Abstract
The transforming growth factor β, Hedgehog, Notch, and Wnt signaling pathways all play critical roles in the development and progression of prostate cancer. It is becoming increasingly apparent that these pathways may intersect with developmentally important transcription factors such as the sex-determining region Y-box 4 (SOX4), homeobox C6, enhancer of zeste 2, and ETS-related gene, which are up-regulated in prostate cancers. For example, identification of the downstream targets of SOX4 and homeobox C6 suggests that these factors may cooperate to activate the Notch pathway and the PI3K/AKT pathway, possibly in response to Wnt signals. PI3K/AKT activation likely occurs indirectly via up-regulation of growth factor receptors, while Notch activation is secondary to up-regulation of Notch pathway components. In addition, SOX4 may affect terminal differentiation via regulation of other transcription factors such as NKX3.1 and MLL, and regulation of components of the microRNA pathway such as Dicer and Argonaute 1. The evidence supporting activation of these pathways in prostate cancer progression suggests that combinations of compounds targeting them may be of benefit to patients with aggressive, metastatic disease.
Prostate cancer is the most prevalent noncutaneous malignancy in American men, with estimates for 2009 at over 192,000 new cases and 27,000 deaths.1 The majority of patients with prostate cancer are clinically asymptomatic with early-stage, organ-confined disease, and in fact, more than 80% of men who reach the age of 80 develop this less aggressive type of prostate cancer. However, a subpopulation of patients with prostate cancer progress to highly invasive, androgen-independent metastatic disease, which is commonly fatal.
In males, the prostate develops in the presence of androgens from obligatory interactions between the urogenital sinus epithelium and the urogenital sinus mesenchyme.2 Several key developmental pathways, including the androgen receptor (AR),2 fibroblast growth factor (FGF),2 transforming growth factor β (TGFβ),3 Hedgehog,4 Notch,5 and Wnt pathways,6 play critical roles in normal prostate development as well as the progression of prostate cancer. Both processes depend on key paracrine effects mediated by stromal-epithelial interactions. For example, FGF7 and FGF10 are secreted by the stroma, while prostate epithelia express the FGF receptor 2 (FGF2R).7 Conversely, prostate epithelia express sonic hedgehog ligand, while the Patched receptor is expressed mainly in the stroma.8 Both FGF and Hedgehog signaling are important for prostatic growth, ductal branching, and differentiation (reviewed in Cunha et al2). Notch signaling is also essential for prostate epithelial proliferation9 and stromal cell survival.10
Many key components of the Hedgehog, TGFβ, and Wnt pathways are up-regulated in embryonic and adult prostate stem cells relative to differentiated prostate epithelial cells.11 Comparison of the expression signatures of fetal and adult prostate stem cells to signatures observed in prostate cancer suggests that these malignancies may activate self-renewal properties via these developmental pathways.11 The potential dependence of prostate cancers on these pathways also suggests that they are ripe for therapeutic intervention, and many teams are actively pursuing novel compounds that target the Notch, Wnt, and Hedgehog pathways.12,13,14
A comprehensive review of the roles of these crucial developmental pathways in prostate cancer is beyond the scope of this review. A short summary of Notch and Wnt signaling in prostate cancer is given below, together with their relationship to key transcriptional regulators in this disease.
The Notch Pathway in Prostate Cancer
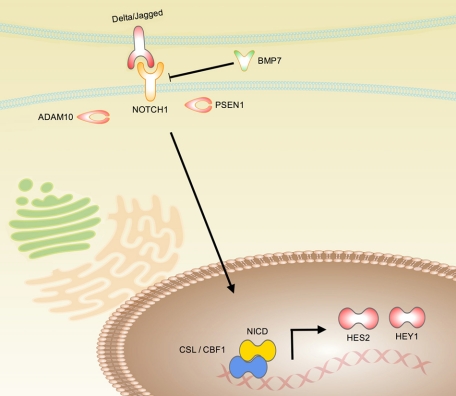
The Notch pathway is an evolutionarily conserved local cell signaling pathway that regulates a host of cellular processes, including cell fate specification, differentiation, proliferation, apoptosis, adhesion, epithelial-mesenchymal transition (EMT), migration, and angiogenesis (reviewed in Bolos et al15). In Notch-mediated neoplasias, Notch can act as an oncogene or as a tumor suppressor, depending on the cellular context and differences in the strength and timing of Notch signals. The Notch receptor is synthesized in the rough endoplasmic reticulum as a single polypeptide precursor and proteolytically cleaved in the trans-golgi network by the furin protease, creating a heterodimeric mature receptor that comprises noncovalently associated extracellular and transmembrane subunits (Figure 1). This assembly travels to the cell surface where it interacts with specific transmembrane ligands, such as jagged and Δ-like 1 (DLL1).16 Following ligand activation and further proteolytic cleavage by the ADAM metallopeptidase proteases (ADAM10 or ADAM17) and γ-secretase complex containing presenilin-1 protease, the Notch intracellular domain is released and translocates to the nucleus where it regulates gene expression, converting repressor complexes into activator complexes (Figure 1). Notch downstream target genes include the hairy-enhancer of split genes (HES1, HES2, and HEY1) among others.
Figure 1.
Schematic of the Notch pathway. The Notch receptor is synthesized in the rough endoplasmic reticulum as a single polypeptide precursor and proteolytically cleaved in the trans-golgi network by the furin protease, creating a heterodimeric mature receptor that comprises noncovalently associated extracellular and transmembrane subunits. This assembly travels to the cell surface where it interacts with specific transmembrane ligands, such as jagged and δ-like 1. Notch activation can be repressed by BMP7 via an unknown mechanism. Following ligand activation and further proteolytic cleavage by ADAM10 and presenilin-1 (PSEN1) proteases, the Notch intracellular domain (NICD) is released and translocates to the nucleus where it regulates gene expression, converting CSL (C-promoter binding factor (CBF-1), suppressor of hairless (Su(H)), lin-12 and glp-1 (Lag-1)) repressor complexes into activator complexes, and inducing expression of the hairy-enhancer of split genes (eg, HES2 and HEY1).
