Abstract
Glycosylation of HIV-1 envelope gp120 determines not only the proper structure, but also the immune responses against this antigen. While glycans may be part of specific epitopes or shield other epitopes from T cells and antibodies, this study provides evidence for a different immunomodulatory function of glycans associated with gp120 residues N230 and N448. These glycans are required for efficient MHC class II-restricted presentation of nearby CD4 T-cell epitopes, even though they are not part of the epitopes. The glycans do not affect CD4 T cell recognition of more distant epitopes, and are not essential for the proper folding and function of gp120. Data on CD4 T-cell recognition of N448 mutants combined with proteolysis analyses and surface electrostatic potential calculation around residue N448 support the notion that N448-glycan near the epitope's C-terminus renders the site to be surface accessible and allows its efficient processing. In contrast, the N230-glycan contributes to the nearby epitope presentation at a step other than the proteolytic processing of the epitope. Hence, N-glycans can determine CD4 T-cell recognition of nearby gp120 epitopes by regulating the different steps in the MHC class II processing and presentation pathway after APCs acquire the intact gp120 antigen exogenously. Modifications of amino acids bearing glycans at the C termini of gp120 helper epitopes may prove to be a useful strategy for enhancing the immunogenicity of HIV-1 envelope gp120.
Keywords: AIDS, HIV envelope gp120, glycosylation, antigen processing/presentation, CD4 T lymphocytes, helper epitopes
Introduction
The HIV-1 envelope glycoprotein made of the exterior gp120 subunit and the transmembrane gp41 subunit is a critical target for protective immune response against HIV-1. The gp120 subunit, which binds to cell receptors CD4 and the chemokine receptors CCR5 or CXCR4, is one of the most heavily glycosylated proteins to be expressed by mammalian viruses as it has on average 25 N-linked glycans per molecule (1). The N-glycans on gp120 are critical for the correct folding and, consequently, the proper structure and function of this envelope glycoprotein (2-5). Many published studies have also demonstrated that a number of the N-glycans on gp120 facilitate virus escape from the host immune system as they block the access of critical epitopes from neutralizing Abs or constrain CTL epitope recognition (6-15). Hence, removal of the N-linked glycans from C2, V1V2, V3 or the silent face of HIV-1 gp120 by site-directed mutagenesis was found to increase the susceptibility of these viruses to neutralizing antibodies (11, 14-16). Similarly, glycosylated gp120 produced in mammalian cells was less efficient than those made in yeast or insect cells in inducing CD8 T cell responses in mice, and treatment of the mammalian cell-derived gp120 with endoglycosidase improved its immunogenicity (12). Nevertheless, N-glycans on gp120 can also be targeted by the immune system (17-19). The broadly neutralizing monoclonal Ab 2G12 recognizes a relatively conserved cluster of oligomannose glycans protruding on the gp120 surface (20, 21). Our previous study identified a specific N-glycan on gp120 that is required for efficient recognition of the neighboring helper epitopes by CD4 T cells (11). Hence, the presence or absence of particular N-linked glycans on HIV-1 gp120 can significantly impact the recognition of this antigen by both humoral and cellular arms of the immune system.
While many studies have reported the effects of various N-glycans on gp120 recognition by antibodies and cytolytic CD8 T cells, very little is understood about how the rich glycosylation of gp120 affects the way this antigen, when provided exogenously, is processed by antigen-presenting cells (APCs) to generate peptide epitopes presented on MHC class II for CD4 T helper cell recognition. Most of the defined human and mouse T helper epitopes are located in distinct regions of gp120, including C1, V2C2, V3, and V4C4 (22), and these epitopes are on sequences exposed on the gp120 surface or near surface accessible flexible loops (22-24). Interestingly, many of the epitopes are also flanked by regions richly decorated with N-linked glycans, although the importance of these glycans has yet to be fully understood. The removal of a glycan by an N-to-Q mutation at residue N448 in the C4 region of gp120 was shown to cause dramatic reduction in CD4 T cell recognition of nearby epitopes, without affecting the more distant epitopes (11). Since the glycan and the N448 residue themselves are located outside the epitopes, we surmise that the failure of CD4 T cells to recognize the C4 epitopes in the N448Q mutant is due to inefficient processing of the C4 region in this gp120 mutant. Indeed, the mutation was associated with increased resistance of the proximal C4 region to proteolysis. This alteration was not apparent when the N448Q mutant was first denatured prior to proteolysis, indicating that the N448Q mutation most likely affects the local C4 conformation and renders the C4 region less accessible to protease. However, the structural requirement dictated by the N448-glycan to allow for efficient processing and generation of the nearby helper epitopes has not been determined. It is also unknown if the processing and generation of helper epitopes from other gp120 regions are likewise affected by nearby N-linked glycans.
In this study, we sought to identify N-linked glycans essential for recognition of helper epitopes from the C2 region of gp120 by introducing N-to-Q mutations at three positions flanking either the amino-terminus or the carboxy-terminus of the epitope-rich C2 region. By comparison with the N448-glycan, we investigated common and unique structural elements critical for the processing of helper epitopes from the C4 and C2 regions. The N448-glycan data led us to postulate that the presence of an N-linked glycan near the C-terminus of a helper epitope is essential for maintaining the surface accessibility of the epitope's C-terminus and allowing its enzymatic processing in order to release the peptide epitope for MHC class II presentation and CD4 T cell recognition. In contrast, the N230-glycan required for presentation of the proximal C2 epitope affects step(s) other than the proteolytic processing of the particular C2 region.
Materials and Methods
Recombinant proteins, synthetic peptides, and monoclonal antibodies
Gp120BH10 proteins with the wild type sequence and with various mutations tested in this study were generated as described (11). Removals of site-specific glycans were verified by alteration in the apparent molecular mass on SDS-PAGE and/or by mass spectroscopy (data not shown). Synthetic gp120 peptides were provided by the NIH AIDS Reagent Repository and the NIBSC Centre for AIDS Reagent, or purchased from Sigma-Genosys. Peptides AQUA-pC4 (AMYAPPISGQI*R) and AQUA-pC2 (VSFEPIPIHYCAPAGFAIL*K) with 13C and 15N-labeled K or R residues were purchased from Sigma-Genosys. Human gp120-specific mAbs were generous gifts from Drs. S. Zolla-Pazner (New York University School of Medicine), J. Robinson (Tulane University School of Medicine), as well as from Drs. H. Katinger (2G12), D. Burton, and C. Barbas (b12) through the NIH AIDS Reagent Repository Program.
Gp120-specific CD4 T cells
Gp120-specific primary CD4 T cell lines PS02 and PS05 were generated from chronically infected HIV-1 patients and maintained as short term cultures (∼ 8 weeks) by in vitro stimulation with irradiated gp120IIIB-treated autologous PBMCs and IL-2 (Roche)(25). Clone DMg26 was generated from a HIV-seronegative volunteer who received HIV-1 gp120W61D vaccine and stimulated with antigen-treated heterologous DR1+ PBMCs (26, 27). These human CD4 T cells recognize MHC class II-restricted epitopes from the C2 or C4 regions of gp120 (Fig. 1). All subjects whose cells were used to establish or stimulate the T cell lines gave informed consent, and the study was reviewed and approved by the Veterans Affairs New York Harbor Healthcare System Institutional Review Board.
FIGURE 1. Site-specific N-linked glycan deletions introduced to gp120 and their positions relative to CD4 T cell epitopes examined in the study.
