Abstract
Bacteria use multiple strategies to bypass the inflammatory responses in order to survive in the host cells. In this review, we discuss the mechanism of the bacerial proteins in inhibiting inflammation. We highlight the anti-inflammatory roles of the type three secretion proteins including Salmonella AvrA, Enteropathogenic Escherichia coli Cif, and Yersinia YopJ, Staphylococcus aureus extracellular adherence protein, and Chlamydia proteins. We also discuss the research progress on the structures of these anti-inflammatory bacterial proteins. The current therapeutic methods for diseases, such as inflammatory bowel diseases, sclerosis, lack influence on the course of chronic inflammation and infection. Therefore, based on the molecular mechanism of the anti-inflammatory bacterial proteins and their 3-Dimension structure, we can design new peptides or non-peptidic molecules that serve as anti-inflammatory drugs without the possible side effect of promoting bacterial infection.
Keywords: Bacteria, type three secretion system, effector, bacterial-tail-specific protease, inflammation, anti-inflammation, acetylation, ubiquitination, phosphorylation, intestine, Salmonella, AvrA
INTRODUCTION
Classical inflammation is a local tissue response to infection or damage. It is a complex process that leads to an elevated blood supply and an increase in vascular permeability in the blood vessels surrounding the affected area [1]. Acute inflammation is a short-term response that usually results in healing: leukocytes infiltrate the damaged region, removing the stimulus and repairing the tissue. Acute inflammation is able to clear the pathogenic bacteria in the host. Whereas bacteria induce inflammation in the host, some bacteria proteins also develop the abilities to inhibit the inflammatory response and to survive in the host. Bacteria have various strategies to attack the inflammatory process including escape of the host defense [2], blockage of leukocyte recruitment to an inflamed area [2, 3], inactivation of anti-microbial peptides [4], stablization of the endogenous inflammatory inhibitors [5, 6], inducation of the expression of the anti-inflammatory cytokine, and cleavage of p65/relA of the NF-κB pathway [7]. Overall, microbial pathogens have applied a variety of mechanisms to manipulate host-cell functions, presumably for their own benefit.
In this review, we discuss the mechanisms of the bacerial proteins in inhibiting inflammation in the host cells. We focus on the anti-inflammatory roles of the type three secretion proteins from Salmonella, Enteropathogenic Escherichia coli (EPEC), and Yersinia, Staphylococcus aureus extracellular adherence protein, and Chlamydia proteins. We also discuss the research progress on the bacterial protein structures. Increasing studies demonstrate that chronic inflammation is associated with modern human diseases [8]. The current therapeutic methods for diseases, such as inflammatory bowel disease, sclerosis, sepsis, lack influence on the course of chronic inflammation and infection. Therefore, insights in the anti-inflammatory mechanisms of the bacterial proteins will provide promising opportunities for therapeutical intervention.
PATHOGENIC BACTERIAL PROTEINS
Pathogenic bacteria have evolved multiple effective “tricks” to hijack the host cellular machinery to creat a suitable niche for their survival and proliferation. One of these strategies is to control the host’s inflammatory responses, which are designed to clear the pathogen in the host. Many bacterial proteins, such as type three secretion effectors, toxins, and extracellular adherence proteins, are known to possess the anti-inflammatory ability, which helps the bacteria bypass the host’s response and prolong the survival of the bacteria in the host cells. In this review, we discuss the following bacterial proteins and their mechanisms to manipulate the inflammatory signaling pathways in the host cells.
Type Three Secretion System (TTSS) and Bacterial Effectors
TTSS is a needle-like protein transport device using by Gram-negative bacteria, including Yersinia, Salmonella, Shigella, EPEC, enterohaemorrhagic E. coli (EHEC), and Pseudomonas [9–13]. The TTSS virulence proteins called effectors are injected into the eukaryotic host cells [14–17]. Delivery of bacterial effectors into mammalian cells by TTSS requires the intimate association of bacteria with target cells. The molecular bases of such intimate association appear to be different in different bacteria involving TTSS components as well as surface determinants not associated with TTSSs [18]. Bacterial effectors display a large repertoire of biochemical activities and modulate the function of crucial host regulatory molecules. These effector proteins paralyze or reprogram the eukaryotic cell to the benefit of the pathogens. The activity of TTSS effectors allows bacteria to invade non-phagocytic cells or to inhibit phagocytosis, to regulate pro-inflammatory responses, to prevent autophagy, or to modulate intracellular trafficking [9, 10, 19]. Clearly, the studies on effectors uncover important mechanisms of regulation in host-bacterial interaction. In this review, we will focus on the research of the TTSS effectors of Salmonella, Yersinia, EPEC, and Chlamydia.
Staphylococcus aureus Extracellular Adherence Protein
Staphylococci (staph) are non-motile Gram-positive spherical bacteria that occur in microscopic clusters resembling grapes. Staphylococcus aureus and Staphylococcus epidermidis are significant in their interactions with humans. S. aureus colonizes mainly the nasal passages, but it may be found regularly in most other anatomical locales, including the skin, oral cavity, and gastrointestinal tract. S epidermidis is an inhabitant of the skin [20]. Staphylococcus aureus is a highly virulent pathogen posing an unabated challenge both in community-acquired as well as nosocomial infections. The organism readily gains access to the tissue through various breaks of the skin or mucosal barrier.
Invading bacteria utilize a variety of molecules to circumvent host defense, including inflammation and tissue regeneration. Among these bacterial proteins, the family of “secreted expanded repertoire adhesion molecules” (SERAM) is of special interest, because these adhesive proteins mediate bacterial colonization as a requirement for further steps in infection. The Staphylococcus aureus extracellular adherence protein (Eap) is one of the SERAMs that inhibit host leukocyte recruitment an anti-inflammatory factor [3].
Chlamydiae Tail-Specific Protease (Tsp)
Bacterial Tsp belongs to the carboxyl-terminal processing proteases (Ctp), which are a group of endoproteases of post-translational protein modification, maturation, and disassembly or degradation. Tail-specific proteases have been identified from bacterial pathogens of medical importance, including Borrelia, Chlamydia, Escherichia, Helicobacter, Pseudomonas, Salmonella, Shigella, Vibrio, and Yersinia [7, 21].
Chlamydiae are parasitic bacterial pathogens that affect over 140 million individuals worldwide. Ocular infection by Chlamydia trachomatis is the leading cause of preventable blindness, and urogenital tract infection by Chlamydia causes sexually transmitted disease, which is the most common cause of sexually transmitted disease in the United States. The annotated Chlamydia genome has two Tsps, CT441 and CT858 (CPAF) [22]. Both proteins target host proteins to interfere with host cellular processes. CPAF degrades regulatory factor X5 (RFX5) and upstream stimulation factor 1 (USF-1), transcription factors required for the expression of the major histocompatibility complex molecules of antigen presentation [23]. Recent study demonstrates that CT441 Tsp is responsible for chlamydial protease activity that cleaves the NF-κB p65 protein, an important regulator of the NF-κB pathway of inflammatory response [7, 21]. Cleavage of the p65 protein can suppress the host immune response against microbial infection.
BACTERIAL PROTEINS’ ANTI-INFLAMMATORY STRATEGIES
The strategies bacteria used to overcome the inflammatory responses in the host cells are various. We discuss the molecular bases of such anti-inflammation effects in the representive bacteria.
1. Salmonella
Salmonella enterica translocates virulent factors into host cells using TTSS to promote host colonization, intracellular bacterial replication and survival, and disease pathogenesis. Salmonella enterica encodes a TTSS within a specific pathogenicity island (SPI) located at centisome 63, which is essential for its pathogenicity [13, 24]. The Salmonella pathogenicity island (SPI)-1 TTSS is a virulence determinant that enables the injection of bacterial effector proteins into the cytosol of eukaryotic cells [25]. The Salmonella pathogenicity island (SPI)-2 is pivotal to the intracellular survival of Salmonella and for virulence in mammals. SPI-2 secretion activity appears to be induced in response to acidification of the vacuole in which it replicates [13, 26, 27]. Bacterial pathogens have evolved a sophisticated arsenal of virulence effectors to modulate host cell biology. Among many Salmonella effectors, we focus on two effectors AvrA and Salmonella secreted factor L (SseL).
AvrA is a Multifunctional Protein with Transacetylase/Deubiquitinase Activities
Salmonella AvrA is a SPI-I effector that is transferred into host cells through TTSS [28]. Recent studies have shown that the AvrA gene is present in 80% of Salmonella enterica serovar typhimurium [29, 30]. Our and others’ publications demonstrate that AvrA is a multifunctional protein that blocks host response to inflammation and influences eukaryotic cell pathways utilizing ubiquitin and acetylation [5, 6, 31].
It is known that AvrA acts as a deubiquitinase to influence eukaryotic cell pathways such as NF-κB and β-catenin that utilize ubiquitin [5]. NF-κB is found in almost all animal cell types and is involved in cellular responses to stimuli such as stress, cytokines, free radicals, ultraviolet irradiation, and bacterial or viral antigens [32–35]. NF-κB plays a key role in regulating the immune response and the expression of many cytokines and chemokines in infection. The inhibitor of κB (IκBα) inhibits the NF-κB activity. IκBα binds to NF-κB to mask the nuclear localization signal so that the NF-κB dimer (p50 & p65) is retained in the cytoplasm. Phosphorylation of IκBα by IκB kinase (IKK) leads to the ubiquitination and degradation of IκBα, resulting in nuclear translocation and activation of NF-κB [36]. β-catenin is another protein which has been shown to be a negative regulator of the proinflammatory NF-κB pathway in epithelial cells [37–39]. This function is in addition to its roles in embryonic development and neoplasia such as colon cancer [40] via enhancement of epithelial cell proliferation.
Recent study has investigated the role of AvrA in intestinal epithelial cells [5]. Target gene and inflammatory cytokine expression, as well as effects on epithelial cell proliferation and apoptosis induced by AvrA-deficient and -sufficient bacterial strains were tested in vivo. AvrA blocks degradation of IκBα and of β-catenin in epithelial cells [5]. AvrA deubiquitinates IκBα which blocks its degradation, leading to the inhibition of NF-κB activation. Correspondingly, target genes of the NF-κB pathway, such as IL-6, were down-regulated during bacterial infection with Salmonella expressing AvrA. AvrA also deubiquitinates and thus blocks degradation of β-catenin. Correspondingly, target genes of the β-catenin pathway, such as c-myc and cyclinD1, were up-regulated with AvrA expression. Increased β-catenin further negatively regulates the NF-κB pathway [5]. These findings suggest an important role for AvrA in regulating host inflammatory responses through the NF-κB and β-catenin pathways.
Interestingly, AvrA also possesses acetyltransferase activity toward specific mitogen-activated protein kinase kinases (MAPKK) and potently inhibits c-Jun N-terminal kinase and NF-κB signaling pathways [31]. The mitogen-activated protein kinase (MAPK) pathways are the key signaling pathways involved in host defenses against microbes. The MAPKs function in a triple kinase sequence that involves the rapid and controlled relay of phosphorylation events to convey “alarm” signals [41]. Perception of diverse threats induces the upstream activators, kinases of the MAPKKK class. These proteins serve to activate the MAPKKs (or MEKKs), which subsequently activate the MAPKs and a downstream battery of immune and cell survival effector systems. MAPKKs such as MKK6 and MKK3 phosphorylate and activate members of the ERK and p38 MAPKs, respectively, which mediate primarily proliferative and cytoprotective responses. In contrast, the MAPKK MKK4/7 activates Jun N-terminal kinase (JNK), which is proinflammatory or, during prolonged activation, potently proapoptotic [42]. Jones et al. have demonstrated that AvrA possesses acetyltransferase activity toward MAPKKs and potently inhibits c-Jun N-terminal kinase (JNK) and NF-kappaB signaling pathways in both transgenic Drosophila and murine models. Furthermore, they found that AvrA dampens the proapoptotic innate immune response to Salmonella at the mouse intestinal mucosa. This activity is consistent with the natural history of Salmonella in mammalian hosts, where the bacteria elicit transient inflammation but do not destroy epithelial cells. These findings suggest that targeting JNK signaling to dampen apoptosis may be a conserved strategy for intracellular pathogens [31].
