Abstract
In humans, four members of the CYP2C subfamily (CYP2C8, CYP2C9, CYP2C18, and CYP2C19) metabolize more than 20% of all therapeutic drugs as well as a number of endogenous compounds. The CYP2C enzymes are found predominantly in the liver, where they comprise ∼20% of the total cytochrome P450. A variety of xenobiotics such as phenobarbital, rifampicin, and hyperforin have been shown to induce the transcriptional expression of CYP2C genes in primary human hepatocytes and to increase the metabolism of CYP2C substrates in vivo in man. This induction can result in drug-drug interactions, drug tolerance, and therapeutic failure. Several drug-activated nuclear receptors including CAR, PXR, VDR, and GR recognize drug responsive elements within the 5′ flanking promoter region of CYP2C genes to mediate the transcriptional upregulation of these genes in response to xenobiotics and steroids. Other nuclear receptors and transcriptional factors including HNF4α, HNF3γ, C/EBPα and more recently RORs, have been reported to regulate the constitutive expression of CYP2C genes in liver. The maximum transcriptional induction of CYP2C genes appears to be achieved through a coordinative cross-talk between drug responsive nuclear receptors, hepatic factors, and coactivators. The transcriptional regulatory mechanisms of the expression of CYP2C genes in extrahepatic tissues has received less study, but these may be altered by perturbations from pathological conditions such as ischemia as well as some of the receptors mentioned above.
Keywords: Human CYP2C, transcription regulation, drug induction, hepatic nuclear receptor, hypoxia
Introduction
The cytochrome P450s (CYP) are a superfamily of enzymes that catalyze the metabolism of xenobiotic drugs and environmental chemicals as well as many endogenous compounds. The human CYP2C subfamily consists of four members clustering at the chromosomal location 10q24 as Cen- CYP2C18-CYP2C19-CYP2C9- and CYP2C8-Tel, and they comprise approximately 20% of the P450 enzymes in the human liver. Except for CYP2C18, which is expressed at the mRNA level but does not appear to be expressed at the protein level in any tissue, the CYP2C proteins are expressed predominantly in the liver (2C9>2C8>2C19). However, they are expressed to variable extents in a number of other extrahepatic tissues such as kidney, gut, brain, heart, aorta, and lung [1, 2]. The CYP2C enzymes are well-known clinically important enzymes that metabolize more than twenty percent of all pharmaceutical drugs. CYP2C substrates include some of the most frequently prescribed drugs, such as the anticoagulant drug coumadin, the anticonvulsant drug phenytoin, the anti-diabetic drugs tolbutamide, glipizide, and rosiglitazone, and numerous nonsteroidal anti-inflammatory drugs such as celecoxib, flurbiprofen, ibuprofen, and diclofenac [3, 4]. CYP2C19 metabolizes the prototype drug S-mephenytoin, antiulcer drugs such as omeprazole and other proton pump inhibitors, diazepam [3, 4], and the platelet inhibitor clopidogrel [5], while CYP2C8 metabolizes rosiglitazone and the anticancer drug paclitaxel [3, 4]. CYP2C8/9 enzymes are also responsible for the hydroxylation of retinoic acid [6], and the CYP2C enzymes are important in the generation of biologically active molecules such as epoxyeicosatrienoic acids (EETs) and hydroxyeicosatrienoic acids (HEETs) from arachidonic acid in both liver and extrahepatic tissues[7].
All of the CYP2C genes exhibit genetic polymorphisms, some of which produce large phenotypic inter-individual variability in the metabolism of certain CYP2C substrates [3, 4, 8-12]. In particular, null polymorphisms of CYP2C19 dramatically affect the metabolism of a number of substrates of this enzyme. When single-nucleotide polymorphisms (SNPs) occur in the coding region, they can result in amino acid changes (some of which alter activity, affinity and turnover number of substrates) or produce premature stop codons, resulting in null (completely nonfunctional) alleles. SNPs can destroy or create new splice sites, producing frame shifts which also produce null alleles. Single or multiple base pair deletions can also cause frame shifts. SNPs also occur in the regulatory regions, and one such SNP produces an ultra-rapid metabolizer allele of CYP2C19 [13]. SNPs of CYP2C9 are well-known to affect dosage and serious bleeding epidsodes of coumadin [4, 14, 15]. A recent report has linked an intron SNP of CYP2C8 to bisphosphonate-related osteonecrosis of the jaw [16]. Moreover, patients treated with clopidogrel who are carriers of the CY2C19 defective alleles have an increase in death from cardiovascular causes and an increase in stent failures [5].
Another factor contributing to inter-individual variability in expression of the CYP2C proteins is their inducibility after exposure of humans to xenobiotics. Studies in vitro in primary human hepatocytes clearly indicate that the expression of CYP2C enzymes is induced by prior exposure to various drugs, including glucocorticoids, rifampicin, paclitaxel and phenobarbital [17, 18]. Moreover, studies in vivo are consistent with changes in the half-life of CYP2C substrates (e.g. tolbutamide, glipizide, S-mephenytoin) in man after prior exposure to drugs such as rifampicin [19-22]. This could potentially result in diminished effectiveness of the drug and possibly therapeutic failure.
Because of the pharmaceutical and physiological significance of the CYP2C enzymes, it is important to understand the transcriptional modulation of the constitutive and inducible expression of CYP2C genes to better understand the basis for inter-individual variability and predict adverse drug-drug interactions. This review will focus on the significant progress over the past few years in unraveling the molecular regulatory mechanisms for both the basal and drug-induced upregulation of human CYP2C genes in liver. The transcriptional regulation of CYP2C genes in extrahepatic tissues as well as in pathological situations is also discussed here.
Induction of CYP2C enzymes by drugs and xenobiotics
A number of clinical reports suggest that the metabolism of CYP2C9, CYP2C8, and CYP2C19 substrates is increased when humans are exposed to a variety of clinical drugs (see Table 1 for references). This induction after prior treatment with drugs results in a faster drug clearance rate, a shorter half-life, and a lower plasma level of drugs that are primarily metabolized by CYP2C enzymes, including coumadin, glyburide, and glipizide (CYP2C9), rosiglitazone and pioglitazone (CYP2C8), and S-mephenytoin and omeprazole (CYP2C19). Administration of some herbal medicines also induces the activity of CYP2C. For example, long-term treatment with St. John's wort, a widely used herbal antidepressant, decreased the plasma concentrations of coumadin and gliclazide (CYP2C9) as well as S-mephenytoin and omeprazole (CYP2C19). Due to clinical concerns resulting from the induction of CYP2C enzymes by drugs, a careful dose increase could be necessary for drugs which are CYP2C substrates to avoid therapeutic failure when co-administered with drugs that are inducers of CYP2C genes.
Table 1. Induction of human CYP2C in liver.
| CYP2C Genes | Transcription Inducers | Clinical Studies | In vitro Studies |
|---|---|---|---|
| CYP2C9 | Phenobarbital | [17, 18, 23] | |
| Rifampicin | [22, 107, 108] | [17, 18, 23, 109] | |
| Hyperforin & St John's wort | [110, 111] | [112] | |
| Avasimibe | [113] | [113] | |
| Ritonavir, Nelfinarir & Lopinavir | [114, 115] | [116] | |
| Dexamethasone | [17, 18] | ||
| Cyclophosphamide or Ifosfamide | [117] | ||
| Nifedipine, Nicardipine, BK8644 & Isradipine | [118] | ||
| Carbamazepine | [119] | ||
| Aprepitant | [120, 121] | ||
| CYP2C8 | Phenobarbital | [122] | [17, 18, 23, 33] |
| Phenytoin | [123] | [33] | |
| Rifampicin | [124-126] | [17, 18, 23, 109] | |
| Hyperforin | [33, 127, 128] | ||
| Dexamethasone | [17, 18, 23, 33] | ||
| Ritonavir & Nelfinarir | [116, 127] | ||
| Cyclophosphamide or Ifosfamide | [117] | ||
| Lithocholic acid | [33] | ||
| Paclitaxel | [129] | [33, 130] | |
| Gemfibrozil, Fenofibric acid & Clofibric acid | [131] | ||
| CYP2C19 | Ritonavir & Nelfinavir | [116] | |
| Rifampicin | [132, 133] | [17, 23, 109] | |
| Hyperforin & St. John's wort | [134, 135] | ||
| Dexamethasone | [17] | ||
| Artemisinin | [136] | [137] |
Information concerning the inducibility of CYP2C genes has been frequently obtained from in vitro induction studies in isolated human primary hepatocytes, which are cited in Table 1. With this cell model, it has been reported that CYP2Cs are induced significantly at the levels of mRNA, protein, and activity by therapeutic reagents, hormones such as glucocorticoid, vitamin D, and the endogenous metabolite lithocholic acid, which was shown to induce CYP2C8. Compared to other CYP genes such as CYP3A4 and CYP2B6, which are strongly induced after exposure to drugs, the CYP2C genes are modestly induced (2-8 fold) [17, 18, 23]. The inducibility of CYP2C genes in liver can be generally ranked as CYP2C8≥ CYP2C9 >CYP2C19. Certain molecules act as inducers for all three CYP2C genes, including phenobarbital, rifampicin, hyperforin (an active ingredient of St. John's wort), and dexamethasone. The induction of CYP2C19 protein and mRNA shows high inter-individual variability in human livers. Polymorphisms in this gene and its low constitutive expression in uninduced liver contribute to this variability in induction.
Nuclear receptor-mediated transcriptional activation of CYP2C genes by drugs in the liver
The transcriptional activation of most P450 genes is mediated by drug responsive nuclear receptors, which are transcriptional factors sensing foreign substances. The nuclear receptors CAR and PXR contain a DNA binding domain (DBD) and a ligand binding domain (LBD). After activation by exposure to xenobiotics, the nuclear receptors bind to the responsive elements as monomers or homo- or hetero-dimers, recruit coactivators to affect chromatin structure, and increase the transcription of target genes [24].
Several nuclear receptors have been identified that mediate the xenobiotic-induced transcriptional activation of the human CYP2C genes (Table 2). The nuclear receptor CAR is responsible for the transcriptional activation of CYP2C9 (in human primary hepatocytes and HepG2 cells), CYP2C8 (in human primary hepatocytes) and CYP2C19 (in HepG2 cells and primary hepatocytes). CAR agonists include drugs such as phenobarbital and artemisinin as well as the chemical CITCO {6-(4-chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehydeO-(3,4-dichlorobenzyl oxime}. CITCO is a high affinity ligand for human CAR (hCAR) which activates hCAR in primary hepatocytes. But it only modestly increases promoter activity in the typical cell-based reporter assays, probably because CAR accumulates in the nucleus in immortalized cells while it is found primarily in the cytoplasm in primary hepatocytes and liver [25]. CAR is constitutively active without ligand, and many xenobiotics (phenobarbital and phenytoin) act primarily by causing its nuclear translocation rather than acting as ligands [26-28]. Another receptor, the human pregnane X receptor (PXR), has been shown to mediate induction of the CYP2C genes by drugs such as rifampicin, artemisinin, and hyperforin, all of which act as ligands for PXR. Dexamethasone, a glucocorticoid mimic drug activates the CYP2C promoters in HepG2 cells via the glucocorticoid receptor (GR). The Vitamin D receptor (VDR) has been reported to produce a modest 2-fold induction of CYP2C9 in human primary hepatocytes by 1α,25-dihydroxyvitamin D3 [29]. It may also mediate the induction of CYP2C8 by lithocholic acid (LCA) in HepG2 cells [30].
