induce RNA looping to sequester the alternative exon from the splicing machinery (Oh et al., 1998; Perez et al., 1997b). However, a later study indicates that PTB exists as a monomer in solution, capable of binding to RNA with high affinity (Amir-Ahmady et al., 2005; Monie et al., 2005); and a NMR study suggests that PTB may use different RRMs (PTB has four) to contact CU-rich RNA elements at different locations to induce RNA looping (Oberstrass et al., 2005). Although no direct experimental evidence is available to demonstrate RNA looping mediated either by PTB dimers or by two RRMs within a single PTB molecule, both models predict extensive PTB-mediated RNA networks during regulated splicing, which is also consistent with the observation that mutating one PTB binding site reduces PTB binding to another site in a model pre-mRNA substrate (Chou et al., 2000).
Although PTB is a well-known splicing repressor, recent splicing array analyses revealed both PTB-dependent exon inclusion and skipping (Boutz et al., 2007; Xing et al., 2008). A recent observation indicates that PTB can promote exon inclusion by antagonizing an inhibitory binding event by a different splicing suppressor (Paradis et al., 2007). However, it is unclear how widely this “suppression-of-suppressor” strategy is used by PTB to regulate alternative splicing. It has also been postulated that PTB may act in a similar fashion to the Nova and Fox families of splicing regulators to promote or suppress splice site selection in a location-dependent manner (Boutz et al., 2007). Genome-wide analysis provides a unique opportunity to directly test this hypothesis, which is key to understand the contribution of PTB to the splicing code in mammals.
Here we employed CLIP-seq to identify direct RNA targets for PTB in HeLa cells, finding that PTB bound to intronic regions near the 5’ or 3’ splice site regardless of whether the site is subject to regulation. About one third of PTB binding events in the human genome are linked to regulated splicing, consistent with PTB being a major splicing regulator in mammals, but the functional outcomes depend on the relative PTB binding frequency on the competing splice sites: Dominant PTB binding near the alternative splice site is correlated with exon skipping whereas overriding PTB binding near a competing constitutive splice site is associated with exon inclusion. We further showed that PTB-mediated exon inclusion could be achieved by inserting a PTB binding site near the flanking constitutive splice sites, thereby elevating the competitiveness of the alternative splice sites. These findings reveal a positional effect of PTB on regulated splicing through modulating the relative strength of competing splice sites, which is fundamentally distinct from the recently elucidated position-dependent activity of the Nova and Fox families of RNA binding proteins in the regulation of alternative splicing.
Results
Evidence for an extensive PTB-RNA interaction network in vivo
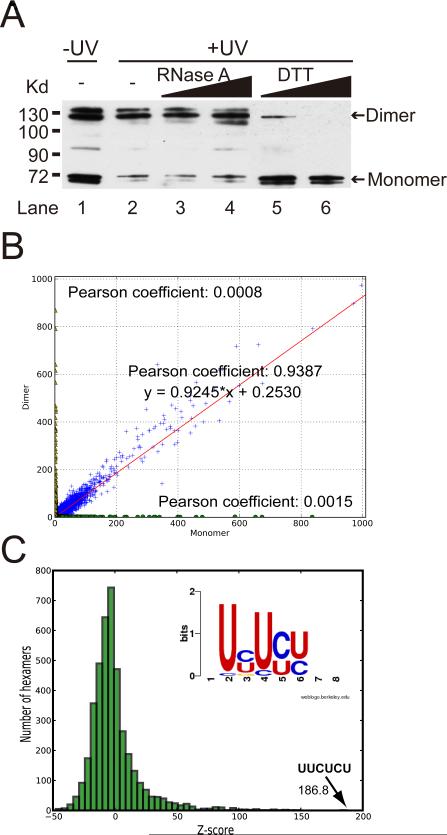
In preparation for genome-wide analysis of PTB binding by CLIP-seq, we first characterized a monoclonal anti-PTB antibody (BB7, described in Chou et al., 2000) on HeLa cells before and after UV treatment by immunoprecipitation/Western blotting. Consistent with a previous study (Perez et al., 1997b), we detected both PTB monomer and dimer under a non-reducing SDS-PAGE condition, but predominantly monomer under a reducing condition (+DTT) (Fig. 1A). UV-treatment dramatically increased the dimeric fraction of PTB. However, the PTB dimer is not tethered by RNA as it is resistant to RNase treatment (Fig. 1A, lanes 3 and 4), but sensitive to DTT (Fig. 1A, lanes 5 and 6), which is consistent with an early observation that the PTB dimer is held together by a specific disulfide bond (Monie et al., 2005).
Figure 1. Extensive PTB-PTB and PTB-RNA interactions in vivo.
(A) Western blotting analysis of immunoprecipitated PTB from mock-treated and UV-treated HeLa cells. The protein was resolved by SDS-PAGE in the presence of either 1 mM (non-reducing) or 10 mM (reducing) DTT as indicated. (B) Overlapping binding with monomeric and dimeric PTB. The Pearson coefficient between monomeric and dimeric tags in a 500 bp window across the whole genome is high (0.9387), which contrasts the lack of correlation between randomized tags and monomeric tags (0.0015) or dimeric tags (0.0008) (See Fig. S5 for further details). (C) Over-represented PTB binding motifs identified by CLIP-seq. Histogram of Z-scores indicates the enrichment of hexamers in CLIP-seq clusters compared to randomly chosen regions of similar sizes in the same genes. Z-score of the top hexamer is indicated. Insert shows the PTB binding consensus calculated from the top 20 enriched hexamers.
The induction of PTB dimerization by UV may be interpreted to indicate that a fraction of PTB might exist as dimer before binding to RNA and UV might catalyze the disulfide bond formation. This would agree with the PTB-PTB interaction detected in the yeast two-hybrid assay (Oh et al., 1998). Alternatively, PTB might bind to RNA as a monomer, but become dimerized upon binding to RNA, which could be enhanced and/or stabilized by UV. This possibility would be consistent with the observation that PTB can bind to RNA as monomer with high affinity (Amir-Ahmady et al., 2005), but PTB binding on one site can influence PTB binding on another site in the same pre-mRNA substrate (Chou et al., 2000). In any case, the ability of PTB to simultaneously engage in protein-protein and protein-RNA interactions suggests that PTB may nucleate extensive RNA-protein interaction networks, which are likely contributed by RNA binding activities of individual RRMs in PTB (Oberstrass et al., 2005; Clerte and Hall, 2009).
By 32P-labeling, we found that both monomeric and dimeric PTB are associated with RNA (Fig. S1). To detect potential functional differences between the monomeric and dimeric forms of PTB, we separately isolated the two protein-RNA complexes and constructed two independent libraries for CLIP-seq analysis. We first determined the quality of the libraries by conventional cloning and sequencing, obtaining 341 and 214 unique tags associated with PTB monomer and dimer, respectively. Most of these tags (~80%) were mapped to introns as expected (Fig. S2) and the average size is ~30nt in length (Fig. S3), which is consistent with the previous report that the minimal PTB binding sequence is 30nt (Amir-Ahmady et al., 2005). This finding suggests that the actual PTB binding sites are likely to reside within, rather than nearby, the sequenced tags in most cases, therefore eliminating the need to computationally extend the tags for subsequent peak finding and motif analysis (Yeo et al., 2009). We further confirmed the specificity of the CLIP assay by performing RIP-PCR analysis on a panel of anti-PTB enriched RNAs (Fig. S4).
Having thoroughly characterized the libraries, we next subjected PCR amplicons to high-throughput sequencing on the Illumina GAII platform, resulting in 2.44 million tags for PTB monomer and 2.37 million for PTB dimer that were uniquely mapped to the human genome (hg18). These high-density reads allowed us to ask first whether PTB monomeric and dimeric tags are differentially distributed in the genome. As shown in Fig. 1B, using a 500bp window, most of the monomeric and dimeric tags are similarly distributed in the genome with a Pearson correlation coefficient of 0.9387 and the coefficient between monomer and dimer increases with the increasing number of tags compared (Fig. S5). We conclude that there are no two separate sets of sites for PTB binding as monomer or dimer in the cell.