Many proteins coordinate distinct signaling pathways in the cells by having multiple functions. Bacterial effectors may have multiple protease activities to modify target proteins, just like the eukaryotic protein A20, which has two enzyme activities [43–45]. Therefore, not surprisingly, Salmonella effector AvrA has multiple protease activities to modify different eukaryotic proteins and to maximize AvrA’s ability to modulate cellular functions.
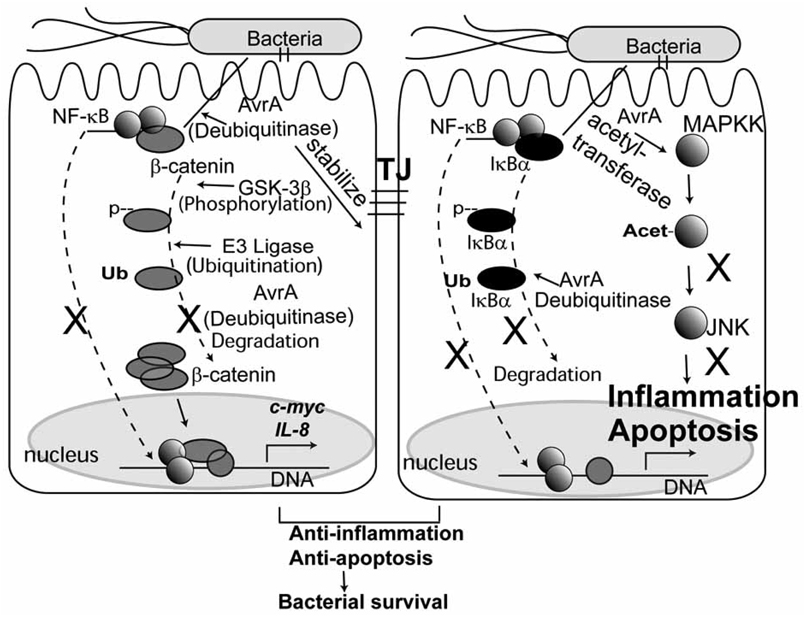
Overall, previous studies on Salmonella and TTSS effector AvrA found that (Fig. 1).
AvrA inhibits the proinflammatory NF-κB pathways [6].
Salmonella can activate the β-catenin pathway in epithelial cells [46].
There is a cross-talk between β-catenin and NF-κB pathways in bacterial-epithelial interaction [39].
β-catenin negatively regulates NF-κB activity in Salmonella-induced inflammation [47].
AvrA protein acts as a deubiquintinase to stabilize β-catenin and IκBα, thus inhibiting NF-κB pathways and activating the β-catenin pathway [5].
AvrA stabilizes intestinal epithelial cell tight Junctions (TJ), even though the other Salmonella TTSS proteins, SopB, SopE, and SopE2, are known to disrupt TJs [48]. AvrA may play a role in stabilizing TJs and balancing the opposing action of other bacterial effectors. Our findings indicate an important role for the bacterial effector AvrA in regulation of intestinal epithelial cell TJs during inflammation. The role of AvrA represents a highly refined bacterial strategy that helps the bacteria survive in the host and dampen the inflammatory response [49].
AvrA activity may account for the self-limited inflammation seen in Salmonellosis. The AvrA protein apparently allows the invading bacterium to dampen innate immune signaling but also prevents the apoptotic elimination in cells that have perceived microbial compromise [5, 31].
AvrA also possesses acetyltransferase activity toward MAPKK and inhibits c-Jun N-terminal kinase and NF-κB signaling pathways [31].
Fig. (1). Schematic model for the anti-inflammatory function of Salmonella effector AvrA in the intestinal epithelial cells.
There is a cross-talk between β-catenin and NF-κB pathways in Salmonella-epithelial interaction. AvrA inhibits the NF-κB pathways. AvrA protein acts as a deubiquintinase to stabilize β-catenin and IκBα, thus inhibiting NF-κB pathways and activating the β-catenin pathway. AvrA stabilizes intestinal epithelial cell tight Junctions (TJ). AvrA also possesses acetyltransferase activity toward MAPKK and inhibits c-Jun N-terminal kinase and NF-κB signaling pathways. The role of AvrA represents a highly refined bacterial strategy that helps the bacteria survive in the host, dampens the inflammatory response, and prevents the apoptotic elimination in cells that have perceived microbial compromise. AvrA may play a role in stabilizing TJs and balancing the opposing action of other bacterial effectors.
However, the exact function and mechanism of AvrA are still not entirely clear. The unanswered questions for the AvrA regulation of the inflammatory pathways in the host include:
How does AvrA play its dual roles as a deubiquitinase and as an acetyltransferase in regulation of the signaling in the host?
What is the functional site of AvrA when it interacts with the host target protein(s)?
What are the biological impacts of AvrA/host interaction?
Therefore, it is important to further elucidate the mechanisms through which AvrA exerts its effects in Salmonella infection and inflammation.
Salmonella Secreted Factor is a Deubiquitinase
The Salmonella TTSS encoded in SPI- 2 translocates a Salmonella-specific protein, designated Salmonella secreted factor L (SseL). Recent studies elucidate that SseL is a putative virulence factor possessing deubiquitinase activity [50]. Expression of SseL in mammalian cells suppresses NF-κB activation downstream of IκBα kinases and impairs IκBα ubiquitination and degradation, but not IκBα phosphorylation. Disruption of the gene encoding SseL in S. enterica serovar typhimurium increases IκBα degradation and ubiquitination, as well as NF-κB activation in infected macrophages, compared with wild-type bacteria. Mice infected with SseL-deficient bacteria mount stronger inflammatory responses, associated with increased production of NF-κB-dependent cytokines [50].
Taken together, AvrA and SseL represent the bacterial deubiquitinases that modulate the host inflammatory response through the NF-κB pathway
2. Yersinia
Yersinia pestis is the infectious agent that caused the Black Death in the Middle Ages, and Yersinia pseudotuberculosis and Yersinia enterocolitica (two closely related food-borne pathogens) are causal agents for gastrointestinal disorders. Yersinia spp. harbor a 70 kb plasmid that encodes Yersinia outer proteins (Yops), also referred to as effectors, and a type III secretion system [51]. To overcome host defenses and inflammatory responses, Yersinia injects specific effector proteins into colonized mammalian cells. Upon translocation of the effectors into the target host cell, these molecules cripple the target cell by blocking phagocytosis, destroying the host defense system [51]. YopJ inhibits the host inflammatory response and induces apoptosis of immune cells by blocking multiple signaling pathways [52], whereas YopH inhibits the PI3-kinase-dependent secretion of IL-2 and proliferation.
Multiple Functional Protease YopJ
Studies on YopJ assigned YopJ multiple functions. Macrophages respond to infection with pathogenic Yersinia by activating MAPK- and NF-κB-signaling pathways. To counteract this response, YopJ deactivates MAPK- and NF-κB-signaling pathways, and induces apoptosis [53]. NF-κB promotes cell survival by up-regulating expression of several apoptosis inhibitor genes. The IκBα superrepressor and A20 are two inhibitors of the NF-κB pathway. To determine whether deactivation of the NF-κB pathway is sufficient for Yersinia-induced apoptosis, IκBα or A20 were expressed in macrophages. Apoptosis levels were substantially higher when active YopJ was delivered into macrophages expressing IκBα A20, suggesting that deactivation of the NF-κB pathway is not sufficient for rapid Yersinia-induced apoptosis. When macrophages expressing A20 were treated with specific inhibitors of MAPKs, similar levels of apoptosis were observed when active or inactive YopJ were delivered during infection. These results suggest that MAPK and NF-κB pathways function together to up-regulate apoptosis inhibitor gene expression in macrophages in response to Yersinia infection and that YopJ deactivates both pathways to promote rapid apoptosis [53].
Early study on YopJ indicates that YopJ is a highly conserved ubiquitin-like molecule, which is covalently added to numerous regulatory proteins [52]. YopJ exerts its pathogenic effect on cells by disrupting the posttranslational modification [52]. Further studies showed that YopJ is actually an acetyltransferase [54, 55]. YopJ uses acetyl-coenzyme A (CoA) to modify the critical serine and threonine residues in the activation loop of MAPKK6 and thereby blocking phosphorylation [54, 55]. YopJ exerts its deleterious effects by catalyzing the acetylation of two serine residues in the activation loop of the MAP kinase kinase, MEK2. This covalent modification prevents the phosphorylation of these serine residues that is required for activation of MEK2 and downstream signal propagation [54, 55]. Studies also showed that YopJ causes acetylation of a threonine residue in the activation loop of both the alpha and beta subunits of the NF-κB pathway kinase, IKK. These results suggest the possibility that serine/threonine acetylation may occur even under nonpathogenic conditions and may be a widespread protein modification regulating protein function in eukaryotic cells [56].
However, some studies demonstrated that YopJ is a deu-biquitinase that negatively regulates signaling by removing ubiquitin moieties from critical proteins, such as tumor necrosis factor receptor-associated factor-2 (TRAF2), TRAF6, and IκBα [57, 58]. Zhou et al. reported that YopJ is a promiscuous deubiquitinating enzyme that negatively regulates signaling by removing ubiquitin moieties from TRAF2, TRAF6, and IκBα. Multi-ubiquitin chains are built by formation of an isopeptide bond between Gly76 of one ubiquitin to the ε-NH2 group of one of the seven potential lysines (K6, K11, K27, K29, K33, K48 or K63) of the preceding ubiquitin. Ployubiqutin chains linked through lysine at posi-ion 48 of ubiquitin (K48) target protein substrates for proteasomal degradation, whereas the K63-polymerized ubiquitin conjugates required for signal transduction [59]. The cylindromatosis tumor suppressor (CYLD) is a deubiquintinase, which attenuates NF-κB signaling by selectively removing K63-linked polyubiquitin chains that activate IκB kinase [60]. In contrast to CYLD, YopJ not only removes K63-link ubiquitins but also cleaves K48-linked chains and thereby inhibits proteasomal degradation of IκBα. A catalytically inactive YopJ mutant loses the ability for deubiquitination of cellular proteins and cleavage both K48- and K63-linked polyubiquitin. Moreover, purified YopJ demonstrates the deubiquitinating activity in vitro [57].
YopJ inhibits TLR-mediated NF-κB and MAPK activation. Additionally, YopJ blocks the induction of the TLR-mediated interferon response. It is shown that YopJ also inhibits interferon regulatory factor 3 (IRF3) signaling [58]. Examination of the NF-κB signaling pathway suggested that YopJ acts at the level of TAK1 (MAP3K7) activation. Further studies revealed a YopJ-dependent decrease in the ubiquitination of TRAF6 and TRAF3. These data support the hypothesis that YopJ is a deubiquitinating protease that acts on TRAF proteins to prevent or remove the K63-polymerized ubiquitin conjugates required for signal transduction [58]. However, these studies do not directly address the alternative hypothesis that YopJ is an acetyltransferase that acts on the activation loop of IKK and MKK proteins.
