Abstract
Pachydermoperiostosis (PDP) is a rare disease characterized by unique phenotypes of the skin and bone, such as thick skin, implying that it may be caused by dysregulation of mesenchymal cells. The aim of this study is to examine the roles of dermal fibroblasts in the pathogenesis of pachydermia in association with Wnt signaling. The numbers of cultured fibroblasts were compared between healthy donors and PDP patients, and mRNA expression profiles in cultured dermal fibroblasts were examined by DNA microarray analysis and real-time reverse transcription-PCR. DKK1 and β-catenin protein expressions were also evaluated by immunohistochemistry in the skin. To evaluate the in vivo roles of DKK1 in mice, DKK1 small interfering RNA was injected to the ears. We found that PDP fibroblasts proliferated more than control fibroblasts and that mRNA expression of a Wnt signaling antagonist, DKK1, was much lower in PDP fibroblasts than in normal ones. Consistently, decreased expression of DKK1 in fibroblasts and enhanced expression of β-catenin were noted in PDP patients. Moreover, recombinant human DKK1 protein decreased the proliferation of dermal fibroblasts. In accord with the above human studies, intradermal injections of DKK1 small interfering RNA into mouse ears increased ear thickness as seen in PDP. Our findings suggest that enhanced Wnt signaling contributes to the development of pachydermia by enhancing dermal fibroblast functions.
Pachydermoperiostosis (PDP), a form of primary hypertrophic osteoarthropathy, is a rare disease1,2,3 diagnosed by the presence of a triad of pachydermia (skin thickening), digital clubbing, and periostosis of long bones. Typically, insidious development of thickening of the fingers and toes, clubbing of the terminal phalanges, enlargement of the hands and feet, hyperhidrosis, increased sebaceous secretion, and velvet coloration of the skin occur mostly in men during adolescence.4 Radiographic signs of bilateral and symmetrical periostosis are frequently observed as a marked irregular periosteal ossification of the tibias and fibulas.3 Touraine et al5 recognized PDP with three clinical presentations or forms: a “complete form” presenting the full-blown phenotype; an “incomplete form” characterized by the phenotype without pachydermia; and a “fruste form” with pachydermia and minimal or absent skeletal changes.
Recently, the incomplete form of PDP, primary osteoarthropathy without pachydermia, was mapped to chromosome 4q33-q34, and gene mutations in HPGD, encoding 15-hydroxyprostaglandin dehydrogenase, the main enzyme of prostaglandin (PG) degradation, were identified.6 Therefore, it has been suggested that the digital clubbing and bone changes are due to elevated PGE2. However, the pathomechanism underlying pachydermia of PDP remains unknown.
Since the major manifestations of complete PDP occur in both skin and bone, the etiology could be related to the dysregulation of bone morphogenetic proteins (BMP), transforming growth factor (TGF)-β, and/or wingless (Wnt) pathways.7,8,9 The Wnt signaling consists of canonical and non-canonical pathways. The canonical pathway involves cytosolic β-catenin stabilization, nuclear translocation and gene regulation, and the non-canonical pathways activate rho, rac, JNK, and protein kinase C.10,11 These signaling pathways are mediated by Wnt protein, which binds to a frizzled Wnt receptor. Wnt signaling is modulated by several different families of secreted down-regulators. Among them, Dickkopf (DKK) is a family of cysteine-rich proteins comprising at least four different forms (DKK1, DKK2, DKK3, and DKK4), which are coordinately expressed in mesodermal lineages. The best studied of these is DKK1, which blocks the canonical Wnt signaling by inducing endocytosis of lipoprotein receptor-related protein 5/6 (LRP5/6) complex12 without affecting the frizzled Wnt receptor.13 DKK1 induces the formation of ectopic heads in Xenopus laevis in the presence of BMP inhibitors14 and modulates apoptosis during vertebrate limb development.15 High mRNA levels of DKK1 in human dermal fibroblasts of the palms and soles inhibit the function and proliferation of melanocytes via the suppression of β-catenin and microphthalmia-associated transcription factor.16,17 In parallel, DKK1 transgenic mice under the control of keratin 14 have no pigmentation on the trunk because of the absence of melanocytes in the inner-follicular epidermis, as well as the lack of hair follicle development.18 These findings suggest that DKK1 is deeply involved in the formation and differentiation of the skin.
Here we investigated two complete cases of PDP using dermal fibroblasts to address the pathogenetic mechanisms. DNA microarray analysis revealed that the proliferation of primary fibroblasts of PDP was increased with decreased expression of DKK1 mRNA in cultured fibroblasts. Consistent with this finding, immunohistochemistry indicated decreased expression of DKK1 in fibroblasts and enhanced expression of β-catenin in the skin of patients with PDP, suggesting that Wnt signaling is enhanced in PDP. The intradermal injection of DKK1 synthetic small interfering RNA (siRNA) increased the ear thickness of mice as seen in PDP. These results suggest that enhanced Wnt signaling contributes to the development of pachydermia.
Materials and Methods
Patients
Case 1
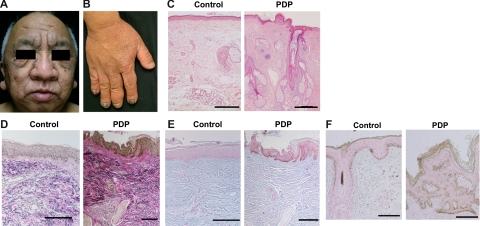
A 50-year-old male was referred to our clinic. The skin on his head and face was thick and oily with a dark velvet color. Naso-labial folds and transverse furrowing of the forehead were prominent (Figure 1A). The hands were enlarged with marked clubbing of the second and fifth digits, as compared with those of an age- and sex-matched healthy donor (Figure 1B). These symptoms developed when he was 18 years old. X-ray examination of the long bones showed major periostosis with cortical thickening and widening of the shafts (data not shown). Histology of the skin showed thickened dermis, and sebaceous and sweat gland enlargement, as compared with that of a healthy control (Figure 1C). Elastica van Gieson staining showed thick and interwoven collagen bundles in some areas of the dermis and also thick and partially fragmented elastic fibers in PDP (Figure 1D). The intensity of mucinous ground substance observed by Alcian blue staining was comparable between a healthy control and a PDP patient (Figure 1E). On the other hand, Fontana Masson staining revealed that the number of melanocytes and the intensity of the staining in the patient with PDP was higher than that in a healthy control (Figure 1F). Neither hepatosplenomegaly nor internal malignancy was found on physical examination or computed tomography scans. Biochemical tests showed normal levels of thyroid-stimulating hormone and growth hormone, which likely rules out thyroid acropathy and acromegaly. Family history was noncontributory. Based on these clinical manifestations and histological findings, the patient was diagnosed as the complete form of PDP.
Figure 1.
Clinical findings and histology of a PDP patient. A, B: Clinical appearances of the face (A) and hands (B) in PDP (case 1) are shown. C: Histology of the face in PDP shows thickened dermis, and hyperplasia of the sebaceous and sweat glands by H&E staining compared with a healthy donor (control). Scale bars = 300 μm. D: The sample of PDP stained with Elastica van Gieson shows thick collagen and elastic fibers in the dermis compared with a healthy donor. E: The intensity of mucinous ground substance observed by Alcian blue staining is comparable between a healthy donor and a PDP patient. F: Melanocytes in the patient with PDP and a healthy control are identified with Fontana Masson staining. Scale bars = 100 μm (D−F).
Case 2
The patient was a 38-year-old male with clinical findings similar to case 1, including pachydermia, digital clubbing, and periostosis. He had no signs or symptoms of hepatosplenomegaly, pulmonary diseases, tumoral syndrome, thyroid acropathy, or acromegaly (data not shown) as reported previously.19
Cell Preparation, Culture, and Reagents
Skin biopsies of the right temple (case 1) and scalp (case 2) were performed for histology and primary culture of fibroblasts. Control donors were matched for age, sex, and biopsy site, and the samples were processed in parallel. Institutional approval and informed consent were obtained from all subjects. The biopsy samples were immersed in Dulbecco’s Modified Eagle Medium (Sigma, St. Louis, MO) containing 10% heat-inactivated fetal calf serum (Invitrogen, Carlsbad, CA), 5 × 10−5 mol/L 2-mercaptoethanol, 2 mmol/L l-glutamine, 25 mmol/L HEPES (Cellgro, Herndon, VA), 1 mmol/L nonessential amino acids, 1 mmol/L sodium pyruvate, 100 units/ml penicillin, and 100 μg/ml streptomycin, with 5% CO2 at 37°C. The fibroblasts were allowed to adhere to the surface of 100-mm plastic tissue culture dishes (Nunc, Roskilde, Denmark). To evaluate the number of fibroblasts, 2 × 105 third-passage fibroblasts were seeded in 1 ml of medium in 24-well dishes and resuspended with trypsin/EDTA 1 week later. The numbers of fibroblasts were evaluated 7 and 14 days after seeding by flow cytometry using FACSCanto (BD Biosciences, San Diego, CA) with standard beads Flow Count (Beckman Coulter, Fullerton, CA) as per the manufacturer’s instructions. The actin bundle formation of cultured fibroblasts from a healthy individual and an individual with PDP were examined by staining with alexa 488-labeled phalloidin antibody (Invitrogen) 5 days after the fourth passage.
