Abstract
Previous studies have suggested that ionizing radiation causes irreparable DNA double-strand breaks in mice and cell lines harboring mutations in any of the three subunits of DNA-dependent protein kinase (DNA-PK) (the catalytic subunit, DNA-PKcs, or one of the DNA-binding subunits, Ku70 or Ku86). In actuality, these mutants vary in their ability to resolve double-strand breaks generated during variable (diversity) joining [V(D)J] recombination. Mutant cell lines and mice with targeted deletions in Ku70 or Ku86 are severely compromised in their ability to form coding and signal joints, the products of V(D)J recombination. It is noteworthy, however, that severe combined immunodeficient (SCID) mice, which bear a nonnull mutation in DNA-PKcs, are substantially less impaired in forming signal joints than coding joints. The current view holds that the defective protein encoded by the murine SCID allele retains enough residual function to support signal joint formation. An alternative hypothesis proposes that DNA-PKcs and Ku perform different roles in V(D)J recombination, with DNA-PKcs required only for coding joint formation. To resolve this issue, we examined V(D)J recombination in DNA-PKcs-deficient (SLIP) mice. We found that the effects of this mutation on coding and signal joint formation are identical to the effects of the SCID mutation. Signal joints are formed at levels 10-fold lower than in wild type, and one-half of these joints are aberrant. These data are incompatible with the notion that signal joint formation in SCID mice results from residual DNA-PKcs function, and suggest a third possibility: that DNA-PKcs normally plays an important but nonessential role in signal joint formation.
V(D)J recombination assembles the variable (V), diversity (D), and joining (J) antigen receptor gene segments within a lymphoid cell, generating a diverse repertoire of receptors in T and B cells (reviewed in ref. 1). Recombination is initiated by the recognition of a pair of signal sequences, which flank each coding element, by the lymphoid-specific recombination activating gene products, RAG1 and RAG2. Site-specific cleavage by the RAG proteins generates recombination intermediates—covalently sealed (hairpin) coding ends and blunt signal ends—which are retained in a postcleavage complex. The ends are differentially processed and joined to form a coding joint (encoding the antigen receptor) and a signal joint. Signal joint formation is typically conservative and precise, without nucleotide loss or addition. In contrast, coding ends are subject to processing so that the coding joint reflects nucleotide loss and/or addition of nontemplated nucleotides.
Mutant mice and cell lines have been described that are defective in both DNA repair and V(D)J recombination, implicating ubiquitous DNA repair proteins in the postcleavage stages of V(D)J recombination (reviewed in ref. 2). Two factors clearly involved in both DNA repair and V(D)J recombination are the XRCC4 protein and DNA-dependent protein kinase (DNA-PK). DNA-PK may serve as a sensor of DNA-damage and/or recruit repair machinery to DNA lesions. The catalytic subunit (DNA-PKcs) is a 460-kDa serine/threonine kinase that exists largely in latent form as a monomer in the absence of DNA damage (reviewed in ref. 3). DNA-PKcs is activated upon association with the regulatory DNA-binding component, called Ku, which has an affinity for DNA lesions such as double-strand breaks. Ku itself is a heterodimer composed of Ku70 and Ku86, which are tightly associated in the cell. Although all three subunits are necessary for optimal activation of DNA-PK, kinase activity is detectable in the absence of Ku under some conditions (4, 5).
The severe combined immunodeficient (SCID) mouse (6) bears a naturally occurring mutation in the DNA-PKcs gene (7–10), resulting in an 83-aa truncation of the C-terminal end of the catalytic subunit of the DNA-PK (11–13). DNA-PKcs protein levels in SCID mice are only about 10% of wild type, and little or no DNA-PK activity is detectable (12, 14), even though the mutation is C-terminal of the kinase domain. SCID mice are sensitive to ionizing radiation (9, 15–17) and are profoundly T and B cell deficient because of a severe defect in coding joint formation (18–21). Levels of coding joints in SCID cell lines (19) and SCID mice (at certain loci) (18, 22, 23) can be reduced as much as 1,000-fold in comparison with wild-type controls. Despite this defect, some coding joint formation is surprisingly efficient (only 10-fold decreased) at “privileged sites” in SCID mice (22, 24–26). On the other hand, careful studies in SCID mice and various DNA-PKcs mutant cell lines demonstrate that signal joint levels are as much as 10-fold reduced and many of these joints display anomalous nucleotide loss that is seldom observed in wild-type controls (19, 27, 28).
