Abstract
Fascin is over-expressed in esophageal squamous cell carcinoma (ESCC) and involved in the proliferation and invasiveness of ESCC cells. In this study, we retrospectively examined the expression of fascin in ESCC samples by immunohistochemistry and revealed that overexpression of fascin was related to poor patient survival. RNAi-mediated knockdown of fascin in ESCC cells significantly inhibited cell proliferation and invasiveness, whereas forced expression of fascin in immortalized esophageal epithelial cells accelerated cell proliferation and invasiveness. To explore the underlying mechanism, cDNA microarray was performed to identify the differential gene expression profiles between a fascin-depleted cell line by RNAi and the corresponding control ESCC cells. Results showed that 296 genes were differentially expressed on fascin depletion. In this study, we focused on two down-regulated genes: CYR61 and CTGF. We found that restored expression of either CYR61 or CTGF led to a recovery of the suppression of cellular proliferation and invasiveness induced by down-regulation of fascin expression; the protein level of CYR61 and CTGF were up-regulated in ESCCs and their expression pattern correlated with fascin overexpression. Finally, analysis of signal transduction revealed that fascin affected the expressions of CYR61 and CTGF through transforming growth factor (TGF)-β pathway. Taken together, we propose that fascin regulates the proliferation and invasiveness of ESCC cells by modulating the expression of CTGF and CYR61 via TGF-β pathway.
Esophageal cancer has the poorest prognosis among the malignant tumors of the digestive tract. Despite advancements in multimodal therapy, the overall 5-year survival rate for patients with esophageal squamous cell carcinoma (ESCC) remains poor.1,2 In an effort to improve prognosis of ESCC, several molecular markers, such as interleukin 6 and matrix metalloproteinase 12, have been identified.3,4 However, the molecular mechanisms underlying esophageal carcinogenesis still remain largely unknown. Fascin, an actin-bundling protein, has recently been implicated in a variety of tumors including ESCC, which promoted our investigation.
Fascin is normally expressed in the mesenchymal tissues and nervous system.5 It is reported to function by forming parallel actin bundles in either lamellipodial or filopodial cell protrusions, which are key cellular structures for environmental guidance and cell migration.6,7 Aberrant fascin expression has been reported in various human carcinomas such as colon cancer and gastric cancer.8,9 Fascin is overexpressed in ESCCs, and its increased expression is associated with a poor prognosis.10 Our previous study reported the overexpression of fascin in the early stage of ESCC progression, which facilitated the proliferation and invasiveness of cancer cells.11,12 However, the molecular basis of fascin function in the progression of ESCC remains largely unknown.
In this study, first, we retrospectively examined the expression of fascin in large amounts of ESCC tissue samples by immunohistochemistry, and showed that overexpression of fascin was related to the poor survival of ESCC patients. RNAi-mediated knockdown of fascin in ESCC cells significantly inhibited cell proliferation and invasiveness, whereas forced expression of fascin in immortalized esophageal epithelial cells accelerated cell proliferation and invasiveness. Then, to explore the mechanism underlying the effects of fascin in ESCCs, we used cDNA microarrays to analyze gene expression profiles in PSC cells (ESCC cells that express high levels of fascin) and PSF8 cells (fascin-depleted ESCC cells).11 Several differentially expressed genes have been identified and categorized based on their biological function. Specially, the proliferation- and invasiveness-related genes were predominantly represented. The data of cDNA array has been validated by real-time RT-PCR, Western blotting, and immunofluorescence analyses. As a result, we focused on two down-regulated genes: CYR61 (Cysteine-rich, angiogenic inducer 61) and CTGF (Connective tissue growth factor), both of which were CCN (CYR61/CTGF/NOV) members.13 Further studies demonstrated that CYR61 and CTGF were directly involved in fascin-mediated proliferation and invasiveness of ESCC cells. Additionally, we also revealed that fascin might affect the expressions of CYR61 and CTGF via TGF-β signaling pathway.
Materials and Methods
Tissue Specimens and Immunohistochemical Staining
Surgically removed tumors embedded in paraffin wax blocks from 198 ESCC cases were retrieved from the archives of the Department of Pathology of the Central Hospital of Shantou City, P.R. China. The cases were received between 1987 and 1997. The cases were selected in this study only if a follow-up was obtained and clinical data were available. Mean age at surgery was 53 years (range 35 to 70), and 138 patients were male and 60 were female. All specimens were fixed in 10% formaldehyde solution, embedded in paraffin blocks, and then cut into 4-μm sections.
Immunohistochemical staining was performed as described.14 The SuperPicTure Polymer Detection kit and the Liquid Substrate kit (Invitrogen, Carlsbad, CA) were used to perform immunohistochemistry according to the manufacturer’s instructions. Each section was independently assessed by two histopathologists without prior knowledge of patient data.
The expression was scored based on intensity and the rate of the positive cells. The intensity was graded as follows: 0, negative; 1, weak; 2, moderate; 3, strong. The rate of positive cells was defined as: 0, <5%; 1, 5% to 25%; 2, 26% to 50%; 3, 51% to 75%; 4, >75%. The final score was calculated by multiplying the intensity grade and the rate of positive cells producing a total range of 0 to 12. For statistical analyses, scores of 0 to 4 were considered −, scores of 5 to 8 were considered +, scores of 9 to 12 were considered ++. + to ++ was defined as positive staining, and ++ was defined as overexpression.
The study was approved by the ethical committee of the Central Hospital of Shantou City and the ethical committee of the Medical College of Shantou University, and written informed consent was obtained from all surgical patients to use resected samples for research.
Cell Lines and Cell Culture
SHEE cells (immortal embryonic esophageal epithelium) and KYSE150 cells (ESCC cell line) were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen) containing 10% fetal calf serum.15,16 The other three human ESCC cell lines, PSF8, PSF10, and PSC, were established in our laboratory.11 These cell lines were derived from EC109 cells, an ESCC cell line with high fascin expression.11,17 The PSF8 and PSF10 cell lines were derived from EC109 cells that were stably transfected with different fascin-siRNAs, and the PSC cell line were derived from EC109 cells stably transfected with empty vector. These cells were maintained in 199 medium (Invitrogen) containing 10% fetal calf serum in the absence or presence of 200 μg/ml G418.
Antibodies and Western Blotting
Total cell lysates were prepared in RIPA buffer, separated by SDS-PAGE and transferred to PVDF membranes (Millipore, Billerica, MA). The membranes were incubated in blocking buffer and then incubated with the indicated antibody. Finally, immunoreactive bands were revealed using luminol reagent (Santa Cruz Biotechnology, DE). Photography and quantitative analyses were done using the FluorChemTMIS-8900 (α Innotech). The following antibodies were used: mouse anti-fascin (DAKO Corporation, Glostrup, Denmark), goat anti-Smad2/3, rabbit anti-p-Smad2/3 (Santa Cruz Biotechnology), mouse anti-β-actin (Sigma, Saint Louis, MO), rabbit anti-CYR61 (Novus, Littleton, CO), mouse anti-CTGF, mouse anti-THBS1 (R&D, Minneapolis, MN), rabbit anti-DSC2 (ARP, Belmont, CA) and rabbit anti-ATF3 (Rockland, Gilbertsville, PA). Detailed information about the antibodies was listed in Table 1.
Table 1.