For treatment with DKK1, fibroblasts were harvested 5 days after a comparable number of passages and cultured again at 1 × 106 cells in one ml of medium with or without recombinant human DKK1 (R&D Systems Inc., Minneapolis, MN) for another 2 days. For treatment with PGE2, fibroblasts were harvested 5 days after a comparable number of passages and cultured again at 1 × 105 cells in two ml of medium with or without PGE2 (Sigma) in the presence of indomethacin (10 μmol/L; Cayman Chemical Co., Ann Arbor, MI) for another 4 days.
Flow Cytometry and Histology
Flow cytometric analysis was performed with doublet discrimination on the FACSCanto20 and FlowJo software (TreeStar, San Carlos, CA).21 Human fibroblasts were treated with cytofix/cytoperm buffer according to the manufacturer’s protocol (BD Biosciences). For cell cycle analysis, fibroblasts were incubated with 7-amino actinomycin D (7-AAD) (BD Biosciences) for 20 minutes at 4°C. After staining with 7-AAD, the DNA contents were analyzed by flow cytometry. For β-catenin staining, fibroblasts were stained with phycoerythrin-labeled β-catenin antibody (H-102, Santa Cruz Biotechnology Inc., Santa Cruz, CA), and mean fluorescence intensity was evaluated by flow cytometry.
For histology, the biopsy samples and the ears of mice were fixed in 10% formaldehyde. Sections of 5-μm thickness were prepared and stained with H&E, Elastica van Gieson, or Alcian blue. Immunohistochemical staining on paraffin-embedded sections was performed using a Vectastain ABC kit (Vector Laboratories, Burlingame, CA).20 Antibodies used were rabbit anti-human polyclonal DKK1 (ab61034, Abcam, Cambridge, UK), mouse monoclonal anti-human β-catenin IgG1 (610153, BD Biosciences, San Diego, CA), and rabbit anti-human polyclonal proliferating cellular nuclear antigen antibodies (SC-7907, Santa Cruz Biotechnology Inc., Santa Cruz, CA). The control antibodies used were rabbit non-immune serum or mouse IgG1 (X0931, Dako, Glostrup, Denmark). The immunoreactivity was visualized by Fast Red or diaminobenzidine (Sigma), and the sections were then counterstained with hematoxylin. Images were acquired on a 600CL-CU cooled charge-coupled device video camera (Pixera, Los Gatos, CA) and processed with InStudio 1.0.0 (Pixera).
Western Blot Analysis
For Western blotting studies, fibroblasts were isolated from a healthy donor. Cytoplasm- and nuclear- proteins were extracted by NucBuster Protein Extraction Kit (Novagen, Darmstadt, Germany). Twenty μg protein samples were electrophoresed by 8% SDS-polyacrylamide gel electrophoresis and electroblotted onto polyvinylidene difluoride membranes for 2 hours at 180 mA. After blocking with 5% skim milk solution, the membranes were incubated with rabbit anti-human β-catenin (SC-7199; 1:1000, Santa Cruz Biotechnology Inc.) polyclonal antibodies or rabbit anti-human glyceraldehyde-3-phosphate dehydrogenase (SC-25778; 1:1000, Santa Cruz Biotechnology Inc.) antibody and detected with horseradish peroxidase-conjugated goat anti-rabbit IgG (Bio-Rad, Hercules, CA). Immunoblots were visualized using the ECL Plus Western Blotting Detection Reagents (GE Health care, Buckinghamshire, UK) according to the manufacturer’s protocol. Bands were quantified by densitometry with the help of a CS Analyzer ver. 2.0 (ATTO, Tokyo, Japan).
Quantitative Reverse Transcription-PCR and Microarray Procedures
Total RNA was extracted from three-passage fibroblasts (case 1 and the control) cultured for 2 days with the RNeasy Mini Kit (QIAGEN, Valencia, CA). cDNA was reverse transcribed from total RNA samples using the TaqMan Reverse Transcription (RT) reagents (Applied Biosystems, Foster City, CA). Human DKK1 (Assay ID: Hs00183740) mRNA expression was quantified using TaqMan Gene Expression Assay (Applied Biosystems) with the ABI PRISM 7700 sequence detection system (Applied Biosystems). As an endogenous reference for these RT-PCR quantification studies, human GAPDH control reagents (Assay ID: Hs99999905) (Applied Biosystems) were used. The relative expression was calculated using the ΔΔ Ct method.22
For DNA microarray analysis, total RNAs were extracted from fibroblasts with the RNeasy Mini Kit (QIAGEN). For transcriptomic profiling, we used an oligonucleotide-based DNA microarray, AceGene (HumanOligoChip30K, DNA Chip Research, Yokohama, Japan). Images were analyzed with DNASIS Array (Hitachi Software Engineering, Tokyo, Japan), according to the manufacturer’s instructions. Mean and SD of background levels were calculated, and genes with intensities less than mean plus 2SD of background levels were excluded from further analysis. The Cy5/Cy3 ratios of all spots on the DNA microarray were normalized by the method of global normalization.
Genetic Analysis for DKK1, TCF, and HPGD
Three healthy controls and two PDP patients (cases 1 and 2) were enrolled and followed up according to local ethical guidelines. Genomic DNA was isolated from primary fibroblasts or peripheral blood leukocytes using proteinase K and the PCI (phenol/chloroform/isoamyl alcohol) extraction procedure. The DKK1 (GenBank: NM012242), TCF7L2 (TCF-4) (GenBank: NM030756), and HPGD (NM000860) genes were amplified via PCR in a thermal cycler (Eppendorf, Hamburg, Germany) using forward and reverse primer pairs (Table 1).
Table 1.
