Abstract
Renal disorders account for a substantial fraction of the budget for health care in many countries. Proteinuria is a frequent manifestation in afflicted patients, but the origin of the proteins varies based on the nature of the disorder. The emerging field of urinary proteomics has the potential to replace kidney biopsy as the diagnostic procedure of choice for patients with some glomerular forms of renal disease. To fully realize this potential, it is vital to understand the basis for the urinary excretion of protein in physiological and pathological conditions. In this review, we discuss the structure of the nephron, the functional unit of the kidney, and the process by which proteins/peptides enter the urine. We discuss several aspects of proteinuria that impact the proteomic analysis of urine of patients with renal diseases.
Keywords: Glomerular Filtration, Nephron, Proteinuria, Proteomics, Renal Tubule
Twenty-four hundred years ago, Hippocrates noted the association between bubbles on the surface of voided urine and kidney disease. This foamy appearance is due to proteinuria, an abnormality oftentimes discovered during routine evaluations in the primary-care setting. The clinical significance of this finding varies widely. Some individuals will be shown to have a benign cause, such as fever, intense activity, exercise, orthostatic proteinuria, or acute illness. Alternatively, serious conditions include a host of ailments intrinsic to the kidney (glomerulonephritis, tubular disorders, interstitial renal disease, and hypertensive renal damage) and various extra-renal disorders (plasma cell dyscrasia, inflammation of the urinary tract, and uroepithelial tumors). To ensure an accurate and timely diagnosis, a knowledgeable approach to the evaluation of proteinuria is critical.
Structure of the nephron
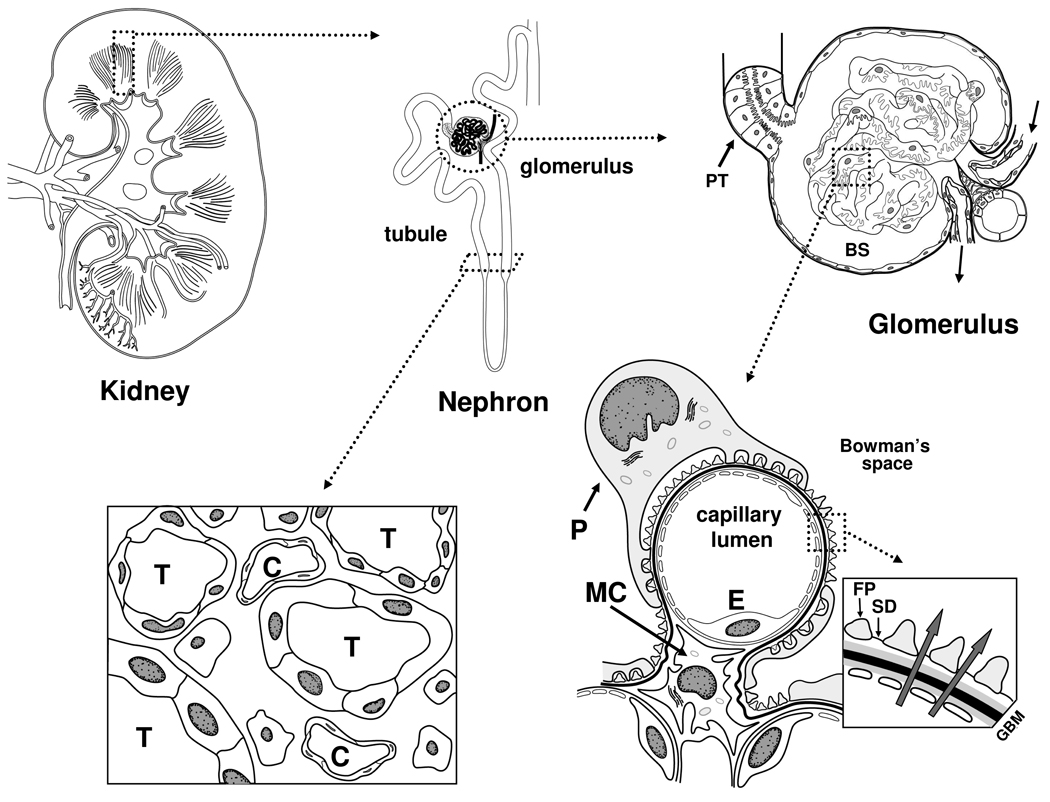
The functional unit of the kidney is the nephron and each normal human kidney contains about 1 × 106 such units. Its essential components are the glomerulus and the tubule (Figure 1). The glomerulus is the site of formation of the primary urine. It is composed of a capillary network lined by a thin layer of endothelial cells separated from overlying epithelial cells (podocytes) by a basement membrane. Bowman’s space is covered by epithelial cells and is the collection site for the primary urine. Upon exiting this space, the urine enters the tubule where its composition is drastically altered before leaving the kidney. The tubule can be divided into several functional segments that differ in their capacities to reabsorb solutes, proteins, and water and secrete various compounds [1]. These structures are interspaced in the interstitium with its small vessels and extracellular matrix.
Figure 1.
Kidney and its structural and functional components. Each human kidney (left top) contains ~1,000,000 functional units or nephrons (middle, top) that span the two regions of the kidney: cortex and medulla. Each nephron is composed of a filtration structure (glomerulus) and a downstream tubule comprised of 11 functional segments. An afferent arteriole delivers blood to the encapsulated glomerulus (right, top) where it enters the capillary network, undergoes ultrafiltration and its residual volume exits by the efferent arteriole to return to the systemic circulation. The ultrafiltrate enters the proximal tubule (PT) as the primary urine and is progressively modified as it flows through the remaining segments of the tubule. Glomeruli are located in the cortex, while the tubular portions of the nephrons span the cortex and medulla. This structural feature creates the necessary concentration gradients for extraction of salts, water and various compounds from the urine in the tubular lumens. In the cross-section of the deep cortex (left, bottom), capillaries (C) and tubuli (T) are depicted, as well as cells in the surrounding tissue, the interstitium. A cross-section of a capillary loop in the glomerulus (right, bottom) reveals the structures and resident cell types responsible for formation of the primary urine: endothelial cell (E) line the capillaries and contain openings (fenestrae) that permit water, salts, and small proteins and low-molecular-weight compounds to filter across the glomerular basement membrane (GBM); arrows in the enlarged inset on the right-hand side depict the flow of ultrafiltrate). These substances then pass through slit diaphragms (SD) that interconnect the interdigitating foot processes (FP), or pedicles, of epithelial cells (podocytes, P) that overlie the capillaries. Upon this entry into Bowman’s space (BS), this fluid is termed primary urine. The mesangium is the centrolobular region of the glomerular tuft that helps to maintain patency of the capillary loops. Mesangial cells (MC) regulate glomerular blood pressure, produce cytokines/chemokines and radical oxygen species, and secrete extracellular matrix proteins necessary for the structural integrity of the glomerulus. Larger circulating substances, such as immune complexes, can more easily enter the mesangium than Bowman’s space because the GBM and slit diaphragms are not present in this area. These processes may culminate in glomerular fibrosis with loss of filtration function of the nephron. In addition, release of various chemokines/cytokines may induce interstitial inflammation and scarring, further damaging the integrity of the nephron to compromise its function.
The glomerular capillary endothelial cells have many fenestrae with diameters ~60–90 nm [2] to allow high permeability to water and small solutes out of the glomerulary capillary bed [2,3]. The diameter of albumin, the most abundant protein in plasma, is ~3.6 nm, but only a tiny fraction of this protein normally reaches Bowman’s space, suggesting that the size of the fenestrae does not substantially contribute to the permselectivity of the glomerular barrier. A high density of fibers in the fenestrae, possibly consisting of negatively charged proteoglycans, apparently influences the permselectivity of the capillary wall [2].
The glomerular basement membrane is composed of an intertwined network of extracellular matrix proteins consisting of collagen type IV, laminin, and nidogen/entactin, with attached proteoglycans such as agrin and perlecan with heparin sulfate chains, as well as glycoproteins. These chains contribute to the selective properties of the filtration barrier [4,5]. Although its role in renal permselectivity has been debated, the glomerular basement membrane is an important part of filtration barrier, as it restricts the flux of fluid. The thickness of basement membrane in the glomerulus (300–350 nm) is twice that of other vascular beds [6]. Abnormalities in the glomerular basement membrane give rise to several pathological conditions characterized by proteinuria and microscopic hematuria [7].
Podocytes on the urinary side of the glomerular basement membrane are terminally differentiated epithelial cells that participate in several glomerular functions, including maintenance of the filtration barrier, regulation of glomerular filtration, support of the capillary tuft, turnover of components of the glomerular basement membrane, production and secretion of vascular endothelial growth factor necessary for the integrity of glomerular endothelial cells, and immunological functions [8–10]. These cells are attached to the underlying glomerular basement membrane via integrins and dystroglycans [11,12]. They are connected to neighboring podocytes by highly specialized gap junctions called slit membranes with ~40 nm-diameter pore-like structures [13]. The glomerular basement membrane and the slit diaphragm constitute the primary barrier for filtration of blood into the urinary compartment, by virtue of their charge and physical characteristics [14–16].
The mesangium is the central region of the glomerulus, comprised of specialized cells with surrounding extracellular matrix that supports the structure and helps to maintain patency of the glomerular capillary bed [17,18]. The only barrier to cross for circulating proteins to reach the mesangium is the endothelial fenestrae. The mesangial matrix consists of collagens, laminin, fibronectin, and proteoglycans with heparin sulfate and chondroitin sulfate chains [19,20]. In some renal diseases, immune complexes from the circulation characteristically attach to mesangial cells. The consequence is cellular activation and proliferation and secretion of cytokines, complement components, and extracellular matrix proteins [19,21]. Activated mesangial cells may also release several growth factors, cytokines, and reactive oxygen species that damage the glomerular basement membrane and podocytes, leading to proteinuria [22–24].
Formation of urine
The kidneys receive about 20–25% of the cardiac output and filtration occurs in the glomerular capillary bed. Glomerular filtration rate (GFR) is the product of [net filtration pressure × hydraulic permeability × filtration area in the glomerular capillaries]. The net ultrafiltration pressure is the difference between the hydrostatic and the osmotic pressures across the capillary loop. In most persons, about 20% of the fluid portion of the blood crosses the filtration barrier to enter Bowman’s space. The cellular elements remain in the capillary lumen to re-enter the systemic circulation. This primary urine has a volume of about 180 liters per day (corresponding to a GFR of 125 ml/min) that contains about 1.5 kg sodium chloride. Proteins with a molecular weight less than 20 kDa easily cross the filtration barrier. As the molecular mass of a protein increases, the fraction that is filtered progressively decreases such that compounds of 60–70 kDa are largely retained in the capillary lumen. The electrical charge of the solutes also influences filtration; negatively charged proteins enter Bowman’s space in amounts far smaller than would be predicted by size criteria.
