Abstract
3,3′-Diindolylmethane (DIM) is an anti-cancer agent that induces cell cycle arrest and apoptosis through unknown mechanisms. Here, we report that DIM selectively induced proteasome-mediated degradation of the class I histone deacetylases (HDAC1, HDAC2, HDAC3, and HDAC8) in colon cancer cells, without affecting the class II HDAC proteins. DIM-induced down-regulation of the class I HDACs was also observed in vivo in tumor xenografts in nude mice. The depletion of the HDACs as the result of DIM treatment caused a reduction of the HDAC activity on the promoters of CDKN1A/WAF1/CIP1 and CDKN1B/KIP1 genes (encoding cyclin-dependent kinase inhibitors p21 and p27, respectively) and significantly increased the expression of p21 and p27, which arrested cells at the G2 phase of the cell cycle. The degradation of the HDACs also caused DNA damage and triggered apoptosis. Thus, DIM acts by selectively targeting the class I HDACs to promote their degradation.
Keywords: 3,3′-Diindolylmethane; histone deacetylase; protein degradation; cell cycle arrest; apoptosis
Introduction
Acetylation and deacetylation of the lysine residues on the histone proteins play a key role in the regulation of gene transcription (1). While histone acetyltransferases catalyze the acetylation of histones and relax the chromatin to increase the accessibility of transcription factors to the promoters of the target genes, histone deacetylases (HDACs) remove the acetyl group from histones and repress transcription (2). Eighteen mammalian HDACs have been identified and they are classified into four groups (3): class I HDACs (HDAC1, −2, −3, and −8) have homology to the yeast protein Rpd3; class II HDACs (HDAC4, −5, −6, −7, −9, and −10) have homology to yeast HDAC Had-1; class III HDACs (sirt1–7) have homology to yeast Sir2 gene; class IV HDAC (HDAC11) does not have sufficient homology with both class I and class II HDACs. Class I HDACs are often overexpressed in various types of cancers comparing to the corresponding normal tissues and their overexpression is correlated with a poor prognosis (4-6). In contrast, the expression of class II HDACs is associated with a better prognosis especially in non-small cell lung cancer and cutaneous T-cell lymphoma. To target the HDAC enzymes, various small molecule inhibitors have been developed and they have been shown to induce differentiation, growth arrest, and apoptosis in cancer cells (7-9). Among the most potent HDAC inhibitors, vorinostat (suberoylanilide hydroxamic acid, SAHA) has been approved for the treatment of cutaneous T-cell lymphoma (10). Other HDAC inhibitors are subjects of extensive clinical evaluations at various stages for treatment of different types of cancers (11-13). Many of these HDAC inhibitors (such as vorinostat) are broad-spectrum inhibitors and inhibit both class I and class II HDACs, while a few are class I-specific inhibitors (14).
3,3′-Diindolylmethane (DIM) is an anti-cancer agent naturally formed during the autolytic breakdown of glucobrassicin which is present in food plants of the Brassica genus, including broccoli, cabbage, Brussels sprouts, and cauliflower (15). The anti-tumor activity of DIM was linked to the down-regulation of androgen receptor (16) and inhibition of mammalian target of rapamycin (mTOR) (17) in prostate cancer cells. DIM was also shown to inhibit mitochondrial H(+)-ATP synthase and induce p21(Cip1/Waf1) expression in breast cancer cells (18). Inhibition of AKT signaling and FLICE-like inhibitory protein by DIM was reported in cholangiocarcinoma cells (19), while inactivation of NF-kappaB (20) and down-regulation of survivin were demonstrated in breast cancer cells treated with DIM (21). DIM also inhibits angiogenesis and invasion by repressing the expression of matrix metalloproteinase MMP-9 and urokinase-type plasminogen activator (uPA) (22). However, it is still not clear how these different effects of DIM eventually lead to cell cycle arrest and apoptosis.
We have recently shown that DIM can enhance the anti-tumor activity of butyrate (an HDAC inhibitor) in a mouse model of colon cancer (23). In this report, we find that DIM selectively induces proteasome-mediated degradation of the class I HDACs (HDAC1, HDAC2, HDAC3, and HDAC8) in colon cancer cells. Down-regulation of HDACs represents a novel mechanism underlying DIM’s ability to induce cell cycle arrest and apoptosis.
Materials and Methods
Cells and Transfection
The colon cancer cell lines HT-29, SW620, RKO, LS174T, and HCT-116 were purchased from American Type Culture Collection. HT-29, LS174T, and RKO cells were cultured in Dulbecco’s Modified Eagle Medium containing 10% fetal bovine serum (FBS). SW620 and HCT-116 cells were cultured in RPMI1640 media containing 10% FBS. For transient transfection, plasmids were transfected into cells using Lipofectamine™Plus Reagent (Invitrogen) following the manufacturer’s protocol.
Drugs and Chemicals
DIM was purchased from LKT Laboratories (St. Paul, MN). MG-132 was purchased from Calbiochem (Gibbstown, NJ). SAHA was purchased from Biovision (Mountain View, CA). Disuccinimidyl suberate was purchased from Pierce (Rockford, IL). Carbobenzoxy-valyl-alanyl-aspartyl-[O-methyl]-fluoromethylketone (z-VAD-fmk) was purchased from R & D Systems (Minneapolis, MN).
Plasmids
Human cDNAs encoding full-length HDAC2 and HDAC8 gene were obtained by PCR amplification using EST clones as templates. These cDNAs were sub-cloned into a pCEP4-Flag vector to express Flag-tagged proteins. Plasmids expressing Flag-tagged HDAC1 and HDAC3 were obtained from Dr. Ed Seto’s lab at H. Lee Moffitt Cancer Center.