PCR and Sequencing Primers
| PCR Primer | Sequence | Tm | Binding site |
|---|---|---|---|
| hDKK1-Exon1, 2 | |||
| Forward | 5′-CGTCTGCTATAACGCTCGCTGGTAG-3′ | 77 | Promoter |
| Reverse | 5′-AATTCATAGACGCTCAAAGGCTGGA-3′ | 73 | Intron2 |
| hDKK1-Exon3, 4 | |||
| Forward | 5′-ACTTGCCCCTACCACAGTTG-3′ | 70 | Intron2 |
| Reverse | 5′-GTTCCTGCCAATCACCAAGT-3′ | 68 | 3′UTR |
| hTCF-4-Exon1 | |||
| Forward | 5′-TGGCTTTTCTTCCTCCTTCA-3′ | 66 | 5′UTR |
| Reverse | 5′-AGAAAAAGAATCGGCGAGGT-3′ | 66 | Intron1 |
| hTCF-4-6 | |||
| Forward | 5′-GCGATTTCTGGCAGGTAGTC-3′ | 70 | Intron7 |
| Reverse | 5′-TAGCGATCCAGGAAGATGCT-3′ | 68 | Intron10 |
| hTCF-4-9 | |||
| Forward | 5′-TTAGTAGGGGTTGGGGGAAG-3′ | 70 | Intron13 |
| Reverse | 5′-TTGGTAGAATCATGAGGTTCTTCTC-3′ | 71 | 3′UTR |
| hHPGD Exon1 | |||
| Forward | 5′-GCTGGCTTGACAGTTTCCTC-3′ | 70 | 5′UTR |
| Reverse | 5′-CAGCCTCAGCTTCAGCAAAT-3′ | 68 | Intron1 |
| hHPGD Exon2 | |||
| Forward | 5′-TTGCTGAAGCTGAGGCTGT-3′ | 68 | Intron1 |
| Reverse | 5′-TCTTGCCTTTCTTTCGGTTT-3′ | 64 | Intron2 |
| hHPGD Exon3 | |||
| Forward | 5′-TCCACAAACCACACATTGAGA-3′ | 67 | Intron2 |
| Reverse | 5′-CCAGCTTTCTGTAACTTCCCTTT-3′ | 70 | Intron3 |
| hHPGD Exon4 | |||
| Forward | 5′-TAGGCAAACCCAAAGAATCC-3′ | 66 | Intron3 |
| Reverse | 5′-CACATGGGAGCAGAGACATC-3′ | 70 | Intron4 |
| hHPGD Exon5 | |||
| Forward | 5′-CCTGGGGAGGCAGAAAAA-3′ | 67 | Intron4 |
| Reverse | 5′-TTTATTTGGTTCTTTATGTGATCTGA-3′ | 67 | Intron5 |
| hHPGD Exon6 | |||
| Forward | 5′-TGCAGAGTTCAGTAGATAAGAGAAGC-3′ | 73 | Intron5 |
| Reverse | 5′-TGCTTGGAATTTAGGCAGAGA-3′ | 67 | Intron6 |
| hHPGD Exon7 | |||
| Forward | 5′-TTGGAAGTAGCAATAGTTTAATGA-3′ | 68 | Intron6 |
| Reverse | 5′-TCACCAAGTGCATGAAGGAA-3′ | 66 | 3′UTR |
| Sequencing Primer | Sequence | Binding site |
|---|---|---|
| hDKK1-Exon1, 2 | ||
| Forward | 5′-CGTCTGCTATAACGCTCGCTGGTAG-3′ | Promoter |
| Reverse | 5′-AATTCATAGACGCTCAAAGGCTGGA-3′ | Intron2 |
| hDKK1-Exon1-S2 | ||
| Forward | 5′-CCACCTTGAACTCGGTTCTC-3′ | Exon1 |
| hDKK1-Exon2-S1 | ||
| Forward | 5′-AGAACGTGCTGAATGTGTGC-3′ | Intron1 |
| hDKK1-Exon3, 4 | ||
| Forward | 5′-ACTTGCCCCTACCACAGTTG-3′ | Intron2 |
| Reverse | 5′-GTTCCTGCCAATCACCAAGT-3′ | 3′UTR |
| hDKK1-Exon3-S1 | ||
| Forward | 5′-CCTTGGATGGGTATTCCAGA-3′ | Exon3 |
| hDKK1-Exon4-S1 | ||
| Forward | 5′-TCATCAGACTGTGCCTCAGG-3′ | Exon4 |
| hDKK1-Exon4-S2 | ||
| Forward | 5′-AAGGTGCTGCACTGCCTATT-3′ | 3′UTR |
| hTCF-4-Exon1 | ||
| Forward | 5′-TGGCTTTTCTTCCTCCTTCA-3′ | 5′UTR |
| Reverse | 5′-AGAAAAAGAATCGGCGAGGT-3′ | Intron1 |
| hTCF-4-Exon9 | ||
| Forward | 5′-GCTTGGGGGTTATGAGACAA-3′ | Intron8 |
| Reverse | 5′-AGACATTCTGCCACCTGACC-3′ | Intron9 |
| hTCF-4-Exon10 | ||
| Forward | 5′-CCTTGGCGTAATGTGTGATG-3′ | Intron9 |
| Reverse | 5′-TAGCGATCCAGGAAGATGCT-3′ | Intron10 |
| hTCF-4-Exon14 | ||
| Forward | 5′-ACATCCCTTAGGTGACCTCA-3′ | Intron13 |
| Reverse | 5′-GGGGGCAAATTAAGAAAAGTG-3′ | 3′UTR |
| hHPGD Exon1 | ||
| Forward | 5′-GCTGGCTTGACAGTTTCCTC-3′ | 5′UTR |
| Reverse | 5′-CAGCCTCAGCTTCAGCAAAT-3′ | Intron1 |
| hHPGD Exon2 | ||
| Forward | 5′-TTGCTGAAGCTGAGGCTGT-3′ | Intron1 |
| Reverse | 5′-TCTTGCCTTTCTTTCGGTTT-3′ | Intron2 |
| hHPGD Exon3 | ||
| Forward | 5′-TCCACAAACCACACATTGAGA-3′ | Intron2 |
| Reverse | 5′-CCAGCTTTCTGTAACTTCCCTTT-3′ | Intron3 |
| hHPGD Exon4 | ||
| Forward | 5′-TAGGCAAACCCAAAGAATCC-3′ | Intron3 |
| Reverse | 5′-CACATGGGAGCAGAGACATC-3′ | intron4 |
| hHPGD Exon5 | ||
| Forward | 5′-CCTGGGGAGGCAGAAAAA-3′ | Intron4 |
| Reverse | 5′-TTTATTTGGTTCTTTATGTGATCTGA-3′ | Intron5 |
| hHPGD Exon6 | ||
| Forward | 5′-TGCAGAGTTCAGTAGATAAGAGAAGC-3′ | Intron5 |
| Reverse | 5′-TGCTTGGAATTTAGGCAGAGA-3′ | Intron6 |
| hHPGD Exon7 | ||
| Forward | 5′-TTGGAAGTAGCAATAGTTTAATGA-3′ | Intron6 |
| Reverse | 5′-TCACCAAGTGCATGAAGGAA-3′ | 3′UTR |
The exons of DKK1, TCF7L2 (TCF-4), and HPGD genes were amplified via PCR in a thermal cycler using the forward and reverse primer pairs indicated in the upper list. Direct sequencing was performed with the BigDye Terminator v3.1 Cycle Sequencing Kit and sequencing primers indicated in the lower list. Binding sites of primers are also indicated.
Amplified products were purified with the QIAquick Gel Extraction Kit (QIAGEN, Valencia, CA) or Wizard SV Gel and PCR Clean-Up System (Promega, Madison, WI) after 1.5% agarose electrophoresis. Direct sequencing was performed with the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) and sequencing primers (Table 1) using capillary electrophoresis (ABI Prism 3130xl Genetic Analyzer; Applied Biosystems), and analyzed with ABI Prism DNA Sequencing Analysis software ver. 5.1 (Applied Biosystems) as previously described.23
Application of Mouse DKK1 siRNA
Mouse DKK1 siRNA (5′-GAA CCA CAC UGA CUU CAA ATT-3′) was purchased from Nippon EGT (Toyama, Japan). siRNA duplexes were generated by mixing sense and antisense single-stranded RNA oligomers equally in an annealing buffer (NIPPON EGT).24 Negative control siRNA (AM4611) was purchased from Ambion (Austin, TX). To impregnate mouse DKK1 siRNA into cationized gelatin microspheres,25 10 μl of PBS solution (pH 7.4) containing 10 μg of mouse DKK1 siRNA was dropped onto 1 mg of the freeze-dried cationized gelatin microspheres, kept overnight at 4°C, and added to 190 μl of PBS. Ten μl of this siRNA solution was injected intradermally into the center of the ears of 8-week-old C57BL/6j female mice (obtained from SLC, Shizuoka, Japan) using a 30-gauge needle four times every 7 days. The same amount of cationized gelatin-conjugated nonsense siRNA was applied as a negative control. The ear thickness was measured before each injection and one week after the last injection using dial-thickness gauge (PG-01, TECLOCK, Okaya, Japan). The injected area was sampled for histology and RT-PCR analysis using 6-mm punch biopsy. Mice were maintained on a 12-hour light/dark cycle under specific pathogen-free conditions. Protocols were approved by the Institutional Animal Care and Use Committee of the University of Occupational and Environmental Health.
Statistical Analysis
Data were analyzed using an unpaired two-tailed t-test. A P value of less than 0.05 was considered to be significant.
Results
Increased S-G2 Phase in Fibroblasts of PDP
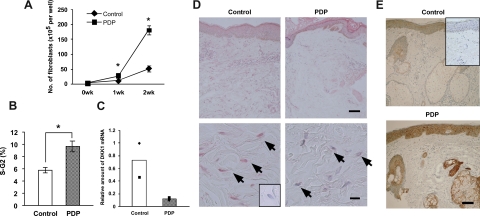
Case 1 had a typical complete form of PDP (Figure 1, A and B) characterized by the triad of pachydermia, digital clubbing, and periostosis.1,2,3 The histology of the skin showed thickened dermis with dense and packed collagen and elastic fibers (Figure 1, C−E), suggesting that the function of fibroblasts was enhanced in PDP. To test the proliferative activity of fibroblasts, we cultured primary fibroblasts from case 1 and a matched control, and monitored their number. As reported previously,26 the number of PDP fibroblasts was significantly higher than that of control fibroblasts (Figure 2A). Similar results were obtained in another typical patient with PDP, case 2 (data not shown). To clarify whether it was due to enhanced cell survival or proliferation, we stained the nuclear contents of fibroblasts with 7-AAD for cell cycle analysis. The ratio of PDP fibroblasts in the cell cycle (S-G2 phase) was higher than that of control fibroblasts (Figure 2B), suggesting that the proliferation of fibroblasts was enhanced in PDP.