Antibodies Used in These Studies
| Antibody to | Mono/polyclonal | Company (catalogue number) | Application (dilution) |
|---|---|---|---|
| Fascin | Mouse monoclonal | DAKO Corporation (M3567) | IHC (1:50) |
| Smad2/3 | Goat polyclonal | Santa Cruz Biotechnology (sc-6033) | Western blotting (1:200) |
| p-Smad2/3 | Rabbit polyclonal | Santa Cruz Biotechnology (sc-11769-R) | Western blotting (1:200) |
| b-actin | Mouse monoclonal | Sigma (A5441) | Western blotting (1:2000) |
| CYR61 | Rabbit polyclonal | Novus (NB100-356) | Western blotting (1:200) IHC (1:50) |
| CTGF | Mouse monoclonal | R&D (MAB660) | Western blotting (1:200) IHC (1:50) |
| THBS1 | Mouse monoclonal | R&D (MAB3074) | Fluorescence staining (1:200) |
| DSC2 | Rabbit polyclonal | ARP (03-610120) | Fluorescence staining (1:200) |
| ATF3 | Rabbit polyclonal | Rockland (600-401-493) | Western blotting (1:200) |
| Anti-mouse IgG-HRP | Goat | Santa Cruz Biotechnology (sc-2031) | Western blotting (1:1000) |
| Anti-rabbit IgG-HRP | Goat | Santa Cruz Biotechnology (sc-2030) | Western blotting (1:1000) |
| Anti-rabbit FITC | Goat | KPL (02-15-06) | Fluorescence staining (1:200) |
| Anti-mouse FITC | Goat | KPL (02-18-06) | Fluorescence staining (1:200) |
RNAi-Mediated Knockdown of Fascin in KYSE150 Cells
Two siRNA expressing plasmids containing different siRNA were used.11 KYSE150 cells were transfected with these plasmids using SuperFect reagent (QIAGEN, Germantown, MD). Cells were harvested 48 hours later and used for further analysis.
Expression Fascin in SHEE Cells and Expression of CYR61 and CTGF in the Fascin-Depleted Cells
To generate the fascin expression vector, the open reading frame of human fascin cDNA was cloned into the eukaryotic expression vector pcDNA3.0 (Invitrogen). The pcDNA3/fascin plasmid or empty pcDNA3 vector was transfected into SHEE cells using SuperFect reagent. Cells were harvested 48 hours later and used for further analysis.
For CYR61 and CTGF expression, PSF8 cells were transfected with the expression plasmid (pcDNA3/CYR61 or pcDNA3/CTGF)18,19 and used for real-time RT-PCR analysis, BrdU incorporation assay, and invasiveness assay 48 hours after transfection.
BrdU Incorporation Assay
BrdU incorporation assay was performed by using the 5-Bromo-2′-deoxy-uridine Labeling and Detection Kit (Roche, Penzberg, Germany) according to the manufacturer’s instructions. Slides for immunostaining were counterstained with the nucleophilic dye 4′,6-diamidino-2-phenyl-indole (DAPI, Sigma). Cells were analyzed by microscopy, and the means of cells with BrdU incorporation were calculated.
Cell Invasiveness Assay and Chamber Migration Assay
Invasiveness assay and migration assay were performed as described before.11 For invasiveness, 1 × 105 cells were seeded onto the top chamber of a 24-well matrigel-coated membrane with 8-μm pores (Millipore), and the bottom chamber was filled with 199 medium with 10% fetal calf serum (in the migration assay, cells were directly seeded on an uncoated chamber). The membranes were fixed and stained by Giemsa reagent, and invasiveness and migration were quantified by counting 10 random fields under a light microscope (400×). The mean value was calculated from data obtained from 3 separate chambers.
cDNA Microarray Analysis
Affymetrix GeneChip Human genome U133 plus 2.0 arrays (Affymetrix, Santa Clara, CA) containing 39,000 known genes and expressed sequence tags were used in this study. Total RNA from PSF8 and PSC cells was isolated.11 RNA of three independent samples of each cell line was mixed as a RNA pool and quantified spectrophotometrically. Preparation of double-stranded cDNAs, biotin-labeled cRNA targets, hybridization, washing, staining, subsequent scanning of the hybridized array, and data processing were performed according to the Affymetrix recommendations. The ratio of the geometric means of the expression intensities for each gene fragment was calculated and reported in terms of the fold change (up or down) relative to the control. The differentially expressed genes were identified from a single experiment and further validated in two fascin-depleted cell lines. The data have been deposited in GEO (accession number: GSE11373).
Fluorescence Staining
Fluorescence staining was performed as described.12 Briefly, cells were fixed and treated with 0.2% (v/v) Triton X-100, and subsequently incubated in goat serum. Primary antibody was then added and incubated overnight at 4°C followed by incubation with FITC-labeled secondary antibody for 30 minutes at 37°C. Finally, the cells were mounted and examined using a fluorescence microscope (Olympus, CKX41).
Real-Time RT-PCR
Total cellular RNA was extracted from cells with TRIzol reagent (Invitrogen) and reverse transcribed to cDNA using the PrimeScript TM RT-PCR kit (TaKaRa, Dalian, China). The real-time RT-PCR assay was performed with the Rotor-Gene 6000 system (Corbett Life Science, Sydney, Australia) using SYBR Premix Ex Taq (TaKaRa) according to the manufacturer’s instructions. All reactions were done in triplicate. The absolute levels of mRNAs of genes were normalized to that of GAPDH mRNA. The relative value from the vehicle-treated control group was considered equal to one arbitrary unit. Primers used in the real-time RT-PCR assay were listed in Table 2.
Table 2.
Primer Sequences for Real-Time RT-PCR
| Genes | Gene ID | Primers |
|---|---|---|
| CTGF | NM_001901 | FP: 5′-CCCAAGGACCCAAACCGTG-3′ |
| RP: 5′-CTAATCATAGTTGGGTCTGGGC-3′ | ||
| THBS1 | NM_003246 | FP: 5′-CGATGGCATTTCCTGATGACAA-3′ |
| RP: 5′-GGCACACTGTCATGGTCAAAATC-3′ | ||
| ATF3 | NM_001030287 | FP: 5′-GCTGCAAAGTGCCGAAACAA-3′ |
| RP: 5′-AGTCAACAGCCCATATGCAGGTC-3′ | ||
| DSC2 | NM_004949 | FP: 5′-AGTGTGAGTTTGTTCATCACAGGTC-3′ |
| RP: 5′-CCATGGCCTCACAGCCTTTA-3′ | ||
| CYR61 | NM_001554 | FP: 5′-ATGAATTGATTGCAGTTGGAAA-3′ |
| RP: 5′-TAAAGGGTTGTATAGGATGCGA-3′ | ||
| GAPDH | NM_002046 | FP: 5′-GCACCGTCAAGGCTGAGAAC-3′ |
| RP: 5′-TGGTGAAGACGCCAGTGGA-3′ |
Treatment with r-hTHBS1 and TGF-β1
EC109 and PSF8 cells were seeded into 6-well plates at 2 × 105 cells per well and cultured for 12 hours. The cells were then serum-starved overnight followed by incubation with r-hTHBS1 (200 or 400 ng/ml, R&D, Minneapolis, MN) or TGF-β1 (10 ng/ml, Invitrogen), respectively. Cells were harvested after different times of treatment and used for further analysis.
Statistical Analysis
Comparisons between data sets were performed using the χ2 test and t test when appropriate. The correlation of fascin expression with CYR61 and CTGF expression was addressed by the Kappa test. Survival was assessed by Kaplan–Meier analysis with log-rank score for determining statistical significance. Relative risk was evaluated by the multivariate Cox proportional hazards model. P < 0.05 was used to determine statistical significance. All statistical tests were performed with SPSS statistic software (SPSS® 13.0 by SPSS Inc).