The V(D)J recombination defect in Ku-deficient mice and embryonic stem (ES) cells severely impairs both coding and signal joint formation (29–34). One interpretation of these results is that Ku70 and Ku86 are necessary for both coding and signal joint formation, whereas DNA-PKcs is necessary for coding joint formation only (9, 35). This model suggests that signal ends normally join independently of DNA-PKcs. Thus, DNA-PKcs, Ku70, and Ku86 might have distinct but overlapping functions in V(D)J recombination. A counterhypothesis proposes that the residual DNA-PKcs function present in SCID lymphocytes is sufficient for signal joint formation (11, 12, 36). The correlation between DNA-PKcs protein levels in SCID thymocytes (10% of wild-type levels) and the 10-fold reduction of signal joint formation is consistent with this view. A corollary to this “partial function” hypothesis further predicts that those coding joints at privileged sites that form with considerable efficiency in SCID mice also depend on the partial function of DNA-PKcs. Results from Ku-deficient mice also show relatively efficient coding joint formation at a privileged site (29–32). This does not refute the partial function hypothesis because DNA-PKcs may function independently of Ku (4, 5) to promote some rearrangements.
Unfortunately, analyses of V(D)J recombination in other DNA-PKcs-defective rodent and human cell lines have not unambiguously supported either one of these hypotheses (see Discussion for details). The only way to resolve these conflicting results is to analyze the effects of a DNA-PKcs null mutation on V(D)J recombination in vivo, in lymphocyte precursors. Jhappan et al. (37) recently described SLIP mice, which bear a null mutation in DNA-PKcs. In these animals, the fortuitous integration of a transgene (in excess of 20 copies) interrupts the first 800 bp of the coding sequence of the DNA-PKcs gene. To reconcile the differences in V(D)J recombination among DNA-PK mutants and to clarify the functions of each component of DNA-PK, we assessed recombination under physiological conditions by analyzing endogenous rearrangements in SLIP mice.
MATERIALS AND METHODS
Mice.
The generation of SLIP mice has been described (37). Adult SLIP mice were transferred from the animal facility at the National Institutes of Health to that of Baylor College of Medicine where they were housed in microisolator cages and daily given antibiotics in their drinking water. Homozygous SLIP matings were maintained; mice were analyzed at 2–4 weeks of age, before onset of tumor formation. BALB/c and SCID (C.B-17 scid/scid) mice were used as controls.
DNA Preparation and Semiquantitative PCR.
DNA was prepared from single-cell suspensions of thymus or bone marrow as described (38). DNA samples were amplified by semiquantitative PCR for the analysis of T cell receptor (TCR)β, TCRδ, and immunoglobulin heavy chain (IgH) recombination products, as described (23, 30). The following primers were used for amplification; probes are indicated. TCR Vβ8-Jβ2.6 coding joints: DR144, DR155 (probe DR154); TCR Dδ2-Jδ1 coding joints: DR123, DR53 (probe DR2); TCR Dδ2-Jδ1 signal joints: DR21, DR162 (probe DR50); IgH D–JH4 coding/hybrid joints: DHL, DR217 (probes DR218 and HYB). PCR products were separated by electrophoresis through 6% polyacrylamide gels (in 1× TBE buffer, 100 mM Tris/90 mM boric acid/1 mM EDTA), electrophoretically transferred in 1/2× TBE onto Gene Screen Plus membranes (DuPont), and hybridized to internal oligonucleotide probes that were 32P end-labeled. Radiolabeled products were analyzed and quantitated using a PhosphorImager (Molecular Dynamics Storm 860). Experiments were performed on numerous samples, and representative data are presented.
RESULTS
Coding Joint Formation Is Severely Impaired in SLIP Mice.
