Abstract
Remodeling of the stromal extracellular matrix and elevated expression of specific proto-oncogenes within the adjacent epithelium represent cardinal features of breast cancer, yet how these events become integrated is not fully understood. To address this question, we focused on tenascin-C (TN-C), a stromal extracellular matrix glycoprotein whose expression increases with disease severity. Initially, nonmalignant human mammary epithelial cells (MCF-10A) were cultured within a reconstituted basement membrane (BM) where they formed three-dimensional (3-D) polarized, growth-attenuated, multicellular acini, enveloped by a continuous endogenous BM. In the presence of TN-C, however, acini failed to generate a normal BM, and net epithelial cell proliferation increased. To quantify how TN-C alters 3-D tissue architecture and function, we developed a computational image analysis algorithm, which showed that although TN-C disrupted acinar surface structure, it had no effect on their volume. Thus, TN-C promoted epithelial cell proliferation leading to luminal filling, a process that we hypothesized involved c-met, a proto-oncogene amplified in breast tumors that promotes intraluminal filling. Indeed, TN-C increased epithelial c-met expression and promoted luminal filling, whereas blockade of c-met function reversed this phenotype, resulting in normal BM deposition, proper lumen formation, and decreased cell proliferation. Collectively, these studies, combining a novel quantitative image analysis tool with 3-D organotypic cultures, demonstrate that stromal changes associated with breast cancer can control proto-oncogene function.
Most contemporary experimental and clinical breast cancer research studies have focused on the gain or loss of function of specific oncogenes or tumor suppressor genes, respectively, as primary transforming events within the mammary epithelium.1 Equally compelling evidence, however, demonstrates that the biochemical and -physical nature of the stromal extracellular matrix (ECM) microenvironment surrounding the epithelium also contributes to breast homeostasis and tumorigenesis.2,3,4 For example, when cultured within a laminin-enriched compliant ECM, normal breast epithelial cells produce an endogenous basement membrane (BM), which directs the formation of polarized, spherical, multicellular acini, each of which contains a single centrally-located lumen.5 On the other hand, blocking exuberrant β1 integrin signaling between malignant human breast cancer cells and their BM microenvironment induces phenotypic reversion and functional normalization of mammary acini.6 In addition, changes within the mammary ECM, such as those associated with postlactational involution, lead to altered signals to normal and malignant epithelium, failing to support duct development in the former while promoting metastasis in the latter.7 Increases in the biophysical stiffness of the normal mammary stromal ECM have also been shown to result in malignant behavior.3 Collectively, these and other studies reinforce the notion that tissue phenotype, specified by the ECM, can exert a dominant effect over gene expression in adjacent epithelial cells.
Three-dimensional (3-D) laminin-based cultures have allowed investigators to elucidate the effects of specific oncogenes on mammary epithelial tissue form and function in an appropriate in vivo–like context.8 For instance, mammary epithelial cells overexpressing either cyclin D1, or the HPV-16 E7 oncogene, form individual acini that possess normal lumens, but that are of a greater size.9 However, overexpression of ErbB2, or coexpression of cyclin D1 and the antiapoptotic protein Bcl-2 in normal mammary epithelial cells, generates a multiacinar hyperproliferative phenotype characterized by luminal filling.9,10 Thus, it appears that different oncogenes exert distinct site-specific effects on 3-D mammary acinar architecture and behavior. Given this, determining how tumor-associated stromal ECM components alter 3-D tissue structure represents a key step in linking changes within the ECM to the activation or repression of specific genes and signaling pathways operating within the epithelium. One approach to this question is to develop computational tools that can accurately measure changes in mammary epithelial tissue structure as a response to alterations in ECM within a pathobiologically-relevant 3-D microenvironment.
Herein, we developed a novel computational model to objectively measure how tenascin-C (TN-C), a stromal ECM glycoprotein induced in experimental and clinical breast cancer,11 affects normal human mammary epithelial 3-D tissue form and function. To accomplish this, we devised an algorithm to quantify acinar surface topography and volume in 3-D cultures of nonmalignant human mammary epithelial (MCF-10A) cells interacting with TN-C. In this way, we discovered that in contrast to control cultures, TN-C–treated cultures were dysmorphic, possessed disrupted endogenous BMs, and acinar interiors filled with proliferating epithelial cells. Because increased expression of the c-met proto-oncogene leads to intraluminal filling and cell proliferation,12 we hypothesized and thereafter demonstrated that c-met contributes to TN-C–dependent loss of normal mammary epithelial tissue architecture by affecting BM integrity, epithelial cell proliferation, and lumen formation. Thus, the quantitative analyses of mammary epithelial tissue architecture presented herein provide a framework for understanding how the tumor stroma controls the function of a proto-oncogene that is amplified in human breast cancer.
Materials and Methods
Reagents
Cell culture medium (Dulbecco’s modified Eagle’s medium, F12/Ham’s), horse serum, 0.25% trypsin-EDTA and penicillin/streptomycin were from Invitrogen (Carlsbad, CA). Epidermal growth factor (EGF) was purchased from Peprotech (Rocky Hill, NJ). Cholera toxin, hydrocortisone, insulin, and normal goat serum were from Sigma-Aldrich (St. Louis, MO). Phenol Red-free, growth factor–reduced Matrigel was from BD Biosciences (San Jose, CA). Human TN-C purified protein (cat.# CC065) was from Chemicon (Temecula, CA). Mouse-anti-human TN-C antibody (cat.# NCL-TENAS-C) was from Novocastra (Newcastle-on-Tyne, UK). Laminin V (human-specific, cat.# MAB19562) and α6 integrin (cat.# MAB1378) antibodies were from Chemicon, and Ki-67 (cat.# 18-0191Z) and cleaved caspase-3 (cat.# 9664) antibodies were from Zymed (Invitrogen; Carlsbad, CA) and Cell Signaling (Danvers, MA), respectively. c-met (cat.# AF276) and HGF (cat.# AF-294-NA) antibodies were from R&D Systems (Minneapolis, MN). Recombinant human hepatocyte growth factor (HGF) (cat.# GF116) was from Chemicon. Goat anti-mouse F(ab′)2 fragments (cat.# 115-006-006) were from Jackson ImmunoResearch (West Grove, PA). Donkey anti-goat–horseradish peroxidase (cat.# sc-2020) secondary antibody was from Santa Cruz Biotechnology (Santa Cruz, CA). All Alexa Fluor-conjugated (Alexa Fluor 488 and 594) secondary antibodies were from Molecular Probes (Invitrogen; Carlsbad, CA). 16% paraformaldehyde was from Electron Microscopy Sciences (Hatfield, PA). The Vectastain ABC and DAB kits, species-specific biotinylated secondary antibodies, and Vectashield mounting medium containing 4′,6 diamidino-2-phenylindole (DAPI) were from Vector Laboratories (Burlingame, CA). Cytoseal and Clear-Rite 3 were from Richard-Allan Scientific (Kalamazoo, MI). Precast 4% to 20% precise protein gels were from Pierce (Rockford, IL).
Immunohistochemistry
Human breast tissue microarrays were from Ambion (Austin, TX), Cybrdi (Frederick, MD), and U.S. Biomax (Rockville, MD). Pathological diagnoses of the paraffin-embedded tissue sections were validated by an experienced pathologist. The histological profiles, ages, and number of patients encompassed in the tissue microarrays are summarized in Table 1. A pronase-based antigen retrieval process was used for TN-C immunohistochemistry,13 and citrate buffer was used for double immunostaining for TN-C and c-met. The primary antibodies were used at the following dilutions: TN-C at 1:50, c-met at 1:50. Sections were then incubated with biotinylated anti-mouse IgG (for TN-C) and biotinylated anti-goat IgG (for c-met) followed by the streptavidin/peroxidase ABComplex. The 3,3′-diaminobenzidine (DAB) chromogenic substrate was used to detect TN-C, whereas the VectorNovaRED substrate was used to detect c-met. Sections were counterstained with hematoxylin QS (for TN-C alone). For sections in which double-immunolabeling was performed, the streptavidin/alkaline phosphatase (AP) ABComplex was used, followed by the Vector Blue AP substrate. Sections coimmunostained for c-met and TN-C were mounted using Vectamount AQ. Images were captured using a Nikon 90i microscope equipped with a Nikon DXM1200 digital camera at 20× magnification (NA 0.75). TN-C immunoreactivity was scored blindly as low or high based on DAB staining intensity (Table 2).