Notch signaling is a key cellular regulator during normal development of many organs, including the prostate9 and bone.17 During prostate differentiation, Notch signaling is absent in stem cells, and highest in the intermediate cells undergoing proliferation, before terminal differentiation.18 Notch signaling is also activated in osteoblast precursor cells17 and prevents osteoblast differentiation.19 Inhibition of Notch signaling is effective at treating mouse models of medulloblastomas that are driven by the Sonic Hedgehog-Smoothened pathway.20 This is particularly relevant to prostate cancer because Hedgehog signaling is activated in advanced prostate cancer21 and targeting of Hedgehog signals inhibits proliferation.22 Cross talk between the Notch and Hedgehog pathways includes Notch regulation of the Gli effectors of the Hedgehog pathway and cooperation between Notch and Hedgehog to activate the Snail family of transcription factors important for mediating the EMT process.23
In the prostate, Notch signaling is required for normal prostate epithelial proliferation and differentiation.9 Normal, primary prostate epithelial cells grown at low cell density without cell-cell contact still contain cleaved Notch receptor, suggesting that prostate progenitor cells may be competent for cell-autonomous Notch1 signaling.24 In addition, Notch signaling is active in intermediate, transit-amplifying prostate cells undergoing rapid proliferation, and Notch inhibition reduces proliferation of primary prostate epithelial cells,18 suggesting that active Notch signaling is a key feature of prostate cancer. Notch signaling is critical for prostate regeneration following castration and hormone replacement, and components of the Notch pathway are important regulators of prostate cancer progression, metastasis, and the EMT.5
Wnt Signaling in Prostate Cancer
The Wnt pathway plays critical roles in the development of many forms of cancer, having been most thoroughly studied in colon and breast cancer. Ectopic expression of Wnt ligands can induce transformation of breast epithelial cells through a Notch-dependent mechanism that requires expression of Notch ligands such as DLL1.25 On binding of Wnt ligands to Frizzled-LRP6 co-receptors,26 the adenomatous polyposis coli (APC)/GSK3β complex is inhibited and β-catenin becomes stabilized, translocates to the nucleus, and activates expression of downstream target genes such as c-myc27 and cyclin D.28 Mutations in the adenomatous polyposis coli gene have been identified as initiating events in familial polyposis coli patients and sporadic colorectal tumors.29
Although less studied, Wnt signaling also appears to be relevant to aggressive prostate cancer and bone metastases. The canonical Wnt ligand Wnt3a can enhance activity of the androgen receptor and growth of LNCaP prostate cancer cells,30 and suppression of Wnt signaling can inhibit proliferation of PC-3 and DU145 cell lines.31 The GSK3β kinase can inhibit activity of AR,32 and AR interacts with β-catenin in vivo in castrate-resistant tumors, but not in noncastrated mice.33 Expression of one negative regulator of Wnt signaling, the dickkopf homolog 1 (DKK1), is reduced in bone metastases and may play a role in the osteoblastic properties of prostate cancer metastases.34 Blocking Wnt activity via stable expression of DKK1 converts osteoblastic C4-2B prostate cancer cells to a highly osteolytic tumor.35 Inhibition of Wnt signaling by using Wnt inhibitory factor 1 (WIF1) reduces Akt kinase activity and induces chemosensitivity in prostate cancer cells with mutations in the PTEN tumor suppressor.36 In our own expression profiling of human prostate cancer specimens, we observed strongly reduced expression of WIF1 that is highly correlated with Gleason score.37 Many negative regulators of the Wnt pathway are shut down during cancer progression via epigenetic silencing and DNA methylation such as secreted frizzled-related protein 1 (SFRP1) and DKK3.38 Loss of DKK3 expression disrupts acinar morphogenesis of RWPE-1 cells in 3D culture models and enhances proliferation.39 SFRP1 inhibits activation of AR in LNCaP cells via a β-catenin-independent mechanism.40 Expression levels of WIF1, DKK3, SFRP1, and SFRP2 are all decreased in primary and metastatic prostate cancers.41,42,43
Developmental Transcription Factors in Prostate Cancer
Several groups have undertaken expression profiling of prostate cancers37,41,44,45,46,47,48,49 and identified genes associated with Gleason grade and prostate cancer progression (reviewed in Hughes et al50). Among the many interesting findings are that several transcription factors that regulate normal embryonic development are associated with prostate cancer progression, including enhancer of zeste 2 (EZH2),51 E. Twenty six (ETS)-family transcription factors,52 homeobox C6 (HOXC6),53 and sex-determining region Y-box 4 (SOX4).37 In fact, SOX4 overexpression has been detected by microarray analysis in no fewer than seven independent studies of PCa.37,41,44,45,46,47,48,49 Overexpression of ETS-family transcription factors ETS-related gene (ERG) and ETS-variant 1 (ETV-1) are driven by chromosomal translocations that fuse the androgen-responsive transmembrane protease, serine 2 (TMPRSS2) promoter to the first exon of these ETS-family genes (reviewed in Kumar-Sinha et al54). Transmembrane protease, serine 2:ERG fusion translocations appear to be a critical genetic event early in the transition from premalignant high-grade prostatic intraepithelial neoplasia to malignant prostate adenocarcinoma and cooperate with PTEN loss to drive cancer progression.55 In silico analysis of prostate cancers that overexpress ERG has shown that up-regulation of Wnt pathway components such as WNT1, adenomatous polyposis coli, AXIN1, and TLE156 is associated with ERG levels. EZH2 is elevated in many cancers57 and is responsible for epigenetic silencing of genes via generation of histone 3 lysine 27 trimethylation marks.58 EZH2, along with several other proteins, is a component of the polycomb complex that is responsible for silencing many developmentally regulated genes,59 including homeobox transcription factors such as HOXC6.60 Increased EZH2 expression is observed later during the disease process during progression to metastatic prostate cancer51 and an EZH2 polycomb repression signature can predict patient outcome in prostate cancer.61
We analyzed the conserved transcription factor binding sites of genes that were overexpressed in prostate cancers by using the CONFAC software that we previously developed.62 CONFAC analysis enables high-throughput analysis of transcription factor binding sites that are evolutionarily conserved between human and mouse genomes for large sets of co-expressed genes that may be co-regulated. Analysis of the promoters of these genes indicated that they were significantly enriched in homeobox transcription factor binding sites. The fact that HOXC6 was overexpressed in prostate cancers and that its expression was strongly correlated with Gleason score led us to further investigate its role in prostate cancer.
The HOXC6 Network
HOX transcription factors are developmentally regulated genes that play crucial roles in tissue patterning.63 However, HOX genes are also expressed in many adult tissues, serving in a variety of roles that ultimately impact cellular differentiation.64 HOX genes are dysregulated in many human cancers including leukemias,65 and solid tumors of the breast, colon, lung, kidney, ovary, and prostate.66 HOXC6 is located on 12q13.3 in humans, and is a relatively small transcription factor that is expressed as two alternatively spliced isoforms 18 kDa and 27 kDa in size. HOXC6 is expressed in osteosarcomas,67 medulloblastomas,68 as well as carcinomas of the breast,69 lung,70 and prostate,53 and is overexpressed in the LNCaP prostate cancer cell line.71 In a recent prostate cancer study, HOXC6 was identified as the gene most strongly correlated with increasing Gleason grade out of a newly identified 16-gene signature.72
We have previously shown that small-interfering RNA knockdown of HOXC6 expression induces apoptosis, and overexpression of HOXC6 results in increased proliferation and decreased apoptosis in LNCaP cells.53 We also identified potential downstream targets of HOXC6 by observing gene expression changes after either increasing or knocking down HOXC6 expression in LNCaP prostate cancer cells. We identified T-cell receptor alternate reading frame protein, insulin-like growth factor binding protein 3 (IGFBP3), and neutral endopeptidase/membrane metallo-endopeptidase as biologically relevant HOXC6 target genes that may influence cell survival and proliferation.
More recently, we have published a comprehensive transcriptional network of genes under the direct control of HOXC6 in prostate cancer.73 To globally identify the direct targets of HOXC6, we performed genome-wide localization chromatin-immunoprecipitation followed by microarray hybridization (ChIP-chip) studies on HOXC6 by using the NimbleGen (Reykjavik, Iceland) 25K promoter array set and identified 468 genes that are bound by HOXC6 in living cells involved in functions such as cell proliferation, development, and apoptosis.73 We identified bone morphogenic protein 7 (BMP7), FGFR2, and platelet-derived growth factor receptor α (PDGFRA), among others, as in vivo direct regulatory targets of HOXC6 in prostate tissue. We went on to show that BMP7 could induce apoptosis in prostate cancer cells, possibly via inhibition of the Notch pathway. We further showed that inhibition of PDGFRA reduces proliferation of prostate cancer cells, and that overexpression of HOXC6 can overcome the effects of PDGFRA inhibition.73
Metastatic prostate cancer is characterized by a strong predisposition for metastasis to the bone.74 It is therefore of interest that HOXC6 directly represses BMP7 expression, a ligand that has osteogenic properties that can produce ectopic bone formation.75 Importantly, BMP7 has recently been shown to inhibit growth of prostate cancer cells in mouse bone suggesting it plays a role in inhibiting prostate cancer bone metastases.76 In addition, BMP7 also appears to counteract TGFβ signaling and inhibit bone metastases of breast cancer cells.77 During normal development, BMP7 represses Notch signaling and HEY1 expression.78
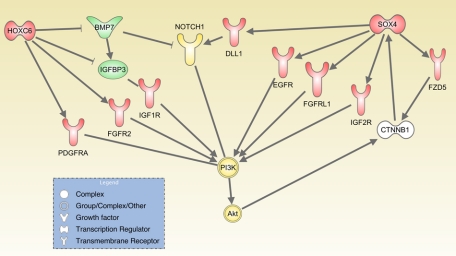
The PI3K/Akt pro-proliferative and survival pathway is influenced by several HOXC6 direct targets such as BMP7,78 IGFBP3,79 and PDGFRA80 (Figure 2), suggesting that one way HOXC6 exerts its pro-survival function is through the PI3K/Akt pathway. HOXC6 activates expression of FGFR2, which is normally expressed in basal epithelial cells of the prostate and binds multiple different FGF ligands,81 supporting the hypothesis that HOXC6 contributes to an undifferentiated cell phenotype.
Figure 2.
Model of the effects of the SOX4 and HOXC6 transcriptional networks on PI3K-AKT pathway activation in prostate cancer. Genes up-regulated by SOX4 or HOXC6 are shown in red, and repression targets are shown in green. Yellow indicates activation.