The downstream tubular portions of the nephron reclaim ~98–99% of the filtered salt and water, and the vast majority of the small proteins in the primary urine. The segments in the tubule vary in their roles in this process, differentially reabsorbing water and solutes [25,26]. More distal segments of the tubule fine-tune the excretion of water, urea, calcium, potassium, and other solutes through reclamation or secretory processes.
Genesis of proteinuria
In the final voided urine, the excreted protein does not normally exceed 150 mg per day, of which albumin accounts for about 20 mg. About half of the protein in the urine of normal individuals is derived from the tubules downstream from the glomerulus or from sources outside of the nephron.
Proteinuria due to glomerular injury
Proteins enter the urinary space through several physiological and pathophysiological mechanisms (Table 1; for further reading, see [27]). Proteins may exit the capillary circulation by crossing the three-layer filtration barrier comprised of the fenestrated endothelial cells, glomerular basement membrane and slit diaphragms of the podocytes. Small proteins (<20 kDa) are freely filtered but are readily reabsorbed downstream by proximal tubular cells. If the podocytes are injured with resultant loss of the organized structure of the actin-rich cytoskeleton [28], undergo changes in the slit-diaphragm proteins, or lose their negative charge on the cell surface, or if the composition and organization of the glomerular basement membrane is altered, they exhibit a characteristic change in appearance: effacement of their foot processes [10]. This alteration is a simplification of the inter-digitating foot processes due to retraction, widening and shortening of the processes of the podocytes. This change is energy-dependent and the result is a flattened elongated appearance of the podocytes and fewer slit diaphragms [10] (Figure 2). In that setting, substantial amounts of moderate-size proteins (e.g., 65-kDa albumin) may enter the primary urine [27]. In some renal diseases, even larger proteins (such as immunoglobulins) leak into the urinary space. The amount of protein excreted by patients with glomerular diseases spans a wide range, from 100 mg to ~20 g per day, depending on the specific disorder. The lower amount is within the normal range, while excretion of greater than 3 g/24h/1.73m2 often leads to nephrotic syndrome, manifested as edema, hypoproteinemia (especially hypoalbuminemia), hypercholesterolemia, and lipiduria.
Table 1.
Classification of proteinuria and main causes.
| Type | Category | Specific diagnosis |
|---|---|---|
| Glomerular | Primary glomerulonephritis | Minimal-change disease Membranous glomerulonephritis Focal segmental glomerulosclerosis Membranoproliferative glomerulonephritis IgA nephropathy |
| Glomerular injury secondary to systemic disorder |
Diabetes mellitus | |
| Systemic lupus erythematosus Amyloidosis Infection (e.g., HIV, hepatitis B and C, streptococcus, syphilis, malaria and endocarditis) Lymphoma Hypertensive nephrosclerosis |
||
| Drug-induced injury | Heroin Non-steroidal anti-inflammatory drugs Gold components Antibiotics Lithium Heavy metals |
|
| Tubular | Tubulointerstitial disease | Uric acid nephropathy Interstitial nephritis Fanconi syndrome Sickle cell disease |
| Drug-induced | Non-steroidal anti-inflammatory drugs Antibiotics Heavy metals |
|
| Overflow | Hemoglobinuria, Myoglobinuria Multiple myeloma Amyloidosis Leukemia Myelodysplastic syndromes |
|
|
Tissue proteinuria |
Acute inflammation of urinary tract | |
| Uroepithelial tumors | ||
| Renal allograft | Cell-mediated rejection Antibody-mediated rejection |
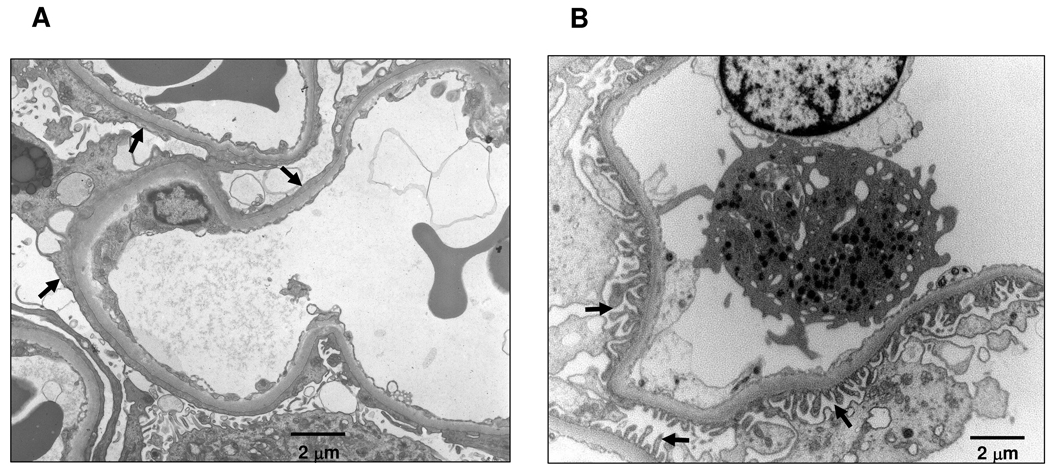
Figure 2.
Electron micrographs of glomerular capillary loops. (A) Glomerulus from a patient with nephrotic-range proteinuria. Black arrows indicate areas of effacement of the foot processes of the epithelial podocyte. Effacement is associated with a substantial decrease in the number of slit diaphragms. (B) Normal glomerulus. Black arrows indicate intact foot processes of the epithelial podocyte overlying the glomerular basement membrane. Slit diaphragms are located between the foot processes. Bars indicate 2 micrometers.
Injury to podocytes, accompanied by foot-process effacement, has been grouped into three classifications: congenital, hereditary and acquired [10]. An example of the first is development of maternal antibodies to fetal neutral endopeptidases that are absent in the mother. Hereditary causes often include genetically determined alterations in podocyte-specific proteins that are often components of the slit diaphragm or the cytoskeleton. Acquired injuries may be subdivided into immune causes (injury due to direct effects of immunoglobulin or immune complexes on the podocytes or indirect effects mediated by alterations in T cells) and non-immune causes (infection with HIV virus, or metabolic or local hypertension arising from diabetes-mediated damage in the mesangium) [10]. Some immune-complex-mediated renal diseases with the initial injury predominantly in the mesangium may damage podocytes through the local release of cytokines or oxidants [29–35].
In a few instances, proteinuria occurs in the absence of effacement of the foot processes of the podocytes [36]. Clinical examples are preeclampsia of pregnancy [37], glomerulonephritis with severe endothelial injury, and some types of familial nephropathy with nephrotic syndrome [38].
Proteinuria due to renal tubular mechanisms
The renal tubules may account for proteinuria by three mechanisms. The first is the fact that excessive filtration may exceed the tubular capacity for reabsorption or degradation. This situation occurs most commonly with plasma cell dyscrasias such as multiple myeloma, amyloidosis, and lymphomas that are associated with monoclonal production of immunoglobulin light chains. These proteins are not only poorly absorbed by tubular cells, but also exert a toxic effect on these cells that leads to scarring and decreased renal clearance function [30–32].
The second mechanism of tubular proteinuria arises from significant injury in the renal interstitium, the tissue surrounding the glomeruli and tubules that includes peritubular capillary and lymphatic networks (Figure 1). Damaged proximal tubular cells may shed brush-border proteins with loss of enzymes, ion channels and transporters, transmembrane glycoproteins and intracellular enzymes. Some proteins enter the urine as a consequence of sloughing of whole tubular epithelial cells [33,34]. This pattern of pathology is common even in diseases in which the inciting event starts in the glomeruli [39]. Inflammation is the most common process to induce interstitial damage and it proceeds in a highly regulated fashion [40,41]. In addition, leukocytes infiltrating the interstitium as a component of the inflammation secrete chemical mediators (e.g., cytokines or chemokines) that alter vascular permeability. These cells may also release other proteins (e.g., IgA) not normally detected in the urine. The effect is often augmentation of the interstitial injury and the outcome is either resolution with return of the histology and function to normal or scarring with fibrosis, tubular atrophy and loss of function [39–41]. Increased interstitial volume due to fibrosis correlates with the degree of impairment of renal function and is a useful histological marker to assess clinical prognosis [40].
A third mechanism for tubular proteinuria is the excretion of exosomes [42]. Their origin may include all of the epithelial cells in the nephron. Whether markers associated with exosomes will be clinically useful in patients with renal disease is not clear, although a recent study of phosphoproteins of urinary exosomes showed absence of the sodium-potassium-chloride co-transporter 2 from urine samples of patients with a genetic renal disease, Bartter syndrome type I [43]. Furthermore, two studies indicated that exosomal proteins may serve as markers of acute kidney injury [44,45].
The quantity of proteins of a tubular source excreted by patients with injured tubules generally does not exceed 1,000 mg per day. As a means to distinguish tubular from glomerular sources of proteinuria, the amount of low-molecular-weight β2-microglobulin relative to albumin is measured [35]. A ratio substantially above the normal value of 0.1 mg/mg characterizes tubular proteinuria.
Excretion of proteins originating outside of the nephron
Excreted proteins may originate at locations outside of the kidney. Small- or middle-molecular-weight proteins entering the circulating blood from other organs may pass through the normal filtration barrier into the proximal tubules in quantities sufficient to escape total reclamation. These proteins may have been secreted into the plasma by any of a wide variety of cells, or be contained within exosomes. Furthermore, proteins may also enter the urine at points downstream from the nephron. These sources include the uroepithelium of the ureter or bladder (e.g., secretory IgA) and, in men, secretions from the prostate gland and vas deferens.
Measurement of proteinuria
The screening test for proteinuria is generally a dipstick urinalysis using a strip of dye-impregnated paper. However, this approach entails several important inherent limitations. Conventional dipsticks detect predominantly albumin in concentrations 20–300 mg/dL and, thus, may not detect concentrations of albumin commonly found in patients with microalbuminuria excreting 30–300 mg albumin per day, considering the usual daily volume of urine is 1–3 liters [27]. Even patients excreting increased amounts of other serum proteins may not test positive by a dipstick analysis that depends on the concentration of protein in the urine sample. A very dilute urine (e.g., specific gravity less than 1.004) may not register the abnormality. In addition, the dipstick is insensitive to immunoglobulins. Alternatively, an alkaline or concentrated urine sample, macroscopic hematuria (visible urinary bleeding) or the presence of some drugs (e.g., cephalosporins and iodinated radiocontrast), mucus, semen or white blood cells may cause a false-positive reading. Contaminants entering the urine during the voiding process, menstrual blood or vaginal secretions, may also lead to such a misleading result.
