The cell nucleus is an intricate and dynamic organelle that is organized into many morphologically distinct regions or “nuclear bodies” (NBs). This compartmentalization represents the spatial and temporal organization of molecular complexes that are important for efficient execution of the essential processes occurring within the nucleus [reviewed in [Zimber et al., 2004]]. While much remains to be understood regarding the structure and function of these NBs, substantial progress has been made in recent years. The rudimentary structures, dynamics, and molecular activities have been investigated in depth for several nuclear bodies including the Cajal body [Morris, 2008], promyelocytic leukemia (PML) nuclear body [Bernardi and Pandolfi, 2007], and nuclear speckles [Lamond and Spector, 2003]. The functional relevance of these NBs is beginning to be revealed. The perinucleolar compartment (PNC), a nuclear body located at the nucleolar periphery, is in comparison much less explored. Here we will highlight the recent advancements in the elucidation of the structure and dynamics of the PNC, and we will speculate potential roles of the PNC in the malignant phenotype.
The PNC
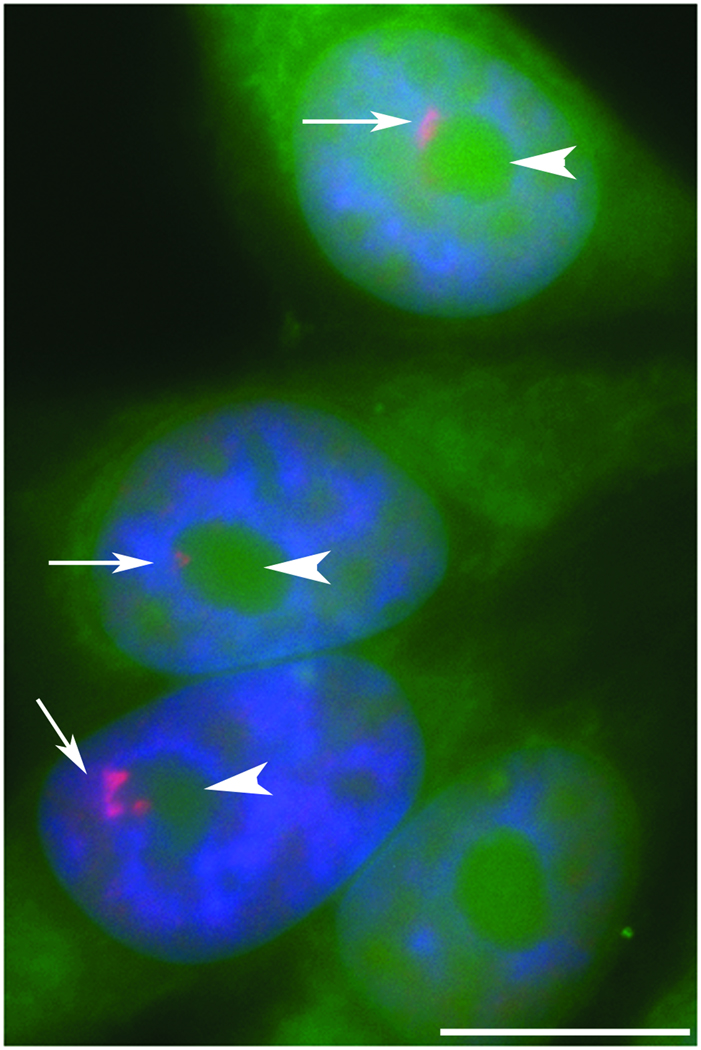
The PNC was first described during the characterization of the polypyrimidine tract binding (PTB) protein, where PTB was shown to localize as a perinucleolar structure [Ghetti et al., 1992]. Subsequently, Matera et al. found that several pol (polymerase) III RNAs are enriched in this structure and termed it the PNC [Matera et al., 1995]. The PNC is an irregularly shaped subnuclear body that ranges in size from .25 up to 4 µm in length ([Matera et al., 1995] and data not published). While the PNC is closely associated with the nucleolus, it is structurally distinct [Huang et al., 1997].