Figure 2.
Characteristics of dermal fibroblasts and histology of the skin in PDP. A: Fibroblasts from a healthy individual (control) and an individual with PDP (case 1) (PDP) were incubated and the numbers of fibroblasts examined. B: The percentages of fibroblasts in S-G2 phase are shown. C: The levels of DKK1 mRNA in fibroblasts from two controls and two PDPs were normalized against GAPDH, and the level of one of the control DKK1 mRNAs is regarded as one. Filled symbols indicate two independent individuals and columns represent the average. D and E: Skin sections were stained with anti-human DKK1 (D) and β-catenin (E) antibodies. Arrows show the perinuclear area of fibroblasts (D). Scale bars: upper panels (150 μm), and lower panels, 10 μm (D), and 100 μm (E). We include that the controls incorporating non-immune serum (D) or mouse IgG1 (E) as insets show no specific reactivity. The student’s t-test was performed (*P < 0.05) (A, B).
Decreased DKK1 Expression in PDP Fibroblasts and Skin
The above results together with the clinical phenotypes involving the skin and bone suggested the possibility that the pathogenesis of PDP is related to dysregulation of BMP, Wnt, and/or TGF-β pathways in mesenchymal cells. To efficiently compare the expression profiles of these genes between PDP fibroblasts (case 1) and matched controls, DNA microarray analysis was performed and the complete array data were deposited in a MIAME-compliant microarray database (GSE17947). Among all genes analyzed, 2573 genes were elevated and 2346 genes were decreased more than twofold in PDP patients compared with a healthy control. The analysis revealed that the mRNA levels of BMP and TGF-β families were comparable between these two groups (Table 2). On the other hand, WNT1, WNT10B, and DKK1 mRNAs were decreased in the patient’s fibroblasts (Table 2). In particular, DKK1 mRNA was markedly decreased. Other molecules, such as levels of LRP5/6, Kremen1, and Kremen2 mRNA were similar between these two groups (Table 2). Moreover, the mRNA levels for collagen families, such as COL4A5, COL9A2, COL10A1, COL11A1, COL12A1, COL14A1, and COL17A1, were elevated, but those for fibronectin and elastin (ELN) families were not (Table 2). These data suggest that the PDP fibroblasts showed enhanced production of several types of collagens in addition to cell proliferation, which might explain the pathogenesis of pachydermia in PDP.
Table 2.
DNA Microarray Analysis
| Gene names | Accession ID | Expression levels
|
Difference | Fold difference | |
|---|---|---|---|---|---|
| Control | PDP | ||||
| BMP2 | NM_001200_1 | 5.72 | 5.87 | 0.16 | − |
| BMP3 | M22491_1 | ND | ND | NA | − |
| BMP4 | NM_001202_1 | 7.20 | 7.46 | 0.26 | − |
| BMP5 | NM_021073_1 | 4.61 | 4.30 | −0.30 | − |
| BMP6 | NM_001718_1 | 4.34 | 4.96 | 0.61 | − |
| BMP7 | NM_001719_1 | ND | ND | NA | − |
| BMP8B | NM_001720_1 | 5.47 | 5.65 | 0.18 | − |
| BMP10 | NM_014482_1 | ND | ND | NA | − |
| BMP15 | NM_005448_1 | ND | ND | NA | − |
| TGFB1 | NM_000660_1 | 8.87 | 7.91 | −0.97 | − |
| TGFB2 | NM_003238_1 | 4.34 | 5.05 | 0.72 | − |
| TGFBR1 | NM_004612_1 | ND | ND | NA | − |
| TGFBR2 | NM_003242_1 | ND | ND | NA | − |
| TGFBR3 | NM_003243_1 | 4.34 | 4.51 | 0.17 | − |
| WNT1 | NM_005430_1 | 6.66 | 5.45 | −1.21 | 0.4 |
| WNT2 | ENSG00000105989 | ND | ND | NA | − |
| WNT2B | NM_024494_1 | 6.36 | 6.95 | 0.60 | − |
| WNT4 | AY009398_1 | 4.33 | 4.35 | 0.03 | − |
| WNT5A | NM_003392_1 | 5.36 | 5.85 | 0.50 | − |
| WNT6 | BC004329_1 | 5.13 | 4.99 | −0.14 | − |
| WNT7A | NM_004625_1 | ND | ND | NA | − |
| WNT8B | NM_003393_1 | 7.27 | 6.88 | −0.39 | − |
| WNT9A | AB060283_1 | 7.36 | 6.92 | −0.44 | − |
| WNT9B | AF028703_1 | 5.69 | 4.47 | −1.22 | 0.4 |
| WNT10A | NM_025216_1 | 4.40 | 5.40 | 1.00 | − |
| WNT10B | NM_003394_1 | 5.73 | 4.48 | −1.25 | 0.4 |
| WNT11 | NM_004626_1 | 6.46 | 5.85 | −0.61 | − |
| WNT16 | NM_016087_1 | 7.63 | 6.80 | −0.83 | − |
| FZD1 | NM_003505_1 | 7.19 | 6.91 | −0.28 | − |
| FZD2 | AB017364_1 | 7.05 | 7.01 | −0.03 | − |
| FZD3 | AJ272427_1 | 7.51 | 6.85 | −0.67 | − |
| FZD3 | NM_017412_1 | ND | ND | NA | − |
| FZD4 | NM_012193_1 | 6.09 | 6.49 | 0.41 | − |
| FZD5 | NM_003468_1 | 5.02 | 5.44 | 0.42 | − |
| FZD6 | NM_003506_1 | 5.95 | 6.37 | 0.42 | − |
| FZD7 | NM_003507_1 | 9.32 | 9.40 | 0.08 | − |
| FZD8 | AB043703_1 | ND | ND | NA | − |
| DKK1 | NM_012242_1 | 11.61 | 8.46 | −3.15 | 0.1 |
| DKK2 | NM_014421_1 | 6.53 | 6.93 | 0.40 | − |
| DKK3 | NM_015881_1 | 9.24 | 9.15 | −0.08 | − |
| KREMEN1 | AB059618_1 | ND | ND | NA | − |
| KREMEN2 | NM_024507_1 | 5.20 | 4.33 | NA | − |
| COL1A1 | K03179_1 | 7.57 | 7.90 | 0.34 | − |
| COL1A2 | NM_000089_1 | 13.54 | 13.98 | 0.44 | − |
| COL2A1 | NM_033150_1 | 9.46 | 10.09 | 0.63 | − |
| COL3A1 | NM_000090_1 | 10.26 | 11.43 | 1.17 | 2.3 |
| COL4A1 | NM_001845_1 | 8.66 | 7.83 | −0.83 | − |
| COL4A2 | X05562_1 | 7.17 | 6.83 | −0.34 | − |
| COL4A3 | U02519_1 | 4.54 | 5.04 | 0.49 | − |
| COL4A4 | NM_000092_1 | 4.73 | 4.30 | −0.43 | − |
| COL4A5 | NM_000495_1 | 4.23 | 5.72 | 1.50 | 2.8 |
| COL4A6 | D63562_1 | 8.41 | 8.68 | 0.28 | − |
| COL5A1 | BC008760_1 | 10.35 | 10.25 | −0.09 | − |
| COL5A3 | NM_015719_1 | 5.57 | 6.20 | 0.62 | − |
| COL6A2 | AY029208_1 | 10.90 | 10.62 | −0.29 | − |
| COL6A3 | NM_004369_1 | 6.64 | 5.37 | −1.27 | 0.4 |
| COL8A1 | NM_001850_1 | 10.47 | 11.09 | 0.61 | − |
| COL8A2 | M60832_1 | 5.43 | 5.55 | 0.11 | − |
| COL9A1 | NM_001851_1 | 6.97 | 6.84 | −0.13 | − |
| COL9A2 | NM_001852_1 | 4.15 | 6.13 | 1.98 | 4 |
| COL9A3 | NM_001853_1 | 4.73 | 5.17 | 0.44 | − |
| COL10A1 | NM_000493_1 | 4.10 | 6.90 | 2.80 | 7 |
| COL11A1 | NM_001854_1 | 6.45 | 9.28 | 2.84 | 7 |
| COL11A2 | J04974_1 | ND | ND | NA | − |
| COL12A1 | NM_004370_1 | 4.98 | 6.26 | 1.27 | 2.5 |
| COL14A1 | Y11711_1 | 4.21 | 5.81 | 1.60 | 3 |
| COL15A1 | NM_001855_1 | ND | ND | NA | − |
| COL17A1 | NM_000494_1 | 4.32 | 6.33 | 2.01 | 4 |
| COL18A1 | NM_030582_1 | 5.73 | 6.03 | 0.30 | − |
| COL19A1 | NM_001858_1 | ND | ND | NA | − |
| FN1 | X07717_1 | 7.10 | 6.69 | −0.41 | − |
| FN5 | NM_020179_1 | 6.41 | 6.71 | 0.30 | − |
| ELN | NM_000501_1 | 7.60 | 7.33 | −0.26 | − |
The upper list of genes related to BMP, TGF-β, and Wnt signaling. The lower list of genes is related to collagens, fibronectins, and elastin. The mRNA expression levels of a healthy donor (control) and the individual with PDP (PDP) are normalized by LOWESS normalization, and indicated by log2. The values in Difference indicate mRNA expression levels of the individual with PDP—those of the healthy individual. The values under ‘Fold Difference’ indicate mRNA expression levels of the individual with PDP/those of the healthy individual, ie, Log2(Difference). The symbol “−” in the Fold Difference indicates non-significant difference between the healthy donor and the individual with PDP. ND, not determined. NA, not applicable.
