Abstract
Cbl is the product of the protooncogene c-cbl and is involved in T cell antigen receptor (TCR)-mediated signaling. To understand the role of Cbl for immune system development and function, we generated a Cbl-deficient mouse strain. In Cbl-deficient mice, positive selection of the thymocytes expressing major histocompatibility complex class II-restricted transgenic TCR was significantly enhanced. Two factors may have contributed to the altered thymic selection. First, Cbl deficiency markedly up-regulated the activity of ZAP-70 and mitogen-activated protein kinases. The mitogen-activated protein kinase pathway was shown previously to be involved in thymic positive selection. Second, Cbl-deficient thymocytes expressed CD3 and CD4 molecules at higher levels, which consequently may increase the avidity of TCR/major histocompatibility complex/coreceptor interaction. Thus, Cbl plays a novel role in modulating TCR-mediated multiple signaling pathways and fine-tunes the signaling threshold for thymic selection.
Thymic positive and negative selections are two major events that form the peripheral T cell repertoire. Both require interactions of T cell antigen receptors (TCR) and coreceptors on thymocytes with major histocompatibility complex (MHC) and other ligands on thymic antigen-presenting cells (1–4). The outcome of the selection is believed to be determined by the strength of intracellular signals delivered by TCR. The current consensus is that thymocytes receiving a moderate-strength TCR signal are positively selected and develop further into mature T cells, whereas cells receiving either too weak or strong signals will be eliminated by “neglect” or negative selection, respectively, during development (5–8). Although the definition of the signal “strength” remains elusive, it probably reflects a combinatory effect of the affinity and avidity of TCR/MHC interactions and the efficiency of intracellular signal transduction determined by the availability of the relevant signal-transducing molecules.
Most of our knowledge concerning the signal transduction during thymic positive and negative selections came from either natural or engineered genetic mutant animals (9). It has been demonstrated that inactivation of the tyrosine kinases Lck and ZAP-70 or the tyrosine phosphatase CD45 abrogates both positive and negative selections (10–13). In contrast, activation of Vav and Ras-Raf-MAP kinase pathway is necessary for positive selection but dispensable for negative selection (14–17). Molecules such as CD5, CD30, IRF-1, and Itk are also known to be involved in thymic selection (18–21) although their place in the TCR-mediated signal transduction pathway is not clear.
c-cbl was identified previously as a cellular homologue of the oncogene v-cbl from Cas-NS1 retrovirus, which causes hematopoietic malignancies in mice (22). While c-cbl transcripts are detected in a variety of tissues, the highest level of expression was observed in thymus and testis (23). The function of Cbl, a protein product of the c-cbl gene, as a signaling protein has been evidenced by its prominent tyrosine phosphorylation upon engagement of various growth factor receptors and antigen receptors (24, 25). Despite lacking any known enzymatic activity, Cbl forms complexes with many different signaling proteins, including Src- and Syk-family tyrosine kinases, phosphatidylinositol-3 kinase (PI-3K), and the adapter proteins such as Grb2 (24, 25). Ota and Samelson reported that Cbl was involved in the control of Syk activity in a mast cell line (26). Genetic studies from Caenorhabditis elegans and Drosophila suggested that Cbl homologues were involved in the regulation of epidermal growth factor receptor-Ras signaling pathway in these organisms (27–29). More recently, T cell signaling was shown to be enhanced in Cbl-deficient mouse thymocytes (30). However, it remains to be determined how the enhanced signaling affects T cell development and function.
MATERIALS AND METHODS
Mice.
All the mice used in this study were bred and maintained at National Institute of Allergy and Infectious Diseases Twinbrook II Animal Facility under specific pathogen-free conditions in accordance with institutional guidelines. To generate the targeted embryonic stem (ES) cell clone, 30 μg of the linearized targeting vector was electroporated into E14.1 ES cells. ES cells were selected with G418 and gancyclovir, and double-resistant clones were screened for homologous recombination by Southern hybridization. Correctly targeted clones were transiently transfected with a Cre recombinase expression vector to remove the neomycin-resistance gene and an exon of the c-cbl gene as indicated in Fig. 1A. Resultant ES clones were injected into blastocysts to obtain chimeric mice. TCR nontransgenic mice used in this study are of mixed C57BL/6 and 129 background (H-2b). Transgenic mice (5C.C7) were selected for the absence of mouse mammary tumor virus (MMTV)-3 and -13 integrants because the 129 background of the ES cells deletes this TCR through superantigen stimulation in the thymus (31). H-Y TCR transgenic mice are of H-2b background.
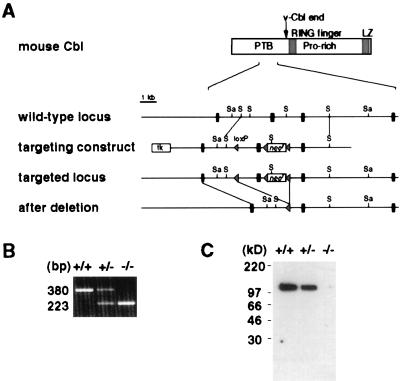
Figure 1.
Generation of Cbl-deficient mice by gene targeting. (A) Partial restriction map of the c-cbl locus, the targeting construct, and the mutated c-cbl locus. The second exon in the figure, which subsequently was deleted by Cre/loxP-mediated DNA recombination, corresponds to nucleotides 681–837 of the published mouse c-cbl cDNA sequence (GenBank accession no. X57111). Black rectangles represent exons, and triangles represent loxP sequence. Sa, SacI; S, SphI. (B) Reverse transcription–PCR analysis of c-cbl transcripts from wild-type (+/+), heterozygous (+/−), and homozygous (−/−) c-cbl mutant thymocytes. The sequences of the primers used in the PCRs correspond to nucleotides 640–663 (sense orientation) and to nucleotides 996-1019 (antisense orientation) of mouse c-cbl cDNA. (C) Immunoblot analysis of the Cbl protein. Thymocyte lysate was immunoblotted with a polyclonal antibody against the C terminus of the Cbl protein.