The debate on the role of YopJ suggests that a bacterial effector has multiple protease activities to modify different eukaryotic proteins. Multiple function can maximize a bacterial effector’s ability to modulate cellular functions in the host.
YopH is a Tyrosine Phosphatase
YopH is a Yersinia TTSS protein that is known to inhibit the phosphatidylinositol 3-kinase (PI 3-kinase)-dependent secretion of IL-2 and proliferation [61]. PI 3-kinase and its target protein kinase B (Akt) are involved in various processes including internalization, chemotaxis, cell survival, and proliferation [62, 63]. Sauvonnet et al. analyzed the activation of Akt in macrophages infected with virulent (pYV+) or avirulent (pYV−) Yersinia enterocolitica [60]. During the early stage of infection with pYV+ and pYV− bacteria, Akt and its targets, glycogen synthase kinase 3 and forkhead transcription factor, became phosphorylated. This phosphorylation induction was inhibited by wortmannin and thus dependent on PI 3-kinase. When infection was carried out with pYV+ bacteria but not with pYV− bacteria, Akt and its targets became dephosphorylated at later time points. The tyrosine phosphatase YopH was responsible for the inactivation of the PI 3-kinase cascade. In macrophages, this inactivation correlated with the downregulation of mRNA coding for monocyte chemoattractant protein 1, suggesting that YopH inhibits recruitment of macrophages to lymph nodes. Consistent with the observation that YopH inactivated the Akt pathway, YopH inhibited PI 3-kinase-dependent secretion of interleukin 2 and proliferation. These data reveal a new effect of YopH in Yersinia pathogenesis [61].
T cell responses are critical to the survival of Yersinia-infected animals. Yersinia has the ability to directly suppress T lymphocyte activation through YopH. Gerke et al. showed that even an average of one Yersinia per T cell is sufficient to inhibit or alter T cell responses [64]. This efficient inhibition is traced to specific targeting by YopH of the adaptor proteins, linker for activation of T cells (LAT) and SH2-domain-containing leukocyte protein of 76 kD (SLP-76), which are crucial for T cell antigen receptor (TCR) signaling. A catalytically inactive YopH translocated via the bacterial TTSS into T cells primarily binds to LAT and SLP-76. Furthermore, among the proteins of the TCR signaling pathway, the tyrosine phosphorylation levels of LAT and SLP-76 are the most affected in T cells exposed to low numbers of Yersinia pseudotuberculosis [64]. YopH’s ability to target the adaptor proteins in the TCR signaling pathway represents a novel strategy by which pathogens efficiently alter T cell-mediated immune responses.
3. Chlamydial Proteins
Chlamydia is a common sexually transmitted disease (STD) caused by the bacterium. A hallmark of chlamydial STD is its asymptomatic nature, although inflammatory cellular response and chronic inflammation are among the underlying mechanisms. Increasing data demonstrate that Chlamydia have the ability to convert a regulatory molecule of host inflammatory response to a dominant negative inhibitor of the same pathway potentially to minimize inflammation. Among these chlamydial proteins, CT441, ChlaDub1, and CPAF have their very own strategies for anti-inflammation.
CT441
NF-κB subunit p65 protein is an important regulator of the NF-κB pathway of inflammatory response. Interestingly, Chlamydia infection does not induce IκBα degradation or NF-κB p65 nuclear translocation. Instead, it promotes p65 cleavage [7, 21]. Chlamydiae causes p65 cleavage into an N terminus-derived p40 fragment and a p22 of the C terminus. The activity is specific because no other protein cleavage or degradation of NF-κB pathway components is detected. Moreover, murine p65 protein is resistant to cleavage by both human and mouse biovars. CT441 Tsp is responsible for Chlamydial protease activity that cleave the NF-κB p65 protein [7, 21].
The essential structure of CT441 as a tail-specific protease in p65 cleavage has been further characterized [21]. The PDZ domain is a common protein–protein interaction structural domain of 80–90 amino-acids found in the signaling proteins [65]. It is found in all kingdoms of life. PDZ is an acronym combining the first letters of three proteins — post synaptic density protein (PSD95), Drosophila disc large tumor suppressor (DlgA), and zonula occludens-1 protein (ZO-1) — which were first discovered to share the domain. PDZ domains are also referred to as DHR (Dlg homologous region) or GLGF (glycine-leucineglycine-phenylalanine) domains. These domains help anchor transmembrane proteins to the cytoskeleton and hold together signaling complexes [65]. CT441 contains a PDZ domain of protein-protein interactions and a Ser/Lys dyad catalytic unit. Mutation at either Ser455 or Lys481 in the active site ablated CT441 activity of p65 cleavage. Additionally, the production of CT441 Tsp, which is detected at the middle and late stages of an infection, correlates with p65 cleavage activity. Thus, CT441 is identified as a tail-specific protease that is capable of interfering with the NF-κB pathway of host antimicrobial and inflammatory responses [21].
ChlaDub1 and ChlaDub2 of Chlamydia trachomatis
In addition to promoting p65 cleavage, Chlamydia sp. also applies other mechanisms to interfere with host inflammatory response. ChlaDub1 and ChlaDub2 of Chlamydia trachomatis are identified to possess both deubiquitinating and deneddylating activities [66]. The genes encoding ChlaDub1 and ChlaDub2 are present in all Chlamydia species except for Chlamydia pneumoniae, and their catalytic domains bear similarity to the catalytic domains of other eukaryotic ubiquitin-like proteases (Ulp). The C. trachomatis DUBs react with activity-based probes and hydrolyse ubiquitinated and neddylated substrates.
Further studies showed that ChlaDub1 of Chlamydia trachomatis suppresses NF-κB activation and inhibits IκBα ubiquitination and degradation [67]. ChlaDub1 acts as a deubiquitinating and deNeddylating protease in infected cells. In transfection experiments, ChlaDub1 suppresses NF-κB activation induced by several pro-inflammatory stimuli and binds the NF-κB inhibitory subunit IκBα, impairing its ubiquitination and degradation. This study provides insight into the mechanism by which C. trachomatis may evade the host inflammatory response by demonstrating that ChlaDub1 is capable of inhibiting IκBα degradation and blocking NF-κB activation [67].
Chlamydial Protease– or Proteasome–like Activity Factor (CPAF)
The CPAF gene is highly conserved among chlamydial strains, but has no significant overall homology with other known genes. CPAF protein is secreted into the host cell cytosol. Chlamydia can escape T lymphocyte immune recognition by degrading host transcription factors required for major histocompatibility complex (MHC) antigen expression. CPAF is necessary and sufficient for the degradation of host transcription factors RFX5 and upstream stimulation factor 1. Thus, CPAF represents a unique secreted protein produced by an obligate intracellular bacterial pathogen to interfere with effective host adaptive immunity [23].
Biochemical studies have demonstrated that cleavage of CPAF into CPAFn and CPAFc is a physiological process required for CPAF proteolytic activity [68]. CPAF was initially synthesized in chlamydia-infected cells as a 70 kDa full-length protein and rapidly cleaved into CPAFn and c fragments. Full-length CPAF expressed via a transgene in mammalian cells remained uncleaved and had no proteolytic activity, whereas CPAF expressed in Escherichia coli cells was processed and possessed RFX5 degradation activity. CPAF mutants deficient in processing even when expressed by E. coli failed to degrade RFX5. More importantly, the RFX5 degradation activity was partially restored when the mutant CPAF was artificially induced to undergo cleavage. These observations together have demonstrated that cleavage of CPAF is necessary and sufficient for CPAF activity [68].
4.Shigella flexneri protein Effector OspF
Shigella flexneri has evolved the capacity to precisely modulate host cell epigenetic 'information' as a strategy for repressing innate immunity [69]. The representative effector is the Shigella type III effector OspF.
OspF is able to inactivate MAPKs. OspF irreversibly removed phosphate groups from the phosphothreonine but not from the phosphotyrosine residue in the activation loop of MAPKs [70]. Mass spectrometry revealed a mass loss of 98 daltons in p-Erk2, due to the abstraction of the alpha proton concomitant with cleavage of the C-OP bond in the phosphothreonine residue. This enzymatic activity, termed phosphothreonine lyase, appeared specific for MAPKs and was shared by other OspF family members [70].
OspF also enabled shigella to block the activation of a subset of NF-κB-responsive genes [69]. Phosphorylation of histone H3 at Ser10 increases chromatin accessibility to transcription factor NF-κB on a subset of genes involved in immune responses. OspF acts as a dually specific phosphatase that dephosphorylated mitogen-activated protein kinases in the nucleus, thus preventing histone H3 phosphorylation at Ser10 in a gene-specific way. The activity of OspF enables shigella to block the activation of a subset of NF-κB-responsive genes, leading to compromised recruitment of polymorphonuclear leukocytes to infected tissues [69].
5.Bordetella Bronchiseptica
TTSS Proteins
Bordetella bronchiseptica establishes respiratory tract infections in laboratory animals with high efficiency. Colonization of Bordetella is persistent and infection is usually asymptomatic in immunocompetent hosts. Recent studies have identified 15 loci that are part of a type III secretion apparatus in B. bronchiseptica and three secreted proteins [71]. Studies showed that type III-secreted products of B. bronchiseptica interact with components of both innate and adaptive immune systems of the host. B. bronchiseptica modulates host immunity by inactivating NF-κB and induces apoptosis in macrophages in vitro and inflammatory cells in vivo. Infection of an epithelial cell line with wild type, but not type III deficient B. bronchiseptica, resulted in rapid aggregation of NF-κB into large complexes in the cytoplasm. NF-κB aggregation was dependent on type III secretion. In summary, the Bordetella TTSS functions to modulate host immune responses during infection [71].
Bordetella pertussis Filamentous Hemagglutinin (FHA)
FHA is a cell-associated and secreted adhesion produced by Bordetella pertussis with pro-apoptotic and pro-inflammatory activity in host cells [72]. Recent studies suggest that longer exposures of host cells to FHA may block NF-κB activation, and perhaps lead to a compromised immune response to this bacterial pathogen [72]. Exposure to FHA resulted in early activation of the NF-κB pathway, as manifested by the IκBα degradation, by NF-κB DNA binding, and by the subsequent secretion of NF-κB-regulated inflammatory cytokines. However, exposure of macrophages and human monocytes to FHA for two hours or more resulted in the accumulation of cytosolic IκBα, and the failure of TNF to activate NF-κB. Thus, these results reveal a complex temporal dynamic and suggest that despite short-term effects to the contrary, longer exposures of host cells to this secreted adhesion may block NF-κB activation and perhaps lead to a compromised immune response to this bacterial pathogen [72].
6. Escherichia coli
Enteropathogenic and enterohemorrhagic Escherichia coli (EPEC and EHEC) use a TTSS to inject microbial proteins into host cells. The cycle inhibiting factor (Cif) was the first cyclomodulin to be identified that is injected into host cells via the type III secretion machinery. Cif provokes cytopathic effects characterized by G(1) and G(2) cell cycle arrests [73], accumulation of the cyclin-dependent kinase inhibitors (CKIs) p21(waf1/cip1), and p27(kip1) [74], and formation of actin stress fibers [75].