We initially confirmed the decreased DKK1 expression using quantitative RT-PCR. Fibroblasts were primarily cultured from two PDP patients (cases 1 and 2) and two matched healthy controls. DKK1 mRNA levels in PDP fibroblasts were consistently lower than those in the control fibroblasts (Figure 2C). We then performed immunohistochemical analysis to evaluate the expression of DKK1 protein in the PDP skin (case 1) and the control. In the normal skin, DKK1 was detected diffusely in the dermis (Figure 2D, upper panels) and notably in the cytoplasm of fibroblasts (Figure 2D, lower panels). The intensity of this expression pattern was substantially decreased in the PDP patient (case 1) (Figure 2D, lower panels). This finding was confirmed with the other PDP patient (case 2) and another matched control (data not shown). We displayed that the controls incorporating non-immune serum (inset, Figure 2D) or mouse IgG1 (inset, Figure 2E) show no specific reactivity.
The decreased expression of DKK1 in PDP suggested that Wnt signaling is enhanced in PDP. Immunohistochemical analysis revealed enhanced β-catenin expression in the PDP skin (case 1), especially around the sebaceous glands, the hair follicles, and the epidermis, and mildly in the dermis, as compared with the control (Figure 2E), supporting the augmented expression of Wnt signaling.
Suppression of Fibroblast Proliferation by DKK1
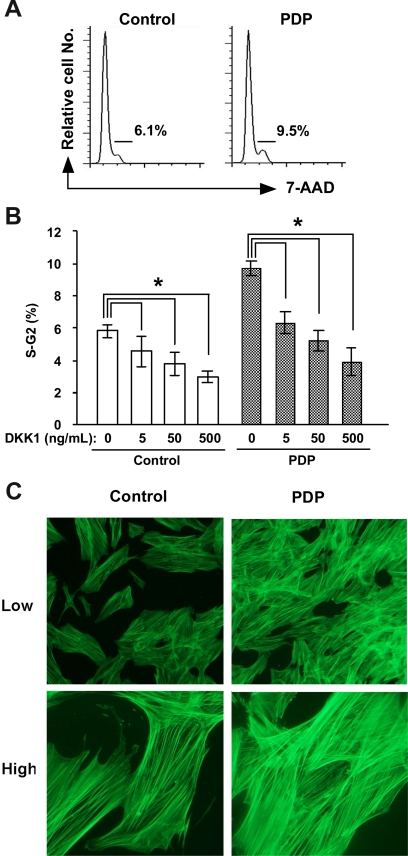
The above results indicated that Wnt signaling is enhanced in PDP through decreased DKK1 expression. However, it was still unknown whether DKK1 directly modulates the function of dermal fibroblasts. To solve this issue, we cultured dermal fibroblasts from a healthy control and the patient with PDP (case 1) in the presence or absence of human recombinant DKK1, and quantitated the DNA contents of fibroblasts by cell cycle analysis with 7-AAD. The ratio of fibroblasts in the cell cycle (S-G2 phase) was higher in the PDP patient than in the control (Figure 3, A and B). In addition, the ratio of fibroblasts with the cell cycle (S-G2 phase) was decreased by treatment with recombinant DKK1 protein in a dose-dependent manner (Figure 3B), implicating the direct involvement of DKK1 in fibroblast proliferation.
Figure 3.
The effect of DKK1 on fibroblast proliferation and actin bundle formation of fibroblasts. A, B: The fibroblasts from a healthy individual (control) and an individual with PDP (PDP) were incubated with or without recombinant human DKK1 protein and the DNA contents of fibroblasts were evaluated with 7-AAD using flow cytometry. Representatives of FACS plots of fibroblasts from a healthy individual (control) and an individual with PDP (PDP) are shown (A). The percentages of fibroblasts in S-G2 phase in triplicated wells are expressed as the mean ± SD (n = 3). The student’s t-test was performed between the indicated groups and *P < 0.05. C: The actin bundle formation of cultured fibroblasts from a healthy individual (control) and an individual with PDP (PDP) were examined by staining with alexa 488-labeled phalloidin antibody 5 days after the fourth passage. Upper panels, low magnification (×10); lower panels, high magnification (×40).
Enhanced Actin Bundle Formation of Fibroblasts in PDP
Wnt signaling is also known to induce cell motility and cytoskeletal rearrangement of NIH3T3, a fibroblast cell line.27 Therefore, we examined the actin bundle formation of cultured fibroblasts with phalloidin staining 5 days after the fourth passage. Fluorescent microscopy showed that the actin bundle formation of PDP fibroblasts is promoted in PDP, as the bundles were thicker and denser than those of control fibroblasts (Figure 3C).
Effect of PGE2 on Fibroblasts
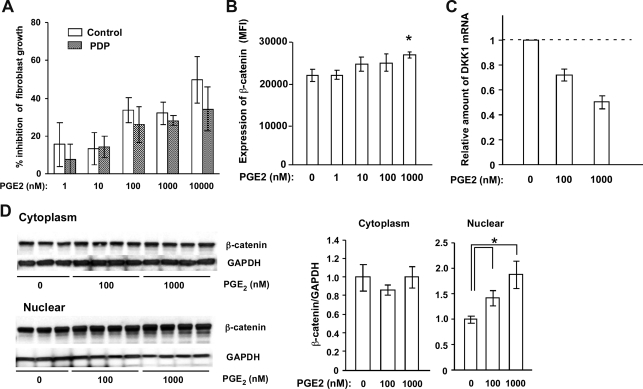
It was recently reported that the incomplete form of PDP is induced by elevated PGE2 due to a mutation in the HPGD gene.6 If this PGE2 alteration also affects pachydermia, PGE2 would be expected to enhance fibroblast function and proliferation. The addition of PGE2 into the cultured medium of fibroblasts decreased the number of dermal fibroblasts from healthy donors in a dose-dependent manner as reported previously28,29 (Figure 4A). A similar effect was observed when PGE2 was added to the culture medium of fibroblasts from the PDP patient (case 2). To examine whether PGE2 affects Wnt signaling in fibroblasts, we measured the amount of β-catenin in fibroblasts after exposure to PGE2 by flow cytometry, and found that β-catenin was significantly increased in fibroblasts by the addition of PGE2 at a dose of 1000 nmol/L (Figure 4B). In addition, the mRNA expression level of DKK1 was significantly decreased by the addition of PGE2 at a dose of 100 and 1000 nmol/L (Figure 4C). Moreover, to determine the effect of PGE2 on β-catenin expression, cytoplasm- and nuclear- protein samples were prepared from fibroblasts treated with or without 0, 100, and 1000 nmol/L PGE2 in the presence of 10 μmol/L indomethacin for 4 days. In the cytoplasm, β-catenin expression was unchanged irrespective of the addition of PGE2. However, β-catenin expression in the nuclei was significantly increased by the treatment with 100 and 1000 nmol/L PGE2 (Figure 4D). These results suggest that PGE2 signaling increases nuclear β-catenin in fibroblasts.