Table 2. The transcriptional regulation of human CYP2C genes by nuclear receptors and their inducers in liver.
| CYP2C Genes | Nuclear Receptors | Inducers |
|---|---|---|
| CYP2C9 | PXR | Rifampicin [32, 34], Hyperforin [34], Phenobarbital [34] |
| CAR | Phenobarbital [31], CITCO [33] | |
| GRα | Glucocorticoid [31] | |
| VDR | Vitamin D [29] | |
| CYP2C8 | PXR | Rifampicin [33], Retonavir [127] |
| CAR | CITCO [33] | |
| GRα | Glucocorticoid [33] | |
| VDR | Lithocholic acid [30] | |
| CYP2C19 | PXR | Rifampicin [35], Artemisinin [137] |
| CAR | Phenobarbital [35], Artemisinin [137] | |
| GRα | Dexamethasone [35] |
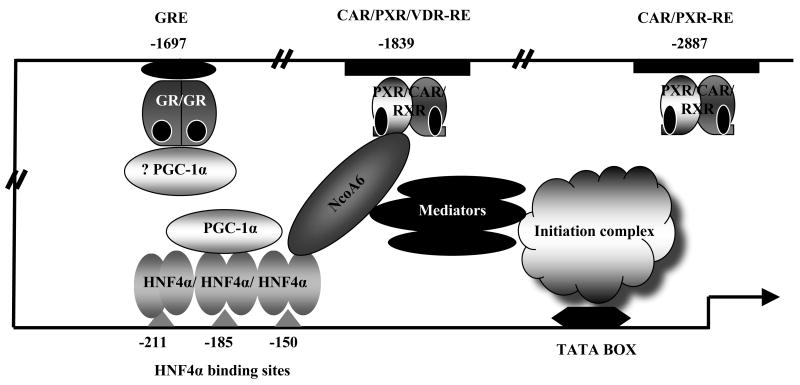
CAR, PXR and VDR form heterodimers with the retinoid X receptor (RXR) while GR forms homodimers which are recognized by specific response elements within the CYP2C promoters. A typical nuclear receptor response element is composed of two half sites related to the hexamer AGGTCA separated by 3-6 bases. Fig (1) presents responsive elements within the CYP2C9, 2C8 and 2C19 upstream promoter regions that have been identified as binding sites for CAR, PXR, GR and VDR in vitro by gel shift assays [29, 31-35]. The response elements of the CYP2C genes exhibit similar but distinct features. Both the CYP2C9 and 2C19 promoters contain a single similar proximal direct repeat spaced with 4 bp nucleotides (DR4) CAR/PXR-RE (located at -1839/-1824 from the translation start site of 2C9 and -1892/-1877 from the translation start of 2C19 respectively), differing by one nucleotide at the 3′ prime end [35]. Both sites showed strong binding to CAR and PXR in vitro, and exchange of these two elements between the two CYP2C promoter constructs did not alter the activation of these two promoters by CAR in a transient transfection assay (Chen, unpublished observations).
Figure 1.
Summary of the known response elements for nuclear receptors regulating the CYP2C8, 2C9, and 2C19 genes. This figure summarizes our current view of the regulatory elements within the three human CYP2C gene promoters. The hexamer sequences are shown in capital letters for each element along with their exact locations within each promoter. The arrows indicate the direct repeat of each element. The elements which bind nuclear receptors in vitro but are identified as nonfunctional by mutagenesis in luciferase promoter studies are shown with grey lines. Newly identified HNF4α sites in CYP2C9 and CYP2C8 are indicated with dark parallel lines (Chen unpublished).
CYP2C9 harbors a second DR5 type CAR/PXR-RE at -2897/-2881 which binds CAR and PXR in gel shift assays [32]. At a similar location in the CYP2C8 promoter there is a DR4 (at -2796/2780) that binds CAR/PXR in gel shift assays but mutation of this element does not affect activation of the CYP2C8 promoter in human hepatocytes by CAR or PXR agonists [33]. In the far upstream region of the 2C8 promoter, another DR4 element was identified at -8805/-8790 that strongly binds to CAR and PXR. Mutation of this element prevents activation of the CYP2C8 in promoter by CAR or PXR agonists in human hepatocytes [33].
Additionally, the three CYP2C promoters harbor a putative DR3 type glucocorticoid response element (GRE) in their proximal regions (at -1697/-1682 for 2C9, -1927/-1913 for 2C8, and -1751/-1737 for 2C19) [31, 33, 35], and the 2C9-GRE was shown to bind hGR in gel shift assays [31]. The core sequences of the GREs are identical for CYP2C9 and 2C19, with a few nucleotides differing in the 5′ flanking region. One base pair in the 5′ half site of the GRE of the CYP2C8 promoter differs from the GREs of 2C9 and 2C19, which results in a change from TGAACT (2C9 and 2C19) to TTAACT. The proximal CAR/PXR-RE of 2C9 has also been shown to bind VDR in vitro.
To evaluate the responsiveness of the CYP2C promoters to induction by xenobiotics and the functionality of putative responsive elements, transient transfection has usually been performed in hepatic carcinoma cell lines such as HepG2 or human primary hepatocytes. CYP2C9 and 2C19 promoters are significantly activated by cotransfection of CAR, PXR (in the presence of rifampicin), and GR (in the presence of dexamethasone) in HepG2 cells [31-35]. Unlike CYP2C9 and 2C19; however, induction of the 2C8 promoter by CAR and PXR ligands was observed in human primary hepatocytes but was not observed in HepG2 cells [33], suggesting the possibility that certain factors that are necessary for CYP2C8 induction in primary hepatocytes are low or absent in HepG2 cells.
Both CAR/PXR-REs appear to contribute to activation of the CYP2C9 promoter by PXR and CAR, but the site at -1839 is more important. For example, mutation of the CAR/PXR-RE at −2897 alone decreased rifampicin/PXR activation by ∼30%, while mutation of the PXR binding site at −1839 bp alone nearly abolished rifampicin/PXR mediated promoter activation [34]. This data suggests that the site at -1839 bp is essential for induction, while the site at -2897 cooperates with the site at -1839 bp. The CAR/PXR-RE at -1839 is further shown to be required for transactivation in the context of a 12kb CYP2C9 promoter by PXR and rifampicin in HepG2 cells [34].
Although activation of the CYP2C19 promoter by CAR and PXR/rifampicin in HepG2 cells was more modest than the activation of the CYP2C9 promoter, mutation of the CAR/PXR-RE at -1892/-1877 completely abolished this activation. Mutation of the CAR/PXR-RE of CYP2C8 at -8805/-8790 completely abolished induction of CYP2C8 promoter activity by CITCO and rifampicin in primary human hepatocytes, but mutation of the putative site at -2796/-2780 had no effect on promoter activation, suggesting that only the distal site is involved in activation of the CYP2C8 gene by CAR and PXR ligands.
Each CYP2C promoter has also been shown to be activated by GR and its ligand dexamethasone via one GRE which is located within the first 2kb of the three promoters. The induction by dexamethasone was much higher for CYP2C9 (>15-fold) than for 2C8 and 2C19 (3-fold and 4-8 fold respectively) in transfection assays in HepG2 cells [31, 33, 35]. Mutation of the GR elements of CYP2C9, CYP2C19, and CYP2C8 abolished dexamethasone induction [31, 33, 35]. The differing extent of dexamethasone induction among the three CYP2C genes is independent of the element itself, since 2C9 and 2C19 share an identical GRE. Possibly promoter context or nucleotides flanking the GRE could play a role. Only the CYP2C9 gene has been examined for upregulation by the VDR ligand 1,25-(OH)2D3 in human primary hepatocytes [29]. The proximal CAR/PXR-RE at -1839/-1824 binds VDR in vitro. When this element was linked to the TK promoter and transfected into HepG2 cells, a modest but reproducible induction (∼1.8 fold) by 1,25-(OH)2D3 was observed in VDR-transfected HepG2 but not in VDR-nontransfected cells. However, the TK promoter is a strong promoter, and the role of this VDR-RE in the induction of CYP2C9 by 1α, 25-dihydroxyvitamin D3 has not been confirmed in the context of the original CYP2C9 promoter.
It is of note that the CAR/PXR-REs in the promoters of all three human CYP2C genes are activated by both CAR or PXR, and gel shift assays confirm that both receptors bind strongly to the identified responsive elements in the human CYP2C gene promoters. These data suggest a symmetrical cross-talk between CAR and PXR in upregulation of human CYP2C genes, though CAR appears much more important for induction of the murine genes Cyp2c29 and Cyp2c37 based on studies in CAR and PXR knockout mice [26, 36]. A similar cross-talk could occur between VDR and CAR/PXR for the expression of CYP2C9, since all three receptors are reported to bind to the proximal CAR/PXR-RE. A corresponding mutual inhibition of induction of the CYP2C9 gene by PXR ligands and vitamin D may occur, as has been observed for CYP3A4, where two PXR binding sites (the distal DR3 motif and proximal everted repeat separated by six base pairs (ER6) bind VDR competitively [29].
Transcriptional regulation of the constitutive expression of CYP2C enzymes in liver and pathological conditions
The human CYP2C enzymes are expressed primarily in the liver, and a number of liver-enriched transcription factors (LETFs) have been shown to regulate the constitutive expression of P450 genes, including the hepatic nuclear factors HNF1α, HNF4α, HNF3γ, HNF6, C/EBP, and DBP as summarized in Table 3. The retinoic acid-related orphan receptors (RORs) have recently been identified as receptors which regulate CYP2C8. [37].
Table 3. The hepatic transcriptional factors of human CYP2C genes.
HNF4α, an orphan nuclear receptor primarily expressed in the liver, kidney, intestine and pancreas, is well known to play a significant role in the regulation of many P450 genes and HNF4α binding sites (described as HepG2-specific P450 2C factor-1 (HPF-1) motifs) were first discovered in rabbit CYP2C genes by Kemper and coworkers [38]. Using adenoviral HNF4α antisense RNAs, Jover et al. [39] were able to reduce endogenous HNF4α and observed a significant (40-45%) reduction of CYP2C9 mRNA content in human primary hepatocytes. A slight but significant decrease in the mRNAs of CYP2C8 and 2C19 was observed with adenoviral siRNA for HNF4α in primary human hepatocytes [40]. These data indicate that HNF4α influences the constitutive expression of all three CYP2C genes. The expression levels of CYP2C8, 2C9, and 2C19 were recently found to be strongly associated with HNF4α content in a study with 20 human liver samples (partial regression analysis), further supporting the role of HNF4α as a predominant regulator for the basal CYP2C gene expression in human liver [41].
HNF4α binds as a homodimer to a DR1 type element and also to the HPF-1 motif (5′ RRRNCAAAGKNCANYY) [42]. These sites are present in the basal promoters of all human CYP2C genes except CYP2C18 (Fig. 1). Both CYP2C9 and 2C19 have two identical HPF-1 motifs located at a similar site in their promoters (at -150/-138 and -185/-173 for 2C9; at -152/-140 and -187/-175 for 2C19) [43, 44]. Gel shift assays show that, both in vitro translated HNF4α protein and nuclear extract from HepG2 cells bind to these sites, with the distal element displaying weaker binding than the proximal one. However, the CYP2C9 basal promoter was significantly activated by HNF4α when cotransfected in human hepatocarcinoma FLC7 and HepG2 cells, while the 2kb of 2C19 basal promoter was not [43]. A chromatin immunoprecipitation (ChIP) study showed that HNF4α associates with the basal CYP2C9 promoter region in vivo in human liver but was not detected in association with the CYP2C19 promoter. Based on these results, it was proposed that CYP2C19 is expressed at lower levels than CYP2C9 in liver due to the lack of sufficient HNF4α binding to the two HNF4 elements within the basal CYP2C19 promoter. However, it is not clear why the basal CYP2C19 promoter is not activated by HNF4α, since it contains two HNF4 sites identical to those found in 2C9. One HPF-1 motif has been also identified in the CYP2C8 promoter at -152/-140 that interacts with HNF4α in vitro. Again, cotransfected HNF4α does not enhance the activity of the CYP2C8 promoter in HepG2 cells, but does transactivate the 2C8 promoter construct in HeLa cells [33]. Recent unpublished studies in our laboratory have identified a second HPF-1 motif in the CYP2C8 promoter.