Electron microscopy demonstrated that the PNC is composed of multiple thick, electron dense-strands, each measuring approximately 80–180 nm in diameter [Huang et al., 1998]. Additionally, a three-dimensional computer reconstruction of the electron microscopic images shows that the PNC forms a reticulated meshwork on the nucleolar surface. Live cell imaging of cells expressing green fluorescent protein-tagged PTB (GFP-PTB) revealed that the PNC is a dynamic structure that makes small, distinct movements along the nucleolar periphery over time. The PNC is detected throughout interphase, is disassembled at the beginning of mitosis, and is reassembled in late telophase in daughter cells. The PNCs in the daughter cells often are similar in number and shape [Huang et al., 1998].
PNC as a Pan-Cancer Marker
The nucleus is a complex organelle in which nucleic acids, transcription factors, and regulatory machinery are highly organized. The compartmentalization of these components may help regulate the coordination of nuclear processes including gene expression, DNA replication, and repair. As all of these processes are significantly changed in cancer cells, a long standing indicator of malignant transformation is the alteration of nuclear morphology [Zaidi et al., 2007]. Nuclear size, shape, and heterogeneous labeling with histological stains have long been used in clinical practices to grade tumors. One specific example of nuclear change associated with cancer cells is the presence of the subnuclear body, the PNC.
Early characterization of the PNC demonstrated that the PNC is predominantly present in cancer cells [Huang et al., 1997]. A broad survey of PNC prevalence (percentage of cells with 1 or more PNC) in over 50 cancerous cell lines and normal cells was conducted to evaluate the tissue and species specificity of the PNC [Norton et al., 2008a]. The results show that PNC prevalence is low in normal cell lines (consistently below 0.5%) and immortalized cell lines (between 0% and 6%). Non-cancerous cell lines are derived from multiple tissue types, including both human and mouse embryonic stem cells. PNC prevalence increases in cancer cells from solid tumors, including carcinomas, blastomas, and sarcomas. However, it does not show significant increases in malignancy of hematopoietic origin. Interestingly, there is a large variation of PNC prevalence among different cancer cell lines (ranging from 5% to near 100%). The heterogeneous nature of PNC prevalence in tumor cells was thought to reflect the malignant potential of the given population. This notion has been supported by additional observations that are outlined below. These findings indicate that PNCs form in a broad range of tumor cells derived from solid tissues, indicating the potential for the PNC to be a pan-cancer marker.
In an effort to determine the association of PNC and malignancy in vivo, PNC prevalence was evaluated in human breast cancer samples of varying clinical stages. It was shown that while PNC prevalence is 0% in normal breast tissue, prevalence increases in parallel with clinical progression of disease, and reaches near 100% in distant metastases [Kamath et al., 2005]. In addition, high PNC prevalence in primary tumors of stage I patients positively correlates with disease relapse in a case-matched study and is predictive of survival in a retrospective 17 year follow-up study [Kamath et al., 2005]. The close correlation between PNC prevalence and metastasis indicates that the PNC containing cells have a metastatic advantage over non-PNC containing cells, and the formation of the PNC may reflect key changes during transformation that associate with metastatic capability. These findings highlight the potential utility of the PNC as a prognostic marker for tumors in breast cancer patients.
The correlation between high PNC prevalence and malignancy was also found in other human tissues and cell lines. Normal, hyperplastic, and malignant uterine smooth muscle tissue samples were examined for PNC, and the results show that PNC prevalence significantly increases in the malignant tissues [Norton et al., 2008a]. In addition, PNC prevalence was also examined in thyroid cell lines derived from papillary, follicular, and anaplastic carcinomas. Similarly, PNC prevalence is significantly increased in the more malignant thyroid cell lines. These results validate the association of the PNC with malignant behavior in cancer cells and affirm the potential utility of the PNC as a prognostic marker for malignancy in solid tissue tumors.