HOXC6 regulates genes with both oncogenic and tumor suppressor activities as well as several genes that are important for prostate branching morphogenesis and metastasis to the bone microenvironment. Interestingly, tumor suppressive genes that were activated by HOXC6 in expression analysis of HOXC6 knockout mice included the hyaluronic acid receptor CD44 and four inhibitors of Wnt signaling: WIF1; DKK3; SFRP1; and SFRP2. Importantly, although HOXC6 activates expression of these genes in normal mouse prostates, all of these genes are silenced by hypermethylation in tumors and cancer cell lines.38,82,83 This hypermethylation may prevent HOXC6 activation of their expression in prostate cancer tissue.
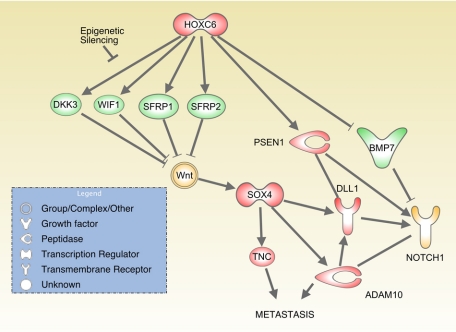
Taken together, these data suggest a model in which HOXC6 is a critical regulator of normal prostate development, directly controlling expression of BMP7, FGFR2, and PDGFRA, indirectly inhibiting Wnt signals and activating Notch signals (Figure 3). However, in prostate cancers, epigenetic silencing of Wnt-suppressing target genes may enable HOXC6 to activate Notch signals without interfering with Wnt signaling.
Figure 3.
Model of the effects of SOX4 and HOXC6 on Notch pathway activation and metastasis. Genes up-regulated by SOX4 or HOXC6 are shown in red, and repression targets are shown in green. Yellow indicates activation. PSEN1 = presenilin-1; TNC = tenascin C.
The SOX4 Network
The SOX4 transcription factor is a developmental transcription factor that regulates progenitor development and Wnt signaling.84,85 SOX4 is a 47 kDa protein that contains a highly conserved high-mobility group DNA-binding domain related to the TCF/LEF family of transcription factors that play important roles in the Wnt pathway. Although the role of SOX4 in the Wnt pathway is still unclear, SOX4 can interact directly with β-catenin and cooperate with β-catenin to activate gene expression.85,86 While SOX4 is not a stem cell marker, as is its relative SOX2,87 it is expressed in intestinal stem cells88 and likely plays a role in the early differentiation and expansion of transit amplifying progenitor cells. Embryonic knockout of SOX4 is lethal around E14 due to cardiac developmental defects and these embryos also show impaired lymphocyte development.89 In adult mice, SOX4 is expressed in the gonads, thymus, T- and pro-B-lymphocyte lineages and to a lesser extent in the lungs, lymph nodes, and heart.90 Tissue specific knockout of SOX4 leads to developmental defects of the pancreas,91 and SOX4 heterozygous mice have impaired bone development,92 whereas prolonged expression of SOX4 inhibits correct neuronal differentiation.93 These studies suggest a crucial role for SOX4 in cell fate decisions and progenitor cell survival.
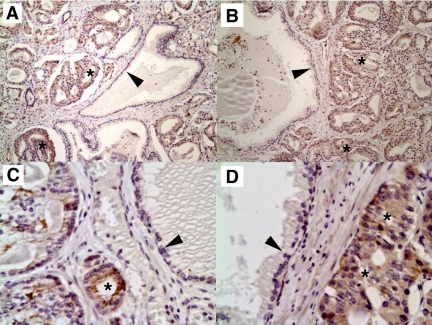
In humans, SOX4 is expressed in the developing breast and osteoblasts, and is up-regulated in response to progestins.94 We recently demonstrated that SOX4 is up-regulated at the mRNA and protein level in prostate cancer and this up-regulation is correlated with Gleason score or tumor grade37 (Figure 4). SOX4 is overexpressed in many other types of human cancers, including leukemias,95 melanomas,96 glioblastomas,97 medulloblastomas,98 and cancers of the bladder99 and lung.100 Furthermore, SOX4 cooperates with the Evi1 transcription factor in mouse models of myeloid leukemias.101 Why is SOX4 overexpressed in so many cancers and what regulates SOX4 expression? Although SOX4 is not induced by androgens,102 it has been shown to be induced by hypoxia and hypoxia-inducible factor 1α (HIF1α),103 angiogenesis,104 progestins,94 estradiol,105 tumor necrosis factor α,106 TGFβ1,107 Wnt signaling,108 and deletion of BMP1 receptor,109 which activates Wnt signaling and activation of the PI3K-AKT pathway. Thus, many signaling pathways that are commonly activated in malignant cells are able to increase SOX4 expression levels.
Figure 4.
Immunohistochemistry of formalin-fixed, paraffin-embedded prostate adenocarcinomas with affinity-purified SOX4 antibodies. SOX4 detection is dark brown; nuclei were counterstained with hematoxylin (blue). Prostate cancer (asterisks) stains much more intensely than benign epithelium (arrowheads). Note the dark staining of some nuclei, consistent with a role of SOX4 in transcription. Negative controls without primary antibody showed no staining of any cell type (not shown). Original magnification: ×400 (A and B); ×600 (C and D).
A metaanalysis examining the transcriptional profiles of human cancers found SOX4 to be one of 64 genes uniquely up-regulated as a general “Cancer Signature,”110 suggesting that it has a fundamental role in multiple tumor types. We found that overexpression of SOX4 in RWPE-1 prostate cells results in anchorage independent growth, implying that in the proper cellular context, SOX4 can act as an oncogene.37 Moreover, SOX4 was recently shown to promote lung metastases of breast cancer cells.111 The SOX4 gene, located on chromosome 6p22.3 is amplified in lung cancers, and SOX4 overexpression increased the transforming ability of the weakly oncogenic RHOA-Q63L mutant.112
Consistent with the suggestion that SOX4 is an oncogene, three independent studies searching for oncogenes have revealed SOX4 to be one of the most common retroviral integration sites, resulting in increased SOX4 mRNA levels.113,114,115 Nevertheless, the precise role that SOX4 plays in cancer progression is not well understood. Although we, and others, have shown that small-interfering RNA knockdown of SOX4 can induce apoptosis,37,116 strong overexpression of SOX4 can also induce apoptosis,117 similar to the c-myc oncogene, suggesting that precise regulation of SOX4 levels is critical for cell survival. Recent evidence suggests this paradox may be due to SOX4’s regulation of p53.118 In response to DNA damage, SOX4 induces p53 stabilization and is critical for transcription of p53 target genes that mediate cell cycle arrest.118 Furthermore, SOX4 can activate expression of PUMA,37 a gene critical for the p53 apoptotic response.119 Interestingly, SOX4 induced colony formation in the p53 compromised RWPE-1 cell line37 but not in a wild-type p53 cell line,120 suggesting that in cells with wild-type p53, SOX4 may act as a tumor suppressor, but in cells lacking wild-type p53, SOX4 may act as an oncogene.
Recently, we have performed a genome-wide promoter analysis by using a ChIP-chip approach to identify those genes that have SOX4 bound at their promoters in human prostate cancer cells.86 We identified 282 genes that are high-confidence direct SOX4 targets, including many genes involved in microRNA (miRNA) processing, transcriptional regulation, developmental pathways, growth factor signaling, and tumor metastasis. SOX4 target genes include regulators of pivotal prostate cancer signaling networks of differentiation, cell survival, and apoptosis.