Tumor Xenografts in Nude Mice
Six to eight weeks old female nude mice (Nu/Nu) were purchased from Charles River (Wilmington, MA). The mice were maintained in sterile conditions using the Innovive IVC System from Innovive (San Diego, CA), following the protocol approved by the Institutional Animal Care and Use Committee of North Dakota State University. Tumor xenografts were established by subcutaneous injection of 2×106 HT-29 cells in the flank area of the mice.
Western Blot Analysis
Cells were lysed in RIPA buffer (1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS in PBS). Complete protease inhibitor cocktail (Roche) was added to lysis buffer before use. Protein concentration was determined by Bio-Rad DC protein assay (Bio-Rad). Protein samples were subjected to SDS-PAGE and transferred to nitrocellulose membrane. The membrane was blocked in 5% non-fat milk in PBS overnight and incubated with primary antibody and subsequently with appropriate horse radish peroxidase-conjugated secondary antibody. Signals were developed with ECL reagents (Pierce) and exposure to X-ray films. Image digitization and quantification were performed with UN-SCAN-IT software from Silk Scientific (Orem, UT). Anti-cleaved caspase-7, anti-cleaved caspase-9, anti-full length caspase-7, anti-HDAC1, anti-HDAC2, anti-HDAC-3, anti-HDAC4, anti-HDAC5, anti-HDAC7, anti-histone H3, anti-acetyl-H3, anti-p21, anti-p27, and anti-SirT2 antibodies were purchased form Cell Signaling Technology. Anti-β-tubulin, anti-Ubc8, anti-HDAC-6, anti-HDAC-8, anti-RLIM, and anti-Bak antibodies were purchased from Santa Cruz Biotechnology. Anti-γH2AX and anti-phospho-KAP1 (S824) polyclonal antibodies were purchased from Bethyl Laboratories. Anti-Hr23b antibody was purchased from Enzo Life Sciences. Anti-Flag-HRP antibody was purchased from Sigma.
Isolation of Ubiquitin-modified Proteins
HCT-116 cells were transfected with plasmids to express the Flag-tagged HDAC proteins. After treatment with DIM, ubiquitin-modified HDAC proteins were isolated from the cells using the ubiquitin affinity resin of the Ubiquitin Enrichment Kit from Pierce (Rockford, IL). HDAC proteins were detected by western blotting with anti-Flag antibody.
Real-time PCR
The mRNA expression was measure by real-time PCR using TaqMan® Gene Expression assays (Cat # Hs02621185_s1 for HDAC1, Hs00231032_m1 for HDAC2, Hs00187320_m1 for HDAC3, Hs00355782_m1 for p21/Cip1, Hs01597588_m1 for p27/kip1) from Applied Biosystems (Foster city, CA). Total RNA was isolated from HT-29 cells using RNeasy® kit (Qiagen). 5 μg of total RNA was used in reverse transcription reaction. The cDNAs were used as templates to perform PCR on an Applied Biosystems 7500 Real-time PCR System following the manufacturer’s protocol.
Chromatin Immunoprecipitation Assay
ChIP assay was performed using the ChIP asay kit from Millipore (Billerica, MA), following the supplied protocol. Immunoprecipitations were performed using anti-HDAC1, anti-HDAC2, anti-HDAC3, anti-H3, anti-acetyl-H3, or control IgG antibodies. PCR was performed with the primers designed from the sequences of the CDKN1A/WAF1/CIP1 promoter (5′-GTGGCTCTGATTGGCTTTCTG-3′ and 5′-CTGAAAACAGGCAGCCCAAG-3′) and CDKN1B/Kip1 promoter (5′-GAGGCGGCGCGCTCGGGAAC-3′ and 5′-AGGGAGGCT GACGAAGAAGA-3′).
Cell cycle analysis
Cell cycle distribution was analyzed using a Accuri C6 Flow Cytometer. Cells were fixed and stained with propidium iodide using the Cell Cycle Phase Determination Kit (Accuri Cytometers) and analyzed following the manufacturer’s protocol.
Immunofluoresence staining
Cells grown in Lab-Tek II chamber slides (Nalge Nunc) were washed with PBS and fixed in 3% paraformaldehyde in PBS for 10 min. The fixed cells were washed and permeabilized with 0.15% Triton X-100 in PBS for 15 min at room temperature. Cells were blocked with 2% bovine serum albumin in PBS for 1h and incubated with anti-phospho-KAP1 (S824) antibody (1:400 dilution) overnight at 4°C. Cells were washed and incubated with Alexa Fluor 488 goat anti-rabbit antibody (Invitrogen) at 1:250 dilution for 1h at room temperature and counterstained with propidium iodide to visualize the nuclei. Cells that were positive for KAP-1 phosphorylation were counted under a fluorescence microscope.
Comet Assay
DNA damage was detected by Comet assay using the Comet Assay Kit from Trevigen. Alkaline electrophoresis was done at pH>13 (200mM NaOH, 1mM EDTA). Images were taken with a fluorescence microscope and analyzed with the CometScore software (TriTek).
Cross-linking Study
Disuccinimidyl suberate (DSS) was dissolved in DMSO at 25 mM concentration. Before protein isolation for western blot, cells were treated with 1 mM DSS in PBS for 30 min at 37°C. The stop solution (1 M Tris, pH 7.5) was then added to a final concentration of 10 mM and incubated for 15 min. Total protein was isolated and western blot was performed as described above.
Detection of Apoptosis
The Cell Death Detection ElisaPLUS kit (Roche) was used to detect apoptosis following the manufacturer’s protocol. This assay determines apoptosis by measuring mono- and oligonucleosomes in the lysates of apoptotic cells. The cell lysates were placed into a streptavidin-coated microplate and incubated with a mixture of anti-histone-biotin and anti-DNA-peroxidase. The amount of peroxidase retained in the immunocomplex was photometrically determined with ABTS as the substrate. Absorbance was measured at 405 nm.