To further address the link between PNC prevalence and metastasis, multiple cell lines of varying metastatic potential have been examined for PNC prevalence. In one experiment, two cell lines were created from a single patient: one from the primary melanoma tumor and one from a distant metastasis. PNC prevalence increases dramatically in cell lines derived from distant metastasis over those derived from the primary tumor, and the same results were observed in a similar experiment using colorectal cell lines. The association of PNC with metastasis was further examined using a well characterized prostate cancer model [Pettaway et al., 1996], in which PC-3 human prostate tumor cells are implanted into a nude mouse prostate and selected according to their ability to metastasize. PNC prevalence is the highest in the cell line enriched with mostly metastatic cells after multiple rounds of selection and is lowest in the localized tumor cells. Additionally, breast cancer cell lines that express a metastatic suppressor gene exhibit a significantly lower prevalence of PNC [Norton et al., 2008a]. These results together support the assertion that PNCs form at advanced stages of transformation and are prominent in cells of high metastatic capacity.
Although the function of the PNC in malignant transformation remains uncertain, it is apparent that the PNC is involved in a process specific to cancer cells, as the PNC does not associate with traits common to both cancer and normal cells. For example, PNC prevalence is not affected by cell proliferation. HeLa cells grown in serum free media exhibit a significantly reduce growth rate, but PNC prevalence does not change. High glucose or low glucose growth conditions do not alter PNC prevalence; demonstrating glycolysis state does not influence PNC structure. Furthermore, changes of differentiation status of cancer cells with various stimulus do not affect PNC prevalence, and PNCs are not present in embryonic stem cells [Norton et al., 2008a]. These findings suggest that the PNC is associated with a process specific to malignancy rather than traits that are common in both cancer and normal cells.
RNA and Protein Components of the PNC
While the complete molecular composition of the PNC has yet to be determined, several protein and RNA components of the PNC have been identified. All known PNC localizing RNAs are non-coding RNAs transcribed by pol III, while all identified PNC-associated proteins are known for their primary role in pol II RNA metabolism. The proteins include CUG-BP [Timchenko et al., 1996], PTB [Ghetti et al., 1992], KSRP [Hall et al., 2004], RAVER1 [Huttelmaier et al., 2001], RAVER2 [Kleinhenz et al., 2005], ROD1, nucleolin [Kopp and Huang, 2005]; RNAs include MRP, RNAse P, hY (1,2,5) [Matera et al., 1995], ALU, and SRP (7SL) [Wang et al., 2003]. For a description of each PNC component see Table 1. While the RNAs that enrich at the PNC are exclusively pol III transcripts, not all pol III RNAs localize to the PNC. For example, U6 and 5S RNA are not detected at the PNC.
Table 1.
Protein and RNA Components of the PNC
| Component Name | Brief Description |
|---|---|
| RNA Components | |
| RNase P RNA | RNA portion of RNase P complex, involved in maturation of 5’ end of tRNA [van Eenennaam et al., 2000] |
| MRP RNA | RNA portion of the MRP complex, involved in mitochondrial DNA replication and pre-ribosomal RNA processing [van Eenennaam et al., 2000] |
| hY RNA | Small RNAs that interact with Ro protein to form Ro ribonucleoproteins (RNPs) [Wolin and Steitz, 1984] |
| Alu RNA | Forms Alu RNA complex, implicated in regulation of transcription and translation [Hasler and Strub, 2006a; Hasler and Strub, 2006b] |
| SRP RNA | RNA portion of the signal recognition particle, involved in secretory protein transport and elongation arrest [Wolin and Walter, 1989] |
| Protein Components | |
| PTB | Protein that prefers pyrimidine rich sequences, involved in RNA splicing and translational regulation [Kozak, 2003; Wagner and Garcia-Blanco, 2001] |
| CUG-BP | RNA binding protein that interacts with polyadenylated RNA and is implicated in alternative splicing and mytonic dystrophy [Savkur et al., 2001; Timchenko et al., 1996] |
| KSRP | Highly expressed in neural cells and involved in RNA splicing and decay [Chen et al., 1997; Min et al., 1997] |
| RAVER1/2 | Both proteins are heterogeneous nuclear RNPs that bind RNA and interact with PTB [Kleinhenz et al., 2005] |
| ROD1 | A homologue of yeast negative regulator of differentiation, shares homology with PTB [Yamamoto et al., 1999] |
| Nucleolin | The only protein component of the PNC also enriched in the nucleolus, involved in rDNA transcription, RNA processing [Ginisty et al., 1999; Tuteja and Tuteja, 1998] |
Although the proteins that localize to the PNC are known to interact with pol II RNAs, no pol II transcripts have been detected in the PNC. In situ hybridization of multiple pol II RNAs did not exhibit enrichment in the PNC [Hall et al., 2004; Kopp and Huang, 2005]. Additionally, inhibition of pol II transcription does not disassemble the structural integrity of the PNC, suggesting that PNC structure is independent of pol II transcription [Huang et al., 1998]. However, the localization the PTB protein to the PNC is dependent upon its RNA binding capacity [Huang et al., 1997]. This suggests that PTB, and perhaps other RNA binding proteins of the PNC, are binding RNA in the PNC.