Each of these genes was bound by SOX4 in ChIP-chip and altered in expression by SOX4 transfection or knockdown. The SOX4 transcriptional network impacts the Notch, Wnt, and PI3K pathways, as well as miRNA processing via regulation of Dicer and Argonaute 1 (AGO1).86
SOX4 and Metastasis
Partial knockdown of SOX4 by short hairpin RNA (shRNA) results in fewer lung metastases by using a xenograft model of breast cancer.111 Our network analysis showed that SOX4 directly regulates a number of genes important for metastasis including epidermal growth factor receptor (EGFR), tenascin C, Integrin αv, Rac1, paxillin, gelsolin, DLL1, ADAM metallopeptidase domain 10 (ADAM10), and growth factor receptor-bound protein 7.86 SOX4 may also promote metastasis and tissue invasion in part by inhibiting terminal differentiation and promoting the EMT process. SOX4 inhibits terminal differentiation via repression of the transcription factor NKX3.1,86 and activation of MLL and MLL3, two histone H3 K4 methyltransferases that induce activation of HOX gene expression.121
MLL methyltransferase complexes can also facilitate E2F activation of S-phase promoters, driving the cell cycle forward, and MLL is a critical oncogene that is often translocated or amplified in myeloid leukemogenesis. Thus, activation of MLL suggests a role for SOX4 in myeloid leukemia122 in which SOX4 may prevent terminal differentiation through activation of MLL and MLL3. SOX4 may also inhibit terminal differentiation via regulation of a host of over 20 other transcription factors, including E74-like factor 5 (ELF5), Serum Response Factor (SRF), nuclear receptor coactivator 4 (NCOA4), retinoblastoma-like 1 p107 (RBL1), zinc finger protein 281 (ZNF281), SOX12, and for khead box A1 (FOXA1).86
The phosphatase PTEN and transcription factor NKX3.1 are prostate cancer tumor suppressors that negatively regulate the PI3K-AKT pathway.120 Mice with prostate-specific compound heterozygous deletions of NKX3.1 and PTEN develop prostate adenocarcinomas and metastases to the lymph node with high frequency,123 implicating the importance of the PI3K-AKT pathway in prostate tumors. SOX4 may promote this effect via simultaneous up-regulation of growth factor receptors such as epidermal growth factor receptor, FGFRL1, and IGF2R, and direct repression of FOXA1 and NKX3.1 (Figure 2).
SOX4 and miRNAs
SOX4 may also impact cellular differentiation via regulation of components of the miRNA pathway. MiRNAs are a small noncoding RNA species that regulate the translation and stability of mRNA messages for hundreds of downstream target genes via partial complementarity to short sequences in the 3′ untranslated regions of mRNAs. The RNA-induced silencing complex, which is composed of AGO1 or AGO2, Tar RNA binding protein (TRBP) and Dicer, processes miRNAs from precursors (pre-miRNA) to their mature form, cleaves target mRNAs, and participates in translational inhibition.124 RNA Helicase A interacts with RNA-induced silencing complex and participates in the loading of small RNAs into the complex.125 SOX4 directly regulates three components of the RNA-induced silencing complex: Dicer; AGO1; and RNA Helicase A.
MiRNAs can act both as tumor suppressors and as oncogenes, depending on the sets of downstream targets that they regulate. Recently, it was found that miR-10b plays a crucial role in promoting metastasis.126 In contrast, miR-335 suppresses metastasis and migration of cancer cells via targeting of tenascin C and SOX4.111 A number of miRNAs are altered in several epithelial tumors, including prostate cancers.127 Moreover, Dicer expression is up-regulated in prostate cancers.127
SOX4 and Developmental Pathways
As noted above, SOX4 plays a role in the Wnt pathway via direct interaction with β-catenin to activate gene expression.85,86 In addition to this role, analysis of the direct targets of SOX4 indicates that it also regulates several Wnt pathway components, including Frizzled receptors (certainly FZD5, and possibly FZD4, FZD6, FZD8, and FZD9) and the ortholog of the groucho repressor, TLE1. In addition, gene set enrichment analysis (GSEA)128 and gene set enrichment analysis Leading Edge analysis129 of gene sets induced by SOX4 determined that these gene sets were enriched in TGFβ-SMAD (Similar to Mothers Against Decapentaplegic) direct target genes such as tenascin C and IGF2R.86 SOX4 is up-regulated by TGFβ-1 treatment,91,107 and SMAD sites are significantly enriched in the SOX4 ChIP-chip peaks, suggesting that SOX4 could interact with SMADs.
Of particular interest is SOX4’s role in activation of the Notch pathway. Preliminary evidence points to SOX4’s ability to activate signaling from the Notch receptor through transcriptional activation of ADAM10, DLL1, and HES2.15 The observations that HOXC6 can repress BMP7 and activate expression of presenilin-1 suggest a model in which HOXC6 and SOX4 may cooperate to activate Notch signaling via targeting multiple components of the Notch pathway (Figure 3), possibly in response to activation of Wnt signals. In addition, SOX4 up-regulation of growth factor receptors such as epidermal growth factor receptor, FGFRL1, and IGF2R could cooperate with HOXC6 up-regulation of PDGFRA, FGFR2, and IGFBP3 to enhance activation of the PI3K-AKT pathway (Figure 2).
Conclusions
The transcriptional networks regulated by SOX4 and HOXC6 suggest crucial roles for these factors in proliferation, survival, and metastasis of prostate cancer cells. Increased expression of these developmental transcription factors may enhance activation of signal transduction pathways such as Notch and PI3K-AKT, contributing to prostate cancer progression and metastasis. These findings suggest that drug combinations that target the Notch and PI3K pathways may be of benefit to patients with metastatic prostate cancer. Moreover, use of compounds that can result in demethylation of DNA such as 5-aza-deoxcycytidine or possibly soy isoflavones may reduce activation of the Wnt pathway and could also be of benefit to this patient population. Future studies will be needed to determine whether SOX4 and/or HOXC6 and their downstream targets represent potential therapeutic targets in prostate cancer.
Acknowledgments
The author would like to thank Dr. Daniel Brat, Dr. Asma Nusrat, and Dr. Debu Tripathy for critical reading of this article. This work was supported by R01 CA106826 from the National Cancer Institute.
Footnotes
Address reprint requests to Carlos S. Moreno, Ph.D., Department of Pathology and Laboratory Medicine, Winship Cancer Institute, Whitehead Research Building, Room 105J, 615 Michael St, Atlanta, GA 30322. E-mail: cmoreno@emory.edu.
Supported by R01 CA106826 from the National Cancer Institute.
The ASIP Cotran Established Investigator Award is given by the American Society for Investigative Pathology to recognize early career investigators with demonstrated excellence as an investigator focusing on the conceptual basis of disease. Carlos S. Moreno, recipient of the 2009 Cotran Established Investigator Award, delivered a lecture entitled “Developmental Pathways and Transcriptional Networks in Prostate Cancer Progression,” on April 21, 2009 at the annual meeting of the American Society for Investigative Pathology in New Orleans, LA.
References
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- Cunha GR, Ricke W, Thomson A, Marker PC, Risbridger G, Hayward SW, Wang YZ, Donjacour AA, Kurita T. Hormonal, cellular, and molecular regulation of normal and neoplastic prostatic development. J Steroid Biochem Mol Biol. 2004;92:221–236. doi: 10.1016/j.jsbmb.2004.10.017. [DOI] [PubMed] [Google Scholar]