In HepG2 cells, where HNF4α is expressed endogenously, the HPF-1 motif at -185/-173 appears to be critical for HNF4α activation of CYP2C9, while mutation of a site at -150/-138 bp leads to a significant reduction in HNF4α activation. However, in the FLC7 cells in which HNF4α is not expressed, Kawashima et al [43] observed that both HNF4α responsive elements contributed equally to activation of the CYP2C9 gene by HNF4α. They also found that the region from -255/-195 bp of the CYP2C9 promoter was necessary for HNF4α to up-regulate the transcription of the CYP2C9 gene and suggested that some other factors might bind to this region and assist HNF4α in this upregulation. Consistent with their results, we recently reported preliminary results identifying a third HNF4α binding element at -211/-199 of the CYP2C9 promoter (Chen, unpublished data). This element aligns in the CYP2C promoter in a reverse orientation with respect to the other two proximal HNF4α sites (Fig. 1) and specifically binds nuclear proteins from HepG2 cells as well as in vitro transcribed and translated HNF4α protein. Three nucleotides in the core motif of a similar element within the CYP2C19 promoter differ from that of CYP2C9, and this difference results in a weaker interaction between this element and HNF4α in gel shift assays. When these three nucleotides were introduced into the CYP2C9 promoter, CYP2C9 activation by HNF4α in HepG2 cells was 50% lower (unpublished data), but these results still do not completely explain the relative unresponsiveness of CYP2C19 to HNF4α compared to that of CYP2C9.
HNF3γ and CCAAT/enhancer-binding protein α (C/EBPα) are two other liver-enriched transcriptional factors implicated in regulating the constitutive expression of the CYP2C genes in the liver[45, 46]. During the isolation and culture of hepatocytes, these two factors have been found to be greatly downregulated, along with a concomitant downregulation of the expression of CYP2C9 [46]. C/EBPs are basic leucine zipper (bZIP) transcription factors with a DNA-binding basic region and a leucine zipper dimerization domain. Homo- or heterodimerized C/EBPs recognize the CCAAT box motif in the promoter region and have been found to regulate the transcription of genes involved in the differentiation of hepatocytes. One factor, C/EBPα, begins to decay in a very early stage of primary hepatocyte culture and continues to decay very rapidly [46]. Moreover, in HepG2 cells the levels of C/EBPα mRNA are only ∼15% of those in human hepatocytes, while the expression of all of three CYP2C genes is much lower in these cells than in liver. The re-expression of this factor in HepG2 cells increased the expression of CYP2C9 (mRNA and protein) while the levels of other liver-enriched factors such as HNF4α were not changed. These data further suggest that C/EBPα may play an important role in maintenance of the expression of CYP2C genes [46]. All of the three CYP2C promoters harbor a CCAAT box in the 5′ flanking region [2, 47], and the deletion of this element significantly decreases the transcriptional activities of the CYP2C9 promoter [48]. It still remains to be established to what extent C/EBPα regulates the constitutive expression of the CYP2C genes.
HNF3γ, a member of the forkhead family of transcription factors, is expressed strongly in adult derivatives of the endoderm posterior to the liver [45]. These transcription factors bind to DNA as monomers and have a distinct conserved winged helix DNA-binding domain that is homologous to the Drosophila homeotic protein called Fork head. This factor also decays rapidly during the culture of human primary hepatocytes, although not as rapidly as C/EBPα, and the level of HNF3γ mRNA in HepG2 cells is only ∼25% of that found in liver [46]. Several putative HNF3γ binding sites have been identified within the 5′ flanking regions of the four human CYP2C genes [45]. The adenoviral expression of ectopic HNF3γ in HepG2 cells resulted in an enhancement in endogenous mRNA levels of CYP2C9 (4.5-fold) and 2C19 (50-fold), as well as 2C8 (20-fold) after cells were treated with a deacetylase inhibitor [45]. Promoter studies in HepG2 cells revealed that HNF3γ activated the promoter activity of CYP2C8 (25-fold), 2C9 (4-fold) and 2C19 (4-fold) [45]. Additional studies are needed to confirm the extent of the regulatory role of HNF3γ in hepatic expression of individual CYP2C genes, such as whether knock-down of endogenous HNF3γ reduces the expression of CYP2C genes, and which putative elements are required for HNF3γ binding and its activation of the CYP2C promoters.
In addition, several other hepatic transcriptional factors have been shown to be implicated in the regulation of hepatic expression of some rodent CYP2C genes, including HNF1, HNF6, C/EBPβ and albumin D-site binding protein (DBP) [38]. The extent to which these factors control the expression of human CYP2C genes remains uncertain.
Recently, we identified retinoid related orphan nuclear receptors (RORs) as new transcriptional regulators for CYP2C8 but not CYP2C9 or CYP2C19 [37]. RORs are constitutively active orphan nuclear receptors. Some natural compound ligands such as cholesterol and all trans-retinoic acid have been found to bind to RORs and modulate their activity [49]. It has been shown that the expression of certain murine P450 genes including Cyp2c70 is altered in ROR knock-out mice [50]. We found that cotransfection of RORα4 and –γ1 significantly increased the promoter activity of a ∼3kb construct of CYP2C8 but not that of CYP2C9 and CYP2C19 in HepG2 cells. Two ROR-REs (at -2289 and -2045 bp within the CYP2C8 promoter region) were identified which bound both RORα4 and –γ1 generated in vitro, but binding of the proximal site was stronger and mutagenesis studies confirmed that the proximal site was the essential one mediating the ROR activation of the CYP2C8 promoter in HepG2 cells. Overexpression of either RORα4 or –γ1 elevated the endogenous CYP2C8 mRNA in HepG2 cells and human primary hepatocytes, while knock-down of either endogenous RORα4 or –γ1 decreased the CYP2C8 expression in HepG2 cells. RORs are also expressed in other extrahepatic tissues including the brain, where CYP2C8 mRNA is preferentially expressed over other CYP2C mRNAs. The role of RORs in regulating CYP2C8 in these extrahepatic tissues is not yet known.
The cooperativity of transcription factors and complexity in transcriptional regulation of human CYP2C genes
In addition to their direct interaction with the responsive element and regulation of the transcription of target genes, nuclear receptors often cooperate with each other or with other factors, such as coactivators and corepressors, to achieve precise modulation of target genes. Moreover, the expression of nuclear receptors can be regulated by endogenous or other receptors exogenous compounds, e.g., glucocorticoids induce the expression of CAR, PXR, and RXR via a direct transactivation mediated by GR and the GR responsive elements within the promoter regions of these nuclear receptors, thus enhancing the expression of target genes including CYP2C9 and CYP2C8 [51, 52]. HNF4α is also known to increase CAR and fetal PXR as well [53-55]. On the other hand, the mRNA expression of CAR, PXR and RXR has been shown to be markedly decreased by the proinflammatory cytokines interleukin (IL)-1β and IL-6. Consistent with these results, the constitutive and inducible mRNA expression of the typical CAR and PXR target genes CYP2C9 and 2C8 are specifically inhibited by these cytokines in human primary hepatocytes [56, 57]. Further studies demonstrated that the inflammatory stimuli by lipopolysaccharides (LPS) and IL-1β caused the nuclear accumulation of NF-κBp65, which acts as an inhibitor of GR and trans-represses the activation of the CAR promoter by glucocorticoid and GR. A ChIP assay also revealed that dexamethasone induced histone H4 acetylation of the proximal CAR gene promoter, while both LPS and IL-1β dramatically inhibited this increased acetylation in human primary hepatocytes [56]. However, recent work shows that the CYP2C genes are downregulated by different inflammatory cytokines in a gene-specific manner in human primary hepatocytes [58].
Recently, transcription factors and coactivators have been found to cooperate in the transcriptional regulation of human CYP2C genes (Fig. 2). A synergy between HNF4α and CAR/PXR was first reported for the CYP3A4 gene, where coexpression of HNF4α and PXR dramatically increased the activity of the CYP3A4 promoter in the presence of PXR ligands [54]. HNF4α has also been shown to synergize with CAR and PXR to enhance the induction of CYP2C9 mediated by these two nuclear receptors in HepG2 cells [44]. This synergy differs from that reported for the CYP3A4 promoter, where the HNF4α binding site essential for the synergy is immediately upstream of two CAR/PXR-REs within the distal XREM. The two HNF4α sites in the CYP2C9 promoter are located at -185 bp and -150 bp, far downstream of the CAR/PXR-RE (at -1839 bp). Mutation of the HNF4α sites essentially abolished the drug induction of CYP2C9 mediated by CAR and PXR, clearly indicating the HNF4α sites are required for the drug responsiveness of the CYP2C9 promoter. In contrast, rifampicin induction of CYP3A4 remained when the HNF4α site was mutated or deleted [54]. Because of the distance between the drug responsive element and HNF4α binding sites in the CYP2C9 promoter, an indirect cross-talk between the receptors was proposed as a likely underlying mechanism for the synergistic activation of the CYP2C9 gene by HNF4α and CAR/PXR. This cross-talk would bridge HNF4α and CAR/PXR via cofactors or other transcriptional factors rather than involving direct interaction between the two nuclear receptors. This hypothesis has gained experimental support from a recent discovery that the nuclear receptor coactivator NCoA6 interacts with CAR and HNF4α and appears to bridge the CAR-RE to the HNF4α sites to cause a synergistic activation of the CYP2C9 promoter in HepG2 cells (Fig. 2) [59]. ChIP analysis showed that NCoA6 interacted with both the HNF4α sites and the CAR sites. Knockdown of NCoA6 disrupted this bridge and decreased the synergistic elevation in expression of CYP2C9 mRNA by CAR and HNF4α.
Figure 2.
Interactions between nuclear receptors and coactivators precisely modulate the transcription of CYP2C9. The coactivator NcoA6 bridges the HNF4α binding site(s) within the basal promoter and the proximal CAR/PXR-RE site by interacting with HNF4α and CAR, producing a synergistic transactivation of the CYP2C9 promoter. The coactivators PGC-1α and SRC-1 also interact with HNF4α, and probably also with GR, to regulate the activity of the CYP2C9 promoter.
A number of coactivators are involved in the indirect modulation of the CYP2C genes. Coactivators are a class of protein factors which do not bind to DNA directly but interact with DNA binding transcription factors and are thus recruited to chromatin. Coactivators recruit histone acetyltransferases and methyltransferases to the promoter region where nuclear receptors bind and facilitate chromatin remodeling, allowing access of general transcriptional machinery to the promoter of the target gene. Two other coactivators have been implicated in the regulation of the CYP2C genes by interacting with the receptor HNF4α: the nuclear receptor coactivator (SRC-1) and the peroxisome proliferator-activated receptor gamma, coactivator 1 alpha (PGC-1α) [60]. Each coactivator activated the CYP2C9 promoter when transfected into human hepatic carcinoma cells. PGC-1α is a known coactivator for HNF4α [61]. Activation of CYP2C9 by PGC-1α may depend largely on the presence of HNF4α in HepG2 cells, since activation of the CYP2C promoter in HeLa cells was dependent on the presence of exogenous HNF4α. The deletion of a short CYP2C9 basal promoter region which harbors the HNF4α sites completely destroyed the activation of the CYP2C9 promoter by HNF4α as well as PGC-1α [60]. It has been proposed that the large reduction of these two cofactors in human carcinoma cells results in a lower expression of CYP2C9 compared to the level in liver or human primary hepatocytes. Consistent with this suggestion, viral-transduced PGC-1α and SRC-1 significantly increased the amount of CYP2C9 mRNA in these cells [60].
Both SRC-1 and PGC-1α have been demonstrated to act as coactivators for other nuclear receptors such as GR, CAR, and PXR, as well as VDR which are known to regulate the induction of human CYP2C genes. They are thus possibly involved in the inducible transcription of CYP2C genes by coactivation of these nuclear receptors. Of note is that the PGC-1α gene is responsive to energy metabolic homeostasis, induced in the liver by fasting and decreased by insulin. This suggests the possibility that target genes such as CYP2C9 could also be regulated by factors that affect energy homeostasis. In fact, CYP2C9 mRNA was decreased in HepG2 cells and human primary hepatocytes treated with insulin [60]. In sum, the transcriptional regulation of CYP2C9 might be subject not only to environmental stimulation by xenobiotic drugs but also affected by various physical conditions such as fasting.