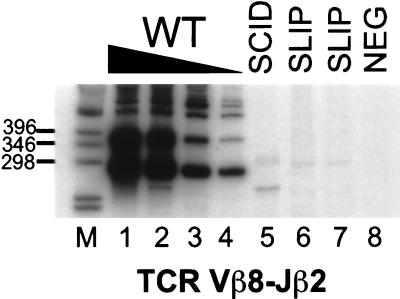
To detect coding joints at the TCRβ locus, semiquantitative PCR of thymocyte DNA was performed for Vβ8-Jβ2.6 rearrangements, which are common in wild-type thymocytes and are detectable in as little as 100 pg of DNA (Fig. 1). Rearrangements in SCID (lane 5) and two SLIP (lanes 6 and 7) thymocyte DNA samples were at least 1,000-fold lower than those from wild-type thymocytes (lanes 1–4). Thus, SLIP mice are clearly defective in coding joint formation, to a degree commensurate with SCID mice.
Figure 1.
Thymocytes from SLIP mice are defective in TCR Vβ8-Jβ2 coding joint formation. Thymocyte DNA samples were subjected to semiquantitative PCR analysis. Titrations with wild-type DNA (100, 10, 1, 0.1 ng; lanes 1–4, respectively) were performed to estimate the abundance of coding joint formation in SCID (lane 5) and two SLIP (lanes 6 and 7) DNA samples (all at 100 ng). Our primers amplify Vβ8 rearrangements to all Jβ2 members. The most prominent band in the wild-type DNA lanes is 276 bp, the expected size for Vβ8-Jβ2.6 (unrearranged DNA is not amplified). A negative PCR control (all reagents without DNA) is shown in lane 8. Relevant sizes of the DNA marker (1-kb ladder; GIBCO/BRL) are indicated adjacent to lane M.
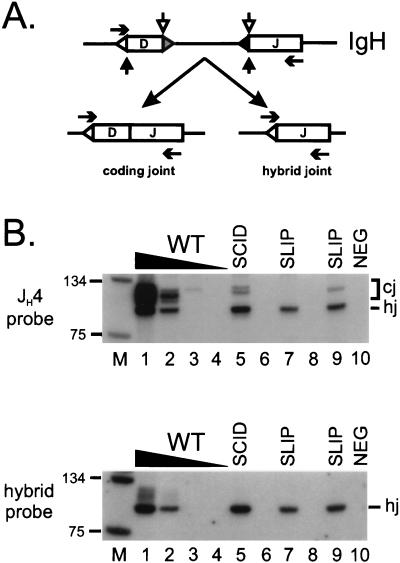
To extend our observations to the B cell lineage, we also assayed for coding joint formation at the IgH chain locus in bone marrow DNA. Two types of IgH D–J products—standard coding joints and nonstandard junctions called hybrid joints—are amplified with the same primer set in this assay, as previously reported (36) and diagrammed in Fig. 2A. Both types of products contain the JH4 coding region and hybridize to a JH4-specific probe. As shown in Fig. 2B (Upper), IgH D–J coding joints (higher molecular weight product) from slip bone marrow DNA were either not detectable (lane 7) or were between 10- and 100-fold less (lane 9) than wild-type mice (lanes 1–4). Variation in coding joint levels has also been observed at this locus in different SCID samples (data not shown). Although coding joint formation is clearly defective in SLIP mice, hybrid joints were formed with considerable efficiency, comparable to levels in wild-type bone marrow DNA (Fig. 2B Upper, lower molecular weight product). To confirm that the lower molecular weight band indeed represents hybrid joints, we stripped and rehybridized the membrane with a hybrid-joint specific probe. As shown in Fig. 2B (Lower), the lower molecular weight band specifically hybridized with this probe, whereas the higher molecular weight coding joints did not. Thus, hybrid joint formation does not require DNA-PKcs, consistent with our proposal that these joints form by RAG-mediated joining (36, 39).
Figure 2.