Table 1.
Summary of Breast Tissue Histology, Patient Age, and Number of Patients
| Histological type | Age range (mean) | Number of patients |
|---|---|---|
| Atypical lobular hyperplasia, ALH | 60 to 67 (63.5) | 2 |
| Ductal carcinoma in situ, DCIS | 58 to 86 (69.5) | 8 |
| Infiltrating ductal carcinoma, IDC | 24 to 88 (55.4) | 189 |
| Lobular carcinoma | 55 | 1 |
| Margin of carcinoma | 31 to 74 (48.6) | 20 |
| Matched benign | 21 to 88 (54) | 25 |
| Medullary carcinoma | 52 to 55 (53.5) | 2 |
| Metastatic adenocarcinoma | 40 to 95 (63.9) | 20 |
| Mucinous adenocarcinoma | 39 to 53 (47.3) | 3 |
| Normal breast tissue | 18 to 69 (46.9) | 104 |
| Paget’s disease | 37 to 51 (46) | 4 |
| Papillary carcinoma | 48 to 67 (57.5) | 2 |
| Sclerosing adenosis | 64 (64) | 2 |
| Tubular carcinoma
|
41
|
1
|
| Total | 18 to 95 (53.2) | 383 |
Table 2.
Stromal TN-C Intensity in Malignant Lesions (IDC), Tumor Margins, Matched Benign, Metastatic, and Normal Tissues
| Histological type | Low TN-C expression (number (%)) | High TN-C expression (number (%)) | Number of patients |
|---|---|---|---|
| Infiltrating ductal carcinoma | 88 (47) | 101 (53)* | 189 |
| Margin of carcinoma | 19 (95) | 1 (5) | 20 |
| Matched benign | 24 (96) | 1 (4) | 25 |
| Metastastic breast carcinoma | 8 (40) | 12 (60)* | 20 |
| Normal breast tissue | 101 (97) | 3 (3)* | 104 |
P < 0.05.
Three-Dimensional Cell Culture
The MCF-10A nonmalignant human mammary epithelial cell line was obtained from the American Tissue Culture Collection (Manassas, VA). MCF-10A cells were maintained in Dulbecco’s modified Eagle’s medium/F12 medium containing horse serum (5%, Invitrogen, Carlsbad, CA), supplemented with EGF (20 ng/ml; Peprotech, Rocky Hill, NJ), hydrocortisone (0.5 μg/ml), cholera toxin (100 ng/ml), insulin (10 μg/ml; all from Sigma-Aldrich, St. Louis, MO), and penicillin/streptomycin (1%) at 37°C and 5% CO2. For 3-D cultures, an overlay method, originally developed by Bissell and colleagues,14,15 was used. Briefly, cells were trypsinized using 0.05% trypsin/0.53 mmol/L EDTA, resuspended in assay medium (Dulbecco’s modified Eagle’s medium/F12, horse serum (2%), supplemented with EGF (10 ng/ml), hydrocortisone (0.5 μg/ml), cholera toxin (100 ng/ml), insulin (10 μg/ml), and penicillin/streptomycin (1%) containing 2% Matrigel (phenol red-free, growth factor–reduced; BD Biosciences, San Jose, CA) and plated on a solidified layer of Matrigel, either with or without purified human TN-C protein (10 μg/ml; Chemicon, Temecula, CA). For c-met blockade studies, c-met antibody (R&D Systems, Minneapolis, MN) or goat IgG (Santa Cruz Biotechnology, Santa Cruz, CA) were used at 0.5 to 2 μg/ml in both the solidified layer of Matrigel as well as in the overlay; the reported phenotypes and analyses were performed using the 1 μg/ml concentration of the antibodies. Overlay assay medium was changed every 2 to 4 days, and 3-D cultures were maintained for 4 to 8 days.
Immunocytochemistry
MCF-10A 3-D cultures (day 4 or day 8) were fixed in 3% paraformaldehyde, rinsed in 1× PBS, permeabilized in 1× PBS containing 0.5% Triton X-100, and stained according to standard methods using primary antibodies diluted in blocking solution (laminin V, 1:100; α6 integrin, 1:200; Ki-67, 1:50; cleaved caspase-3, 1:100; c-met, 1:50). This was followed by incubation in Alexa Fluor–conjugated species-specific secondary antibodies diluted in IF buffer (130 mmol/L NaCl, 7 mmol/L Na2HPO4, 3.5 mmol/L NaH2PO4, 7.7 mmol/L NaN3, 0.1% bovine serum albumin, 0.2% Triton X-100, 0.05% Tween-20) containing 10% normal goat serum (either Alexa Fluor 488 or 594 conjugated antibodies, 1:100). 3-D cultures were washed with IF Buffer and 1× PBS, and mounted with Vectashield medium containing DAPI. Z-stacks of images were obtained at 1- or 2-μm intervals using a Zeiss LSM 510 laser scanning confocal microscope (oil-immersion 25× objective, NA 0.8).
Nuclear Counts
Quantification of cellular proliferation (Ki-67 immunoreactivity) and apoptosis (caspase-3 immunoreactivity) was achieved using images obtained at 5-μm intervals from confocal Z-stacks of acini costained with DAPI. ImageJ software (http://rsb.info.nih.gov/ij/) was used to adjust color in the red, green, and blue channels, achieve optimal brightness and contrast for the channel of interest, to convert image stacks to 8-bit files, to perform edge detection, and to threshold the images to delineate nuclei. Ki-67 or cleaved caspase-3–positive nuclei, as well as total nuclei (DAPI), were manually counted in thresholded image Z-stacks at 5-μm intervals. % Ki-67– or cleaved caspase-3–positive nuclei refers to the percent ratio of positive/total nuclei per individual acinus as measured for 17 to 100 acini per treatment condition.
Computational Biology
Image Preprocessing
Z-stacks of confocal images derived from laminin V– or α6 integrin–immunostained acini were imported into Matlab (The MathWorks; Natick, MA), and stored as a 3-D matrix volume, V = (vi,j,k)X×Y×Z, where there were Z-confocal slices each of resolution X×Y. Immunostaining intensity for a particular 3-D pixel (voxel) was thus described by V(i,j,k). These data were histogram-normalized to the full range of intensity values 0 to 255, such that low and high values corresponded to unstained and more strongly stained areas, respectively. A median filter was used to process each section of the Z-stack images (a particular value of k) to remove pixel noise.
Active Contour Optimization
To trace the edges of individual acini, an active surface was used. This is a 3-D version of an active contour16,17 or ‘snake,’18 which is defined as a discrete set of points S = s(p,q) = (xp,q,yp,q,zp,q),19 where each point in s represented a vector of length 3, containing its x, y, z coordinates. Here, s(p,q) represents a 2-D grid of points that is wrapped around to form a cylinder, q is the index for a slice number (corresponding to a specific confocal image slice at a particular z-depth), and there are Z slices in total.19 For each slice q, the active contour formed a two-dimensional (2-D) outline consisting of P discrete points, typically in the order of 200. The active contour was constrained to be a cylinder by setting s(P,q) = s(1,q) (for all values of q).19 We did not constrain the active contour to be a closed sphere as the confocal data for the extreme polar regions of the acini was either too noisy or not present to allow for accurate fitting.
To fit the x, y, z positions of the active surface S to volumetric data V, the equatorial section was manually traced in an approximate fashion. The equatorial slice was the only slice manually traced, which was subsequently optimized using the procedure described below. Briefly, all of the remaining slices above and below the equatorial plane were sequentially and independently fit using the previous slice fit as the starting point. Active contour optimization was restricted in the z axis as previously described because of limited resolution in the z dimension.20
For any particular section, the contour was optimized using an iterative procedure to minimize the energy function, Etotal = Eimage+Espacing. As with all active contour models, the final contour was balanced between low-level image and high-level shape properties.18 The low-level properties promoted points in the active contour to move toward regions of highest local immunostaining intensity. Optimization based solely on low-level features typically resulted in jagged and unsatisfactory contours, which could get ‘caught’ or ‘snagged’ on highly stained debris. For this reason, high-level features were also included and these took the form of prior knowledge regarding a certain degree of smoothness of the contour.