Results
Overexpression of Fascin Was Related to Poor Survival of ESCC Patients
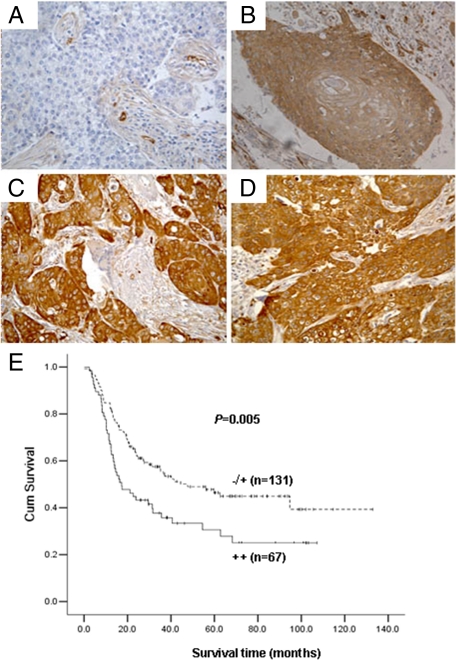
Our previous study has shown that fascin was up-regulated in ESCC.12 Here, we further confirmed the overexpression of fascin (++) in ESCCs by immunohistochemical staining (Figure 1, A, B, C, and D). Kaplan–Meier survival analysis was used to evaluate the association between fascin expression and the overall survival. Result of the statistical analysis demonstrated that overexpression of fascin was associated with decreased overall survival. Patients without fascin overexpression had a 5-year survival of 46.5%, compared with 30.6% for fascin overexpressing patients (P = 0.005; Figure 1E). Moreover, we performed multivariate analysis to assess the independent predictive value of fascin expression for overall survival, and the risk ratio for fascin overexpressing patients was 1.795 (P = 0.004; Table 3).
Figure 1-8375.
Fascin expression in ESCCs and its impact on patient survival. A through D: Immunohistochemical staining of fascin in ESCCs (400×). A: A negative-staining case. B: A case with 1+ staining. C and D: Two cases with 2+ staining. E: Kaplan–Meier estimate of the survival by fascin expression status.
Table 3.
Multivariate Cox Regression Analysis by Fascin Overexpression
| Variate | Risk ratio | 95% confidence interval | P value |
|---|---|---|---|
| Sex | 1.298 | 0.858 to 1.964 | 0.217 |
| Age | 0.887 | 0.574 to 1.371 | 0.590 |
| Histologic grade | 1.258 | 0.718 to 2.201 | 0.422 |
| Primary tumor | 1.263 | 0.722 to 2.211 | 0.413 |
| Regional lymph nodes | 2.289 | 1.487 to 3.524 | 0.000* |
| Distant metastasis | 1.455 | 0.640 to 3.310 | 0.371 |
| Tumor size | 1.483 | 0.954 to 2.304 | 0.08* |
| Location | — | 1.674 to 1.075 | 0.023* |
| Fascin overexpression | 1.795 | 1.201 to 2.684 | 0.004* |
Significant at the 0.05 level.
Effects of Fascin Expression on Cell Proliferation and Invasiveness
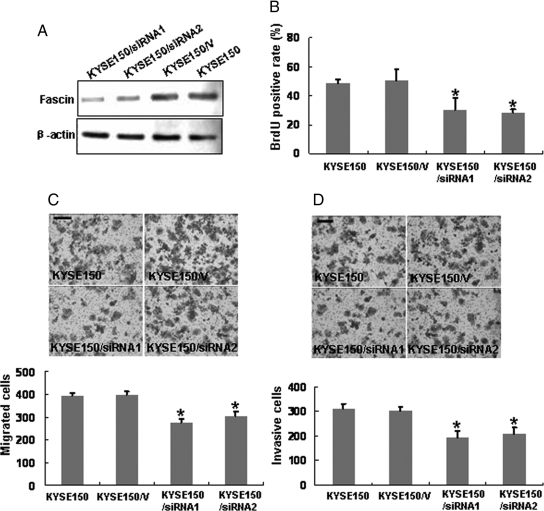
Our previous work has shown that siRNA-mediated knockdown of fascin in EC109 cells (an ESCC cell line) led to decreased cellular proliferation and invasiveness.11 Here we further confirmed these results by RNAi in KYSE150 cells, another ESCC cell line. Western blotting results revealed that two siRNA-treated cell clones showed markedly decrease of fascin expression level compared with the control cells (Figure 2A). The two clones were then used for further studies. The effect of fascin depletion on cell proliferation was evaluated by BrdU incorporation assay. We found that siRNA-treated KYSE150 cells showed less BrdU incorporation than control cells, indicating that depletion of fascin in ESCC cells inhibited cell proliferation (Figure 2B). The motility of siRNA-treated clones was examined by migration assay, and the migration rates of siRNA-treated cells were decreased as compared with the control (Figure 2C). Furthermore, Fascin depletion also dramatically reduced the invasive capability of ESCC cells (Figure 2D).
Figure 2-8375.
Inhibition of cell proliferation and invasiveness by RNAi-mediated knockdown of fascin in KYSE150 cells. KYSE150 cells were transfected with fascin siRNA-expressing plasmid (KYSE150/siRNA) or empty vector (KYSE150/V). A: Western blot analysis was used to show RNAi-mediated knockdown of fascin. Equal loading was ascertained using β-actin as an internal control. B: Cell proliferation rate was determined by BrdU incorporation assay. Modified Boyden chamber assay (C) and invasiveness assay (D) were used to determine the effects of fascin knockdown on cell migration and invasiveness. Migrated and invasive cells were fixed and stained, and representative fields were photographed (bar, 100 μm). For quantification, the cells were counted in 10 random fields under a light microscope (400×). Each experiment was performed in triplicate, and results represent the mean ± SD of three experiments. *P < 0.05.
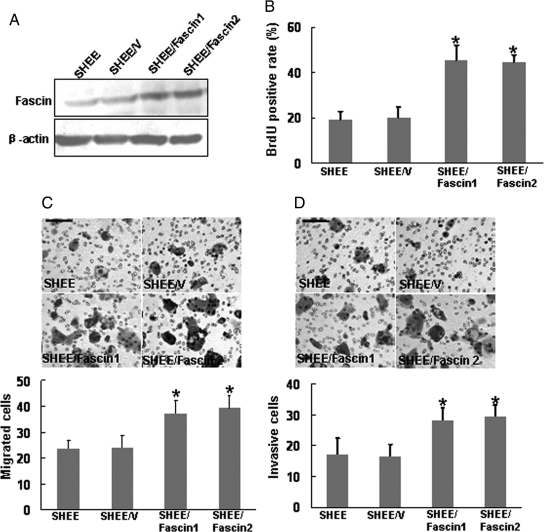
In addition, we overexpressed fascin in SHEE cells, an immortalized esophageal epithelial cell line which had low fascin level,20 and then determined the effect exerted by forced expression of fascin on cell proliferation and invasiveness. We identified two fascin-transfected SHEE clones with high expression of fascin by Western blotting (Figure 3A). Further analysis by performing BrdU incorporation assay, migration assay, and invasiveness assay revealed that forced expression of fascin dramatically accelerated the proliferation, migration, and invasiveness of SHEE cells (Figure 3, B, C, and D).
Figure 3-8375.
Forced expression of fascin in SHEE cells accelerated cell proliferation and invasiveness. SHEE cells were transfected with either the empty pcDNA3 vector (SHEE/V) or the fascin expression vector (SHEE/Fascin). A: Two SHEE/Fascin clones were selected for their high expression of fascin as shown by Western blot analysis. BrdU incorporation assay (B), modified Boyden chamber assay (C), and invasiveness assay (D) were used to determine the effects of fascin on cell proliferation, migration, and invasiveness. Representative fields were photographed (scale bar, 100 μm). *P < 0.05.
Genes Differentially Expressed on Fascin Knockdown
To explore the mechanism through which fascin influences the proliferation and invasiveness of ESCC cells, we selected PSF8 (fascin-depleted) and PSC (fascin high-expressed), two ESCC cell lines recently established in our laboratory for cDNA microarray analysis, and identified genes that were differentially expressed on fascin knockdown. Among 39,000 genes examined, 296 were differentially expressed to a significant degree (≥2-fold difference between PSF8 and PSC samples). Specifically, 78 genes were down-regulated and 218 genes were up-regulated on fascin knockdown. The intensities of gene expression signals revealed that 46 genes showed ≥10-fold difference in expression levels between the PSF8 set and the control set, 41 gene fragments demostrated ≥5-fold to 10-fold difference, and 209 gene fragments had ≥2-fold to 5-fold difference in expression levels between the two cells.