Antibodies and Flow Cytometry.
Anti-CD3ɛ (145–2C11), anti-CD4 (RM4–5), anti-CD5 (53–7.3), anti-CD8 (53–6.7), anti-CD28 (37.51), anti-CD69 (H1.2F3), anti-TCR Vα11 (RR8–1), and anti-TCR Vβ3 (KJ25) mAbs were obtained from PharMingen.
Flow cytometry was performed with FACScan (Becton Dickinson). Data were analyzed with flowjo analysis program (Tree Star, San Carlos, CA).
Detection of MMTV Genomes.
Genomic DNA was analyzed by PCR for the presence of MMTV-3 and -13. Primers were designed to amplify MMTV sequences specific for Vβ3-deleting virus integrants (32). The 129 strain does not have MMTV-1 or -6. Primer sequences are: sense, 5′-AAA AAG GGG AAA GAG GAG TGC GCT TGT CAA-3′, and antisense, 5′-ATT ATA TGT CTT TTG GCC TCC TCT CTA CTG-3′.
Peripheral T Cell Proliferation Assay.
CD4+ T cells were purified by depleting CD8+, B220+, and Mac1+ cells from mesenteric lymph node cells by using magnetic beads (PerSeptive Biosystems, Framingham, MA). Purity of CD4+ cells was 85–95% as assessed by flow cytometry. After stimulating with plate-bound anti-CD3 antibody for 42 hr, the culture was pulsed with 5 μCi/ml of [3H]thymidine and incubated for an additional 12 hr. [3H]Thymidine incorporation was measured by a scintillation counter, and the values were normalized against those of cells stimulated with PMA and ionomycin.
Western Blotting, Immunoprecipitation, and PI-3K Assay.
For all biochemical analyses, thymocytes were isolated and cultured ex vivo overnight before the experiment. Splenocytes were used fresh. Cells (5 × 107) were stimulated with 10 μg/ml of biotinylated anti-CD3ɛ, CD4, or CD28 antibodies followed by crosslinking with 25 μg/ml of streptavidin at 37°C. Stimulated cells were lysed in 1% Triton X-100 containing buffer with protease and phosphatase inhibitors and immunoprecipitated with indicated antibodies as described previously (33). Protein was separated by SDS/PAGE, electrotransferred to polyvinylidene difluoride membranes, and blotted with indicated antibodies.
The following antibodies were used: antiphosphotyrosine (4G10, Upstate Biotechnology, Lake Placid, NY), anti-Cbl and anti-Lck (Santa Cruz Biotechnology), anti-ZAP-70 (Transduction Laboratories, Lexington, KY), and anti-MAP kinase (Promega).
For the determination of PI-3K activity, cell lysate was immunoprecipitated with 4G10 and the associated PI-3K was assayed in vitro by 32P incorporation into phosphatidylinositol. The resulting phosphatidylinositol 3-phosphate was resolved by TLC in chloroform/methanol/water/ammonium hydroxide as described (34). 32P-labeled spots were visualized by autoradiography and quantitated by a densitometer.
RESULTS
Generation of Cbl-Deficient Mice.
Mice deficient in Cbl were generated by using a gene-targeting strategy depicted in Fig. 1A. The targeting vector was designed to delete one exon of the c-cbl gene and introduce a reading-frame shift in the resulting c-cbl transcripts (Fig. 1A). In c-cbl mutant mice, absence of aberrant transcripts of the c-cbl gene was confirmed by reverse transcription–PCR, and the Cbl protein was not detected by an antibody against the C terminus of the protein (Fig. 1 B and C). Homozygous c-cbl mutant (c-cbl−/−) mice are generally healthy and fertile. Inspection of various tissues in the adult c-cbl−/− animals (up to 4 months old) revealed no gross abnormality except a mild splenomegaly. Spleens from c-cbl−/− mice were two to three times larger than those of their wild-type (wt) littermates, primarily because of erythroid hyperplasia (data not shown).
Cbl Deficiency Elevates Cell Surface Molecules Related to Thymic Positive Selection.
Thymus is one of the adult organs in which Cbl is most highly expressed (23). We therefore examined whether the thymocyte development was affected by Cbl deficiency. Although the total number of thymocytes and the ratio of CD4/CD8 double-positive (DP), CD4, and CD8 single-positive (SP) thymocytes were almost the same in c-cbl−/− and wt mice, an increased proportion of thymocytes from c-cbl−/− mutants expressed higher levels of TCRαβ/CD3, CD5, and CD69 molecules than those from wt mice (Fig. 2). In DP cells, the expression of CD4 molecule was elevated moderately in c-cbl−/− mice (CD4 mean fluorescence intensity of wt and c-cbl−/− DP cells are 48.6 and 54.9, respectively). Since the up-regulation of TCR, CD5, and CD69 molecules are associated with thymocytes undergoing positive selection (35–40), these results suggested that Cbl deficiency might have influenced the TCR signaling and thymic selection.
Figure 2.
Flow-cytometric analysis of thymocytes. (A) CD4 and CD8 staining of thymocytes from c-cbl+/+ and c-cbl−/− mice. (B) Expression of surface markers on thymocytes. Shaded patterns and the thick lines represent c-cbl wt and homozygous mutant cells, respectively.