IgH recombination products in SLIP mice. (A) Coding joints (D–J) and hybrid joints (5′ D recombination signal sequence to J) are amplified from the same primers (horizontal arrows). Coding elements are depicted by boxes and recombination signal sequences are represented by triangles adjacent to the coding elements. Both types of joints depend on RAG-mediated cleavage at the recombination signal sequence 5′ of the J coding element. Normal coding joints are generated when cleavage also occurs 3′ of the D coding element (open vertical arrows) and hybrid joint formation depends on cleavage on the 5′ side of this element (solid vertical arrows). (B) Coding joint formation is defective in slip bone marrow. Bone marrow DNA samples were subjected to semiquantitative PCR analysis; titrations with wild-type DNA (100, 10, 1, 0.1 ng; lanes 1–4, respectively), SCID (lane 5), and two SLIP (lanes 7 and 9) DNA samples (all at 100 ng). Both coding (cj) and hybrid joints (hj) are detected by a JH4-specific probe (Upper); expected sizes are ≈100 bp for coding joints [numerous sizes caused by amplification of several D region family members by a degenerate primer (DHL)] and 83 bp for a perfect hybrid joint. The membrane was stripped and hybridized to an oligonucleotide probe representing a perfect hybrid joint (Lower). Relevant sizes of the DNA marker (lane M) are indicated.
TCR Dδ2-Jδ1 Coding Joint Formation in SCID Mice Does Not Depend on Residual DNA-PKcs Function.
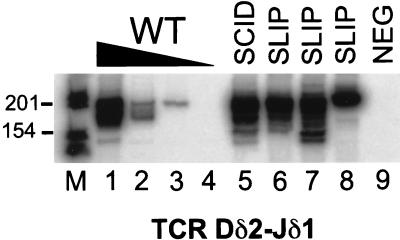
To determine whether “privileged site” coding joints are formed independently of DNA-PKcs or whether a residual function of the defective DNA-PKcs protein in SCID mice is responsible for these types of joints, we analyzed coding joint formation at the TCRδ locus in SLIP thymocytes. Our analysis of coding joint formation at the TCRδ locus is shown in Fig. 3. As expected, TCR Dδ2-Jδ1 coding joints were detected at a relatively high frequency in SCID thymocytes (lane 5) compared with wild type (lanes 1–4) (22, 24, 26). TCR Dδ2-Jδ1 rearrangements were also observed at significant levels (similar to SCID) in several SLIP thymocyte DNA samples (lanes 6–8). These data indicate that TCR Dδ2-Jδ2 coding joint formation in SCID mice is largely DNA-PKcs-independent and suggests that these rearrangements might normally occur in a DNA-PKcs-independent manner.
Figure 3.
Privileged site coding joint formation in SCID mice does not depend on residual DNA-PKcs functions. Thymocyte DNA samples were subjected to semiquantitative PCR analysis for TCR Dδ2-Jδ1 coding joints. Titrations with wild-type DNA (100, 10, 1, 0.1 ng; lanes 1–4, respectively) were performed to estimate the abundance of coding joint formation in SCID (lane 5) and three SLIP (lanes 6–8) DNA samples (all at 100 ng). The expected size is 199 bp. A negative PCR control (all reagents without DNA) is shown in lane 9. Relevant sizes of the DNA marker are indicated adjacent to lane M.
Signal Joint Formation in SLIP Mice.
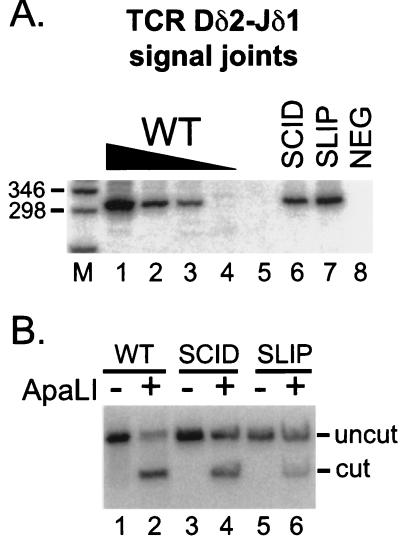
To explore further the role of DNA-PKcs in V(D)J recombination, we searched for signal joints in SLIP mice. As shown in Fig. 4, TCR Dδ2-Jδ1 signal joints were readily detectable in wild-type (lanes 1–4) and SCID thymocytes (lane 6). Signal joint formation in SCID mice is about 10-fold lower than in wild type, consistent with reduced levels reported in mice (38) and cell lines (19). Signal joints also were easily detected in SLIP thymocyte DNA (lane 7) at levels comparable to SCID. Thus, signal joints in DNA-PKcs null mice form at one-tenth the frequency of those in wild-type thymocytes.