Low-level image features were calculated as Eimage = −w.Gσ*|∇V(i,j,k)| in which Gσ was a 2-D Gaussian with SD, σ. |∇V(i,j,k)| is the gradient of the immunostaining intensity V at the point (i,j,k) and w was used to set the relative importance of low-level image features. The dx, dy maps referred to below represent this gradient. High-level shape properties were defined by equal spacing of surface points in each section Espacing = Σ(d(a,b) − d(b,c)), where d(a,b) and d(b,c) represented Euclidian distances between the contiguous points ab and bc, respectively. In practice, Espacing was minimized by calculating a vector for each point directed toward the nearest point on the bisector of the two neighboring points.21 This approach resulted in Espacing = 0 when all points were equally spaced. By decreasing the value of w in Eimage, the contribution of Espacing was increased, resulting in smoother fits.
A sequential technique for fitting the surface S to the acinar surface was used. Initially, the entire surface was fitted using a larger Gaussian blur of the immunostaining intensity (typically σ = 6 pixels), which had the effect of smoothing over extraneous debris, and producing a good overall approximate fit after 400 iterations. A sequentially better fit was obtained by decreasing blur (smaller σ), calculating 400 iterations, and repeating until a minimum σ of 2 pixels was reached.
Morphological Measures: Surface Roughness, Volume, and Mercator Projections
A morphological measure was calculated based on the Euclidian difference between the 3-D surface of the acinus (delineated by the active mesh) and a best fitting ellipsoid model. The ellipsoid was chosen as an accurate representation of a smooth surface, which closely resembled the shape of spherical acini that became increasingly ellipsoidal on squash introduced by coverslip placement. An ellipsoid, defined as x2a2 + y2b2 + z2c2 = 1 where a, b, and c are parameters defining the scale of the ellipsoid along each dimension, was fitted to the active surface using additional parameters to capture the center location, and orientation around the z axis only. Best fitting ellipsoid parameters (one ellipsoid for each acinus) were calculated in spherical coordinates to minimize the root-mean-square (RMS) error between the ellipsoid and the acinar surface data. Obtaining a reliable surface roughness measure was clearly dependent on the ellipsoid, and so it was crucial to find the globally optimal ellipsoid parameters. To accomplish this, we optimized 100 separate ellipsoid models, each with different starting parameters. From these, the best fitting model was taken as a starting point and a further 100 estimates were produced, with slightly perturbed parameter values. The best fitting set of parameter values was chosen at this point. This ellipsoid parameter fitting process was applied to each acinus. It was not possible in all cases to directly use the active surface to accurately measure acinar volume because polar regions of some acini were absent or noisy. We found estimation of acinar volume on the ellipsoid fit, which is given by V = 4/3πabc to be a satisfactory and consistent method, given the data at polar regions. Mercator projection maps were generated by plotting the difference between the acinar surface and the best ellipsoid model at each spherical coordinate. This effectively ‘unwraps’ the 3-D approximately spherical structure for easier visualization. Protrusions and indentations of different magnitudes of the acini are shown as positive and negative values, respectively, in the Mercator maps.
Western Analysis
Cells were extracted from Matrigel using 1× PBS containing 0.5 mmol/L EDTA, and lysed in 1× Cell Lysis Buffer (20 mmol/L Tris-HCl (pH 7.5), 150 mmol/L NaCl, 1 mmol/L EDTA, 1 mmol/L EGTA, 1% Triton X-100, 2.5 mmol/L sodium pyrophosphate, 1 mmol/L β-glycerophosphate, 1 mmol/L sodium vanadate, 1 μg/ml leupeptin) containing phosphatase and protease inhibitors. Conditioned medium was concentrated before Western blotting. Cell lysates and conditioned medium were separated by SDS-PAGE on 4% to 20% gels (Pierce, Rockford, IL), transferred to polyvinylidene difluoride membranes and probed with anti–c-met, -HGF, -GAPDH (Cell Signaling, Danvers, MA), or –β-actin (Sigma-Aldrich, St. Louis, MO) antibodies. Horseradish peroxidase–conjugated secondary antibodies and enhanced chemiluminescence (ECL) were used to visualize proteins using the Fuji LAS-3000 system. ImageGauge V4.22 software was used for densitometric quantification.
RNA Extraction and Semiquantitative RT-PCR
RNA extraction was performed using the RNeasy Minikit under methods provided by Qiagen (Valencia, CA). Synthesis of cDNA was performed using random hexamers and Superscript II reverse transcriptase (Invitrogen) at 42°C for 50 minutes starting with 1 to 5 μg of total RNA. One μl of cDNA was then used for PCR, implementing PCR beads per the manufacturer’s instructions (GE Health care, Piscataway, NJ). The sequences of primers used in PCR amplification are as follow: for TN-C, the forward primer: 5′-GCTCTCGATGGTCCATCTGGC-3′, and the reverse primer: 5′-TCGTAGCAGTGGTAGAACTC-3′. Quantum RNA 18S classic internal standards (cat# 1716) were from Ambion (Austin, TX). PCR analyses were done using a thermocycler for 32 cycles. Products were separated in 1.2% agarose gels. Bands were visualized by ethidium bromide or Vistra Green staining (GE Health care).
Quantitative RT-PCR
RNA extraction (from day 8 cultures) was performed as described above. Synthesis of cDNA was performed using the iScript cDNA Synthesis Kit (cat.# 170-8891, Bio-Rad, Hercules, CA) according to the manufacturer’s instructions, at 42°C for 30 minutes starting with 0.2 to 1 μg of total RNA. One μl of cDNA was then used for PCR, implementing the iQ SYBR Green Supermix (cat.# 170-8880, Bio-Rad), along with 500 nmol/L of the following primers: c-met: forward: 5′-CAGGCAGTGCAGCATGTAGTG-3′ and reverse: 5′-TAAGGTGGGGCTCCTCTTGTCA-3′. Stratagene QPCR Human Reference Total RNA (cat. #750500, Stratagene, La Jolla, CA) was used as a positive control. PCR reactions were performed in triplicate. PCR analyses were performed and detected using the MyiQ2 Two-Color Real-Time PCR Detection System (Bio-Rad).
Statistics
Statistics were performed using an unpaired Student t test. Error bars show standard error of the mean (SEM). Significant P values were <0.05 (* or **) where indicated. In cases where a sample size of n is indicated, n refers to the number of individual acini. All experiments were performed at least in triplicate.
Results
TN-C Promotes Mammary Epithelial Cell Proliferation and Disrupts 3-D Tissue Organization
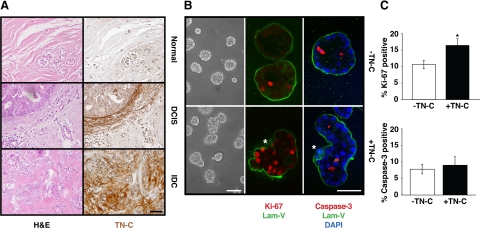
Immunohistochemical staining of human breast tissue microarrays, encompassing 383 individuals, showed that although TN-C expression was negligible within normal breast tissue (Figure 1A, top right), it appeared at the stromal-epithelial interface in premalignant lesions, including ductal carcinoma in situ (DCIS; Figure 1A, middle right), and at higher levels in the stroma of infiltrating ductal carcinoma (IDC; Figure 1A, bottom right). Elevated expression of stromal TN-C within tumors directly correlated with breast disease and progressive loss of epithelial tissue architecture (Figure 1A). Whereas only 3% to 5% of normal, matched benign, and carcinoma margin tissues expressed high levels of TN-C, strong stromal TN-C staining was evident in 53% of IDCs and 60% of breast cancer metastases (Table 2).
Figure 1.
Stromal TN-C alters normal 3-D mammary epithelial tissue architecture. A: Tissue sections from normal human mammary gland (top), DCIS (middle), and IDC (lower) stained with hematoxylin and eosin (H&E) (left) or TN-C (right). Scale bar, 50 μm. B: Morphology of MCF-10A acini generated in Matrigel in the absence (top) or presence (bottom) of TN-C for 8 days (phase contrast, left). Confocal immunofluorescence staining with laminin-V (green; middle and right), Ki-67 (red; middle), and cleaved caspase-3 (red; right) in 8 day cultures. Asterisk indicates loss of a continuous BM (middle) and position of cells residing outside of the BM zone (right) in the presence of TN-C. Scale bars, 50 μm. C: Quantification of Ki-67 immunoreactivity in MCF-10A acini revealed a 1.6-fold increase in proliferation (n = 66 acini, *P < 0.009), yet no differences in apoptosis (quantification of cleaved caspase-3 immunoreactivity; n = 40 acini, P = 0.68) in response to TN-C.