Results
DIM Selectively Decreases the Protein Levels of the Class I HDACs
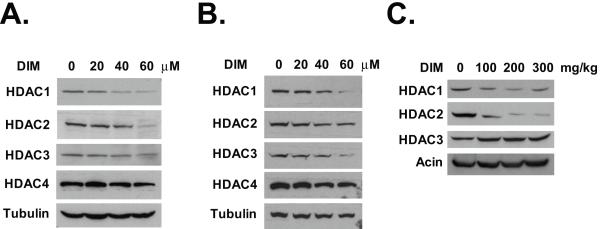
We have recently shown that DIM can enhance the antitumor activity of HDAC inhibitor butyrate (23). To determine if DIM has a direct effect on the levels of HDAC proteins, we performed western blot analysis on colon cancer HT-29 cells treated with various doses of DIM. As shown in Fig. 1A, exposure to DIM selectively decreased the levels of class I HDACs (HDAC1, HDAC2, and HDAC3). However, DIM had no effect on the levels of the class II HDACs (HDAC4, HDAC5, HDAC6, and HDAC7) or the class III HDACs (SIRT2) (Fig. 1A, Supplemental Fig. 1B, and data not shown). We have examined the effects of DIM on another colon cancer cell line SW620. DIM decreased the levels of the class I HDACs in SW620 cells, but had no effects on the class II HDACs (Fig. 1B). The other class I HDAC, HDAC8, was not expressed in HT-29 or SW620 cells. In HCT-116 colon cancer cells, DIM induced significant decrease in the levels of all four class I HDACs, including HDAC8 (Supplemental Fig. 1A). To determine if the effect of DIM on the HDACs can be applied in general to other colon cancer cell lines, we repeated the experiment using RKO and LS174T cells. DIM was able to down-regulate the class I HDACs in all these colon cancer cell lines (data not shown). Since both HT-29 and SW620 cells express mutant p53, the effects of DIM on the HDAC proteins appear to be p53-independent. Importantly, we found that DIM was able to down-regulate both HDAC1 and HDAC2 in vivo in HT-29 xenografts in nude mice (Fig. 1C). DIM treatment did not decrease HDAC3 in the tumor xenografts, reflecting a more complicated environment in the in vivo situation. Previously, it has been shown that an oral dose of 250 mg/kg of DIM produced plasma DIM concentration of 19 μg/ml in mice, which equals to about 77 μM (24). We did not observe any toxicity to the mice even at the highest dose used (300 mg/kg/day).
Fig. 1.
DIM decreases the levels of the class I HDACs. (A) HT-29 cells were treated with various doses of DIM for 24 hours. Western blotting was done using the indicated antibodies. (B) SW620 cells were treated with various doses of DIM for 24 hours. Western blotting was done using the indicated antibodies. (C) Nude mice bearing HT-29 xenografts were treated with DIM at oral doses of 100, 200, 300 mg/kg/day for two days. Tumor samples were collected 24h after the second dose and analyzed by western blotting. Representative results from three independent blots were shown.
Down-regulation of HDACs by DIM is a Result of Proteasomal Degradation
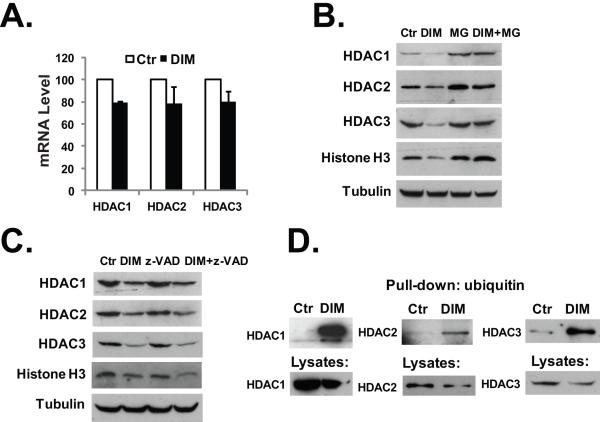
To understand the mechanism of DIM-induced decrease of the HDAC proteins, we first determined if DIM could affect the mRNA expression of the HDACs. Using real-time PCR analysis, we found that treatment with DIM had only minimal effect (causing ~20% decreases) on the levels of the mRNAs of class I HDACs (Fig. 2A). On the other hand, DIM treatment did not inhibit the translation of the mRNAs of the class I HDACs (Supplemental Fig. 2A). We next examined if the proteasome-mediated protein degradation was involved in the down-regulation of HDACs after DIM treatment. As shown in Fig. 2B, treatment with DIM in the presence of proteasome inhibitor MG-132 did not result in the decrease of HDAC1, HDAC2, and HDAC3 proteins. DIM treatment also induced a decrease in the level of histone H3 protein in HT-29 cells, which was also blocked by MG-132 (Fig. 2B). In contrast, co-treatment of HT-29 cells with a broad spectrum caspase inhibitor z-VAD had no effects on DIM-induced decreases in the HDACs and histone H3 protein (Fig. 2C), indicating that the decreases of these proteins were not the result of the activation of apoptosis. Furthermore, by isolating ubiquitin-modified proteins in DIM-treated cells, we demonstrated that DIM treatment significantly induced the formation of the ubiquitin-labeled HDAC1, HDAC2, HDAC3, and HDAC8 (Fig. 2D and Supplemental Fig. 1D). Thus, ubiquitylation of the class I HDACs is involved in the proteasome-mediated degradation of these proteins. It has been shown that the E2 ubiquitin conjugase Ubc8 and the E3 ubiquitin ligase RLIM are involved in the protein degradation of HDAC2 (25). However, we did not detect the expression of Ubc8 in HT-29 cells (data not shown) and RLIM expression was not affected by DIM treatment (Supplemental Fig. 1B). Furthermore, we observed a decrease of the interaction between RLIM and HDAC2 after DIM treatment, indicating that RLIM may not act as the E3 ubiquitin ligase for HDAC2 in this situation (Supplemental Fig. 1C). Recently, a genome wide screening identified HR23B as a critical factor for HDAC inhibitor-induced apoptosis (26). HR23B shuttles ubiquitinated proteins to the proteasome. However, we did not observe an induction of HR23B by DIM in HT-29 cells (Supplemental Fig. 2B).