Transcriptional regulation of CYP2C genes in extrahepatic tissues and pathological conditions
Human CYP2C enzymes are widely distributed in a variety of extrahepatic tissues, but the level of human CYP2C transcripts and proteins in these tissues is lower than that in liver [1, 2]. Moreover, the pattern of expression of the individual CYP2C enzymes and transcripts differ in these organs, suggesting that the regulatory control of the CYP2C genes differs in various extra-hepatic tissues. However, the regulatory control of the CYP2Cs in extrahepatic tissues has received less study than that of liver.
In the human intestine, CYP2Cs are the second most abundant subfamily of P450 enzymes (25% of total immunoquantified P450s) [62]. Treatment with the PXR ligand rifampicin in healthy humans significantly increases the mRNA and protein level of CYP2C9, 2C8 and 2C19 as well as their enzymatic activity in the small intestine [63-65]. The order of inducibility is similar to that in hepatic CYP2Cs: 2C8>2C9>2C19 [64], but the induction response is reported to be weaker (about 1.5-fold at the mRNA level) in the small intestine than in the liver, as quantified using intestinal biopsies [65]. Notably, CAR, PXR, and HNF4α are also expressed in the small intestine.
In the kidney, CYP2Cs are well known renal arachidonic acid epoxygenases, and their metabolites, EETs, play an antihypertensive role [66]. In human kidneys, the mRNAs and proteins of CYP2C9 and CYP2C8 have been detected [2, 67], and CYP2C8 has been suggested to be responsible for the generation of active renal vasodilatory epoxygenases (EETs) [68]. The expression and activity of murine renal Cyp2c44 and rat CYP2C23 are decreased in several hypertension animal models (salt-sensitive [66], diet-induced [69], and obesity-related albuminuria [70]). Several lines of experimental evidence have suggested that increased levels of cytokines such as TGF-β and TNFα might be involved in downregulation of the renal CYP2C genes [71, 72]. Recently, EETs have been identified as potent ligands for human PPARα and -γ in vitro and shown to transactivate both receptors in human hepatic carcinoma cells [73]. The expression of murine renal Cyp2c44 was increased by ligands for PPARα [74, 75]. However, there is no human equivalent of Cyp2c44 [76, 77], and at present there are no reports as to whether renal CYP2C8 or 2C9 can be modulated by PPAR agonists.
In the brain, CYP2C8 mRNA is expressed at a higher level than other CYP2C mRNAs, and CYP2C8 mRNA is expressed at higher levels in brain than any other extrahepatic tissues we tested (Delozier and Goldstein, unpublished). Low levels of CYP2C9 and 2C19 mRNAs were reported in the whole brain [2, 78, 79], where these enzymes could be implicated in the local metabolism of psychoactive drugs and xenobiotics as well as possibly in the regulation of the cerebral blood flow through production of EETs. mRNAs of CYP2C subfamily members such as CYP2C8 and 2C9 have also been identified in human astrocytoma cells [80]. Cocaine treatment reduced mRNAs or proteins of CYP2C8 and 2C9 in human U373 MG astrocytoma cells, along with a simultaneous downregulation of CAR and GR, two nuclear receptors which could be involved in this decrease [80]. RORs are newly identified as transcriptional regulators of CYP2C8 in HepG2 cells [81]. RORα and -β are well expressed in different regions of the brain, where they play a role in the control of circadian rhythm. It would be of interest to examine whether ROR and CYP2C8 are colocalized in the brain, and whether CYP2C8 is upregulated by RORs in the brain.
Of note is the expression of CYP2C8 and 2C9 in human endothelial cells (ECs) [82, 83], where they metabolize endogenous arachidonic acid into vasoreactive EETs. CYP2C9 appears to be predominant in the heart, aorta, and cardiac vessels, while CYP2C8 is found in the heart [1]. EETs play critical roles in vascular homeostasis as endothelial-derived hyperpolarizing factors (EDHF) [84]. More importantly, they act as signal molecules that elicit multiple cellular activities, including promotion of endothelial cell proliferation, migration and angiogenesis [85]. Because of the cardioprotective role of EETs in cardiovascular disease [86], it is important to understand the regulation of the expression and activity of CYP2C genes in ECs. Accumulating evidence has demonstrated that the expression of the CYP2C genes in ECs is affected by multiple stimuli, such as hemodynamic and physio-chemical forces [87] as well as the glucocorticoid cortisol [88]. A dramatic enhancement in the expression of the CYP2C genes was reported to be elicited by the Ca2+ antagonist nifedipine in human umbilical endothelial cells (mRNA) and porcine coronary arteries (mRNA & protein) [89]. Some HMG-CoA reductase inhibitors, such as cerivastatin, fluvastatin, and lovastatin, have been found to induce the expression of CYP2C mRNA and protein in native and cultured endothelial cells, but not that of the CYP3A or CYP2J genes [90, 91]. The mechanisms underlying the induction of the CYP2C genes by nifedipine remain to be elucidated. There have been suggestions that the induction by certain statins may be mediated by CAR. For example, CAR has been shown to be activated by statins including cerivastatin, fluvastatin, and atorvastatin in hepatocellular carcinoma FLC7 cells stably transfected with hCAR [92].
Chronic hypoxia has been reported to induce human CYP2C genes and angiogenesis in human endothelial cells [93]. It is also known that hepatic expression of CYP2C genes is markedly increased (including mRNA, protein, and activity) in sudden infant death syndrome (SIDS) patients with an unchanged overall hepatic P450 content although the mechanism is unknown [94, 95]. Recently, the expression of CYP2C8 or 2C9 mRNA and production of EETs were found to be augmented in human endothelial cells upon exposure to hypoxia [93]. The activity of the CYP2C9 promoter was also reported to be modestly enhanced by hypoxia treatment in human endothelial cells [93].The mechanism of this proposed upregulation of the transcription of the CYP2C genes has not been defined. The vascular endothelial growth factor (VEGF) plays a key regulatory role in physiological and pathological angiogenesis. Hypoxia induces its expression by stabilizing the hypoxia inducible factor-1 (HIF-1), which binds to the hypoxia response element (HRE) within the VEGF promoter and strongly enhances its transcription. VEGF was recently reported to activate the promoter of CYP2C9 and enhance the expression of CYP2C8 mRNA and protein in endothelial cells [96]. This enhanced expression is dependent upon the phosphorylation of the AMP-activated protein kinase (AMPK), an energy sensor which is activated under stress situations such as hypoxia. Over-expression of wild type AMPK increased CYP2C expression without VEGF, while the dominant negative AMPK mutant prevented induction of CYP2C expression by VEGF. In the liver, AMPK is activated by PB and some PB-type drugs [97] and this induced AMPK activity has been reported to be essential for the PB induction of P450 in human hepatoma cells and primary hepatocytes [98-100]. Intracellular production of the mitochondrial reactive oxygen species (ROS) triggered by PB seems to be necessary for AMPK activation and induction of P450s by PB since interference with ROS production diminished the phosphorylation of AMPK and decreased PB induction of P450 genes in male leghorn chick hepatoma LMH cells [99].
Interestingly, the CYP2C8/9 metabolite 11, 12-EET was recently shown to increase the level of HIF-1α in human umbilical artery endothelial cells (HUAEC) and human hepatoma cells (Hep3B), possibly via the stabilization of HIF-1α [101]. Induction of VEGF mRNA by hypoxia was enhanced by overexpression of CYP2C8 but efficiently inhibited in HUAEC by sulfaphenazole (10 μM), a high affinity inhibitor of CYP2C9. Although sulphaphenazole also inhibits CYP2C8, the IC50 for CYPC8 is two orders of magnitude lower (IC50 130 μM) than for CYP2C9 (IC50 0.6 μM) [102]. However, the activity of the luciferase promoter containing the hypoxia response element (HRE) from the VEGF promoter as an enhancer was induced by exogenous 11, 12- EET but suppressed by 10 μM sulfaphenazole under hypoxia in HUAEC [101].
Although a self-positive feedback mechanism could be proposed for the induction of CYP2C by hypoxia, it is unclear how EETs increase HIF-1α protein and how phosphorylated AMPK activates the transcription of the CYP2C genes. mRNA of HIF-1α was not increased by EETs [101]; therefore the observed enhancement in induction of HIF-1α proteins by EETs under hypoxia is not due to augmented transcription. EETs have been shown to activate the PI3K/Akt pathway to promote tube formation [103], while this pathway has been shown to be required for protection of HIF-1α from degradation [104]. Possibly EETs could stabilize HIF-1α via activation of the PI3K/Akt pathway to induce the expression of VEGF. More research is needed to clarify the possible effects of hypoxia on CYP2C genes and the mechanism(s) involved.
Conclusions
Human CYP2C enzymes metabolize 20% of clinical drugs and also metabolize arachidonic acid to produce EETs, important endogenous signal molecules that regulate many physiological processes such as vasodilation and angiogenesis. The expression of CYP2C genes is transcriptionally upregulated by exposure to xenobiotics. Drug-responsive nuclear receptors as well as hepatic transcriptional factors bind to cis elements within CYP2C gene promoters to regulate the transcription of CYP2C genes. HNF4α is probably the most important receptor for upregulating the constitutive expression of the CYP2Cs in liver. Variability in expression of the CYP2C enzymes has been shown to correlate with levels of HNF4α in human liver. Moreover, cross-talk between HNF4α sites and PXR/CAR sites appears to be necessary for optimal induction in response to drugs. Other regulatory factors, such as coactivators, corepressors, and signal pathways indirectly modulate the expression of human CYP2C genes. Very little progress has yet been made on the transcriptional regulation of the extrahepatic CYP2Cs.
Animals carrying both transgenic human nuclear receptors and human CYP2Cs would be a promising experimental model for better understanding the transcriptional regulation of human CYP2C genes in vivo [105], due to the lack of direct orthologs for human CYP2C genes in animals. There are also ligand/agonist differences between rodent and human nuclear receptors such as PXR and CAR; therefore, it would be beneficial to use mice with humanized nuclear receptors. For example, Scheer and coworkers [106] have established murine lines with human PXR and human CAR. These mice could be used to establish human CYP2C models. Human primary hepatocytes grown under different matrices also remain an in vitro model for answering some of these questions. Future studies will undoubtedly address how pathological/physiological conditions and stresses perturb CYP2C expression.
Acknowledgments
We thank Drs. Masahiko Negishi and Tatsuya Sueyoshi (NIEHS) for comments regarding the manuscript and Anne Motten of NIEHS for editing this manuscript. We thank Dr. Sailesh Surapureddi for assistance with formatting for submission. This study was supported by the Intramural Research Program of NIH, National Institute of Environmental Health Sciences under NIH intramural project number Z01ES02124.