To begin to determine whether stromal TN-C directly affects the structure and function of the normal human mammary epithelium, TN-C–deficient MCF-10A cells (See Supplemental Figure S1 at http://ajp.amjpathol.org) were cultivated for 4 to 8 days in the presence of a reconstituted basement membrane (ie, Matrigel), either with or without addition of exogenous purified human TN-C protein. Although control and TN-C–treated cultures both formed 3-D acini, their gross appearance differed: control acini were mostly smooth and spherical, whereas acini exposed to TN-C appeared to be more disorganized, accounting for 66.3% of total acini (See Supplemental Figure S2 panel A at http://ajp.amjpathol.org and Figure 1B, left panels). To further assess these differences, we evaluated endogenous BM organization: whereas control acini were surrounded by a smooth, continuous, laminin-V–rich BM, the BM appeared to contain numerous gaps and folds when TN-C was present (Figure 1B, middle and right panels). In fact, BM disruption was twice as common in TN-C–containing cultures when compared with controls (56% versus 28.5%; See Supplemental Figure S2 panel B at http://ajp.amjpathol.org). In addition, in some fields, epithelial cells often appeared to be transiting through gaps in the BM into the surrounding TN-C–enriched microenvironment. Because it appeared that TN-C played a role in BM disorganization and possibly contributed to cells transitioning through BM gaps, we postulated that TN-C may be involved in negatively regulating BM assembly, or in potentiating BM degradation. Despite these observations, the mechanism for which is still under investigation, the most striking difference between the control and TN-C–treated cultures was the apparent size and overall organization of the acini.
Because TN-C is known to support cell proliferation and survival in remodeling human tissues during development and disease,22 its ability to alter normal 3-D mammary epithelial tissue architecture may occur by increasing proliferation or by decreasing apoptosis. Ki-67 immunostaining revealed a significant 1.6-fold increase in epithelial cell proliferation in response to TN-C (Figure 1B, middle & Figure 1C, left; 16.4 ± 2.0% versus 10.6 ± 1.3%; P < 0.009). In contrast, levels of apoptosis within acini, as determined by immunostaining for cleaved caspase-3, were identical in control and TN-C–treated cultures, (Figure 1B, right & Figure 1C, right; −TN-C: 7.8 ± 1.5%, +TN-C: 9.0 ± 2.5%; P = 0.34).
TN-C Affects Acinar Surface Architecture, but not Volume
Because TN-C promotes epithelial cell proliferation without affecting the levels of apoptosis, this might culminate in the formation of larger acini. Alternatively, increased proliferation may lead to intraluminal filling, without affecting acinar volume. An objective test of these ideas is critical, because different cancer-associated genes are known to exert different effects on 3-D mammary acinar organization.23 3-D ECM-based cultures represent the current state-of-the-art method for examining cell behavior in an appropriate in vivo–like tissue context. However, they remain difficult to measure because of their relatively large size and heterogeneous nature (Figure 1B, left). To redress this, we developed an image analysis tool to objectively quantify surface topographical features and volumes of 3-D mammary acini in mass cultures. Surface roughness, evaluated by visualizing the endogenous BM, was chosen as an initial metric, because it is accepted that BM alterations not only reflect the state of tumorigenesis,24 but that perturbation or loss of this feature in and of itself may contribute to tumorigenesis.25
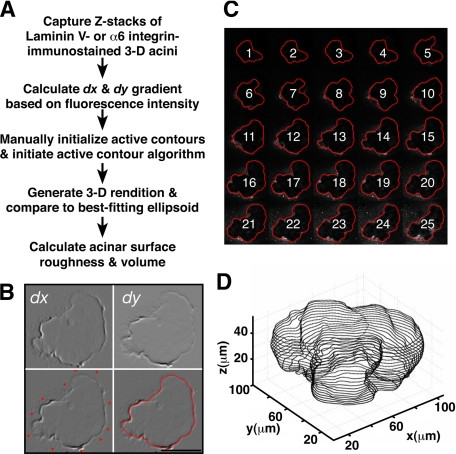
To trace the exterior surface of mammary acini, an active contours-based algorithm was devised (Figure 2A). Individual 2-D image slices acquired at 1 to 2 μm intervals, as confocal Z-stacks of laminin-V or α6 integrin-stained acini, were used for this procedure. Next, the green channel (representing laminin-V or α6 integrin immunoreactivity) in a 2-D equatorial plane confocal image was converted to an 8-bit grayscale image. After noise removal, blurred image gradients (dx and dy) were obtained (Figure 2B, top). Subsequently, the active contour for the equatorial Z plane was initiated by manually selecting numerous points close to the edge of the acinus (Figure 2B, lower left), which were then finalized automatically using the active contour algorithm to create an accurate outline of the outer acinar edge (Figure 2B, lower right). This initial equatorial contour was used as a point of reference for automatic tracing of the remaining image slices in the Z stack for each acinus, resulting in a montage of images (Figure 2C). Traced montages were then compared with the original images to check for any errors in tracing, and each montage was then used to construct a 3-D rendition of each acinus (Figure 2D).
Figure 2.
Reconstructing 3-D mammary epithelial tissue architecture. A: Schema delineating the computational steps used to measure acinar surface roughness and volume. B: In this example, the equatorial 2-D confocal slice from a laminin-V immunostained acinus, treated previously with TN-C for 8 days, was subjected to dx and dy pre-processing based on fluorescence intensity (top). Active contours were manually initiated by selecting multiple points close to the edge of the acinus (lower left). After active contour fitting, a final trace of the edge was obtained automatically (lower right). C: The trace obtained in B was extrapolated to all subsequent remaining 2-D slices of the acinus resulting in the automated generation of a montage of traces. D: Montages described in C were used to render a 3-D projection.
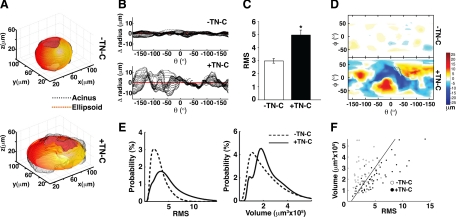
Using this method, we examined the effects of TN-C on tissue architecture in 103 individual 3-D acinar renditions generated in more than three independent experiments. Outer surface roughness was measured by comparing the 3-D rendition to a customized, best-fitting ellipsoid, which represents a ‘perfectly smooth’ surface (Figure 3A). Even before calculating surface roughness, 3-D reconstructions of individual acini suggested that TN-C has a profound effect on normal mammary epithelial tissue architecture. Specifically, when compared with control cultures, numerous protrusions and indentations, reminiscent of branched structures, were apparent (Figure 3A). Further analysis of these features confirmed that TN-C promotes a greater degree of radial deviation from the perfect ellipsoid when compared with a control acinus (Figure 3B). To measure surface roughness in control and TN-C-treated acini, we calculated a RMS value based on the difference between the Euclidian distance of a 3-D acinar rendition versus a best-fitting ellipsoid. Smooth acini possessed lower RMS values, whereas acini with a rougher surface produced higher RMS values (Figure 3C). TN-C–treated cultures exhibited a significant 1.7-fold increase in mean RMS value, indicative of increased surface roughness. In addition to quantifying acinar surface roughness, other features were explored. By ‘unwrapping’ 3-D renditions, Mercator projections, which display acinar surface topography in a map-like format, were generated, permitting visualization and comparison of the finer surface features of control and TN-C-treated acini (Figure 3D). This analysis revealed that control acini possess a relatively “flat” surface landscape, whereas acini exposed to TN-C harbor numerous protrusions (orange to red colors) and indentations (light to deep blue colors).
Figure 3.