Recent studies identify a family of proteins, homologous to the type III effector Cif from EPEC, in pathogens including Yersinia, Photorhabdus, and Burkholderia that contain functional TTSSs. Although these Cif homologs are remarkably divergent in primary sequence, the catalytic triad is strictly conserved and was shown to be crucial for cell cycle arrest, cytoskeleton reorganization, accumulation of cyclin-dependent kinase inhibitors, and formation of actin stress fibers [76]. Bacterial Cifs interfere with the eukaryotic cell cycle and block mitosis. These might constitute powerful weapons for immune evasion by inhibiting expansion of lymphocytes. Cell-cycle inhibitors might also impair epithelial-barrier integrity allowing the entry of pathogenic bacteria into the body or prolonging their local existence by blocking the shedding of epithelia [77].
7. Staphylococcus aureus Eap Inhibits Host Leukocyte Recruitment
Staphylococcus aureus has the capacity to infect and disseminate into the bloodstream and distant sites in the host when skin or mucosal barriers are breached or when host immunity is affected. The range of diseases caused by S. aureus is broad and includes endocarditis, osteomyelitis, and septic shock [20, 78]. S. aureus produces a number of cell surfacelocalized proteins that are responsible for binding to fibronectin, collagen, fibrinogen, and vitronectin, among others. These extracellular matrixbinding proteins have been proposed to contribute to successful colonization and persistence at various sites in the host [78]. Among these Staphylococcus aureus, Eap acts as a anti-inflammatory factor that inhibits host leukocyte recruitment [3]. As an immediate response towards bacterial infection or injury, leukocytes migrate from the blood stream into extravascular sites of inflammation. This coordinated sequence of adhesion and locomotion steps requires the expression and upregulation of various adhesion receptors on the surface of mobile and stationary vascular cells. Chavakis, et al. reportesd that Eap disrupted beta(2)-integrin and urokinase receptor mediated leukocyte adhesion due to its direct interactions with the host adhesive proteins intercellular adhesion molecule 1 (ICAM-1), fibrinogen or vitronectin in vitro. Eap-expressing S. aureus induced a 2 to 3-fold lower neutrophil recruitment in bacterial peritonitis in mice as compared with an Eap-negative strain, isolated Eap prevented beta(2)-integrin-dependent neutrophil recruitment. Thus, the specific interactions with ICAM-1 and extracellular matrix proteins render Eap a potent anti-inflammatory factor. This inhibition of leukocyte recruitment by Eap may therefore explain the impaired wound healing frequently seen in S. aureus-infected chronic wounds [3].
However, recent studies from Scriba et al showed that Eap activates expression of proinflammatory Cytokines [79]. They found that the Eap protein can induce both human and murine cells to produce copious amounts of IL-6 and TNF. The effects of Eap are likely to be highly variable depending on both the body site and the levels of expression of both ICAM-1 and Eap [79].
In summary, S. aureus virulence factors may have differential effects, depending on bacterial density and the stage of infection. During the early stage of infection, Eap is present at levels that correspond to the stimulatory concentration. This would, together with the action of other bacterial factors, result in an expansion and proliferation of human immune cells and, consequently, bacterial clearance. In contrast, during a later stage of infection, Eap, along with other secreted Staphylococcal products, may accumulate to reach levels in the extracellular environment that are capable of triggering apoptosis and necrosis. This generates an immunosuppressive state in the host, which benifit bacteria and allow the establishment of a long-term persistence and, therefore, relapsing or chronic infections [78].
KEY INFLAMMATORY CELL-SIGNALING PATHWAYS INVOVLED IN BACTERIA-HOST INTERACTIONS
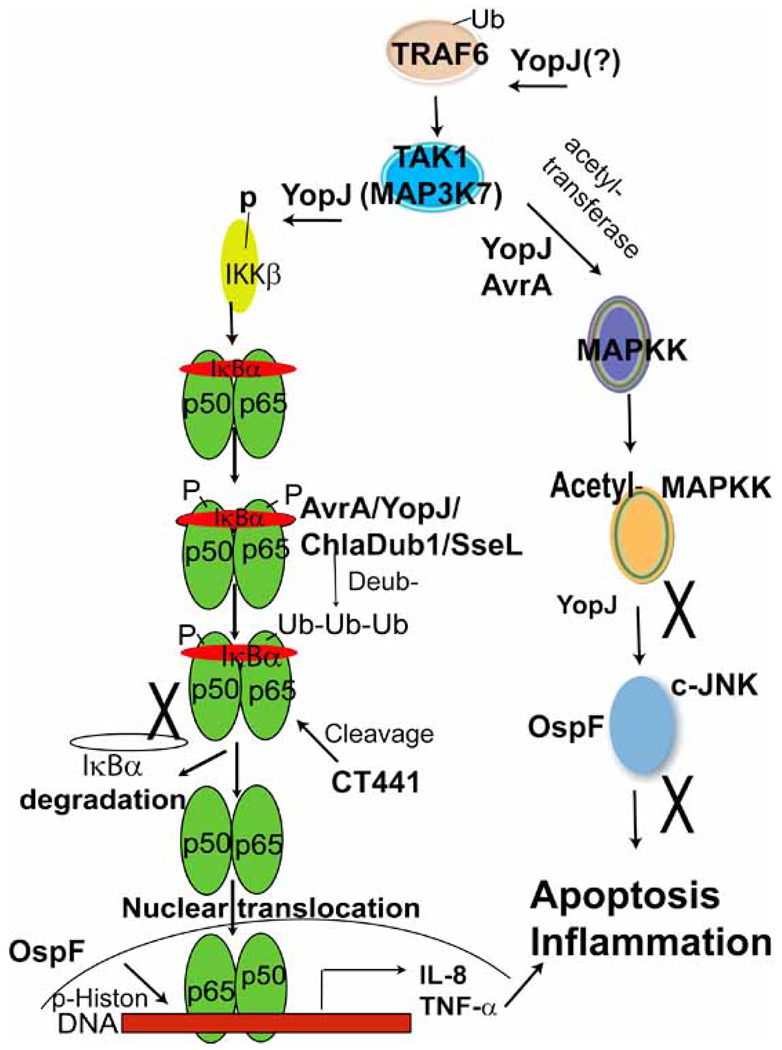
Bacterial pathogens evolve the capacities to prevent or reduce the upregulation of the inflammatory response, thus blocking the influx of phagocytic leukocytes, and/or to activate or augment apoptotic pathways in immunoregulatory cells such as macrophages—both events likely permissive for bacterial proliferation and dissemination [31, 42]. We note that the NF-κB and MAPKs pathways are the key inflammatory cell-signaling pathways involved in the bacterial-host interaction. Given the critical role NF-κB and MAPKs pathways in antimicrobial signaling, it is not surprising that bacterial proteins display a large repertoire of biochemical activities and modulate the function of crucial host regulatory molecules to surmount their effects (Fig. 2). The simplified MAPK signaling through sequential steps involves TRAF6, TAK1, MKK6, and MAPK. A parallel NF-κB pathway involves TRAF6, TAK1, IKKβ, IκBα phosphorylation, ubiquitination, NF-κB translocation, and resulting in activation of NF-κB. Bacterial TTSS effectors including Salmonella AvrA, SseL, Yersinia YopJ, and Chlamydia ChlaDub1/2 are reported as deubiquitinases to target the ubiquitination of IκBα, thus inhibiting the NF-κB activity. Chlamydia protein CT441 has a unique strategy to cleave the NF-κB subunit p65 and to attenuate the NF-κB activity. Additionally, Shigela OspF is able to modify phosphorylation of histone H3 at Ser10 increasing chromatin accessibility to transcription factor NF-κB on a subset of genes involved in immune responses. The MAPK pathway is modulated by AvrA/YopJ through acetylation. AvrA and YopJ inhibit the c-JNK pathway, attenuate the inflammatory responses, and interfere with the cell apoptotic process. OspF acts as a dually specific phosphatase that dephosphorylated MAPK, thus preventing histone H3 phosphorylation at Ser10 in a gene-specific way [69].
Fig. (2). Inactivation of NF-κB and MAPK pathways (simplified version) by the bacterial proteins.
As shown here, MAPK signaling through sequential steps involves TRAF6, TAK1, and MAPKK. A parallel NF-κB pathway involves TRAF6, TAK1, IKKβ, IκBα phosphorylation (P), ubiquitination (Ub), NF-κB translocation, activation of NF-κB transcription, and target gene expression. Bacterial TTSS effectors including Salmonella AvrA, SseL, Yersinia YopJ, and Chlamydia ChlaDub1/2 act as deubiquitinases to target the ubiquitination of IκBα, thus inhibiting the NF-κB activity. Chlamydia protein CT441 cleaves the NF-κB subunit p65 and attenuate the NF-κB activity. Additionally, Shigela OspF modifies phosphorylation of histone H3 at Ser10 increases chromatin accessibility to transcription factor NF-κB. Acting as a broad-specificity deubiquitinase, YopJ removes Ub from proteins, including TRAF6 and IκBα. MAPKK and IKKβ are activated by phosphorylation of serine or threonine residues in their activation loops. YopJ inactivates these pathways. The MAPK pathway is modulated by AvrA through acetylation. AvrA, YopJ, and OspF inhibit the c-JNK pathway, attenuate the inflammatory responses, and interfere with the cell apoptotic process.
Bacterial toxins are also known to interfere with the MAPK pathway. Anthrax lethal toxin, produced by the bacterium Bacillus anthracis, is the major cause of death in animals infected with anthrax. Lethal factor (LF) is shown as a protease that cleaves the amino terminus of MAPKK1 and MAPKK2. This cleavage inactivates MAPKK1 and inhibits the MAPK signal transduction pathway [80]. We also note that bacterial toxins exert their function by introducing covalent, non-reversible modifications of their target host cell proteins, whereas bacterial TTSS effectors mimick the function of host cell proteins [26]. Many eukaryotic proteins coordinate distinct signaling pathways in the cells by having multiple functions. Not surprisingly, many bacterial effectors discussed in this review also have multiple protease activities. These activities allow bacteria to modify different eukaryotic proteins and maximize bacterial effector’s ability to modulate cellular functions.
STRUCTURES OF THE ANTI-INFLAMMATORY BACTERIAL PROTEINS
Structure-based biochemical and enzymatic analysis allow us to propose a general mechanism for the anti-inflammatory bacterial proteins. In this section, the representative structures of the bacterial proteins are discussed.
1. OspF Family of Phosphothreonine Lyase
The OspF family of phosphothreonine lyase, including SpvC from Salmonella, irreversibly inactivates the dual-phosphorylated host MAPKs (pT-X-pY) through beta elimination. Recently, crystal structures of SpvC and its complex with a phosphopeptide substrate were determined [81]. SpvC adopts a unique fold of alpha/beta type. The disordered N terminus harbors a canonical D motif for MAPK substrate docking. The enzyme-substrate complex structure indicates that recognition of the phosphotyrosine followed by insertion of the threonine phosphate into an arginine pocket places the phosphothreonine into the enzyme active site. This requires the conformational flexibility of pT-X-pY, which suggests that p38 (pT-G-pY) is likely the preferred physiological substrate. Structure-based biochemical and enzymatic analysis indicate a general acid/base mechanism for beta elimination reaction catalyzed by the phosphothreonine lyase. The mechanism provides a structural understanding of MAPK inactivation by a family of pathogenic effectors conserved in plant and animal systems [81].