Figure 4.
Effect of PGE2 on fibroblasts. A: The % inhibition of the number of fibroblasts from a healthy donor and a PDP patient by the addition of PGE2 was evaluated as (Number of fibroblasts without PGE2 − Number of fibroblasts with PGE2)/Number of fibroblasts without PGE2 × 100. The growth inhibitory effect of PGE2 is dose-dependent and comparable between these two groups. The values are expressed as the mean ± SD (n = 3) and are representative of two independent experiments. B, C: The effects of PGE2 on β-catenin expression and DKK1 mRNA levels in fibroblasts were evaluated. The mean fluorescent intensity (MFI) of β-catenin (B) and DKK1 mRNA (C) in fibroblasts after exposure to PGE2 is shown. The amount of DKK1 mRNA relative to GAPDH mRNA without the addition of PGE2 is regarded as one. The values are expressed as the mean ± SD (n = 3) and *P < 0.05. D: Cytoplasm- (right panel) and nuclear- (left panel) protein samples from fibroblasts treated with or without 0, 100, and 1000 nmol/L PGE2 for 4 days were used to determine the effect of PGE2 on β-catenin expression. The values are expressed as the mean ± SD (n = 3 to 4) and *P < 0.05.
Genetic Analysis for DKK1, TCF-4, and HPGD Genes
To address the cause of PDP, we initially analyzed the sequences of HPGD, and found no mutation including single nucleotide polymorphism that was different among three healthy donors and two PDP patients (data not shown). Rather, our current results suggest that the pathogenesis of the complete form of PDP may be attributable to enhanced Wnt signaling secondary to decreased DKK1 expression. Moreover, it remains uncertain how DKK1 expression is reduced in PDP. One possible mediator is TCF7L2 (TCF-4), which binds to the DKK1 promoter, thus enhancing activity of DKK1.30 Hence, we further analyzed the sequence of DKK1 and TCF-4. However, sequence analyses of the coding sequences of DKK1 and TCF-4, including exon-intron boundaries revealed no mutation (data not shown). In addition, the primers used in this study sequenced all exon-intron boundaries of DKK1, TCF4, and HPGD, but no mutation was found.
DKK1 siRNA Enhances Ear Thickness in Mice
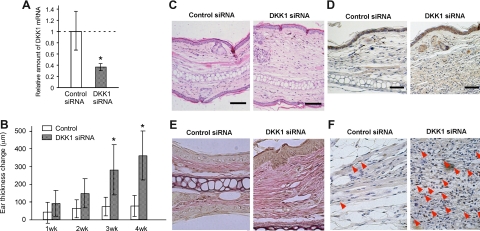
Finally, we used mice to pursue direct evidence for DKK1 involvement in pachydermia. We injected a solution of mouse DKK1 siRNA or control siRNA intradermally into the ears of mice four times every 7 days. Quantitative RT-PCR analysis revealed that this procedure successfully suppressed the expression of DKK1 mRNA in the skin by about 60% (Figure 5A). The ear thickness was measured every week before each injection and 1 week after the last injection. The ear thickness was significantly augmented by the application of DKK1 siRNA (Figure 5B). The histological findings showed that the dermis was thickened with increased fibroblasts (Figure 5, C−F). Consistent with these findings, enhanced β-catenin expression was observed diffusely in the dermis treated with DKK1 siRNA. (Figure 5D).
Figure 5.
The effect of mouse DKK1 siRNA on pachydermia. A: Mouse DKK1 siRNA and control-scrambled siRNA (control siRNA) solutions were injected intradermally into the ears of mice four times every 7 days. The expression of DKK1 mRNA in the skin 1 week after the last siRNA injection was evaluated by quantitative RT-PCR analysis. The mean value of DKK1 mRNA relative to GAPDH mRNA treated with scrambled siRNA is regarded as one (n = 5). B: The ear thickness was measured every week before each injection and one week after the last injection. Columns show the mean ± SD (n = 8, each group) from two independent experiments. The student’s t-test was performed between the indicated groups and *P < 0.05. C−D: The skin from the ears one week after the last siRNA injection was fixed and stained with H&E (C). In addition, skin sections were stained with anti-β-catenin antibody by immunohistochemistry. Enhanced β-catenin expression is visible diffusely in the dermis of the skin treated with DKK1 siRNA (D). Scale bars = 100 μm. E: The samples stained with Elastica van Gieson shows the thick collagen and elastic fibers in the dermis of mice treated with DKK1 siRNA. F: The samples are stained with proliferating cellular nuclear antigen (PCNA). Red arrowheads depict PCNA positive cells.
Discussion
We showed that Wnt/DKK1 plays a key role in the development of pachydermia in several aspects. Firstly, proliferation of fibroblasts from the PDP patients was promoted with a higher ratio in the cell cycle than compared with normal fibroblasts, and human recombinant DKK1 protein decreased their proliferation. Secondly, the expression levels of DKK1 mRNA in PDP fibroblasts and DKK1 protein in PDP skin were lower than those in healthy controls. Thirdly, β-catenin intensity in the skin from PDP was pronounced by immunohistochemistry. Finally, application of mouse DKK1 siRNA increased the thickness of the skin in accordance with the elevated β-catenin levels. These results suggest that enhanced Wnt signaling is related to the development of pachydermia.
Pachydermia is one of the clinical manifestations of the complete form of PDP, which involves both skin and bone. For example, BMP, TGF-β, and Wnt families are the possible molecules responsible for the changes in both organs. There are several congenital diseases related to both organs, such as basal cell nevus syndrome, synovitis acne pustulosis hyperostosis ostitis syndrome, Klippel-Trenaunay-Weber syndrome, and Buschke-Ollendorff syndrome.31,32,33 Bushchke-Ollendorff syndrome, in which osteopoikilosis is associated with connective tissue nevi, is particularly of note, since mutations in LEMD3, a gene implicated in BMP signaling, are candidates for its pathogenesis.34 However, we could not detect a significant difference in mRNA expression for BMP or TGF-β families between PDP and control fibroblasts by DNA microarray analysis.
Recently, the incomplete form of PDP was attributed to elevated PGE2 due to the mutation of HPGD. The skeletal phenotype of PDP, particularly clubbing and periostosis, can clearly be explained by elevated PGE2, since it is well known that PGE2 stimulates the activity of both osteoclasts and osteoblasts,35 leading to bone deposition (periostosis) and resorption (acro-osteolysis), respectively. However, we could not detect a mutation in HPGD. In addition, the level of serum PGE2 from one of our PDP cases (case 1) was within the normal range (data not shown). In fact, long-term therapeutic administration of exogenous PGE2 for skin ulcers secondary to systemic sclerosis, arteriosclerosis obliterans, and Buerger diseases does not induce pachydermia, sebaceous hyperplasia, or velvet coloration of the skin as adverse effects. Moreover, the addition of PGE2 into the fibroblast culture did not induce proliferation. Therefore, it remains unknown how the skin manifestations of PDP are induced.
Here we focused on Wnt signaling in the development of pachydermia. Fibroblasts from PDP skin and bone marrow-derived fibroblasts of PDP patients are known to grow faster than those of healthy donors.26,36 The transfection of DKK1 into cultured mouse fibroblasts, NIH3T3, blocked WNT2-induced cell growth and the WNT2-induced increase in uncomplexed β-catenin.37 WNT3a induced motility and cytoskeletal rearrangement of NIH3T3 cells.27 These previous reports suggest that enhanced Wnt/β-catenin signaling promotes fibroblast proliferation and cytoskeletal rearrangement. In fact, we found that the frequency of PDP fibroblasts in cycle was increased, and that actin bundle formation was more pronounced in PDP fibroblasts. Moreover, the addition of human recombinant DKK1 consistently suppressed the fibroblast proliferation.