Dipstick proteinuria should be confirmed by a colorimetric or turbidometric assay for total protein. More precise determinations of proteinuria are derived from measurements in a 24-hour collection or a random “spot” sample to calculate the protein/creatinine ratio. About 85% of urinary creatinine is derived from the circulation as a result of filtration across the glomerular basement membrane, with about 15% originating from secretion by renal tubular cells. Thus, excretion of creatinine is used as a gauge of GFR in patients with reasonably well preserved clearance function. However, it is often difficult for individuals to collect a timed sample of urine correctly. Although the rate of protein excretion varies during the day, due to differences in posture, physical activity, intake of dietary protein and hemodynamic factors [46], several studies have found that the urinary protein/creatinine ratio in a spot urine sample closely correlated with the 24-hour excretion [46–48]. Because of the excellent correlation between these two approaches and its less cumbersome collection technique, use of a random sample has gained increasing favor in the clinic (for details on other current laboratory tests for urinary protein analysis, please see [49]). The National Kidney Foundation of the United States has concluded that a random sample suffices for the quantitative measurement of proteinuria. A normal urinary protein/creatinine ratio is less than 0.1 g/g [50,51].
Proteinuria in patients with renal disease
Most physicians interpret the importance of proteinuria in the context of GFR. The most frequently used estimate of GFR in clinical practice is creatinine clearance. The amount of creatinine excreted per day depends on the muscle mass of the individual and, to some degree, on physical activity. These factors vary with age, race and body composition. GFR declines with age starting in the fourth decade, by as much as 8 mL/min/1.73m2/decade [52]. Use of creatinine clearance becomes increasingly problematic as damage to the nephron worsens and GFR declines because the amount of creatinine entering the urine as a result of tubular secretion is proportionally greater. Some investigators have favored measurement of another compound, cystatin C, as a better measure of GFR [53]. The use of iothalamate or inulin can overcome this problem, because both are freely filtered in the glomerulus and neither is secreted to a significant degree. However, use of either compound requires an intravenous infusion and, thus, this approach is not practical for clinical purposes. To minimize the complicating factors in the use of creatinine clearance to assess renal clearance function, the National Kidney Foundation of the United States has endorsed two equations that take age, sex, and ethnicity into account:
- Cockcroft-Gault creatinine clearance [54], mL/min:
Above formula is for men; multiply by 0.85 to obtain the result for women - Modification of Diet in Renal Disease study (MDRD) estimated GFR, abbreviated version [55], mL/min/1.73m2:
- Estimated GFR = 186 × [serum creatinine concentration]−1.154 × [age]−0.203 × [0.742, if female] × [1.210, if black]
- The four-variable MDRD calculation or use of an extended equation that includes two more clinical parameters, blood urea nitrogen (BUN) and serum albumin concentrations, can be done easily at the website http://newtech.kidney.org/professionals/kdoqi/gfr_calculator.cfm.
Both formulas entail some inaccuracies at the higher range of GFR and the current recommendation is to report a specific value for estimated GFR only if the result is less than 60 mL/min/1.73m2. Evidence of kidney damage is defined as pathologic abnormalities or markers of damage, including abnormalities in blood or urine, or by imaging studies. Chronic kidney disease is defined as either evidence of kidney damage or GFR <60mL/min/1.73m2.
Nearly all (99%) normal persons excrete less than 150 mg protein per day. The major exception to this criterion applies to normal pregnancy. Due to an increase in GFR that begins early in gestation (perhaps due to hormonally mediated vasodilatation) and continues through the third trimester, the upper limit for normal increases to 300 mg per day. Overt proteinuria, an amount that is easily detectable by routine screening methods, generally ranges 300 – 500 mg per day. To satisfy the standard for chronic kidney disease, overt proteinuria should be documented on several occasions over a three-month interval. Such a persistent abnormality must be distinguished from transient proteinuria that may be detected in the normal persons with short-term losses due to exercise or a febrile illness or in diabetic patients with poor glycemic control.
Overt proteinuria usually precedes decline in GFR, particularly in patients with diseases initially damaging the filtration barrier, and is generally asymptomatic. As the glomerular injury progresses, or in patients with diseases affecting the other components of the nephron, proteinuria may worsen due to scarring of the glomerular basement membrane or damage of the renal epithelial cells. For these patients, a spot urine protein/creatinine ratio measurement is at least as reliable as a 24-hour urinary protein collection in predicting progression of renal disease [48]. Patients whose inciting injury is confined to the tubular and interstitial compartments of the kidney (e.g., nephrotoxins or vascular insufficiency) often have proteinuria of a modest degree. Assays for particular peptides (e.g., β2-microglobulin and kidney injury molecule-1) are used to confirm the tubular source of the proteinuria and they have been proposed as biomarkers for acute kidney injury (for review, see [56]).
In an effort to detect glomerular renal injury at earlier stages, investigators have turned their attention to the excretion of albumin. Microalbuminuria (excretion of 30–300 mg albumin per day or per g creatinine) may indeed be more sensitive than overt proteinuria in this regard. For example, in patients with type 1 diabetes mellitus, microalbuminuria may occur in as few as five years after the onset of insulin dependency. Compared to normoalbuminuric patients, individuals with persistent microalbuminuria have a three- to four-fold greater risk to progress to overt proteinuria and loss of clearance function [57,58]. From another perspective, among microalbuminuric patients with type 1 diabetes mellitus, 20–45% progress to overt proteinuria over the next 10 years, 30–60% remain microalbuminuric, and the rest return to normoalbuminuria [59,60]. Some investigators contend that microalbuminuria is not the first clinical sign of renal disease in patients with type 1 diabetes mellitus, but this opinion remains controversial [60].
Clinically, proteinuria is classified as "selective" when albumin constitutes a substantial majority of the urinary protein or “nonselective” when the profile of the excreted protein reflects that of the proteins in the circulation. When the urinary protein losses exceed 3 g per day, the serum albumin concentration usually decreases due the liver’s inability to synthesize sufficient albumin to compensate for the urinary losses, and edema frequently develops. Proteinuria in most adults with glomerular disease is non-selective. In contrast, in patients with orthostatic proteinuria (manifests only while the individual is upright, and usually carries a benign prognosis), the pattern is highly selective [61]. Many clinical studies support the association of worsening proteinuria with progressive damage to the nephron and loss of clearance function [62–64]. The simplest hypothesis for this observation is that increasingly severe proteinuria triggers a downstream inflammatory cascade around epithelial cells of the renal tubules, leading to interstitial injury, fibrosis, and tubular atrophy. Because albumin is an abundant polyanion in circulating blood and binds a variety of cytokines, chemokines, and lipid mediators [65–69], it is plausible that in patients with glomerular proteinuria these small molecules initiate interstitial inflammation. Furthermore, glomerular injury may add activated mediators to the filtrate or alter the balance of cytokine inhibitors and activators, leading to a critical level of activated cytokines that damage downstream tubular epithelial cells. However, after uptake of albumin, epithelial cells lining the proximal tubules release an array of cytokines and chemokines that contribute to the inflammation in the interstitial compartment. As the inflammation heals, the resultant scarring can substantially decrease glomerular filtration. Indeed, some investigators have indicated that the degree of interstitial scarring is a better marker for prognosis than glomerular scarring in patients with some forms of glomerulonephritis [70]. Studies have shown that patients excreting substantial amounts of β2-microglobulin (proximal tubular source) and IgG or albumin (glomerular source) have an unfavorable clinical course [71,72]. High concentrations of IgG or albumin may damage the podocyte cytoskeleton [73].
Diagnosis of proteinuric renal disease
Although some patients with proteinuria due to structural abnormalities of the kidney (polycystic disease, congenital dysplasia, or reflux nephropathy) may be diagnosed by non-invasive imaging techniques such as magnetic resonance or ultrasound, the gold standard for most patients with proteinuria due to glomerulus-based disorders is the renal biopsy. Using a percutaneous approach, often during localization with real-time ultrasound, a small sample of renal tissue is removed with a cutting needle. Evaluation of the tissue includes light microscopy, immunofluorescence studies and electron microscopy, and the diagnostic yield is usually much better for glomerular diseases than for other types of renal disorders. Patients with glomerular disease (glomerulonephritis, termed an “inflammation” of the glomerulus) usually have morphological alterations in the glomeruli and clinically exhibit microscopic hematuria with varying degrees of proteinuria, generally more than 1 g per day (Table 1). For patients with glomerular renal diseases, the biopsy generally provides sufficient details to allow diagnosis and to address the approach for treatment. Patients with diseases for which therapy may include use of immunomodulatory drugs for years, such as lupus nephritis, renal vasculitis or IgA nephropathy, repetitive biopsy is frequently undertaken for monitoring the histological response and adjusting treatment. Serious complications of renal biopsy due to bleeding are not uncommon [74,75]. The procedure is more difficult in young children for whom conscious sedation is required.
Proteomic analysis of urine
As can be appreciated from the above discussion, urine contains a wide variety of compounds, ranging from small-molecular-mass molecules, such as metabolites, to peptides and polypeptides and finally to macromolecular complexes, such as exosomes [76,77]. Analysis of each of these different components requires specific approaches, and, consequently, specialized fields such as metabolomics, proteomics, and peptidomics have emerged. Each approach has contributed to discipline of urinary biomarker research. Although these fields differ, many common considerations apply, being dictated by general requirements of clinically-related studies [78]. In the sections below, we review some of these aspects, with emphasis on proteomics and peptidomics.
Technical aspects of proteomics and peptidomics
Before a sample of body fluid is analyzed in a clinical laboratory for specific biomarker(s) of a disease, several aspects of the methodology must be taken into account. First, collection and storage of the sample must be tested for reliability and, if necessary, optimized. While sampling renal tissue that is directly affected by the disease process would be ideal, this procedure is invasive and quite expensive and entails well-documented risk (as discussed in greater detail in the accompanying Viewpoint in this issue). Proteins/peptides may be shed or secreted into the local circulation. Unfortunately, collection of blood from young children can be a traumatic experience for the patient and parents. But even in older children and adults, sampling of blood has significant shortfalls. While blood is a rich source of proteins and peptides, potential biomarkers present in modest quantities may escape detection because of the difficulties encountered in the process of separating them during the preparation steps or analytical assays from more abundant material of about the same charge or size. Second, the handling and preparation of the sample should be kept to a minimum. A protocol that includes immediately freezing the sample, such as serum with its abundant enzymatic activity that may degrade the biomarker of interest, is a challenging requirement in a standard clinical setting. Third, a validated platform and, if possible and applicable, even an FDA-approved method of detection should be used and performed under reproducible conditions with appropriate controls. Fourth, the results should be reported in an easy-to-understand format with appropriate reference values. All of these facets assume that a specific biomarker or panel of biomarkers was identified and characterized with suitable methods for detection. For analysis of urinary proteomic biomarkers of renal diseases, most researchers have indicated that the second-void sample of the day provides the most reproducible results [79,80]. Use of the second-void sample shortens exposure of compounds to proteases in the urine and potentially lessens the risk for reduction in the concentration or alteration of the composition of a potential biomarker. However, some prefer a first-void sample if the search is for markers of bladder cancer [81,82] due to the potentially greater amount excreted. Several recent publications summarize the experience and suggestions for sample handling and provide a detailed discussion of other considerations relevant to urinary proteome analysis in clinical settings [78,83,84]. Urine samples can be stored frozen, ideally in aliquots to prevent repeated freeze-thaw cycles [84]. The temperature should be −20°C or lower. If the sample is to be stored for more than five years, a temperature of −80°C is preferable [84]. Preparation of urine specimens for analysis is entirely dependent on the particular markers being assayed and on the methodology used. In principle, proteomic techniques usually require two-step protocols, separation and detection, that are frequently coupled with a specific method for sample preparation. The optimal clinical assay should require minimal handling of the sample, to reduce cost and risk of possible artifacts.