TN-C increases surface roughness, but not acinar volume. A: Examples of 3-D renditions of individual acini (black) generated in the absence (top) or presence (bottom) of TN-C in 8 day cultures. A customized ellipsoid (red-yellow) was designed and fitted to each individual acinus. B: Change in radius for each acinar slice (black) denotes distance away from the perfect ellipsoid for the acini depicted in A; the red horizontal line at ‘0′ represents the perfect ellipsoid. In the absence of TN-C (top), each slice did not deviate more than a few μms from the perfect ellipsoid, whereas in the presence of TN-C (bottom), deviations exceeded 10 μm. C: 3-D acinar structure was quantified by measuring root mean square (RMS; absolute difference from perfect ellipsoid) values for acini cultured for 8 days in the absence or presence of TN-C. TN-C evoked a 1.7-fold increase in RMS (−TN-C = 2.98 ± 0.18, +TN-C = 4.96 ± 0.38, n = 103; P = 0.024). D: Mercator projections of 3-D acini: protrusions are displayed in yellow and red while indentations appear blue. E: Distribution of RMS values (left) and volumes (right) for acini generated in the absence (dashed line) or presence (solid line) of TN-C for 8 days. (F) Plotting RMS versus volume yields 2 distinct groups of acini with the line showing the best linear classification using logistic regression at a performance of 79.5%.
The distribution of RMS values (acinar surface roughness) and acinar volumes in both control and TN-C–treated conditions was then analyzed. Using logistic regression analysis, we calculated performance in terms of percent correct classification of a condition obtained via a bootstrap technique.26 RMS measurements alone resulted in good discrimination performance of 69.8 ± 4.8% (Figure 3E, left), ie, the control and TN-C–treated cultures were clearly separable into 2 populations. Taking volume measurements alone into account resulted in a lower performance of 56.6 ± 4.7% (Figure 3E, right), which was insufficient to separate the two populations. Thus, TN-C did not change acinar volume in the 103 acini analyzed. Nevertheless, on taking both RMS and volume into account, the control and TN-C–exposed acini could be segregated with approximately 80% accuracy (Figure 3F). Therefore, although TN-C had no significant effect on acinar volume, on consideration of both volume and surface roughness, it became possible to predict the culture conditions of an individual acinus, ie, whether they were cultured in the absence or presence of TN-C.
TN-C Promotes Luminal Filling via c-Met
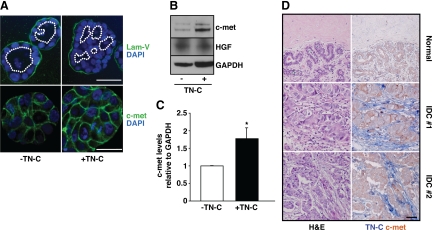
Overall, because TN-C had no effect on acinar volume, while increasing net cell proliferation, we hypothesized that stromal TN-C promotes epithelial cell proliferation leading to intraluminal filling. To test this, we re-evaluated nuclear staining within confocal Z-stacks of acini: this analysis revealed that whereas control acini possess a central lumen lined by a single layer of epithelial cells (Figure 4A, upper left), TN-C–treated acini contain numerous lumens surrounded by multiple cell layers (Figure 4A, upper right). Based on these findings, we then explored the relationship between TN-C and c-met (ie, the HGF receptor), a proto-oncogene that is amplified or overexpressed, but not mutated, in human breast cancer, where it has been postulated to promote cell proliferation, migration and intraluminal filling.27 Given these prior observations, we tested the hypothesis that TN-C regulates c-met function to promote cell proliferation and subsequent intraluminal filling. Consistent with this, immunofluorescence staining of 3-D acini showed increased epithelial c-met levels in the presence of TN-C (Figure 4A, lower). Western immunoblotting confirmed these findings, revealing that TN-C significantly upregulates c-met expression in 3-D cultures (Figure 4, B and C). Although HGF secretion is generally considered to be a hallmark of stromal cells, we nevertheless measured HGF in both the assay medium containing Matrigel (±TN-C), either with or without epithelial cells. These studies showed that HGF levels were negligible in cell-free media (See Supplemental Figure S3 at http://ajp.amjpathol.org), that MCF10-A cells represent a source of this ligand (Figure 4B), and that TN-C has no effect on HGF production (Figure 4B). Thus, TN-C appears to promote increased expression of c-met while having no effect on the level of its ligand. To address whether TN-C enhances c-met at the transcriptional or posttranslational level, real-time PCR was performed, which showed a 1.35-fold increase in c-met transcript levels in the presence of TN-C (data not shown).
Figure 4.
TN-C promotes luminal filling and upregulates c-met. A: Confocal immunofluorescence staining with laminin-V (green; upper) and DAPI (nuclei, blue) revealed changes in lumen structure (white dotted lines) in acini cultured with or without TN-C. C-met staining intensity is increased in the presence of TN-C (lower right) when compared with control (lower left). Scale bars, upper panels, 50 μm; lower panels, 25 μm. B: Western blot analysis confirmed that c-met levels are increased in the presence of TN-C, whereas secreted HGF levels remain unchanged. GAPDH is the loading control. C: Densitometric analyses of c-met levels relative to GAPDH reveal a significant 1.8-fold increase in the presence of TN-C. D: Tissue sections from the normal human mammary gland (top) and grade 2 IDCs (middle and bottom) stained with H&E (left) or c-met and TN-C (red and blue, respectively; right). Scale bar, 50 μm.
Next, to explore the potential relationship between TN-C–dependent increases in c-met expression in vivo, we co-immunostained human breast cancer tissue sections for these proteins. In normal tissue, the majority of the samples (43%) were negative for both stromal TN-C and epithelial c-met (Figure 4D, upper). However, in IDC sections, stromal TN-C expression was evident adjacent to the c-met–positive epithelial compartment in 60% of the cases, with only 5% of the tissues exhibiting no expression of either protein (Figure 4D, middle and lower).
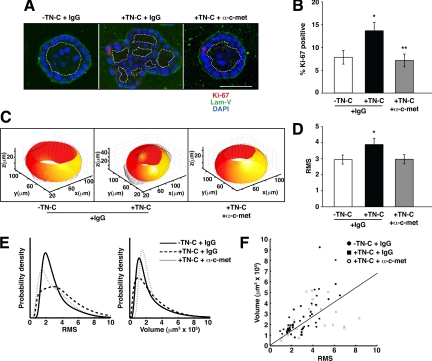
These results indicated that a TN-C/c-met signaling axis may also be relevant to human breast cancer, yet these studies did not address directly the functional nature of this connection. To explore this link, we inhibited c-met function in TN-C–treated 3-D mammary acini using a function-blocking c-met antibody. When compared with IgG-treated controls, c-met inhibition (at 1 μg/ml) blocked the TN-C phenotype resulting in the formation of a continuous BM, accompanied by a 52% decrease in epithelial cell proliferation, and the appearance of single lumens within individual acini (Figure 5, A and B). Furthermore, by quantifying changes in acinar architecture after c-met blockade using the active contours-based algorithm (Figure 5C), we demonstrated that RMS values (acinar surface roughness) were reduced to control levels (Figure 5D), fully supporting the notion that TN-C generates a c-met-dependent phenotype. The distribution of RMS values was similar for control and c-met blockade groups, whereas it was higher for TN-C–treated (+IgG) cultures, whereas volumes were not significantly changed between all three groups (Figure 5E). Interestingly, we were once again able to segregate the TN-C–treated acini from control and c-met blockade acini by taking both the RMS and volume measurements into account, with approximately 72% accuracy (Figure 5F). This finding further supports the predictive value of both RMS and volume measurements in objectively assessing individual acini, and their subsequent placement into either control or TN-C–treated groups.
Figure 5.
Blocking c-met function reverses the TN-C–dependent phenotype. A: Representative confocal immunofluorescence photomicrographs showing reversion of the TN-C–induced phenotype on introduction of a function-blocking c-met antibody. B: C-met blockade significantly decreases the proliferation of MCF-10A acini treated with TN-C (−TN-C + IgG, 7.82 ± 1.49; +TN-C + IgG, 13.65 ± 1.82; +TN-C + α-c-met, 7.12 ± 1.41; n = 60; *P = 0.017; **P = 0.0079). C: Examples of 3-D renditions of individual acini (black), generated in the absence of TN-C (left), presence of TN-C (middle), and presence of TN-C and c-met blocking antibody (right), along with customized ellipsoids (red-yellow). D: C-met blockade restores normal acinar architecture as measured by RMS (−TN-C + IgG, 2.95 ± 0.28; +TN-C + IgG, 3.89 ± 0.39; +TN-C + α-c-met, 2.97 ± 0.29; n = 74; *P = 0.036). E: Distribution of RMS values (left) and volumes (right) for acini generated in the absence of TN-C (solid line), presence of TN-C (horizontally dashed line), and in the presence of TN-C and c-met blocking antibody (vertically dashed line) for 8 days. F: Plotting volume versus RMS results in clustering of the 3 groups of acini into 2 distinct populations, (ie, the absence of TN-C and presence of TN-C and c-met blocking antibody form one cluster, whereas the presence of TN-C and IgG form a different cluster), with the line showing the best linear classification using logistic regression at a performance of 72%.