The source of DKK1 and how it works in the skin are issues that remain to be clarified. According to our immunohistochemical analysis, the major source of DKK1 in the skin seems to be fibroblasts, because the DKK1 expression in fibroblasts was low in PDP. Since DKK1 is a secreted antagonist and may affect bystander cells in the vicinity of fibroblasts, the dysregulated production of DKK1 possibly modulates the functions of not only fibroblasts but also other cells, such as keratinocytes and melanocytes. It was reported that high DKK1 expression by dermal fibroblasts in the palms and soles inhibits the function of melanocytes via suppression of β-catenin and microphthalmia-associated transcription factor, and enhances keratinocyte proliferation.16,17,38 Mice with an overexpression of DKK1 in skin consistently lacked formation of appendages, such as hair follicles, and the mice had no skin pigmentation on the trunk.18
The role of DKK1 has been more extensively studied in bone than in the skin. DKK1 is known to inhibit osteoblast differentiation, and the overproduction of DKK1 was noted in osteolytic bone lesions of patients with multiple myeloma.39 The elevated DKK1 levels in bone marrow plasma and peripheral blood from the patients were correlated with the presence of focal bone lesions. Recombinant human DKK1 inhibited the differentiation of osteoblast precursor cells in vitro.40,41 These previous observations could explain the periostosis in PDP possibly secondary to decreased DKK1 expression. Since fibroblasts and osteoblasts are derived from mesenchymal origin, they seem to share in common the mechanism of differentiation and proliferation. Although we did not address the relationship between DKK1 and the skeletal phenotype in PDP, it would be of interest to analyze the function of osteoblasts in PDP.
The next question is how Wnt signaling is enhanced. One possibility provided by our present study is the suppression of DKK1 expression in fibroblasts. The mechanism by which DKK1 is down-regulated in PDP remains to be elucidated. It can be hypothesized that there is a mutation in DKK1 or molecules controlling DKK1 expression, such as TCF-4. However, no mutation was detected in either exons of DKK1 or TCF-4 genes. Therefore, in the present study, we could not determine the genetic mechanism responsible for the complete form of PDP and/or pachydermia. Given the defect in PDP appears to altered expression of DKK1, it will be of interest in future studies to analyze the regulatory regions of DKK1, especially around the TCF binding sites, an issue which remains to be clarified.
On the other hand, the Wnt/β-catenin pathway is known to increase DKK1 mRNA and protein, thus initiating a negative feedback loop.42 It can be hypothesized that this negative feedback regulation might be dysregulated in PDP. Moreover, due to this negative feedback system, DKK1 can work as a tumor suppressor gene in some types of neoplasia.42,43 Hypertrophic osteoarthritis is occasionally induced by a variety of thoracoabdominal, sometimes malignant, conditions. The relationship between decreased DKK1 expression and secondary hypertrophic osteoarthritis in association with malignancy may be an interesting issue to pursue.
It still remains unclear whether PDP in our cases could be attributed to the mutation in HPGD or not. Of note is that our cases were diagnosed as the complete form of PDP including pachydermia and adolescent onset, but that the cases with HPGD mutation had the incomplete form of PDP without pachydermia and with early onset (within the first year of their lives). The onset of the PDP is bimodal. The first peak is during the first year of the life and the second at the age of 15 years.3,44 Therefore, the pathogenesis of PDP might be subdivided into at least two groups. However, further clinical studies in combination with HPGD mutation analysis will be required to clarify this.
In PDP, clinical cutaneous manifestations include pachydermia, seborrhea, and velvet colored skin. At present, we could not show direct evidence that all of the phenotypes of PDP were induced by enhanced Wnt signaling secondary to the suppressed expression of DKK1. In addition, the number of cases in our study was limited. However, our findings, together with those of previous studies suggest that the Wnt signaling pathway was promoted in accordance with decreased DKK1 expression, leading to increased fibroblast proliferation, enhanced pigmentation of the skin, and adnexal hyperplasia.
Acknowledgments
We thank Ms. Rie Murase and Dr. Yosuke Okada for technical assistance, and Dr. Tatsuya Ishibe for discussion.
Footnotes
Address reprint requests to Dr. Kenji Kabashima, Department of Dermatology, Kyoto University Graduate School of Medicine, 54 Shogoin Kawara-cho, Sakyo-ku, Kyoto 606-8507, Japan. E-mail: kaba@kuhp.kyoto-u.ac.jp.
Supported in part by grants from the Ministry of Education, Culture, Sports, Science and Technology and the Ministry of Health, Labour and Welfare of Japan.
K.K. and J.S. contributed equally to this work.
References
- Vogl A, Goldfischer S. Pachydermoperiostosis: primary or idiopathic hypertrophic osteoarthropathy. Am J Med. 1962;33:166–187. doi: 10.1016/0002-9343(62)90016-5. [DOI] [PubMed] [Google Scholar]
- Shawarby K, Ibrahim MS. Pachydermoperiostosis. A review of literature and report on four cases. Br Med J. 1962;1:763–766. doi: 10.1136/bmj.1.5280.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimoin DL. Pachydermoperiostosis (idiopathic clubbing and periostosis): genetic and physiologic considerations. N Engl J Med. 1965;272:923–931. doi: 10.1056/NEJM196505062721801. [DOI] [PubMed] [Google Scholar]
- Jajic I, Jajic Z, Grazio S. Minor but important symptoms and signs in primary hypertrophic osteoarthropathy. Clin Exp Rheumatol. 2001;19:357–358. [PubMed] [Google Scholar]
- Touraine A, Solente G, Gole A. Un syndrome osteodermopathique: la pachydermie plicaturee avec pachyperiostose des extremites. Presse Med. 1935;43:1820–1824. [Google Scholar]
- Uppal S, Diggle CP, Carr IM, Fishwick CW, Ahmed M, Ibrahim GH, Helliwell PS, Latos-Bielenska A, Phillips SE, Markham AF, Bennett CP, Bonthron DT. Mutations in 15-hydroxyprostaglandin dehydrogenase cause primary hypertrophic osteoarthropathy. Nat Genet. 2008;40:789–793. doi: 10.1038/ng.153. [DOI] [PubMed] [Google Scholar]
- Castori M, Sinibaldi L, Mingarelli R, Lachman RS, Rimoin DL, Dallapiccola B. Pachydermoperiostosis: an update. Clin Genet. 2005;68:477–486. doi: 10.1111/j.1399-0004.2005.00533.x. [DOI] [PubMed] [Google Scholar]
- Kornak U, Mundlos S. Genetic disorders of the skeleton: a developmental approach. Am J Hum Genet. 2003;73:447–474. doi: 10.1086/377110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchkarev VA, Sharov AA. BMP signaling in the control of skin development and hair follicle growth. Differentiation. 2004;72:512–526. doi: 10.1111/j.1432-0436.2004.07209005.x. [DOI] [PubMed] [Google Scholar]
- Zorn AM. Wnt signalling: antagonistic Dickkopfs. Curr Biol. 2001;11:R592–595. doi: 10.1016/s0960-9822(01)00360-8. [DOI] [PubMed] [Google Scholar]
- Kawano Y, Kypta R. Secreted antagonists of the Wnt signalling pathway. J Cell Sci. 2003;116:2627–2634. doi: 10.1242/jcs.00623. [DOI] [PubMed] [Google Scholar]
- Mao B, Wu W, Davidson G, Marhold J, Li M, Mechler BM, Delius H, Hoppe D, Stannek P, Walter C, Glinka A, Niehrs C. Kremen proteins are Dickkopf receptors that regulate Wnt/beta-catenin signalling. Nature. 2002;417:664–667. doi: 10.1038/nature756. [DOI] [PubMed] [Google Scholar]
- Mao B, Wu W, Li Y, Hoppe D, Stannek P, Glinka A, Niehrs C. LDL-receptor-related protein 6 is a receptor for Dickkopf proteins. Nature. 2001;411:321–325. doi: 10.1038/35077108. [DOI] [PubMed] [Google Scholar]