Proteomic techniques rely most frequently on gel or column separation, followed by an on-line or off-line detection method, such as mass spectrometry (MS) or immunodetection. Table 2 compiles the commonly used proteomic separation and detection techniques for analysis of urinary biomarkers. As noted, each technique has its advantages and disadvantages that must be considered for the clinical application. In addition, factors such as turn-around time, expense of equipment, training of laboratory personnel, reproducibility, calibration and standardization, and central laboratory vs. portable instrument should be considered. For biomarker identification, parameters such as reproducibility of sample preparation and sample analysis must be considered as well as statistical analysis applied for identification of relevant biomarkers [76,78].
Table 2.
Examples of proteomic separation and detection techniques for analysis of biomarkers.
| Technique | Separation | Detection | Comments relevant to potential use in a clinical laboratory (references) |
|---|---|---|---|
| 2-D PAGE | Isoelectric focusing in 1st dimension, followed by SDS-PAGE in 2nd dimension |
Western blotting or MS | High resolution, suitable for proteins 10–200 kDa Quantitative assessments and reproducibility not easy to perform |
| Post-translational modifications result in multiple spots for each protein ([136,137]) |
|||
| SELDI-MS | Differential retention on surface plate |
TOF MS | Selective technique with high throughput |
| Reproducibility and ion identification not easy to achieve ([158,159]) |
|||
| LC-MS and MS/MS |
HPLC (usually reverse- phase column) |
MS or MS/MS | Performance dependent on LC column |
| Low throughput | |||
| Can provide amino acid sequence in MS/MS ([160]) | |||
| MRM/SID LC- MS/MS |
LC | MS/MS (transition states for specific peptide ions monitored) |
Quantitative and specific, can assess many analytes in each sample |
| For quantitation, SID is necessary ([161]) | |||
| CE-MS | CE | TOF, FT ICR, or Orbitrap MS | High resolution, applicable to peptides and small proteins, not suitable for proteins >10 kDa |
| FT and Orbitrap can analyze post-translational modifications ([75,131,132,135]) |
Validation of the identification of the peaks shown by MS is vital in establishing confidence in the process that defined a biomarker of interest. It is also important to ascertain additional data that may be practical for expanding the clinical usefulness of the biomarker. More detailed information about the composition of the biomarker may clarify the pathophysiology of the disease process and lead to better treatment. Furthermore, such precise particulars may guide development of an independent, rapid, highly accurate and less expensive assay, such as one using an isotope-labeled standard or antibody. One approach to characterize a peptide biomarker is to establish its sequence.
Protein sequencing takes advantage of MS-based dissociation of the target ion. Commonly, these ions are fragmented by collision-induced dissociation (CID) with atoms of an inert gas [85,86]. After dissociation, the masses of the produced fragments are recorded (MS/MS scan) and, by subtracting consecutive signals, adjacent residues are determined. Repetition of this process can determine the amino acid sequence of the entire peptide. CID has some important inherent limitations: (1) fragmentation often fails with peptides with a high content of Pro, Arg, and/or Lys, (2) post-translational modifications (PTMs) complicate interpretation of spectra, and (3) longer peptides (≥15–18 amino acids) yield poor information. However, with use of an ion trap MS, CID can produce several spectra per second for many types of peptides and, thus, can sequence thousands of peptides in a day. To enhance the effectiveness of CID for larger peptides and proteins, additional steps prior to MS analysis are included, such as proteolytic digests (using site-specific proteases such as trypsin or chymotrypsin) sometimes combined with removal of PTMs (e.g., deglycosylation of glycoproteins).
In 1998, a new method for peptide/protein ion fragmentation was introduced, electron capture dissociation (ECD), using a Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometer [87]. In contrast to CID, ECD does not preferentially cleave PTMs from modified peptide ions and more randomly cleaves the backbone bonds of large peptides and even whole protein ions. This feature enables, in many instances, direct characterization of whole proteins (top-down approach) [88–106].
As the ECD fragmentation is limited to FT-ICR MS instruments, investigators have sought to develop a similarly effective fragmentation technique using a more commonly available instrument. These efforts produced a new ion dissociation method, electron transfer dissociation (ETD), that can be performed using a bench-top mass spectrometer with an ion trap [85,86,107–110]. This approach uses negatively charged ions to deliver the electron. With an appropriate electron-transfer reagent, the transferred electron induces fragmentation of the peptide backbone. Both ECD and ETD fragment cations of peptides and proteins in a relatively sequence-independent fashion, can be used for identification of PTMs, and can be performed on a chromatographic time-scale.
Application of FT-ICR MS has permitted analysis of various PTMs, including the aberrancies in the O-glycosylation of IgA1 hinge-region glycopeptides [111,112] that play a central role in the pathogenesis of IgA nephropathy, as discussed below. FT-ICR MS enables heterogeneity profiling of glycopeptides as well as detailed sequence-specific studies due to the availability of MS/MS techniques with FT-ICR MS (fragmentation using CID, ECD, ETD, and infrared-multiphoton dissociation, IRMPD). These approaches facilitate identification of the sites of attachment of glycans on IgA1 and are applicable to other types of PTMs on other proteins in various organisms [85,99,102,113–119].
Clinical examples
It is not necessary that patients exhibit clinical proteinuria or even have renal disease before urinary proteomics may be considered as a potentially useful tool in diagnosis. As has been shown for patients with coronary disease [120] or urological cancers [121,122], urinary proteomics can detect biomarkers of disease in the absence of abnormal proteinuria.
Several centers have addressed the question of whether urinary proteomics is helpful in the identification of the type of renal disease or in the assessment of the prognosis. While several challenges must be addressed before for urinary proteomics can be adapted for the clinic (please see the accompanying Viewpoint in this issue) preliminary findings suggest that this approach has the potential to be developed into diagnostic assays that would be less expensive and safer than the current methods. One example with substantial clinical impact is patients with IgA nephropathy, the most common form of glomerulonephritis worldwide [123]. This disease is a leading cause of end-stage renal failure [124] and apparently arises due to mesangial deposition of circulating immune complexes [125,126] containing galactose-deficient IgA1 [127,128]. While the serum level of this aberrant immunoglobulin [129] and the urinary excretion of IgA-containing immune complexes [130,131] are elevated in patients with IgA nephropathy [129], renal biopsy is still necessary for an accepted diagnosis [76,78,130–143]. A recent study used CE-MS to identify a panel of polypeptide biomarkers that distinguished patients with IgA nephropathy from those with other renal disorders with a 90% sensitivity and 90% specificity [144]. The origin of the fragments distinctive for IgA nephropathy remains unknown, but unique proteases in inflamed glomeruli or in the urine may generate disease-specific polypeptides [145]. Similar evidence for apparently disease-specific urinary biomarkers has been reported for patients with other common renal diseases, including membranous glomerulonephritis, focal segmental glomerulosclerosis, systemic lupus erythematosus, minimal-change disease and diabetic nephropathy [135,146], using SELDI-TOF MS [78,132,135–137,141,144–149]. Otu et al. found a urinary 12-peak protein signature in Pima Indians with type 2 diabetes mellitus and normoalbuminuria that predicted development of diabetic nephropathy 10 years later [150]. If validated, these findings will lead to a more focused approach to treatment, and hopefully a reduction in the prevalence of diabetic nephropathy. For patients with other types of renal disorders, the diagnosis requires careful refinement before proceeding with investigations of the urinary proteome. For example, a cohort of patients with the clinical diagnosis (without a renal biopsy) of hypertensive renal disease may have a spectrum of renal pathology that differs greatly from nephrosclerosis due to primary hypertension. Indeed, in a study of patients presenting for transplantation for renal failure attributed hypertensive renal disease, many patients had clinical evidence of proteinuria suggestive of undiagnosed glomerular disease [151]. Embarking on a search for disease-specific urinary biomarker (pattern) using proteomic methods in this setting would be futile, or worse, misleading.
Urinary proteomics may nevertheless prove to be valuable for the clinic because the currently applied diagnostic methods are expensive and entail risks for morbidity, and diagnosis in the earliest stages of renal injury provides an opportunity to therapeutically intervene for the best outcome. It is necessary to validate findings in large prospective multi-center studies that permit assessment of the specificity and sensitivities of the different approaches. Irrespective of the technology, certain rules and approaches should be followed to maximize the clinical impact of each study [78]. Along these lines, a publicly accessible database of the urinary peptidome was published in 2008, providing a new tool for studies focused on urinary biomarkers [135,152–157].
Summary
Renal diseases affect a substantial portion of the world’s population and account for a significant fraction of the costs of health care. Afflicted individuals manifest a wide spectrum of clinical and laboratory features. For patients with the largest subset of renal diseases, i.e. glomerular disorders, renal biopsy is the current standard for diagnosis, and for directing and monitoring therapy. Unfortunately, this test is invasive and expensive. These aspects frequently limit the applicability of the biopsy procedure, often denying patients an opportunity for early diagnosis and treatment before irreversible renal injury has developed. Recent advances in urinary proteomic analyses have the potential to significantly enhance the care of patients with renal disease, as well as other disorders, and to alleviate some of the financial burden on the health care system. To fully realize the potential of these emerging techniques, it is vital that clinicians and basic scientists understand the processes by which proteins are excreted in the urine. By building on this foundation, investigators will more likely minimize the pitfalls that often slow the pace of progress in medical research.
Acknowledgements
The authors thank Mr. David Fisher for his artistic help with Figure 1. BAJ, SH, and JN were supported in part by NIH grants DK078244, DK080301, DK071802, DK077279, DK061525, and DK064400, General Clinical Research Center of the University of Alabama at Birmingham (M01 RR00032), and by CCTS grant 1UL1RR025777-01.
Abbreviations
- GFR
glomerular filtration rate
- IgA
immunoglobulin A
- MDRD
modified diet in renal disease
- MRM
multiple reaction monitoring
- SID
stable isotope dilution
Footnotes
Conflict of interest statement
The authors do not have any financial/commercial conflicts of interest to declare.