To determine roles of these differentially expressed genes, they were categorized based on biological functions, including cell adhesion, invasiveness, apoptosis and proliferation, transcription, and cell signaling (Table 4).
Table 4.
Representative Cancer-Related Genes with Altered Relative Expression upon Fascin Knockdown
| Probe set | Gene title | Fold change (+, up; −, down*) | UniGene ID |
|---|---|---|---|
| Cell proliferation- and apoptosis-related genes | |||
| 219478_at | WAP four-disulfide core domain 1 (WFDC1) | −2.06 | Hs.36688 |
| 202934_at | Hexokinase 2 (HK2) | +2.00 | Hs.406266 |
| 205302_at | Insulin-like growth factor binding protein 1 (IGFBP1) | +2.03 | Hs.401316 |
| 200632_s_at | N-myc downstream regulated gene 1 (NDRG1) | +2.09 | Hs.372914 |
| 216115_at | Neurofibromin 1 (NF1) | +2.12 | Hs.113874 |
| 213236_at | SAM and SH3 domain containing 1 (SASH1) | +2.21 | Hs.193133 |
| 204237_at | GULP, engulfment adaptor PTB domain containing 1 (GULP1) | +7.00 | Hs.470887 |
| Cell adhesion- and invasiveness-related genes | |||
| 209101_at | Connective tissue growth factor (CTGF) | −3.83 | Hs.410037 |
| 201289_at | Cysteine-rich, angiogenic inducer 61 (CYR61) | −2.00 | Hs.8867 |
| 206295_at | Interleukin 18 (IL-18) | −2.00 | Hs.83077 |
| 201110_s_at | Thrombospondin 1 (THBS1) | −2.00 | Hs.164226 |
| 202672_s_at | Activating transcription factor 3 (ATF3) | +2.00 | Hs.460 |
| 244632_at | Contactin 5 (CNTN5) | +2.00 | Hs.268707 |
| 226817_at | Desmocollin 2 (DSC2) | +2.14 | Hs.95612 |
| 219251_s_at | PTPRF interacting protein, binding protein 1 (PPFIBP1) | +2.77 | Hs.389945 |
| 211488_s_at | Integrin, beta 8 (ITGB8) | +4.79 | Hs.547130 |
| Cell transcription-related genes | |||
| 225753_at | Zinc finger protein 513 (ZNF513) | −10.22 | Hs.515872 |
| 244140_at | Wolf-Hirschhorn syndrome candidate 1 (WHSC1) | −8.45 | Hs.113876 |
| 205966_at | AF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor (TAF13) | −5.86 | Hs.502508 |
| 208989_s_at | F-box and leucine-rich repeat protein 11 (FBXL11) | −5.04 | Hs.124147 |
| 207826_s_at | Inhibitor of DNA binding 3 (ID3) | −2.90 | Hs.76884 |
| 203543_s_at | Kruppel-like factor 9 (KLF9) | +2.50 | Hs.150557 |
| 230332_at | Zinc finger, CCHC domain containing 7 (ZCCHC7) | +2.82 | Hs.371588 |
| 209189_at | V-fos FBJ murine osteosarcoma viral oncogene homolog (FOS) | +3.69 | Hs.25647 |
| 239619_at | Zinc finger protein 395 (ZNF395) | +4.05 | Hs.435535 |
| 1564963_x_at | Zinc finger protein 92 (ZNF92) | +5.30 | Hs.9521 |
| Signal-related genes | |||
| 206002_at | G protein-coupled receptor 64 (GPR64) | −6.64 | Hs.184942 |
| 209288_s_at | CDC42 effector protein 3 (CDC42EP3) | −3.64 | Hs.260024 |
| 213094_at | G protein–coupled receptor 126 (GPR126) | −3.37 | Hs.44197 |
| 218353_at | Regulator of G-protein signaling 5 (RGS5) | +2.92 | Hs.274368 |
| 207446_at | Toll-like receptor 6 (TLR6) | +18.2 | Hs.227105 |
| 1552691_at | ADP-ribosylation factor-like 11 (ARL11) | +25.00 | Hs.348334 |
All of the fold changes, P < 0.05.
Validation of the Microarray Data
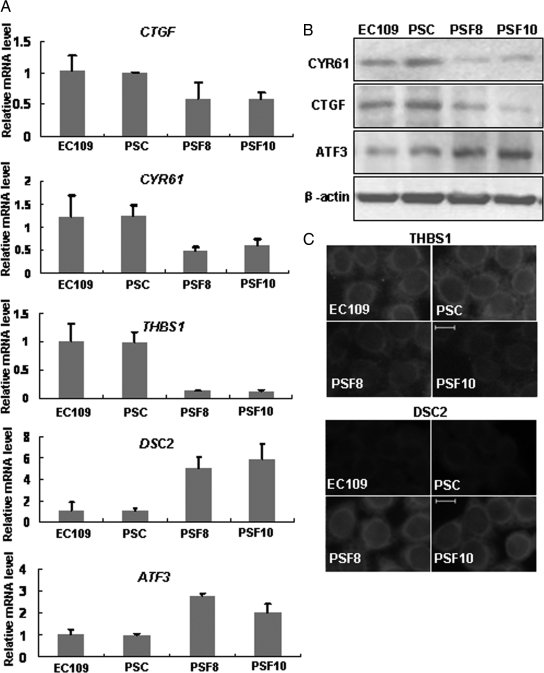
To confirm the microarray data, real-time RT-PCR was used to analyze the expression levels of five genes, including CTGF, CYR61, THBS1, ATF3, and DSC2. The examination were performed in EC109, PSC, PSF8 cells, and PSF10 cells, another fascin-depleted cell line established by our laboratory.11 Consistent with the microarray data, the expression levels of CTGF, CYR61, and THBS1 were down-regulated, whereas ATF3 and DSC2 were up-regulated in PSF8 and PSF10 cells compared with PSC cells and to EC109 control cells, validating the microarray analysis (Figure 4A). Western blotting and fluorescence staining were also used to confirm the gene expression pattern, and the results of these assays corresponded to the data of real-time RT-PCR and microarray analysis (Figure 4, B and C).
Figure 4-8375.
Real-time RT-PCR, Western blotting, and immunofluorescence analysis validate the microarray data. A: Real-time RT-PCR analysis was performed to analyze the expression levels of five genes: CTGF, CYR61, THBS1, ATF3, and DSC2. The expression levels were normalized by the level of GAPDH and represented as fold change over PSC cells. B: Western blot analysis was performed to determine the protein levels of CTGF, CYR61, and ATF3. C: Immunofluorescence analysis was performed to compare THBS1 and DSC2 expression. Representative images show localization of THBS1 and DSC2. Scale bar, 10 μm. EC109 is a human ESCC cell line that expresses high levels of fascin (control). PSC is an EC109 cell line transfected with nonspecific siRNA (control). PSF8 and PSF10 are two different PSC-based fascin-depleted cell lines prepared using RNAi method.
Based on these analyses, we focused on two proliferation- and invasiveness-related genes, CTGF and CYR61 because of the following reasons. First, both CYR61 and CTGF are TGF-β–targeted genes21,22 that were reported to participate in the proliferation and invasiveness of various cancer cells.23,24 Second, both of them are secreted proteins belonging to CCN family.13
Correlation between Fascin Expression and the Protein Levels of CYR61 and CTGF in ESCCs
To explore whether there is a correlation between the expression of fascin and CYR61 or CTGF, the expression patterns of their protein products were determined by immunohistochemical staining of 80 ESCC specimens and 50 normal esophageal mucosas.