Abbreviation
- AMPK
AMP activated protein kinase
- CAR
the constitutive androstane receptor
- C/EBPα
CCAAT/enhancer-binding protein α
- CITCO
[6-(4-Chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl)oxime]
- DR
direct repeat
- EETs
epoxyeicosatrienoic acids
- ER
everted repeat
- GR
the glucocorticoid receptor
- HEETs
hydroxyeicosatrienoic acids
- HIF-1
hypoxia inducible factor-1
- HNF
hepatic nuclear factor
- HPF-1
HepG2-specific P450 2C factor-1
- HRE
hypoxia responsive element
- LCA
lithocholic acid
- PGC-1α
the peroxisome proliferator-activated receptor gamma coactivator-1α
- PI3K/Akt
phosphoinositide 3-kinase/protein kinase B
- PPAR
the peroxisome proliferator-activated receptor
- PXR
the pregnane X receptor
- RE
responsive element
- ROR
Retinoic acid related-related orphan receptors
- ROS
reactive oxygen species
- RXR
the retinoid X receptor
- siRNA
silencing RNA
- SRC-1
the nuclear receptor coactivator-1
- VDR
the vitamin D receptor
- VEGF
vascular endothelial growth factor
- XREM
xenobiotic response element
References
- 1.Delozier TC, Kissling GE, Coulter SJ, Dai D, Foley JF, Bradbury JA, Murphy E, Steenbergen C, Zeldin DC, Goldstein JA. Detection of human CYP2C8, CYP2C9, and CYP2J2 in cardiovascular tissues. Drug Metab Dispos. 2007;35(4):682–688. doi: 10.1124/dmd.106.012823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klose TS, Blaisdell JA, Goldstein JA. Gene structure of CYP2C8 and extrahepatic distribution of the human CYP2Cs. J Biochem Mol Toxicol. 1999;13(6):289–295. doi: 10.1002/(sici)1099-0461(1999)13:6<289::aid-jbt1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 3.Goldstein JA. Clinical relevance of genetic polymorphisms in the human CYP2C subfamily. Br J Clin Pharmacol. 2001;52(4):349–355. doi: 10.1046/j.0306-5251.2001.01499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee CR, Goldstein JA, Pieper JA. Cytochrome P450 2C9 polymorphisms: a comprehensive review of the in-vitro and human data. Pharmacogenetics. 2002;12(3):251–263. doi: 10.1097/00008571-200204000-00010. [DOI] [PubMed] [Google Scholar]
- 5.Mega JL, Close SL, Wiviott SD, Shen L, Hockett RD, Brandt JT, Walker JR, Antman EM, Macias W, Braunwald E, Sabatine MS. Cytochrome p-450 polymorphisms and response to clopidogrel. N Engl J Med. 2009;360(4):354–362. doi: 10.1056/NEJMoa0809171. [DOI] [PubMed] [Google Scholar]
- 6.Marill J, Cresteil T, Lanotte M, Chabot GG. Identification of human cytochrome P450s involved in the formation of all-trans-retinoic acid principal metabolites. Mol Pharmacol. 2000;58(6):1341–1348. doi: 10.1124/mol.58.6.1341. [DOI] [PubMed] [Google Scholar]
- 7.Fleming I, Michaelis UR, Bredenkotter D, Fisslthaler B, Dehghani F, Brandes RP, Busse R. Endothelium-derived hyperpolarizing factor synthase (Cytochrome P450 2C9) is a functionally significant source of reactive oxygen species in coronary arteries. Circ Res. 2001;88(1):44–51. doi: 10.1161/01.res.88.1.44. [DOI] [PubMed] [Google Scholar]
- 8.de Morais SM, Wilkinson GR, Blaisdell J, Nakamura K, Meyer UA, Goldstein JA. The major genetic defect responsible for the polymorphism of S-mephenytoin metabolism in humans. J Biol Chem. 1994;269(22):15419–15422. [PubMed] [Google Scholar]
- 9.Dai D, Zeldin DC, Blaisdell JA, Chanas B, Coulter SJ, Ghanayem BI, Goldstein JA. Polymorphisms in human CYP2C8 decrease metabolism of the anticancer drug paclitaxel and arachidonic acid. Pharmacogenetics. 2001;11(7):597–607. doi: 10.1097/00008571-200110000-00006. [DOI] [PubMed] [Google Scholar]
- 10.de Morais SM, Wilkinson GR, Blaisdell J, Meyer UA, Nakamura K, Goldstein JA. Identification of a new genetic defect responsible for the polymorphism of (S)-mephenytoin metabolism in Japanese. Mol Pharmacol. 1994;46(4):594–598. [PubMed] [Google Scholar]
- 11.Klotz U, Schwab M, Treiber G. CYP2C19 polymorphism and proton pump inhibitors. Basic Clin Pharmacol Toxicol. 2004;95(1):2–8. doi: 10.1111/j.1600-0773.2004.pto950102.x. [DOI] [PubMed] [Google Scholar]
- 12.Sullivan-Klose TH, Ghanayem BI, Bell DA, Zhang ZY, Kaminsky LS, Shenfield GM, Miners JO, Birkett DJ, Goldstein JA. The role of the CYP2C9-Leu359 allelic variant in the tolbutamide polymorphism. Pharmacogenetics. 1996;6(4):341–349. doi: 10.1097/00008571-199608000-00007. [DOI] [PubMed] [Google Scholar]
- 13.Sim SC, Risinger C, Dahl ML, Aklillu E, Christensen M, Bertilsson L, Ingelman-Sundberg M. A common novel CYP2C19 gene variant causes ultrarapid drug metabolism relevant for the drug response to proton pump inhibitors and antidepressants. Clin Pharmacol Ther. 2006;79(1):103–113. doi: 10.1016/j.clpt.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 14.Limdi NA, Arnett DK, Goldstein JA, Beasley TM, McGwin G, Adler BK, Acton RT. Influence of CYP2C9 and VKORC1 on warfarin dose, anticoagulation attainment and maintenance among European-Americans and African-Americans. Pharmacogenomics. 2008;9(5):511–526. doi: 10.2217/14622416.9.5.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Limdi NA, McGwin G, Goldstein JA, Beasley TM, Arnett DK, Adler BK, Baird MF, Acton RT. Influence of CYP2C9 and VKORC1 1173C/T genotype on the risk of hemorrhagic complications in African-American and European-American patients on warfarin. Clin Pharmacol Ther. 2008;83(2):312–321. doi: 10.1038/sj.clpt.6100290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarasquete M, Gonzalez M, San Miguel J, Garcia-Sanz R. Bisphosphonate-related osteonecrosis: genetic and acquired risk factors. Oral Dis. 2009 doi: 10.1111/j.1601-0825.2009.01568.x. [DOI] [PubMed] [Google Scholar]
- 17.Raucy JL, Mueller L, Duan K, Allen SW, Strom S, Lasker JM. Expression and induction of CYP2C P450 enzymes in primary cultures of human hepatocytes. J Pharmacol Exp Ther. 2002;302(2):475–482. doi: 10.1124/jpet.102.033837. [DOI] [PubMed] [Google Scholar]
- 18.Gerbal-Chaloin S, Pascussi JM, Pichard-Garcia L, Daujat M, Waechter F, Fabre JM, Carrere N, Maurel P. Induction of CYP2C genes in human hepatocytes in primary culture. Drug Metab Dispos. 2001;29(3):242–251. [PubMed] [Google Scholar]
- 19.Zilly W, Breimer DD, Richter E. Induction of drug metabolism in man after rifampicin treatment measured by increased hexobarbital and tolbutamide clearance. Eur J Clin Pharmacol. 1975;9(23):219–227. doi: 10.1007/BF00614021. [DOI] [PubMed] [Google Scholar]
- 20.Zhou HH, Anthony LB, Wood AJ, Wilkinson GR. Induction of polymorphic 4′-hydroxylation of S-mephenytoin by rifampicin. Br J Clin Pharmacol. 1990;30(3):471–475. doi: 10.1111/j.1365-2125.1990.tb03799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williamson KM, Patterson JH, McQueen RH, Adams KF, Jr, Pieper JA. Effects of erythromycin or rifampin on losartan pharmacokinetics in healthy volunteers. Clin Pharmacol Ther. 1998;63(3):316–323. doi: 10.1016/S0009-9236(98)90163-1. [DOI] [PubMed] [Google Scholar]
- 22.Niemi M, Backman JT, Neuvonen M, Neuvonen PJ, Kivisto KT. Effects of rifampin on the pharmacokinetics and pharmacodynamics of glyburide and glipizide. Clin Pharmacol Ther. 2001;69(6):400–406. doi: 10.1067/mcp.2001.115822. [DOI] [PubMed] [Google Scholar]
- 23.Madan A, Graham RA, Carroll KM, Mudra DR, Burton LA, Krueger LA, Downey AD, Czerwinski M, Forster J, Ribadeneira MD, Gan LS, LeCluyse EL, Zech K, Robertson P, Jr, Koch P, Antonian L, Wagner G, Yu L, Parkinson A. Effects of prototypical microsomal enzyme inducers on cytochrome P450 expression in cultured human hepatocytes. Drug Metab Dispos. 2003;31(4):421–431. doi: 10.1124/dmd.31.4.421. [DOI] [PubMed] [Google Scholar]
- 24.Handschin C, Meyer UA. Induction of drug metabolism: the role of nuclear receptors. Pharmacol Rev. 2003;55(4):649–673. doi: 10.1124/pr.55.4.2. [DOI] [PubMed] [Google Scholar]
- 25.Maglich JM, Parks DJ, Moore LB, Collins JL, Goodwin B, Billin AN, Stoltz CA, Kliewer SA, Lambert MH, Willson TM, Moore JT. Identification of a novel human constitutive androstane receptor (CAR) agonist and its use in the identification of CAR target genes. J Biol Chem. 2003;278(19):17277–17283. doi: 10.1074/jbc.M300138200. [DOI] [PubMed] [Google Scholar]
- 26.Jackson JP, Ferguson SS, Negishi M, Goldstein JA. Phenytoin induction of the cyp2c37 gene is mediated by the constitutive androstane receptor. Drug Metab Dispos. 2006;34(12):2003–2010. doi: 10.1124/dmd.106.012005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawana K, Ikuta T, Kobayashi Y, Gotoh O, Takeda K, Kawajiri K. Molecular Mechanism of Nuclear Translocation of an Orphan Nuclear Receptor, SXR. Mol Pharmacol. 2003;63(3):524–531. doi: 10.1124/mol.63.3.524. [DOI] [PubMed] [Google Scholar]
- 28.Zelko I, Sueyoshi T, Kawamoto T, Moore R, Negishi M. The peptide near the C terminus regulates receptor CAR nuclear translocation induced by xenochemicals in mouse liver. Mol Cell Biol. 2001;21(8):2838–2846. doi: 10.1128/MCB.21.8.2838-2846.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drocourt L, Ourlin JC, Pascussi JM, Maurel P, Vilarem MJ. Expression of CYP3A4, CYP2B6, and CYP2C9 is regulated by the vitamin D receptor pathway in primary human hepatocytes. J Biol Chem. 2002;277(28):25125–25132. doi: 10.1074/jbc.M201323200. [DOI] [PubMed] [Google Scholar]
- 30.Makishima M, Lu TT, Xie W, Whitfield GK, Domoto H, Evans RM, Haussler MR, Mangelsdorf DJ. Vitamin D receptor as an intestinal bile acid sensor. Science. 2002;296(5571):1313–1316. doi: 10.1126/science.1070477. [DOI] [PubMed] [Google Scholar]
- 31.Gerbal-Chaloin S, Daujat M, Pascussi JM, Pichard-Garcia L, Vilarem MJ, Maurel P. Transcriptional regulation of CYP2C9 gene. Role of glucocorticoid receptor and constitutive androstane receptor. J Biol Chem. 2002;277(1):209–217. doi: 10.1074/jbc.M107228200. [DOI] [PubMed] [Google Scholar]
- 32.Ferguson SS, LeCluyse EL, Negishi M, Goldstein JA. Regulation of human CYP2C9 by the constitutive androstane receptor: discovery of a new distal binding site. Mol Pharmacol. 2002;62(3):737–746. doi: 10.1124/mol.62.3.737. [DOI] [PubMed] [Google Scholar]