Furthermore, PNC structure is sensitive to RNase treatment, signifying that RNA is important for PNC structural integrity. These observations post at least two possible explanations: that these proteins are either interacting with pol II transcripts that have yet to be detected in the PNC, or previously uncharacterized interactions are occurring between the PNC-associated proteins and the pol III RNAs. Research is ongoing to establish the complete composition of the PNC, and to determine the protein-RNA interactions occurring in association with the PNC.
PNC and Pol III Transcription
The importance of RNA in PNC stability was first observed when RNase, but not DNase, treatment in permeabilized cells eliminated PNC [Huang et al., 1998]. Since all RNA detected in the PNC are of pol III origin, the significance of pol III transcription in PNC stability was evaluated. The results demonstrated that inhibition of polymerase III causes disassembly of the PNC [Wang et al., 2003]. Interestingly it is not polymerase III activity, but rather the continuous production of pol III transcripts themselves that are important for PNC structure. Overexpression of one of the PNC-associated RNAs, RNase MRP RNA, from a pol II promoter was able to partially overcome the PNC disassociation caused by pol III inhibition [Wang et al., 2003]. This evidence supports the importance of pol III RNA to the PNC.
It is likely that the RNAs composing the PNC are newly transcribed. Five minute pulse labeling with BrU shows a concentration of newly synthesized RNA in the PNC [Huang et al., 1998]. Furthermore, while pol III inhibition leads to rapid disassociation of the PNC, the mature pol III RNAs in their functional complexes remain intact at similar time points [Wang et al., 2003]. These observations demonstrate that the PNC is enriched with newly synthesized pol III RNA that are not in their known functional complex, and these RNAs are critical to the structural integrity of the PNC. Although the PNC is associated with newly transcribed RNAs, it is not the site of transcription for several of the RNAs identified in the PNC. In situ hybridization of the genes encoding four of the PNC associated RNAs show no association with the PNC [Kopp and Huang, 2005; Matera et al., 1995]. In addition, it is also unlikely that the PNC acts as an assembly point for the newly transcribed RNAs into their prospective ribonucleoprotein complexes, because the protein subunits of these complexes are not detected in the PNC [Hall et al., 2004; Kopp and Huang, 2005]. Therefore, the PNC may represent a transitional depot between the newly synthesized pol III RNA and the assembly of their final functional complex.