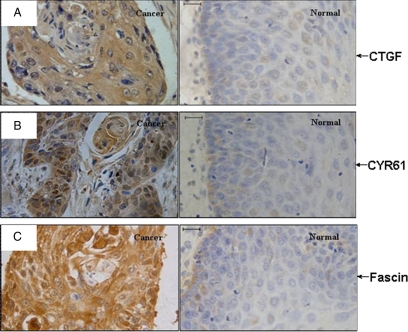
Positive cytoplasmic immunostaining of CTGF and CYR61 protein was observed in both cancerous and normal mucosa of the esophagus. In ESCC samples, most of the cancer cells showed intense immunostaining signals of the two proteins, whereas only negligible or weak positive signals were detected in the normal epithelium of the esophagus (Figure 5A-C). Positive rate (+ to ++) of CTGF and CYR61 proteins was higher in ESCC samples than in normal mucosa (Table 5).
Figure 5-8375.
Increased expressions of CYR61 and CTGF in ESCC tissues. Photomicrographs are shown to compare immunohistochemical staining of (A) CTGF, (B) CYR61, and (C) fascin in normal esophageal epithelium (normal) and ESCC tissues (cancer). Scale bars, 20 μm.
Table 5.
IHC Staining of ESCC Tumors and Normal Esophageal Epithelium Tissues
| Diagnosis | Fascin |
CYR61 |
CTGF |
||||||
|---|---|---|---|---|---|---|---|---|---|
| − | + to ++ | P | − | + to ++ | P | − | + to ++ | P | |
| Normal (n = 50) | 40 | 10 | <0.01 | 37 | 9 | <0.01 | 42 | 8 | <0.01 |
| Carcinoma (n = 80) | 8 | 72 | 37 | 43 | 17 | 63 | |||
We also identified a positive correlation between the protein level of fascin and CTGF (r = 0.557; P < 0.01), or CYR61 (r = 0.376; P < 0.05) in ESCC samples, which was consistent with the microarray data. Furthermore, Kaplan–Meier analysis suggested that overexpression of CYR61 or CTGF was also related to poor survival of patient with ESCC (Data shown in another submitted manuscript, Supplementary Figure S1, see http://ajp.amjpathol.org).
CYR61 and CTGF Were Involved in the Fascin-Mediated Cell Proliferation and Invasiveness of ESCC Cells
The data from clinical studies suggested that CYR61 and CTGF might mediate the function of fascin in ESCC. To further address this issue, we explore the function of the two genes in ESCC cells.
Our previous study reported that CTGF was involved in the tumorigenicity of ESCC cells19; however, the role of CYR61 in ESCC remained unknown. By using RNAi method, we found that CYR61 knockdown led to decreased proliferation and invasiveness of ESCC cells (Supplemental Figure S2, see http://ajp.amjpathol.org).
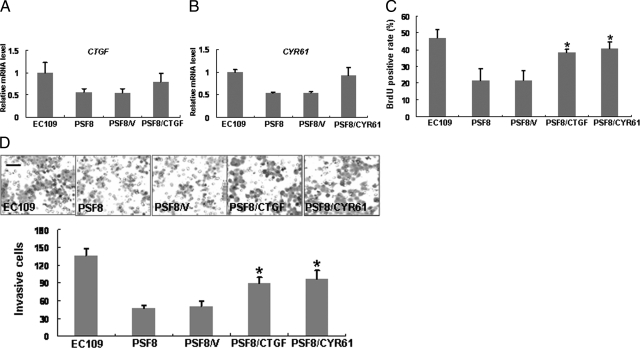
Then, to find whether CYR61 and CTGF were directly involved in fascin-induced alterations in cell proliferation and invasiveness, we transfected PSF8 cells with CYR61-expressing or CTGF-expressing plasmid, respectively. Real-time RT-PCR analysis showed that the expressions of both CTGF and CYR61 were restored in the transfected PSF8 cells (Figure 6, A and B). Moreover, BrdU incorporation assay and invasiveness assay revealed that the elevated expression of CTGF or CYR61 resulted in partial recoveries of cellular proliferation and invasiveness. Cell proliferated rates of CYR61- and CTGF-restored cells recovered by about 67% and 51%, and their invasiveness recovered by about 70% and 53%, respectively (Figure 6C and D).
Figure 6-8375.
Restored expression of CTGF or CYR61 resulted in a recovery of the suppression of cellular proliferation and invasiveness induced by down-regulation of fascin expression. A and B: PSF8 cells (fascin-depleted) were transfected with either the empty pcDNA3 vector (PSF8/V), the CTGF expression vector (PSF8/CTGF), or the CYR61 expression vector (PSF8/CYR61). Restored expression of CTGF (A) or CYR61 (B) was shown by real-time RT-PCR analysis. C and D: Effect of CTGF or CYR61 restored-expression on cell proliferation and invasiveness assay was determined by using BrdU incorporation assay (C) and invasiveness assay (D). Invasive cells were fixed and stained, and representative fields were photographed (Scale bar, 100 μm). *P < 0.05. Cell lines were as described in Figure 4.
Fascin Affected the Expressions of CYR61 and CTGF via TGF-β Pathway
It has been reported that CYR61 and CTGF were target genes of TGF-β.21,22 To examine whether this is the same case in ESCC cells, we treated EC109 cells with TGF-β1, and found that the level of p-Smad2/3, CTGF and CYR61 were up-regulated in a time-dependent manner (Figure 7, A and B). Based on this result, we proposed that fascin might affect the expressions of CTGF and CYR61 via TGF-β pathway. Thus we examined the activation status of this pathway by detecting the phosphorylation of Smad2/3 in fascin-depleted cells by Western blotting and found that the level of p-Smad2/3 was decreased in the fascin-depleted cells (Figure 7C). These results suggested that fascin might affect the expressions of CYR61 and CTGF via TGF-β pathway.
Figure 7-8375.
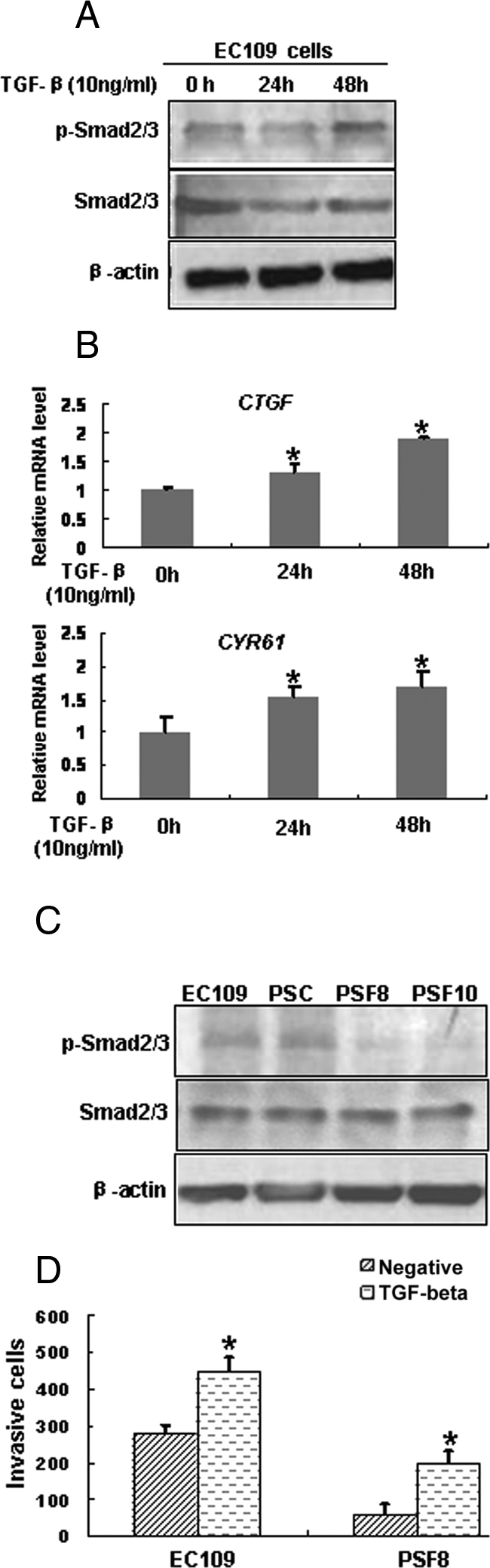
Fascin affected the expressions of CTGF and CYR61 via TGF-β pathway. Induced expressions of p-Smad2/3 (A), CTGF, and CYR61 (B) by addition of exogenous TGF-β1 protein in EC109 cells were addressed by Western blotting or real-time RT-PCR. C: Smad2/3 phosphorylation in whole-cell extracts was determined by Western blotting to evaluate the TGF-β signaling pathway. D: TGF-β1 treatment led to a recovery of the fascin-mediated alteration of cell invasiveness. *P < 0.05. Cell lines were as described in Figure 4.