Figure 4.
Signal joint formation in SLIP mice. (A) TCR Dδ2-Jδ1 signal joints were amplified from thymocyte DNA preparations (the same DNA concentrations as in Fig. 3): wild-type titration (lanes 1–4), SCID (lane 6), and SLIP (lane 7). The expected size is 301 bp. A negative PCR control (all reagents without DNA) is shown in lane 8. Relevant sizes of the DNA marker (lane M) are indicated. (B) Signal joint PCR products were subjected to digestion with ApaLI. Wild type (lanes 1 and 2), SCID (lanes 3 and 4), and SLIP (lanes 5 and 6); undigested (lanes 1, 3, and 5), and ApaLI treated (lanes 2, 4, and 6). The expected sizes after digestion are 167 and 134 bp; only the larger product hybridizes to the probe.
To ascertain the fidelity of signal joint formation in SLIP mice, we subjected the remainder of the PCR products to digestion with the restriction enzyme ApaLI. Only perfect signal joints, formed without loss or addition of nucleotides, are digested by this endonuclease. Fig. 4B shows that, unlike signal joints formed in wild-type thymocyte DNA, which are largely ApaLI sensitive (lane 2), more of the signal joints derived from SLIP thymocyte DNA are ApaLI resistant (lane 6), indicating that many of the signal joints formed were imperfect. Signal joints from SCID thymocyte DNA were also largely imperfect (lane 4), as reported (19, 27, 28). On average, signal joints from additional SCID and SLIP samples were 50 and 55% ApaLI-resistant compared with 5–15% of ApaLI-resistant signal joints in wild-type mice [quantitated by PhosphorImager; data not shown and ref. (25)], indicating that roughly one-half of the signal joints are joined imprecisely in DNA-PKcs-deficient mice. We conclude that signal joints from SLIP mice are formed with frequency and fidelity similar to those of SCID mice.
DISCUSSION
Effects on Coding Joint Formation.
Our data show that DNA-PKcs-deficient SLIP mice are impaired in coding joint formation at multiple loci in both the T and B cell lineages. Levels of coding joints in SLIP mice are virtually indistinguishable from those in SCID mice and are similar to those in Ku86-deficient mice (29, 30). DNA-PKcs is thus required for efficient coding joint formation, and the residual DNA-PKcs protein found in SCID thymocytes does not assist this reaction. Recent analysis of DNA-PKcs null mice generated by gene targeting (40) supports these conclusions.
Hairpin coding ends accumulate in the thymocytes of SCID (38), Ku86-deficient (30), and DNA-PKcs-deficient mice (40). Our data show that these ends are used for efficient formation of hybrid joints in the absence of DNA-PKcs. These ends are thus available for some joining reactions. In vivo and biochemical experiments have found that hybrid joints can form by RAG proteins without the participation of Ku86 or XRCC4 (36, 39, 41, 42). Together, these data support a model in which the hairpin coding ends remain associated with the RAG proteins in a postcleavage complex that requires DNA-PKcs and other DNA repair factors for further processing (30, 43).
Although coding joint formation is severely compromised in SCID, Ku, and DNA-PKcs null mice, some coding joints can be detected in these animals (this work and refs. 31 and 40), suggesting the presence of a DNA-PK-independent bypass pathway for coding joint formation (36). Certain rearrangements, such as TCR Dδ2-Jδ1, appear to be much less affected by these mutations. Previous studies in SCID and Ku-deficient mice could not determine definitively whether these rearrangements require DNA-PKcs, because residual DNA-PKcs function could be present in these animals (4, 5, 12). Our data establish that these rearrangements are DNA-PKcs-independent. What is responsible for the relatively high levels of DNA-PK-independent rearrangements at these “privileged” sites? Recent data have shown that the TCRβ enhancer can specifically augment formation of certain TCRβ coding joints (44). Although the molecular basis of this phenomenon remains unknown, such enhancer effects could account for the ability of rearrangements at certain sites to bypass the normal requirement for DNA-PKcs.