- 33.Ferguson SS, Chen Y, LeCluyse EL, Negishi M, Goldstein JA. Human CYP2C8 is transcriptionally regulated by the nuclear receptors constitutive androstane receptor, pregnane X receptor, glucocorticoid receptor, and hepatic nuclear factor 4alpha. Mol Pharmacol. 2005;68(3):747–757. doi: 10.1124/mol.105.013169. [DOI] [PubMed] [Google Scholar]
- 34.Chen Y, Ferguson SS, Negishi M, Goldstein JA. Induction of human CYP2C9 by rifampicin, hyperforin, and phenobarbital is mediated by the pregnane X receptor. J Pharmacol Exp Ther. 2004;308(2):495–501. doi: 10.1124/jpet.103.058818. [DOI] [PubMed] [Google Scholar]
- 35.Chen Y, Ferguson SS, Negishi M, Goldstein JA. Identification of constitutive androstane receptor and glucocorticoid receptor binding sites in the CYP2C19 promoter. Mol Pharmacol. 2003;64(2):316–324. doi: 10.1124/mol.64.2.316. [DOI] [PubMed] [Google Scholar]
- 36.Jackson JP, Ferguson SS, Moore R, Negishi M, Goldstein JA. The constitutive active/androstane receptor regulates phenytoin induction of Cyp2c29. Mol Pharmacol. 2004;65(6):1397–1404. doi: 10.1124/mol.65.6.1397. [DOI] [PubMed] [Google Scholar]
- 37.Chen Y, Coulter S, Jetten AM, Goldstein JA. Identification of Human CYP2C8 as a Retinoic Acid Receptor-Related Orphan Receptor (ROR) Target Gene. J Pharmacol Exp Ther. 2009 doi: 10.1124/jpet.108.148916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akiyama TE, Gonzalez FJ. Regulation of P450 genes by liver-enriched transcription factors and nuclear receptors. Biochim Biophys Acta. 2003;1619(3):223–234. doi: 10.1016/s0304-4165(02)00480-4. [DOI] [PubMed] [Google Scholar]
- 39.Jover R, Bort R, Gomez-Lechon MJ, Castell JV. Cytochrome P450 regulation by hepatocyte nuclear factor 4 in human hepatocytes: a study using adenovirus-mediated antisense targeting. Hepatology. 2001;33(3):668–675. doi: 10.1053/jhep.2001.22176. [DOI] [PubMed] [Google Scholar]
- 40.Kamiyama Y, Matsubara T, Yoshinari K, Nagata K, Kamimura H, Yamazoe Y. Role of human hepatocyte nuclear factor 4alpha in the expression of drug-metabolizing enzymes and transporters in human hepatocytes assessed by use of small interfering RNA. Drug Metab Pharmacokinet. 2007;22(4):287–298. doi: 10.2133/dmpk.22.287. [DOI] [PubMed] [Google Scholar]
- 41.Wortham M, Czerwinski M, He L, Parkinson A, Wan YJ. Expression of constitutive androstane receptor, hepatic nuclear factor 4 alpha, and P450 oxidoreductase genes determines interindividual variability in basal expression and activity of a broad scope of xenobiotic metabolism genes in the human liver. Drug Metab Dispos. 2007;35(9):1700–1710. doi: 10.1124/dmd.107.016436. [DOI] [PubMed] [Google Scholar]
- 42.Venepally P, Chen D, Kemper B. Transcriptional regulatory elements for basal expression of cytochrome P450IIC genes. J Biol Chem. 1992;267(24):17333–17338. [PubMed] [Google Scholar]
- 43.Kawashima S, Kobayashi K, Takama K, Higuchi T, Furihata T, Hosokawa M, Chiba K. Involvement of hepatocyte nuclear factor 4alpha in the different expression level between CYP2C9 and CYP2C19 in the human liver. Drug Metab Dispos. 2006;34(6):1012–1018. doi: 10.1124/dmd.106.009365. [DOI] [PubMed] [Google Scholar]
- 44.Chen Y, Kissling G, Negishi M, Goldstein JA. The nuclear receptors constitutive androstane receptor and pregnane X receptor cross-talk with hepatic nuclear factor 4alpha to synergistically activate the human CYP2C9 promoter. J Pharmacol Exp Ther. 2005;314(3):1125–1133. doi: 10.1124/jpet.105.087072. [DOI] [PubMed] [Google Scholar]
- 45.Bort R, Gomez-Lechon MJ, Castell JV, Jover R. Role of hepatocyte nuclear factor 3 gamma in the expression of human CYP2C genes. Arch Biochem Biophys. 2004;426(1):63–72. doi: 10.1016/j.abb.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 46.Jover R, Bort R, Gomez-Lechon MJ, Castell JV. Re-expression of C/EBP alpha induces CYP2B6, CYP2C9 and CYP2D6 genes in HepG2 cells. FEBS Lett. 1998;431(2):227–230. doi: 10.1016/s0014-5793(98)00746-7. [DOI] [PubMed] [Google Scholar]
- 47.de Morais SM, Schweikl H, Blaisdell J, Goldstein JA. Gene structure and upstream regulatory regions of human CYP2C9 and CYP2C18. Biochem Biophys Res Commun. 1993;194(1):194–201. doi: 10.1006/bbrc.1993.1803. [DOI] [PubMed] [Google Scholar]
- 48.Ibeanu GC, Goldstein JA. Transcriptional regulation of human CYP2C genes: functional comparison of CYP2C9 and CYP2C18 promoter regions. Biochemistry (Mosc) 1995;34(25):8028–8036. doi: 10.1021/bi00025a008. [DOI] [PubMed] [Google Scholar]
- 49.Jetten AM, Joo JH. Retinoid-related Orphan Receptors (RORs): Roles in Cellular Differentiation and Development. Adv Dev Biol. 2006;16:313–355. doi: 10.1016/S1574-3349(06)16010-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang HS, Angers M, Beak JY, Wu X, Gimble JM, Wada T, Xie W, Collins JB, Grissom SF, Jetten AM. Gene expression profiling reveals a regulatory role for ROR alpha and ROR gamma in phase I and phase II metabolism. Physiol Genomics. 2007;31(2):281–294. doi: 10.1152/physiolgenomics.00098.2007. [DOI] [PubMed] [Google Scholar]
- 51.Pascussi JM, Gerbal-Chaloin S, Fabre JM, Maurel P, Vilarem MJ. Dexamethasone enhances constitutive androstane receptor expression in human hepatocytes: consequences on cytochrome P450 gene regulation. Mol Pharmacol. 2000;58(6):1441–1450. doi: 10.1124/mol.58.6.1441. [DOI] [PubMed] [Google Scholar]
- 52.Pascussi JM, Drocourt L, Fabre JM, Maurel P, Vilarem MJ. Dexamethasone induces pregnane X receptor and retinoid X receptor-alpha expression in human hepatocytes: synergistic increase of CYP3A4 induction by pregnane X receptor activators. Mol Pharmacol. 2000;58(2):361–372. doi: 10.1124/mol.58.2.361. [DOI] [PubMed] [Google Scholar]
- 53.Kamiya A, Inoue Y, Gonzalez FJ. Role of the hepatocyte nuclear factor 4alpha in control of the pregnane X receptor during fetal liver development. Hepatology. 2003;37(6):1375–1384. doi: 10.1053/jhep.2003.50212. [DOI] [PubMed] [Google Scholar]
- 54.Tirona RG, Lee W, Leake BF, Lan LB, Cline CB, Lamba V, Parviz F, Duncan SA, Inoue Y, Gonzalez FJ, Schuetz EG, Kim RB. The orphan nuclear receptor HNF4alpha determines PXR- and CAR-mediated xenobiotic induction of CYP3A4. Nat Med. 2003;9(2):220–224. doi: 10.1038/nm815. [DOI] [PubMed] [Google Scholar]
- 55.Ding X, Lichti K, Kim I, Gonzalez FJ, Staudinger JL. Regulation of constitutive androstane receptor and its target genes by fasting, cAMP, hepatocyte nuclear factor alpha, and the coactivator peroxisome proliferator-activated receptor gamma coactivator-1alpha. J Biol Chem. 2006;281(36):26540–26551. doi: 10.1074/jbc.M600931200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Assenat E, Gerbal-Chaloin S, Larrey D, Saric J, Fabre JM, Maurel P, Vilarem MJ, Pascussi JM. Interleukin 1beta inhibits CAR-induced expression of hepatic genes involved in drug and bilirubin clearance. Hepatology. 2004;40(4):951–960. doi: 10.1002/hep.20387. [DOI] [PubMed] [Google Scholar]
- 57.Pascussi JM, Gerbal-Chaloin S, Pichard-Garcia L, Daujat M, Fabre JM, Maurel P, Vilarem MJ. Interleukin-6 negatively regulates the expression of pregnane X receptor and constitutively activated receptor in primary human hepatocytes. Biochem Biophys Res Commun. 2000;274(3):707–713. doi: 10.1006/bbrc.2000.3219. [DOI] [PubMed] [Google Scholar]
- 58.Aitken AE, Morgan ET. Gene-specific effects of inflammatory cytokines on cytochrome P450 2C, 2B6 and 3A4 mRNA levels in human hepatocytes. Drug Metab Dispos. 2007;35(9):1687–1693. doi: 10.1124/dmd.107.015511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Surapureddi S, Rana R, Reddy JK, Goldstein JA. Nuclear receptor coactivator 6 mediates the synergistic activation of human cytochrome P-450 2C9 by the constitutive androstane receptor and hepatic nuclear factor-4alpha. Mol Pharmacol. 2008;74(3):913–923. doi: 10.1124/mol.108.048983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martinez-Jimenez CP, Castell JV, Gomez-Lechon MJ, Jover R. Transcriptional activation of CYP2C9, CYP1A1, and CYP1A2 by hepatocyte nuclear factor 4alpha requires coactivators peroxisomal proliferator activated receptor-gamma coactivator 1alpha and steroid receptor coactivator 1. Mol Pharmacol. 2006;70(5):1681–1692. doi: 10.1124/mol.106.025403. [DOI] [PubMed] [Google Scholar]
- 61.Rhee J, Ge H, Yang W, Fan M, Handschin C, Cooper M, Lin J, Li C, Spiegelman BM. Partnership of PGC-1alpha and HNF4alpha in the regulation of lipoprotein metabolism. J Biol Chem. 2006;281(21):14683–14690. doi: 10.1074/jbc.M512636200. [DOI] [PubMed] [Google Scholar]
- 62.Paine MF, Hart HL, Ludington SS, Haining RL, Rettie AE, Zeldin DC. The human intestinal cytochrome P450 “pie”. Drug Metab Dispos. 2006;34(5):880–886. doi: 10.1124/dmd.105.008672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lapple F, von Richter O, Fromm MF, Richter T, Thon KP, Wisser H, Griese EU, Eichelbaum M, Kivisto KT. Differential expression and function of CYP2C isoforms in human intestine and liver. Pharmacogenetics. 2003;13(9):565–575. doi: 10.1097/00008571-200309000-00005. [DOI] [PubMed] [Google Scholar]
- 64.Glaeser H, Drescher S, Eichelbaum M, Fromm MF. Influence of rifampicin on the expression and function of human intestinal cytochrome P450 enzymes. Br J Clin Pharmacol. 2005;59(2):199–206. doi: 10.1111/j.1365-2125.2004.02265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oscarson M, Burk O, Winter S, Schwab M, Wolbold R, Dippon J, Eichelbaum M, Meyer UA. Effects of rifampicin on global gene expression in human small intestine. Pharmacogenet Genomics. 2007;17(11):907–918. doi: 10.1097/FPC.0b013e3280143dfc. [DOI] [PubMed] [Google Scholar]
- 66.Capdevila JH, Falck JR, Imig JD. Roles of the cytochrome P450 arachidonic acid monooxygenases in the control of systemic blood pressure and experimental hypertension. Kidney Int. 2007;72(6):683–689. doi: 10.1038/sj.ki.5002394. [DOI] [PubMed] [Google Scholar]
- 67.Enayetallah AE, French RA, Thibodeau MS, Grant DF. Distribution of soluble epoxide hydrolase and of cytochrome P450 2C8, 2C9, and 2J2 in human tissues. J Histochem Cytochem. 2004;52(4):447–454. doi: 10.1177/002215540405200403. [DOI] [PubMed] [Google Scholar]