- Glinka A, Wu W, Delius H, Monaghan AP, Blumenstock C, Niehrs C. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature. 1998;391:357–362. doi: 10.1038/34848. [DOI] [PubMed] [Google Scholar]
- Grotewold L, Ruther U. The Wnt antagonist Dickkopf-1 is regulated by Bmp signaling and c-Jun and modulates programmed cell death. EMBO J. 2002;21:966–975. doi: 10.1093/emboj/21.5.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Itami S, Watabe H, Yasumoto K, Abdel-Malek ZA, Kubo T, Rouzaud F, Tanemura A, Yoshikawa K, Hearing VJ. Mesenchymal-epithelial interactions in the skin: increased expression of dickkopf1 by palmoplantar fibroblasts inhibits melanocyte growth and differentiation. J Cell Biol. 2004;165:275–285. doi: 10.1083/jcb.200311122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Passeron T, Watabe H, Yasumoto K, Rouzaud F, Hoashi T, Hearing VJ. The effects of dickkopf 1 on gene expression and Wnt signaling by melanocytes: mechanisms underlying its suppression of melanocyte function and proliferation. J Invest Dermatol. 2007;127:1217–1225. doi: 10.1038/sj.jid.5700629. [DOI] [PubMed] [Google Scholar]
- Andl T, Reddy ST, Gaddapara T, Millar SE. WNT signals are required for the initiation of hair follicle development. Dev Cell. 2002;2:643–653. doi: 10.1016/s1534-5807(02)00167-3. [DOI] [PubMed] [Google Scholar]
- Masuda K, Moriwaki S, Takigawa M, Furukawa F, Higashishiba T, Fukamizu H. A case of pachydermoperiostosis. Rinsho Hifuka (Japanese) 2000;54:398–401. [Google Scholar]
- Kabashima K, Banks TA, Ansel KM, Lu TT, Ware CF, Cyster JG. Intrinsic lymphotoxin-beta receptor requirement for homeostasis of lymphoid tissue dendritic cells. Immunity. 2005;22:439–450. doi: 10.1016/j.immuni.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Kabashima K, Haynes NM, Xu Y, Nutt SL, Allende ML, Proia RL, Cyster JG. Plasma cell S1P1 expression determines secondary lymphoid organ retention versus bone marrow tropism. J Exp Med. 2006;203:2683–2690. doi: 10.1084/jem.20061289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2[-Delta Delta C(T)] method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Kabashima K, Sakabe J, Yamada Y, Tokura Y. “Nagashima-type” keratosis as a novel entity in the palmoplantar keratoderma category. Arch Dermatol. 2008;144:375–379. doi: 10.1001/archderm.144.3.375. [DOI] [PubMed] [Google Scholar]
- Maekawa M, Yamamoto T, Tanoue T, Yuasa Y, Chisaka O, Nishida E. Requirement of the MAP kinase signaling pathways for mouse preimplantation development. Development. 2005;132:1773–1783. doi: 10.1242/dev.01729. [DOI] [PubMed] [Google Scholar]
- Kushibiki T, Matsumoto K, Nakamura T, Tabata Y. Suppression of tumor metastasis by NK4 plasmid DNA released from cationized gelatin. Gene Ther. 2004;11:1205–1214. doi: 10.1038/sj.gt.3302285. [DOI] [PubMed] [Google Scholar]
- Matucci-Cerinic M, Sacerdoti L, Perrone C, Carossino A, Cagnoni ML, Jajic I, Lotti T. Pachydermoperiostosis (primary hypertrophic osteoarthropathy): in vitro evidence for abnormal fibroblast proliferation. Clin Exp Rheumatol. 1992;10 Suppl 7:57–60. [PubMed] [Google Scholar]
- Kim SE, Choi KY. EGF receptor is involved in WNT3a-mediated proliferation and motility of NIH3T3 cells via ERK pathway activation. Cell Signal. 2007;19:1554–1564. doi: 10.1016/j.cellsig.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Fine A, Goldstein RH. The effect of PGE2 on the activation of quiescent lung fibroblasts. Prostaglandins. 1987;33:903–913. doi: 10.1016/0090-6980(87)90117-1. [DOI] [PubMed] [Google Scholar]
- Korn JH, Halushka PV, LeRoy EC. Mononuclear cell modulation of connective tissue function: suppression of fibroblast growth by stimulation of endogenous prostaglandin production. J Clin Invest. 1980;65:543–554. doi: 10.1172/JCI109698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niida A, Hiroko T, Kasai M, Furukawa Y, Nakamura Y, Suzuki Y, Sugano S, Akiyama T. DKK1, a negative regulator of Wnt signaling, is a target of the beta-catenin/TCF pathway. Oncogene. 2004;23:8520–8526. doi: 10.1038/sj.onc.1207892. [DOI] [PubMed] [Google Scholar]
- Kahn MF, Chamot AM. SAPHO syndrome. Rheum Dis Clin North Am. 1992;18:225–246. [PubMed] [Google Scholar]
- Oduber CE, van der Horst CM, Hennekam RC. Klippel-Trenaunay syndrome: diagnostic criteria and hypothesis on etiology. Ann Plast Surg. 2008;60:217–223. doi: 10.1097/SAP.0b013e318062abc1. [DOI] [PubMed] [Google Scholar]
- Ehrig T, Cockerell CJ. Buschke-Ollendorff syndrome: report of a case and interpretation of the clinical phenotype as a type 2 segmental manifestation of an autosomal dominant skin disease. J Am Acad Dermatol. 2003;49:1163–1166. doi: 10.1016/s0190-9622(03)00481-x. [DOI] [PubMed] [Google Scholar]
- Hellemans J, Preobrazhenska O, Willaert A, Debeer P, Verdonk PC, Costa T, Janssens K, Menten B, Van Roy N, Vermeulen SJ, Savarirayan R, Van Hul W, Vanhoenacker F, Huylebroeck D, De Paepe A, Naeyaert JM, Vandesompele J, Speleman F, Verschueren K, Coucke PJ, Mortier GR. Loss-of-function mutations in LEMD3 result in osteopoikilosis. Buschke-Ollendorff syndrome and melorheostosis. Nat Genet. 2004;36:1213–1218. doi: 10.1038/ng1453. [DOI] [PubMed] [Google Scholar]
- Raisz LG, Pilbeam CC, Fall PM. Prostaglandins: mechanisms of action and regulation of production in bone. Osteoporos Int. 1993;3 Suppl 1:136–140. doi: 10.1007/BF01621888. [DOI] [PubMed] [Google Scholar]
- Fontenay-Roupie M, Dupuy E, Berrou E, Tobelem G, Bryckaert M. Increased proliferation of bone marrow-derived fibroblasts in primitive hypertrophic osteoarthropathy with severe myelofibrosis. Blood. 1995;85:3229–3238. [PubMed] [Google Scholar]
- Fedi P, Bafico A, Nieto Soria A, Burgess WH, Miki T, Bottaro DP, Kraus MH, Aaronson SA. Isolation and biochemical characterization of the human Dkk-1 homologue, a novel inhibitor of mammalian Wnt signaling. J Biol Chem. 1999;274:19465–19472. doi: 10.1074/jbc.274.27.19465. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Passeron T, Hoashi T, Watabe H, Rouzaud F, Yasumoto K, Hara T, Tohyama C, Katayama I, Miki T, Hearing VJ. Dickkopf 1 (DKK1) regulates skin pigmentation and thickness by affecting Wnt/beta-catenin signaling in keratinocytes. FASEB J. 2008;22:1009–1020. doi: 10.1096/fj.07-9475com. [DOI] [PubMed] [Google Scholar]
- Qiang YW, Endo Y, Rubin JS, Rudikoff S. Wnt signaling in B-cell neoplasia. Oncogene. 2003;22:1536–1545. doi: 10.1038/sj.onc.1206239. [DOI] [PubMed] [Google Scholar]
- Tian E, Zhan F, Walker R, Rasmussen E, Ma Y, Barlogie B, Shaughnessy JD., Jr The role of the Wnt-signaling antagonist DKK1 in the development of osteolytic lesions in multiple myeloma. N Engl J Med. 2003;349:2483–2494. doi: 10.1056/NEJMoa030847. [DOI] [PubMed] [Google Scholar]
- Morvan F, Boulukos K, Clement-Lacroix P, Roman Roman S, Suc-Royer I, Vayssiere B, Ammann P, Martin P, Pinho S, Prognonec P, Mollat P, Niehrs C, Baron R, Rawadi G. Deletion of a single allele of the Dkk1 gene leads to an increase in bone formation and bone mass. J Bone Miner Res. 2006;21:934–945. doi: 10.1359/jbmr.060311. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Sancho JM, Aguilera O, Garcia JM, Pendas-Franco N, Pena C, Cal S, Garcia de Herreros A, Bonilla F, Munoz A. The Wnt antagonist DICKKOPF-1 gene is a downstream target of beta-catenin/TCF and is downregulated in human colon cancer. Oncogene. 2005;24:1098–1103. doi: 10.1038/sj.onc.1208303. [DOI] [PubMed] [Google Scholar]
- Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- Matucci-Cerinic M, Lotti T, Jajic I, Pignone A, Bussani C, Cagnoni M. The clinical spectrum of pachydermoperiostosis (primary hypertrophic osteoarthropathy). Medicine (Baltimore) 1991;70:208–214. doi: 10.1097/00005792-199105000-00005. [DOI] [PubMed] [Google Scholar]