- Assinder SJ, Dong Q, Kovacevic Z, Richardson DR. The TGF-beta, PI3K/Akt and PTEN pathways: established and proposed biochemical integration in prostate cancer. Biochem J. 2009;417:411–421. doi: 10.1042/BJ20081610. [DOI] [PubMed] [Google Scholar]
- Shaw A, Bushman W. Hedgehog signaling in the prostate. J Urol. 2007;177:832–838. doi: 10.1016/j.juro.2006.10.061. [DOI] [PubMed] [Google Scholar]
- Leong KG, Gao WQ. The Notch pathway in prostate development and cancer. Differentiation. 2008;76:699–716. doi: 10.1111/j.1432-0436.2008.00288.x. [DOI] [PubMed] [Google Scholar]
- Robinson DR, Zylstra CR, Williams BO. Wnt signaling and prostate cancer. Curr Drug Targets. 2008;9:571–580. doi: 10.2174/138945008784911831. [DOI] [PubMed] [Google Scholar]
- Thomson AA. Role of androgens and fibroblast growth factors in prostatic development. Reproduction. 2001;121:187–195. doi: 10.1530/rep.0.1210187. [DOI] [PubMed] [Google Scholar]
- Freestone SH, Marker P, Grace OC, Tomlinson DC, Cunha GR, Harnden P, Thomson AA. Sonic hedgehog regulates prostatic growth and epithelial differentiation. Dev Biol. 2003;264:352–362. doi: 10.1016/j.ydbio.2003.08.018. [DOI] [PubMed] [Google Scholar]
- Wang XD, Leow CC, Zha J, Tang Z, Modrusan Z, Radtke F, Aguet M, de Sauvage FJ, Gao WQ. Notch signaling is required for normal prostatic epithelial cell proliferation and differentiation. Dev Biol. 2006;290:66–80. doi: 10.1016/j.ydbio.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Orr B, Grace OC, Vanpoucke G, Ashley GR, Thomson AA. A role for notch signaling in stromal survival and differentiation during prostate development. Endocrinology. 2009;150:463–472. doi: 10.1210/en.2008-0383. [DOI] [PubMed] [Google Scholar]
- Blum R, Gupta R, Burger PE, Ontiveros CS, Salm SN, Xiong X, Kamb A, Wesche H, Marshall L, Cutler G, Wang X, Zavadil J, Moscatelli D, Wilson EL. Molecular signatures of prostate stem cells reveal novel signaling pathways and provide insights into prostate cancer. PLoS One. 2009;4:e5722. doi: 10.1371/journal.pone.0005722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi M, Nguyen C, Lee SC, Kahn M. ICG-001, a novel small molecule regulator of TCF/beta-catenin transcription. Med Chem. 2005;1:467–472. doi: 10.2174/1573406054864098. [DOI] [PubMed] [Google Scholar]
- Feldmann G, Fendrich V, McGovern K, Bedja D, Bisht S, Alvarez H, Koorstra JB, Habbe N, Karikari C, Mullendore M, Gabrielson KL, Sharma R, Matsui W, Maitra A. An orally bioavailable small-molecule inhibitor of Hedgehog signaling inhibits tumor initiation and metastasis in pancreatic cancer. Mol Cancer Ther. 2008;7:2725–2735. doi: 10.1158/1535-7163.MCT-08-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, O'Neil J, Liberator CD, Hardwick JS, Dai X, Zhang T, Tyminski E, Yuan J, Kohl NE, Richon VM, Van der Ploeg LH, Carroll PM, Draetta GF, Look AT, Strack PR, Winter CG. Inhibition of NOTCH signaling by gamma secretase inhibitor engages the RB pathway and elicits cell cycle exit in T-cell acute lymphoblastic leukemia cells. Cancer Res. 2009;69:3060–3068. doi: 10.1158/0008-5472.CAN-08-4295. [DOI] [PubMed] [Google Scholar]
- Bolos V, Grego-Bessa J, de la Pompa JL. Notch signaling in development and cancer. Endocr Rev. 2007;28:339–363. doi: 10.1210/er.2006-0046. [DOI] [PubMed] [Google Scholar]
- Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131:965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- Nobta M, Tsukazaki T, Shibata Y, Xin C, Moriishi T, Sakano S, Shindo H, Yamaguchi A. Critical regulation of bone morphogenetic protein-induced osteoblastic differentiation by Delta1/Jagged1-activated Notch1 signaling. J Biol Chem. 2005;280:15842–15848. doi: 10.1074/jbc.M412891200. [DOI] [PubMed] [Google Scholar]
- Ceder JA, Jansson L, Helczynski L, Abrahamsson PA. Delta-like 1 (Dlk-1), a novel marker of prostate basal and candidate epithelial stem cells, is downregulated by notch signalling in intermediate/transit amplifying cells of the human prostate. Eur Urol. 2008;54:1344–1353. doi: 10.1016/j.eururo.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Sciaudone M, Gazzerro E, Priest L, Delany AM, Canalis E. Notch 1 impairs osteoblastic cell differentiation. Endocrinology. 2003;144:5631–5639. doi: 10.1210/en.2003-0463. [DOI] [PubMed] [Google Scholar]
- Hallahan AR, Pritchard JI, Hansen S, Benson M, Stoeck J, Hatton BA, Russell TL, Ellenbogen RG, Bernstein ID, Beachy PA, Olson JM. The SmoA1 mouse model reveals that notch signaling is critical for the growth and survival of sonic hedgehog-induced medulloblastomas. Cancer Res. 2004;64:7794–7800. doi: 10.1158/0008-5472.CAN-04-1813. [DOI] [PubMed] [Google Scholar]
- Sheng T, Li C, Zhang X, Chi S, He N, Chen K, McCormick F, Gatalica Z, Xie J. Activation of the hedgehog pathway in advanced prostate cancer. Mol Cancer. 2004;3:29. doi: 10.1186/1476-4598-3-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez P, Hernandez AM, Stecca B, Kahler AJ, DeGueme AM, Barrett A, Beyna M, Datta MW, Datta S, Ruiz i Altaba A. Inhibition of prostate cancer proliferation by interference with sonic hedgehog-GLI1 signaling. Proc Natl Acad Sci USA. 2004;101:12561–12566. doi: 10.1073/pnas.0404956101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17:548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Dalrymple S, Antony L, Xu Y, Uzgare AR, Arnold JT, Savaugeot J, Sokoll LJ, De Marzo AM, Isaacs JT. Role of notch-1 and E-cadherin in the differential response to calcium in culturing normal versus malignant prostate cells. Cancer Res. 2005;65:9269–9279. doi: 10.1158/0008-5472.CAN-04-3989. [DOI] [PubMed] [Google Scholar]
- Ayyanan A, Civenni G, Ciarloni L, Morel C, Mueller N, Lefort K, Mandinova A, Raffoul W, Fiche M, Dotto GP, Brisken C. Increased Wnt signaling triggers oncogenic conversion of human breast epithelial cells by a Notch-dependent mechanism. Proc Natl Acad Sci USA. 2006;103:3799–3804. doi: 10.1073/pnas.0600065103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bafico A, Liu G, Yaniv A, Gazit A, Aaronson SA. Novel mechanism of Wnt signalling inhibition mediated by Dickkopf-1 interaction with LRP6/Arrow. Nat Cell Biol. 2001;3:683–686. doi: 10.1038/35083081. [DOI] [PubMed] [Google Scholar]
- He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- Verras M, Brown J, Li X, Nusse R, Sun Z. Wnt3a growth factor induces androgen receptor-mediated transcription and enhances cell growth in human prostate cancer cells. Cancer Res. 2004;64:8860–8866. doi: 10.1158/0008-5472.CAN-04-2370. [DOI] [PubMed] [Google Scholar]
- Lu W, Tinsley HN, Keeton A, Qu Z, Piazza GA, Li Y. Suppression of Wnt/beta-catenin signaling inhibits prostate cancer cell proliferation. Eur J Pharmacol. 2009;602:8–14. doi: 10.1016/j.ejphar.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas TR, Kim J, Vakar-Lopez F, Sabichi AL, Troncoso P, Jenster G, Kikuchi A, Chen SY, Shemshedini L, Suraokar M, Logothetis CJ, DiGiovanni J, Lippman SM, Menter DG. Glycogen synthase kinase-3 beta is involved in the phosphorylation and suppression of androgen receptor activity. J Biol Chem. 2004;279:19191–19200. doi: 10.1074/jbc.M309560200. [DOI] [PubMed] [Google Scholar]
- Wang G, Wang J, Sadar MD. Crosstalk between the androgen receptor and beta-catenin in castrate-resistant prostate cancer. Cancer Res. 2008;68:9918–9927. doi: 10.1158/0008-5472.CAN-08-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall CL, Kang S, MacDougald OA, Keller ET. Role of Wnts in prostate cancer bone metastases. J Cell Biochem. 2006;97:661–672. doi: 10.1002/jcb.20735. [DOI] [PubMed] [Google Scholar]