Effects on Signal Joint Formation.
Two hypotheses have been proposed to explain why the murine SCID mutation predominantly affects coding joint formation, whereas null mutations in the other components of DNA-PK affect both coding and signal joints. According to the prevailing view, signal joints formed in SCID cells result from residual DNA-PKcs function (11, 12, 36). Our current data refute this hypothesis, because virtually identical levels of signal joints are found in SLIP mice. The recent analysis of DNA-PKcs knockout mice (40) supports this conclusion as well.
The second hypothesis posits that signal joint formation does not depend on DNA-PKcs (9, 35). Strictly speaking, this is confirmed by our results and by analysis of DNA-PKcs knockout thymocytes and ES cells (40), because significant levels of signal joints are observed in these situations. However, the observations outlined below lead us to suggest a third possibility: that DNA-PKcs normally plays an important, but nonessential, role in signal joint formation. We find that the abundance of signal joints is consistently reduced (up to 10-fold) in DNA-PKcs-deficient mice. Similar reductions have been observed in thymocytes of SCID (38) and targeted DNA-PKcs null mice (40), as well as in various DNA-PKcs-deficient cell lines (19, 45, 46). Furthermore, up to one-half of the signal joints formed in the absence of DNA-PKcs are abnormal. Signal joints formed in wild-type cells are generally sensitive to ApaLI, indicating that the junctions are perfect; without nucleotide loss or addition (this work and refs. 9, 25, 27, 28, and 34). In particular, the loss of nucleotides from signal joints is extremely rare (19). Together these observations indicate that DNA-PKcs plays an important role in signal joint formation, increasing both the efficiency and the fidelity of this reaction.
The aforementioned considerations suggest that there may be at least two pathways for signal joint formation: an efficient, faithful, DNA-PKcs-dependent pathway and an inefficient, error-prone, DNA-PKcs-independent pathway. Although the mechanisms involved remain unknown, the following model is consistent with the available data. After cleavage, signal ends are thought to be protected from nucleotide loss by continued association with the RAG proteins in a postcleavage complex (30, 47, 48). DNA-PKcs may facilitate faithful and efficient joining of the signal ends, perhaps by recruiting the joining machinery to the postcleavage complex. A similar role for DNA-PKcs has been suggested for coding joint formation, which would involve recruitment of the hairpin opening nuclease (9, 43). According to this view, in the absence of DNA-PKcs, both coding and signal ends would be left to join by less efficient alternative pathways. In the case of signal ends, joining could occur by simple blunt-end ligation. Of course, if this reaction occurred outside of the postcleavage complex the ends would be freely available to nuclease activities. It is striking that the precision of signal joint formation in SLIP mice (≈50%) closely resembles the precision with which blunt-ended DNA molecules microinjected into nuclei of mammalian cells are ligated (also ≈50%) (49). The much more severe defect in coding joint formation in the absence of DNA-PKcs could reflect less efficient alternative pathways for joining the hairpin coding ends.
Our suggestion that an alternative pathway may be responsible for signal joint formation in the absence of functional DNA-PKcs is supported further by analysis of DNA-PKcs null ES cells. Surprisingly, although fibroblasts from the DNA-PKcs null mice exhibit a SCID-like hypersensitivity to ionizing radiation, the corresponding ES cells do not (40). To explain these observations, Gao et al. (40) proposed the existence of an alternate DNA-PKcs-independent double-strand break repair pathway that is active in ES cells. Interestingly, signal joint formation, which is decreased ≈10-fold in thymocytes from DNA-PKcs-deficient mice, is more normal in the corresponding ES cells, with all joints formed precisely and only a 2.5-fold decrease in signal joint levels (average from three sets of experiments) (40). Thus, a DNA-PKcs-independent pathway might be responsible for both radioresistance and signal joint formation. Recent biochemical work has identified efficient DNA-PK-independent end joining pathways in extracts from rodent cells (50).