Positive Selection of CD4+ T Cells Is Enhanced in Cbl-Deficient Mice.
The apparent normal cellularity in c-cbl−/− thymus prompted us to speculate that changes of the thymocyte development caused by Cbl deficiency might have been masked by the heterogeneity of the TCR repertoire in normal mice. To explore this possibility, we analyzed the positive and negative selection of T cells expressing a defined transgenic TCR in the absence of Cbl.
We tested thymic positive selection in 5C.C7 TCR transgenic mice that express a Vα11/Vβ3 TCR transgene specific for a pigeon cytochrome c peptide presented by the MHC class II Ek molecules (41). During development, thymocytes expressing the transgenic TCR are positively selected in mice expressing Ek, whereas the development of the same cells is arrested at DP stage in the absence of this selecting class II molecule. This is shown clearly by the presence of a large number of CD4 SP cells in Eb/k and Ek/k mice and the nearly complete absence of CD4 SP T cells in Eb/b mice (Fig. 3A and Table 1). Additionally, the degree of positive selection is strongly affected by the dosage of the ligand for TCR (42); in wt Ek/k mice (with two Ek alleles) there were significantly more CD4 SP T cells (63.7%) and fewer DP thymocytes (26.6%) than in the Eb/k (one Ek allele) littermate (47.8% SP CD4 and 43.3% DP thymocytes, Fig. 3 and Table 1). Comparison of thymocytes from c-cbl−/− and wt Eb/b mice revealed similar frequencies of DP, CD4, and CD8 SP T cells (Fig. 3A and Table 1), indicating that positive selection of the mutant CD4 SP T cells still requires the interaction of transgenic TCR and its ligand, Ek. However, in c-cbl−/− Eb/k mice, positive selection of CD4 SP T cells apparently was enhanced to a level equivalent to that seen in wt Ek/k mice, as the percentage of CD4 SP T cells increased to 70.4% and that of DP thymocytes decreased to 20.3% while the total number of thymocytes and CD8 SP T cells remained comparable to that in the wt Eb/k mice (Fig. 3A and Table 1). The enhancement of positive selection also was observed in Ek/k mice, where there was a further increase of CD4 SP cells and a reduction of DP thymocytes in c-cbl−/− mice as compared with wt mice (Fig. 3A and Table 1). As shown in Table 1, this observation was reproducible in multiple independent experiments.
Figure 3.
Positive selection in Cbl-deficient mice. (A) Positive selection of 5C.C7 TCR transgenic thymocytes. Freshly isolated thymocytes of indicated genotypes were stained with antibodies and analyzed by flow cytometry. The percentage of each cell population is indicated in the 5% contour profiles. More than 95% of thymocytes expressed transgenic TCR α and β chains. Total thymocyte numbers are: Eb/b wt, 2.3 × 108; Eb/b c-cbl−/−, 1.7 × 108; Eb/k wt, 4.7 × 108; Eb/k c-cbl−/−, 2.5 × 108; Ek/k wt, 1.8 × 108; Ek/k c-cbl−/−, 0.9 × 108. There was no statistically significant difference in total cell numbers between groups. (B) Positive selection of H-Y TCR transgenic thymocytes in female mice. Total thymocyte numbers of the wt and c-cbl−/− mice are 1.0 × 108 and 1.3 × 108, respectively. More than five pairs of mice were analyzed, and a representative experiment is shown here. There was no statistically significant difference in total thymocyte numbers.
Table 1.
Positive selection of 5C.C7 TCR transgenic thymocytes
| Experiment | DP
|
CD4 SP
|
CD8 SP
|
||||
|---|---|---|---|---|---|---|---|
| +/+ or +/− | −/− | +/+ or +/− | −/− | +/+ or +/− | −/− | ||
| Eb/b | 1* | 80.0% | 79.8 | 7.2 | 9.2 | 1.7 | 2.2 |
| 2 | 87.1 | 87.6 | 2.4 | 3.7 | 2.9 | 2.1 | |
| 3 | 79.7 | 67.5 | 3.9 | 4.8 | 2.7 | 3.0 | |
| Eb/k | 1* | 43.3 | 20.3 | 47.8 | 70.4 | 4.1 | 4.0 |
| 2 | 50.3 | 25.5 | 37.2 | 61.3 | 7.5 | 4.3 | |
| 3 | 45.4 | 24.9 | 44.3 | 65.2 | 6.6 | 4.2 | |
| Ek/k | 1* | 26.6 | 16.0 | 63.7 | 68.5 | 3.9 | 6.0 |
| 2 | 24.8 | 12.2 | 62.0 | 69.3 | 3.5 | 3.1 | |
Thymocytes from 7- to 8-week-old mice of indicated genotypes were analyzed by flow cytometry. +/+, +/−, and −/− refer to c-cbl genotype.
These experiments are shown in Fig. 3A.
Selection of Thymocytes with MHC Class I-Restricted TCR Is Not Affected in the Absence of Cbl.
To examine whether the c-cbl−/− mutation influences selection of thymocytes with MHC class I-restricted TCR, we crossed the c-cbl−/− mice to a transgenic line expressing a TCR specific for the H-Y antigen (43). In this model, thymocytes with the transgenic TCR are positively selected in female but deleted by negative selection in male mice (43). As shown in Figs. 3B and 4A, the ratio of DP to CD8 SP thymocytes was comparable between wt and c-cbl−/− female mice, and DP thymocytes were almost completely absent in both wt and c-cbl−/− male mice. Therefore, we concluded that in the absence of Cbl neither positive nor negative selection was affected in H-Y TCR transgenic model.
Figure 4.