- Hall CL, Bafico A, Dai J, Aaronson SA, Keller ET. Prostate cancer cells promote osteoblastic bone metastases through Wnts. Cancer Res. 2005;65:7554–7560. doi: 10.1158/0008-5472.CAN-05-1317. [DOI] [PubMed] [Google Scholar]
- Ohigashi T, Mizuno R, Nakashima J, Marumo K, Murai M. Inhibition of Wnt signaling downregulates Akt activity and induces chemosensitivity in PTEN-mutated prostate cancer cells. Prostate. 2005;62:61–68. doi: 10.1002/pros.20117. [DOI] [PubMed] [Google Scholar]
- Liu P, Ramachandran S, Ali-Seyed M, Scharer CD, Laycock N, Dalton WB, Williams H, Karanam S, Datta MW, Jaye D, Moreno CS. Sex-determining region Y box 4 is a transforming oncogene in human prostate cancer cells. Cancer Res. 2006;46:4011–4019. doi: 10.1158/0008-5472.CAN-05-3055. [DOI] [PubMed] [Google Scholar]
- Lodygin D, Epanchintsev A, Menssen A, Diebold J, Hermeking H. Functional epigenomics identifies genes frequently silenced in prostate cancer. Cancer Res. 2005;65:4218–4227. doi: 10.1158/0008-5472.CAN-04-4407. [DOI] [PubMed] [Google Scholar]
- Kawano Y, Kitaoka M, Hamada Y, Walker MM, Waxman J, Kypta RM. Regulation of prostate cell growth and morphogenesis by Dickkopf-3. Oncogene. 2006;25:6528–6537. doi: 10.1038/sj.onc.1209661. [DOI] [PubMed] [Google Scholar]
- Kawano Y, Diez S, Uysal-Onganer P, Darrington RS, Waxman J, Kypta RM. Secreted Frizzled-related protein-1 is a negative regulator of androgen receptor activity in prostate cancer. Br J Cancer. 2009;100:1165–1174. doi: 10.1038/sj.bjc.6604976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K, Pienta KJ, Rubin MA, Chinnaiyan AM. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412:822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- Varambally S, Yu J, Laxman B, Rhodes DR, Mehra R, Tomlins SA, Shah RB, Chandran U, Monzon FA, Becich MJ, Wei JT, Pienta KJ, Ghosh D, Rubin MA, Chinnaiyan AM. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, McDonald C, Thomas R, Dhir R, Finkelstein S, Michalopoulos G, Becich M, Luo JH. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Barrette TR, Rubin MA, Ghosh D, Chinnaiyan AM. Meta-analysis of microarrays: interstudy validation of gene expression profiles reveals pathway dysregulation in prostate cancer. Cancer Res. 2002;62:4427–4433. [PubMed] [Google Scholar]
- Ernst T, Hergenhahn M, Kenzelmann M, Cohen CD, Bonrouhi M, Weninger A, Klaren R, Grone EF, Wiesel M, Gudemann C, Kuster J, Schott W, Staehler G, Kretzler M, Hollstein M, Grone HJ. Decrease and gain of gene expression are equally discriminatory markers for prostate carcinoma: a gene expression analysis on total and microdissected prostate tissue. Am J Pathol. 2002;160:2169–2180. doi: 10.1016/S0002-9440(10)61165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe J, Li C, Higgins JP, Van De Rijn M, Bair E, Montgomery K, Ferrari M, Egevad L, Rayford W, Bergerheim U, Ekman P, DeMarzo AM, Tibshirani R, Botstein D, Brown PO, Brooks JD, Pollack JR. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci USA. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Duggan DJ, Chen Y, Sauvageot J, Ewing CM, Bittner ML, Trent JM, Isaacs WB. Human prostate cancer and benign prostatic hyperplasia: molecular dissection by gene expression profiling. Cancer Res. 2001;61:4683–4688. [PubMed] [Google Scholar]
- Magee JA, Araki T, Patil S, Ehrig T, True L, Humphrey PA, Catalona WJ, Watson MA, Milbrandt J. Expression profiling reveals hepsin overexpression in prostate cancer. Cancer Res. 2001;61:5692–5696. [PubMed] [Google Scholar]
- Welsh JB, Sapinoso LM, Su AI, Kern SG, Wang-Rodriguez J, Moskaluk CA, Frierson HF, Jr, Hampton GM. Analysis of gene expression identifies candidate markers and pharmacological targets in prostate cancer. Cancer Res. 2001;61:5974–5978. [PubMed] [Google Scholar]
- Hughes C, Murphy A, Martin C, Sheils O, O'Leary J. Molecular pathology of prostate cancer. J Clin Pathol. 2005;58:673–684. doi: 10.1136/jcp.2002.003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, Menon A, Jing X, Cao Q, Han B, Yu J, Wang L, Montie JE, Rubin MA, Pienta KJ, Roulston D, Shah RB, Varambally S, Mehra R, Chinnaiyan AM. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- Ramachandran S, Liu P, Young AN, Yin-Goen Q, Lim SD, Laycock N, Amin MB, Carney JK, Marshall FF, Petros JA, Moreno CS. Loss of HOXC6 expression induces apoptosis in prostate cancer cells. Oncogene. 2005;24:188–198. doi: 10.1038/sj.onc.1207906. [DOI] [PubMed] [Google Scholar]
- Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, Alimonti A, Nardella C, Varmeh S, Scardino PT, Cordon-Cardo C, Gerald W, Pandolfi PP. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–624. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iljin K, Wolf M, Edgren H, Gupta S, Kilpinen S, Skotheim RI, Peltola M, Smit F, Verhaegh G, Schalken J, Nees M, Kallioniemi O. TMPRSS2 fusions with oncogenic ETS factors in prostate cancer involve unbalanced genomic rearrangements and are associated with HDAC1 and epigenetic reprogramming. Cancer Res. 2006;66:10242–10246. doi: 10.1158/0008-5472.CAN-06-1986. [DOI] [PubMed] [Google Scholar]
- Bachmann IM, Halvorsen OJ, Collett K, Stefansson IM, Straume O, Haukaas SA, Salvesen HB, Otte AP, Akslen LA. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J Clin Oncol. 2006;24:268–273. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- Saramaki OR, Tammela TL, Martikainen PM, Vessella RL, Visakorpi T. The gene for polycomb group protein enhancer of zeste homolog 2 (EZH2) is amplified in late-stage prostate cancer. Genes Chromosomes Cancer. 2006;45:639–645. doi: 10.1002/gcc.20327. [DOI] [PubMed] [Google Scholar]
- Cooper CS, Foster CS. Concepts of epigenetics in prostate cancer development. Br J Cancer. 2009;100:240–245. doi: 10.1038/sj.bjc.6604771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Gong Y, Yue J, Qiang B, Yuan J, Peng X. Cooperation between EZH2: NSPc1-mediated histone H2A ubiquitination and Dnmt1 in HOX gene silencing. Nucleic Acids Res. 2008;36:3590–3599. doi: 10.1093/nar/gkn243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Rhodes DR, Tomlins SA, Cao X, Chen G, Mehra R, Wang X, Ghosh D, Shah RB, Varambally S, Pienta KJ, Chinnaiyan AM. A polycomb repression signature in metastatic prostate cancer predicts cancer outcome. Cancer Res. 2007;67:10657–10663. doi: 10.1158/0008-5472.CAN-07-2498. [DOI] [PubMed] [Google Scholar]
- Karanam S, Moreno CS. CONFAC: automated application of comparative genomic promoter analysis to DNA microarray datasets. Nucleic Acids Res. 2004;32:W475–W484. doi: 10.1093/nar/gkh353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veraksa A, Del Campo M, McGinnis W. Developmental patterning genes and their conserved functions: from model organisms to humans. Mol Genet Metab. 2000;69:85–100. doi: 10.1006/mgme.2000.2963. [DOI] [PubMed] [Google Scholar]
- Cillo C, Cantile M, Faiella A, Boncinelli E. Homeobox genes in normal and malignant cells. J Cell Physiol. 2001;188:161–169. doi: 10.1002/jcp.1115. [DOI] [PubMed] [Google Scholar]
- Celetti A, Barba P, Cillo C, Rotoli B, Boncinelli E, Magli MC. Characteristic patterns of HOX gene expression in different types of human leukemia. Int J Cancer. 1993;53:237–244. doi: 10.1002/ijc.2910530211. [DOI] [PubMed] [Google Scholar]
- Abate-Shen C. Deregulated homeobox gene expression in cancer: cause or consequence? Nat Rev Cancer. 2002;2:777–785. doi: 10.1038/nrc907. [DOI] [PubMed] [Google Scholar]
- Bodey B, Bodey B, Jr, Siegel SE, Luck JV, Kaiser HE. Homeobox B3, B4, and C6 gene product expression in osteosarcomas as detected by immunocytochemistry. Anticancer Res. 2000;20:2717–2721. [PubMed] [Google Scholar]