Comparison with Other DNA-PKcs Mutants.
Our results are easily reconciled with data from most of the previously reported DNA-PKcs mutants, which display a range of V(D)J recombination defects, as summarized in Table 1. Regardless of the presence or absence of defective DNA-PKcs protein, most of the mutant cell lines exhibit severe defects in coding joint formation, with less severe deficiencies in formation of signal joints, much as we see in SCID and SLIP mice. However, a Chinese hamster ovary cell line (XR-C1) bearing an unidentified mutation believed to affect DNA-PKcs is clearly deficient in both signal and coding joints (54). Furthermore, analysis of a spontaneous mutation causing immunodeficiency in Arabian foals (equine SCID) also revealed severe defects in formation of both coding and signal joints (55). It is not clear why these particular mutations give phenotypes that differ from those observed in SCID and DNA-PKcs-deficient mice. Perhaps these mutations create dominant negative alleles of DNA-PKcs that inhibit signal joint formation. Analysis of DNA-PKcs expression in equine SCID cells argues against this possibility, because this mutation deletes about 900 amino acids, resulting in an unstable protein product that is not detectable (55). In the case of the XR-C1 cell line, both coding and signal joints were rescued by microcell transfer of human chromosome 8. Although the human DNA-PKcs gene resides on this chromosome, the mutation in XR-C1 cells has not been precisely mapped to the SCID locus, and it is conceivable that there might be multiple mutations. Finally, the alternative joining pathway that we have suggested could be absent or less efficient in one or both of these contexts. Recent work provides a precedent for interspecies differences in DNA-PK-independent end joining pathways, because robust DNA-PK-independent joining is observed in extracts from rodent, but not human, cells (50).
Table 1.
Effects on V(D)J recombination in DNA-PKcs mutants
| Mutants | Mutation* | DNA-PKcs†
|
V(D)J‡
|
sj fidelity, % | Refs. | ||||
|---|---|---|---|---|---|---|---|---|---|
| Level | Binding | Activity | cj | sj | |||||
| Cell lines§ | |||||||||
| SCGR11 | SCID(m) | pt4045 | — | — | — | Defective | Normal | 80 | 9, 28 |
| fibrobl | SCID(m) | pt4045 | ND | ND | ND | Defective | Normal | 80 | 27 |
| pre-B | SCID(m) | pt4045 | ND | ND | ND | Defective | Reduced | 50 | 19 |
| SX-9 | MC(m) | L3192P | + | + | — | Defective | 10X↓¶ | 12 | 45, 51 |
| irs-20 | CHO | E4124K | ++ | + | — | Defective | 8X↓ | 75 | 46, 51, 52 |
| MO59J | MG(h) | nd | — | ND | — | Defective | Normal | 95 | 35, 53 |
| V3 | CHO | pt4024 | — | — | — | Defective | 2-10X↓ | 47–80 | 8, 9, 27, 46, 51 |
| DNA-PKcsN/N | ES(m) | null | — | ND | — | Defective | 2.5X↓ | 100 | 40 |
| XR-C1 | CHO | nd | — | ND | — | Defective | 60X↓ | — | 54 |
| Animals | |||||||||
| SCID | equ | pt3160 | — | ND | — | Defective | Defective | — | 55, 56 |
| SCID | mur | pt4045 | + | + | ±‖ | Defective | 10X↓ | ∼50 | 11–14, 38, this work |
| DNA-PKcsN/N | mur | null | — | ND | — | Defective | 10X↓ | ∼100 | 40 |
| SLIP | mur | null | — | ND | ND | Defective | 10X↓ | ∼50 | 37, this work |
ND, not done.
pt, premature termination; amino acid substitutions indicated by amino acid number preceded by wild-type amino acid (single-letter abbreviation) followed by substitution. The irs-20 mutation also was reported as E4120K (46). Note that the kinase domain is between amino acids 3719 and 4127.