It has long been known that pol III transcription is deregulated in cancer (reviewed in [White, 2004]). Early experiments demonstrated that pol III is hyperactive in mouse melanomas [Schwartz et al., 1974], and more recent investigations support these findings in human cell lines and tissues [Liebhaber et al., 1978; Winter et al., 2000]. During the experimental examination of the role of pol III in tumorigenesis, a recent study showed that the overexpression of the pol III transcripts, tRNA and 5S rRNA, is sufficient to induce increased proliferation and oncogenic transformation [Marshall et al., 2008]. These findings demonstrate that increased pol III transcription not only correlates with malignancy, but may play a direct role in the development or progression of cancer. The link between pol III transcription and cancer becomes relevant when examining the role of the PNC in malignancy. While the pol III transcripts that localize to the PNC do not individually play a known role in the development of cancer, the concentration of pol III transcripts in the PNC could be indicative of the deregulation of pol III transcription in cancer cells. Additionally, it raises the question of whether the nucleation of pol III transcripts to the PNC is not merely a consequence of malignancy but rather a factor involved in regulating the function of these RNAs in disease progression and maintenance. Studies are underway to analyze the complexes the pol III RNA form in association with the PNC and their functional relevance to malignancy.
PNC Association with DNA Locus
During early characterization of the PNC it was observed that the PNC is a heritable structure, where daughter cells have the same number of PNC as parental cells, and the PNC are often spatially arranged as mirror images [Huang et al., 1997; Norton et al., 2008b]. These early observations suggested a possible link between the PNC and DNA. Recent chemical and cell biology studies have provided further evidence that the PNC is associated with a DNA locus.
Chemical biology studies have shown that PNC structure can be disassembled by a large array of genome toxic drugs. The disassembly of the PNC is not due to the DNA-damage response from treated cells, but rather DNA damage itself. While not all types of DNA damage disrupt the PNC, DNA intercalators and cross linkers do, suggesting that the base-pairing capacity of DNA is critical to the structural of the PNC. Further evidence to support a PNC-DNA association came from cell biology studies using a cdk1 conditional mutant. At non-permissive conditions, the mutant is defective in cell division but allows for continuous endoreplication. If the PNC is associated with a DNA locus, it should interact with the newly replicated DNA, and PNC prevalence would increase. As was predicted, PNC prevalence was shown to increase in unison with replication cycle, providing further support that PNC is associated with DNA [Norton et al., 2008b]. Additionally, treatment with camptothecin, a topo I inhibitor and blocker of cell cycle at S/G2 phase, blocks cells and increases DNA content in each cell. Again, PNC prevalence was shown to increase with the enhanced amount of DNA produced upon camptothecin treatment. These experiments provide strong evidence that the PNC is associated with DNA.
Examination of synchronized HeLa cells shows that during S phase, a majority of PNCs split into a doublet form, but then reformed during G2 phase [Norton et al., 2008b]. This is consistent with the behavior of a DNA locus and suggests that the PNC associated locus replicates at mid S phase. To evaluate how chromatin structure could impact the PNC, cells were treated with a histone deacetylase (HDAC) inhibitor to enhance the acetylation state of the chromatin. Upon HDAC treatment, the structure of the PNC changed from a dense, round structure to an extended, fibrous structure, indicating that the PNC associated locus is responsive to the epigenetic regulations. Taken together, these studies strongly support the association of the PNC with a DNA locus. It is thought that the nucleation of PNC on a DNA locus may represent at least two possibilities: the locus may be a site of transcription of yet to be identified PNC associated RNA, or the PNC may regulate gene expression at this locus. Studies are underway to identify the DNA locus that the PNC is nucleated upon, which will provide clues to the potential function of the PNC.