Discussion
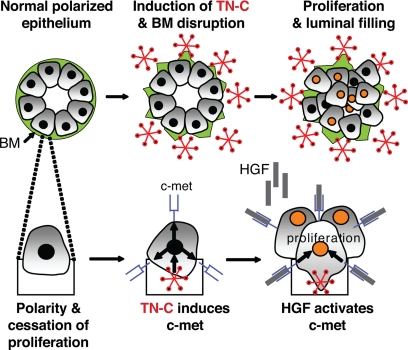
Although it is fully accepted that alterations in the ECM microenvironment25 and transformation of the adjacent epithelium by specific proto-oncogenes1 contribute to cancer initiation and progression, how these events converge to impact overall normal breast epithelial structure and function is less well understood. To this end, the novel quantitative image analyses described herein allowed us to connect a pathologically-relevant breast cancer stromal component (ie, TN-C) with the regulation and function of a proto-oncogene (ie, c-met) known to be overexpressed in human breast cancer (Figure 6). Importantly, this tool and associated methodologies may now be applied to understand more precisely how other extrinsic or intrinsic factors control 3-D mammary epithelial tissue organization and function.
Figure 6.
Hypothetical schema delineating how stromal TN-C promotes epithelial cell proliferation and intraluminal filling within the mammary epithelium via up-regulation of c-met.
TN-C is a multifunctional context-dependent ECM glycoprotein that has been postulated to contribute to pathological tissue remodeling via its ability to impact genome stability,28 cell cycle progression,29 production of matrix metalloproteinases (MMPs),30 expression of vascular endothelial growth factor,31 as well as cross-modulation of receptor tyrosine kinases.11 Even so, the precise manner in which TN-C controls mammary epithelial tissue behavior remained obscure. In this study, we not only show that TN-C upregulates c-met, but that this pathway has functional consequences on both tissue form and function. Because TN-C and c-met have both been postulated to represent reliable markers for poor prognosis in breast cancer,11,32 these studies indicate that TN-C and its downstream effectors represent a potentially useful target in breast cancer treatment.33 Consistent with this, albeit in different cancer tumor types, TN-C radioimmuno-, RNAi-, and aptamer-based strategies are already being evaluated at the clinical level for targeting malignant glioma and non-Hodgkins lymphoma.34,35,36 In addition, growth and angiogenesis are reduced using HGF and c-met antagonists in an in vivo human breast cancer model.37,38 Given these findings, targeting breast cancer via TN-C may represent a viable novel treatment option for breast cancer.
In 3-D mammary epithelial cultures treated with TN-C, we recorded deficits in the production, assembly, and/or stability of the endogenous BM. Furthermore, we have preliminary evidence of cellular trans-migration through the points of BM loss into the TN-C–enriched ECM. Analyses of various cells and tissues have also shown that TN-C protein is associated with a migratory phenotype in vivo and in tissue culture. For example, activated focal adhesion kinase promotes fibroblast migration via its ability to control TN-C expression at the transcriptional level.39 Importantly, both focal adhesion kinase– and TN-C-null mice exhibit migration and wound healing defects, respectively40,41 and in vivo knockdown of TN-C expression in avian embryos attenuates neural crest cell migration.42 Once deposited in the ECM, TN-C may also control migration by disassembling stable focal adhesions,43 or by modulating the strength of cell binding interactions with other ECM molecules, including fibronectin,44 which is also induced in the breast tumor stroma.45 As well, based on our current results, it is possible that TN-C promotes cell motility via induction of c-met, given that this receptor appears to control this function in both embryonic development and cancer.27
As well, it is plausible that TN-C disrupts endogenous BM by regulating the expression or activity of MMPs, an effect that has already been described for synovial fibroblasts, which produce increased levels of MMP-9 when interacting with mixed substrates of TN-C and fibronectin.30 However, gelatin substrate zymography did not reveal differences in MMP-9 levels or activity in response to TN-C (data not shown). Nonetheless, TN-C may stimulate MMPs that are not detectable via gelatin zymography, including MMP-12, which is upregulated by TN-C in 3-D cultures of gliomas,46 MMP-3, which is activated in the involuting breast in concert with TN-C,47 and MMP-14/MT1-MMP, an MMP identified as critical for trans-migration across in vivo–derived BMs.48 Alternatively, it is possible that TN-C disrupts mammary epithelial tissue architecture in an MMP-independent manner through regulation of other ECM-degrading enzymes, such as the serine proteinase, urokinase plasminogen activator, another reliable prognostic marker in breast cancer,49 and one that has been found to be expressed in the tumor stroma together with TN-C.50 It is also possible that TN-C leads to disruption of the BM in a protease-independent manner. For example, recent studies have shown that oral squamous cell carcinoma invasion and metastasis is associated with co-deposition of laminin-V and TN-C, and that these molecules form a physical complex.51 Thus, TN-C may sterically interfere with normal BM deposition in tumors.
A central finding of this study is that TN-C promotes intraluminal filling of acini, a process that can occur when increases in cell proliferation are coupled with deficits in apoptosis within the acinar interior. For example, luminal filling has been noted when cyclin D1 is overexpressed with the anti-apoptotic protein Bcl-2.23 In our studies, however, apoptosis was not regulated by TN-C, suggesting that TN-C influences a different pathway. Accordingly, we focused on c-met, a proto-oncogene that is not only required for normal breast development,52 but also for increased cell proliferation, invasion through the ECM,53 and luminal filling.12 Furthermore, and in common with TN-C, overexpression of c-met has been linked with poor prognosis and a high risk of metastasis.54 At a histological level, c-met overexpression occurs in invasive ductal breast tumors bearing luminal aberrations,55 and activation of c-met by HGF has been shown to stimulate extensive development of branched structures by mammary epithelial cells cultured in 3-D.56 Thus, it is plausible that TN-C–dependent induction of c-met promotes a branched phenotype that may manifest itself in our system as increased surface roughness. In support of this idea, TN-C is expressed around the neck of the breast bud, and surrounding portions of the branching ducts during mammary gland development.57 Our findings are also consistent with recent reports that showed that overexpression of c-met in MCF-10A cells and in the mouse mammary gland leads to increased branching and intraluminal filling, respectively.12,58 Furthermore, a link between TN-C and c-met has been previously demonstrated in colon cancer–associated myofibroblasts, which secrete both TN-C and HGF, and have been shown to promote colon cancer cell invasion.59
Mechanistically, c-met can collaborate with c-myc in promoting breast adenocarcinomas.12 This may be relevant in the context of the present findings, because TN-C has already been shown to induce c-myc expression in 3-D cultures of differentiated mouse mammary epithelial cells.29 Future studies will focus on how TN-C regulates c-met expression in the human mammary epithelium, and whether it also controls other pathways that collaborate with c-met. To this end, we have recently discovered that c-met forms a complex with activated epidermal growth factor receptors (EGFRs) in 2-D cultures of MCF-10A cells (Taraseviciute and Jones, unpublished). Because TN-C has the ability to potentiate the activity of EGFRs60 and EGFRs cross-modulate c-met activity,61 it is plausible that TN-C not only upregulates c-met expression but that it indirectly controls its activity via interactions with EGFRs.
In summary, the combined use of a 3-D organotypic culture system with a novel computational image analysis tool has allowed us to objectively measure how TN-C compromises human mammary epithelial tissue structure, gene expression and function. These studies reinforce the idea that microenvironmental cues originating within the tumor stroma can act in both a dominant and paracrine fashion to control the expression and function of epithelial genes already associated with the development and progression of breast cancer.
Acknowledgments
We thank members of the Jones laboratory, the Institute for Medicine and Engineering, Dr. Paul Jedlicka (University of Colorado Denver), for providing pathology expertise regarding the pathological diagnoses and grade information in human breast tissue microarrays and Roland Baddeley and Eleanor Martin for their helpful comments.
Footnotes
Address reprint requests to Peter Lloyd Jones, Ph.D., University of Pennsylvania, 1010 Vagelos Research Laboratories, 3340 Smith Walk, Philadelphia, PA 19104-6383. E-mail: jonespl@mail.med.upenn.edu.