- 68.Zeldin DC, DuBois RN, Falck JR, Capdevila JH. Molecular cloning, expression and characterization of an endogenous human cytochrome P450 arachidonic acid epoxygenase isoform. Arch Biochem Biophys. 1995;322(1):76–86. doi: 10.1006/abbi.1995.1438. [DOI] [PubMed] [Google Scholar]
- 69.Wang MH, Smith A, Zhou Y, Chang HH, Lin S, Zhao X, Imig JD, Dorrance AM. Downregulation of renal CYP-derived eicosanoid synthesis in rats with diet-induced hypertension. Hypertension. 2003;42(4):594–599. doi: 10.1161/01.HYP.0000090123.55365.BA. [DOI] [PubMed] [Google Scholar]
- 70.Dey A, Williams RS, Pollock DM, Stepp DW, Newman JW, Hammock BD, Imig JD. Altered kidney CYP2C and cyclooxygenase-2 levels are associated with obesity-related albuminuria. Obes Res. 2004;12(8):1278–1289. doi: 10.1038/oby.2004.162. [DOI] [PubMed] [Google Scholar]
- 71.Luo P, Zhou Y, Chang HH, Zhang J, Seki T, Wang CY, Inscho EW, Wang MH. Glomerular 20-HETE, EETs, and TGF-{beta}1 in diabetic nephropathy. Am J Physiol Renal Physiol. 2009;296(3):F556–563. doi: 10.1152/ajprenal.90613.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Elmarakby AA, Quigley JE, Pollock DM, Imig JD. Tumor necrosis factor alpha blockade increases renal Cyp2c23 expression and slows the progression of renal damage in salt-sensitive hypertension. Hypertension. 2006;47(3):557–562. doi: 10.1161/01.HYP.0000198545.01860.90. [DOI] [PubMed] [Google Scholar]
- 73.Ng VY, Huang Y, Reddy LM, Falck JR, Lin ET, Kroetz DL. Cytochrome P450 eicosanoids are activators of peroxisome proliferator-activated receptor alpha. Drug Metab Dispos. 2007;35(7):1126–1134. doi: 10.1124/dmd.106.013839. [DOI] [PubMed] [Google Scholar]
- 74.Nakagawa K, Holla VR, Wei Y, Wang WH, Gatica A, Wei S, Mei S, Miller CM, Cha DR, Price E, Jr, Zent R, Pozzi A, Breyer MD, Guan Y, Falck JR, Waterman MR, Capdevila JH. Salt-sensitive hypertension is associated with dysfunctional Cyp4a10 gene and kidney epithelial sodium channel. J Clin Invest. 2006;116(6):1696–1702. doi: 10.1172/JCI27546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Muller DN, Theuer J, Shagdarsuren E, Kaergel E, Honeck H, Park JK, Markovic M, Barbosa-Sicard E, Dechend R, Wellner M, Kirsch T, Fiebeler A, Rothe M, Haller H, Luft FC, Schunck WH. A peroxisome proliferator-activated receptor-alpha activator induces renal CYP2C23 activity and protects from angiotensin II-induced renal injury. Am J Pathol. 2004;164(2):521–532. doi: 10.1016/s0002-9440(10)63142-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nelson DR, Zeldin DC, Hoffman SM, Maltais LJ, Maltais LJ, Wain HM, Nebert DW. Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes, and alternative-splice variants. Pharmacogenetics. 2004;14(1):35–18. doi: 10.1097/00008571-200401000-00001. [DOI] [PubMed] [Google Scholar]
- 77.Wang H, Zhao Y, Bradbury JA, Graves JP, Foley J, Blaisdell JA, Goldstein JA, Zeldin DC. Cloning, expression, and characterization of three new mouse cytochrome p450 enzymes and partial characterization of their Fatty Acid oxidation activities. Mol Pharmacol. 2004;65(5):1148–1158. doi: 10.1124/mol.65.5.1148. [DOI] [PubMed] [Google Scholar]
- 78.Knupfer H, Knupfer MM, Hotfilder M, Preiss R. P450-expression in brain tumors. Oncol Res. 1999;11(1112):523–528. [PubMed] [Google Scholar]
- 79.McFayden MC, Melvin WT, Murray GI. Regional distribution of individual forms of cytochrome P450 mRNA in normal adult human brain. Biochem Pharmacol. 1998;55(6):825–830. doi: 10.1016/s0006-2952(97)00516-9. [DOI] [PubMed] [Google Scholar]
- 80.Malaplate-Armand C, Ferrari L, Masson C, Visvikis-Siest S, Lambert H, Batt AM. Down-regulation of astroglial CYP2C, glucocorticoid receptor and constitutive androstane receptor genes in response to cocaine in human U373 MG astrocytoma cells. Toxicol Lett. 2005;159(3):203–211. doi: 10.1016/j.toxlet.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 81.Chen Y, Jetten A, Goldstein J. The retinoic acid receptor-related orphan receptor (ROR) regulates human CYP2C8. The FASEB Journal. 2007;21:886.813. [Google Scholar]
- 82.Hillig T, Krustrup P, Fleming I, Osada T, Saltin B, Hellsten Y. Cytochrome P450 2C9 plays an important role in the regulation of exercise-induced skeletal muscle blood flow and oxygen uptake in humans. J Physiol. 2003;546(Pt 1):307–314. doi: 10.1113/jphysiol.2002.030833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lin JHC, Kobari Y, Zhu Y, Stemerman MB, Pritchard KAJ. Human umbilical vein endothelial cells express P450 2C8 mRNA:cloning of endothelial P450 epoxygenase. Endothelium. 1996;4:219–229. [Google Scholar]
- 84.Fleming I. Cytochrome p450 and vascular homeostasis. Circ Res. 2001;89(9):753–762. doi: 10.1161/hh2101.099268. [DOI] [PubMed] [Google Scholar]
- 85.Fleming I. DiscrEET regulators of homeostasis: epoxyeicosatrienoic acids, cytochrome P450 epoxygenases and vascular inflammation. Trends Pharmacol Sci. 2007;28(9):448–452. doi: 10.1016/j.tips.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 86.Chehal MK, Granville DJ. Cytochrome p450 2C (CYP2C) in ischemic heart injury and vascular dysfunction. Can J Physiol Pharmacol. 2006;84(1):15–20. doi: 10.1139/y05-139. [DOI] [PubMed] [Google Scholar]
- 87.Fisslthaler B, Popp R, Michaelis UR, Kiss L, Fleming I, Busse R. Cyclic stretch enhances the expression and activity of coronary endothelium-derived hyperpolarizing factor synthase. Hypertension. 2001;38(6):1427–1432. doi: 10.1161/hy1201.096532. [DOI] [PubMed] [Google Scholar]
- 88.Bauersachs J, Christ M, Ertl G, Michaelis UR, Fisslthaler B, Busse R, Fleming I. Cytochrome P450 2C expression and EDHF-mediated relaxation in porcine coronary arteries is increased by cortisol. Cardiovasc Res. 2002;54(3):669–675. doi: 10.1016/s0008-6363(02)00257-2. [DOI] [PubMed] [Google Scholar]
- 89.Fisslthaler B, Hinsch N, Chataigneau T, Popp R, Kiss L, Busse R, Fleming I. Nifedipine increases cytochrome P4502C expression and endothelium-derived hyperpolarizing factor-mediated responses in coronary arteries. Hypertension. 2000;36(2):270–275. doi: 10.1161/01.hyp.36.2.270. [DOI] [PubMed] [Google Scholar]
- 90.Fisslthaler B, Michaelis UR, Randriamboavonjy V, Busse R, Fleming I. Cytochrome P450 epoxygenases and vascular tone: novel role for HMG-CoA reductase inhibitors in the regulation of CYP 2C expression. Biochim Biophys Acta. 2003;1619(3):332–339. doi: 10.1016/s0304-4165(02)00492-0. [DOI] [PubMed] [Google Scholar]
- 91.Bertrand-Thiebault C, Masson C, Siest G, Batt AM, Visvikis-Siest S. Effect of HMGCoA reductase inhibitors on cytochrome P450 expression in endothelial cell line. J Cardiovasc Pharmacol. 2007;49(5):306–315. doi: 10.1097/FJC.0b013e31803e8756. [DOI] [PubMed] [Google Scholar]
- 92.Kobayashi K, Yamanaka Y, Iwazaki N, Nakajo I, Hosokawa M, Negishi M, Chiba K. Identification of HMG-CoA reductase inhibitors as activators for human, mouse and rat constitutive androstane receptor. Drug Metab Dispos. 2005;33(7):924–929. doi: 10.1124/dmd.104.002741. [DOI] [PubMed] [Google Scholar]
- 93.Michaelis UR, Fisslthaler B, Barbosa-Sicard E, Falck JR, Fleming I, Busse R. Cytochrome P450 epoxygenases 2C8 and 2C9 are implicated in hypoxia-induced endothelial cell migration and angiogenesis. J Cell Sci. 2005;118(23):5489–5498. doi: 10.1242/jcs.02674. [DOI] [PubMed] [Google Scholar]
- 94.Treluyer JM, Cheron G, Sonnier M, Cresteil T. Cytochrome P-450 expression in sudden infant death syndrome. Biochem Pharmacol. 1996;52(3):497–504. doi: 10.1016/0006-2952(96)00253-5. [DOI] [PubMed] [Google Scholar]
- 95.Treluyer JM, Benech H, Colin I, Pruvost A, Cheron G, Cresteil T. Ontogenesis of CYP2C-dependent arachidonic acid metabolism in the human liver: relationship with sudden infant death syndrome. Pediatr Res. 2000;47(5):677–683. doi: 10.1203/00006450-200005000-00020. [DOI] [PubMed] [Google Scholar]
- 96.Webler AC, Michaelis UR, Popp R, Barbosa-Sicard E, Murugan A, Falck JR, Fisslthaler B, Fleming I. Epoxyeicosatrienoic acids are part of the VEGF-activated signaling cascade leading to angiogenesis. Am J Physiol Cell Physiol. 2008;295(5):C1292–1301. doi: 10.1152/ajpcell.00230.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shindo S, Numazawa S, Yoshida T. A physiological role of AMP-activated protein kinase in phenobarbital-mediated constitutive androstane receptor activation and CYP2B induction. Biochem J. 2007;401(3):735–741. doi: 10.1042/BJ20061238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rencurel F, Foretz M, Kaufmann MR, Stroka D, Looser R, Leclerc I, da Silva Xavier G, Rutter GA, Viollet B, Meyer UA. Stimulation of AMP-activated protein kinase is essential for the induction of drug metabolizing enzymes by phenobarbital in human and mouse liver. Mol Pharmacol. 2006;70(6):1925–1934. doi: 10.1124/mol.106.029421. [DOI] [PubMed] [Google Scholar]
- 99.Blattler SM, Rencurel F, Kaufmann MR, Meyer UA. In the regulation of cytochrome P450 genes, phenobarbital targets LKB1 for necessary activation of AMP-activated protein kinase. Proc Natl Acad Sci U S A. 2007;104(3):1045–1050. doi: 10.1073/pnas.0610216104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rencurel F, Stenhouse A, Hawley SA, Friedberg T, Hardie DG, Sutherland C, Wolf CR. AMP-activated protein kinase mediates phenobarbital induction of CYP2B gene expression in hepatocytes and a newly derived human hepatoma cell line. J Biol Chem. 2005;280(6):4367–4373. doi: 10.1074/jbc.M412711200. [DOI] [PubMed] [Google Scholar]
- 101.Suzuki S, Oguro A, Osada-Oka M, Funae Y, Imaoka S. Epoxyeicosatrienoic acids and/or their metabolites promote hypoxic response of cells. J Pharmacol Sci. 2008;108(1):79–88. doi: 10.1254/jphs.08122fp. [DOI] [PubMed] [Google Scholar]
- 102.Marques-Soares C, Dijols S, Macherey AC, Wester MR, Johnson EF, Dansette PM, Mansuy D. Sulfaphenazole derivatives as tools for comparing cytochrome P450 2C5 and human cytochromes P450 2Cs: identification of a new high affinity substrate common to those enzymes. Biochemistry (Mosc) 2003;42(21):6363–6369. doi: 10.1021/bi027391+. [DOI] [PubMed] [Google Scholar]