Amino acid substitutions were introduced into gp120BH10 to remove N-glycans linked with residues 197, 230, 234, 406, 448, or 463 (marked by circles). The mutated proteins were tested for recognition by gp120-specific CD4 T cell lines PS02, DMg26, and PS05. Locations of the epitopes recognized by these T cell lines were highlighted by circles of different colors: pink for PS02 and blue for DMg26 in the C2 region (overlap was highlighted with both pink and blue), and red for PS05 in the C4 region. This diagram (modified from Leonard et al. (46)) shows the relatively conserved (C1-C5) and variable (V1-V5) regions of the gp120 protein, while the 30-amino acid signal peptide at the N terminus was omitted.
Construction and production of gp120 mutants
The construction of PCR template gp120BH10-pUC19 for use in site-directed mutagenesis was described previously (11) and primers used to introduce mutations are described in the Supplemental Table 1. Site directed mutagenesis was performed with the Quick Change XL Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer's instructions. The entire gp120 gene of each mutant was sequenced to confirm that the specific mutation(s) were introduced successfully without any other changes. After the mutations were introduced, the gp120BH10 cassette was then removed from pUC19 and inserted to the expression vector pEE14 for transfection and expression of soluble gp120 in mammalian cells. Transfection of CHO-L761h cells with the pEE14 expression vector bearing WT or mutated gp120 was performed by calcium phosphate precipitation, and stable clones selected by glutamine depletion (11). To express mutants with different substitutions at residues 448 and 230, a transient expression system was established using 293T cells and the ProFection Mammalian Transfection System (Promega). Gp120 proteins were purified from the culture supernatants by affinity chromatography using the anti-V3 mAb 694 (11). Purity of each gp120 preparation was assessed by SDS-PAGE and Coomassie blue staining, and their concentrations were determined with the NanoDrop 1000 spectrophotometer.
Binding of gp120 mutants to mAbs and CD4
ELISA to detect mAb or soluble CD4 (Perkins Elmer) binding to gp120 proteins were done as previously described (28). Gp120 proteins were captured onto ELISA wells by sheep anti-C5 antibodies (Cliniqa) and reacted with different human anti-gp120 mAbs. Detection of the mAb binding was done using alkaline phosphatase-conjugated anti-human IgG (Sigma). CD4 binding was detected similarly using the mouse anti-CD4 mAb OKT4 and alkaline phosphatase-conjugated anti-mouse IgG. The optical density was read at 405nm with an ELISA plate reader. Each condition was tested in triplicates, and each experiment was performed independently two or more times with comparable results.
T cell proliferation assay
T cell proliferation was assessed by the standard 3H-thymidine incorporation assay. PBMCs from autologous or heterologous healthy donors with matched MHC class II alleles were irradiated (12,000 RADS) and treated with antigens for 18-22 hrs prior to use as APCs in the assay. T cell response to APCs alone in the absence of any antigen was determined in each assay as background proliferation. Each experimental condition was tested in triplicates, and all experiments were performed at least twice.
Quantitative MALDI-TOF MS and LC- ESI MS/MS analyses
The generation of peptide pC4 (AMYAPPISGQIR) from trypsin digestion of gp120 was detected by MALDI-TOF MS as described (11). In brief, gp120 proteins (0.5 μg/μl) were digested with trypsin (0.1 μg/μl) in 25mM ammonium bicarbonate overnight at 37°C. The digested products were then mixed 1:1(vol) with 5pmol/μl of AQUA-pC4 and analyzed by MALDI-TOF MS (TofSpec 2E, Waters-Micromass, MA). The amounts of gp120 peptides in the digestion reactions were calculated according to the following equation: AreaAQUA/ConcentrationAQUA = Areasample/Concentrationsample, with AreaAQUA and Areasample representing the areas under the monoisotopic peaks for the AQUA or sample peptides in the in the MALDI-TOF spectra.
LC-ESI MS/MS was used to detect DMg26 epitope-containing peptide pC2 (VSFEPIPIHYCAPAGFAILK), as this peptide was not detectable with MALDI-TOF MS. After trypsin digestion of the gp120 WT or mutants (2 pmol each), the digestion reactions were reduced with dithiothreitol, alkylated with iodoacetamide, and then mixed with 2 pmol of the AQUA-pC2 peptide. The samples were then loaded for LC-ESI MS/MS analyses with 20% acetonitrile in 0.1% trifluoroacetic acid as the sample buffer. The amount of pC2 generated from trypsin digestion of gp120 samples were calculated as described above for pC4.
Circular Dichroism analysis of gp120 proteins
Circular dichroism analyses were performed on 2.5μM of gp120 WT or mutants in 10 mM sodium phosphate buffer pH 7 at 25°C. The sample compartment was purged with nitrogen gas and a 1 mm curvette was used. Data points were measured between 250 and 190 nm using a Jasco J-810 Spectrophotometer. Triplicate scans were averaged, and the spectrum for the phosphate background was subtracted.
Statistical analysis
Statistical analyses were performed using one-way ANOVA with Dunnett's post tests or two-way ANOVA with Bonferroni's post tests (GraphPad Prism 4).
Results
HIV gp120 mutants lacking N-glycans in the C2 region bind to CD4 and Abs
We have observed the importance of a single glycan linked to residue N448 in the C4 region of the HIV-1 envelope gp120 for the generation and recognition of CD4 T cell epitopes that are located close to but do not encompass residue N448 (Fig.1 and (11)). The requirement for this N448-glycan was further verified in Fig. 2A by the failure of gp120 mutants lacking this specific glycan, due to either an N-to-Q substitution at position 448 (N448Q) or a T-to-A substitution at position 450 (T450A), to be recognized efficiently by CD4 T cells specific for the nearby PS05 epitope (Fig 2A). The contribution of the N448-glycan was specific since removal of two other glycans in the V4 and C4 regions (N406Q and N463Q, respectively) had no effect on the PS05 epitope recognition (Fig. 2A and (11)). To determine if other N-glycans in the gp120 are also critical for efficient processing and recognition of the neighboring CD4 T cell epitopes, we examined three glycans in the C2 region of gp120 linked to N197, N230, and N234 (Fig. 1). These N-glycans were selected because they flank a cluster of epitopes frequently recognized by human and mouse gp120-specific CD4 T cells (22). Indeed, two of the CD4 T cell lines (DMg26 and PS02) that we have been studying over the past decade are specific for epitopes in this particular region (25, 26) and were tested in this study. Gp120 lacking glycans linked to N197, N230, or N234 were constructed by introducing N-to-Q mutations at these specific positions individually. Recombinant proteins were expressed in CHO cells and purified by affinity chromatography as previously described (11).
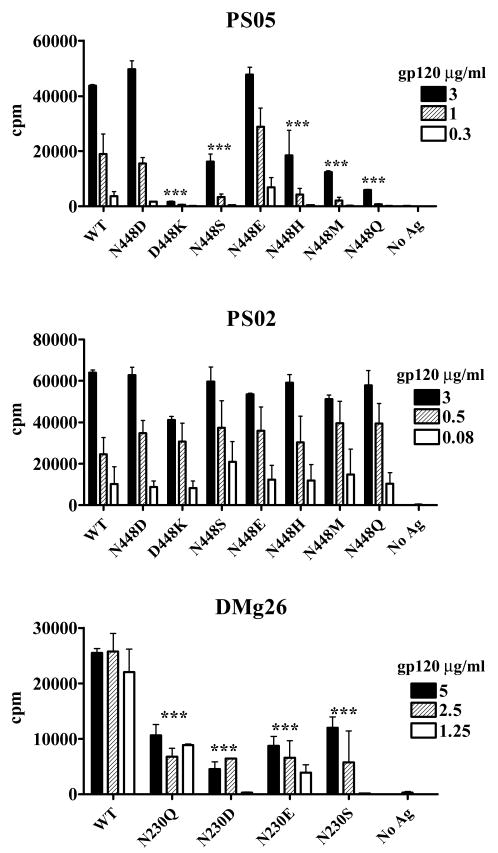
FIGURE 2. Recognition of gp120 mutants lacking specific N-glycans by CD4 T cells and mAbs.