We therefore used combined tags to determine over-represented motifs in PTB binding clusters (see Methods), finding that CU-rich hexamers are highly enriched (Fig. 1C and Fig. S6A). About 21% clusters (5.17% for random, p-value=0) contain the top-scored motif UUCUCU (Z-score, 186.5). The top 20 motifs are all CU-enriched; 83.56% of total clusters obtained contains at least one of top 20 motifs (38.56% for random, p-value=0) (Fig. S6B), and the consensus generated from the top 20 hexamers is UYUYU (insert in Fig. 1C). In fact, the C/U percentage is broadly elevated surrounding the PTB binding sites, but not among randomly selected background sequences (Fig. S7), which fully corroborates with biochemcially defined PTB binding characteristics (Ashiya and Grabowski, 1997; Perez et al., 1997a). Interestingly, ~90% of identified PTB binding sites overlap with those predicted by an algorithm based on the biochemical properties of PTB (Gama-Carvalho et al., 2006), but the number of the experimentally detected sites represents only ~1% of ~5 million sites predicted in the human genome by the algorithm (data not shown). Thus, biochemically deduced consensus may not be sufficient to predict true binding sites because the recognition of some consensus motifs may be obstructed by competition of other RNA binding proteins or by certain RNA secondary structures. Indeed, we did note a few under-represented, A/G-rich motifs (Fig. S6A), indicating that depletion of A/G-rich sequences may help maximize the single-strandedness of PTB binding sites by minimizing potential stem-loop structures due to basepairings between C/U and A/G rich sequences. These observations suggest that experimentally validated binding sites coupled with critical features in local genomic context will help further improve prediction algorithms for RNA binding splicing factors.
Genomic landscape of PTB binding
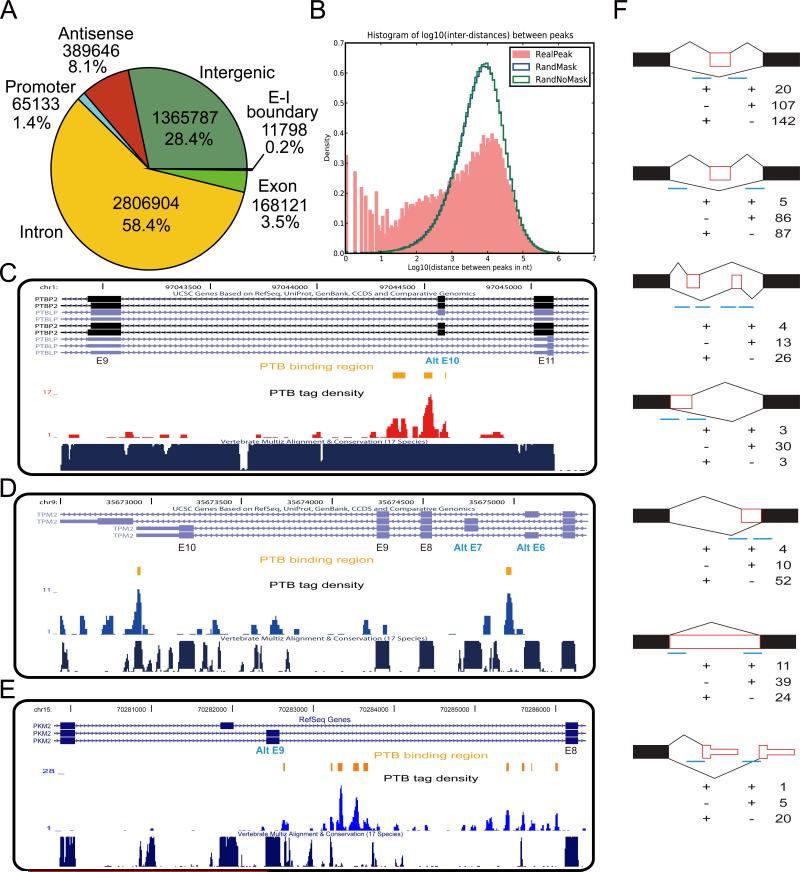
By mapping the sequenced tags to the knownGene set from the UCSC genome database, we found that 58.4% of the tags are localized in introns (Fig. 2A), with the relative density (counts per kb) 17-fold higher in introns than exons, indicating that most of the tags are derived from pre-mRNAs (a fraction of PTB binding events may also be derived from excised lariats). A sizable fraction of tags were mapped to antisense transcripts (8.1%) and intergenic regions (28.4%), implying that PTB may also bind to many non-coding RNA and/or unannotated transcripts, which is subject to future studies.
Figure 2. Genomic landscape of PTB binding.
(A) The distribution of PTB tags in the human genome (hg18). (B) The distribution of PTB binding clusters relative to one another in the same genes. (C) Screen shot of PTB binding around the well-characterized nPTB exon 10. (D) Screen shot of PTB binding around TPM2 exon 7 and the two alternative polyadenylation sites. (E) Screen shot of PTB binding in the intron proceeding the regulated exon 9 in PKM2 gene. (F) PTB binding clusters associated with six major alternative RNA processing modes. The patterns of PTB binding in 250nt intronic and 30nt exonic regions around splice sites were counted. The filled black boxes indicate constitutive exons or exonic regions, whereas empty red boxes show alternative exons or exonic regions. The short blue lines mark the regions where PTB binding clusters were present (+) or absent (-). The number is the total events of PTB binding at each location.
We next focused on clustered PTB binding events by identifying peaks above the gene-specific, randomized background as previously described (Yeo et al., 2009). The resulting 64,314 peaks were further merged to 51,394 clusters by placing PTB peaks within a 50nt window. Interestingly, while more than half (56.5%) of PTB binding clusters are separated by 1 to 10kb, as expected from independent binding events, a significant fraction (43.5%) of PTB clusters appears to be more closely positioned (<1 kb) (Fig. 2B), likely reflecting a concert action of multiple PTB binding events in regulated splicing. Further analysis revealed that PTB binds to 10,372 out of the 24,378 annotated human genes (30,986/66,803 knownGene transcripts). This number might be an underestimate because our current sequencing density has not yet reached saturation according to power analysis (data not shown). Given the fact that most sequence tags contain PTB binding consensus, indicating that contamination with other non-specific RNA is minimal, this binding profile suggests that PTB is a major RNA binding protein that may be widely involved in RNA metabolism in mammals.
Association of PTB binding with alternative splicing events
We next explored how frequently PTB binding is linked to annotated alternative splicing events. We separately examined PTB association with several major modes of alternative splicing, including cassette exon, alternative 5’ splice site, alternative 3’ splice site, and retained intron, based on the knownAlt track of the UCSC genome browser (Karolchik et al., 2008). This analysis revealed that 28.3% of PTB binding events are associated with annotated alternative splicing, and 22.2% of all annotated alternative splicing events are linked to PTB binding, thus suggesting a prevalent role of PTB as a splicing regulator in the human genome. PTB is involved in all common modes of alternative splicing (Table 1), with cassette exons being the most frequent targets for PTB regulation (z-score 14.11, compared to 100 trials of randomly placed clusters). Many PTB binding events are also found on “constitutive” introns and exons, which might be associated with alternative splicing events that have not yet been annotated. Alternatively, PTB may function to repress decoy splicing signals within constitutively spliced genes, which deserves a close look in future studies.
Table 1.