Fig. 2.
DIM induces proteasome-mediated degradation of the HDACs. (A) HT-29 cells were treated with 40 μM DIM for 24 hours, total RNA was isolated and real-time PCR analysis was done as described in Materials and Methods. (B) HT-29 cells were treated with 40 μM DIM, 10 μM of MG-132, or 40 μM DIM plus 10 μM of MG-132 for 24h. Western blotting was performed with the indicated antibodies. (C) HT-29 cells were treated with 40 μM DIM, 20 μM of z-VAD, or 40 μM DIM plus 20 μM of z-VAD for 24h. Western blotting was performed with the indicated antibodies. (D) HCT-116 cells were transfected with plasmids to express Flag-tagged HDAC proteins. 24h after transfection, cells were treated with 40 μM DIM for additional 24 hours. Cell lysates in RIPA buffer were collected and ubiquitin-modified proteins were isolated as described in Materials and Methods, followed by western blot analysis. The experiments have been repeated three times.
Degradation of HDACs by DIM Activates p21 and p27 Expression
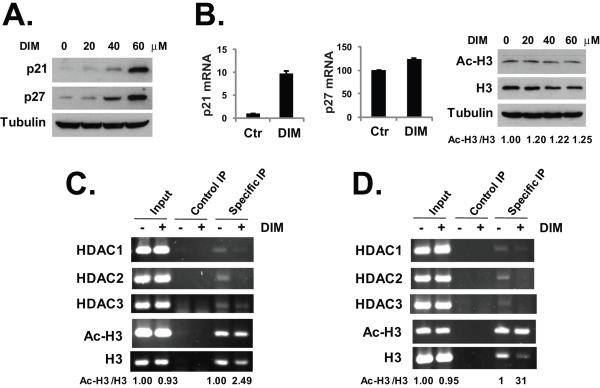
The class I HDACs have been linked to the transcriptional repression of the cyclin dependent kinase inhibitor p21 (27) and the HDAC inhibitors have been shown to induce cell cycle arrest by activating the expression of p21 and p27 (28,29). We hypothesize that DIM-induced depletion of the class I HDACs can activate the expression of p21 and p27, because the HDAC repression on the promoters of these genes will be removed. As shown in Fig. 3A, we found that treatment with DIM had induced significant increases in the protein levels of both p21 and p27 in HT-29 cells. Notably, DIM also induced p21 expression in vivo in the HT-29 xenografts in nude mice (Fig. 5B). DIM had no effects on p27 in the tumor xenografts (data not shown). To determine if the increase of p21 and p27 expression occurred at transcription level, we used real-time PCR to measure the mRNA levels of p21 and p27 in HT-29 cells. As shown in Fig. 3B, exposure to DIM resulted in about ten-fold increase in the p21 mRNA and about 23% increase in p27 mRNA expression. The global effects of DIM-induced HDAC depletion on cellular histone acetylation were examined by western blot analysis. As shown in Fig. 3B, while DIM caused a decrease in the total protein level of histone H3, the percentage of acetylated H3 was increased by about 20–25%. More importantly, we examined the effects of DIM on the levels of HDAC proteins bound to the p21 and p27 promoters in HT-29 cells. Using chromatin immunoprecipitation (ChIP) assays, we found that DIM caused significant decreases (if not a complete loss) in the levels of HDAC1–3 associated with the p21 and p27 promoters (Figs. 3C and 3D). While DIM also induced a decrease in the amount of total histone H3 bound to the p21 and p27 promoters, the relative levels of acetylated H3 over total H3 on the p21 and p27 promoters were increased by 2.49 and 31 folds, respectively (Figs. 3C and 3D). The discrepancy between the effects of DIM on H3 acetylation within the p21 and p27 promoters and DIM-induced p21 and p27 expression levels indicates that histone acetylation is not the only factor regulating the expression of these genes. Other factors may also regulate the expression of p21 and p27, for example the recruitment of histone methyltransferases and histone methylation, which may be affected by the DIM-induced depletion of HDAC proteins.
Fig. 3.
DIM activates p21 and p27 expression. (A) HT-29 cells were treated with various doses of DIM for 24 hours. Western blotting was performed with the indicated antibodies. (B) HT-29 cells were treated with 40 μM DIM for 24 hours, total RNA was isolated and real-time PCR analysis was done as described in Materials and Methods. For western blot analysis, HT-29 cells were treated with various doses of DIM for 24 hours. Western blotting was performed with an anti-acetylated H3 and anti-H3 antibodies and anti-tubulin antibody. (C) HT-29 cells were treated with 40 μM DIM for 24 hours. ChIP assay was performed as described in Materials and Methods, using primers specific for the p21 promoter and the indicated antibodies. (D) HT-29 cells were treated with 40 μM DIM for 24 hours. ChIP assay was performed as described in Materials and Methods, using primers specific for p27 promoter and the indicated antibodies. Relative protein levels and DNA band signals were quantified and shown under the gels. The experiments have been repeated three times.