A. N448-glycan is required for CD4 T cell recognition of the neighboring PS05 epitope. CD4 T cell recognition of gp120 mutants lacking glycans at N448 (N448Q or T450A), N406 (N406Q), or N463 (N463Q) was evaluated using a C4-specific CD4 T cell line PS05. T cell proliferation was measured in the standard 3H-thymidine incorporation assay. Autologous human PBMCs were used as APCs after irradiation and treatment with wild type and mutant gp120 proteins at the indicated concentrations. All experiments were in triplicates and representative results of at least two independent experiments were shown. Average counts per minute (cpm) and standard deviations from triplicate wells were shown. ***, p<0.001 compared with the WT.
B. Removal of N-glycans from the C2 region of gp120 does not abrogate CD4 binding and mAb reactivities. The CD4-binding capacity and gp120-specific mAb reactivity of the mutated gp120 proteins lacking glycans in the C2 region (N197Q, N230Q or N234Q) were determined by ELISA. Mutant and wild type gp120 proteins were captured onto ELISA plates and incubated with the designated concentrations of soluble CD4 (sCD4) or human anti-gp120 mAbs (b12, 447, 2G12, C11, and EH21). Means and standard deviations of OD405 values from triplicate wells are shown. The data are representative results from one of two independent experiments. *, p<0.05 compared with WT at the same concentration.
C. N230-glycan is required for CD4 T cell recognition of the nearby DMg26 epitope. Recognition of gp120 mutants lacking glycans at N197, N230, or N234 was assessed using CD4 T cell lines specific for C2 epitopes (DMg26 and PS02) or a C4 epitope (PS05) in T cell proliferation assays. All experiments were performed in triplicates and representative results of at least two independent experiments were shown. Average counts per minute (cpm) and standard deviations from triplicate wells were shown. ***, p<0.001 compared with the WT.
To assess whether any of these mutations affect gp120 function and antigenicity, we first determined the capacity of the mutants to bind soluble CD4 (sCD4) and a panel of gp120-specific mAbs. Each of the three mutants displayed similar CD4-binding activities as WT (Fig. 2B). All three mutants were also reactive with each of the anti-gp120 mAbs tested, including mAbs specific for the conformation-dependent CD4-binding site (b12), a conformation-dependent epitope involving the N- and C-termini of gp120 (C11), the V3 loop (447), and the N-terminus C1 region (EH21). As expected, these mutants remained reactive with mAb 2G12 specific for a mannose-dependent epitope involving glycans at residues 332, 339 and 392 (20) that are distant from the C2 glycans examined here. Nevertheless, small but significant differences in mAb reactivities were noted with mAbs b12 and 447 which had lower reactivities with mutant N234Q, and with mAb EH21 which displayed lower reactivities with N230Q and N234Q (Fig. 2B). On the other hand, the reactivities of mAbs 2G12 and C11 with the mutants were slightly enhanced, at least at the highest concentration of mAbs tested (1 μg/ml). Since none of the mutations abrogated mAb reactivity and CD4 binding activity, the changes observed are most likely due to localized perturbations and do not reflect global alterations affecting the overall conformation and function of the gp120 proteins.
Similar to the N448-glycan mutants, HIV gp120 mutant lacking N230-glycan is poorly recognized by CD4 T cells specific for a nearby epitope
To examine if any of the three N-glycans in the C2 region was critical for CD4 T cell recognition of neighboring epitopes, we tested the three gp120 mutants for recognition by two different C2-specific CD4 T cell lines, DMg26 and PS02, as well as by the PS05 cell line specific for a more distant C4 epitope. The T cell line DMg26 poorly recognized the N230Q mutant, while recognition of the other two mutants, N197Q and N234Q, was comparable to that of WT (Fig 2C). Of note, the N230Q mutation is only three amino acids away from the carboxyl terminus of the DMg26 epitope, whereas N197Q and N234Q mutations are more distant from the DMg26 epitope (Fig. 1). The other C2-specific line, PS02, recognized N230Q as well as N197Q, N234Q, and WT. The PS02 epitope overlaps with the amino-terminus of DMg26 epitope but its carboxyl-terminus is 10 amino acids away from the N230Q mutation. The T cell line PS05, which is specific for an epitope in the distant C4 region of gp120, also recognized all three mutants as well as the WT. Hence, the N-to-Q mutation at position 230 in the gp120 protein affected specifically CD4 T cell recognition of the nearest DMg26 epitope but had no affect on the more distant epitopes. These data indicate that the uptake and intracellular transport of the N230Q mutant by APCs are likely to proceed normally, but the subsequent step(s) in the processing and presentation of the specific epitope near N230 may be hindered. The findings are reminiscent of our data showing that the loss of N448-glycan due to N448Q or T450A mutations also diminished CD4 T cell recognition of the nearby epitopes in the C4 region and especially the most adjacent PS05 epitope (11) (see Fig. 1 and Fig. 2A).
We have now identified two glycans, linked to gp120 residues N230 and N448, that are critical for the recognition of the adjacent CD4 T cell epitopes. The N448 and N230 residues and their associated glycans are not parts of the PS05 and DMg26 epitopes, respectively, as both epitopes can be represented entirely by synthetic peptides devoid of any glycans and lacking these N residues. In fact, the minimal epitopes for these two CD4 T cell lines have been mapped using nested peptides and do not encompass N448 or N230 (Fig. 1 and data not shown).
N230 and N448 share common features in relation of their respective nearby epitopes
Although N230-glycan and N448-glycan were clearly essential for the efficient recognition of CD4 T cell epitopes near these respective glycans, the reason was unknown. We searched for common structural features at the two gp120 regions that may shed light on the role of these glycans. Both N448 and N230 are outside the CD4 T cell epitopes of interest, but are located only two or three amino acids away from the C-termini of PS05 or DMg26 epitopes, respectively (Fig. 1). Near the C termini of these epitopes, there are also cysteine residues that form disulphide bonds (C228-C239 for DMg26 and C378-C445 for PS05), and again are only two or three amino acids away from N230-glycan or N448-glycan, respectively. Considering that antigenic processing is needed for MHC class II presentation and T cell recognition of DMg26 and PS05 epitopes (11, 26), mutations and glycan removals at N230 and N448 are likely to cause alterations in the local structures near the C-termini of the epitopes that limit the accessibility of these regions to enzymes needed for generating the peptide epitopes, including endopeptidases that cleave the C termini and reductases that break the disulfide bonds.
As each gp120 molecule contains >20 N-linked glycans, we also examined the 3D structure of gp120 for the potential interactions of N230-glycan and N448-glycan with other glycans protruding from the same gp120 surfaces. The crystal structure of CD4 and mAb-bound gp120 core showing the proximal sugar moities (29) was used to map the glycan locations on the gp120 protein surface. The most recent cryo-electron tomography of HIV-1 virus particles demonstrated that this liganded gp120 structure, but not the unliganded SIV gp120 structure, fit best into the electron density of the native envelope glycoproteins on the HIV virion surface (30). The three C2 glycans studied here (linked to N197, N230, and N234) flank the DMg26 epitope at either amino or carboxyl ends in the gp120 linear sequence, but only N230-glycan and N234-glycan are in close proximity to each other and to the DMg26 epitope, while the N197-glycan is considerably distant in the primary gp120 sequence and is located at the opposite end of the gp120 surface in the tertiary gp120 structure (Fig. 3A). However, in addition to N234-glycan, a glycan linked to N241 is also found near N230, such that N230-glycan is flanked on two sides by N234- and N241-glycans to form a triple sugar complex near the C terminus of DMg26 epitope. Of the three glycans, two glycans linked to N230 and N234 project out toward the same direction, while one glycan linked to N241 has a different orientation (Fig 3A). Similarly, the N448-glycan is also flanked on two sides by glycans linked to N262 and N295 to form three sugar towers, all of which raise up in the same orientation from the gp120 surface around the C terminus of PS05 epitope (Fig. 3B). Since neighboring glycans can interact to provide structural support to each other, removal of one N-glycan from the middle of such glycan complexes may weaken the vertical support of the sugar towers, rendering the remaining glycans to collapse and cover the sites otherwise accessible for the processing and generation of T cell epitopes.