2. Cyclomodulin Cif is a Structural Member of the Cysteine Protease Superfamily
The TTSS effector Cif produced by EPEC and EHEC is able to block host eukaryotic cell-cycle progression [73, 82]. Cif is composed of a C-terminal active domain (residues 21– 282) and an exchangeable N-terminal translocation signal encoded by the first ∼20 amino acids [83]. A crystal structure of Cif reveals that Cif is a divergent member of the superfamily of enzymes including cysteine proteases, transglutaminases, and acetyltransferases that share a common catalytic triad. The putative catalytic triad of Cif consists of cysteine, histidine, and glutamine residues. These studies showed that these residues are essential for the ability of Cif to cause the cytopathic effect when introduced into cells and during pathogenic E. coli infection [82].
Interestedly, Cif shares structural homology with Pseudomonas syringae AvrPphB (PDB ID code 1ukf) with a Z-score of 4.0 and root-mean-square deviation (RMSD) of 3.8 over 88 residues [84]. AvrPphB is a member of a superfamily of related enzymes containing cysteine proteases, acetyltransferases, and transglutaminases. While there is considerable divergence across this superfamily in the overall fold, a core anti-parallel β-sheet and an N-terminal helix, which packs against the β-strands, are always present [82]. Mutation of the conserved active site residues abolishes the ability of Cif to block cell-cycle progression. However, irreversible cysteine protease inhibitors do not abolish the Cif cytopathic effect, suggesting that another enzymatic activity may underlie the biological activity of this virulence factor [82].
Structure of Cif homolog in Burkholderia pseudomallei (CHBP) reveals a papain-like fold and a conserved Cys-His-Gln catalytic triad despite the lack of primary sequence identity. For CHBP and Cif, only the putative catalytic Cys is susceptible to covalent modification by E-64, a specific inhibitor of papain-like cysteine proteases. Unlike papain-like enzymes where the S2 site is the major determinant of cleavage-site specificity, CHBP has a characteristic negatively charged pocket occupying surface areas corresponding to the S1/S1' site in papain-like proteases. The negative charge is provided by a conserved aspartate, and the pocket best fits an arginine, as revealed by molecular docking analysis. Mutation analysis establishes the essential role of the catalytic triad and the negatively charged pocket in inducing cell cycle arrest in host cells. Overall, these results demonstrate that bacterial pathogens have evolved a unique papain-like hydrolytic activity to block the normal host cell cycle progression [85].
The X-ray crystal structure studies further discover four Cif homologs encoded by different pathogenic or symbiotic bacteria isolated from vertebrates or invertebrates. Cif homologs from the enterobacteria Yersinia pseudotuberculosis, Photorhabdus luminescens, Photorhabdus asymbiotica, and the beta-proteobacterium Burkholderia pseudomallei all induce cytopathic effects identical to those observed with E. coli Cif. Although these Cif homologs are remarkably divergent in primary sequence, the catalytic triad is strictly conserved and is crucial for cell cycle arrest, cytoskeleton reorganization and CKIs accumulation. These results reveal that Cif proteins form a growing family of bacterial cyclo-modulins that interact with very distinct hosts including insects, nematodes and humans [76].
Recently, Crow et al. also report that X-ray crystal structure of Cif from Photorhabdus luminescens and Burk-holderia pseudomallei [85]. They found that both of these proteins adopt an overall fold similar to the papain subfamily of cysteine proteases, as originally identified in the structure of a truncated form of Cif from EPEC, despite sharing only limited sequence identity. The structure of an N-terminal region, referred to here as the 'tail-domain' (absent in the EPEC Cif structure), suggests a surface likely to be involved in host-cell substrate recognition. The conformation of the Cys-His-Gln catalytic triad is retained, and the essential cysteine is exposed to solvent and addressable by small molecule reagents [86].
3. Structural Requirements for Yersinia YopJ Inhibition of MAP Kinase Pathways
In the early study, the activity of a protein is predicted based on the secondary structure. Comparing the predicted secondary structure of YopJ with all of the known secondary structures derived from crystallographic data, 52]. Recent study on YopJ showed the structural requirements for YopJ inhibition of MAPK pathways [87]. YopJ uses the unique activity of Ser/Thr acetylation to inhibit the activation of the MAPKK and prevent activation by phosphorylation. YopJ is also able to block yeast MAPK signaling pathways using this mechanism. Hao et al. performed a genetic screen to isolate mutants in the yeast MAPKK, Pbs2, that suppress YopJ inhibition [87]. They found that one suppressor contains a mutation in a conserved tyrosine residue and bypasses YopJ inhibition by increasing the basal activity of Pbs2. Mutations on the hydrophobic face of the conserved G alpha-helix in the kinase domain prevent both binding and acetylation by YopJ. Corresponding mutants in human MAPKKs showed that they are conserved not only structurally, but also functionally. These studies reveal a conserved binding site found on the superfamily of MAPKKs while providing insight into the molecular interactions required for YopJ inhibition [87].
4. Structural Basis for Chlamydia protease CPAF
CPAF degrades host proteins, enabling Chlamydia to evade host defenses and replicate. Using a computational approach to search the protein databank for structures that are compatible with the CPAF amino acid sequence, Chen et al. reveal that CPAF possesses a fold similar to that of the catalytic domains of the tricorn protease from Thermoplasma acidophilum, and that CPAF residues H105, S499, and E558 are structurally analogous to the tricorn protease catalytic triad residues H746, S965, and D1023 [88]. Substitution of these putative CPAF catalytic residues blocked CPAF from degrading substrates in vitro, while the wild type and a non-catalytic control mutant of CPAF remained cleavage-competent. Substrate cleavage is also correlated with processing of CPAF into N-terminal (CPAFn) and C-terminal (CPAFc) fragments, suggesting that these putative catalytic residues may also be required for CPAF maturation [88].
Studies further demonstrate that CPAF degrades many host molecules and plays a major role in Chlamydia pathogenesis. CPAF is a homodimer of the catalytic domains, each of which comprises two distinct subunits [89]. Dormancy of the CPAF zymogen is maintained by an internal inhibitory segment that binds the CPAF active site and blocks its homodimerization. CPAF activation is initiated by trans-autocatalytic cleavage, which induces homodimerization and conformational changes that assemble the catalytic triad. This assembly leads to two autocatalytic cleavages and removal of the inhibitory segment, enabling full CPAF activity. CPAF is covalently bound and inhibited by the proteasome inhibitor lactacystin [89].
POTENTIAL APPLICATION OF BACTERIAL COMPONENTS IN ANTI-INFLAMAMTION THERA-PEUTICAL INTERVENTION
The studies on the action of bacterial proteins uncover the new facets of bacterial-host interactions. Bacterial proteins display a large repertoire of biochemical activities and modulate the function of crucial host regulatory molecules such as NF-κB. Because microbial pathogens have developed various strategies for modulating the host’s innate and adaptive immunity, bacterial proteins can be viewed as potential therapeutic agents for human immunopathological diseases.
1. Staphylococcus aureus Eap and Its Therapeutic Applications
S. aureus may gain access to other capillaries, arterioles and venules, attach to the vessel wall and start disseminating from focal sites of infection, after passing through the endothelial cell layer and/or the extracellular matrix stroma [90]. S. aureus Eap may inhibit vascular cell proliferation and angiogenesis possibly via direct interference with agonist-stimulated endothelial functions. Eap also interferes with T-cells function shifting towards a Th2-cell response and reducing delayed-type hypersensitivity reactions. Thus, Eap as a potent anti-inflammatory and anti-angiogenic factor is a promising candidate for new therapeutic modalities in inflammatory vascular diseases [90].
Severe inflammatory response syndrome occurs when the body's response to an overwhelming infection becomes uncontrolled. The immune response means to clear the pathogen and its toxins actually cause damage to the host's own tissues when too many immune signals are released. Thus, the immune response must be carefully controlled to prevent damage to the host. Due to the lack of influence on the course of infection, isolated Eap could be used as a lead substance in designing new peptides or non-peptidic molecules that could serve as anti-inflammatory drugs without the possible side effect of promoting S. aureus infection. Non-regulated adhesiveness of leukocytes, of circulating tumor cells and/or endothelial cells results in uncontrolled cellular extravasation and causes atherosclerosis and rheumatoid arthritis, or leads to tumor metastasis. In such pathological processes, Eap-derived molecules could be devised as ICAM-1 blocking agents to achieve an antiadhesive, anti-inflammatory potential during therapeutic interventions [91].
2. Staphylococcus aureus Eap Inhibits Inflammation in Sclerosis
Multiple sclerosis (MS) is a devastating inflammatory disorder of the central nervous system. A major hallmark of MS is the infiltration of T cells reactive against myelin components [92]. T cell infiltration is mediated by the interaction of integrins of the beta1 and beta2 family expressed by lymphocytes with their endothelial counter-receptors, vascular cell adhesion molecule 1 and ICAM-1. The ability of Eap to modulate T-cell and prevent leukocyte extravasation [3, 91] suggested that that Eap may be used to treat multiple sclerosis [93]. It is reported that Eap inhibits experimental autoimmune encephalomyelitis (EAE) in mice. Eap reduced adhesion of peripheral blood T cells to immobilized ICAM-1 as well as their adhesion and transmigration of TNF-activated human endothelium under static and shear flow conditions. In a delayed-type hypersensitivity model, both T cell infiltration and the corresponding tissue edema were significantly reduced by Eap. In addition, Eap administration prevented the development of EAE and markedly decreased infiltration of inflammatory cells into the CNS. Strikingly, intervention with Eap after the onset of EAE suppressed the disease. Collectively, these findings indicate that Eap represents an attractive treatment for autoimmune neuroinflammatory disorders such as MS [93]. Thus, the specific interactions with ICAM-1 and extracellular matrix proteins render Eap a potent anti-inflammatory factor, which may serve as a new therapeutic substance to block leukocyte extravasation in patients with hyperinflammatory pathologies [3].
3. The Anti-Inflammatory Activities of Staphylococcus aureus Chemotaxis Inhibitory Protein in Systemic Amyloidosis, Alzheimer's, and Prion Disease
Chemotaxis inhibitory protein from S. aureus(CHIPS) impaires the neutrophil responses to FPR-like1 (FPRL1) agonists [2]. Interestingly, FPRL1 is used by at least three amyloidogenic ligands: serum amyloid A [94], the 42-aa form of β amyloid (Aβ 1–42) [95], and the prion protein fragment PrP106–126 [96]. The activation of FPRL1 by Aβ 1–42 or PrP106–126 may be responsible for accumulation and activation of mononuclear phagocytes) as well as fibrillar formation that is associated with the pathogenesis of Alzheimer’s disease and prion diseases [97]. The Alzheimer’s patient will benefit from a combination of different drugs and the development of FPRL1-specific antagonists may have promising therapeutic potential in retarding the progression of the disease [2]. As a novel described FPRL1 antagonist, chemotaxis inhibitory protein of S. aureus might lead to the development of therapeutic agents in FPRL1-mediated inflammatory components of diseases such as systemic amyloidosis, Alzheimer's, and prion disease.
4. Anti-Inflammatory Components of Y. pseudotuberculosis and Salmonella in Colitis Treatment
Elevated levels of activated NF-κB and a number of NF-κB-induced proinflammatory mediators have been implicated in inflammatory bowel diseases (IBD) and are found in the intestinal mucosa of IBD patients. Increased intestinal permeability is also found in IBD patients [98, 99]. Inhibitors of the NF-κB pathway are viewed as potential therapeutic agents. Because bacteria have developed various strategies for modulating the host’s innate and adaptive immunity, bacterial proteins can be applied to the development of drugs for IBD treatment.