To assess whether the suppression of TGF-β pathway mediated by fascin down-regulation was responsible for the decreased invasive capability of PSF8 cells, we tested the effects of TGF-β1 on the invasive capability of EC109 cells and PSF8 cells. By invasiveness assay, we found that treatment with TGF-β1 not only significantly facilitated the invasiveness ability of EC109 cells, but also resulted in the recovery of the invasiveness ability of PSF8 cells (Figure 7D). However, the invasiveness of PSF8 cells was still lower than that of EC109 cells, even in the presence of TGF-β1. Thus, we reasoned that fascin-mediated alteration in cell invasiveness was related to activation of TGF-β pathway.
Fascin Altered the Activation of TGF-β Pathway through THBS1
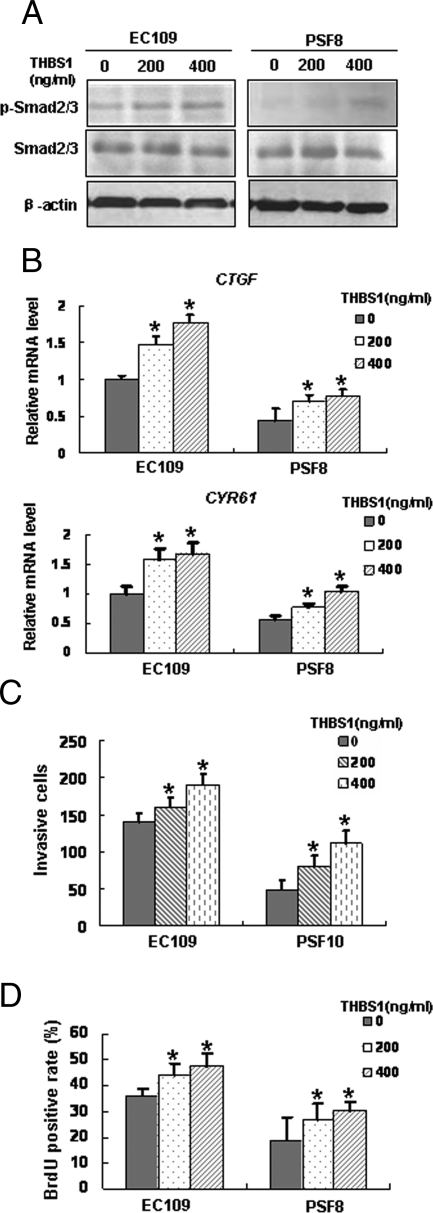
Our microarray data showed that THBS1, an activator of the TGF-β pathway,25 was down-regulated in the fascin-depleted cells (Figure 4, A and C). To address its role in fascin-mediated functions in ESCC cells, EC109 cells and PSF8 cells were treated with exogenous THBS1 protein and the expression levels of p-Smad2/3, CTGF, and CYR61 were examined by Western blotting or real-time RT-PCR. Results showed that on THBS1 treatment, levels of p-Smad2/3, CTGF, and CYR61 were increased in both EC109 cells and PSF8 cells, suggesting that THBS1 might induce the expressions of CTGF and CYR61 through TGF-β pathway (Figure 8, A and B). Furthermore, the results of invasiveness assay and BrdU incorporation assay revealed that THBS1 treatment resulted in increased proliferation and invasiveness of both EC109 cells and PSF8 cells (Figure 8, C and D).
Figure 8-8375.
Impact of fascin on the TGF-β pathway was possibly mediated by the expression of THBS1. Induced expressions of p-Smad2/3 (A), CTGF, and CYR61 (B) by addition of exogenous THBS1 protein were determined by Western blotting or real-time RT-PCR. Proliferation (BrdU incorporation assay, C) and invasiveness (invasiveness assay, D) of EC109 and PSF8 cells were addressed under control conditions or pretreated with THBS1. *P < 0.05. Cell lines were as described in Figure 4.
Discussion
The involvement of fascin in cellular proliferation and invasiveness has been reported in a variety of cancers including ESCC.8,9,10,11,12 In this study, the retrospective analysis of a large amount of ESCC samples further proved that fascin was over-expressed in ESCCs and revealed that over-expression of fascin was related to poor survival. At the cellular level, we showed that RNAi-mediated knockdown of fascin in ESCC cells significantly inhibited cellular proliferation and invasiveness, whereas forced expression of fascin in immortalized esophageal epithelial cells accelerated cellular proliferation and invasiveness. Combined with our previous findings of fascin,11,12 all of the results strongly suggested that fascin was involved in the development and progression of ESCC.
It has been reported that the roles of fascin in carcinoma cells are mediated either by increasing the formation or functional effectiveness of cell protrusions, or by the binding of phosphorylated fascin to active Protein Kinase C (PKC).26 However, the mechanism underlying the oncogenic effect of fascin has not been fully understood. Here, we demonstrated that on fascin knockdown, the expression of several cell invasiveness–related genes and cell proliferation–related genes was altered, implicating they might mediate the effect of fascin in cell proliferation and invasiveness. Further analysis suggested that fascin depletion led to down-regulation of certain genes that stimulate/promote cellular invasiveness and proliferation, such as CTGF, CYR61, and THBS1,23,27 as well as up-regulation of certain genes that inhibit invasiveness or proliferation, such as ATF3 and DSC2.28,29 Moreover, expression of certain transcription factors and signal molecules such as ID3 and FOS were also altered in the fascin-depleted cells, indicating that fascin might affect a complex network that contributes to tumorigenisis and progression.
Among these differentially expressed genes, CTGF and CYR61 caught our attention. Both CTGF and CYR61 belong to the CCN family.13,23 Members of this family stimulate mitosis, adhesion, extracellular matrix production, growth, and migration in multiple cell types through their interaction with protein partners distributed in extracellular matrix and cell surface.22,23 Dysregulation of this family has been shown in a number of cancers, such as breast and ovarian cancers.30,31 However, genes in this family appear to have different or even opposing functions in different cancers.32 Previous studies showed that CTGF was overexpressed in ESCCs and involved in the tumorigenicity of ESCC cells.19 Our results confirmed up-regulation of CTGF in ESCCs, and we also found that CYR61 was overexpressed in ESCCs and participated in the regulation of proliferation and invasiveness. Moreover, restored expression of CYR61 or CTGF led to a partial recovery of the suppression of cellular proliferation and invasiveness induced by down-regulation of fascin expression. Interestingly, overexpression of CTGF or CYR61 was also related to poor survival of patients with ESCC (data not shown). These results suggested that CTGF and CYR61 might be involved in the function of fascin in ESCC.
It was reported that CTGF and CYR61 were target genes of both TGF-β pathway and Wnt/β-catenin pathway.33 We reported here that TGF-β, rather than Wnt pathway (data not shown), played a predominant role in the fascin-mediated alterations of CTGF and CYR61. Despite an established role in suppressing tumor formation, the TGF-β pathway can also stimulate tumor invasion.34 In ESCC, TGF-β pathway was disturbed and the expression of TGF-β1 was up-regulated, which might be responsible for the high invasive capability of ESCC cells.35,36,37 Our results showed that the activity of TGF-β pathway was decreased on fascin depletion. CTGF and CYR61, downstream effectors of TGF-β action, were also down-regulated, and might influence the composition of microenvironment and cell-matrix interactions, which contributed to the alterations of cellular proliferation and invasiveness.