FIGURE 3. Triple N-glycan towers on the gp120 surfaces near the C termini of DMg26 and PS05 epitopes.
A. The glycans linked to residues N230, N234 and N241 form triple glycan towers with the middle N230-glycan flanked by N234 and N241 glycans on each side. The sugar towers protrude from the surface of gp120 inner domain near the C terminus of DMg26 epitope (blue). In contrast, the N197-linked glycan is found on the opposite side of gp120.
B. The N448-linked glycan is also flanked by two glycans linked to N262 and N295, and the three glycans make triple sugar towers on the surface of gp120 outer domain close to the C terminus of PS05 epitope (red). Asparagine residues associated with the glycans are shown in grey, whereas the proximal sugar units resolved in the gp120 crystal structure (29) are in orange.
Removal of additional glycans near N230 and N448 did not fully restore CD4 T cell recognition of the neighboring epitopes
To test the proposed sugar tower hypothesis, we constructed one gp120 mutant with N-to-Q substitutions that remove all three glycans clustering in the C2 region (ΔGC2T with mutations at positions 230, 234 and 241) and another mutant lacking the three glycans towering over the C terminus of PS05 in the C4 region (ΔGC4T with mutations at positions 448, 262 and 295). Removal of the additional glycans near N230 and N448 did not fully restore CD4 T cell recognition of the neighboring epitopes (Fig. 4A). Moreover, the triple mutant ΔGC4T decreased the recognition of a distant PS02 epitope (Fig. 4A), suggesting that these mutations induce more global alterations in the antigenicity of gp120. This observation corresponded with ELISA data showing that the ΔGC4T mutant failed to bind CD4 and lost reactivities with most of the anti-gp120 mAbs tested (Fig. 4B). The ΔGC2T mutant also displayed a substantial decrease in CD4 binding capacity and mAb reactivities. Only anti-V3 mAb 694 remained reactive with ΔGC4T and ΔGC2T at the WT level. Conformational changes in ΔGC4T and ΔGC2T were also evident from the circular dichroism (CD) analyses, in which both ΔGC2T and ΔGC4T showed slight but significant shifts in their CD spectra (Fig. 4C), indicative of the presence of more random coil in these mutants. Hence, removal of the three glycan clusters by ΔGC2T or ΔGC4T mutations could partially improve the recognition of neighboring CD4 T cell epitopes, indicating the importance of inter-glycan interactions in modulating recognition of T cell epitopes from a richly glycosylated gp120 antigen. However, the triple mutations also substantially altered the core gp120 structure, affecting the overall antigenicity of gp120 beyond the CD4 T cell epitopes of interest.
FIGURE 4. Antigenicity and conformation changes of the triple gp120 mutants ΔGC2T and ΔGC4T.
A. CD4 T cell recognition of the ΔGC2T and ΔGC4T mutants. The triple mutant ΔGC2T (lacking glycans linked to N230, N234 and N241) was tested along with the single N230Q mutant and wild type (WT) gp120 for recognition by the C2-specific CD4 T cell line DMg26 in a standard 3H-thymidine incorporation assay (left panel). By contrast, the triple mutant ΔGC4T (lacking glycans linked to N448, N262 and N295) was tested in comparison with the single N448Q mutant and WT gp120 for recognition by CD4 T cell lines specific for the nearby C4 epitope (PS05; middle panel) or the C2 epitope (PS02; right panel). Irradiated autologous or heterologous DR-matched PBMCs were treated with gp120 proteins at the indicated concentrations and used as APCs in the T cell proliferation assay. 3H-thymidine incorporation in the absence of any antigen was recorded as background and subtracted from responses to the tested antigens to obtain Δ cpm. Average Δ cpm and standard deviation from triplicate wells are shown. *** p<0.001, ** p<0.01 compared with WT and N448Q.
B. CD4 and mAb binding of the ΔGC2T (left graphs) and ΔGC4T (right graphs) mutants. ELISA was used to assess the capacity of the triple mutants to bind CD4 and different anti-gp120 mAbs. Each triple mutant was tested in parallel with the relevant single mutant and WT gp120. Average OD405 and standard deviations from triplicate wells were shown. ***, p<0.001 compared with WT at the same sCD4 or mAb concentrations.
C. Circular dichroism spectra of the ΔGC2T and ΔGC4T proteins. The circular dichroism spectra of triple mutants ΔGC2T and ΔGC4T were compared with WT gp120 and the relevant single C2 (N230Q) or C4 (N448Q) mutants. Averages of triplicate scans are shown.
Substitution of N448 to a negatively charged amino acid fully restores CD4 T cell recognition of the neighboring C4 epitope
To investigate further the mechanisms by which N448 glycan and N230 glycan influence the recognition of the respective PS05 and DMg26 epitopes, we tested the effects of various amino acid substitutions at positions 448 and 230 and defined the nature of amino acids that could replace the N residues and their associated glycans without incurring the loss of CD4 T cell recognition of the neighboring epitopes. Except for the N448M substitution, the amino acids introduced were selected from those naturally expressed at positions 448 and 230 by HIV-1 subtype B isolates recorded in the Los Alamos HIV-1 sequence database. The database searches revealed that residue N448 and a glycosylation motif N448-I-T were found in as many as 425 (93.6%) of 454 HIV-1 subtype B envelopes, while the remaining envelopes contain one of the following residues: K, S, I, Q, H, D, Y, or E at position 448. On the other hand, the most prevalent amino acid at position 230 is D (66.5%, 302/454), whereas residue N230 and the glycosylation motif N230-X-T/S are expressed by only 24.5% (111/454) of the isolates. Residues E, Q, and S were also found at position 230. Hence, a panel of gp120 mutants was constructed to express amino acids K, S, H, D, E or M at position 448 or amino acids D, E, or S at position 230.
The panel of N448 mutants was tested in comparison with WT and the N448Q mutant for recognition by the C4-specific CD4 T cell line PS05 (Fig. 5). Notably, two of the mutants, N448D and N448E, were recognized by PS05 at comparable levels as WT. Mutants N448S, N448H, and N448M were poorly recognized, similar to N448Q, whereas mutant N448K abrogated PS05 recognition to the levels below those of N448Q. As expected, all of the mutants were recognized as well as WT by the T cell line PS02 specific for a distant C2 epitope. It is of interest to point out that based on surface electrostatic potential prediction, the local area around residue N448 is anticipated to be positively charged (Fig. 6), and thus, a basic K residue would be highly de-stabilizing while a negatively charged amino acid D or E in place of N448 and its glycan could have stabilizing effects on this gp120 surface. The N230 mutants were also tested for CD4 T cell recognition by DMg26 cells specific for a C2 epitope adjacent to N230. However, similar to N230Q, the other three mutants examined (N230D, N230E and N230S) were not recognized efficiently by DMg26 cells. Hence, no amino acid residues were yet defined that could replace N230 for the DMg26 epitope recognition, but residue N448 and its associated glycan could be substituted with a negatively charged residue D or E to allow the CD4 T cell recognition of PS05 epitope.