Study shows that anti-inflammatory components produced by Y. pseudotuberculosis are effective in reducing experimental colitis in mice [100]. Rectal instillation of trinitrobenzene sulfonic acid (TNBS) induces acute colitis in the mouse. The efficacy of Yersinia pseudotuberculosis anti-inflammatory components in preventing TNBS-triggered colitis was tested. Animals were orally inoculated with virulence-attenuated Yersinia cells prior to TNBS administration. Under these experimental conditions, colonic lesions and tumor necrosis factor alpha mRNA levels were significantly reduced [100]. These observations suggest that anti-inflammatory bacterial components might be potential drugs for the treatment of IBD.
Salmonella AvrA protein is translocated into host cells by a type three secretion system. Current studies on AvrA revealed that 1) AvrA protein expression is finely tuned in the Salmonella enterica serovars; 2) Physiologically, AvrA inhibits the activity of NF-κB, a key regulator of inflammation and host; 3) One of the mechanisms on how AvrA overcomes the host inflammatory responses is to act as a deubiquitinase, thus resulting in stabilization of IκBα and β-catenin, the negative regulators of proinflammatory NF-κB pathway; and 4) AvrA must alter many very effective host defense mechanisms to attenuate the inflammatory response, thus enhancing infection in the host. Specifically, we have significant data to support the proposed hypothesis that AvrA can attenuate NF-κB activity and increase epithelial integrity [5, 49]. Because inhibition of the NF-κB pathway and maintenance of epithelial cell integrity are viewed as potential therapeutic strategies, the effects of AvrA on both anti-inflammation and the tight junction structure make AvrA a promising candidate for IBD therapy.
5. Development of Vaccines/Therapies Against Pathogens
An important virulence strategy evolved by bacterial pathogens to overcome host defenses is the modulation of host cell death. Understanding the mechanism of the anti-inflammatory bacterial proteins has novel implications for the development of vaccines/therapies against bacterial pathogens. For example, engineered cytotoxic Y. pestis strain can induce very rapid, effective and long-lasting protection against bubonic and pneumonic plague. Y. pestis endowed with increased cytotoxicity is avirulent in a bubonic plague model and induces rapid protection against pneumonic plague [101]. Y. pestis restricted capacity to induce cell death in macrophages due to ineffective translocation of kYopJ, as opposed to the readily translocated YopP, the YopJ homologue of the enteropathogen Y. enterocolitica Oratio8. When YopJ was replaced with YopP, the YopP-expressing Y. pestis strain exhibited high cytotoxic activity against macrophages. Following subcutaneous infection, this strain had reduced ability to colonize internal organs, was unable to induce septicemia and exhibited reduction in virulence. Animals were protected against septicemic or primary pneumonic plague three days after subcutaneous administration of this strain. These findings indicate that an inverse relationship exists between the cytotoxic potential of Y. pestis and its virulence following subcutaneous infection. These observations have implications for the development of vaccines/therapies against Y. pestis [101].
CONCLUSION AND FUTURE STUDIES ON THE BACTERIAL PROTEINS IN MODULATING HOST INFLAMMATION
For a bacterium to be a successful vertebrate pathogen, it must overcome or alter many very effective host defense mechanisms [102]. Recently, an array of bacterial proteins has been identified and characterized in their functions and mechanisms to inhibit the inflammatory responses in the host. Here, we focus on the anti-inflammatory TTSS effectors of Salmonella, EPEC, Chlamydia, and Yersinia, Staphylococcus aureus extracellular adherence protein, and Chlamydia proteins. The bacterial proteins we discussed in this review are essential for the death of infected host cells and can block host proinflammatory responses by inhibiting both the NF-κB and MAPK pathways. These pathways are important for evasion of the host immune response and aid in establishing a systemic infection. The effector AvrA we are exploiting also uses remarkable strategies to regulate eukaryotic signaling pathways by modulation both NF-κB and β-catenin pathways.
Acute enteritis caused by food borne bacteria is the most common cause of infectious gastroenteritis in the United States. These disorders are a public health concern in the developed nations, and are a great cause of morbidity and mortality in developing countries. Therefore, it is important to understand how the bacterial effectors work. The studies on the action of bacterial effectors will discover the new facets of bacteria-host interaction and will lead to the development of new therapeutic drugs against important human pathogens.
Chronic inflammatory diseases are known to be involved in the misregulation of the NF-κB and MAKP pathway. Anti-inflammaory bacterila proteins also target these similar signaling pathways. Therefore, understanding bacterial proteins will not only lead to insights into the pathophysiology of inflmmation and infection, to the new anti-microbioal strategies, but also provide the plateform for the design therapies in chronic inflammatory diseases. The therapeutic implications of bacterial proteins in anti-inflammation will be an exciting and promising field of translational studies.
ACKNOWLEDGEMENT
I thank all present and former members of my group for their contribution to our own work on the bacterial-epithelial interactions and Salmonella TTSS effector AvrA in intestinal inflammation.
GRANTS
This work was supported by NIDDK KO1 DK075386 and American Society Research Scholar Grant RSG-09-075-01-MBC to J. S.
ABBREVIATION
- Cif
Cycle inhibiting factor
- CoA
Acetyl-coenzyme A
- CPAF
Chlamydial protease– or proteasome–like activity factor
- EAE
Experimental autoimmune encephalomyelitis
- Eap
Extracellular adherence protein
- EPEC
Enteropathogenic Escherichia coli
- FPR
Formyl peptide receptor
- FKHRL1
Forkhead transcription factor
- GSK-3
Glycogen synthase kinase 3
- IBD
Inflammatory bowel diseases
- ICAM
Intercellular adhesion molecule
- IκBα
Inhibitor of NF-κB
- IKK
IκB kinase
- JNK
JUN-NH2-terminal kinase
- MAPK
Mitogen-activated protein kinase
- MAKK
MAK kinase kinase
- MCP-1
Monocyte chemoattractant protein 1
- NF-κB
Nuclear factor κB
- SseL
Salmonella secreted factor L
- Tsp
Tail-specific protease
- TRAF6
Tumor necrosis factor receptor-associated factor-6
- TNBS
Trinitrobenzene sulfonic acid
- Yops
Yersinia outer proteins
REFERENCES
- 1.Weiss U. Inflammation. Nature. 2008;454(7203):427. doi: 10.1038/454427a. [DOI] [PubMed] [Google Scholar]
- 2.Prat C, Bestebroer J, de Haas CJ, van Strijp JA, van Kessel KP. A new staphylococcal anti-inflammatory protein that antagonizes the formyl peptide receptor-like 1. J. Immunol. 2006;177(11):8017–8026. doi: 10.4049/jimmunol.177.11.8017. [DOI] [PubMed] [Google Scholar]
- 3.Chavakis T, Hussain M, Kanse SM, Peters G, Bretzel RG, Flock JI, Herrmann M, Preissner KT. Staphylococcus aureus extracellular adherence protein serves as anti-inflammatory factor by inhibiting the recruitment of host leukocytes. Nat Med. 2002;8(7):687–693. doi: 10.1038/nm728. [DOI] [PubMed] [Google Scholar]
- 4.Peschel A. How do bacteria resist human antimicrobial peptides? Trends Microbiol. 2002;10(4):179–186. doi: 10.1016/s0966-842x(02)02333-8. [DOI] [PubMed] [Google Scholar]
- 5.Ye Z, Petrof EO, Boone D, Claud EC, Sun J. Salmonella effector AvrA regulation of colonic epithelial cell inflammation by deubiquitination. Am. J. Pathol. 2007;171(3):882–892. doi: 10.2353/ajpath.2007.070220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collier-Hyams LS, Zeng H, Sun J, Tomlinson AD, Bao ZQ, Chen H, Madara JL, Orth K, Neish AS. Cutting edge: Salmonella AvrA effector inhibits the key proinflammatory, anti-apoptotic NF-kappa B pathway. J. Immunol. 2002;169(6):2846–2850. doi: 10.4049/jimmunol.169.6.2846. [DOI] [PubMed] [Google Scholar]
- 7.Lad SP, Li J, da Silva Correia J, Pan Q, Gadwal S, Ulevitch RJ, Li E. Cleavage of p65/RelA of the NF-kappaB pathway by Chlamydia. Proc. Natl. Acad. Sci. USA. 2007;104(8):2933–2938. doi: 10.1073/pnas.0608393104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454(7203):428–435. doi: 10.1038/nature07201. [DOI] [PubMed] [Google Scholar]
- 9.Mattoo S, Lee YM, Dixon JE. Interactions of bacterial effector proteins with host proteins. Curr. Opin. Immunol. 2007;19(4):392–401. doi: 10.1016/j.coi.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 10.Schlumberger MC, Hardt WD. Salmonella type III secretion effectors: pulling the host cell's strings. Curr. Opin. Microbiol. 2006;9(1):46–54. doi: 10.1016/j.mib.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 11.Patel JC, Rossanese OW, Galan JE. The functional interface between Salmonella and its host cell: opportunities for therapeutic intervention. Trends Pharmacol. Sci. 2005;26(11):564–570. doi: 10.1016/j.tips.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 12.Patel JC, Hueffer K, Lam TT, Galan JE. Diversification of a Salmonella virulence protein function by ubiquitin-dependent differential localization. Cell. 2009;137(2):283–294. doi: 10.1016/j.cell.2009.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galan JE. Salmonella interactions with host cells: type III secretion at work. Annu. Rev. Cell. Dev. Biol. 2001;17:53–86. doi: 10.1146/annurev.cellbio.17.1.53. [DOI] [PubMed] [Google Scholar]
- 14.Mota LJ, Cornelis GR. The bacterial injection kit: type III secretion systems. Ann. Med. 2005;37(4):234–249. doi: 10.1080/07853890510037329. [DOI] [PubMed] [Google Scholar]
- 15.Coburn B, Sekirov I, Finlay BB. Type III secretion systems and disease. Clin. Microbiol. Rev. 2007;20(4):535–549. doi: 10.1128/CMR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coburn B, Li Y, Owen D, Vallance BA, Finlay BB. Salmonella enterica serovar Typhimurium pathogenicity island 2 is necessary for complete virulence in a mouse model of infectious enterocolitis. Infect. Immun. 2005;73(6):3219–3227. doi: 10.1128/IAI.73.6.3219-3227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Staskawicz BJ, Mudgett MB, Dangl JL, Galan JE. Common and contrasting themes of plant and animal diseases. Science. 2001;292(5525):2285–2289. doi: 10.1126/science.1062013. [DOI] [PubMed] [Google Scholar]