References
- 1.Briggs JP, Kriz W, Schnermann JB. In: Primer on Kidney Disease. Greenberg A, editor. Philadelphia: Elsevier Saunders; 2005. pp. 2–19. [Google Scholar]
- 2.Rostgaard J, Qvortrup K. Sieve plugs in fenestrae of glomerular capillaries--site of the filtration barrier? Cells Tissues Organs. 2002;170:132–138. doi: 10.1159/000046186. [DOI] [PubMed] [Google Scholar]
- 3.Deen WM, Lazzara MJ, Myers BD. Structural determinants of glomerular permeability. Am. J. Physiol. Renal Physiol. 2001;281:F579–F596. doi: 10.1152/ajprenal.2001.281.4.F579. [DOI] [PubMed] [Google Scholar]
- 4.Hassell JR, Robey PG, Barrach HJ, Wilczek J, et al. Isolation of a heparan sulfate-containing proteoglycan from basement membrane. Proc. Natl. Acad. Sci. USA. 1980;77:4494–4498. doi: 10.1073/pnas.77.8.4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanwar YS, Linker A, Farquhar MG. Increased permeability of the glomerular basement membrane to ferritin after removal of glycosaminoglycans (heparan sulfate) by enzyme digestion. J. Cell. Biol. 1980;86:688–693. doi: 10.1083/jcb.86.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonsib SM. In: Heptinstall's Pathology of the Kidney. Jennette JC, Olson JL, Schwartz MM, Silva FG, editors. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 1–70. [Google Scholar]
- 7.Barker DF, Hostikka SL, Zhou J, Chow LT, et al. Identification of mutations in the COL4A5 collagen gene in Alport syndrome. Science. 1990;248:1224–1227. doi: 10.1126/science.2349482. [DOI] [PubMed] [Google Scholar]
- 8.Hauser PV, Collino F, Bussolati B, Camussi G. Nephrin and endothelial injury. Curr. Opin. Nephrol. Hypertens. 2009;18:3–8. doi: 10.1097/MNH.0b013e32831a4713. [DOI] [PubMed] [Google Scholar]
- 9.Mundel P, Kriz W. Structure and function of podocytes: an update. Anat. Embryol. (Berl) 1995;192:385–397. doi: 10.1007/BF00240371. [DOI] [PubMed] [Google Scholar]
- 10.Shankland SJ. The podocyte's response to injury: role in proteinuria and glomerulosclerosis. Kidney Int. 2006;69:2131–2147. doi: 10.1038/sj.ki.5000410. [DOI] [PubMed] [Google Scholar]
- 11.Sever S, Altintas MM, Nankoe SR, Moller CC, et al. Proteolytic processing of dynamin by cytoplasmic cathepsin L is a mechanism for proteinuric kidney disease. J. Clin. Invest. 2007;117:2095–2104. doi: 10.1172/JCI32022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ronco P. Proteinuria: is it all in the foot? J. Clin. Invest. 2007;117:2079–2082. doi: 10.1172/JCI32966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wartiovaara J, Ofverstedt LG, Khoshnoodi J, Zhang J, et al. Nephrin strands contribute to a porous slit diaphragm scaffold as revealed by electron tomography. J. Clin. Invest. 2004;114:1475–1483. doi: 10.1172/JCI22562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asanuma K, Yanagida-Asanuma E, Takagi M, Kodama F, et al. The role of podocytes in proteinuria. Nephrology (Carlton) 2007;12 Suppl 3:S15–S20. doi: 10.1111/j.1440-1797.2007.00876.x. [DOI] [PubMed] [Google Scholar]
- 15.Kriz W. Progressive renal failure--inability of podocytes to replicate and the consequences for development of glomerulosclerosis. Nephrol. Dial. Transplant. 1996;11:1738–1742. [PubMed] [Google Scholar]
- 16.Yu D, Petermann A, Kunter U, Rong S, et al. Urinary podocyte loss is a more specific marker of ongoing glomerular damage than proteinuria. J. Am. Soc. Nephrol. 2005;16:1733–1741. doi: 10.1681/ASN.2005020159. [DOI] [PubMed] [Google Scholar]
- 17.Michael AF. The glomerular mesangium. Contrib. Nephrol. 1984;40:7–16. [PubMed] [Google Scholar]
- 18.Inkyo-Hayasaka K, Sakai T, Kobayashi N, Shirato I, et al. Three-dimensional analysis of the whole mesangium in the rat. Kidney Int. 1996;50:672–683. doi: 10.1038/ki.1996.364. [DOI] [PubMed] [Google Scholar]
- 19.Mene P, Simonson MS, Dunn MJ. Physiology of the mesangial cell. Physiol. Rev. 1989;69:1347–1424. doi: 10.1152/physrev.1989.69.4.1347. [DOI] [PubMed] [Google Scholar]
- 20.Ohyama K, Seyer JM, Raghow R, Kang AH. Extracellular matrix phenotype of rat mesangial cells in culture. Biosynthesis of collagen types I, III, IV, and V and a low molecular weight collagenous component and their regulation by dexamethasone. J. Lab. Clin. Med. 1990;116:219–227. [PubMed] [Google Scholar]
- 21.Schlondorff D. The glomerular mesangial cell: an expanding role for a specialized pericyte. FASEB J. 1987;1:272–281. doi: 10.1096/fasebj.1.4.3308611. [DOI] [PubMed] [Google Scholar]
- 22.Abboud HE, Poptic E, DiCorleto P. Production of platelet-derived growth factorlike protein by rat mesangial cells in culture. J. Clin. Invest. 1987;80:675–683. doi: 10.1172/JCI113121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aron DC, Rosenzweig JL, Abboud HE. Synthesis and binding of insulin-like growth factor I by human glomerular mesangial cells. J. Clin. Endocrinol. Metab. 1989;68:585–591. doi: 10.1210/jcem-68-3-585. [DOI] [PubMed] [Google Scholar]
- 24.Zoja C, Wang JM, Bettoni S, Sironi M, et al. Interleukin-1 β and tumor necrosis factor-α induce gene expression and production of leukocyte chemotactic factors, colonystimulating factors, and interleukin-6 in human mesangial cells. Am. J. Pathol. 1991;138:991–1003. [PMC free article] [PubMed] [Google Scholar]
- 25.Evan AP, Gattone VH, 2nd, Connors BA. Ultrastructural features of the rabbit proximal tubule. Arch. Histol. Cytol. 1992;55 Suppl:139–145. doi: 10.1679/aohc.55.suppl_139. [DOI] [PubMed] [Google Scholar]
- 26.Brenner BM, Falchuk KH, Keimowitz RI, Berliner RW. The relationship between peritubular capillary protein concentration and fluid reabsorption by the renal proximal tubule. J. Clin. Invest. 1969;48:1519–1531. doi: 10.1172/JCI106118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glassock RJ. In: Primer on Kidney Disease. Greenberg A, editor. Philadelphia: Elsevier Saunders; 2005. pp. 36–46. [Google Scholar]
- 28.Hsu HH, Hoffmann S, Endlich N, Velic A, et al. Mechanisms of angiotensin II signaling on cytoskeleton of podocytes. J. Mol. Med. 2008;86:1379–1394. doi: 10.1007/s00109-008-0399-y. [DOI] [PubMed] [Google Scholar]
- 29.Lai KN, Leung JC, Chan LY, Saleem MA, et al. Podocyte injury induced by mesangial-derived cytokines in IgA nephropathy. Nephrol. Dial. Transplant. 2009;24:62–72. doi: 10.1093/ndt/gfn441. [DOI] [PubMed] [Google Scholar]
- 30.Lee YJ, Han HJ. Albumin-stimulated DNA synthesis is mediated by Ca2+/PKC as well as EGF receptor-dependent p44/42 MAPK and NF-kappaB signal pathways in renal proximal tubule cells. Am. J. Physiol. Renal Physiol. 2008;294:F534–F541. doi: 10.1152/ajprenal.00408.2007. [DOI] [PubMed] [Google Scholar]
- 31.Batuman V. Proximal tubular injury in myeloma. Contrib. Nephrol. 2007;153:87–104. doi: 10.1159/000096762. [DOI] [PubMed] [Google Scholar]
- 32.Pote A, Zwizinski C, Simon EE, Meleg-Smith S, et al. Cytotoxicity of myeloma light chains in cultured human kidney proximal tubule cells. Am. J. Kidney Dis. 2000;36:735–744. doi: 10.1053/ajkd.2000.17620. [DOI] [PubMed] [Google Scholar]
- 33.Oliver J, Macdowell M, Lee YC. Cellular mechanisms of protein metabolism in the nephron. I. The structural aspects of proteinuria; tubular absorption, droplet formation, and the disposal of proteins. J. Exp. Med. 1954;99:589–604. doi: 10.1084/jem.99.6.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zoja C, Benigni A, Remuzzi G. Cellular responses to protein overload: key event in renal disease progression. Curr. Opin. Nephrol. Hypertens. 2004;13:31–37. doi: 10.1097/00041552-200401000-00005. [DOI] [PubMed] [Google Scholar]
- 35.Schardijn GH, Statius van Eps LW. Beta 2-microglobulin: its significance in the evaluation of renal function. Kidney Int. 1987;32:635–641. doi: 10.1038/ki.1987.255. [DOI] [PubMed] [Google Scholar]
- 36.Kalluri R. Proteinuria with and without renal glomerular podocyte effacement. J. Am. Soc. Nephrol. 2006;17:2383–2389. doi: 10.1681/ASN.2006060628. [DOI] [PubMed] [Google Scholar]
- 37.Pirani CL, Pollak VE, Lannigan R, Folli G. The Renal Glomerular Lesions of Pre-Eclampsia: Electron Microscopic Studies. Am. J. Obstet. Gynecol. 1963;87:1047–1070. doi: 10.1016/0002-9378(63)90100-5. [DOI] [PubMed] [Google Scholar]
- 38.Branten AJ, van den Born J, Jansen JL, Assmann KJ, et al. Familial nephropathy differing from minimal change nephropathy and focal glomerulosclerosis. Kidney Int. 2001;59:693–701. doi: 10.1046/j.1523-1755.2001.059002693.x. [DOI] [PubMed] [Google Scholar]
- 39.Nangaku M. Mechanisms of tubulointerstitial injury in the kidney: final common pathways to end-stage renal failure. Intern. Med. 2004;43:9–17. doi: 10.2169/internalmedicine.43.9. [DOI] [PubMed] [Google Scholar]
- 40.Strutz F, Muller GA. Interstitial pathomechanisms underlying progressive tubulointerstitial damage. Kidney Blood Press. Res. 1999;22:71–80. doi: 10.1159/000025911. [DOI] [PubMed] [Google Scholar]
- 41.Palmer BF. The renal tubule in the progression of chronic renal failure. J. Investig. Med. 1997;45:346–361. [PubMed] [Google Scholar]