FIGURE 5. CD4 T cell recognition of gp120 proteins with different amino acids at residues 448 or 230.
CD4 T cell recognition of the different gp120 mutants was tested in 3H-thymidine incorporation assays. The panel of N448 mutants was tested for recognition by CD4 T cells specific for a C4 epitope near residue 448 (PS05; top) or a distant C2 epitope (PS02; middle). The panel of N230 mutants was tested for recognition by DMg26 cells specific for a C2 epitope nearest to residue 230. Irradiated PBMCs were treated with the different gp120 antigens at the indicated concentrations and used as APCs. Average counts per minute (cpm) and standard deviations from triplicate wells are shown. ***, p<0.001 compared with WT.
Figure 6. Electrostatic surface of gp120 near residue N488.
The electrostatic potential at the solvent-accessible surface is shown and colored according to the local electrostatic potential, ranging from blue (positive) to red (negative). The location of N448 was also labeled. The electrostatic potential calculation was done using the ICM software (Molsoft LLC.)
Failure of C4-specific CD4 T cells to recognize N448 mutants is associated with increased resistance of the C4 region to proteolytic processing
We have shown previously that the failure of the N448Q mutant to stimulate CD4 T cell line PS05 recognizing a nearby C4 epitope was associated with an increased resistance of the C4 region in the N448Q mutant to proteolysis needed for releasing the peptide epitopes from this gp120 region (11). In this study, we confirmed that indeed trypsin digestion of N448Q yielded a significantly lower amount of peptide pC4 (AMYAPPISGQIR) which overlaps with PS05 epitope than digestion of WT (Fig. 7A). The quantitative analyses of peptide pC4 generated from in vitro trypsin digestion of different gp120 proteins were done using mass spectrometry (MALDI-TOF MS) with an internal AQUA peptide standard as performed previously (11). Since we identified that N448D and N448E mutations removed N448-glycan without affecting CD4 T cell recognition of the C4 epitope, one of these mutants (N448D) was examined in a parallel trypsin digestion experiment to assess how the C4 region of this mutant differed from those of N448Q and WT in its susceptibility to trypsin cleavage. The data show that digestion of the N448D mutant yielded a comparable amount of peptide pC4 to that of WT and this pC4 yield was significantly higher than the amount obtained from N448Q digestion (p<0.05). From 0.5 pmol/μl of each protein, on average 0.52 and 0.45 pmol/μl of peptide pC4 were obtained from N448D and WT, respectively. In contrast, only 0.32 pmol/μl (71% of that of WT) of peptide pC4 was produced upon digestion of the N448Q mutant. These data clearly show a correlation between the relative susceptibility of the C4 region as expressed in the different gp120 proteins to proteolysis and the recognition of the respective gp120 proteins by C4-specific CD4 T cells. The finding further attests that the key step affected by mutations introduced at residue 448 is the processing of gp120 antigen for generating the nearby C4 epitope.
FIGURE 7. Quantitative analyses of peptides pC4 and pC2 produced from trypsin digestion of the different gp120 proteins.
A. The amounts of peptide pC4 generated from trypsin digestion of WT and mutated gp120 (N448Q and N448D) were determined by MOLDI-TOF MS with the internal standard AQUA-pC4 peptide. Gp120 proteins (0.5 μg/μl) were digested with trypsin and mixed with 1:1 with 0.5 pmol/μl AQUA-pC4. The pC4 amount from each sample was calculated by comparing the ion counts of pC4 with that of AQUA-pC4. The average amount of pC4 from WT (0.45 pmol/μl) was normalized to 100% and the relative yields of pC4 produced from the mutants were calculated. Averages and standard deviations from triple digestions were shown. *, p<0.05 compared with WT and N448D.
B. The amounts of peptide pC2 generated from trypsin digestion of the C2 mutants and WT gp120 were quantified by LC-ESI MS/MS using the AQUA-pC2 peptide as an internal standard. The pC2 peptide is recognized by CD4 T cell line DMg26. Gp120 proteins (2 pmol each) was digested with trypsin and AQUA-pC2 (2 pmol) was added prior to LC-ESI MS/MS analysis. The ion counts of pC2 and AQUA-pC2 in each sample were determined, and the amounts of pC2 generated from the mutants were calculated relative to that from WT. Averages and standard deviations from two independent experiments are shown. No statistical differences were observed among these C2 mutants and between the mutants and WT gp120.
We subsequently analyzed whether the N230Q mutation may also hinder the processing and generation of the adjacent DMg26 epitope from the C2 region of gp120. To assess the sensitivity of the C2 region to enzymatic processing in general, trypsin digestion assays were performed on the mutant and WT proteins. The digestion products were then subjected to reduction and alkylation, and analyzed by liquid chromatography-electrospray ionization tandem mass spectrometry (LC-ESI MS/MS). A tryptic fragment VSFEPIPIHYCAPAGFAILK (designated pC2), which overlaps with the DMg26 epitope and is recognized by the DMg26 cells, was generated as a result of cleavage of the C2 region. The yield of the pC2 peptide was quantified from the LC-ESI MS/MS spectra by comparison with a specified amount of the AQUA pC2 peptide added to the analyzed samples (11). Digestion of the N230Q mutant produced a similar amount of pC2 compared to WT and the two other mutants (N197Q and N234Q) (Fig. 7B). Hence, unlike N448, the amino acid substitution and glycan removal at N230 did not appear to have a direct effect on the proteolytic processing and generation of the nearby CD4 T cell epitope. Taken together with differences observed on the effects of amino acid substitutions at N230 vs. N448 (Fig. 5), the data further indicate a distinct role of N230-glycan from that of N448-glycan in determining the CD4 T cell recognition of the nearby epitopes.
Discussion
HIV-1 envelope glycoprotein gp120 is an important target for effective immune responses against the virus. However, broadly reactive and protective immune responses to gp120 are rarely elicited in HIV-1-infected subjects and among healthy recipients of HIV-1 envelope-based vaccines. (31-35). Many reasons can account for non-protective responses against gp120, including the presence of immunogenic but highly variable regions and >20 N–glycans decorating the protein surface (36-38), and a better understanding about how the different gp120 components shape the immune responses to this antigen is needed in order to design more effective gp120 immunogens. In this study, we have examined the role of N-glycans in influencing gp120 antigen processing for MHC class II presentation and CD4 T cell recognition. Very little is known about how the numerous N-glycans on gp120 affect the way this glycoprotein, when exogenously provided to APCs, is processed into peptide epitopes that stimulate the helper CD4 T cells necessary for establishing CD8 T cell- or antibody-mediated effector functions. It has been noted that N-glycans are found to flank each of the four hot-spots where the vast majority of human and mouse T helper epitopes cluster (22), but the relevance of these glycans is not fully understood. Similar to their shielding effects on Ab and CD8 T cell epitopes, the glycans were believed to also obstruct the processing and presentation of CD4 T cell epitopes. Surprisingly, our data reveal that certain N-glycans on gp120 are actually needed for recognition of the nearby CD4 T cell epitopes. The N448-glycan was previously identified to be critical for recognition of two proximal epitopes in the C4 region. In this study, the glycan associated with residue N230 in the C2 region was found to be required for an epitope located three amino acids upstream, indicating that this finding may be applicable to many of the glycans often found to flank many CD4 T cell epitopes in HIV-1 gp120. The critical involvement of N448-glycan in T cell recognition of nearby epitopes provided an explanation for earlier findings by Sjolander et al. (39) showing that immunization of mice with gp120 mutants lacking N448-glycan, along with other glycans linked to N406 and N463, resulted in the loss of T cell responses to a specific epitope located in the C4 region. Interestingly, substitution of N448 with a D or an E, but not with other amino acids, was found to permit efficient recognition of the C4 helper epitope, indicating that in the absence of N448-glycan, an acidic amino acid side chain could serve a similar function as the sugar moieties in facilitating the presentation and recognition of this epitope. Altogether these results demonstrate the important role of N-glycans flanking gp120 helper epitopes in modulating CD4 T cell responses to gp120 and suggest that modification of N-glycans and other amino acids flanking the four helper epitope-rich areas on gp120 may be one strategy that can be utilized to improve the immunogenicity of gp120 antigens for HIV envelope-based vaccines.