PTB binding clusters associated with different modes of alternative splicing. Column 1 shows the total number of events in each mode extracted from the UCSC knownAlt track. The observed number of PTB clusters associated with each mode is the count of PTB clusters within the region covering the alternative exon, the flanking intron(s) and constitutive exons. The expected number is the averaged number of association in 100 trials (random placement of PTB clusters). The column z-score shows the significance of association.
| Alt event | # Total events | # PTB cluster associated | Z-score (100 random trials) | |
|---|---|---|---|---|
| Observed | expected | |||
| Cassette exon | 7449 | 5824 | 5053 | 14.11 |
| Alt Terminal | 909 | 815 | 661 | 8.56 |
| Retained intron | 1446 | 147 | 96 | 6.59 |
| Mulx exon | 522 | 662 | 581 | 4.04 |
| Alt5Prime | 1970 | 582 | 524 | 3.24 |
| Alt3Prime | 3207 | 805 | 748 | 2.70 |
We next determined how PTB binding might affect splice site selection on both known and newly identified PTB target genes. As expected, a significant number of tags were mapped to the PTBP2 (also known as nPTB) gene, a well-known PTB target in which the alternative exon 10 is repressed by PTB. Notably, PTB binds preferentially to the sequences upstream of the 3’ splice site of exon 10 in nPTB as previously characterized (Boutz et al., 2007; Spellman et al., 2007). We also identified a binding cluster near the downstream 5’ splice site and some distributive PTB binding in the upstream intron (Fig. 2C), suggesting that PTB binds to multiple locations surrounding the regulated exon, which may collectively contribute to PTB-mediated exon repression, a situation similar to the well-characterized c-Src N1 exon (Sharma et al., 2005).
In another example (Fig. 2D), we identified two PTB binding clusters between the two mutually exclusive exons (exon 6 and 7) in the TPM2 gene, which is consistent with the observed repression of exon 7 in non-muscle cells (Sauliere et al., 2006; Spellman et al., 2007). We also detected prevalent PTB binding near the polyadenylation site for E10 in agreement with the observed utilization of the E11 polyadenylation site in non-muscle cells. Interestingly, we note multiple PTB binding events between the regulated exons and polyadenylation sites, suggesting a potential RNA network that may underlie the coordinated regulation of both events as reported (Spellman et al., 2007).
The high-quality PTB-RNA interaction map also help assign PTB as a regulator to previously uncharacterized alternative splicing events. For example, the pyruvate kinase 2 (PKM2) gene expresses two mutually exclusive isoforms, and such regulated splicing appears to be critical for cancer metabolism and tumor growth (Christofk et al., 2008). Although PTB has been implicated in the regulation of PKM2 splicing, critical cis-acting regulatory elements has remained undefined (Spellman et al., 2007). We found extensive PTB binding clusters in the intron preceding the alternative exon 9 (Fig. 2E) and RT-PCR confirmed PTB-dependent repression of PKM2 exon 9 in HeLa cells (data not shown). This finding raises the possibility that PTB may contribute to certain cancer phenotype by regulating the alternative splicing of PKM2.
PTB binding appears to associate with regulated cassette exons more significantly than other modes of alternative splicing (Table 1). On the well-characterized c-Src gene, PTB binds to both sides of the regulated exon N1 (Amir-Ahmady et al., 2005). To estimate how frequently PTB binds to both sides of alternative exons or exonic sequences, we analyzed a large number of annotated alternative splicing/polyadenylation events in comparison with mapped PTB binding events (Fig. 2F). This analysis revealed several interesting trends: First, while the bracket binding mode of PTB is clearly associated with many regulated RNA processing events, PTB appears to bind to either up- or downstream of the alternative splice site in the majority of cases. Secondly, among regulated cassette exons, PTB has the same tendency to bind to one of the competing (constitutive versus alternative) splice sites, implying that PTB does not always target the alternative splice site, which have distinct functional consequences (see below). Thirdly, in most cases of regulated 5’ and 3’ splice site choices, PTB appears to prefer binding on the intronic side, predicting that PTB may favor the distal splice site by repressing the proximal site in general.
PTB-dependent repression or enhancement of alternative splicing in vivo
Most minigene-based analysis focused on the consensus PTB binding motif near a regulated exon(s), which leaves a general impression that PTB preferentially targets alternative splice sites for regulation. The PTB CLIP-seq data now offer an unbiased view on the actual location of PTB binding on PTB-regulated genes. We found both known and new locations for PTB binding on all 13 previously documented PTB-regulated exons. This prompted us to examine additional candidates based on prevalent PTB binding events. Of 32 targets assayed, we found 22 altered splicing in response to PTB knockdown by RNAi (>5% absolute change), and among these, 10 showed PTB-dependent inclusion and 12 exhibited PTB-dependent exon skipping (supplementary Table S1). This finding confirmed the previous observation that PTB regulates both exon inclusion and skipping in vivo (Boutz et al., 2007; Xing et al., 2008).
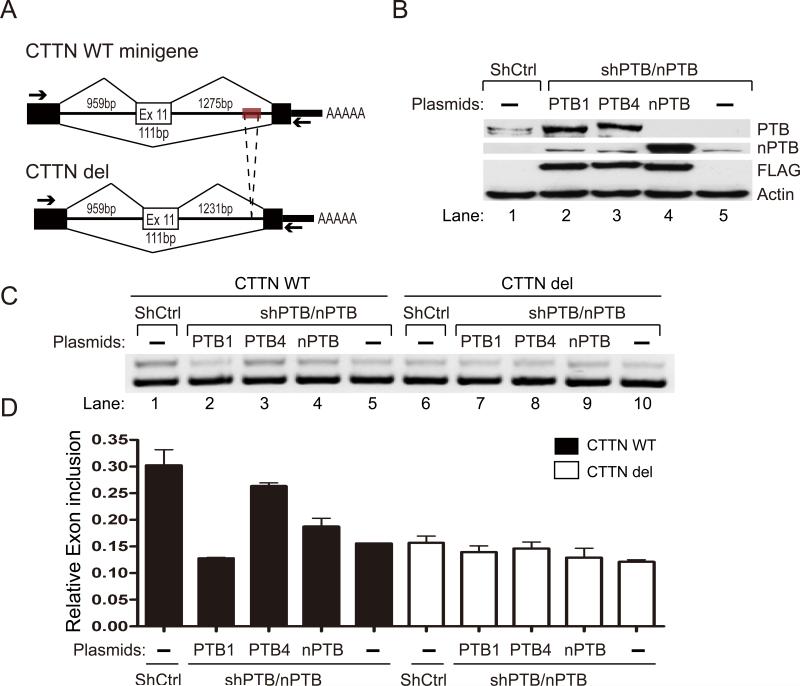
While the mechanism for PTB-mediated exon skipping is well characterized on multiple minigene models in literature, it has been unclear how PTB enhances exon inclusion. We first wished to establish sequence-dependent regulation of exon inclusion by PTB. For this purpose, we selected the CTTN gene that showed positive regulation by PTB to construct a minigene for analysis in transfected cells. The minigene, which contains the cassette exon 11 and flanking introns and exons, was expressed from the CMV promoter in pcDNA3 (Fig. 3A; note that this minigene was spliced less efficiently than the endogenous gene (see Fig. 4B), likely because the minigene might miss some positive regulatory elements in the construct and/or impair efficient transcription/splicing coupling as on the endogenous gene). In response to simultaneous knockdown of PTB and nPTB by shRNAs (Fig. 3B, lane 5), we detected a significant reduction of exon 11 inclusion in cells co-transfercted with the minigene reporter (Fig. 3C, compare lane 1 treated with control shRNA with lane 5 treated with combined shRNAs against PTB and nPTB).
Figure 3. PTB-dependent inclusion of alternative exon.
(A) Schematic representation of the CTTN minigene constructs, showing both wild-type and the mutant that lacks the 44nt PTB-binding cluster. The mapped PTB binding site is marked in grey and the PCR primers used to detect alternatively spliced products are indicated by arrows. (B) Exogenously expressed PTB isoforms and nPTB in double PTB/nPTB knockdown cells. (C) Semi-quantitative RT-PCR analysis of wt and mutant CTTN pre-mRNA splicing in response to PTB/nPTB knockdown with or without complementation with exogenously expressed PTB isoforms or nPTB. (D) Quantification of the data as in (C) based on three independent experiments. Error bars are based on SEM; the statistical significance is determined by student t-test (P-value<0.05).