- Bodey B, Bodey B, Jr, Siegel SE, Kaiser HE. Immunocytochemical detection of the homeobox B3, B4, and C6 gene products in childhood medulloblastomas/primitive neuroectodermal tumors. Anticancer Res. 2000;20:1769–1780. [PubMed] [Google Scholar]
- Dressman HK, Hans C, Bild A, Olson JA, Rosen E, Marcom PK, Liotcheva VB, Jones EL, Vujaskovic Z, Marks J, Dewhirst MW, West M, Nevins JR, Blackwell K. Gene expression profiles of multiple breast cancer phenotypes and response to neoadjuvant chemotherapy. Clin Cancer Res. 2006;12:819–826. doi: 10.1158/1078-0432.CCR-05-1447. [DOI] [PubMed] [Google Scholar]
- Bodey B, Bodey B, Jr, Groger AM, Siegel SE, Kaiser HE. Immunocytochemical detection of homeobox B3, B4, and C6 gene product expression in lung carcinomas. Anticancer Res. 2000;20:2711–2716. [PubMed] [Google Scholar]
- Miller GJ, Miller HL, van Bokhoven A, Lambert JR, Werahera PN, Schirripa O, Lucia MS, Nordeen SK. Aberrant HOXC expression accompanies the malignant phenotype in human prostate. Cancer Res. 2003;63:5879–5888. [PubMed] [Google Scholar]
- Bibikova M, Chudin E, Arsanjani A, Zhou L, Garcia EW, Modder J, Kostelec M, Barker D, Downs T, Fan JB, Wang-Rodriguez J. Expression signatures that correlated with Gleason score and relapse in prostate cancer. Genomics. 2007;89:666–672. doi: 10.1016/j.ygeno.2007.02.005. [DOI] [PubMed] [Google Scholar]
- McCabe CD, Spyropoulos DD, Martin D, Moreno CS. Genome-wide analysis of the homeobox C6 transcriptional network in prostate cancer. Cancer Res. 2008;68:1988–1996. doi: 10.1158/0008-5472.CAN-07-5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeneman KS, Yeung F, Chung LW. Osteomimetic properties of prostate cancer cells: a hypothesis supporting the predilection of prostate cancer metastasis and growth in the bone environment. Prostate. 1999;39:246–261. doi: 10.1002/(sici)1097-0045(19990601)39:4<246::aid-pros5>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Franceschi RT, Wang D, Krebsbach PH, Rutherford RB. Gene therapy for bone formation: in vitro and in vivo osteogenic activity of an adenovirus expressing BMP7. J Cell Biochem. 2000;78:476–486. doi: 10.1002/1097-4644(20000901)78:3<476::aid-jcb12>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Buijs JT, Rentsch CA, van der Horst G, van Overveld PG, Wetterwald A, Schwaninger R, Henriquez NV, Ten Dijke P, Borovecki F, Markwalder R, Thalmann GN, Papapoulos SE, Pelger RC, Vukicevic S, Cecchini MG, Lowik CW, van der Pluijm G. BMP7, a putative regulator of epithelial homeostasis in the human prostate, is a potent inhibitor of prostate cancer bone metastasis in vivo. Am J Pathol. 2007;171:1047–1057. doi: 10.2353/ajpath.2007.070168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buijs JT, Henriquez NV, van Overveld PG, van der Horst G, ten Dijke P, van der Pluijm G. TGF-beta and BMP7 interactions in tumour progression and bone metastasis. Clin Exp Metastasis. 2007;24:609–617. doi: 10.1007/s10585-007-9118-2. [DOI] [PubMed] [Google Scholar]
- Grishina IB, Kim SY, Ferrara C, Makarenkova HP, Walden PD. BMP7 inhibits branching morphogenesis in the prostate gland and interferes with Notch signaling. Dev Biol. 2005;288:334–347. doi: 10.1016/j.ydbio.2005.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricort JM, Binoux M. Insulin-like growth factor-binding protein-3 activates a phosphotyrosine phosphatase: effects on the insulin-like growth factor signaling pathway. J Biol Chem. 2002;277:19448–19454. doi: 10.1074/jbc.M200439200. [DOI] [PubMed] [Google Scholar]
- van der Geer P, Hunter T, Lindberg RA. Receptor protein-tyrosine kinases and their signal transduction pathways. Annu Rev Cell Biol. 1994;10:251–337. doi: 10.1146/annurev.cb.10.110194.001343. [DOI] [PubMed] [Google Scholar]
- Cotton LM, O'Bryan MK, Hinton BT. Cellular signaling by fibroblast growth factors (FGFs) and their receptors (FGFRs) in male reproduction. Endocr Rev. 2008;29:193–216. doi: 10.1210/er.2007-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissmann C, Wild PJ, Kaiser S, Roepcke S, Stoehr R, Woenckhaus M, Kristiansen G, Hsieh JC, Hofstaedter F, Hartmann A, Knuechel R, Rosenthal A, Pilarsky C. WIF1, a component of the Wnt pathway, is down-regulated in prostate, breast, lung, and bladder cancer. J Pathol. 2003;201:204–212. doi: 10.1002/path.1449. [DOI] [PubMed] [Google Scholar]
- Zou H, Molina JR, Harrington JJ, Osborn NK, Klatt KK, Romero Y, Burgart LJ, Ahlquist DA. Aberrant methylation of secreted frizzled-related protein genes in esophageal adenocarcinoma and Barrett’s esophagus. Int J Cancer. 2005;116:584–591. doi: 10.1002/ijc.21045. [DOI] [PubMed] [Google Scholar]
- Cheung M, Abu-Elmagd M, Clevers H, Scotting PJ. Roles of Sox4 in central nervous system development. Brain Res Mol Brain Res. 2000;79:180–191. doi: 10.1016/s0169-328x(00)00109-1. [DOI] [PubMed] [Google Scholar]
- Sinner D, Kordich JJ, Spence JR, Opoka R, Rankin S, Lin SC, Jonatan D, Zorn AM, Wells JM. Sox17 and Sox4 differentially regulate beta-catenin/T-cell factor activity and proliferation of colon carcinoma cells. Mol Cell Biol. 2007;27:7802–7815. doi: 10.1128/MCB.02179-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharer CD, McCabe CD, Ali-Seyed M, Berger MF, Bulyk ML, Moreno CS. Genome-wide promoter analysis of the SOX4 transcriptional network in prostate cancer cells. Cancer Res. 2009;69:709–717. doi: 10.1158/0008-5472.CAN-08-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Fevr T, Robine S, Louvard D, Huelsken J. Wnt/beta-catenin is essential for intestinal homeostasis and maintenance of intestinal stem cells. Mol Cell Biol. 2007;27:7551–7559. doi: 10.1128/MCB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ya J, Schilham MW, de Boer PA, Moorman AF, Clevers H, Lamers WH. Sox4-deficiency syndrome in mice is an animal model for common trunk. Circ Res. 1998;83:986–994. doi: 10.1161/01.res.83.10.986. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Oosterwegel M, van Norren K, Clevers H. Sox-4, an Sry-like HMG box protein, is a transcriptional activator in lymphocytes. EMBO J. 1993;12:3847–3854. doi: 10.1002/j.1460-2075.1993.tb06063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson ME, Yang KY, Kalousova A, Lau J, Kosaka Y, Lynn FC, Wang J, Mrejen C, Episkopou V, Clevers HC, German MS. The HMG box transcription factor Sox4 contributes to the development of the endocrine pancreas. Diabetes. 2005;54:3402–3409. doi: 10.2337/diabetes.54.12.3402. [DOI] [PubMed] [Google Scholar]
- Nissen-Meyer LS, Jemtland R, Gautvik VT, Pedersen ME, Paro R, Fortunati D, Pierroz DD, Stadelmann VA, Reppe S, Reinholt FP, Del Fattore A, Rucci N, Teti A, Ferrari S, Gautvik KM. Osteopenia, decreased bone formation and impaired osteoblast development in Sox4 heterozygous mice. J Cell Sci. 2007;120:2785–2795. doi: 10.1242/jcs.003855. [DOI] [PubMed] [Google Scholar]
- Potzner MR, Griffel C, Lutjen-Drecoll E, Bosl MR, Wegner M, Sock E. Prolonged Sox4 expression in oligodendrocytes interferes with normal myelination in the central nervous system. Mol Cell Biol. 2007;27:5316–5326. doi: 10.1128/MCB.00339-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt SM, Clarke CL. Expression and hormonal regulation of the Sox4 gene in mouse female reproductive tissues. Biol Reprod. 1999;61:476–481. doi: 10.1095/biolreprod61.2.476. [DOI] [PubMed] [Google Scholar]
- Andersson A, Ritz C, Lindgren D, Eden P, Lassen C, Heldrup J, Olofsson T, Rade J, Fontes M, Porwit-Macdonald A, Behrendtz M, Hoglund M, Johansson B, Fioretos T. Microarray-based classification of a consecutive series of 121 childhood acute leukemias: prediction of leukemic and genetic subtype as well as of minimal residual disease status. Leukemia. 2007;21:1198–1203. doi: 10.1038/sj.leu.2404688. [DOI] [PubMed] [Google Scholar]