- 42.Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. USA. 2004;101:13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gonzales PA, Pisitkun T, Hoffert JD, Tchapyjnikov D, et al. Large-scale proteomics and phosphoproteomics of urinary exosomes. J. Am. Soc. Nephrol. 2009;20:363–379. doi: 10.1681/ASN.2008040406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou H, Cheruvanky A, Hu X, Matsumoto T, et al. Urinary exosomal transcription factors, a new class of biomarkers for renal disease. Kidney Int. 2008;74:613–621. doi: 10.1038/ki.2008.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou H, Pisitkun T, Aponte A, Yuen PS, et al. Exosomal Fetuin-A identified by proteomics: a novel urinary biomarker for detecting acute kidney injury. Kidney Int. 2006;70:1847–1857. doi: 10.1038/sj.ki.5001874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodby RA, Rohde RD, Sharon Z, Pohl MA, et al. The urine protein to creatinine ratio as a predictor of 24-hour urine protein excretion in type 1 diabetic patients with nephropathy. The Collaborative Study Group. Am. J. Kidney Dis. 1995;26:904–909. doi: 10.1016/0272-6386(95)90054-3. [DOI] [PubMed] [Google Scholar]
- 47.Schwab SJ, Christensen RL, Dougherty K, Klahr S. Quantitation of proteinuria by the use of protein-to-creatinine ratios in single urine samples. Arch. Intern. Med. 1987;147:943–944. [PubMed] [Google Scholar]
- 48.Ruggenenti P, Gaspari F, Perna A, Remuzzi G. Cross sectional longitudinal study of spot morning urine protein:creatinine ratio, 24 hour urine protein excretion rate, glomerular filtration rate, and end stage renal failure in chronic renal disease in patients without diabetes. BMJ. 1998;316:504–509. doi: 10.1136/bmj.316.7130.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barratt J, Topham P. Urine proteomics: the present and future of measuring urinary protein components in disease. CMAJ. 2007;177:361–368. doi: 10.1503/cmaj.061590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hutchison AS, O'Reilly DS, MacCuish AC. Albumin excretion rate, albumin concentration, and albumin/creatinine ratio compared for screening diabetics for slight albuminuria. Clin. Chem. 1988;34:2019–2021. [PubMed] [Google Scholar]
- 51.Mattix HJ, Hsu CY, Shaykevich S, Curhan G. Use of the albumin/creatinine ratio to detect microalbuminuria: implications of sex and race. J. Am. Soc. Nephrol. 2002;13:1034–1039. doi: 10.1681/ASN.V1341034. [DOI] [PubMed] [Google Scholar]
- 52.Hoang K, Tan JC, Derby G, Blouch KL, et al. Determinants of glomerular hypofiltration in aging humans. Kidney Int. 2003;64:1417–1424. doi: 10.1046/j.1523-1755.2003.00207.x. [DOI] [PubMed] [Google Scholar]
- 53.Dharnidharka VR, Kwon C, Stevens G. Serum cystatin C is superior to serum creatinine as a marker of kidney function: a meta-analysis. Am. J. Kidney Dis. 2002;40:221–226. doi: 10.1053/ajkd.2002.34487. [DOI] [PubMed] [Google Scholar]
- 54.Cockcroft DW, Gault MH. Prediction of creatinine clearance from serum creatinine. Nephron. 1976;16:31–41. doi: 10.1159/000180580. [DOI] [PubMed] [Google Scholar]
- 55.K/DOQI clinical practice guidelines for chronic kidney disease: evaluation, classification, and stratification. Am. J. Kidney Dis. 2002;39:S1–S266. [PubMed] [Google Scholar]
- 56.Han WK, Waikar SS, Johnson A, Betensky RA, et al. Urinary biomarkers in the early diagnosis of acute kidney injury. Kidney Int. 2008;73:863–869. doi: 10.1038/sj.ki.5002715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mann JF, Yi QL, Gerstein HC. Albuminuria as a predictor of cardiovascular and renal outcomes in people with known atherosclerotic cardiovascular disease. Kidney Int. 2004:S59–S62. doi: 10.1111/j.1523-1755.2004.09215.x. [DOI] [PubMed] [Google Scholar]
- 58.de Jong PE, Gansevoort RT. Prevention of chronic kidney disease: the next step forward! Nephrology (Carlton) 2006;11:240–244. doi: 10.1111/j.1440-1797.2006.00575.x. [DOI] [PubMed] [Google Scholar]
- 59.Caramori ML, Fioretto P, Mauer M. The need for early predictors of diabetic nephropathy risk: is albumin excretion rate sufficient? Diabetes. 2000;49:1399–1408. doi: 10.2337/diabetes.49.9.1399. [DOI] [PubMed] [Google Scholar]
- 60.Caramori M, Mauer M. Primer on Kidney Disease. Philadelphia: Elsevier Saunders; 2005. pp. 241–248. [Google Scholar]
- 61.Wingo CS, Clapp WL. Proteinuria: potential causes and approach to evaluation. Am. J. Med. Sci. 2000;320:188–194. doi: 10.1097/00000441-200009000-00010. [DOI] [PubMed] [Google Scholar]
- 62.Yamagata K, Iseki K, Nitta K, Imai H, et al. Chronic kidney disease perspectives in Japan and the importance of urinalysis screening. Clin. Exp. Nephrol. 2008;12:1–8. doi: 10.1007/s10157-007-0010-9. [DOI] [PubMed] [Google Scholar]
- 63.Palmer BF. Proteinuria as a therapeutic target in patients with chronic kidney disease. Am. J. Nephrol. 2007;27:287–293. doi: 10.1159/000101958. [DOI] [PubMed] [Google Scholar]
- 64.Jafar TH, Stark PC, Schmid CH, Landa M, et al. Proteinuria as a modifiable risk factor for the progression of non-diabetic renal disease. Kidney Int. 2001;60:1131–1140. doi: 10.1046/j.1523-1755.2001.0600031131.x. [DOI] [PubMed] [Google Scholar]
- 65.Zoja C, Morigi M, Figliuzzi M, Bruzzi I, et al. Proximal tubular cell synthesis and secretion of endothelin-1 on challenge with albumin and other proteins. Am. J. Kidney Dis. 1995;26:934–941. doi: 10.1016/0272-6386(95)90058-6. [DOI] [PubMed] [Google Scholar]
- 66.Wang Y, Chen J, Chen L, Tay YC, et al. Induction of monocyte chemoattractant protein-1 in proximal tubule cells by urinary protein. J. Am. Soc. Nephrol. 1997;8:1537–1545. doi: 10.1681/ASN.V8101537. [DOI] [PubMed] [Google Scholar]
- 67.Zoja C, Donadelli R, Colleoni S, Figliuzzi M, et al. Protein overload stimulates RANTES production by proximal tubular cells depending on NF-κB activation. Kidney Int. 1998;53:1608–1615. doi: 10.1046/j.1523-1755.1998.00905.x. [DOI] [PubMed] [Google Scholar]
- 68.Tang S, Leung JC, Abe K, Chan KW, et al. Albumin stimulates interleukin-8 expression in proximal tubular epithelial cells in vitro and in vivo. J. Clin. Invest. 2003;111:515–527. doi: 10.1172/JCI16079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nakajima H, Takenaka M, Kaimori JY, Hamano T, et al. Activation of the signal transducer and activator of transcription signaling pathway in renal proximal tubular cells by albumin. J. Am. Soc. Nephrol. 2004;15:276–285. doi: 10.1097/01.asn.0000109672.83594.02. [DOI] [PubMed] [Google Scholar]
- 70.Coppo R, D'Amico G. Factors predicting progression of IgA nephropathies. J. Nephrol. 2005;18:503–512. [PubMed] [Google Scholar]
- 71.D'Amico G, Bazzi C. Pathophysiology of proteinuria. Kidney Int. 2003;63:809–825. doi: 10.1046/j.1523-1755.2003.00840.x. [DOI] [PubMed] [Google Scholar]
- 72.Branten AJ, Wetzels JF. Influence of albumin infusion on the urinary excretion of beta2-microglobulin in patients with proteinuria. Nephron. 1999;81:329–333. doi: 10.1159/000045301. [DOI] [PubMed] [Google Scholar]
- 73.Morigi M, Buelli S, Angioletti S, Zanchi C, et al. In response to protein load podocytes reorganize cytoskeleton and modulate endothelin-1 gene: implication for permselective dysfunction of chronic nephropathies. Am. J. Pathol. 2005;166:1309–1320. doi: 10.1016/S0002-9440(10)62350-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Madaio MP. Renal biopsy. Kidney Int. 1990;38:529–543. doi: 10.1038/ki.1990.236. [DOI] [PubMed] [Google Scholar]
- 75.Parrish AE. Complications of percutaneous renal biopsy: a review of 37 years' experience. Clin. Nephrol. 1992;38:135–141. [PubMed] [Google Scholar]
- 76.Fliser D, Novak J, Thongboonkerd V, Argilés À, et al. Advances in urinary proteome analysis and biomarker discovery. J. Am. Soc. Nephrol. 2007;18:1057–1071. doi: 10.1681/ASN.2006090956. [DOI] [PubMed] [Google Scholar]
- 77.Hewitt SM, Dear J, Star RA. Discovery of protein biomarkers for renal diseases. J. Am. Soc. Nephrol. 2004;15:1677–1689. doi: 10.1097/01.asn.0000129114.92265.32. [DOI] [PubMed] [Google Scholar]
- 78.Mischak H, Apweiler R, Banks RE, Conaway M, et al. Clinical Proteomics: a need to define the field and to begin to set adequate standards. Proteomics-Clin. Appl. 2007;1:148–156. doi: 10.1002/prca.200600771. [DOI] [PubMed] [Google Scholar]
- 79.Goodsaid F, Bandow J, Mischak H. REPORT - Grand Rounds in Proteomics at the FDA (White Oak, Silver Spring, MD, USA, April 3, 2007) Proteomics Clin. Appl. 2007;1:1526–1531. doi: 10.1002/prca.200700575. [DOI] [PubMed] [Google Scholar]
- 80.HKUPP. 2nd HKUPP Workshop"Towards Standards for Urine Proteomics"; November 1, 2006; San Francisco, CA, USA: 2006. [Google Scholar]
- 81.Theodorescu D. Molecular pathogenesis of urothelial bladder cancer. Histol. Histopathol. 2003;18:259–274. doi: 10.14670/HH-18.259. [DOI] [PubMed] [Google Scholar]
- 82.Schiffer E, Mischak H, Theodorescu D, Vlahou A. Challenges of using mass spectrometry as a bladder cancer biomarker discovery platform. World J. Urol. 2008;26:67–74. doi: 10.1007/s00345-007-0234-z. [DOI] [PubMed] [Google Scholar]
- 83.Lee RS, Monigatti F, Briscoe AC, Waldon Z, et al. Optimizing sample handling for urinary proteomics. J. Proteome Res. 2008 doi: 10.1021/pr800301h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thongboonkerd V. Practical points in urinary proteomics. J. Proteome Res. 2007;6:3881–3890. doi: 10.1021/pr070328s. [DOI] [PubMed] [Google Scholar]