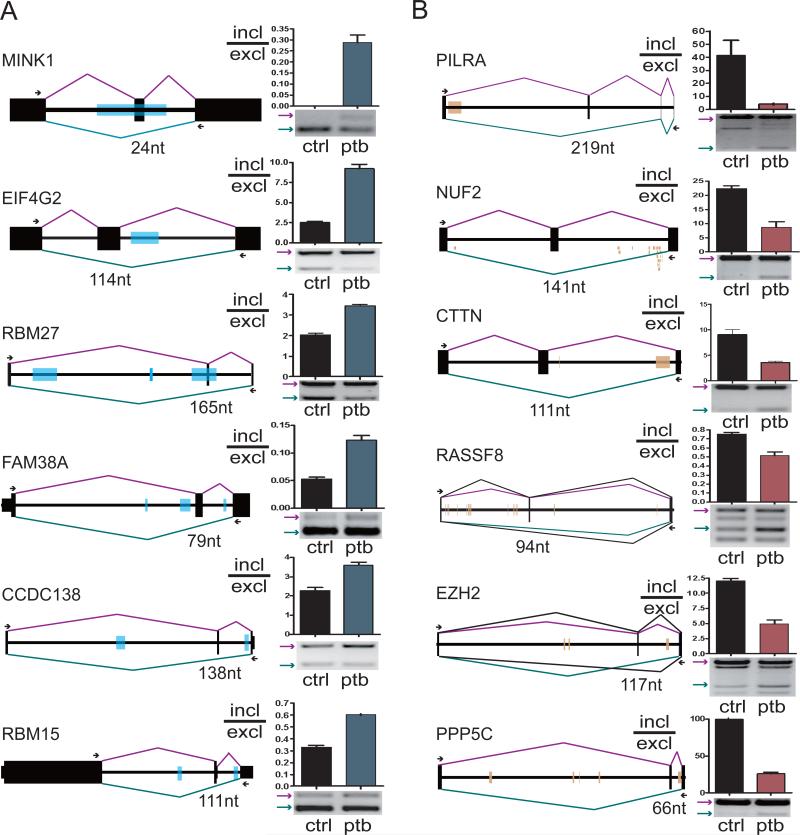
Figure 4. PTB can either represses or enhances alternative splicing in vivo.
(A) Examples of PTB-dependent exon skipping. Each is schematically diagramed (exon: black box, intron: black line) with mapped PTB binding clusters as marked by blue boxes. PTB RNAi induced splicing changes are shown on the right. Error bars are based on SEM from three independent experiments. All detected changes are significant as determined by the Student's t-test (P-value<0.05). (B) Examples of PTB-dependent exon inclusion with mapped PTB binding clusters marked by brown boxes. For NUF2, PTB tags (not clusters) are shown under the intron line.
We next attempted to rescue the splicing defect by co-transfecting the cell with a plasmid expressing PTB or nPTB, each of which contains a synonymous mutation that disrupts the shRNA target. By Western blotting, these exogenous genes were robustly expressed (Fig. 3B). We observed that the full-length PTB (PTB4, see below) was able to fully rescue the inclusion of the alternative exon 11 (Fig. 3C, lane 3). nPTB was also capable of rescuing exon 11 inclusion to a significant degree (Fig. 3C and 3D). Previous studies showed that the PTB gene expresses two major isoforms PTB4 and PTB1, which differ by the presence or absence of the alternative exon 9 (Wollerton et al., 2001). We found that PTB1 had little activity in rescuing the inclusion of CTTN exon 11 in comparison with the exon 9-containing PTB4, even though both proteins were expressed at comparable levels in transfected cells. This observation is consistent with the previous study that reported a stronger activity of PTB4 than PTB1 in regulated splicing (Wollerton et al., 2001). In these rescue experiments, we did not detect further increase in exon 11 inclusion even though the exogenous PTB or nPTB was overexpressed, indicating that PTB or nPTB is involved in the regulation but is not the only regulator(s) for this alternative splicing event (as a result, it is no longer a rate limiting factor in PTB-overexpressed cells).
To determine whether the regulation is dependent on the mapped PTB binding site in the intron, we deleted the 44nt PTB binding site in the reporter and found that the mutation abolished the response to exogenous PTB or nPTB (Fig. 3C and 3D). Deletion of the PTB binding site renders levels of exon inclusion in the CTTN minigene similar to those caused in the wild type by depletion of PTB/nPTB, further supporting the involvement of these proteins in regulation. Deletion of the PTB binding sites also abolished the functional rescue by any PTB isoforms. We conclude from these experiments that PTB/nPTB is also directly involved in regulated exon inclusion in addition to its widely perceived role in exon skipping.
Mechanistic insights into PTB-regulated alternative splicing
In order to understand the mechanisms for PTB-dependent exon inclusion or skipping, we analyzed the PTB binding pattern with respect to the functional consequence of alternative splicing and realized some general trends for PTB-regulated splicing (Fig. 4). Among PTB-mediated exon repression events, we note that PTB binding typically takes place near the alternative exon. This is clearly the case with both the MINK1 and EIF4G2 genes (Fig. 4A, row 1 and 2). However, PTB also binds to other intronic locations besides around the alternative exon as seen on the RBM27 and FAM38A gene (Fig. 4A, row 3 and 4). The remaining two examples (CCDC138, and RBM15, row 5 and 6 in Fig. 4A) illustrate PTB binding on both sides of the regulated exon, although the upstream PTB binding sites appear to vary in distance from the regulated exon. In these cases, we notice a relatively short intron after the alternative exon, indicating that PTB binding in the intron might obstruct the intron definition process (Fox-Walsh et al., 2005), thus resulting in PTB-dependent skipping of the alternative exon. These examples agree in general with the established principle of PTB-dependent exon skipping where PTB appears to mainly act to interfere with the recognition of the splice sites associated with the alternative exon.
PTB-dependent exon inclusion seems to exhibit a different trend. As illustrated in Fig. 4B, the first three examples exhibited PTB binding events that are far away from the alternative exon and close to the competing constitutive 5’ (RILRA, row 1) or 3’ splice site (NUF2, row 2). This trend may also be applicable to the CTTN gene (Fig. 4B, row 3), despite of a minor PTB binding site near the alternative exon, and our mutagenesis study showed that the major site near the downstream constitutive 3’ splice site was responsible for PTB-dependent exon inclusion (Fig. 3). The remaining three examples (RASSF8, EZH2 and PPP5C, row 4 to 6) are not clear-cut. PTB clearly binds to both sides of the alternative exon in each case, which is similar to the situation with PTB-dependent exon skipping events. However, both PTB binding sites appear closer to the competing constitutive 5’ and 3’ splice sites than to the alternative exon. Together, these examples appear to point to the trend that the PTB binding sites associated with PTB-dependent exon inclusion events are associated with competing constitutive splice sites.
To generalize the trend for both PTB-regulated exon inclusion and skipping, we collected a number of PTB-regulated exons, including 22 identified in the present study and 11 that has been previously reported in humans (supplementary Table S1). In addition, we found that the CLIP tags are generally mapped to the conserved regions on PTB-regulated mouse genes as reported previously (Boutz et al., 2007), which strongly implicates similar regulation between mice and humans. We directly tested a subset of the human orthologs of several PTB-regulated genes previously characterized on mouse cells, including SPAG9, PTB, nPTB, TPM1, KTN1, TPM2 and MINK1, and found that these genes all similarly responded to PTB knockdown in HeLa cells. We therefore included additional PTB-regulated splicing events in mouse cells (supplementary Table S1), resulting in a total of 55 PTB-regulated splicing events (41 PTB-dependent exon skipping and 14 PTB-dependent exon inclusion) for further analysis. As controls, we selected 100 groups of randomly sampled constitutive exons (each group contains 50 exons) for similar analysis.