Supported by a DOD predoctoral award to P.L.J. and A.T. (BC050720), an EPSRC grant to B.T.V. (GR/S47953/01[P]), and by University of Pennsylvania Academic Enrichment funds to P.L.J.
A.G. and B.T.V. contributed equally to this work.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Bissell MJ, Radisky D. Putting tumours in context. Nat Rev Cancer. 2001;1:46–54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Schedin P, Mitrenga T, McDaniel S, Kaeck M. Mammary ECM composition and function are altered by reproductive state. Mol Carcinog. 2004;41:207–220. doi: 10.1002/mc.20058. [DOI] [PubMed] [Google Scholar]
- Aggeler J, Ward J, Blackie LM, Barcellos-Hoff MH, Streuli CH, Bissell MJ. Cytodifferentiation of mouse mammary epithelial cells cultured on a reconstituted basement membrane reveals striking similarities to development in vivo, J Cell Sci. 1991;99 (Pt 2):407–417. doi: 10.1242/jcs.99.2.407. [DOI] [PubMed] [Google Scholar]
- Weaver VM, Petersen OW, Wang F, Larabell CA, Briand P, Damsky C, Bissell MJ. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel SM, Rumer KK, Biroc SL, Metz RP, Singh M, Porter W, Schedin P. Remodeling of the mammary microenvironment after lactation promotes breast tumor cell metastasis. Am J Pathol. 2006;168:608–620. doi: 10.2353/ajpath.2006.050677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer. 2005;5:675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- Debnath J, Mills KR, Collins NL, Reginato MJ, Muthuswamy SK, Brugge JS. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell. 2002;111:29–40. doi: 10.1016/s0092-8674(02)01001-2. [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- Jones PL. Extracellular matrix and tenascin-C in pathogenesis of breast cancer. Lancet. 2001;357:1992–1994. doi: 10.1016/S0140-6736(00)05133-3. [DOI] [PubMed] [Google Scholar]
- Welm AL, Kim S, Welm BE, Bishop JM. MET and MYC cooperate in mammary tumorigenesis, Proc Natl Acad Sci USA. 2005;102:4324–4329. doi: 10.1073/pnas.0500470102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PL, Chapados R, Baldwin HS, Raff GW, Vitvitsky EV, Spray TL, Gaynor JW. Altered hemodynamics controls matrix metalloproteinase activity and tenascin-C expression in neonatal pig lung. Am J Physiol Lung Cell Mol Physiol. 2002;282:L26–L35. doi: 10.1152/ajplung.00171.2001. [DOI] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Li ML, Aggeler J, Farson DA, Hatier C, Hassell J, Bissell MJ. Influence of a reconstituted basement membrane and its components on casein gene expression and secretion in mouse mammary epithelial cells, Proc Natl Acad Sci USA. 1987;84:136–140. doi: 10.1073/pnas.84.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Gonzalez R, Barcellos-Hoff MH, Ortiz-de-Solorzano C. Quantitative image analysis in mammary gland biology. J Mammary Gland Biol Neoplasia. 2004;9:343–359. doi: 10.1007/s10911-004-1405-9. [DOI] [PubMed] [Google Scholar]
- McInerney T, Terzopoulos D. Deformable models in medical image analysis: a survey. Med Image Anal. 1996;1:91–108. doi: 10.1016/s1361-8415(96)80007-7. [DOI] [PubMed] [Google Scholar]
- Kass M, Witkin A, Terzopoulos D. Snakes: active contour models. Int J Comp Vision. 1988;1:321–331. [Google Scholar]
- Takanashi I, Muraki S, Doi A, Kaufman A. San Jose CA: SPIE-Int. Soc. Opt. Eng,; Three-dimensional active net for volume extraction. 1998:p. 184–193. [Google Scholar]
- Cohen LD, Cohen I. Finite-element methods for active contour models and balloons for 2-D and 3-D images. IEEE Transactions on Pattern Analysis and Machine Intelligence. 1993;15:1131–1147. [Google Scholar]
- Perrin DP, Smith CE. Kauai HI: IEEE Comput. Soc,; Rethinking classical internal forces for active contour models. 2001:p. 615–620. [Google Scholar]
- Jones FS, Jones PL. The tenascin family of ECM glycoproteins: structure, function, and regulation during embryonic development and tissue remodeling. Dev Dyn. 2000;218:235–259. doi: 10.1002/(SICI)1097-0177(200006)218:2<235::AID-DVDY2>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Reginato MJ, Mills KR, Becker EB, Lynch DK, Bonni A, Muthuswamy SK, Brugge JS. Bim regulation of lumen formation in cultured mammary epithelial acini is targeted by oncogenes. Mol Cell Biol. 2005;25:4591–4601. doi: 10.1128/MCB.25.11.4591-4601.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen OW, Ronnov-Jessen L, Howlett AR, Bissell MJ. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells, Proc Natl Acad Sci USA. 1992;89:9064–9068. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell MJ, Rizki A, Mian IS. Tissue architecture: the ultimate regulator of breast epithelial function. Curr Opin Cell Biol. 2003;15:753–762. doi: 10.1016/j.ceb.2003.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B, Tibshirani R. New York: Chapman & Hall,; An Introduction to the Bootstrap. 1993 [Google Scholar]
- Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Met, metastasis, motility and more. Nat Rev Mol Cell Biol. 2003;4:915–925. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- Orend G, Chiquet-Ehrismann R. Tenascin-C induced signaling in cancer. Cancer Lett. 2006;244:143–163. doi: 10.1016/j.canlet.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Boudreau N, Werb Z, Bissell MJA-b. Suppression of apoptosis by basement membrane requires three-dimensional tissue organization and withdrawal from the cell cycle, Proc Natl Acad Sci USA. 1996;93:3509–3513. doi: 10.1073/pnas.93.8.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremble P, Chiquet-Ehrismann R, Werb Z. The extracellular matrix ligands fibronectin and tenascin collaborate in regulating collagenase gene expression in fibroblasts. Mol Biol Cell. 1994;5:439–453. doi: 10.1091/mbc.5.4.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Hiraiwa N, Hashimoto H, Yamazaki Y, Kusakabe M. Tenascin-C regulates angiogenesis in tumor through the regulation of vascular endothelial growth factor expression. Int J Cancer. 2004;108:31–40. doi: 10.1002/ijc.11509. [DOI] [PubMed] [Google Scholar]
- Ghoussoub RA, Dillon DA, D'Aquila T, Rimm EB, Fearon ER, Rimm DL. Expression of c-met is a strong independent prognostic factor in breast carcinoma. Cancer. 1998;82:1513–1520. doi: 10.1002/(sici)1097-0142(19980415)82:8<1513::aid-cncr13>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Ioachim E, Charchanti A, Briasoulis E, Karavasilis V, Tsanou H, Arvanitis DL, Agnantis NJ, Pavlidis N. Immunohistochemical expression of extracellular matrix components tenascin, fibronectin, collagen type IV and laminin in breast cancer: their prognostic value and role in tumour invasion and progression. Eur J Cancer. 2002;38:2362–2370. doi: 10.1016/s0959-8049(02)00210-1. [DOI] [PubMed] [Google Scholar]
- Zukiel R, Nowak S, Wyszko E, Rolle K, Gawronska I, Barciszewska MZ, Barciszewski J. Suppression of human brain tumor with interference RNA specific for tenascin-C. Cancer Biol Ther. 2006;5:1002–1007. doi: 10.4161/cbt.5.8.2886. [DOI] [PubMed] [Google Scholar]
- Reardon DA, Akabani G, Coleman RE, Friedman AH, Friedman HS, Herndon JE, 2nd, McLendon RE, Pegram CN, Provenzale JM, Quinn JA, Rich JN, Vredenburgh JJ, Desjardins A, Gururangan S, Badruddoja M, Dowell JM, Wong TZ, Zhao XG, Zalutsky MR, Bigner DD. Salvage radioimmunotherapy with murine iodine-131-labeled antitenascin monoclonal antibody 81C6 for patients with recurrent primary and metastatic malignant brain tumors: phase II study results. J Clin Oncol. 2006;24:115–122. doi: 10.1200/JCO.2005.03.4082. [DOI] [PubMed] [Google Scholar]