Negative selection in Cbl-deficient mice. (A) Negative selection of self-reactive T cells in H-Y male mice. Thymocytes from age-matched wt and c-cbl−/− male mice with transgenic H-Y TCR were compared. Total thymocyte numbers of the wt and c-cbl−/− male mice are 1.7 × 107 and 2.4 × 107, respectively. A total of four pairs of mice were analyzed, and there was no significant difference in total thymocyte numbers between wt and c-cbl−/− mice. (B) Clonal deletion by superantigen. Peripheral blood lymphocytes were stained with anti-CD4, anti-CD8, and anti-Vβ3 antibodies and analyzed by flow cytometry. Integration of MMTV-3 and/or -13 genomes was detected by PCR (PCR does not distinguish between MMTV-3 and -13). The percentage of Vβ3+ cells out of total T cells is plotted.
Superantigen-Mediated Negative Selection Is Not Altered in Cbl-Deficient Mice.
It has been proposed that viral superantigen-mediated negative selection might have a different signaling requirement compared with MHC/peptide-mediated negative selection (19). Therefore, we examined clonal deletion of T cells reactive to endogenous viral superantigen in c-cbl−/− mice. Normally, T cells expressing TCR with Vβ3 element normally are deleted in the 129 mouse strain, which carries endogenous superantigens encoded by MMTV-3 and -13 (31). In both wt and c-cbl−/− mice, the percentage of Vβ3+ peripheral T cells was equally reduced in the presence of these viral superantigens (Fig. 4B). From these results we conclude that both the MHC/peptide- and superantigen-mediated negative selections are not impaired in the absence of Cbl.
Cbl Deficiency Affects Various Signaling Pathways in Thymocytes.
Phosphorylation of multiple signaling proteins is the earliest event during TCR-mediated T cell activation. We first compared the protein phosphorylation in c-cbl−/− and wt thymocytes after stimulation with anti-CD3ɛ antibody. To reduce the variations introduced by different levels of TCR expression between wt and c-cbl−/− thymocytes, both wt and mutant cells were incubated overnight at 37°C before stimulation. Such a treatment up-regulated TCR/CD3 on wt thymocytes to a level comparable to that on mutant cells (data not shown). In both wt and c-cbl−/− thymocytes, multiple protein bands became tyrosine-phosphorylated 1 min after stimulation (Fig. 5A). However, the level of tyrosine phosphorylation was elevated significantly on almost all phosphoprotein bands in c-cbl−/− thymocytes. In addition, two unidentified tyrosine phosphoprotein bands, pp110 and pp140, were observed reproducibly only in stimulated c-cbl−/− thymocytes. These results collectively indicate that in normal thymocytes, Cbl provides an inhibitory signal and negatively regulates tyrosine phosphorylation of multiple cellular proteins.
Figure 5.
Altered TCR-mediated signal transduction in Cbl-deficient thymocytes. (A) Tyrosine phosphorylation of cellular proteins upon CD3ɛ stimulation. Thymocytes were stimulated with anti-CD3ɛ antibody for indicated time periods. Equal amount of protein from cell lysate was separated by SDS/PAGE and immunoblotted with 4G10 antiphosphotyrosine antibody. The positions of Cbl and molecular size standards are indicated. (B) Analysis of individual signaling molecules. Thymocytes were stimulated for 2 min with antibodies against CD3ɛ and CD4 as indicated. Equal amount of protein from cell lysate was immunoprecipitated (IP) with indicated antibodies and immunoblotted with antibodies specific for phosphotyrosine (pY), Lck, ZAP-70, or PLCγ1. For MAP kinase (MAPK), total cell lysate was immunoblotted with antibodies specific for active MAP kinase or total MAP kinase. (C) PI-3K activity of thymocytes. Thymocytes were stimulated with anti-CD3ɛ and anti-CD28 antibodies for 2 min. PI-3K activity was measured as described previously (34). The data are presented as fold increase over control (unstimulated) and represent the average ± range from two separate experiments.
Hyperphosphorylation of multiple proteins in the stimulated c-cbl−/− thymocytes suggested that the absence of Cbl had disregulated the upstream components of the TCR signaling cascade. Therefore, we next examined the phosphorylation of two tyrosine kinases, Lck and ZAP-70, that have been shown to transduce signals at the proximal end of the TCR signaling pathway (44–46). Phosphorylation of Lck was comparable between c-cbl−/− and wt thymocytes after stimulation with either anti-CD3ɛ or anti-CD3ɛ/CD4 antibodies (Fig. 5B). In contrast, stimulation with anti-CD3ɛ or anti-CD3ɛ/CD4 antibodies led to an approximately 10-fold increase in tyrosine phosphorylation of ZAP-70 in c-cbl−/− thymocytes compared with wt cells (Fig. 5B). These results, together with the observation that ZAP-70 associates with Cbl (47, 48), strongly suggest that ZAP-70 is the most upstream component of the tyrosine kinase cascade affected by Cbl deficiency.
Activation of tyrosine kinases through TCR leads to activation of multiple signaling pathways including the Ras-Raf-MAP kinase cascade, PI-3K, and phospholipase C (PLC)-γ (44–46, 49). Therefore, we investigated whether these downstream signals were affected in c-cbl−/− thymocytes. Mitogen-activated protein (MAP) kinase activity in total cell lysate was determined with antibodies against the active form of ERK1 and ERK2 (50). We repeatedly detected approximately three times more active ERK1 and ERK2 in c-cbl−/− thymocytes than in wt thymocytes after stimulation with anti-CD3ɛ or anti-CD3ɛ/CD4 antibodies (Fig. 5B). This result clearly indicates that MAP kinase activity is negatively regulated by Cbl.