Concluding Remarks
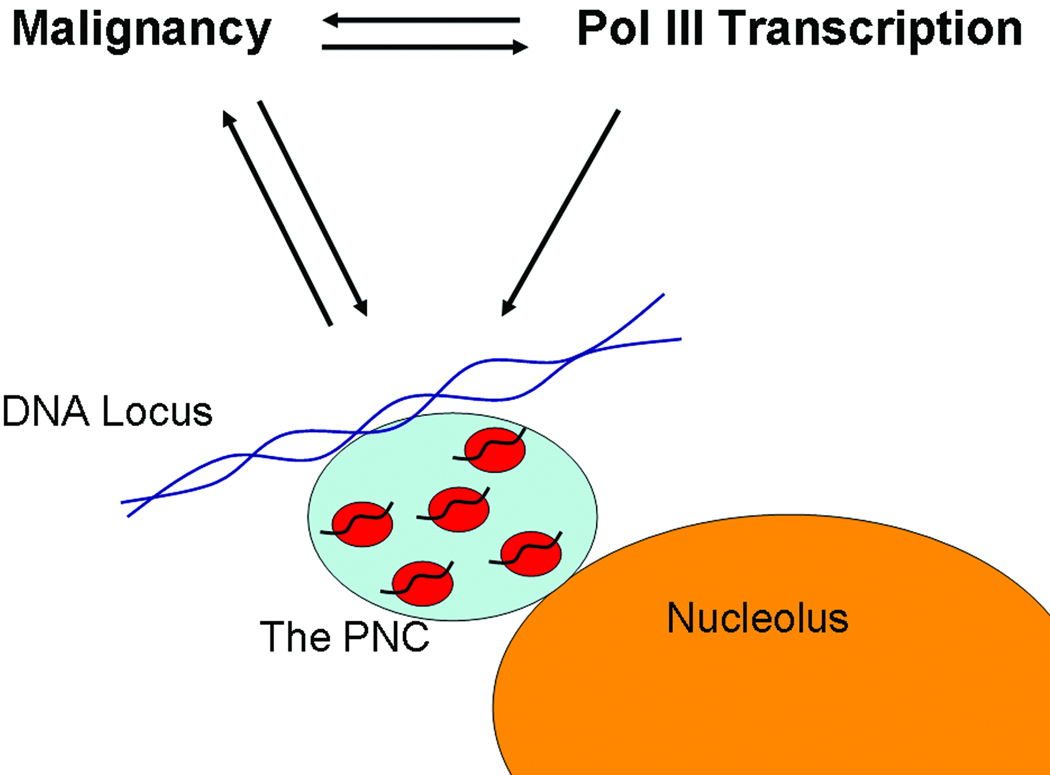
Much progress has been made in the characterization of the PNC since its discovery in 1992. The PNC is a dynamic nuclear body that correlates with cancer and metastasis and has potential as a prognostic marker for solid tissue tumors. The PNC is enriched with a mixture of pol III RNAs and RNA binding proteins that are primarily implicated in pol II transcription. The structural integrity of the PNC is dependent upon continuous transcription of pol III RNA and intact DNA structure, and the PNC is associated with an undefined DNA locus. Since pol III transcription is deregulated in cancer cells and directly contributes to the transformed phenotype, the formation of the PNC may play a role in the maintenance and/or promotion of malignancy. Based on these observations, our current working model (Figure 3) is that newly transcribed pol III RNA nucleates on a DNA locus to form the PNC. These RNAs may interact with PNC associated proteins to form previously uncharacterized complexes. In this way, the PNC may serve as a transitional depot where these newly synthesized pol III RNAs are regulated. Future studies should focus on identifying the molecular complexes and the DNA locus that are associated with the PNC. These advancements will work to further the understanding of the functional relevance of PNCs in cancer cells.
Figure 1.
Figure 2.
Acknowledgements
We thank Kelly Kopp for providing PNC images and Paul Schook for his critical review of the manuscript. S.H. is funded by 1 R01 GM078555-01A1, and C.B.P is funded by the T32 CA080621-06A2 training grant.