- Hicke BJ, Stephens AW, Gould T, Chang YF, Lynott CK, Heil J, Borkowski S, Hilger CS, Cook G, Warren S, Schmidt PG. Tumor targeting by an aptamer. J Nucl Med. 2006;47:668–678. [PubMed] [Google Scholar]
- Martin TA, Parr C, Davies G, Watkins G, Lane J, Matsumoto K, Nakamura T, Mansel RE, Jiang WG. Growth and angiogenesis of human breast cancer in a nude mouse tumour model is reduced by NK4, a HGF/SF antagonist. Carcinogenesis. 2003;24:1317–1323. doi: 10.1093/carcin/bgg072. [DOI] [PubMed] [Google Scholar]
- Jiang WG, Grimshaw D, Martin TA, Davies G, Parr C, Watkins G, Lane J, Abounader R, Laterra J, Mansel RE. Reduction of stromal fibroblast-induced mammary tumor growth, by retroviral ribozyme transgenes to hepatocyte growth factor/scatter factor and its receptor, c-MET. Clin Cancer Res. 2003;9:4274–4281. [PubMed] [Google Scholar]
- McKean DM, Sisbarro L, Ilic D, Kaplan-Alburquerque N, Nemenoff R, Weiser-Evans M, Kern MJ, Jones PL. FAK induces expression of Prx1 to promote tenascin-C-dependent fibroblast migration. J Cell Biol. 2003;161:393–402. doi: 10.1083/jcb.jcb.200302126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilic D, Furuta Y, Kanazawa S, Takeda N, Sobue K, Nakatsuji N, Nomura S, Fujimoto J, Okada M, Yamamoto T. Reduced cell motility and enhanced focal adhesion contact formation in cells from FAK-deficient mice. Nature. 1995;377:539–544. doi: 10.1038/377539a0. [DOI] [PubMed] [Google Scholar]
- Matsuda A, Yoshiki A, Tagawa Y, Matsuda H, Kusakabe M. Corneal wound healing in tenascin knockout mouse. Invest Ophthalmol Vis Sci. 1999;40:1071–1080. [PubMed] [Google Scholar]
- Tucker RP. Abnormal neural crest cell migration after the in vivo knockdown of tenascin-C expression with morpholino antisense oligonucleotides. Dev Dyn. 2001;222:115–119. doi: 10.1002/dvdy.1171. [DOI] [PubMed] [Google Scholar]
- Murphy-Ullrich JE, Lightner VA, Aukhil I, Yan YZ, Erickson HP, Hook M. Focal adhesion integrity is downregulated by the alternatively spliced domain of human tenascin. J Cell Biol. 1991;115:1127–1136. doi: 10.1083/jcb.115.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotz MM, Burdsal CA, Erickson HP, McClay DR. Cell adhesion to fibronectin and tenascin: quantitative measurements of initial binding and subsequent strengthening response. J Cell Biol. 1989;109:1795–1805. doi: 10.1083/jcb.109.4.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koukoulis GK, Howeedy AA, Korhonen M, Virtanen I, Gould VE. Distribution of tenascin, cellular fibronectins and integrins in the normal, hyperplastic and neoplastic breast. J Submicrosc Cytol Pathol. 1993;25:285–295. [PubMed] [Google Scholar]
- Sarkar S, Nuttall RK, Liu S, Edwards DR, Yong VW. Tenascin-C stimulates glioma cell invasion through matrix metalloproteinase-12. Cancer Res. 2006;66:11771–11780. doi: 10.1158/0008-5472.CAN-05-0470. [DOI] [PubMed] [Google Scholar]
- Werb Z, Ashkenas J, MacAuley A, Wiesen JF. Extracellular matrix remodeling as a regulator of stromal-epithelial interactions during mammary gland development, involution and carcinogenesis. Braz J Med Biol Res. 1996;29:1087–1097. [PubMed] [Google Scholar]
- Hotary K, Li XY, Allen E, Stevens SL, Weiss SJ. A cancer cell metalloprotease triad regulates the basement membrane transmigration program. Genes Dev. 2006;20:2673–2686. doi: 10.1101/gad.1451806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy MJ. Urokinase plasminogen activator and its inhibitor. PAI-1, as prognostic markers in breast cancer: from pilot to level 1 evidence studies, Clin Chem. 2002;48:1194–1197. [PubMed] [Google Scholar]
- Jahkola T, Toivonen T, von Smitten K, Virtanen I, Wasenius VM, Blomqvist C. Cathepsin-D, urokinase plasminogen activator and type-1 plasminogen activator inhibitor in early breast cancer: an immunohistochemical study of prognostic value and relations to tenascin-C and other factors. Br J Cancer. 1999;80:167–174. doi: 10.1038/sj.bjc.6690336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz M, Hansen T, Richter P, Borsi L, Bohmer FD, Hyckel P, Schleier P, Katenkamp D, Zardi L, Kosmehl H, Berndt A. Complex formation of the laminin-5 gamma2 chain and large unspliced tenascin-C in oral squamous cell carcinoma in vitro and in situ: implications for sequential modulation of extracellular matrix in the invasive tumor front. Histochem Cell Biol. 2006;126:125–131. doi: 10.1007/s00418-005-0126-5. [DOI] [PubMed] [Google Scholar]
- Niranjan B, Buluwela L, Yant J, Perusinghe N, Atherton A, Phippard D, Dale T, Gusterson B, Kamalati T. HGF/SF: a potent cytokine for mammary growth, morphogenesis and development. Development. 1995;121:2897–2908. doi: 10.1242/dev.121.9.2897. [DOI] [PubMed] [Google Scholar]
- Peruzzi B, Bottaro DP. Targeting the c-Met signaling pathway in cancer. Clin Cancer Res. 2006;12:3657–3660. doi: 10.1158/1078-0432.CCR-06-0818. [DOI] [PubMed] [Google Scholar]
- Beviglia L, Matsumoto K, Lin CS, Ziober BL, Kramer RH. Expression of the c-Met/HGF receptor in human breast carcinoma: correlation with tumor progression. Int J Cancer. 1997;74:301–309. doi: 10.1002/(sici)1097-0215(19970620)74:3<301::aid-ijc12>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Lengyel E, Prechtel D, Resau JH, Gauger K, Welk A, Lindemann K, Salanti G, Richter T, Knudsen B, Vande Woude GF, Harbeck N. C-Met overexpression in node-positive breast cancer identifies patients with poor clinical outcome independent of Her2/neu. Int J Cancer. 2005;113:678–682. doi: 10.1002/ijc.20598. [DOI] [PubMed] [Google Scholar]
- Soriano JV, Pepper MS, Nakamura T, Orci L, Montesano R. Hepatocyte growth factor stimulates extensive development of branching duct-like structures by cloned mammary gland epithelial cells. J Cell Sci. 1995;108 (Pt 2):413–430. doi: 10.1242/jcs.108.2.413. [DOI] [PubMed] [Google Scholar]
- Osin PP, Anbazhagan R, Bartkova J, Nathan B, Gusterson BA. Breast development gives insights into breast disease. Histopathology. 1998;33:275–283. doi: 10.1046/j.1365-2559.1998.00479.x. [DOI] [PubMed] [Google Scholar]
- Wrobel CN, Debnath J, Lin E, Beausoleil S, Roussel MF, Brugge JS. Autocrine CSF-1R activation promotes Src-dependent disruption of mammary epithelial architecture. J Cell Biol. 2004;165:263–273. doi: 10.1083/jcb.200309102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wever O, Nguyen QD, Van Hoorde L, Bracke M, Bruyneel E, Gespach C, Mareel M. Tenascin-C and SF/HGF produced by myofibroblasts in vitro provide convergent pro-invasive signals to human colon cancer cells through RhoA and Rac. FASEB J. 2004;18:1016–1018. doi: 10.1096/fj.03-1110fje. [DOI] [PubMed] [Google Scholar]
- Jones PL, Crack J, Rabinovitch M. Regulation of tenascin-C, a vascular smooth muscle cell survival factor that interacts with the alpha v beta 3 integrin to promote epidermal growth factor receptor phosphorylation and growth. J Cell Biol. 1997;139:279–293. doi: 10.1083/jcb.139.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom JD, Westermark B, Heldin NE. Epidermal growth factor receptor signaling activates met in human anaplastic thyroid carcinoma cells. Exp Cell Res. 2000;259:293–299. doi: 10.1006/excr.2000.4967. [DOI] [PubMed] [Google Scholar]