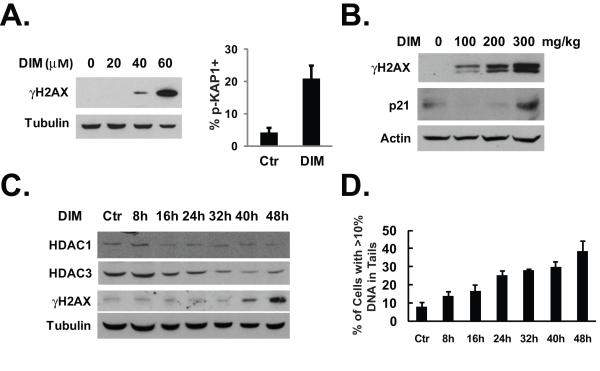
Fig. 5.
DIM induces DNA damage. (A) Left, HT-29 cells were treated with various doses of DIM for 24 hours. Western blotting was done using indicated antibodies. Right, HT-29 cells were cultured in chamber slides and treated with 40 μM of DIM for 48 hours. Immunofluoresence staining with anti-phospho-KAP1 antibody was done as described in Materials and Methods. The percentage of cells that are positive for KAP1 phosphorylation was determined after counting approximately 100 cells in each experiment. The average percentage of three independent experiments was shown. (B) Nude mice treatment was described in Fig. 1C. Tumor samples were analyzed by western blotting with indicated antibodies. (C) HT-29 cells were treated with 40 μM of DIM for various lengths of time as indicated. Western blotting was done using the indicated antibodies. (D) HT-29 cells were treated with 40 μM of DIM for various lengths of time as indicated. Comet assay was performed as described in Materials and Methods. The average results from two independent experiments were shown.
DIM Induces G2 Cell Cycle Arrest
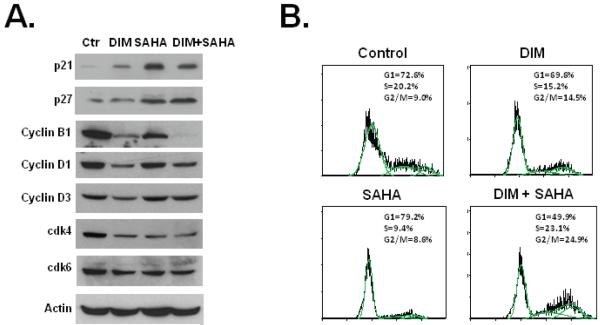
p21 and p27 can arrest cells at both the G1 and G2 phases (30-33). The effects of DIM on p21 and p27 expression led us to further examine how cell cycle regulatory proteins and cell cycle progression itself were affected by DIM treatment. Comparing to HDAC inhibitor SAHA, DIM had different effects on the protein levels of cyclins and cyclin-dependent kinases (Fig. 4A). While DIM significantly decreased the levels of cyclin B1, cyclin D1, and cyclin D3, SAHA did not affect the expression of these proteins. Both DIM and SAHA decreased cdk4 expression without significantly affecting the level of cdk6 (Fig. 4A). We found that DIM induced a G2 phase cell cycle arrest in HT-29 cells (Fig. 4B). While SAHA induced a G1 phase cell cycle arrest, the combination of DIM and SAHA induced a more significant G2 phase arrest than when DIM was used alone (Fig. 4B). SAHA was known to induce G1 or G2/M arrests depending the concentrations used (34).
Fig. 4.
DIM induces G2 cell cycle arrest. (A) HT-29 cells were treated with 40 μM DIM for 24h, 2 μM SAHA for 24h, or 40 μM DIM for 24h followed by 2 μM SAHA for additional 24h. Western blot were performed using the indicated antibodies. (B) HT-29 cells were treated with 40 μM DIM for 24h, 2 μM SAHA for 24h, or 40 μM DIM for 24h followed by 2 μM SAHA for additional 24h. Cell cycle distribution was analyzed as described in Materials and Methods. The experiments have been repeated three times.
DIM Treatment Causes DNA Damage
Studies in the embryonic fibroblasts from HDAC3 knockout mouse have shown that depletion of HDAC3 can cause DNA damage that is associated with defective DNA double-strand break repair (35). We hypothesize that by depleting all class I HDACs, DIM can cause DNA damage in colon cancer cells. Phosphorylated H2AX (also named as γH2AX) has been used as a reliable indicator to measure DNA double-strand breaks (36). We found that treatment with DIM caused a significant increase in the levels of γH2AX protein (Fig. 5A, left). To further confirm the occurrence of DNA damage after DIM treatment, we examined an additional marker of DNA damage response. KAP-1 (KRAB-associated protein) is phosphorylated on serine 824 in response to DNA damage, which relaxes the chromatin (37). Using immunofluoresence staining, we observed a significant increase in the number of cells positively stained with anti-phospho-KAP1 antibody in response to DIM treatment (Fig. 5A, right). Induction of γH2AX expression was also observed in vivo in HT-29 xenografts in nude mice after DIM treatment (Fig. 5B). We determined the time courses of DIM-induced HDAC degradation and found that the decrease of HDACs occurred about 24 hours ahead of the appearance of γH2AX (Fig. 5C). Using quantitative Comet assay, we measured the time course of DIM-induced DNA damage. As shown in Fig. 5D, the appearance of DNA damage in cells correlated with the decrease of the levels of the HDAC proteins.