- 18.Lara-Tejero M, Galan JE. Salmonella enterica serovar typhimurium pathogenicity island 1-encoded type III secretion system translocases mediate intimate attachment to nonphagocytic cells. Infect. Immun. 2009;77(7):2635–2642. doi: 10.1128/IAI.00077-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galan JE. SnapShot: effector proteins of type III secretion systems. Cell. 2007;130(1):192. doi: 10.1016/j.cell.2007.06.042. [DOI] [PubMed] [Google Scholar]
- 20.Lowy FD. Staphylococcus aureus infections. N. Engl. J. Med. 1998;339(8):520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 21.Lad SP, Yang G, Scott DA, Wang G, Nair P, Mathison J, Reddy VS, Li E. Chlamydial CT441 is a PDZ domain-containing tail-specific protease that interferes with the NF-kappaB pathway of immune response. J. Bacteriol. 2007;189(18):6619–6625. doi: 10.1128/JB.00429-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, Koonin EV, Davis RW. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282(5389):754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 23.Zhong G, Fan P, Ji H, Dong F, Huang Y. Identification of a chlamydial protease-like activity factor responsible for the degradation of host transcription factors. J. Exp. Med. 2001;193(8):935–942. doi: 10.1084/jem.193.8.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kubori T, Galan JE. Salmonella type III secretion-associated protein InvE controls translocation of effector proteins into host cells. J. Bacteriol. 2002;184(17):4699–4708. doi: 10.1128/JB.184.17.4699-4708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garner MJ, Hayward RD, Koronakis V. The Salmonella pathogenicity island 1 secretion system directs cellular cholesterol redistribution during mammalian cell entry and intracellular trafficking. Cell. Microbiol. 2002;4(3):153–165. doi: 10.1046/j.1462-5822.2002.00181.x. [DOI] [PubMed] [Google Scholar]
- 26.Galan JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444(7119):567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- 27.Salcedo SP, Holden DW. Bacterial interactions with the eukaryotic secretory pathway. Curr. Opin. Microbiol. 2005;8(1):92–98. doi: 10.1016/j.mib.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 28.Hardt WD, Galan JE. A secreted Salmonella protein with homology to an avirulence determinant of plant pathogenic bacteria. Proc. Natl. Acad. Sci. USA. 1997;94(18):9887–9892. doi: 10.1073/pnas.94.18.9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ben-Barak Z, Streckel W, Yaron S, Cohen S, Prager R, Tschape H. The expression of the virulence-associated effector protein gene avrA is dependent on a Salmonella enterica-specific regulatory function. Int. J. Med. Microbiol. 2006;296(1):25–38. doi: 10.1016/j.ijmm.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 30.Streckel W, Wolff AC, Prager R, Tietze E, Tschape H. Expression profiles of effector proteins SopB, SopD1, SopE1, and AvrA differ with systemic, enteric, and epidemic strains of Salmonella enterica. Mol. Nutr. Food Res. 2004;48(7):496–503. doi: 10.1002/mnfr.200400035. [DOI] [PubMed] [Google Scholar]
- 31.Jones RM, Wu H, Wentworth C, Luo L, Collier-Hyams L, Neish AS. Salmonella AvrA Coordinates Suppression of Host Immune and Apoptotic Defenses via JNK Pathway Blockade. Cell. Host Microbe. 2008;3(4):233–244. doi: 10.1016/j.chom.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 32.Perkins ND. Integrating cell-signalling pathways with NF-kappaB IKK function. Nat. Rev Mol. Cell Biol. 2007;8(1):49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 33.Gilmore TD. Introduction to NF-kappaB: players, pathways, perspectives. Oncogene. 2006;25(51):6680–6684. doi: 10.1038/sj.onc.1209954. [DOI] [PubMed] [Google Scholar]
- 34.Brasier AR. The NF-kappaB regulatory network. Cardiovasc. Toxicol. 2006;6(2):111–130. doi: 10.1385/ct:6:2:111. [DOI] [PubMed] [Google Scholar]
- 35.Gilmore TD, Herscovitch M. Inhibitors of NF-kappaB signaling: 785 and counting. Oncogene. 2006;25(51):6887–6899. doi: 10.1038/sj.onc.1209982. [DOI] [PubMed] [Google Scholar]
- 36.Bonizzi G, Karin M. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004;25(6):280–288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 37.Deng J, Miller SA, Wang HY, Xia W, Wen Y, Zhou BP, Li Y, Lin SY, Hung MC. beta-catenin interacts with and inhibits NF-kappa B in human colon and breast cancer. Cancer Cell. 2002;2(4):323–334. doi: 10.1016/s1535-6108(02)00154-x. [DOI] [PubMed] [Google Scholar]
- 38.Deng J, Xia W, Miller SA, Wen Y, Wang HY, Hung MC. Crossregulation of NF-kappaB by the APC/GSK-3beta/beta-catenin pathway. Mol. Carcinog. 2004;39(3):139–146. doi: 10.1002/mc.10169. [DOI] [PubMed] [Google Scholar]
- 39.Sun J, Hobert ME, Duan Y, Rao AS, He TC, Chang EB, Madara JL. Crosstalk between NF-kappaB and beta-catenin pathways in bacterial-colonized intestinal epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2005;289(1):G129–G137. doi: 10.1152/ajpgi.00515.2004. [DOI] [PubMed] [Google Scholar]
- 40.Morin PJ. beta-catenin signaling and cancer. Bioessays. 1999;21(12):1021–1030. doi: 10.1002/(SICI)1521-1878(199912)22:1<1021::AID-BIES6>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 41.Roux PP, Blenis J. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol. Mol. Biol. Rev. 2004;68(2):320–344. doi: 10.1128/MMBR.68.2.320-344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weston CR, Davis RJ. The JNK signal transduction pathway. Curr. Opin. Cell Biol. 2007;19(2):142–149. doi: 10.1016/j.ceb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 43.Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, Tsui C, Hurley P, Chien M, Chai S, Hitotsumatsu O, McNally E, Pickart C, Ma A. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat. Iimmunol. 2004;5(10):1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 44.Hitotsumatsu O, Ahmad RC, Tavares R, Wang M, Philpott D, Turer EE, Lee BL, Shiffin N, Advincula R, Malynn BA, Werts C, Ma A. The ubiquitin-editing enzyme A20 restricts nucleotide-binding oligomerization domain containing 2-triggered signals. Immunity. 2008;28(3):381–390. doi: 10.1016/j.immuni.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, Ma A, Koonin EV, Dixit VM. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430(7000):694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 46.Sun J, Hobert ME, Rao AS, Neish AS, Madara JL. Bacterial activation of beta-catenin signaling in human epithelia. Am. J. Physiol. Gastrointest. Liver Physiol. 2004;287(1):G220–G227. doi: 10.1152/ajpgi.00498.2003. [DOI] [PubMed] [Google Scholar]
- 47.Duan Y, Liao AP, Kuppireddi S, Ye Z, Ciancio MJ, Sun J. beta-Catenin activity negatively regulates bacteria-induced inflammation. Lab. Invest. 2007;87(6):613–624. doi: 10.1038/labinvest.3700545. [DOI] [PubMed] [Google Scholar]
- 48.Boyle EC, Brown NF, Finlay BB. Salmonella enterica serovar Typhimurium effectors SopB, SopE, SopE2 and SipA disrupt tight junction structure and function. Cell Microbiol. 2006;8(12):1946–1957. doi: 10.1111/j.1462-5822.2006.00762.x. [DOI] [PubMed] [Google Scholar]
- 49.Liao AP, Petrof EO, Kuppireddi S, Zhao Y, Xia Y, Claud EC, Sun J. Salmonella type III effector AvrA stabilizes cell tight junctions to inhibit inflammation in intestinal epithelial cells. PLoS ONE. 2008;3(6):e2369. doi: 10.1371/journal.pone.0002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Le Negrate G, Faustin B, Welsh K, Loeffler M, Krajewska M, Hasegawa P, Mukherjee S, Orth K, Krajewski S, Godzik A, Guiney DG, Reed JC. Salmonella secreted factor L deubiquitinase of Salmonella typhimurium inhibits NF-kappaB, suppresses IkappaBalpha ubiquitination and modulates innate immune responses. J. Immunol. 2008;180(7):5045–5056. doi: 10.4049/jimmunol.180.7.5045. [DOI] [PubMed] [Google Scholar]
- 51.Orth K. Function of the Yersinia effector YopJ. Curr. Opin. Microbiol. 2002;5(1):38–43. doi: 10.1016/s1369-5274(02)00283-7. [DOI] [PubMed] [Google Scholar]
- 52.Orth K, Xu Z, Mudgett MB, Bao ZQ, Palmer LE, Bliska JB, Mangel WF, Staskawicz B, Dixon JE. Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science. 2000;290(5496):1594–1597. doi: 10.1126/science.290.5496.1594. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Y, Ting AT, Marcu KB, Bliska JB. Inhibition of MAPK and NF-kappa B pathways is necessary for rapid apoptosis in macrophages infected with Yersinia. J. Immunol. 2005;174(12):7939–7949. doi: 10.4049/jimmunol.174.12.7939. [DOI] [PubMed] [Google Scholar]
- 54.Mukherjee S, Keitany G, Li Y, Wang Y, Ball HL, Goldsmith EJ, Orth K. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science. 2006;312(5777):1211–1214. doi: 10.1126/science.1126867. [DOI] [PubMed] [Google Scholar]
- 55.Mukherjee S, Hao YH, Orth K. A newly discovered post-translational modification--the acetylation of serine and threonine residues. Trends Biochem. Sci. 2007;32(5):210–216. doi: 10.1016/j.tibs.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 56.Mittal R, Peak-Chew SY, McMahon HT. Acetylation of MEK2 and I kappa B kinase (IKK) activation loop residues by YopJ inhibits signaling. Proc. Natl. Acad. Sci. USA. 2006;103(49):18574–18579. doi: 10.1073/pnas.0608995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou H, Monack DM, Kayagaki N, Wertz I, Yin J, Wolf B, Dixit VM. Yersinia virulence factor YopJ acts as a deubiquitinase to inhibit NF-kappa B activation. J. Exp. Med. 2005;202(10):1327–1332. doi: 10.1084/jem.20051194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sweet CR, Conlon J, Golenbock DT, Goguen J, Silverman N. YopJ targets TRAF proteins to inhibit TLR-mediated NF-kappaB, MAPK and IRF3 signal transduction. Cell Microbiol. 2007;9(11):2700–2715. doi: 10.1111/j.1462-5822.2007.00990.x. [DOI] [PubMed] [Google Scholar]
- 59.Bhoj VG, Chen ZJ. Ubiquitylation in innate and adaptive immunity. Nature. 2009;458(7237):430–437. doi: 10.1038/nature07959. [DOI] [PubMed] [Google Scholar]
- 60.Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature. 2003;424(6950):793–796. doi: 10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- 61.Sauvonnet N, Lambermont I, van der Bruggen P, Cornelis GR. YopH prevents monocyte chemoattractant protein 1 expression in macrophages and T-cell proliferation through inactivation of the phosphatidylinositol 3-kinase pathway. Mol. Microbiol. 2002;45(3):805–815. doi: 10.1046/j.1365-2958.2002.03053.x. [DOI] [PubMed] [Google Scholar]
- 62.Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378(6559):785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 63.Vanhaesebroeck B, Alessi DR. The PI3K-PDK1 connection: more than just a road to PKB. Biochem. J. 2000;346(Pt 3):561–576. [PMC free article] [PubMed] [Google Scholar]