- 85.Coon JJ, Syka JE, Shabanowitz J, Hunt DF. Tandem mass spectrometry for peptide and protein sequence analysis. Biotechniques. 2005;38:519–523. doi: 10.2144/05384TE01. [DOI] [PubMed] [Google Scholar]
- 86.Coon JJ, Ueberheide B, Syka JE, Dryhurst DD, et al. Protein identification using sequential ion/ion reactions and tandem mass spectrometry. Proc. Natl. Acad. Sci. USA. 2005;102:9463–9468. doi: 10.1073/pnas.0503189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zubarev RA, Kelleher NL, McLafferty FW. Electron capture dissociation of multiply charged protein cations. A nonergodic process. J. Am. Chem. Soc. 1998;120:3265–3266. [Google Scholar]
- 88.Farrugia JM, O'Hair RAJ, Reid GE. Do all b(2) ions have oxazolone structures? Multistage mass spectrometry and ab initio studies on protonated N-acyl amino acid methyl ester model systems. Int. J. Mass Spectrom. 2001;210:71–87. [Google Scholar]
- 89.Valaskovic GA, Kelleher NL, McLafferty FW. Attomole protein characterization by capillary electrophoresis-mass spectrometry. Science. 1996;273:1199–1202. doi: 10.1126/science.273.5279.1199. [DOI] [PubMed] [Google Scholar]
- 90.Reid GE, Shang H, Hogan JM, Lee GU, et al. Gas-phase concentration, purification, and identification of whole proteins from complex mixtures. J. Am. Chem. Soc. 2002;124:7353–7362. doi: 10.1021/ja025966k. [DOI] [PubMed] [Google Scholar]
- 91.Stephenson JL, McLuckey SA, Reid GE, Wells JM, et al. Ion/ion chemistry as a top-down approach for protein analysis. Curr. Opin. Biotechnol. 2002;13:57–64. doi: 10.1016/s0958-1669(02)00285-9. [DOI] [PubMed] [Google Scholar]
- 92.Davey CA, Sargent DF, Luger K, Maeder AW, et al. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 A resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 93.Sze SK, Ge Y, Oh H, McLafferty FW. Top-down mass spectrometry of a 29-kDa protein for characterization of any posttranslational modification to within one residue. Proc. Natl. Acad. Sci. USA. 2002;99:1774–1779. doi: 10.1073/pnas.251691898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.VerBerkmoes NC, Bundy JL, Hauser L, Asano KG, et al. Integrating "top-down" and "bottom-up" mass spectrometric approaches for proteomic analysis of Shewanella oneidensis. J. Proteome Res. 2002;1:239–252. doi: 10.1021/pr025508a. [DOI] [PubMed] [Google Scholar]
- 95.Hogan JM, Pitteri SJ, McLuckey SA. Phosphorylation site identification via ion trap tandem mass spectrometry of whole protein and peptide ions: bovine alpha-crystallin A chain. Anal. Chem. 2003;75:6509–6516. doi: 10.1021/ac034410s. [DOI] [PubMed] [Google Scholar]
- 96.He M, McLuckey SA. Charge permutation reactions in tandem mass spectrometry. J. Mass Spectrom. 2004;39:1231–1259. [Google Scholar]
- 97.Ge Y, Lawhorn BG, ElNaggar M, Sze SK, et al. Detection of four oxidation sites in viral prolyl-4-hydroxylase by top-down mass spectrometry. Protein Sci. 2003;12:2320–2326. doi: 10.1110/ps.03244403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ge Y, Lawhorn BG, ElNaggar M, Strauss E, et al. Top down characterization of larger proteins (45 kDa) by electron capture dissociation mass spectrometry. J. Am. Chem. Soc. 2002;124:672–678. doi: 10.1021/ja011335z. [DOI] [PubMed] [Google Scholar]
- 99.Zabrouskov V, Giacomelli L, van Wijk KJ, McLafferty FW. A new approach for plant proteomics: characterization of chloroplast proteins of Arabidopsis thaliana by top-down mass spectrometry. Mol. Cell. Proteomics. 2003;2:1253–1260. doi: 10.1074/mcp.M300069-MCP200. [DOI] [PubMed] [Google Scholar]
- 100.Amunugama R, Hogan JM, Newton KA, McLuckey SA. Whole protein dissociation in a quadrupole ion trap: identification of an a priori unknown modified protein. Anal. Chem. 2004;76:720–727. doi: 10.1021/ac034900k. [DOI] [PubMed] [Google Scholar]
- 101.Newton KA, Pitteri SJ, Laskowski M, Jr, McLuckey SA. Effects of single amino acid substitution on the collision-induced dissociation of intact protein ions: Turkey ovomucoid third domain. J. Proteome Res. 2004;3:1033–1041. doi: 10.1021/pr049910w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Forbes AJ, Patrie SM, Taylor GK, Kim YB, et al. Targeted analysis and discovery of posttranslational modifications in proteins from methanogenic archaea by top-down MS. Proc. Natl. Acad. Sci. USA. 2004;101:2678–2683. doi: 10.1073/pnas.0306575101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kelleher NL. Top-down proteomics. Anal. Chem. 2004;76:197A–203A. [PubMed] [Google Scholar]
- 104.Pesavento JJ, Kim YB, Taylor GK, Kelleher NL. Shotgun annotation of histone modifications: a new approach for streamlined characterization of proteins by top down mass spectrometry. J. Am. Chem. Soc. 2004;126:3386–3387. doi: 10.1021/ja039748i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Geer LY, Markey SP, Kowalak JA, Wagner L, et al. Open mass spectrometry search algorithm. J. Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 106.Oh HB, Lin C, Hwang HY, Zhai H, et al. Infrared photodissociation spectroscopy of electrosprayed ions in a Fourier transform mass spectrometer. J. Am. Chem. Soc. 2005;127:4076–4083. doi: 10.1021/ja040136n. [DOI] [PubMed] [Google Scholar]
- 107.Coon JJ, Shabanowitz J, Hunt DF, Syka JE. Electron transfer dissociation of peptide anions. J. Am. Soc. Mass Spectrom. 2005;16:880–882. doi: 10.1016/j.jasms.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 108.Chi A, Coon JJ, Syka JEP, Bai D, et al. Efficient, large-scale phosphoproteome analysis with electron transfer dissociation mass spectrometry. Proceedings of the 53rd ASMS Conference; San Antonio, TX. 2005. p. 2005. [Google Scholar]
- 109.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, et al. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. USA. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Coon JJ, Syka JEP, Schwartz JC, Shabanowitz J, et al. Anion dependence in the partitioning between proton and electron transfer in ion/ion reactions. Int. J. Mass Spectrom. 2004;236:33–42. [Google Scholar]
- 111.Renfrow MB, MacKay CL, Chalmers MJ, Julian BA, et al. Analysis of O-glycan heterogeneity in IgA1 myeloma proteins by Fourier transform ion cyclotron resonance mass spectrometry: Implications for IgA nephropathy. Anal. Bioanal. Chem. 2007;389:1397–1407. doi: 10.1007/s00216-007-1500-z. [DOI] [PubMed] [Google Scholar]
- 112.Renfrow MB, Cooper HJ, Tomana M, Kulhavy R, et al. Determination of aberrant O-glycosylation in the IgA1 hinge region by electron capture dissociation Fourier transform-ion cyclotron resonance mass spectrometry. J. Biol. Chem. 2005;280:19136–19145. doi: 10.1074/jbc.M411368200. [DOI] [PubMed] [Google Scholar]
- 113.Chalmers MJ, Hakansson K, Johnson R, Smith R, et al. Protein kinase A phosphorylation characterized by tandem Fourier transform ion cyclotron resonance mass spectrometry. Proteomics. 2004;4:970–981. doi: 10.1002/pmic.200300650. [DOI] [PubMed] [Google Scholar]
- 114.Håkansson K, Cooper HJ, Emmett MR, Costello CE, et al. Electron capture dissociation and infrared multiphoton dissociation MS/MS of an N-glycosylated tryptic peptic to yield complementary sequence information. Anal. Chem. 2001;73:4530–4536. doi: 10.1021/ac0103470. [DOI] [PubMed] [Google Scholar]
- 115.Håkansson K, Chalmers MJ, Quinn JP, McFarland MA, et al. Combined electron capture and infrared multiphoton dissociation for multistage MS/MS in a Fourier transform ion cyclotron resonance mass spectrometer. Anal. Chem. 2003;75:3256–3262. doi: 10.1021/ac030015q. [DOI] [PubMed] [Google Scholar]
- 116.Kelleher RL, Zubarev RA, Bush K, Furie B, et al. Localization of labile posttranslational modifications by electron capture dissociation: The case of gamma-carboxyglutamic acid. Anal. Chem. 1999;71:4250–4253. doi: 10.1021/ac990684x. [DOI] [PubMed] [Google Scholar]
- 117.Marshall AG, Hendrickson CL, Jackson GS. Fourier transform ion cyclotron resonance mass spectrometry: A primer. Mass Spectrom. Rev. 1998;17:1–35. doi: 10.1002/(SICI)1098-2787(1998)17:1<1::AID-MAS1>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 118.Mirgorodskaya E, Roepstorff P, Zubarev RA. Localization of O-glycosylation sites in peptides by electron capture dissociation in a Fourier transform mass spectrometer. Anal. Chem. 1999;71:4431–4436. doi: 10.1021/ac990578v. [DOI] [PubMed] [Google Scholar]
- 119.Mormann M, Paulsen H, Peter-Katalinic J. Electron capture dissociation of O-glycosylated peptides: radical site-induced fragmentation of glycosidic bonds. Eur. J. Mass Spectrom. 2005;11:497–511. doi: 10.1255/ejms.738. [DOI] [PubMed] [Google Scholar]
- 120.Zimmerli LU, Schiffer E, Zurbig P, Good DM, et al. Urinary proteomic biomarkers in coronary artery disease. Mol. Cell. Proteomics. 2008;7:290–298. doi: 10.1074/mcp.M700394-MCP200. [DOI] [PubMed] [Google Scholar]
- 121.Rasmussen HH, Orntoft TF, Wolf H, Celis JE. Towards a comprehensive database of proteins from the urine of patients with bladder cancer. J. Urol. 1996;155:2113–2119. [PubMed] [Google Scholar]