Funded By:
Contract grant sponsor: NIH-GM; Contract grant number: R01 GM078555-01A1 for SH
Contract grant sponsor: NCI; Contract grant number: T32 CA080621-06A2 for CP
References
- Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- Chen W, Bocker W, Brosius J, Tiedge H. Expression of neural BC200 RNA in human tumours. J Pathol. 1997;183:345–351. doi: 10.1002/(SICI)1096-9896(199711)183:3<345::AID-PATH930>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Ghetti A, Pinol-Roma S, Michael WM, Morandi C, Dreyfuss G. hnRNP I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 1992;20:3671–3678. doi: 10.1093/nar/20.14.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginisty H, Sicard H, Roger B, Bouvet P. Structure and functions of nucleolin. J Cell Sci. 1999;112(Pt 6):761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- Hall MP, Huang S, Black DL. Differentiation-induced colocalization of the KH-type splicing regulatory protein with polypyrimidine tract binding protein and the c-src pre-mRNA. Mol Biol Cell. 2004;15:774–786. doi: 10.1091/mbc.E03-09-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasler J, Strub K. Alu elements as regulators of gene expression. Nucleic Acids Res. 2006a;34:5491–5497. doi: 10.1093/nar/gkl706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasler J, Strub K. Alu RNP and Alu RNA regulate translation initiation in vitro. Nucleic Acids Res. 2006b;34:2374–2385. doi: 10.1093/nar/gkl246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Deerinck TJ, Ellisman MH, Spector DL. The dynamic organization of the perinucleolar compartment in the cell nucleus. J Cell Biol. 1997;137:965–974. doi: 10.1083/jcb.137.5.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Deerinck TJ, Ellisman MH, Spector DL. The perinucleolar compartment and transcription. J Cell Biol. 1998;143:35–47. doi: 10.1083/jcb.143.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttelmaier S, Illenberger S, Grosheva I, Rudiger M, Singer RH, Jockusch BM. Raver1, a dual compartment protein, is a ligand for PTB/hnRNPI and microfilament attachment proteins. J Cell Biol. 2001;155:775–786. doi: 10.1083/jcb.200105044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RV, Thor AD, Wang C, Edgerton SM, Slusarczyk A, Leary DJ, Wang J, Wiley EL, Jovanovic B, Wu Q, Nayar R, Kovarik P, Shi F, Huang S. Perinucleolar compartment prevalence has an independent prognostic value for breast cancer. Cancer Res. 2005;65:246–253. [PubMed] [Google Scholar]
- Kleinhenz B, Fabienke M, Swiniarski S, Wittenmayer N, Kirsch J, Jockusch BM, Arnold HH, Illenberger S. Raver2, a new member of the hnRNP family. FEBS Lett. 2005;579:4254–4258. doi: 10.1016/j.febslet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Kopp K, Huang S. Perinucleolar compartment and transformation. J Cell Biochem. 2005;95:217–225. doi: 10.1002/jcb.20403. [DOI] [PubMed] [Google Scholar]
- Kozak M. Alternative ways to think about mRNA sequences and proteins that appear to promote internal initiation of translation. Gene. 2003;318:1–23. doi: 10.1016/s0378-1119(03)00774-1. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- Liebhaber SA, Wolf S, Schlessinger D. Differences in rRNA metabolism of primary and SV40-transformed human fibroblasts. Cell. 1978;13:121–127. doi: 10.1016/0092-8674(78)90143-5. [DOI] [PubMed] [Google Scholar]
- Marshall L, Kenneth NS, White RJ. Elevated tRNA(iMet) synthesis can drive cell proliferation and oncogenic transformation. Cell. 2008;133:78–89. doi: 10.1016/j.cell.2008.02.035. [DOI] [PubMed] [Google Scholar]
- Matera AG, Frey MR, Margelot K, Wolin SL. A perinucleolar compartment contains several RNA polymerase III transcripts as well as the polypyrimidine tract-binding protein, hnRNP I. J Cell Biol. 1995;129:1181–1193. doi: 10.1083/jcb.129.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min H, Turck CW, Nikolic JM, Black DL. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 1997;11:1023–1036. doi: 10.1101/gad.11.8.1023. [DOI] [PubMed] [Google Scholar]