THBS1 is an extracellular matrix glycoprotein, which is thought to be a potential regulator of tumor growth and metastasis.38 THBS1 is capable of activating TGF-β signal pathway, leading to transcription of Cyr61 and CTGF.39,40 Methylation-induced silencing of THBS1 expression correlated with impaired TGF-β signaling as indicated by decreased Smad2 phosphorylation and nuclear localization.41 Specifically, we reported here down-regulation of THBS1 in the fascin-depleted cells. We also suggested that effects of fascin down-expression on the TGF-β pathway were possibly mediated by the decreased expression of THBS1. Furthermore, we observed that expression of THBS1 is up-regulated in cancerous tissues of ESCC and its expression pattern correlated with fascin overexpression, consistent with a potential involvement in the function of fascin (data not shown). Previous studies had shown that fascin was a critical component that was required for actin cytoskeletal organization and cell migration responses to THBS1.42,43 However, how fascin affected the expression of THBS1 needed further study. Other than the regulation of THBS1 expression, fascin might influence TGF-β pathway by interaction with certain key component, and down-regulation of fascin altered the activation status of TGF-β pathway through its decreased interaction with this molecule. Further study to identify the hypothetical interaction is required.
In summary, our study reported fascin was up-regulated in ESCCs and overexpression of fascin was related to poor survival. We also found that fascin might affect the expressions of CYR61 and CTGF through TGF-β pathway, and CYR61 and CTGF were directly involved in the fascin-mediated alterations of cellular proliferation and invasiveness. Taken together, we propose that fascin regulates the proliferation and invasiveness of ESCC cells by affecting the expressions of CYR61 and CTGF via TGF-β pathway.
Footnotes
Address reprint requests to En-Min Li, Ph.D., Department of Biochemistry and Molecular Biology, Medical College of Shantou University, Shantou 515041, China; or Li-Yan Xu, Ph.D., Institute of Oncologic Pathology, the Key Immunopathology Laboratory of Guangdong Province, Medical College of Shantou University, Shantou 515041, China; or Prof. Dong Xie, Ph.D., Laboratory of Molecular Oncology, Institute for Nutritional Sciences, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, 294 Tai Yuan Road, Shanghai 200031, China. E-mail: nmli@stu.edu.cn, liyanxu1130@yahoo.com.cn, or dxie@sibs.ac.cn.
Supported by grants from the National High Technology Research and Development Program of China (No. 2006AA02A403), National Natural Science Foundation of China (No. 30672376, No. 30772485), National Natural Science Foundation of China-Guangdong Joint Fund (No. U0932001), National Natural Science Funds for Distinguished Young Scholar (No. 30725010), and Key Program of National Natural Science Foundation of China (No.30930023).
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Mariette C, Balon JM, Piessen G, Fabre S, Van Seuningen I, Triboulet JP. Pattern of recurrence following complete resection of esophageal carcinoma and factors predictive of recurrent disease. Cancer. 2003;97:1616–1623. doi: 10.1002/cncr.11228. [DOI] [PubMed] [Google Scholar]
- Law SY, Fok M, Cheng SW, Wong J. A comparison of outcome after resection for squamous cell carcinomas and adenocarcinomas of the esophagus and cardia. Surg Gynecol Obstet. 1992;175:107–112. [PubMed] [Google Scholar]
- Wang LS, Chow KC, Wu CW. Expression and up-regulation of interleukin-6 in oesophageal carcinoma cells by n-sodium butyrate. Br J Cancer. 1999;80:1617–1622. doi: 10.1038/sj.bjc.6690571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Shimada Y, Gorrin-Rivas MJ, Itami A, Li Z, Hong T, Maeda M, Komoto I, Kawabe A, Kaganoi J, Imamura M. Clinicopathological significance of human macrophage metalloelastase expression in esophageal squamous cell carcinoma. Oncology. 2002;63:378–384. doi: 10.1159/000066231. [DOI] [PubMed] [Google Scholar]
- Adams JC. Formation of stable microspikes containing actin and the 55 kDa actin bundling protein, fascin, is a consequence of cell adhesion to thrombospondin-1: implications for the anti-adhesive activities of thrombospondin-1. J Cell Sci. 1995;108:1977–1990. doi: 10.1242/jcs.108.5.1977. [DOI] [PubMed] [Google Scholar]
- Kureishy N, Sapountzi V, Prag S, Anilkumar N, Adams JC. Fascins, and their roles in cell structure and function. Bioessays. 2002;24:350–361. doi: 10.1002/bies.10070. [DOI] [PubMed] [Google Scholar]
- Adams JC. Roles of fascin in cell adhesion and motility. Curr Opin Cell Biol. 2004;16:590–596. doi: 10.1016/j.ceb.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Jawhari AU, Buda A, Jenkins M, Shehzad K, Sarraf C, Noda M, Farthing MJ, Pignatelli M, Adams JC. Fascin, an actin-bundling protein, modulates colonic epithelial cell invasiveness and differentiation in vitro. Am J Pathol. 2003;162:69–80. doi: 10.1016/S0002-9440(10)63799-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Shimada Y, Kawamura J, Yamasaki S, Imamura M. The prognostic relevance of fascin expression in human gastric carcinoma. Oncology. 2004;67:262–270. doi: 10.1159/000081327. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Ito T, Inoue H, Okumura T, Tanaka E, Tsunoda S, Higashiyama M, Watanabe G, Imamura M, Shimada Y. Prognostic significance of fascin overexpression in human esophageal squamous cell carcinoma. Clin Cancer Res. 2005;11:2597–2605. doi: 10.1158/1078-0432.CCR-04-1378. [DOI] [PubMed] [Google Scholar]
- Xie JJ, Xu LY, Zhang HH, Cai WJ, Mai RQ, Xie YM, Yang ZM, Niu YD, Shen ZY, Li EM. Role of fascin in the proliferation and invasiveness of esophageal carcinoma cells. Biochem Biophys Res Commun. 2005;337:355–362. doi: 10.1016/j.bbrc.2005.09.055. [DOI] [PubMed] [Google Scholar]
- Zhang H, Xu L, Xiao D, Xie J, Zeng H, Cai W, Niu Y, Yang Z, Shen Z, Li E. Fascin is a potential biomarker for early-stage oesophageal squamous cell carcinoma. J Clin Pathol. 2006;59:958–964. doi: 10.1136/jcp.2005.032730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigstock DR. The CCN family: a new stimulus package. J Endocrinol. 2003;178:169–175. doi: 10.1677/joe.0.1780169. [DOI] [PubMed] [Google Scholar]
- Zeng H, Xu L, Xiao D, Zhang H, Wu X, Zheng R, Li Q, Niu Y, Shen Z, Li E. Altered expression of ezrin in esophageal squamous cell carcinoma. J Histochem Cytochem. 2006;54:889–896. doi: 10.1369/jhc.5A6881.2006. [DOI] [PubMed] [Google Scholar]
- Shen ZY, Xu LY, Li EM, Cai WJ, Shen J, Chen MH, Cen S, Tsao SW, Zeng Y. The multistage process of carcinogenesis in human esophageal epithelial cells induced by human papillomavirus. Oncol Rep. 2004;11:647–654. [PubMed] [Google Scholar]