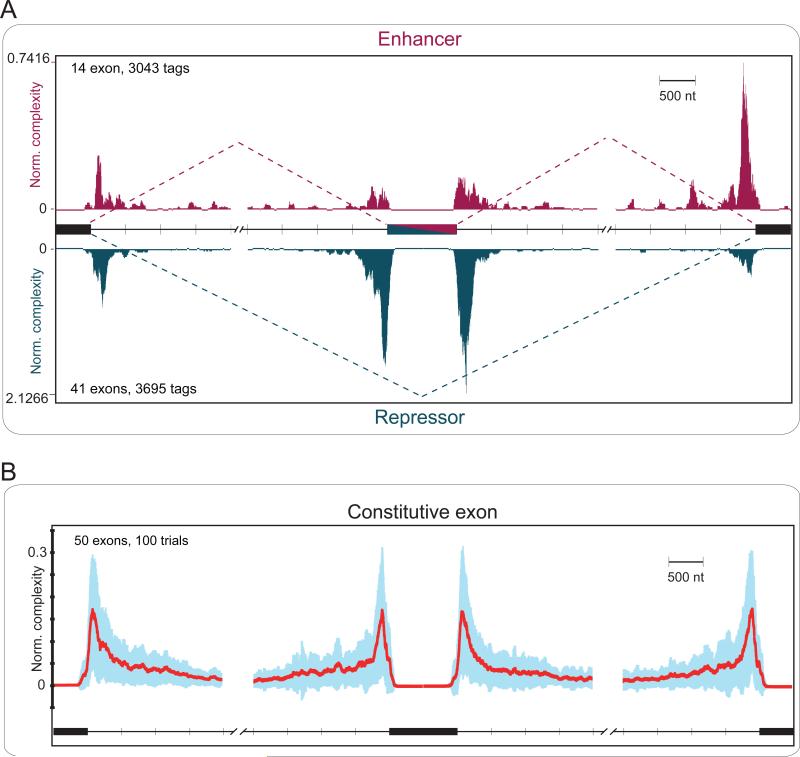
By integrating all PTB binding events, we generated an RNA map associated with PTB-repressed, PTB-enhanced, and PTB-nonregulated (constitutive) exons on a scaled pre-mRNA model, an approach that has been recently used for analysis of position-dependent activities of Nova (Licatalosi et al., 2008). Interestingly, the map revealed that PTB binds to both the 5’ and 3’ splice sites of constitutive exons as well as to both the 5’ and 3’ splice sites of alternative exons (Fig. 5). While PTB binding to the 3’ splice site is expected (because of the polypyrimdine tract as part of the splicing signal at the 3’ splice site), we were surprised by equally frequent PTB binding at the 5’ splice site. Most PTB-dependent exon skipping events (bottom portion of Fig. 5A) are associated with PTB binding near either side of the alternative exon, which is fully consistent with functional studies conducted so far on model minigenes. In contrast, the RNA map associated with PTB-dependent exon inclusion events (top portion of Fig. 5A) suggests that PTB binds prevalently to the flanking constitutive splice sites, especially at the downstream constitutive 3’ splice site (Fig. 5A). This most likely reflects PTB interference with the recognition of the competing constitutive 3’ splice site, therefore in favor of the selection of the upstream alternative exon. On non-regulated exons, we found no clear bias in PTB binding to intronic regions near any upstream or downstream splice sites (Fig. 5B). Together, these findings formally suggest a PTB-mediated splice site titration mechanism by which the relative binding frequency near the competing constitutive and alternative splice sites dictates the functional outcome, which appears to be neutralized on non-regulated constitutive exons (see further in Discussion).
Figure 5. Composite functional map of PTB-regulated splicing.
(A) PTB-regulated cassette exons are collected from previously reported cases and those that are validated in the present study. Among 55 PTB-regulated splicing events compiled, 14 exhibited PTB-depended splicing inclusion and 41 showed PTB-depended exon skipping. (B) RNA map on constitutive exons. Red line shows the average of normalized complexity of 100 sets (each contains 50 randomly selected exons) of constitutive exons, the upper and lower light blue boundaries show the one standard deviation (see Methods for further details).
Induction of exon inclusion by engineered PTB binding sites
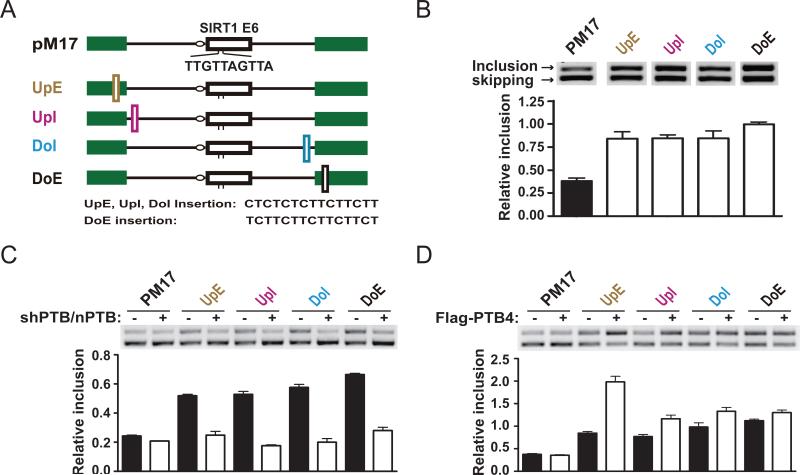
We demonstrated that the PTB binding site near the constitutive 3’ splice site of the CTTN gene is responsible for PTB-dependent inclusion of the upstream alternative exon (Fig. 3). To further test the hypothesis that PTB induces the inclusion of the alternative exon by weakening the competing constitutive splice site(s), we engineered a different minigene containing a SIRT1 exon (Fig. 6A), which was previously used to screen for cis-acting splicing suppressors (Wang et al., 2004). To improve the PTB response range of the reporter, we made a minor modification on the sequence in the SIRT1 exon to reduce its inclusion level and selected 4 regions to insert a PTB binding site (Fig. 6B). In order not to directly interfere with U1 binding, the positions for insertion in the upstream exon (UpE) or intron (UpI) are both ~15nt away from the constitutive 5’ splice site. To avoid obstruction of 3’ splicing signals, the position for insertion in the downstream intron (DoI) is 15nt upstream of the branchpoint whereas the position for insertion in the downstream exon (DoE) is 10nt from the 3’ AG di-nucleotide.
Figure 6. Mechanism of PTB-dependent exon inclusion.
(A) The reporter construct. pM17 is derived from pZW8-ESS17 (Wang et al, 2004) with a cis-acting regulatory element disrupted by the inserted sequence in the alternative SIRT1 exon. Four positions are selected for inserting a PTB binding site as diagrammed. (B) Splicing of the parental and mutant reporters in transfected HeLa cells was determined by RT-PCR (representative gel images shown as inserts) and quantified. (C and D) PTB-dependent exon inclusion through the inserted PTB binding sites. The effect on exon inclusion was diminished by the shRNAs against PTB and nPTB in co-transfected HeLa cells (C). The effect on exon inclusion could be further enhanced by PTB overexpression (D). Together, these results demonstrate that PTB is directly involved in enhancing exon inclusion through the inserted binding sites. Error bars are based on SEM derived from three independent experiments.
We transfected the parental splicing reporter and the PTB site insertion derivatives into HeLa cell and analyzed the splicing products by semi-quantitative RT-PCR. As shown in Fig. 6B, insertion of a PTB binding site near either the 5’ or 3’ constitutive splice site significantly enhanced the inclusion of the alternative exon. RNAi knockdown of PTB and nPTB completely abolished the exon inclusion induced by inserted PTB binding sites (Fig. 6C), and overexpression of PTB4 further enhanced exon inclusion in a PTB binding site-dependent manner (Fig. 6D). These observations provide an unequivocal support to the splice site titration mechanism for PTB-dependent exon inclusion where weakening the constitutive 5’ or 3’ splice site enhances the competitiveness of the alternative 5’ or 3’ splice site. These findings have therefore documented a new positional effect for a general splicing repressor to positively regulate alternative splicing in mammalian cells.
Discussion
Our global analysis of PTB-RNA interactions in the human genome provides mechanistic insights into PTB-regulated RNA processing. Besides competing directly with U2AF65 binding to interfere with 3’ splice site recognition (Lin and Patton, 1995; Sauliere et al., 2006; Singh et al., 1995), PTB has been shown to use multiple mechanisms to regulate alternative RNA processing by binding to regions other than the core splicing signals on minigene models (Izquierdo et al., 2005; Sharma et al., 2008; Spellman and Smith, 2006). We have now generalized and significantly extended these findings at the genome level.