- Talantov D, Mazumder A, Yu JX, Briggs T, Jiang Y, Backus J, Atkins D, Wang Y. Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin Cancer Res. 2005;11:7234–7242. doi: 10.1158/1078-0432.CCR-05-0683. [DOI] [PubMed] [Google Scholar]
- Sun L, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, Passaniti A, Menon J, Walling J, Bailey R, Rosenblum M, Mikkelsen T, Fine HA. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Lee CJ, Appleby VJ, Orme AT, Chan WI, Scotting PJ. Differential expression of SOX4 and SOX11 in medulloblastoma. J Neurooncol. 2002;57:201–214. doi: 10.1023/a:1015773818302. [DOI] [PubMed] [Google Scholar]
- Aaboe M, Birkenkamp-Demtroder K, Wiuf C, Sorensen FB, Thykjaer T, Sauter G, Jensen KM, Dyrskjot L, Orntoft T. SOX4 expression in bladder carcinoma: clinical aspects and in vitro functional characterization. Cancer Res. 2006;66:3434–3442. doi: 10.1158/0008-5472.CAN-05-3456. [DOI] [PubMed] [Google Scholar]
- Friedman R, Bangur C, Zasloff E, Fan L, Wang T, Watanabe Y, Kalos M. Molecular and immunological evaluation of the transcription factor SOX-4 as a lung tumor vaccine antigen. J Immunol. 2004;172:3319–3327. doi: 10.4049/jimmunol.172.5.3319. [DOI] [PubMed] [Google Scholar]
- Boyd KE, Xiao YY, Fan K, Poholek A, Copeland NG, Jenkins NA, Perkins AS. Sox4 cooperates with Evi1 in AKXD-23 myeloid tumors via transactivation of proviral LTR. Blood. 2006;107:733–741. doi: 10.1182/blood-2003-05-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco AM, Gillis KA, Li Y, Brown EL, Sadler TM, Achilleos M, Greenberger LM, Frost P, Bai W, Zhang Y. Identification and validation of novel androgen-regulated genes in prostate cancer. Endocrinology. 2004;145:3913–3924. doi: 10.1210/en.2004-0311. [DOI] [PubMed] [Google Scholar]
- Manalo DJ, Rowan A, Lavoie T, Natarajan L, Kelly BD, Ye SQ, Garcia JG, Semenza GL. Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood. 2005;105:659–669. doi: 10.1182/blood-2004-07-2958. [DOI] [PubMed] [Google Scholar]
- van Beijnum JR, Dings RP, van der Linden E, Zwaans BM, Ramaekers FC, Mayo KH, Griffioen AW. Gene expression of tumor angiogenesis dissected: specific targeting of colon cancer angiogenic vasculature. Blood. 2006;108:2339–2348. doi: 10.1182/blood-2006-02-004291. [DOI] [PubMed] [Google Scholar]
- Harvell DM, Richer JK, Allred DC, Sartorius CA, Horwitz KB. Estradiol regulates different genes in human breast tumor xenografts compared with the identical cells in culture. Endocrinology. 2006;147:700–713. doi: 10.1210/en.2005-0617. [DOI] [PubMed] [Google Scholar]
- Banno T, Gazel A, Blumenberg M. Effects of tumor necrosis factor-alpha (TNF-alpha) in epidermal keratinocytes revealed using global transcriptional profiling. J Biol Chem. 2004;279:32633–32642. doi: 10.1074/jbc.M400642200. [DOI] [PubMed] [Google Scholar]
- Ruebel KH, Leontovich AA, Tanizaki Y, Jin L, Stilling GA, Zhang S, Coonse K, Scheithauer BW, Lombardero M, Kovacs K, Lloyd RV. Effects of TGFbeta1 on gene expression in the HP75 human pituitary tumor cell line identified by gene expression profiling. Endocrine. 2008;33:62–76. doi: 10.1007/s12020-008-9060-3. [DOI] [PubMed] [Google Scholar]
- Lowry WE, Blanpain C, Nowak JA, Guasch G, Lewis L, Fuchs E. Defining the impact of beta-catenin/Tcf transactivation on epithelial stem cells. Genes Dev. 2005;19:1596–1611. doi: 10.1101/gad.1324905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobielak K, Stokes N, de la Cruz J, Polak L, Fuchs E. Loss of a quiescent niche but not follicle stem cells in the absence of bone morphogenetic protein signaling. Proc Natl Acad Sci USA. 2007;104:10063–10068. doi: 10.1073/pnas.0703004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc Natl Acad Sci USA. 2004;101:9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina PP, Castillo SD, Blanco S, Sanz-Garcia M, Largo C, Alvarez S, Yokota J, Gonzalez-Neira A, Benitez J, Clevers HC, Cigudosa JC, Lazo PA, Sanchez-Cespedes M. The SRY-HMG box gene: SOX4, is a target of gene amplification at chromosome 6p in lung cancer. Hum Mol Genet. 2009;18:1343–1352. doi: 10.1093/hmg/ddp034. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Shen H, Akagi K, Morse HC, Malley JD, Naiman DQ, Jenkins NA, Copeland NG. New genes involved in cancer identified by retroviral tagging. Nat Genet. 2002;32:166–174. doi: 10.1038/ng949. [DOI] [PubMed] [Google Scholar]
- Lund AH, Turner G, Trubetskoy A, Verhoeven E, Wientjens E, Hulsman D, Russell R, DePinho RA, Lenz J, van Lohuizen M. Genome-wide retroviral insertional tagging of genes involved in cancer in Cdkn2a-deficient mice. Nat Genet. 2002;32:160–165. doi: 10.1038/ng956. [DOI] [PubMed] [Google Scholar]
- Shin MS, Fredrickson TN, Hartley JW, Suzuki T, Agaki K, Morse HC., 3rd High-throughput retroviral tagging for identification of genes involved in initiation and progression of mouse splenic marginal zone lymphomas. Cancer Res. 2004;64:4419–4427. doi: 10.1158/0008-5472.CAN-03-3885. [DOI] [PubMed] [Google Scholar]
- Pramoonjago P, Baras AS, Moskaluk CA. Knockdown of Sox4 expression by RNAi induces apoptosis in ACC3 cells. Oncogene. 2006;25:5626–5639. doi: 10.1038/sj.onc.1209566. [DOI] [PubMed] [Google Scholar]
- Ahn SG, Kim HS, Jeong SW, Kim BE, Rhim H, Shim JY, Kim JW, Lee JH, Kim IK. Sox-4 is a positive regulator of Hep3B and HepG2 cells’ apoptosis induced by prostaglandin (PG)A(2) and delta(12)-PGJ(2). Exp Mol Med. 2002;34:243–249. doi: 10.1038/emm.2002.34. [DOI] [PubMed] [Google Scholar]
- Pan X, Zhao J, Zhang WN, Li HY, Mu R, Zhou T, Zhang HY, Gong WL, Yu M, Man JH, Zhang PJ, Li AL, Zhang XM. Induction of SOX4 by DNA damage is critical for p53 stabilization and function. Proc Natl Acad Sci USA. 2009;106:3788–3793. doi: 10.1073/pnas.0810147106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J, MacLean KH, Han J, Chittenden T, Ihle JN, McKinnon PJ, Cleveland JL, Zambetti GP. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 2003;4:321–328. doi: 10.1016/s1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- Lei Q, Jiao J, Xin L, Chang CJ, Wang S, Gao J, Gleave ME, Witte ON, Liu X, Wu H. NKX3.1 stabilizes p53, inhibits AKT activation, and blocks prostate cancer initiation caused by PTEN loss. Cancer Cell. 2006;9:367–378. doi: 10.1016/j.ccr.2006.03.031. [DOI] [PubMed] [Google Scholar]
- Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- Chowdhury T, Brady HJ. Insights from clinical studies into the role of the MLL gene in infant and childhood leukemia. Blood Cells Mol Dis. 2008;40:192–199. doi: 10.1016/j.bcmd.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Abate-Shen C, Banach-Petrosky WA, Sun X, Economides KD, Desai N, Gregg JP, Borowsky AD, Cardiff RD, Shen MM. Nkx3.1; Pten mutant mice develop invasive prostate adenocarcinoma and lymph node metastases. Cancer Res. 2003;63:3886–3890. [PubMed] [Google Scholar]
- Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- Robb GB, Rana TM. RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol Cell. 2007;26:523–537. doi: 10.1016/j.molcel.2007.04.016. [DOI] [PubMed] [Google Scholar]
- Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- Ambs S, Prueitt RL, Yi M, Hudson RS, Howe TM, Petrocca F, Wallace TA, Liu CG, Volinia S, Calin GA, Yfantis HG, Stephens RM, Croce CM. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68:6162–6170. doi: 10.1158/0008-5472.CAN-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for gene set enrichment analysis. Bioinformatics. 2007;23:3251–3253. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]