DIM Induces Apoptosis and Enhances HDAC Inhibitor-induced Apoptosis
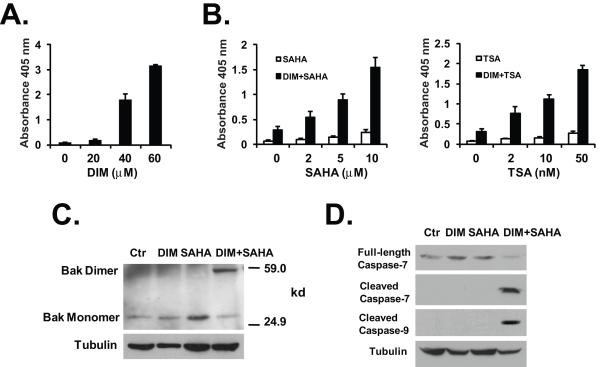
Since DIM caused significant decreases in the levels of HDAC proteins as well as DNA damage, we reasoned that DIM could induce apoptosis in colon cancer cells and enhance the effects of HDAC inhibitors on HDAC inhibition and induction of apoptosis. We found that treatment with DIM alone induced apoptosis in HT-29 cells in a dose-dependent manner (Fig. 6A). Pre-treatment with low dose of DIM also significantly enhanced SAHA-induced apoptosis in HT-29 cells (Fig. 6B, left). Similarly, DIM treatment significantly potentiated apoptosis induced by another HDAC inhibitor, trichostatin A (Fig. 6B, right). Using the CalcuSyn software, we demonstrated that the combinations of DIM/SAHA and DIM/Trichostatin A were synergistic (CI < 1) in apoptosis induction at most doses (Supplemental Fig. 3A). A combination of low doses of DIM and SAHA treatment triggered the formation of dimerized pro-apoptotic protein Bak (Fig. 6C), representing the activated Bak (38,39). The dimerization of Bak was not seen in cells treated with either DIM or SAHA alone. We also found that caspase-7 and caspase-9 were activated by the DIM/SAHA combination treatment (indicated by the appearance of cleaved caspase-7 and cleaved caspase-9), but not by the individual agent alone (Fig. 6D). To determine the role of down-regulation of individual HDAC protein in the enhancement of SAHA-induced apoptosis, we knocked-down the individual class I HDAC protein in HCT-116 cell by siRNA (Supplemental Fig. 3B), and analyzed the effects of siRNA knock-down on responses to SAHA. Knock-down of HDAC2 or HDAC3 by siRNA exhibited most significant enhancement of SAHA-induced apoptosis in these cells (Supplemental Fig. 3C). The data suggest that DIM-induced degradation of the class I HDACs (especially HDAC2 and HDAC3) can directly enhance the activity of an HDAC inhibitor in apoptosis induction.
Fig. 6.
DIM induces apoptosis and enhances HDAC inhibitor-induced apoptosis. (A) HT-29 cells were treated with various doses of DIM for 24h. Apoptosis was analyzed using the Cell Death Detection ElisaPLUS kit as described in Materials and Methods. (B) HT-29 cells were treated with or without 20 μM of DIM for 24 hours, followed by various doses of SAHA or Trichostatin A (TSA) for additional 24h. Apoptosis was analyzed as described in (A). The average results from three independent experiments were shown. (C) DIM/SAHA treatment induced Bak activation. HT-29 cells were treated with 20 μM DIM for 24h, 2 μM SAHA for 24h, or 20 μM DIM for 24h followed by 2 μM SAHA for additional 24h. Bak dimerization was detected by cross-linking experiments as described in Materials and Methods. Protein lysates were analyzed by western blotting. (D) HT-29 cells were treated as described in (C) and western blot was performed using the indicated antibodies.
Discussion
DIM has been intensively evaluated as an anti-tumor agent in various types of cancers (40). However, the mechanism of its activity in inducing cell cycle arrest and apoptosis has not been defined. In this study, we demonstrate that DIM selectively induces proteasomal degradation of the class I HDACs (HDAC1, HDAC2, HDAC3, and HDAC8). Down-regulation of the HDACs by DIM not only activates the expression of p21 and p27 to cause cell cycle arrest, but also induces DNA damage to trigger apoptosis in colon cancer cells. Thus, our data establish a mechanistic basis for the anti-tumor activity of DIM. Since the cells used in our experiments express mutant p53, our results also provide a mechanism for the p53-independent activation of p21 and p27 by DIM. Reduction of the levels of the HDAC proteins that are bound to the p21 and p27 promoters derepresses the transcription of these genes.
It is recently reported that DIM induces p21 expression by inhibiting the mitochondrial H+-ATP synthase and causing oxidative stress in human breast cancer cells (18). We did observe increased ROS production in DIM-treated HT-29 cells, which was suppressed by coincubation with antioxidants (vitamin E plus vitamin C). However, the antioxidants did not block DIM-mediated down-regulation of the class I HDACs or the induction of γH2AX and p21 (Supplemental Fig. 2C), indicating that ROS production was not a critical mechanism in these cells.
The HDAC inhibitors act by inhibiting the catalytic activity of HDAC enzymes. These agents are often broad spectrum inhibitors that target many of the class I, II and IV HDAC isoforms (for example, vorionostat, panobinostat, belinostat, etc). In contrast, DIM selectively induces protein degradation of the class I HDACs. Very few agents have ever been found to possess this type of activity, i.e., inducing HDAC degradation. While valproic acid was shown to induce HDAC2 degradation (25), tumor necrosis factor-α was reported to deplete HDAC1 (41). However, these agents only cause the degradation of a single HDAC enzyme but not the rest of class I HDACs. In comparison, DIM selectively induces the degradation of all class I HDACs. Class I HDACs are often overexpressed in various types of cancers and the elevation of these proteins correlates with a poor prognosis. On the other hand, the expression of the class II HDACs (HDAC4, 5, 6, 7, and 10) is associated with a better prognosis (14). This suggests that class II HDACs should be avoided in therapy targeting the HDAC enzymes. By selectively targeting class I HDACs for degradation, DIM offers an advantage over the pan-HDAC inhibitors in cancer treatment.
Histone deacetylases are components of high molecular weight multisubunit complexes of co-repressor proteins that are recruited by transcription factors to the promoters to regulate gene expression. DIM-induced degradation of the class I HDACs will result in a disruption of the protein-protein interactions among the HDACs and other proteins in these co-repressor complexes. Such effects could not be achieved by HDAC inhibitors, which may not significantly affect the protein interactions. Because of this, DIM may produce different anti-tumor activities from those of the HDAC inhibitors.