- 103.Webler AC, Popp R, Korff T, Michaelis UR, Urbich C, Busse R, Fleming I. Cytochrome P450 2C9-induced angiogenesis is dependent on EphB4. Arterioscler Thromb Vasc Biol. 2008;28(6):1123–1129. doi: 10.1161/ATVBAHA.107.161190. [DOI] [PubMed] [Google Scholar]
- 104.Zhou J, Schmid T, Frank R, Brune B. PI3K/Akt is required for heat shock proteins to protect hypoxia-inducible factor 1alpha from pVHL-independent degradation. J Biol Chem. 2004;279(14):13506–13513. doi: 10.1074/jbc.M310164200. [DOI] [PubMed] [Google Scholar]
- 105.Lofgren S, Baldwin RM, Hiratsuka M, Lindqvist A, Carlberg A, Sim SC, Schulke M, Snait M, Edenro A, Fransson-Steen R, Terelius Y, Ingelman-Sundberg M. Generation of mice transgenic for human CYP2C18 and CYP2C19: characterization of the sexually dimorphic gene and enzyme expression. Drug Metab Dispos. 2008;36(5):955–962. doi: 10.1124/dmd.107.019349. [DOI] [PubMed] [Google Scholar]
- 106.Scheer N, Ross J, Rode A, Zevnik B, Niehaves S, Faust N, Wolf CR. A novel panel of mouse models to evaluate the role of human pregnane X receptor and constitutive androstane receptor in drug response. J Clin Invest. 2008;118(9):3228–3239. doi: 10.1172/JCI35483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Heimark LD, Gibaldi M, Trager WF, O'Reilly RA, Goulart DA. The mechanism of the warfarin-rifampin drug interaction in humans. Clin Pharmacol Ther. 1987;42(4):388–394. doi: 10.1038/clpt.1987.168. [DOI] [PubMed] [Google Scholar]
- 108.Niemi M, Backman JT, Fromm MF, Neuvonen PJ, Kivisto KT. Pharmacokinetic interactions with rifampicin : clinical relevance. Clin Pharmacokinet. 2003;42(9):819–850. doi: 10.2165/00003088-200342090-00003. [DOI] [PubMed] [Google Scholar]
- 109.Rae JM, Johnson MD, Lippman ME, Flockhart DA. Rifampin is a selective, pleiotropic inducer of drug metabolism genes in human hepatocytes: studies with cDNA and oligonucleotide expression arrays. J Pharmacol Exp Ther. 2001;299(3):849–857. [PubMed] [Google Scholar]
- 110.Jiang X, Williams KM, Liauw WS, Ammit AJ, Roufogalis BD, Duke CC, Day RO, McLachlan AJ. Effect of St John's wort and ginseng on the pharmacokinetics and pharmacodynamics of warfarin in healthy subjects. Br J Clin Pharmacol. 2004;57(5):592–599. doi: 10.1111/j.1365-2125.2003.02051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xu H, Williams KM, Liauw WS, Murray M, Day RO, McLachlan AJ. Effects of St John's wort and CYP2C9 genotype on the pharmacokinetics and pharmacodynamics of gliclazide. Br J Pharmacol. 2008;153(7):1579–1586. doi: 10.1038/sj.bjp.0707685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Komoroski BJ, Zhang S, Cai H, Hutzler JM, Frye R, Tracy TS, Strom SC, Lehmann T, Ang CY, Cui YY, Venkataramanan R. Induction and inhibition of cytochromes p450 by the St. John's wort constituent hyperforin in human hepatocyte cultures. Drug Metab Dispos. 2004;32(5):512–518. doi: 10.1124/dmd.32.5.512. [DOI] [PubMed] [Google Scholar]
- 113.Sahi J, Stern RH, Milad MA, Rose KA, Gibson G, Zheng X, Stilgenbauer L, Sadagopan N, Jolley S, Gilbert D, LeCluyse EL. Effects of avasimibe on cytochrome P450 2C9 expression in vitro and in vivo. Drug Metab Dispos. 2004;32(12):1370–1376. doi: 10.1124/dmd.104.000208. [DOI] [PubMed] [Google Scholar]
- 114.Fulco PP, Zingone MM, Higginson RT. Possible antiretroviral therapy-warfarin drug interaction. Pharmacotherapy. 2008;28(7):945–949. doi: 10.1592/phco.28.7.945. [DOI] [PubMed] [Google Scholar]
- 115.Lim ML, Min SS, Eron JJ, Bertz RJ, Robinson M, Gaedigk A, Kashuba AD. Coadministration of lopinavir/ritonavir and phenytoin results in two-way drug interaction through cytochrome P-450 induction. J Acquir Immune Defic Syndr. 2004;36(5):1034–1040. doi: 10.1097/00126334-200408150-00006. [DOI] [PubMed] [Google Scholar]
- 116.Dixit V, Hariparsad N, Li F, Desai P, Thummel KE, Unadkat JD. Cytochrome P450 enzymes and transporters induced by anti-human immunodeficiency virus protease inhibitors in human hepatocytes: implications for predicting clinical drug interactions. Drug Metab Dispos. 2007;35(10):1853–1859. doi: 10.1124/dmd.107.016089. [DOI] [PubMed] [Google Scholar]
- 117.Chang TK, Yu L, Maurel P, Waxman DJ. Enhanced cyclophosphamide and ifosfamide activation in primary human hepatocyte cultures: response to cytochrome P-450 inducers and autoinduction by oxazaphosphorines. Cancer Res. 1997;57(10):1946–1954. [PubMed] [Google Scholar]
- 118.Drocourt L, Pascussi JM, Assenat E, Fabre JM, Maurel P, Vilarem MJ. Calcium channel modulators of the dihydropyridine family are human pregnane X receptor activators and inducers of CYP3A, CYP2B, and CYP2C in human hepatocytes. Drug Metab Dispos. 2001;29(10):1325–1331. [PubMed] [Google Scholar]
- 119.Spina E, Pisani F, Perucca E. Clinically significant pharmacokinetic drug interactions with carbamazepine. An update. Clin Pharmacokinet. 1996;31(3):198–214. doi: 10.2165/00003088-199631030-00004. [DOI] [PubMed] [Google Scholar]
- 120.Depre M, Van Hecken A, Oeyen M, De Lepeleire I, Laethem T, Rothenberg P, Petty KJ, Majumdar A, Crumley T, Panebianco D, Bergman A, de Hoon JN. Effect of aprepitant on the pharmacokinetics and pharmacodynamics of warfarin. Eur J Clin Pharmacol. 2005;61(56):341–346. doi: 10.1007/s00228-005-0907-8. [DOI] [PubMed] [Google Scholar]
- 121.Shadle CR, Lee Y, Majumdar AK, Petty KJ, Gargano C, Bradstreet TE, Evans JK, Blum RA. Evaluation of potential inductive effects of aprepitant on cytochrome P450 3A4 and 2C9 activity. J Clin Pharmacol. 2004;44(3):215–223. doi: 10.1177/0091270003262950. [DOI] [PubMed] [Google Scholar]
- 122.Kutt H. Phenobarbital: interactions with other drugs. In: Levy R, Mattson RH, Meldrum BS, editors. Antiepileptic drugs. 4th. Raven Press; New York: 1995. pp. 389–400. [Google Scholar]
- 123.Kutt H. Phenytoin: interactions with other drugs. Part 1. Clinical aspects. In: Levy R, Mattson RH, Meldrum BS, editors. Antiepileptic drugs. 4th. Raven press; New York: 1995. pp. 315–328. [Google Scholar]
- 124.Jaakkola T, Backman JT, Neuvonen M, Laitila J, Neuvonen PJ. Effect of rifampicin on the pharmacokinetics of pioglitazone. Br J Clin Pharmacol. 2006;61(1):70–78. doi: 10.1111/j.1365-2125.2005.02515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Niemi M, Backman JT, Neuvonen PJ. Effects of trimethoprim and rifampin on the pharmacokinetics of the cytochrome P450 2C8 substrate rosiglitazone. Clin Pharmacol Ther. 2004;76(3):239–249. doi: 10.1016/j.clpt.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 126.Park JY, Kim KA, Kang MH, Kim SL, Shin JG. Effect of rifampin on the pharmacokinetics of rosiglitazone in healthy subjects. Clin Pharmacol Ther. 2004;75(3):157–162. doi: 10.1016/j.clpt.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 127.Dussault I, Lin M, Hollister K, Wang EH, Synold TW, Forman BM. Peptide mimetic HIV protease inhibitors are ligands for the orphan receptor SXR. J Biol Chem. 2001;276(36):33309–33312. doi: 10.1074/jbc.C100375200. [DOI] [PubMed] [Google Scholar]
- 128.Komoroski BJ, Parise RA, Egorin MJ, Strom SC, Venkataramanan R. Effect of the St. John's wort constituent hyperforin on docetaxel metabolism by human hepatocyte cultures. Clin Cancer Res. 2005;11(19 Pt 1):6972–6979. doi: 10.1158/1078-0432.CCR-04-2488. [DOI] [PubMed] [Google Scholar]
- 129.Gustafson DL, Long ME, Bradshaw EL, Merz AL, Kerzic PJ. P450 induction alters paclitaxel pharmacokinetics and tissue distribution with multiple dosing. Cancer Chemother Pharmacol. 2005;56(3):248–254. doi: 10.1007/s00280-004-0988-6. [DOI] [PubMed] [Google Scholar]
- 130.Synold TW, Dussault I, Forman BM. The orphan nuclear receptor SXR coordinately regulates drug metabolism and efflux. Nat Med. 2001;7(5):584–590. doi: 10.1038/87912. [DOI] [PubMed] [Google Scholar]
- 131.Prueksaritanont T, Richards KM, Qiu Y, Strong-Basalyga K, Miller A, Li C, Eisenhandler R, Carlini EJ. Comparative effects of fibrates on drug metabolizing enzymes in human hepatocytes. Pharm Res. 2005;22(1):71–78. doi: 10.1007/s11095-004-9011-5. [DOI] [PubMed] [Google Scholar]
- 132.Feng HJ, Huang SL, Wang W, Zhou HH. The induction effect of rifampicin on activity of mephenytoin 4′-hydroxylase related to M1 mutation of CYP2C19 and gene dose. Br J Clin Pharmacol. 1998;45(1):27–29. doi: 10.1046/j.1365-2125.1998.00643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhou HH, Anthony LB, Wood AJ, Wilkinson GR. Induction of polymorphic 4′-hydroxylation of S-mephenytoin by rifampicin. Br J Clin Pharmacol. 1990;30(3):471–475. doi: 10.1111/j.1365-2125.1990.tb03799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang LS, Zhou G, Zhu B, Wu J, Wang JG, Abd El-Aty AM, Li T, Liu J, Yang TL, Wang D, Zhong XY, Zhou HH. St John's wort induces both cytochrome P450 3A4-catalyzed sulfoxidation and 2C19-dependent hydroxylation of omeprazole. Clin Pharmacol Ther. 2004;75(3):191–197. doi: 10.1016/j.clpt.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 135.Wang LS, Zhu B, Abd El-Aty AM, Zhou G, Li Z, Wu J, Chen GL, Liu J, Tang ZR, An W, Li Q, Wang D, Zhou HH. The influence of St John's Wort on CYP2C19 activity with respect to genotype. J Clin Pharmacol. 2004;44(6):577–581. doi: 10.1177/0091270004265642. [DOI] [PubMed] [Google Scholar]
- 136.Svensson US, Ashton M, Trinh NH, Bertilsson L, Dinh XH, Nguyen VH, Nguyen TN, Nguyen DS, Lykkesfeldt J, Le DC. Artemisinin induces omeprazole metabolism in human beings. Clin Pharmacol Ther. 1998;64(2):160–167. doi: 10.1016/S0009-9236(98)90149-7. [DOI] [PubMed] [Google Scholar]
- 137.Burk O, Arnold KA, Nussler AK, Schaeffeler E, Efimova E, Avery BA, Avery MA, Fromm MF, Eichelbaum M. Antimalarial artemisinin drugs induce cytochrome P450 and MDR1 expression by activation of xenosensors pregnane X receptor and constitutive androstane receptor. Mol Pharmacol. 2005;67(6):1954–1965. doi: 10.1124/mol.104.009019. [DOI] [PubMed] [Google Scholar]