- Shimada Y, Imamura M, Wagata T, Yamaguchi N, Tobe T. Characterization of 21 newly established esophageal cancer cell lines. Cancer. 1992;69:277–284. doi: 10.1002/1097-0142(19920115)69:2<277::aid-cncr2820690202>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Department of Cell Biology of CICAMS Establishment of a cell line from human esophageal carcinoma. Chin Med J (Engl) 1976;2:357–364. [PubMed] [Google Scholar]
- Tong X, Xie D, O'Kelly J, Miller CW, Muller-Tidow C, Koeffler HP. Cyr61, a member of CCN family, is a tumor suppressor in non-small cell lung cancer. J Biol Chem. 2001;276:47709–47714. doi: 10.1074/jbc.M107878200. [DOI] [PubMed] [Google Scholar]
- Deng YZ, Chen PP, Wang Y, Yin D, Koeffler HP, Li B, Tong XJ, Xie D. Connective tissue growth factor is overexpressed in esophageal squamous cell carcinoma and promotes tumorigenicity through beta-catenin-T-cell factor/Lef signaling. J Biol Chem. 2007;282:36571–36581. doi: 10.1074/jbc.M704141200. [DOI] [PubMed] [Google Scholar]
- Rong J, Xu LY, Cai WJ, Xiong XD, Li JT, Fang WK, Shen ZY, Li EM. Expression of fascin 1 gene in the process of the immortalized esophageal carcinoma carcinogenesis. Ai Zheng. 2004;23:243–248. [PubMed] [Google Scholar]
- Chen Y, Du XY. Functional properties and intracellular signaling of CCN1/Cyr61. J Cell Biochem. 2007;100:1337–4135. doi: 10.1002/jcb.21194. [DOI] [PubMed] [Google Scholar]
- Grotendorst GR, Okochi H, Hayashi N. A novel transforming growth factor beta response element controls the expression of the connective tissue growth factor gene. Cell Growth Differ. 1996;7:469–480. [PubMed] [Google Scholar]
- Leask A, Abraham DJ. All in the CCN family: essential matricellular signaling modulators emerge from the bunker. J Cell Sci. 2006;119:4803–4810. doi: 10.1242/jcs.03270. [DOI] [PubMed] [Google Scholar]
- Leivonen SK, Kähäri VM. Transforming growth factor-beta signaling in cancer invasion and metastasis. Int J Cancer. 2007;121:2119–2124. doi: 10.1002/ijc.23113. [DOI] [PubMed] [Google Scholar]
- Breitkopf K, Sawitza I, Westhoff JH, Wickert L, Dooley S, Gressner AM. Thrombospondin 1 acts as a strong promoter of transforming growth factor beta effects via two distinct mechanisms in hepatic stellate cells. Gut. 2005;54:673–681. doi: 10.1136/gut.2004.042911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Skacel M, Adams JC. Roles of fascin in human carcinoma motility and signaling: prospects for a novel biomarker? Int J Biochem Cell Biol. 2005;37:1787–1804. doi: 10.1016/j.biocel.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Jo WS, Mizukami Y, Duerr EM, Zukerberg LR, Chung DC. Wnt signaling can repress thrombospondin-1 expression in colonic tumorigenesis. Cancer Biol Ther. 2005;4:1361–1366. doi: 10.4161/cbt.4.12.2201. [DOI] [PubMed] [Google Scholar]
- Bottone FG, Jr, Moon Y, Kim JS, Alston-Mills B, Ishibashi M, Eling TE. The anti-invasive activity of cyclooxygenase inhibitors is regulated by the transcription factor ATF3. Mol Cancer Ther. 2005;4:693–703. doi: 10.1158/1535-7163.MCT-04-0337. [DOI] [PubMed] [Google Scholar]
- Khan K, Hardy R, Haq A, Ogunbiyi O, Morton D, Chidgey M. Desmocollin switching in colorectal cancer. Br J Cancer. 2006;95:1367–1370. doi: 10.1038/sj.bjc.6603453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D, Nakachi K, Wang H, Elashoff R, Koeffler HP. Elevated levels of connective tissue growth factor WISP-1, and CYR61 in primary breast cancers associated with more advanced features. Cancer Res. 2001;61:8917–8923. [PubMed] [Google Scholar]
- Gery S, Xie D, Yin D, Gabra H, Miller C, Wang H, Scott D, Yi WS, Popoviciu ML, Said JW, Koeffler HP. Ovarian carcinomas: cCN genes are aberrantly expressed and CCN1 promotes proliferation of these cells. Clin Cancer Res. 2005;11:7243–7254. doi: 10.1158/1078-0432.CCR-05-0231. [DOI] [PubMed] [Google Scholar]
- Jiang WG, Watkins G, Fodstad O, Douglas-Jones A, Mokbel K, Mansel RE. Differential expression of the CCN family members Cyr61 CTGF and Nov in human breast cancer. Endocr Relat Cancer. 2004;11:781–791. doi: 10.1677/erc.1.00825. [DOI] [PubMed] [Google Scholar]
- Labbé E, Lock L, Letamendia A, Gorska AE, Gryfe R, Gallinger S, Moses HL, Attisano L. Transcriptional cooperation between the transforming growth factor-beta and Wnt pathways in mammary and intestinal tumorigenesis. Cancer Res. 2007;67:75–84. doi: 10.1158/0008-5472.CAN-06-2559. [DOI] [PubMed] [Google Scholar]
- Pardali K, Moustakas A. Actions of TGF-beta as tumor suppressor and pro-metastatic factor in human cancer. Biochim Biophys Acta. 2007;1775:21–62. doi: 10.1016/j.bbcan.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Rees JR, Onwuegbusi BA, Save VE, Alderson D, Fitzgerald RC. In vivo and in vitro evidence for transforming growth factor-beta1-mediated epithelial to mesenchymal transition in esophageal adenocarcinoma. Cancer Res. 2006;66:9583–9590. doi: 10.1158/0008-5472.CAN-06-1842. [DOI] [PubMed] [Google Scholar]
- Von Rahden BH, Stein HJ, Feith M, Pühringer F, Theisen J, Siewert JR, Sarbia M. Overexpression of TGF-beta1 in esophageal (Barrett’s) adenocarcinoma is associated with advanced stage of disease and poor prognosis. Mol Carcinog. 2006;45:786–794. doi: 10.1002/mc.20259. [DOI] [PubMed] [Google Scholar]
- Miller AV, Alvarez SE, Spiegel S, Lebman DA. Sphingosine kinases and sphingosine-1-phosphate are critical for transforming growth factor beta-induced extracellular signal-regulated kinase 1 and 2 activation and promotion of migration and invasion of esophageal cancer cells. Mol Cell Biol. 2008;28:4142–4151. doi: 10.1128/MCB.01465-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargiannidou I, Zhou J, Tuszynski GP. The role of thrombospondin-1 in tumor progression. Exp Biol Med. 2001;226:726–733. doi: 10.1177/153537020222600803. [DOI] [PubMed] [Google Scholar]
- Crawford SE, Stellmach V, Murphy-Ullrich JE, Ribeiro SM, Lawler J, Hynes RO, Boivin GP, Bouck N. Thrombospondin-1 is a major activator of TGF-beta1 in vivo. Cell. 1998;93:1159–1170. doi: 10.1016/s0092-8674(00)81460-9. [DOI] [PubMed] [Google Scholar]
- Annes JP, Munger JS, Rifkin DB. Making sense of latent TGF beta activation. J Cell Sci. 2003;116:217–224. doi: 10.1242/jcs.00229. [DOI] [PubMed] [Google Scholar]
- Rojas A, Meherem S, Kim YH, Washington MK, Willis JE, Markowitz SD, Grady WM. The aberrant methylation of TSP1 suppresses TGF-beta1 activation in colorectal cancer. Int J Cancer. 2008;123:14–21. doi: 10.1002/ijc.23608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams JC. Characterization of cell-matrix adhesion requirements for the formation of fascin microspikes. Mol Biol Cell. 1997;8:2345–2363. doi: 10.1091/mbc.8.11.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams JC, Schwartz MA. Stimulation of fascin spikes by thrombospondin-1 is mediated by the GTPases Rac and Cdc42. J Cell Biol. 2000;150:807–822. doi: 10.1083/jcb.150.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]