- 122.Sarto C, Deon C, Doro G, Hochstrasser DF, et al. Contribution of proteomics to the molecular analysis of renal cell carcinoma with an emphasis on manganese superoxide dismutase. Proteomics. 2001;1:1288–1294. doi: 10.1002/1615-9861(200110)1:10<1288::AID-PROT1288>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 123.Julian BA, Waldo FB, Rifai A, Mestecky J. IgA nephropathy, the most common glomerulonephritis worldwide. A neglected disease in the United States? Am. J. Med. 1988;84:129–132. doi: 10.1016/0002-9343(88)90019-8. [DOI] [PubMed] [Google Scholar]
- 124.Wyatt RJ, Julian BA, Bhathena DB, Mitchell BL, et al. IgA nephropathy: presentation, clinical course, and prognosis in children and adults. Am. J. Kidney Dis. 1984;4:192–200. doi: 10.1016/s0272-6386(84)80071-2. [DOI] [PubMed] [Google Scholar]
- 125.Berger J, Hinglais N. Les depots intercapillaires d'IgA-IgG (Intercapillary deposits of IgA-IgG) J. Urol. Nephrol. 1968;74:694–695. [PubMed] [Google Scholar]
- 126.Jennette JC. The immunohistology of IgA nephropathy. Am. J. Kidney Dis. 1988;12:348–352. doi: 10.1016/s0272-6386(88)80022-2. [DOI] [PubMed] [Google Scholar]
- 127.Hiki Y, Odani H, Takahashi M, Yasuda Y, et al. Mass spectrometry proves under-O-glycosylation of glomerular IgA1 in IgA nephropathy. Kidney Int. 2001;59:1077–1085. doi: 10.1046/j.1523-1755.2001.0590031077.x. [DOI] [PubMed] [Google Scholar]
- 128.Allen AC, Bailey EM, Brenchley PEC, Buck KS, et al. Mesangial IgA1 in IgA nephropathy exhibits aberrant O-glycosylation: Observations in three patients. Kidney Int. 2001;60:969–973. doi: 10.1046/j.1523-1755.2001.060003969.x. [DOI] [PubMed] [Google Scholar]
- 129.Moldoveanu Z, Wyatt RJ, Lee J, Tomana M, et al. Patients with IgA nephropathy have increased serum galactose-deficient IgA1 levels. Kidney Int. 2007;71:1148–1154. doi: 10.1038/sj.ki.5002185. [DOI] [PubMed] [Google Scholar]
- 130.Matousovic K, Novak J, Tomana M, Kulhavy R, et al. IgA1-containing immune complexes in the urine of IgA nephropathy patients. Nephrol. Dial. Transplant. 2006;21:2478–2484. doi: 10.1093/ndt/gfl240. [DOI] [PubMed] [Google Scholar]
- 131.Julian BA, Wittke S, Novak J, Good DM, et al. Electrophoretic methods for analysis of urinary polypeptides in IgA-associated renal diseases. Electrophoresis. 2007;28:4469–4483. doi: 10.1002/elps.200700237. [DOI] [PubMed] [Google Scholar]
- 132.Haubitz M, Wittke S, Weissinger EM, Walden M, et al. Urine protein patterns can serve as diagnostic tools in patients with IgA nephropathy. Kidney Int. 2005;67:2313–2320. doi: 10.1111/j.1523-1755.2005.00335.x. [DOI] [PubMed] [Google Scholar]
- 133.Haubitz M, Mischak H, Julian BA, Novak J. Urinary protein patterns as a diagnostic tool in patients with IgA nephropathy. European Renal Disease - Touch Briefings 2008. In Press. [Google Scholar]
- 134.Good DM, Zürbig P, Bauer HW, Behrens G, et al. Naturally occurring human urinary peptides for use in diagnosis of chronic kidney disease. Mol. Cell. Proteomics. 2008 doi: 10.1074/mcp.M110.001917. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Coon JJ, Zürbig P, Dakna M, Dominiczak AF, et al. CE-MS analysis of the human urinary proteome for biomarker discovery and disease diagnostics. Proteomics-Clin. Appl. 2008;2:964–973. doi: 10.1002/prca.200800024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Park MR, Wang EH, Jin DC, Cha JH, et al. Establishment of a 2-D human urinary proteomic map in IgA nephropathy. Proteomics. 2006;6:1066–1076. doi: 10.1002/pmic.200500023. [DOI] [PubMed] [Google Scholar]
- 137.Rocchetti MT, Centra M, Papale M, Bortone G, et al. Urine protein profile of IgA nephropathy patients may predict the response to ACE-inhibitor therapy. Proteomics. 2008;8:206–216. doi: 10.1002/pmic.200700492. [DOI] [PubMed] [Google Scholar]
- 138.Novak J, Wittke S, Haubitz M, Walden M, et al. Urinary protein analyses reveal patterns and biomarkers specific for IgA nephropathy (IgAN) J. Am. Soc. Nephrol. 2005;16:155. [Google Scholar]
- 139.Mischak H, Julian BA, Novak J. High-resolution proteome/peptidome analysis of peptides and low-molecular-weight proteins in urine. Proteomics-Clin. Appl. 2007;1:792–804. doi: 10.1002/prca.200700043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Julian BA, Wyatt RJ, Matousovic K, Moldoveanu Z, et al. IgA nephropathy: a clinical overview. Contrib. Nephrol. 2007;157:19–26. doi: 10.1159/000102284. [DOI] [PubMed] [Google Scholar]
- 141.Schiffer E, Mischak H, Novak J. High resolution proteome/peptidome analysis of body fluids by capillary electrophoresis coupled with mass spectrometry. Proteomics. 2006;6:5615–5627. doi: 10.1002/pmic.200600230. [DOI] [PubMed] [Google Scholar]
- 142.Galla JH, Spotswood MF, Harrison LA, Mestecky J. Urinary IgA in IgA nephropathy and Henoch-Schoenlein purpura. J. Clin. Immunol. 1985;5:298–306. doi: 10.1007/BF00918248. [DOI] [PubMed] [Google Scholar]
- 143.Halling SFE, Söderberg MP, Berg UB. Henoch-Schönlein nephritis: clinical findings related to renal function and morphology. Pediatr. Nephrol. 2005;20:46–51. doi: 10.1007/s00467-004-1650-6. [DOI] [PubMed] [Google Scholar]
- 144.Julian BA, Wittke S, Novak J, Zürbig P, et al. Urinary polypeptide biomarkers of IgA-associated renal diseases. J. Am. Soc. Nephrol. 2007;18 doi: 10.1007/s00345-007-0192-5. 782A. [DOI] [PubMed] [Google Scholar]
- 145.Candiano G, Musante L, Bruschi M, Petretto A, et al. Repetitive fragmentation products of albumin and α1-antitrypsin in glomerular diseases associated with nephrotic syndrome. J. Am. Soc. Nephrol. 2006;17:3139–3148. doi: 10.1681/ASN.2006050486. [DOI] [PubMed] [Google Scholar]
- 146.Rossing K, Mischak H, Dakna M, Zurbig P, et al. Urinary proteomics in diabetes and CKD. J. Am. Soc. Nephrol. 2008;19:1283–1290. doi: 10.1681/ASN.2007091025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Neuhoff N, Kaiser T, Wittke S, Krebs R, et al. Mass spectrometry for the detection of differentially expressed proteins: a comparison of surface-enhanced laser desorption/ionization and capillary electrophoresis/mass spectrometry. Rapid Commun. Mass Spectrom. 2004;18:149–156. doi: 10.1002/rcm.1294. [DOI] [PubMed] [Google Scholar]
- 148.Mischak H, Kaiser T, Walden M, Hillmann M, et al. Proteomic analysis for the assessment of diabetic renal damage in humans. Clin. Sci. (Lond) 2004;107:485–495. doi: 10.1042/CS20040103. [DOI] [PubMed] [Google Scholar]
- 149.Kolch W, Neususs C, Pelzing M, Mischak H. Capillary electrophoresis-mass spectrometry as a powerful tool in clinical diagnosis and biomarker discovery. Mass Spectrom. Rev. 2005;24:959–977. doi: 10.1002/mas.20051. [DOI] [PubMed] [Google Scholar]
- 150.Otu HH, Can H, Spentzos D, Nelson RG, et al. Prediction of diabetic nephropathy using urine proteomic profiling 10 years prior to development of nephropathy. Diabetes Care. 2007;30:638–643. doi: 10.2337/dc06-1656. [DOI] [PubMed] [Google Scholar]
- 151.Schlessinger SD, Tankersley MR, Curtis JJ. Clinical documentation of end-stage renal disease due to hypertension. Am. J. Kidney Dis. 1994;23:655–660. doi: 10.1016/s0272-6386(12)70275-5. [DOI] [PubMed] [Google Scholar]
- 152.Wolfe RA, Ashby VB, Milford EL, Ojo AO, et al. Comparison of mortality in all patients on dialysis, patients on dialysis awaiting transplantation, and recipients of a first cadaveric transplant. N. Engl. J. Med. 1999;341:1725–1730. doi: 10.1056/NEJM199912023412303. [DOI] [PubMed] [Google Scholar]
- 153.Ivanyi B. A primer on recurrent and de novo glomerulonephritis in renal allografts. Nat. Clin. Prac. Nephrol. 2008;4:446–457. doi: 10.1038/ncpneph0854. [DOI] [PubMed] [Google Scholar]
- 154.Clarke W, Silverman BC, Zhang Z, Chan DW, et al. Characterization of renal allograft rejection by urinary proteomic analysis. Ann. Surg. 2003;237:660–664. doi: 10.1097/01.SLA.0000064293.57770.42. discussion 664–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wittke S, Haubitz M, Walden M, Rohde F, et al. Detection of acute tubulointerstitial rejection by proteomic analysis of urinary samples in renal transplant recipients. Am. J. Transplant. 2005;5:2479–2488. doi: 10.1111/j.1600-6143.2005.01053.x. [DOI] [PubMed] [Google Scholar]
- 156.Schaub S, Rush D, Wilkins J, Gibson IW, et al. Proteomic-based detection of urine proteins associated with acute renal allograft rejection. J. Am. Soc. Nephrol. 2004;15:219–227. doi: 10.1097/01.asn.0000101031.52826.be. [DOI] [PubMed] [Google Scholar]
- 157.Schaub S, Wilkins JA, Nickerson P. Proteomics and renal transplantation: searching for novel biomarkers and therapeutic targets. Contrib. Nephrol. 2008;160:65–75. doi: 10.1159/000125934. [DOI] [PubMed] [Google Scholar]
- 158.Trocme C, Marotte H, Baillet A, Pallot-Prades B, et al. Apolipoprotein A-I and platelet factor 4 are biomarkers for Infliximab response in rheumatoid arthritis. Ann. Rheum. Dis. 2008 doi: 10.1136/ard.2008.093153. In Press, PMID: 18664547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhang X, Jin M, Wu H, Nadasdy T, et al. Biomarkers of lupus nephritis determined by serial urine proteomics. Kidney Int. 2008;74:799–807. doi: 10.1038/ki.2008.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Domon B, Aebersold R. Mass spectrometry and protein analysis. Science. 2006;312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- 161.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat. Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]