Interference of splice site recognition and communication by PTB-mediated RNA networks
RNA looping has been proposed as one of the mechanisms for PTB-mediated splicing repression to sequester the alternative exon from the splicing machinery (Chou et al., 2000; Wagner and Garcia-Blanco, 2001). PTB dimerization was initially postulated to facilitate RNA looping (Oh et al., 1998; Perez et al., 1997b), but a later structural analysis suggests potential induction of RNA looping via RRM3 and RRM4 in the same PTB molecule to simultaneously bind to cis-acting RNA elements (Oberstrass et al., 2005). However, these two modes of RNA looping induced by inter- or intra-molecular interactions do not have to be mutually exclusive. Although purified PTB exists predominantly as a monomer in solution, which can bind to RNA with high affinity, it has been suggested that PTB binding to RNA may create the spatial proximity for enhanced PTB-PTB interactions on RNA, which may be stabilized by the induced formation of a disulfide bond (Amir-Ahmady et al., 2005; Monie et al., 2005; Oberstrass et al., 2005). Our data are fully consistent with PTB binding to RNA as monomer and subsequent disulfide bond formation on closely spaced PTB molecules on target RNA. Interestingly, we found that UV can further enhance or stabilize PTB-PTB interactions. Importantly, our data indicate that there are no separate sets of binding sties for monomeric and dimeric PTB in the human genome. However, this does not undermine the potential synergy between protein-protein and protein-RNA interactions that may be critical for induced RNA looping surrounding PTB-regulated exons as previously proposed (Wagner and Garcia-Blanco, 2001).
Given frequent PTB binding in multiple locations in a single intron in many cases (e.g. Fig. S6B), we may envision an extensive RNA network nucleated by PTB, which may be the underlying mechanism for the observed interference of both exon definition and the transition from exon definition to intron definition during spliceosome assembly (Izquierdo et al., 2005; Sharma et al., 2005; Sharma et al., 2008). As there is no reason to believe that PTB-mediated RNA network has to be restricted within a single regulatory unit, we may further speculate that the network may spread on multiple intronic and exonic locations in the same pre-mRNA molecule, thereby allowing a coordinated regulation of multiple RNA processing events as evidenced on the TPM2 gene (Spellman et al., 2007). Such network may be more prevalent than what we can image at this point because initial PTB-RNA interactions may induce additional PTB binding to other sites that may not even contain a motif for high affinity binding by PTB, which may be further enhanced by other PTB co-factors, such as Raver1 (Gromak et al., 2003).
Mechanisms for positive and negative regulation of splice site selection by PTB
The PTB-RNA interaction map also suggests a potential mechanism for positive and negative regulation of splice site selection by PTB, depending its binding relative to competing constitutive and alternative splice sites. Interestingly, PTB not only binds to intronic locations near the 3’ splice site, but also to sites closer to the 5’ splice site regardless of whether the splice site is subjected to alternative choices. If predominant PTB binding occurs near a constitutive splice site, it may weaken the site, thereby raising the competitiveness of the competing alternative site. A minor modulation of splice site recognition may be translated into a major functional consequence as demonstrated by a recent kinetic analysis of splice site competition (Yu et al., 2008). This principle may be generally applicable to RNA binding splicing regulators to give rise to either a positive or negative functional outcome that depends on where the factor binds.
A positional effect has clearly emerged from recent genome-wide studies of splicing regulators (Licatalosi et al., 2008; Yeo et al., 2009). However, the positional effect we observed with PTB-regulated splicing appears to be fundamentally distinct from that exerted by Nova and Fox2. In those cases, Nova and Fox2 binding to their cis-acting elements upstream and downstream of the alternative exon generally represses or enhances the selection of the exon, respectively, but it is presently unclearly how such opposite effects on splice site selection are achieved. In contrast, PTB appears to be sampling multiple intronic locations in a pre-mRNA to exert a negative effect on the selection of the nearby splice site. PTB binding close to intronic region near the 3’ and 5’ alternative splice site likely results in skipping of the alternative exon whereas PTB binding to sequences adjacent to constitutive exons tends to induce the inclusion of the alternative exon.
It is important to point out that potential composite effects may count for some apparent exceptions to this general trend. For example, the PTB binding pattern was similar on both the CCDC138 and EZH2 genes, but PTB knockdown had opposite effects on these two genes. The PTB binding events on the CCDC138 gene are both far away from the regulated exon, yet the net effect is PTB-dependent exon skipping, perhaps because the exon skipping effect due to strong PTB binding on both sides of the alternative exon might be dominant over its influence on the downstream constitutive exon. Therefore, the final functional outcomes in many cases may be determined by the sum of those competing binding events. In addition, most alternative splicing events are likely subjected to regulation by multiple different splicing regulators, which may act synergistically or antagonistically. Therefore, the possibility that other regulators may override the effect of PTB binding on certain regulated exons may be account for various exceptions to the position effect observed, thus emphasizing the combinatory control of alternative splicing that likely operate in mammalian cells.
Material and Methods
Cell culture, plasmids, and RNAi
HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM) with 10% newborn bovine serum plus 100 U penicillin /streptomycin (Gibico) at 37°C in 5% CO2. Oligofectamine and Lipo2000 (both from Invitrogen) were used for siRNA and plasmid transfection, respectively, according to manufacturer's instructions. To construct the expression plasmids for PTB1, PTB4 and nPTB, flag-tagged primer sets were used to amplify the coding region of individual genes and the PCR products were inserted into pcDNA3 between the EcoR1 and Not1 sites. The CTTN minigene was constructed by amplifying Exon 10 to Exon 12 regions, which were inserted into pcDNA3 between the EcoR1 and Xho1 sites. Validated siRNAs against PTB and nPTB were purchased from Ribbobio.
CLIP-seq, RIP-PCR and validation of PTB-regulated splicing
HeLa cells were UV-irradiated at 400 mj and collected by scraping the cells from 15-cm plates. The CLIP procedure was performed as described (Yeo et al., 2009). Immunoprecipitated RNA was extracted using Trizol, and after DNase I (Promega) treatment, RNA was reverse transcribed using MMLV with N9 random primer at 37°C followed by inactivation of the reverse transcriptase at 70°C for 15min. The resulting cDNA was analysis by PCR. Supplementary Table S2 lists PCR primers for validation of PTB-dependent splicing.
Bioinformatics analysis
The sequenced tags longer than 18nt were mapped to the human genome sequence by allowing 2 mismatches, 2 insertions or deletions, and only those with maximal identity (≥90%) were kept. To analyze genomic distribution of tags, known human genes (knownGene track from the UCSC genome browser) were chosen as the gene set, which contained 66,803 entries. We arbitrarily defined promoter regions as 5kb upstream of the transcriptional start site of the gene. For genes having multiple isoforms, the transcript with the longest length, the largest number of exons, the longest CDS, or the maximum length of all exons was chosen to analyze the tag distribution in exon, intron, 5’ UTR, CDS and 3’ UTR.
The PTB binding sites enriched of tags were detected by using the similar strategy as previously described (Yeo et al., 2009). The differences were: 1) tags were not extended; 2) peak identification was independently done for each gene cluster of 25,179 clusters, which were grouped by using the program clusterGenes on all known genes; 3) consecutive positions with same height was counted only once; 4) control tags were randomly placed on gene cluster with repetitive-elements masked; 5) For each height level h, the p-value was assigned as the ratio of the number of heights higher than h divided by the total number of heights in 100 random placements, and the p-values for all heights were adjusted by using Bonferroni correction to account for multiple hypotheses testing. The smallest height that gave an FDR<0.001 was defined as the threshold height. Consecutive nucleotide positions with height higher than the threshold were identified as significant PTB binding peak. If multiple peaks were detected less than 50nt from one another, they were merged to represent a single PTB binding site or cluster.