In contrast to the enhanced ZAP-70 and MAP kinase activation, both PI-3K and PLCγ1 activities were reduced in c-cbl−/− thymocytes. PI-3K activity in c-cbl−/− thymocytes was approximately half of that in wt cells when cells were stimulated with either anti-CD3ɛ or anti-CD3ɛ/CD28 antibodies (Fig. 5C). Similar to PI-3K, the level of PLCγ1 phosphorylation was reduced in c-cbl−/− thymocytes after CD3ɛ/CD4 cross-linking (Fig. 5B), indicating that the absence of Cbl prevented full activation of PLCγ1 in the mutant cells.
Peripheral T Cells from Cbl-Deficient Mice Are Less Responsive to Antigenic Stimulation.
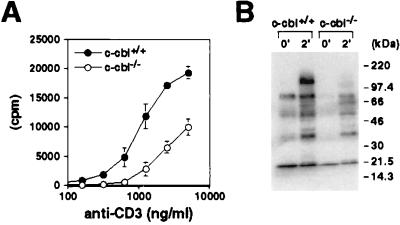
How does the altered thymic selection affect the function of mature T cells? We first examined in vitro T cell proliferative responses upon stimulation through TCR. Purified peripheral CD4+ T cells from 5C.C7 transgenic mice (Eb/k background) were stimulated with anti-CD3 antibodies, and cell proliferation was measured by [3H]thymidine incorporation. As shown in Fig. 6A, cells from both wt and c-cbl−/− mice responded to anti-CD3 stimulation in a dose-dependent manner. However, cells from the mutant mice incorporated only half as much thymidine compared with those from wt mice at each antibody concentration.
Figure 6.
Function of Cbl-deficient peripheral T cells. (A) Proliferative response of peripheral T cells. Purified CD4+ T cells from 5C.C7 transgenic mice were stimulated with plate-bound anti-CD3 ɛ antibody. Culture was pulsed with [3H]thymidine for the last 12 hr of incubation. (B) Total tyrosine phosphorylation of splenocytes. Splenocytes were enriched for T cells by passing through nylon wool columns. T cells were stimulated with anti-CD3ɛ and anti-CD4 antibodies for 2 min. Equal amount of protein from total cell lysate was separated by SDS/PAGE and immunoblotted with 4G10 antibody.
To study whether the lower responsiveness of T cells from c-cbl−/− mice was reflected at the biochemical level, we analyzed protein tyrosine phosphorylation in peripheral T cells in response to TCR stimulation. Consistent with the above results, the level of protein tyrosine phosphorylation in the c-cbl−/− peripheral T cells was reduced significantly compared with that in the wt T cells (Fig. 6B).
DISCUSSION
Cbl has been shown to be involved in the signal transduction of various growth-factor receptors and antigen receptors. However, the physiological significance of this molecule for the immune system remained elusive. The results presented here demonstrate that Cbl is involved in positive selection of T cells in thymus. In the absence of Cbl, thymocytes with MHC class II-restricted transgenic TCR were more efficiently selected. Preliminary experiments using another class II-restricted transgenic TCR mouse strain showed similar results (unpublished data).
Previous experiments revealed that loss of CD5 molecule also enhanced thymic positive selection in some TCR transgenic mice (18). However, intracellular signals affected in CD5- and Cbl-deficient mouse models are different. In CD5-deficient thymocytes, phosphorylation of PLCγ1 and Vav was enhanced. In contrast, Cbl deficiency led to a marked increase of tyrosine phosphorylation on multiple proteins and elevated ZAP-70 and MAP kinase activities while PLCγ1 phosphorylation was reduced (Fig. 5). These results indicate that Cbl is a negative regulator of thymic positive selection but its mechanism of action is distinct from that of CD5.
Stimulation of T cells through TCR leads to a concerted activation of multiple signaling pathways. We demonstrated that ZAP-70 and MAP kinase were hyperactivated in Cbl-deficient thymocytes. On the other hand, activity of PLCγ and PI-3K was reduced in the absence of Cbl. Therefore, Cbl may function as an organizer to coordinate the action of multiple signaling pathways during thymocyte activation. Current available data suggest that different mechanisms may be employed by Cbl to modulate each signaling pathway. For instance, Syk, a homologue of ZAP-70, has been shown to bind to Cbl directly, and its kinase activity was reduced when Cbl was overexpressed (26). Because Cbl is known to associate with ZAP-70 as well (47, 48), Cbl may regulate ZAP-70 activity in a similar manner. As for the MAP kinase pathway, because no direct interaction between Cbl and MAP kinases has been demonstrated, the regulation of MAP kinase activity by Cbl may be mediated by intermediary molecules. Cbl may control MAP kinase activation by inhibiting the tyrosine kinases (such as ZAP-70) that are linked with MAP kinase activation in T cells (44–46, 49). Alternatively, SOS, a Ras GDP release factor, and Cbl bind to the same Src homology 3 domain of Grb2 (25, 46). Therefore, Cbl may compete with SOS for the binding sites on Grb2 and consequently reduces the number of Grb2 molecules available for SOS-mediated activation of Ras-MAP kinase pathway. Activation of PI-3K requires the recruitment of its regulatory subunit, p85, to the membrane, and Cbl is known to be the major phosphoprotein associated with the p85 subunit in T cells activated through TCR (49, 51). Therefore, the reduction of PI-3K activity was most likely caused by the failure of the membrane localization of the p85 subunit mediated by Cbl. The mechanism of PLCγ1 phosphorylation remains unclear at present. One may postulate that Cbl regulates PLCγ phosphorylation by establishing a bridge between PLCγ and its tyrosine kinase.