The findings from this study also imply that a vast array of mutations can occur on gp120 to result in the loss, addition, or repositioning of different N-glycans on the gp120 surface that lead to dramatic modulations of helper T cell responses to this antigen. As the glycans are outside the helper epitopes, these mutations would facilitate immune escape without any alterations on epitopes themselves. Indeed, three of the four hotspots known to contain most of helper epitopes are in the relatively conserved constant regions (C1, C2, and C4) of gp120, but the preservation of these epitopes do not necessarily correspond to their successful recognition by the T cells. As our study shows, the C2 recognition is determined not only by the amino acid sequence within the epitope itself, but also by the presence of N230-glycan near its C-terminus. Among HIV-1 subtype B, the C2 epitope recognized by DMg26 is conserved among 62% (282/454) of viruses, but the glycosylation motif at N230 is found only in a quarter of the viruses. Similarly, PS05 recognition of the C4 epitope requires a factor outside the epitope itself, i.e. the presence of an N-glycan or an acidic residue D or E at position 448 flanking the C terminus of the epitope. When position 448 is occupied by Q, S, H, M, or K, or when mutations occur at position 450 to eliminate the glycosylation motif N-X-T/S, recognition of this T cell epitope is hampered or completely blocked.
In this study, we further investigated the reasons as to why N-glycans at positions 230 and 448 are required for CD4 T cell recognition of nearby helper epitopes. First we evaluated common features shared between N230Q and N448Q mutations. Both mutations abrogated specifically CD4 T cell recognition of the neighboring epitopes without affecting more distant epitopes, indicating that the uptake and intracellular transport of the mutant proteins into the endolysosomes of APCs are not impeded. The N230- and N448-glycans are located only two or three amino acids outside the C termini of the affected epitopes, each of which also bears a disulfide bond in the vicinity (Fig. 1). This observation raises a possibility that these glycan affects enzymatic processing of the epitope C termini, including endoproteolytic cleavage and disulfide bond reduction needed to release the epitopes for MHC class II presentation (40-42). Experimental data from the trypsin digestion, which serves as a surrogate for probing the relative sensitivity of specific gp120 fragments to enzymatic processing, support this idea, albeit only for N448 (Fig 7). The N448Q mutation, which abrogated CD4 T cell recognition of the nearby C4 epitope, retarded trypsin cleavage from the C4 region of gp120. In comparison, the N448D mutation had no effect on the C4 epitope recognition and the C4 region of this mutant was digested as efficiently as that of WT. By contrast, the N230Q mutation did not appear to have any effects on proteolysis of the neighboring C2 region, suggesting that unlike the N448-glycan, the N230-glycan is likely to be involved in other steps of the MHC class II processing and presentation pathway. Nevertheless, the exact processes and enzymes necessary for generating the different gp120 helper epitopes are not at all understood and the step(s) influenced by the N230 glycan needs to be investigated in future studies.
N-glycans have been implicated to play a role in shielding antibodies and CD8 T cells from accessing critical epitopes on gp120. Thus, it is seemingly counterintuitive that the presence of an N-glycan actually facilitates the proteolytic processing of the nearby helper epitopes while its removal interferes with this process. The importance of the glycan moiety associated with N448, and not the amino acid N itself, is confirmed by poor CD4 T cell recognition of the T450A mutant, which retains residue N448 but lacks its associated glycan (Fig 2A). The question remains as to how a glycan at position 448 contributes to the processing of the nearby C4 epitope. The N448-glycan protrudes from the outer domain surface of gp120 core and, on its own, is dispensable for proper folding of the gp120 protein as HIV-1 with envelope lacking this glycan is still infectious (11, 14). Comparison of the crystal structures of CD4-bound and unliganded gp120 reveals that the position of N448 at the C terminus of the β22 strand is relatively unchanged (13, 43), indicating the rigidity of this particular gp120 region. Interestingly, while the PS05 epitope which encompasses loop F and β22 and is upstream of N448 is mostly surface exposed, amino acids immediately after N448 (forming part of β23) is solvent inaccessible (13). Based on this observation, we postulate that the presence of N448-glycan is necessary to maintain the surface accessibility of the C terminal region of β22 and the PS05 epitope, and that removal of this glycan may cause this strand to cave along with β23. In the absence of N448-glycan, two neighboring glycans could also collapse and bury this site further (Fig. 3B). When we removed all three glycans towering the site, the PS05 epitope recognition was restored to a small degree, but the overall gp120 conformation was seriously disrupted. The importance of an N-glycan for maintaining the stability of a local area on a protein is also evident from a report by Wyss et al demonstrating that the single N-linked glycan in the human CD2 protein forms hydrogen bonds and van der Wall contacts with neighboring amino acid residues in order to provide a counterbalance for an unfavorable clustering of five positively charged K residues (44, 45). Notably, this glycan could be removed when a negatively charge amino acid was introduced to the middle of the K cluster. The electrostatic potential analyses of gp120 surface also indicate that a large area of the protein surface around N448 is positively charged (Fig. 6), and substitution of N448-glycan with a negatively charge amino acid D or E enabled CD4 T cell recognition of the PS05 epitope. In contrast, substitution to an uncharged residue or to a positively charge residue K failed to restore PS05 epitope recognition (Fig. 5). The successful substitution of N448-glycan with an acidic residue confirms that the N448 glycan is not part of the PS05 epitope and is not absolutely necessary for the epitope recognition. Rather, the glycan or a negatively charge amino acid at residue 448 is required for maintaining the conformational stability of the gp120 surface at the carboxy end of PS05 epitope that allows efficient processing to release this particular epitope from the gp120 core protein. Similar to N-glycans, flexible loops are also frequently found to flank gp120 helper epitopes, especially at their C termini (23). Considering the high surface accessibility of N-glycans and flexible loops, both of these structures are likely to contribute to efficient cleavage of the epitope C termini.
In summary, CD4 T cell recognition of HIV-1 gp120 is dictated not only by the epitopes to be presented by the relevant MHC class II alleles, but also by factors outside the epitopes themselves, including N-glycans or amino acid residues that influence the local gp120 surfaces and determine the epitope accessibility to the processing machinery of the APCs. Hence, vast arrays of mutations affecting N-glycans or amino acids outside the T cell epitopes can facilitate HIV-1 escape from immune recognition.
Supplementary Material
Acknowledgments
The authors thank the volunteers for their participation in this study, Ms. Anne Dwyer for coordinating the volunteer visits and blood donation, N. Kalaya Steede for assistance with CD analyses, Drs. Xiang-Peng Kong (New York University School of Medicine) and Max Totrov (Molsoft Inc.) for helpful discussions and critiques, and Dr. Jennifer Fuller for editing the manuscript.