The sequences extracted from genome according to PTB binding clusters were used to detect overrepresented motifs (Defrance et al., 2008). For each cluster, it was extended to the two sides by 25nt, as some clusters had small lengths. Background sequences include those from randomly selected intervals in genes or random sequences generated with respect to order 0 and 1 Markov models (same single and di-nucleotide frequencies) built from knownGenes (Ponty et al., 2006). Identification of overrepresented k-mers (k=2, 3, 4, 5, 6 and 7) was based on random intervals. The pictogram was plotted according to WebLogo (http://weblogo.berkeley.edu/), which is based on the alignment of the top 20 motifs by ClustalW (http://www.ebi.ac.uk/Tools/clustalw/).
Normalized complexity map of PTB–RNA interactions was generated as described in (Licatalosi et al., 2008). The composite pre-mRNA was made by joining the longest upstream exon, upstream intron, middle exon (PTB-regulated exon), downstream intron and downstream exon. The tags around PTB-regulated individual exons were mapped to the composite pre-mRNA according to their positions relative to the nearest splice site. The tags in one transcript were first normalized to their number across the region covering the PTB-regulated exon and flanking introns and exons, and then to the number of different transcripts with tags at a given position as described (Licatalosi et al., 2008). For comparison, we extracted 6460 sets of three constitutive internal exons that are associated with PTB binding from knownGene set (hg18). Normalized complexity map was similarly created on 50 randomly selected constitutive exons to deduce both averaged PTB binding events with standard deviation.
The analysis used the programs from Jim Kent's source code (http://www.soe.ucsc.edu/~kent), bx-python library (http://bitbucket.org/james_taylor/bx-python/), pygr libray (http://code.google.com/p/pygr/), and homemade python codes.
Supplementary Material
Figure S1. Analysis of PTB and associated RNA after Crosslinking Immunoprecipitation (CLIP). Left panel: PTB-RNA complex with the associated RNA labeled with [γ-32P] ATP after SDS-PAGE separation and transfer to nitrocellulose. Right panel: After autoradiography, the same membrane was processed for Western.
Figure S2. Pie-chart of the distribution of PTB binding tags identified by direct cloning/sequencing the human genome. Tags associated with PTB monomer (A) and dimer (B) show a similar genomic distribution.
Figure S3. The length distribution of sequenced cloned tags. About 900 clones containing the cDNA reverse-transcribed from the PTB-bound RNA tags were sequenced. After filtering, unique 341 monomeric (A) and 211 dimeric (B) PTB-associated tags (≥16 nt) were obtained and plotted against the tag length.
Figure S4. Validation of anti-PTB enriched by RIP-PCR. (A) Determination of the IP efficiency of the monoclonal BB7 anti-PTB antibody. (B) RT-PCR analysis of anti-PTB enriched RNA using specific primers designed around identified tag sequences. The major band in SERINC2 sample corresponds to primer dimer.
Figure S5. The Pearson correlation coefficient between monomeric and dimeric PTB tags. The genome was partitioned into a series of bins by a specific window size, the number of tags in each bin was counted for both monomeric and dimeric tags, and then the correlation efficient in all bins was calculated. The coefficient, indicating the similarity of the binding profile of monomeric and dimeric tags, increases as the number of tags increases, and also increases as the window size becomes larger.
Figure S6. Enriched and depleted motifs among PTB CLIP tags. (A) List of top 20 enriched motifs and 5 under-represented motifs. (B) The percentage of PTB binding sites that contain top enriched motifs.
Figure S7. The C/U content of the sequences associated with PTB binding peaks in comparison with background. Colored shade represents the standard derivation in each case. The results clearly illustrate significant elevation of the C/U content surrounding the PTB binding peaks.
Supplementary Table S1. List of PTB-regulated splicing events used to construct the RNA map. kgID: knownGene ID in UCSC genome database (hg18). The “Fold change” value means the change of relative inclusion level between PTB knockdown and normal condition, NA: not available, INF: Infinite. In Up/down column, ‘+’ and ‘-’ mean the inclusion level of the exon is reduced or enhanced under PTB knockdown, respectively. In the src (data source) column: a) validated by Xue in this study; number is the literature id listed under the table below.
Supplementary Table S2. List of PCR primers used to validate PTB-regulated splicing events. IN: inclusion, SK: skipping.
Reference:
1. Xing, Y., Stoilov, P., Kapur, K., Han, A., Jiang, H., Shen, S., Black, D.L., and Wong, W.H. (2008). MADS: a new and improved method for analysis of differential alternative splicing by exon-tiling microarrays. RNA 14, 1470-1479.
2. Boutz, P.L., Stoilov, P., Li, Q., Lin, C.H., Chawla, G., Ostrow, K., Shiue, L., Ares, M., Jr., and Black, D.L. (2007). A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev 21, 1636-1652.
3. Wollerton, M.C., Gooding, C., Wagner, E.J., Garcia-Blanco, M.A., and Smith, C.W. (2004). Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol Cell 13, 91-100.
4. Spellman, R., Llorian, M., and Smith, C.W. (2007). Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol Cell 27, 420-434
5. Cote, J., Dupuis, S., Jiang, Z., and Wu, J.Y. (2001). Caspase-2 pre-mRNA alternative splicing: Identification of an intronic element containing a decoy 3' acceptor site. Proc Natl Acad Sci U S A 98, 938-943.
6. Izquierdo, J.M., Majos, N., Bonnal, S., Martinez, C., Castelo, R., Guigo, R., Bilbao, D., and Valcarcel, J. (2005). Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol Cell 19, 475-484.
Acknowledgements
The authors are grateful to Chris Smith, Zefeng Wang, and Reuven Agami for sending us PTB expression plasmids and other regents, and Miriam Llorian for advice on PTB RNAi, and Alain Denise for suggestions in computational analysis. We are also indebted to members of the Yi Zhang lab for cooperation and discussion during this course of this investigation. This work is supported by the China 863 program (2007AA02Z112) to Y.Z., the China 973 program (2005CB724604) to Y.Z. and X.D.F., and by US NIH grants (GM049369, HG004659, GM084317) to D.L.B., G.Y., and X.D.F.