Our results demonstrated that loss of Cbl enhanced positive selection of CD4 but not CD8 lineage T cells, suggesting that Cbl might be involved in the control of cell lineage determination. The function of Cbl as a negative regulator of MAP kinase pathway is compatible with genetic evidence from C. elegans and Drosophila and indicates that the function of Cbl is evolutionarily conserved (27–29). Previous reports indicated that thymic positive but not negative selection was impaired in transgenic mice expressing either a dominant-negative form of Ras or Raf protein or a catalytically inactive Mek-1 (a MAP kinase kinase) (15–17). Sharp et al. (52) demonstrated that transgenic expression of the gain-of-function mutant of ERK2 favored CD4 commitment while a MAP kinase inhibitor could direct thymocytes into the CD8 lineage. Another recent study showed that in the absence of Csk, a negative regulator of Src-family tyrosine kinases, CD4 but not CD8 lineage cells were generated without TCR engagement (53). These reports collectively indicate that positive selection requires the activation of the MAP kinase pathway, that CD4/CD8 lineage determination may require different intracellular signals, and that the activation of Src-family kinases and/or the MAP kinase pathway may selectively favor CD4 lineage commitment or survival. We showed that in Cbl-deficient thymocytes, both tyrosine kinase and MAP kinase activities were increased. This may explain why positive selection of thymocytes with MHC class II-restricted TCR in c-cbl−/− mice was enhanced whereas class I-restricted H-Y female thymocytes did not show a comparable change in thymic selection in the absence of Cbl.
One interesting observation in c-cbl−/− mice is that thymocytes expressed an increased level of cell surface markers such as TCR/CD3, CD4, CD5, and CD69. The increased expression of TCR, CD5, and CD69 molecules has been associated with the thymocytes undergoing positive selection in normal mice (35–40). However, up-regulation of TCR in Cbl-deficient mice appears to be independent of the TCR triggering, because in 5C.C7 TCR transgenic thymocytes the high-level expression of the transgenic TCR was maintained even in the Eb/b or MHC class II-deficient environment (unpublished data). Cbl is a RING-finger domain containing protein (25). It has been reported recently that the RING-finger domain may bind to the membrane lipid phosphatidylinositol-3-phosphate and regulate endocytic/vacuolar membrane traffic (54, 55). Therefore, it is conceivable that Cbl deficiency impairs the trafficking of membrane proteins in thymocytes and alters the expression of certain surface molecules. Based on these considerations, we propose that the negative regulatory role of Cbl for thymic positive selection may operate at two levels: at the cytoplasmic level, Cbl directly inhibits intracellular signaling machinery downstream of TCR, and at the membrane level, it may down-regulate the expression of cell surface TCR complexes and coreceptors and consequently reduce the avidity of TCR/MHC interactions.
Present data also indicated that peripheral T cells from c-cbl−/− mice responded rather poorly to TCR stimulation. Considering that Cbl is expressed at a much lower level in wt mature T cells (less than 5% of that in thymocytes; unpublished data), it may not play a significant regulatory role in mature T cells. The level of total tyrosine phosphorylation in mature c-cbl−/− T cells upon TCR stimulation was reduced compared with that in wt cells (Fig. 6B). Since the absence of Cbl enhanced overall tyrosine phosphorylation in c-cbl−/− thymocytes, it seems unlikely that the same molecule has complete opposite functions in thymus and periphery and that the absence of Cbl directly inhibited proliferation and phosphorylation of the stimulated mutant peripheral T cells. A more likely explanation is that Cbl deficiency promoted positive selection of T cells with lower responsiveness to TCR-mediated signals. Since the expression level of molecules involved in TCR signaling may vary among individual thymocytes, wt thymocytes expressing a smaller amount of signaling molecules should be less sensitive to TCR triggering and may eventually be eliminated by “neglect” during thymic selection, whereas in c-cbl−/− mice, the same weak selection signal may be amplified to a level above the threshold for positive selection and consequently rescue these lower responders from death and allow them to become mature T cells.
Taken together, our results indicate that Cbl plays a novel role in thymic positive selection by fine-tuning the signaling threshold of the developing thymocyte. In the absence of Cbl, DP thymocytes can be more efficiently selected even when they receive a relatively weak signal from TCR/MHC interaction. Selection as such may lead to the enrichment of T cells with reduced responsiveness to TCR triggering in the mature T cell pool and ultimately attenuate the peripheral T cell response in the mutant mice. Thus, regulation of TCR signals by Cbl provides a mechanism that ensures selection of a T cell pool with a proper responsiveness to antigen stimulation.
Acknowledgments
We thank B. J. Fowlkes, R. N. Germain, M. J. Lenardo, W. E. Paul, and L. E. Samelson for helpful discussions and critical review of the manuscript.