Footnotes
This study was supported by funds from a Merit Review Award and the Research Enhancement Award Program of the US Department of Veterans Affairs, the New York University Center for AIDS Research Immunology Core (AI-27742), the Training Program in TB and HIV Prevention and Treatment (D43 TWO1409), the Louisiana Vaccine Center and South Louisana Institute for Infectious Disease Research (LVC/SLIIDR), and by NIH grant AI-48371. The mass spectrometry analysis was supported in part by NIH grants NS-050276 and RR-14662 to Dr. Thomas A. Neubert, Skirball Institute, New York University School of Medicine.
Publisher's Disclaimer: This is an author-produced version of a manuscript accepted for publication in The Journal of Immunology (The JI). The American Association of Immunologists, Inc. (AAI), publisher of The JI, holds the copyright to this manuscript. This version of the manuscript has not yet been copyedited or subjected to editorial proofreading by The JI; hence, it may differ from the final version published in The JI (online and in print). AAI (The JI) is not liable for errors or omissions in this author-produced version of the manuscript or in any version derived from it by the U.S. National Institutes of Health or any other third party. The final, citable version of record can be found at www.jimmunol.org.
References
- 1.Zhang M, Gaschen B, Blay W, Foley B, Haigwood N, Kuiken C, Korber B. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology. 2004;14:1229–1246. doi: 10.1093/glycob/cwh106. [DOI] [PubMed] [Google Scholar]
- 2.Fischer PB, Karlsson GB, Butters TD, Dwek RA, Platt FM. N-butyldeoxynojirimycin-mediated inhibition of human immunodeficiency virus entry correlates with changes in antibody recognition of the V1/V2 region of gp120. J Virol. 1996;70:7143–7152. doi: 10.1128/jvi.70.10.7143-7152.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fischer PB, Karlsson GB, Dwek RA, Platt FM. N-butyldeoxynojirimycin-mediated inhibition of human immunodeficiency virus entry correlates with impaired gp120 shedding and gp41 exposure. J Virol. 1996;70:7153–7160. doi: 10.1128/jvi.70.10.7153-7160.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y, Luo L, Rasool N, Kang CY. Glycosylation is necessary for the correct folding of human immunodeficiency virus gp120 in CD4 binding. J Virol. 1993;67:584–588. doi: 10.1128/jvi.67.1.584-588.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morikawa Y, Moore JP, Jones IM. HIV-1 envelope protein gp120 expression by secretion in E. coli: assessment of CD4 binding and use in epitope mapping. J Virol Methods. 1990;29:105–113. doi: 10.1016/0166-0934(90)90013-6. [DOI] [PubMed] [Google Scholar]
- 6.Duenas-Decamp MJ, Peters P, Burton D, Clapham PR. Natural resistance of human immunodeficiency virus type 1 to the CD4bs antibody b12 conferred by a glycan and an arginine residue close to the CD4 binding loop. J Virol. 2008;82:5807–5814. doi: 10.1128/JVI.02585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Granados-Gonzalez V, Claret J, Berlier W, Vincent N, Urcuqui-Inchima S, Lucht F, Defontaine C, Pinter A, Genin C, Riffard S. Opposite immune reactivity of serum IgG and secretory IgA to conformational recombinant proteins mimicking V1/V2 domains of three different HIV type 1 subtypes depending on glycosylation. AIDS Res Hum Retroviruses. 2008;24:289–299. doi: 10.1089/aid.2007.0187. [DOI] [PubMed] [Google Scholar]
- 8.Ho YS, Abecasis AB, Theys K, Deforche K, Dwyer DE, Charleston M, Vandamme AM, Saksena NK. HIV-1 gp120 N-linked glycosylation differs between plasma and leukocyte compartments. Virol J. 2008;5:14. doi: 10.1186/1743-422X-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mori K, Sugimoto C, Ohgimoto S, Nakayama EE, Shioda T, Kusagawa S, Takebe Y, Kano M, Matano T, Yuasa T, Kitaguchi D, Miyazawa M, Takahashi Y, Yasunami M, Kimura A, Yamamoto N, Suzuki Y, Nagai Y. Influence of glycosylation on the efficacy of an Env-based vaccine against simian immunodeficiency virus SIVmac239 in a macaque AIDS model. J Virol. 2005;79:10386–10396. doi: 10.1128/JVI.79.16.10386-10396.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mori K, Yasutomi Y, Ohgimoto S, Nakasone T, Takamura S, Shioda T, Nagai Y. Quintuple deglycosylation mutant of simian immunodeficiency virus SIVmac239 in rhesus macaques: robust primary replication, tightly contained chronic infection, and elicitation of potent immunity against the parental wild-type strain. J Virol. 2001;75:4023–4028. doi: 10.1128/JVI.75.9.4023-4028.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Chien PC, Jr, Tuen M, Visciano ML, Cohen S, Blais S, Xu CF, Zhang HT, Hioe CE. Identification of an N-linked glycosylation in the C4 region of HIV-1 envelope gp120 that is critical for recognition of neighboring CD4 T cell epitopes. J Immunol. 2008;180:4011–4021. doi: 10.4049/jimmunol.180.6.4011. [DOI] [PubMed] [Google Scholar]
- 12.Doe B, Steimer KS, Walker CM. Induction of HIV-1 envelope (gp120)-specific cytotoxic T lymphocyte responses in mice by recombinant CHO cell-derived gp120 is enhanced by enzymatic removal of N-linked glycans. Eur J Immunol. 1994;24:2369–2376. doi: 10.1002/eji.1830241017. [DOI] [PubMed] [Google Scholar]
- 13.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCaffrey RA, Saunders C, Hensel M, Stamatatos L. N-linked glycosylation of the V3 loop and the immunologically silent face of gp120 protects human immunodeficiency virus type 1 SF162 from neutralization by anti-gp120 and anti-gp41 antibodies. J Virol. 2004;78:3279–3295. doi: 10.1128/JVI.78.7.3279-3295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ly A, Stamatatos L. V2 loop glycosylation of the human immunodeficiency virus type 1 SF162 envelope facilitates interaction of this protein with CD4 and CCR5 receptors and protects the virus from neutralization by anti-V3 loop and anti-CD4 binding site antibodies. J Virol. 2000;74:6769–6776. doi: 10.1128/jvi.74.15.6769-6776.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolk T, Schreiber M. N-Glycans in the gp120 V1/V2 domain of the HIV-1 strain NL4-3 are indispensable for viral infectivity and resistance against antibody neutralization. Med Microbiol Immunol. 2006;195:165–172. doi: 10.1007/s00430-006-0016-z. [DOI] [PubMed] [Google Scholar]
- 17.Perdomo MF, Levi M, Sallberg M, Vahlne A. Neutralization of HIV-1 by redirection of natural antibodies. Proc Natl Acad Sci U S A. 2008;105:12515–12520. doi: 10.1073/pnas.0805777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sirois S, Touaibia M, Chou KC, Roy R. Glycosylation of HIV-1 gp120 V3 loop: towards the rational design of a synthetic carbohydrate vaccine. Curr Med Chem. 2007;14:3232–3242. doi: 10.2174/092986707782793826. [DOI] [PubMed] [Google Scholar]
- 19.Wang LX. Toward oligosaccharide- and glycopeptide-based HIV vaccines. Curr Opin Drug Discov Devel. 2006;9:194–206. [PubMed] [Google Scholar]