- 64.Gerke C, Falkow S, Chien YH. The adaptor molecules LAT and SLP-76 are specifically targeted by Yersinia to inhibit T cell activation. J. Exp. Med. 2005;201(3):361–371. doi: 10.1084/jem.20041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ranganathan R, Ross EM. PDZ domain proteins: scaffolds for signaling complexes. Curr. Biol. 1997;7(12):R770–R773. doi: 10.1016/s0960-9822(06)00401-5. [DOI] [PubMed] [Google Scholar]
- 66.Misaghi S, Balsara ZR, Catic A, Spooner E, Ploegh HL, Starnbach MN. Chlamydia trachomatis-derived deubiquitinating enzymes in mammalian cells during infection. Mol. Microbiol. 2006;61(1):142–150. doi: 10.1111/j.1365-2958.2006.05199.x. [DOI] [PubMed] [Google Scholar]
- 67.Le Negrate G, Krieg A, Faustin B, Loeffler M, Godzik A, Krajewski S, Reed JC. ChlaDub1 of Chlamydia trachomatis suppresses NF-kappaB activation and inhibits IkappaBalpha ubiquitination and degradation. Cell Microbiol. 2008;10(9):1879–1892. doi: 10.1111/j.1462-5822.2008.01178.x. [DOI] [PubMed] [Google Scholar]
- 68.Dong F, Pirbhai M, Zhong Y, Zhong G. Cleavage-dependent activation of a chlamydia-secreted protease. Mol. Microbiol. 2004;52(5):1487–1494. doi: 10.1111/j.1365-2958.2004.04072.x. [DOI] [PubMed] [Google Scholar]
- 69.Arbibe L, Kim DW, Batsche E, Pedron T, Mateescu B, Muchardt C, Parsot C, Sansonetti PJ. An injected bacterial effector targets chromatin access for transcription factor NF-kappaB to alter transcription of host genes involved in immune responses. Nat. Immunol. 2007;8(1):47–56. doi: 10.1038/ni1423. [DOI] [PubMed] [Google Scholar]
- 70.Li H, Xu H, Zhou Y, Zhang J, Long C, Li S, Chen S, Zhou JM, Shao F. The phosphothreonine lyase activity of a bacterial type III effector family. Science. 2007;315(5814):1000–1003. doi: 10.1126/science.1138960. [DOI] [PubMed] [Google Scholar]
- 71.Yuk MH, Harvill ET, Cotter PA, Miller JF. Modulation of host immune responses, induction of apoptosis and inhibition of NF-kappaB activation by the Bordetella type III secretion system. Mol. Microbiol. 2000;35(5):991–1004. doi: 10.1046/j.1365-2958.2000.01785.x. [DOI] [PubMed] [Google Scholar]
- 72.Abramson T, Kedem H, Relman DA. Modulation of the NF-kappaB pathway by Bordetella pertussis filamentous hemagglutinin. PLoS ONE. 2008;3(11):e3825. doi: 10.1371/journal.pone.0003825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marches O, Ledger TN, Boury M, Ohara M, Tu X, Goffaux F, Mainil J, Rosenshine I, Sugai M, De Rycke J, Oswald E. Enteropathogenic and enterohaemorrhagic Escherichia coli deliver a novel effector called Cif, which blocks cell cycle G2/M transition. Mol. Microbiol. 2003;50(5):1553–1567. doi: 10.1046/j.1365-2958.2003.03821.x. [DOI] [PubMed] [Google Scholar]
- 74.Samba-Louaka A, Nougayrede JP, Watrin C, Jubelin G, Oswald E, Taieb F. Bacterial cyclomodulin Cif blocks the host cell cycle by stabilizing the cyclin-dependent kinase inhibitors p21 and p27. Cell Microbiol. 2008;10(12):2496–2508. doi: 10.1111/j.1462-5822.2008.01224.x. [DOI] [PubMed] [Google Scholar]
- 75.Nougayrede JP, Boury M, Tasca C, Marches O, Milon A, Oswald E, De Rycke J. Type III secretion-dependent cell cycle block caused in HeLa cells by enteropathogenic Escherichia coli O103. Infect. Iimmun. 2001;69(11):6785–6795. doi: 10.1128/IAI.69.11.6785-6795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jubelin G, Chavez CV, Taieb F, Banfield MJ, Samba-Louaka A, Nobe R, Nougayrede JP, Zumbihl R, Givaudan A, Escoubas JM, Oswald E. Cycle inhibiting factors (CIFs) are a growing family of functional cyclomodulins present in invertebrate and mammal bacterial pathogens. PLoS ONE. 2009;4(3):e4855. doi: 10.1371/journal.pone.0004855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nougayrede JP, Taieb F, De Rycke J, Oswald E. Cyclomodulins: bacterial effectors that modulate the eukaryotic cell cycle. Trends Microbiol. 2005;13(3):103–110. doi: 10.1016/j.tim.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 78.Haggar A, Shannon O, Norrby-Teglund A, Flock JI. Dual effects of extracellular adherence protein from Staphylococcus aureus on peripheral blood mononuclear cells. J. Infect. Dis. 2005;192(2):210–217. doi: 10.1086/430948. [DOI] [PubMed] [Google Scholar]
- 79.Scriba TJ, Sierro S, Brown EL, Phillips RE, Sewell AK, Massey RC. The Staphyloccous aureus Eap protein activates expression of proinflammatory cytokines. Infect. Immun. 2008;76(5):2164–2168. doi: 10.1128/IAI.01699-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Duesbery NS, Webb CP, Leppla SH, Gordon VM, Klimpel KR, Copeland TD, Ahn NG, Oskarsson MK, Fukasawa K, Paull KD, Vande Woude GF. Proteolytic inactivation of MAP-kinase-kinase by anthrax lethal factor. Science. 1998;280(5364):734–737. doi: 10.1126/science.280.5364.734. [DOI] [PubMed] [Google Scholar]
- 81.Zhu Y, Li H, Long C, Hu L, Xu H, Liu L, Chen S, Wang DC, Shao F. Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase. Mol. Cell. 2007;28(5):899–913. doi: 10.1016/j.molcel.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 82.Hsu Y, Jubelin G, Taieb F, Nougayrede JP, Oswald E, Stebbins CE. Structure of the cyclomodulin Cif from pathogenic Escherichia coli. J. Mol. Biol. 2008;384(2):465–477. doi: 10.1016/j.jmb.2008.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Charpentier X, Oswald E. Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector Cif, using TEM-1 beta-lactamase as a new fluorescence-based reporter. J. Bacteriol. 2004;186(16):5486–5495. doi: 10.1128/JB.186.16.5486-5495.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 1995;20(11):478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 85.Yao Q, Cui J, Zhu Y, Wang G, Hu L, Long C, Cao R, Liu X, Huang N, Chen S, Liu L, Shao F. A bacterial type III effector family uses the papain-like hydrolytic activity to arrest the host cell cycle. Proc. Natl. Acad. Sci. USA. 2009;106(10):3716–3721. doi: 10.1073/pnas.0900212106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Crow A, Race PR, Jubelin G, Varela Chavez C, Escoubas JM, Oswald E, Banfield MJ. Crystal structures of Cif from bacterial pathogens Photorhabdus luminescens and Burkholderia pseudomallei. PLoS ONE. 2009;4(5):e5582. doi: 10.1371/journal.pone.0005582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hao YH, Wang Y, Burdette D, Mukherjee S, Keitany G, Goldsmith E, Orth K. Structural requirements for Yersinia YopJ inhibition of MAP kinase pathways. PLoS ONE. 2008;3(1):e1375. doi: 10.1371/journal.pone.0001375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen D, Chai J, Hart PJ, Zhong G. Identifying catalytic residues in CPAF, a Chlamydia-secreted protease. Arch. Biochem. Biophys. 2009;485(1):16–23. doi: 10.1016/j.abb.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huang Z, Feng Y, Chen D, Wu X, Huang S, Wang X, Xiao X, Li W, Huang N, Gu L, Zhong G, Chai J. Structural basis for activation and inhibition of the secreted chlamydia protease CPAF. Cell Host Microbe. 2008;4(6):529–542. doi: 10.1016/j.chom.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 90.Chavakis T, Wiechmann K, Preissner KT, Herrmann M. Staphylococcus aureus interactions with the endothelium: the role of bacterial "secretable expanded repertoire adhesive molecules" (SERAM) in disturbing host defense systems. Thromb. Haemost. 2005;94(2):278–285. doi: 10.1160/TH05-05-0306. [DOI] [PubMed] [Google Scholar]
- 91.Chavakis T, Preissner KT, Herrmann M. The anti-inflammatory activities of Staphylococcus aureus. Trends Immunol. 2007;28(9):408–418. doi: 10.1016/j.it.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 92.tHart BA, Amor S. The use of animal models to investigate the pathogenesis of neuroinflammatory disorders of the central nervous system. Curr. Opin. Neurol. 2003;16(3):375–383. doi: 10.1097/01.wco.0000073940.19076.43. [DOI] [PubMed] [Google Scholar]
- 93.Xie C, Alcaide P, Geisbrecht BV, Schneider D, Herrmann M, Preissner KT, Luscinskas FW, Chavakis T. Suppression of experimental autoimmune encephalomyelitis by extracellular adherence protein of Staphylococcus aureus. J. Exp. Med. 2006;203(4):985–994. doi: 10.1084/jem.20051681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Su SB, Gong W, Gao JL, Shen W, Murphy PM, Oppenheim JJ, Wang JM. A seven-transmembrane, G protein-coupled receptor, FPRL1, mediates the chemotactic activity of serum amyloid A for human phagocytic cells. J. Exp. Med. 1999;189(2):395–402. doi: 10.1084/jem.189.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Le Y, Gong W, Tiffany HL, Tumanov A, Nedospasov S, Shen W, Dunlop NM, Gao JL, Murphy PM, Oppenheim JJ, Wang JM. Amyloid (beta)42 activates a G-protein-coupled chemoattractant receptor, FPR-like-1. J. Neurosci. 2001;21(2):RC123. doi: 10.1523/JNEUROSCI.21-02-j0003.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Le Y, Yazawa H, Gong W, Yu Z, Ferrans VJ, Murphy PM, Wang JM. The neurotoxic prion peptide fragment PrP(106–126) is a chemotactic agonist for the G protein-coupled receptor formyl peptide receptor-like 1. J. Immunol. 2001;166(3):1448–14451. doi: 10.4049/jimmunol.166.3.1448. [DOI] [PubMed] [Google Scholar]
- 97.Cui Y, Le Y, Yazawa H, Gong W, Wang JM. Potential role of the formyl peptide receptor-like 1 (FPRL1) in inflammatory aspects of Alzheimer's disease. J. Leukoc. Biol. 2002;72(4):628–635. [PubMed] [Google Scholar]
- 98.Sartor RB. Mechanisms of disease: pathogenesis of Crohn's disease and ulcerative colitis. Nat. Clin. Pract. 2006;3(7):390–407. doi: 10.1038/ncpgasthep0528. [DOI] [PubMed] [Google Scholar]
- 99.Sartor RB, Muehlbauer M. Microbial host interactions in IBD: implications for pathogenesis and therapy. Curr. Gastroenterol. Reports. 2007;9(6):497–507. doi: 10.1007/s11894-007-0066-4. [DOI] [PubMed] [Google Scholar]
- 100.Marceau M, Dubuquoy L, Caucheteux-Rousseaux C, Foligne B, Desreumaux P, Simonet M. Yersinia pseudotuberculosis anti-inflammatory components reduce trinitrobenzene sulfonic acid-induced colitis in the mouse. Infect. Immun. 2004;72(4):2438–2441. doi: 10.1128/IAI.72.4.2438-2441.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zauberman A, Tidhar A, Levy Y, Bar-Haim E, Halperin G, Flashner Y, Cohen S, Shafferman A, Mamroud E. Yersinia pestis endowed with increased cytotoxicity is avirulent in a bubonic plague model and induces rapid protection against pneumonic plague. PLoS ONE. 2009;4(6):e5938. doi: 10.1371/journal.pone.0005938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Finlay BB, McFadden G. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell. 2006;124(4):767–782. doi: 10.1016/j.cell.2006.01.034. [DOI] [PubMed] [Google Scholar]