ABBREVIATIONS
- TCR
T cell antigen receptor
- MHC
major histocompatibility complex
- PI-3K
phosphatidylinositol-3 kinase
- PLC
phospholipase C
- MMTV
mammary tumor virus
- wt
wild type
- SP
single positive
- DP
double positive
References
- 1.Robey E, Fowlkes B J. Annu Rev Immunol. 1994;12:675–705. doi: 10.1146/annurev.iy.12.040194.003331. [DOI] [PubMed] [Google Scholar]
- 2.Jameson S C, Hogquist K A, Bevan M J. Annu Rev Immunol. 1995;13:93–126. doi: 10.1146/annurev.iy.13.040195.000521. [DOI] [PubMed] [Google Scholar]
- 3.Kisielow P, von Boehmer H. Adv Immunol. 1995;58:87–209. doi: 10.1016/s0065-2776(08)60620-3. [DOI] [PubMed] [Google Scholar]
- 4.Benoist C, Mathis D. Curr Opin Immunol. 1997;9:245–249. doi: 10.1016/s0952-7915(97)80143-4. [DOI] [PubMed] [Google Scholar]
- 5.Ashton-Rickardt P G, Van Kaer L, Schumacher T N, Ploegh H L, Tonegawa S. Cell. 1993;73:1041–1049. doi: 10.1016/0092-8674(93)90281-t. [DOI] [PubMed] [Google Scholar]
- 6.Ashton-Rickardt P G, Bandeira A, Delaney J R, Van Kaer L, Pircher H P, Zinkernagel R M, Tonegawa S. Cell. 1994;76:651–663. doi: 10.1016/0092-8674(94)90505-3. [DOI] [PubMed] [Google Scholar]
- 7.Hogquist K A, Jameson S C, Heath W R, Howard J L, Bevan M J, Carbone F R. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 8.Sebzda E, Wallace V A, Mayer J, Yeung R S, Mak T W, Ohashi P S. Science. 1994;263:1615–1618. doi: 10.1126/science.8128249. [DOI] [PubMed] [Google Scholar]
- 9.Fischer A, Malissen B. Science. 1998;280:237–243. doi: 10.1126/science.280.5361.237. [DOI] [PubMed] [Google Scholar]
- 10.Hashimoto K, Sohn S J, Levin S D, Tada T, Perlmutter R M, Nakayama T. J Exp Med. 1996;184:931–943. doi: 10.1084/jem.184.3.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Penninger J M, Wallace V A, Molina T, Mak T W. J Immunol. 1996;157:5359–5366. [PubMed] [Google Scholar]
- 12.Negishi I, Motoyama N, Nakayama K, Senju S, Hatakeyama S, Zhang Q, Chan A C, Loh D Y. Nature (London) 1995;376:435–438. doi: 10.1038/376435a0. [DOI] [PubMed] [Google Scholar]
- 13.Byth K F, Conroy L A, Howlett S, Smith A J, May J, Alexander D R, Holmes N. J Exp Med. 1996;183:1707–1718. doi: 10.1084/jem.183.4.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turner M, Mee P J, Walters A E, Quinn M E, Mellor A L, Zamoyska R, Tybulewicz V L. Immunity. 1997;7:451–460. doi: 10.1016/s1074-7613(00)80367-2. [DOI] [PubMed] [Google Scholar]
- 15.Swan K A, Alberola-Ila J, Gross J A, Appleby M W, Forbush K A, Thomas J F, Perlmutter R M. EMBO J. 1995;14:276–285. doi: 10.1002/j.1460-2075.1995.tb07001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Shea C C, Crompton T, Rosewell I R, Hayday A C, Owen M J. Eur J Immunol. 1996;26:2350–2355. doi: 10.1002/eji.1830261012. [DOI] [PubMed] [Google Scholar]
- 17.Alberola-Ila J, Forbush K A, Seger R, Krebs E G, Perlmutter R M. Nature (London) 1995;373:620–623. doi: 10.1038/373620a0. [DOI] [PubMed] [Google Scholar]
- 18.Tarakhovsky A, Kanner S B, Hombach J, Ledbetter J A, Muller W, Killeen N, Rajewsky K. Science. 1995;269:535–537. doi: 10.1126/science.7542801. [DOI] [PubMed] [Google Scholar]
- 19.Amakawa R, Hakem A, Kundig T M, Matsuyama T, Simard J J, Timms E, Wakeham A, Mittruecker H W, Griesser H, Takimoto H, et al. Cell. 1996;84:551–562. doi: 10.1016/s0092-8674(00)81031-4. [DOI] [PubMed] [Google Scholar]
- 20.Penninger J M, Sirard C, Mittrucker H W, Chidgey A, Kozieradzki I, Nghiem M, Hakem A, Kimura T, Timms E, Boyd R, et al. Immunity. 1997;7:243–254. doi: 10.1016/s1074-7613(00)80527-0. [DOI] [PubMed] [Google Scholar]
- 21.Liao X C, Littman D R. Immunity. 1995;3:757–769. doi: 10.1016/1074-7613(95)90065-9. [DOI] [PubMed] [Google Scholar]
- 22.Langdon W Y, Hartley J W, Klinken S P, Ruscetti S K, Morse H C., III Proc Natl Acad Sci USA. 1989;86:1168–1172. doi: 10.1073/pnas.86.4.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langdon W Y, Hyland C D, Grumont R J, Morse H C., III J Virol. 1989;63:5420–5424. doi: 10.1128/jvi.63.12.5420-5424.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langdon W Y. Aust N Z J Med. 1995;25:859–864. doi: 10.1111/j.1445-5994.1995.tb02892.x. [DOI] [PubMed] [Google Scholar]
- 25.Miyake S, Lupher M L, Jr, Andoniou C E, Lill N L, Ota S, Douillard P, Rao N, Band H. Crit Rev Oncog. 1997;8:189–218. doi: 10.1615/critrevoncog.v8.i2-3.30. [DOI] [PubMed] [Google Scholar]
- 26.Ota Y, Samelson L E. Science. 1997;276:418–420. doi: 10.1126/science.276.5311.418. [DOI] [PubMed] [Google Scholar]