- 20.Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, Kunert R, Zhu P, Wormald MR, Stanfield RL, Roux KH, Kelly JW, Rudd PM, Dwek RA, Katinger H, Burton DR, Wilson IA. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300:2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- 21.Sanders RW, Venturi M, Schiffner L, Kalyanaraman R, Katinger H, Lloyd KO, Kwong PD, Moore JP. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J Virol. 2002;76:7293–7305. doi: 10.1128/JVI.76.14.7293-7305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Surman S, Lockey TD, Slobod KS, Jones B, Riberdy JM, White SW, Doherty PC, Hurwitz JL. Localization of CD4+ T cell epitope hotspots to exposed strands of HIV envelope glycoprotein suggests structural influences on antigen processing. Proc Natl Acad Sci U S A. 2001;98:4587–4592. doi: 10.1073/pnas.071063898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mirano-Bascos D, Tary-Lehmann M, Landry SJ. Antigen structure influences helper T-cell epitope dominance in the human immune response to HIV envelope glycoprotein gp120. Eur J Immunol. 2008;38:1231–1237. doi: 10.1002/eji.200738011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarkar S, Kalia V, Murphey-Corb M, Montelaro RC. Detailed analysis of CD4+ Th responses to envelope and Gag proteins of simian immunodeficiency virus reveals an exclusion of broadly reactive Th epitopes from the glycosylated regions of envelope. J Immunol. 2002;168:4001–4011. doi: 10.4049/jimmunol.168.8.4001. [DOI] [PubMed] [Google Scholar]
- 25.Cohen S, Tuen M, Hioe CE. Propagation of CD4+ T cells specific for HIV type 1 envelope gp120 from chronically HIV type 1-infected subjects. AIDS Res Hum Retroviruses. 2003;19:793–806. doi: 10.1089/088922203769232593. [DOI] [PubMed] [Google Scholar]
- 26.Jones GJ, von Hoegen P, Weber J, Rees AD. Immunization with human immunodeficiency virus type 1 rgp120W61D in QS21/MPL adjuvant primes T cell proliferation and C-C chemokine production to multiple epitopes within variable and conserved domains of gp120W61D. J Infect Dis. 1999;179:558–566. doi: 10.1086/314626. [DOI] [PubMed] [Google Scholar]
- 27.Hioe CE, Jones GJ, Rees AD, Ratto-Kim S, Birx D, Munz C, Gorny MK, Tuen M, Zolla-Pazner S. Anti-CD4-binding domain antibodies complexed with HIV type 1 glycoprotein 120 inhibit CD4+ T cell-proliferative responses to glycoprotein 120. AIDS Res Hum Retroviruses. 2000;16:893–905. doi: 10.1089/08892220050042837. [DOI] [PubMed] [Google Scholar]
- 28.Tuen M, Visciano ML, Chien PC, Jr, Cohen S, Chen PD, Robinson J, He Y, Pinter A, Gorny MK, Hioe CE. Characterization of antibodies that inhibit HIV gp120 antigen processing and presentation. Eur J Immunol. 2005;35:2541–2551. doi: 10.1002/eji.200425859. [DOI] [PubMed] [Google Scholar]
- 29.Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zolla-Pazner S. Identifying epitopes of HIV-1 that induce protective antibodies. Nat Rev Immunol. 2004;4:199–210. doi: 10.1038/nri1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown BK, Wieczorek L, Sanders-Buell E, Rosa Borges A, Robb ML, Birx DL, Michael NL, McCutchan FE, Polonis VR. Cross-clade neutralization patterns among HIV-1 strains from the six major clades of the pandemic evaluated and compared in two different models. Virology. 2008;375:529–538. doi: 10.1016/j.virol.2008.02.022. [DOI] [PubMed] [Google Scholar]
- 33.Yuste E, Sanford HB, Carmody J, Bixby J, Little S, Zwick MB, Greenough T, Burton DR, Richman DD, Desrosiers RC, Johnson WE. Simian immunodeficiency virus engrafted with human immunodeficiency virus type 1 (HIV-1)-specific epitopes: replication, neutralization, and survey of HIV-1-positive plasma. J Virol. 2006;80:3030–3041. doi: 10.1128/JVI.80.6.3030-3041.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mascola JR, Snyder SW, Weislow OS, Belay SM, Belshe RB, Schwartz DH, Clements ML, Dolin R, Graham BS, Gorse GJ, Keefer MC, McElrath MJ, Walker MC, Wagner KF, McNeil JG, McCutchan FE, Burke DS. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. The National Institute of Allergy and Infectious Diseases AIDS Vaccine Evaluation Group. J Infect Dis. 1996;173:340–348. doi: 10.1093/infdis/173.2.340. [DOI] [PubMed] [Google Scholar]
- 35.Moore JP, Cao Y, Qing L, Sattentau QJ, Pyati J, Koduri R, Robinson J, Barbas CF, 3rd, Burton DR, Ho DD. Primary isolates of human immunodeficiency virus type 1 are relatively resistant to neutralization by monoclonal antibodies to gp120, and their neutralization is not predicted by studies with monomeric gp120. J Virol. 1995;69:101–109. doi: 10.1128/jvi.69.1.101-109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pinter A. Roles of HIV-1 Env variable regions in viral neutralization and vaccine development. Curr HIV Res. 2007;5:542–553. doi: 10.2174/157016207782418470. [DOI] [PubMed] [Google Scholar]
- 37.Hu SL, Stamatatos L. Prospects of HIV Env modification as an approach to HIV vaccine design. Curr HIV Res. 2007;5:507–513. doi: 10.2174/157016207782418542. [DOI] [PubMed] [Google Scholar]
- 38.Pantophlet R, Burton DR. GP120: target for neutralizing HIV-1 antibodies. Annu Rev Immunol. 2006;24:739–769. doi: 10.1146/annurev.immunol.24.021605.090557. [DOI] [PubMed] [Google Scholar]
- 39.Sjolander S, Bolmstedt A, Akerblom L, Horal P, Olofsson S, Morein B, Sjolander A. N-linked glycans in the CD4-binding domain of human immunodeficiency virus type 1 envelope glycoprotein gp160 are essential for the in vivo priming of T cells recognizing an epitope located in their vicinity. Virology. 1996;215:124–133. doi: 10.1006/viro.1996.0015. [DOI] [PubMed] [Google Scholar]
- 40.Watts C. The exogenous pathway for antigen presentation on major histocompatibility complex class II and CD1 molecules. Nat Immunol. 2004;5:685–692. doi: 10.1038/ni1088. [DOI] [PubMed] [Google Scholar]
- 41.Li P, Gregg JL, Wang N, Zhou D, O'Donnell P, Blum JS, Crotzer VL. Compartmentalization of class II antigen presentation: contribution of cytoplasmic and endosomal processing. Immunol Rev. 2005;207:206–217. doi: 10.1111/j.0105-2896.2005.00297.x. [DOI] [PubMed] [Google Scholar]
- 42.Bryant PW, Lennon-Dumenil AM, Fiebiger E, Lagaudriere-Gesbert C, Ploegh HL. Proteolysis and antigen presentation by MHC class II molecules. Adv Immunol. 2002;80:71–114. doi: 10.1016/S0065-2776(02)80013-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, Harrison SC. Determining the structure of an unliganded and fully glycosylated SIV gp120 envelope glycoprotein. Structure. 2005;13:197–211. doi: 10.1016/j.str.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 44.Wyss DF, Wagner G. The structural role of sugars in glycoproteins. Curr Opin Biotechnol. 1996;7:409–416. doi: 10.1016/s0958-1669(96)80116-9. [DOI] [PubMed] [Google Scholar]
- 45.Wyss DF, Choi JS, Li J, Knoppers MH, Willis KJ, Arulanandam AR, Smolyar A, Reinherz EL, Wagner G. Conformation and function of the N-linked glycan in the adhesion domain of human CD2. Science. 1995;269:1273–1278. doi: 10.1126/science.7544493. [DOI] [PubMed] [Google Scholar]
- 46.Leonard CK, Spellman MW, Riddle L, Harris RJ, Thomas JN, Gregory TJ. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J Biol Chem. 1990;265:10373–10382. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.