Supplementary Material
Acknowledgements
We thank Jodie Haring for providing help with real-time PCR analysis and Dr. Ed Seto (H. Lee Moffitt Cancer Center) for providing the HDAC1 and HDAC3 expression vectors.
This research was supported by NIH Grants CA130062 (Bin Guo) and RR015566.
References
- 1.Clayton AL, Hazzalin CA, Mahadevan LC. Enhanced histone acetylation and transcription: a dynamic perspective. Mol Cell. 2006;23(3):289–96. doi: 10.1016/j.molcel.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 2.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 3.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10(1):32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weichert W, Roske A, Gekeler V, et al. Association of patterns of class I histone deacetylase expression with patient prognosis in gastric cancer: a retrospective analysis. Lancet Oncol. 2008;9(2):139–48. doi: 10.1016/S1470-2045(08)70004-4. [DOI] [PubMed] [Google Scholar]
- 5.Weichert W, Roske A, Gekeler V, et al. Histone deacetylases 1, 2 and 3 are highly expressed in prostate cancer and HDAC2 expression is associated with shorter PSA relapse time after radical prostatectomy. Br J Cancer. 2008;98(3):604–10. doi: 10.1038/sj.bjc.6604199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weichert W, Roske A, Niesporek S, et al. Class I histone deacetylase expression has independent prognostic impact in human colorectal cancer: specific role of class I histone deacetylases in vitro and in vivo. Clin Cancer Res. 2008;14(6):1669–77. doi: 10.1158/1078-0432.CCR-07-0990. [DOI] [PubMed] [Google Scholar]
- 7.Marks P, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK. Histone deacetylases and cancer: causes and therapies. Nat Rev Cancer. 2001;1(3):194–202. doi: 10.1038/35106079. [DOI] [PubMed] [Google Scholar]
- 8.Johnstone RW, Licht JD. Histone deacetylase inhibitors in cancer therapy: is transcription the primary target? Cancer Cell. 2003;4(1):13–8. doi: 10.1016/s1535-6108(03)00165-x. [DOI] [PubMed] [Google Scholar]
- 9.Marks PA, Richon VM, Rifkind RA. Histone deacetylase inhibitors: inducers of differentiation or apoptosis of transformed cells. J Natl Cancer Inst. 2000;92(15):1210–6. doi: 10.1093/jnci/92.15.1210. [DOI] [PubMed] [Google Scholar]
- 10.Grant S, Easley C, Kirkpatrick P. Vorinostat. Nat Rev Drug Discov. 2007;6(1):21–2. doi: 10.1038/nrd2227. [DOI] [PubMed] [Google Scholar]
- 11.Garber K. HDAC inhibitors overcome first hurdle. Nat Biotechnol. 2007;25(1):17–9. doi: 10.1038/nbt0107-17. [DOI] [PubMed] [Google Scholar]
- 12.Paris M, Porcelloni M, Binaschi M, Fattori D. Histone deacetylase inhibitors: from bench to clinic. J Med Chem. 2008;51(6):1505–29. doi: 10.1021/jm7011408. [DOI] [PubMed] [Google Scholar]
- 13.Dokmanovic M, Clarke C, Marks PA. Histone deacetylase inhibitors: overview and perspectives. Mol Cancer Res. 2007;5(10):981–9. doi: 10.1158/1541-7786.MCR-07-0324. [DOI] [PubMed] [Google Scholar]
- 14.Balasubramanian S, Verner E, Buggy JJ. Isoform-specific histone deacetylase inhibitors: The next step? Cancer Lett. 2009;280(2):211–21. doi: 10.1016/j.canlet.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 15.Aggarwal BB, Ichikawa H. Molecular targets and anticancer potential of indole-3-carbinol and its derivatives. Cell Cycle. 2005;4(9):1201–15. doi: 10.4161/cc.4.9.1993. [DOI] [PubMed] [Google Scholar]
- 16.Bhuiyan MM, Li Y, Banerjee S, et al. Down-regulation of androgen receptor by 3,3′-diindolylmethane contributes to inhibition of cell proliferation and induction of apoptosis in both hormone-sensitive LNCaP and insensitive C4-2B prostate cancer cells. Cancer Res. 2006;66(20):10064–72. doi: 10.1158/0008-5472.CAN-06-2011. [DOI] [PubMed] [Google Scholar]
- 17.Kong D, Banerjee S, Huang W, et al. Mammalian target of rapamycin repression by 3,3′-diindolylmethane inhibits invasion and angiogenesis in platelet-derived growth factor-D-overexpressing PC3 cells. Cancer Res. 2008;68(6):1927–34. doi: 10.1158/0008-5472.CAN-07-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gong Y, Sohn H, Xue L, Firestone GL, Bjeldanes LF. 3,3′-Diindolylmethane is a novel mitochondrial H(+)-ATP synthase inhibitor that can induce p21(Cip1/Waf1) expression by induction of oxidative stress in human breast cancer cells. Cancer Res. 2006;66(9):4880–7. doi: 10.1158/0008-5472.CAN-05-4162. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y, Xu J, Jhala N, et al. Fas-mediated apoptosis in cholangiocarcinoma cells is enhanced by 3,3′-diindolylmethane through inhibition of AKT signaling and FLICE-like inhibitory protein. Am J Pathol. 2006;169(5):1833–42. doi: 10.2353/ajpath.2006.060234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rahman KM, Ali S, Aboukameel A, et al. Inactivation of NF-kappaB by 3,3′-diindolylmethane contributes to increased apoptosis induced by chemotherapeutic agent in breast cancer cells. Mol Cancer Ther. 2007;6(10):2757–65. doi: 10.1158/1535-7163.MCT-07-0336. [DOI] [PubMed] [Google Scholar]