- Morris GE. The Cajal body. Biochim Biophys Acta. 2008;1783:2108–2115. doi: 10.1016/j.bbamcr.2008.07.016. [DOI] [PubMed] [Google Scholar]
- Norton JT, Pollock CB, Wang C, Schink JC, Kim JJ, Huang S. Perinucleolar compartment prevalence is a phenotypic pancancer marker of malignancy. Cancer. 2008a;113:861–869. doi: 10.1002/cncr.23632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton JT, Wang C, Gjidoda A, Henry RW, Huang S. The perinucleolar compartment is directly associated with DNA. J Biol Chem. 2008b doi: 10.1074/jbc.M807255200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettaway CA, Pathak S, Greene G, Ramirez E, Wilson MR, Killion JJ, Fidler IJ. Selection of highly metastatic variants of different human prostatic carcinomas using orthotopic implantation in nude mice. Clin Cancer Res. 1996;2:1627–1636. [PubMed] [Google Scholar]
- Savkur RS, Philips AV, Cooper TA. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- Schwartz LB, Sklar VE, Jaehning JA, Weinmann R, Roeder RG. Isolation and partial characterization of the multiple forms of deoxyribonucleic acid-dependent ribonucleic acid polymerase in the mouse myeloma, MOPC 315. J Biol Chem. 1974;249:5889–5897. [PubMed] [Google Scholar]
- Timchenko LT, Miller JW, Timchenko NA, DeVore DR, Datar KV, Lin L, Roberts R, Caskey CT, Swanson MS. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja R, Tuteja N. Nucleolin: a multifunctional major nucleolar phosphoprotein. Crit Rev Biochem Mol Biol. 1998;33:407–436. doi: 10.1080/10409239891204260. [DOI] [PubMed] [Google Scholar]
- van Eenennaam H, Jarrous N, van Venrooij WJ, Pruijn GJ. Architecture and function of the human endonucleases RNase P and RNase MRP. IUBMB Life. 2000;49:265–272. doi: 10.1080/15216540050033113. [DOI] [PubMed] [Google Scholar]
- Wagner EJ, Garcia-Blanco MA. Polypyrimidine tract binding protein antagonizes exon definition. Mol Cell Biol. 2001;21:3281–3288. doi: 10.1128/MCB.21.10.3281-3288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Politz JC, Pederson T, Huang S. RNA polymerase III transcripts and the PTB protein are essential for the integrity of the perinucleolar compartment. Mol Biol Cell. 2003;14:2425–2435. doi: 10.1091/mbc.E02-12-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RJ. RNA polymerase III transcription and cancer. Oncogene. 2004;23:3208–3216. doi: 10.1038/sj.onc.1207547. [DOI] [PubMed] [Google Scholar]
- Winter AG, Sourvinos G, Allison SJ, Tosh K, Scott PH, Spandidos DA, White RJ. RNA polymerase III transcription factor TFIIIC2 is overexpressed in ovarian tumors. Proc Natl Acad Sci U S A. 2000;97:12619–12624. doi: 10.1073/pnas.230224097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin SL, Steitz JA. The Ro small cytoplasmic ribonucleoproteins: identification of the antigenic protein and its binding site on the Ro RNAs. Proc Natl Acad Sci U S A. 1984;81:1996–2000. doi: 10.1073/pnas.81.7.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin SL, Walter P. Signal recognition particle mediates a transient elongation arrest of preprolactin in reticulocyte lysate. J Cell Biol. 1989;109:2617–2622. doi: 10.1083/jcb.109.6.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Tsukahara K, Kanaoka Y, Jinno S, Okayama H. Isolation of a mammalian homologue of a fission yeast differentiation regulator. Mol Cell Biol. 1999;19:3829–3841. doi: 10.1128/mcb.19.5.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Javed A, Pratap J, Montecino M, van Wijnen A, Lian JB, Stein JL, Stein GS. Nuclear microenvironments in biological control and cancer. Nat Rev Cancer. 2007;7:454–463. doi: 10.1038/nrc2149. [DOI] [PubMed] [Google Scholar]
- Zimber A, Nguyen QD, Gespach C. Nuclear bodies and compartments: functional roles and cellular signalling in health and disease. Cell Signal. 2004;16:1085–1104. doi: 10.1016/j.cellsig.2004.03.020. [DOI] [PubMed] [Google Scholar]