References
- Amir-Ahmady B, Boutz PL, Markovtsov V, Phillips ML, Black DL. Exon repression by polypyrimidine tract binding protein. RNA. 2005;11:699–716. doi: 10.1261/rna.2250405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashiya M, Grabowski PJ. A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart. RNA. 1997;3:996–1015. [PMC free article] [PubMed] [Google Scholar]
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares M, Jr., Black DL. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cama-Carvalho M, Barbosa-Morais N, Brodsky AS, Silver PA, Carmo-Fonseca M. Genome-wide identification of functionally distinct subsets of cellular mRNAs associated with two nucleocytoplasmic-shuttling mammalian splicing factors. Genome Biol. 2006;7:R113. doi: 10.1186/gb-2006-7-11-r113. 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou MY, Underwood JG, Nikolic J, Luu MH, Black DL. Multisite RNA binding and release of polypyrimidine tract binding protein during the regulation of c-src neural-specific splicing. Mol Cell. 2000;5:949–957. doi: 10.1016/s1097-2765(00)80260-9. [DOI] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Clerte C, Hall KB. The domains of polypyrimidine tract binding protein have distinct RNA structural preferences. Biochem. 2009 doi: 10.1021/bi8016872. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defrance M, Janky R, Sand O, van Helden J. Using RSAT oligo-analysis and dyad-analysis tools to discover regulatory signals in nucleic sequences. Nat Protoc. 2008;3:1589–1603. doi: 10.1038/nprot.2008.98. [DOI] [PubMed] [Google Scholar]
- Fox-Walsh KL, Dou Y, Lam BJ, Hung SP, Baldi PF, Hertel KJ. The architecture of pre-mRNAs affects mechanisms of splice-site pairing. Proc Natl Acad Sci U S A. 2005;102:16176–16181. doi: 10.1073/pnas.0508489102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XD. Towards a splicing code. Cell. 2004;119:736–738. doi: 10.1016/j.cell.2004.11.039. [DOI] [PubMed] [Google Scholar]
- Gromak N, Rideau A, Southby J, Scadden AD, Gooding C, Huttelmaier S, Singer RH, Smith CW. The PTB interacting protein raver1 regulates alpha-tropomyosin alternative splicing. EMBO J. 2003;22:6356–6364. doi: 10.1093/emboj/cdg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo JM, Majos N, Bonnal S, Martinez C, Castelo R, Guigo R, Bilbao D, Valcarcel J. Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol Cell. 2005;19:475–484. doi: 10.1016/j.molcel.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Karolchik D, Kuhn RM, Baertsch R, Barber GP, Clawson H, Diekhans M, Giardine B, Harte RA, Hinrichs AS, Hsu F, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Patton JG. Regulation of alternative 3' splice site selection by constitutive splicing factors. RNA. 1995;1:234–245. [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- Monie TP, Hernandez H, Robinson CV, Simpson P, Matthews S, Curry S. The polypyrimidine tract binding protein is a monomer. RNA. 2005;11:1803–1808. doi: 10.1261/rna.2214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, et al. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- Oh YL, Hahm B, Kim YK, Lee HK, Lee JW, Song O, Tsukiyama-Kohara K, Kohara M, Nomoto A, Jang SK. Determination of functional domains in polypyrimidine-tract-binding protein. Biochem J. 1998;331(Pt 1):169–175. doi: 10.1042/bj3310169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis C, Cloutier P, Shkreta L, Toutant J, Klarskov K, Chabot B. hnRNP I/PTB can antagonize the splicing repressor activity of SRp30c. RNA. 2007;13:1287–1300. doi: 10.1261/rna.403607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez I, Lin CH, McAfee JG, Patton JG. Mutation of PTB binding sites causes misregulation of alternative 3' splice site selection in vivo. RNA. 1997a;3:764–778. [PMC free article] [PubMed] [Google Scholar]
- Perez I, McAfee JG, Patton JG. Multiple RRMs contribute to RNA binding specificity and affinity for polypyrimidine tract binding protein. Biochemistry. 1997b;36:11881–11890. doi: 10.1021/bi9711745. [DOI] [PubMed] [Google Scholar]
- Ponty Y, Termier M, Denise A. GenRGenS: software for generating random genomic sequences and structures. Bioinformatics. 2006;22:1534–1535. doi: 10.1093/bioinformatics/btl113. [DOI] [PubMed] [Google Scholar]
- Sauliere J, Sureau A, Expert-Bezancon A, Marie J. The polypyrimidine tract binding protein (PTB) represses splicing of exon 6B from the beta-tropomyosin pre-mRNA by directly interfering with the binding of the U2AF65 subunit. Mol Cell Biol. 2006;26:8755–8769. doi: 10.1128/MCB.00893-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Falick AM, Black DL. Polypyrimidine tract binding protein blocks the 5' splice site-dependent assembly of U2AF and the prespliceosomal E complex. Mol Cell. 2005;19:485–496. doi: 10.1016/j.molcel.2005.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Kohlstaedt LA, Damianov A, Rio DC, Black DL. Polypyrimidine tract binding protein controls the transition from exon definition to an intron defined spliceosome. Nat Struct Mol Biol. 2008;15:183–191. doi: 10.1038/nsmb.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Valcarcel J, Green MR. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- Spellman R, Llorian M, Smith CW. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol Cell. 2007;27:420–434. doi: 10.1016/j.molcel.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman R, Smith CW. Novel modes of splicing repression by PTB. Trends Biochem Sci. 2006;31:73–76. doi: 10.1016/j.tibs.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Wagner EJ, Garcia-Blanco MA. Polypyrimidine tract binding protein antagonizes exon definition. Mol Cell Biol. 2001;21:3281–3288. doi: 10.1128/MCB.21.10.3281-3288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Wollerton MC, Gooding C, Robinson F, Brown EC, Jackson RJ, Smith CW. Differential alternative splicing activity of isoforms of polypyrimidine tract binding protein (PTB). RNA. 2001;7:819–832. doi: 10.1017/s1355838201010214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Stoilov P, Kapur K, Han A, Jiang H, Shen S, Black DL, Wong WH. MADS: a new and improved method for analysis of differential alternative splicing by exon-tiling microarrays. RNA. 2008;14:1470–1479. doi: 10.1261/rna.1070208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Maroney PA, Denker JA, Zhang XH, Dybkov O, Luhrmann R, Jankowsky E, Chasin LA, Nilsen TW. Dynamic regulation of alternative splicing by silencers that modulate 5' splice site competition. Cell. 2008;135:1224–1236. doi: 10.1016/j.cell.2008.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Analysis of PTB and associated RNA after Crosslinking Immunoprecipitation (CLIP). Left panel: PTB-RNA complex with the associated RNA labeled with [γ-32P] ATP after SDS-PAGE separation and transfer to nitrocellulose. Right panel: After autoradiography, the same membrane was processed for Western.
Figure S2. Pie-chart of the distribution of PTB binding tags identified by direct cloning/sequencing the human genome. Tags associated with PTB monomer (A) and dimer (B) show a similar genomic distribution.
Figure S3. The length distribution of sequenced cloned tags. About 900 clones containing the cDNA reverse-transcribed from the PTB-bound RNA tags were sequenced. After filtering, unique 341 monomeric (A) and 211 dimeric (B) PTB-associated tags (≥16 nt) were obtained and plotted against the tag length.
Figure S4. Validation of anti-PTB enriched by RIP-PCR. (A) Determination of the IP efficiency of the monoclonal BB7 anti-PTB antibody. (B) RT-PCR analysis of anti-PTB enriched RNA using specific primers designed around identified tag sequences. The major band in SERINC2 sample corresponds to primer dimer.
Figure S5. The Pearson correlation coefficient between monomeric and dimeric PTB tags. The genome was partitioned into a series of bins by a specific window size, the number of tags in each bin was counted for both monomeric and dimeric tags, and then the correlation efficient in all bins was calculated. The coefficient, indicating the similarity of the binding profile of monomeric and dimeric tags, increases as the number of tags increases, and also increases as the window size becomes larger.
Figure S6. Enriched and depleted motifs among PTB CLIP tags. (A) List of top 20 enriched motifs and 5 under-represented motifs. (B) The percentage of PTB binding sites that contain top enriched motifs.
Figure S7. The C/U content of the sequences associated with PTB binding peaks in comparison with background. Colored shade represents the standard derivation in each case. The results clearly illustrate significant elevation of the C/U content surrounding the PTB binding peaks.
Supplementary Table S1. List of PTB-regulated splicing events used to construct the RNA map. kgID: knownGene ID in UCSC genome database (hg18). The “Fold change” value means the change of relative inclusion level between PTB knockdown and normal condition, NA: not available, INF: Infinite. In Up/down column, ‘+’ and ‘-’ mean the inclusion level of the exon is reduced or enhanced under PTB knockdown, respectively. In the src (data source) column: a) validated by Xue in this study; number is the literature id listed under the table below.
Supplementary Table S2. List of PCR primers used to validate PTB-regulated splicing events. IN: inclusion, SK: skipping.
Reference:
1. Xing, Y., Stoilov, P., Kapur, K., Han, A., Jiang, H., Shen, S., Black, D.L., and Wong, W.H. (2008). MADS: a new and improved method for analysis of differential alternative splicing by exon-tiling microarrays. RNA 14, 1470-1479.
2. Boutz, P.L., Stoilov, P., Li, Q., Lin, C.H., Chawla, G., Ostrow, K., Shiue, L., Ares, M., Jr., and Black, D.L. (2007). A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev 21, 1636-1652.
3. Wollerton, M.C., Gooding, C., Wagner, E.J., Garcia-Blanco, M.A., and Smith, C.W. (2004). Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol Cell 13, 91-100.
4. Spellman, R., Llorian, M., and Smith, C.W. (2007). Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol Cell 27, 420-434
5. Cote, J., Dupuis, S., Jiang, Z., and Wu, J.Y. (2001). Caspase-2 pre-mRNA alternative splicing: Identification of an intronic element containing a decoy 3' acceptor site. Proc Natl Acad Sci U S A 98, 938-943.
6. Izquierdo, J.M., Majos, N., Bonnal, S., Martinez, C., Castelo, R., Guigo, R., Bilbao, D., and Valcarcel, J. (2005). Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol Cell 19, 475-484.