- 21.Rahman KW, Li Y, Wang Z, Sarkar SH, Sarkar FH. Gene expression profiling revealed survivin as a target of 3,3′-diindolylmethane-induced cell growth inhibition and apoptosis in breast cancer cells. Cancer Res. 2006;66(9):4952–60. doi: 10.1158/0008-5472.CAN-05-3918. [DOI] [PubMed] [Google Scholar]
- 22.Kong D, Li Y, Wang Z, Banerjee S, Sarkar FH. Inhibition of angiogenesis and invasion by 3,3′-diindolylmethane is mediated by the nuclear factor-kappaB downstream target genes MMP-9 and uPA that regulated bioavailability of vascular endothelial growth factor in prostate cancer. Cancer Res. 2007;67(7):3310–9. doi: 10.1158/0008-5472.CAN-06-4277. [DOI] [PubMed] [Google Scholar]
- 23.Bhatnagar N, Li X, Chen Y, Zhou X, Garrett SH, Guo B. 3,3′-diindolylmethane enhances the efficacy of butyrate in colon cancer prevention through down-regulation of survivin. Cancer Prev Res (Phila Pa) 2009;2(6):581–9. doi: 10.1158/1940-6207.CAPR-08-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderton MJ, Manson MM, Verschoyle R, et al. Physiological modeling of formulated and crystalline 3,3′-diindolylmethane pharmacokinetics following oral administration in mice. Drug Metab Dispos. 2004;32(6):632–8. doi: 10.1124/dmd.32.6.632. [DOI] [PubMed] [Google Scholar]
- 25.Kramer OH, Zhu P, Ostendorff HP, et al. The histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2. Embo J. 2003;22(13):3411–20. doi: 10.1093/emboj/cdg315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fotheringham S, Epping MT, Stimson L, et al. Genome-wide loss-of-function screen reveals an important role for the proteasome in HDAC inhibitor-induced apoptosis. Cancer Cell. 2009;15(1):57–66. doi: 10.1016/j.ccr.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 27.Wilson AJ, Byun DS, Popova N, et al. Histone deacetylase 3 (HDAC3) and other class I HDACs regulate colon cell maturation and p21 expression and are deregulated in human colon cancer. J Biol Chem. 2006;281(19):13548–58. doi: 10.1074/jbc.M510023200. [DOI] [PubMed] [Google Scholar]
- 28.Archer SY, Meng S, Shei A, Hodin RA. p21(WAF1) is required for butyrate-mediated growth inhibition of human colon cancer cells. Proc Natl Acad Sci U S A. 1998;95(12):6791–6. doi: 10.1073/pnas.95.12.6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chun SG, Zhou W, Yee NS. Combined targeting of histone deacetylases and hedgehog signaling enhances cytoxicity in pancreatic cancer. Cancer Biol Ther. 2009;8(14):1328–39. doi: 10.4161/cbt.8.14.8633. [DOI] [PubMed] [Google Scholar]
- 30.Medema RH, Klompmaker R, Smits VA, Rijksen G. p21waf1 can block cells at two points in the cell cycle, but does not interfere with processive DNA-replication or stress-activated kinases. Oncogene. 1998;16(4):431–41. doi: 10.1038/sj.onc.1201558. [DOI] [PubMed] [Google Scholar]
- 31.Niculescu AB, 3rd, Chen X, Smeets M, Hengst L, Prives C, Reed SI. Effects of p21(Cip1/Waf1) at both the G1/S and the G2/M cell cycle transitions: pRb is a critical determinant in blocking DNA replication and in preventing endoreduplication. Mol Cell Biol. 1998;18(1):629–43. doi: 10.1128/mcb.18.1.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee J, Kim JA, Barbier V, Fotedar A, Fotedar R. DNA damage triggers p21WAF1-dependent Emi1 down-regulation that maintains G2 arrest. Mol Biol Cell. 2009;20(7):1891–902. doi: 10.1091/mbc.E08-08-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sgambato A, Cittadini A, Faraglia B, Weinstein IB. Multiple functions of p27(Kip1) and its alterations in tumor cells: a review. J Cell Physiol. 2000;183(1):18–27. doi: 10.1002/(SICI)1097-4652(200004)183:1<18::AID-JCP3>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 34.Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene. 2007;26(37):5541–52. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- 35.Bhaskara S, Chyla BJ, Amann JM, et al. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol Cell. 2008;30(1):61–72. doi: 10.1016/j.molcel.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fernandez-Capetillo O, Nussenzweig A. Linking histone deacetylation with the repair of DNA breaks. Proc Natl Acad Sci U S A. 2004;101(6):1427–8. doi: 10.1073/pnas.0307342101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ziv Y, Bielopolski D, Galanty Y, et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8(8):870–6. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 38.Wei MC, Lindsten T, Mootha VK, et al. tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c. Genes Dev. 2000;14(16):2060–71. [PMC free article] [PubMed] [Google Scholar]
- 39.Upreti M, Chu R, Galitovskaya E, Smart SK, Chambers TC. Key role for Bak activation and Bak-Bax interaction in the apoptotic response to vinblastine. Mol Cancer Ther. 2008;7(7):2224–32. doi: 10.1158/1535-7163.MCT-07-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarkar FH, Li Y, Wang Z, Kong D. Cellular signaling perturbation by natural products. Cell Signal. 2009;21(11):1541–7. doi: 10.1016/j.cellsig.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vashisht Gopal YN, Arora TS, Van Dyke MW. Tumour necrosis factor-alpha depletes histone deacetylase 1 protein through IKK2. EMBO Rep. 2006;7(3):291–6. doi: 10.1038/sj.embor.7400613. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.