- 27.Yoon C H, Lee J, Jongeward G D, Sternberg P W. Science. 1995;269:1102–1105. doi: 10.1126/science.7652556. [DOI] [PubMed] [Google Scholar]
- 28.Hime G R, Dhungat M P, Ng A, Bowtell D D. Oncogene. 1997;14:2709–2719. doi: 10.1038/sj.onc.1201223. [DOI] [PubMed] [Google Scholar]
- 29.Meisner H, Daga A, Buxton J, Fernandez B, Chawla A, Banerjee U, Czech M P. Mol Cell Biol. 1997;17:2217–2225. doi: 10.1128/mcb.17.4.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy M A, Schnall R G, Venter D J, Barnett L, Bertoncello I, Thien C B F, Langdon W Y, Bowtell D D L. Mol Cell Biol. 1998;18:4872–4882. doi: 10.1128/mcb.18.8.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomonari K, Fairchild S, Rosenwasser O A. Immunol Rev. 1993;131:131–168. doi: 10.1111/j.1600-065x.1993.tb01534.x. [DOI] [PubMed] [Google Scholar]
- 32.Pullen A M, Choi Y, Kushnir E, Kappler J, Marrack P. J Exp Med. 1992;175:41–47. doi: 10.1084/jem.175.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiest D L, Ashe J M, Howcroft T K, Lee H M, Kemper D M, Negishi I, Singer D S, Singer A, Abe R. Immunity. 1997;6:663–671. doi: 10.1016/s1074-7613(00)80442-2. [DOI] [PubMed] [Google Scholar]
- 34.Kole H K, Garant M J, Kole S, Bernier M. J Biol Chem. 1996;271:14302–14307. doi: 10.1074/jbc.271.24.14302. [DOI] [PubMed] [Google Scholar]
- 35.Fowlkes B J, Edison L, Mathieson B J, Chused T M. J Exp Med. 1985;162:802–822. doi: 10.1084/jem.162.3.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohashi P S, Pircher H, Burki K, Zinkernagel R M, Hengartner H. Nature (London) 1990;346:861–863. doi: 10.1038/346861a0. [DOI] [PubMed] [Google Scholar]
- 37.Shortman K, Vremec D, Egerton M. J Exp Med. 1991;173:323–332. doi: 10.1084/jem.173.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bendelac A, Matzinger P, Seder R A, Paul W E, Schwartz R H. J Exp Med. 1992;175:731–742. doi: 10.1084/jem.175.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kearse K P, Takahama Y, Punt J A, Sharrow S O, Singer A. J Exp Med. 1995;181:193–202. doi: 10.1084/jem.181.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lucas B, Germain R N. Immunity. 1996;5:461–477. doi: 10.1016/s1074-7613(00)80502-6. [DOI] [PubMed] [Google Scholar]
- 41.Seder R A, Paul W E, Davis M M, Fazekas de St. Groth B. J Exp Med. 1992;176:1091–1098. doi: 10.1084/jem.176.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berg L J, Frank G D, Davis M M. Cell. 1990;60:1043–1053. doi: 10.1016/0092-8674(90)90352-f. [DOI] [PubMed] [Google Scholar]
- 43.Kisielow P, Bluthmann H, Staerz U D, Steinmetz M, von Boehmer H. Nature (London) 1988;333:742–746. doi: 10.1038/333742a0. [DOI] [PubMed] [Google Scholar]
- 44.Alberola-Ila J, Takaki S, Kerner J D, Perlmutter R M. Annu Rev Immunol. 1997;15:125–154. doi: 10.1146/annurev.immunol.15.1.125. [DOI] [PubMed] [Google Scholar]
- 45.Weiss A, Littman D R. Cell. 1994;76:263–274. doi: 10.1016/0092-8674(94)90334-4. [DOI] [PubMed] [Google Scholar]
- 46.Wange R L, Samelson L E. Immunity. 1996;5:197–205. doi: 10.1016/s1074-7613(00)80315-5. [DOI] [PubMed] [Google Scholar]
- 47.Fournel M, Davidson D, Weil R, Veillette A. J Exp Med. 1996;183:301–306. doi: 10.1084/jem.183.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lupher M L, Jr, Reedquist K A, Miyake S, Langdon W Y, Band H. J Biol Chem. 1996;271:24063–24068. doi: 10.1074/jbc.271.39.24063. [DOI] [PubMed] [Google Scholar]
- 49.Cantrell D. Annu Rev Immunol. 1996;14:259–274. doi: 10.1146/annurev.immunol.14.1.259. [DOI] [PubMed] [Google Scholar]
- 50.Payne D M, Rossomando A J, Martino P, Erickson A K, Her J H, Shabanowitz J, Hunt D F, Weber M J, Sturgill T W. EMBO J. 1991;10:885–892. doi: 10.1002/j.1460-2075.1991.tb08021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hartley D, Corvera S. J Biol Chem. 1996;271:21939–21943. doi: 10.1074/jbc.271.36.21939. [DOI] [PubMed] [Google Scholar]
- 52.Sharp L L, Schwarz D A, Bott C M, Marshall C J, Hedrick S M. Immunity. 1997;7:609–618. doi: 10.1016/s1074-7613(00)80382-9. [DOI] [PubMed] [Google Scholar]
- 53.Schmedt C, Saijo K, Niidome T, Kuhn R, Aizawa S, Tarakhovsky A. Nature (London) 1998;394:901–904. doi: 10.1038/29802. [DOI] [PubMed] [Google Scholar]
- 54.Gaullier J M, Simonsen A, D’Arrigo A, Bremnes B, Stenmark H, Aasland R. Nature (London) 1998;394:432–433. doi: 10.1038/28767. [DOI] [PubMed] [Google Scholar]
- 55.Patki V, Lawe D C, Corvera S, Virbasius J V, Chawla A. Nature (London) 1998;394:433–434. doi: 10.1038/28771. [DOI] [PubMed] [Google Scholar]