Level, relative DNA-PKcs protein levels compared with wild type at +++; Binding, DNA-PK binding to DNA; Activity, DNA-PKcs activity.
cj, coding joint; sj, signal joint. Fold reductions are included as reported or calculated by us from reported data. Defective, low to undetectable; normal, wild-type levels; reduced, less than normal, but not as severe as defective.
(m), murine; (h), human; MC, mammary carcinoma; MG, malignant glioma; CHO, Chinese hamster ovary; ES, embryonic stem cell.
Although SX9 was reported to be defective in both coding and signal joint formation, the data show that the signal joint defect is only 10-fold below wild type.
Nonoverlapping Roles for DNA-PKcs, Ku, and XRCC4.
To date, four factors have been found to be critical for double-strand break repair and V(D)J recombination: Ku70, Ku86, DNA-PKcs, and XRCC4 (reviewed in refs. 1 and 2). All four of these proteins are required for coding joint formation. Although signal joint formation is quite dependent on Ku (29–31) and XRCC4 (41, 57), our data show that this reaction is clearly less dependent on DNA-PKcs. We have suggested that signal joint formation in DNA-PKcs-deficient cells may result from a DNA-PK-independent bypass pathway. The much more severe effect of mutations in the other three factors on signal joint formation could simply reflect their involvement in both the primary and the bypass pathways for signal joint formation. One possibility is that XRCC4 and Ku are instrumental in disassembling or remodeling the postcleavage complex, so that in their absence the ends are prevented from entering the bypass pathway. This suggestion is supported by the observation that coding and signal ends are not joined, but rather are protected from degradation in XRCC4-deficient cell lines and Ku-deficient cells and mice (30, 39, 41). An alternative or additional possibility is that Ku and XRCC4 may be required for joining of ends by the alternative pathway. This is supported by recent biochemical data showing that Ku brings DNA termini together and stimulates their ligation (58), and XRCC4 interacts with and stimulates the activity of DNA ligase IV (59, 60).
DNA-PKcs, Ku, and XRCC4 clearly have nonoverlapping roles in other aspects of DNA metabolism. For example, Ku86- and Ku70-deficient mice exhibit growth retardation that is not observed in SCID or DNA-PKcs-deficient animals (29, 31, 37, 40). Furthermore, although Ku-deficient yeast show defects in telomere maintenance (61–63), yeast bearing a targeted disruption of the XRCC4 homolog, LIF-1, do not (64). These data suggest that Ku has functions that do not involve DNA-PKcs and are not shared by XRCC4 and that the Ku heterodimer might interact with other important protein complexes.
Acknowledgments
We thank Fred Alt for sharing unpublished data on DNA-PKcs knockout mice. We greatly appreciate the efforts of Jeff Lin and Mary Lowe for their excellent technical and secretarial skills, respectively. We are also grateful to Vicky Brandt for critical review of the manuscript. This work was supported by grants from the National Institutes of Health (AI-36420 and CA-76317).
ABBREVIATIONS
- DNA-PK
DNA-dependent protein kinase
- IgH
Immunoglobulin heavy chain
- RAG
recombination activating gene
- TCR
T cell receptor
- V(D)J
variable (diversity) joining
- SCID
severe combined immunodeficient
- ES
embryonic stem
Note
After submission of this manuscript, Taccioli et al. (65) reported an additional targeted deletion of DNA-PKcs in mice. Although no quantitative analysis was shown, signal joint formation was reported to be “close to normal frequency,” but only 30% of the signal joints were perfect. The results of our study are in agreement with this report (65) and that of Gao et al. (40). Thus, all three studies of DNA-PKcs-deficient mice show abnormal formation of signal joints. We conclude that DNA-PKcs is important for faithful and efficient joining of signal ends.
References
- 1.Gellert M. Adv Immunol. 1997;64:39–64. doi: 10.1016/s0065-2776(08)60886-x. [DOI] [PubMed] [Google Scholar]
- 2.Jeggo P A. Adv Genet. 1998;38:185–218. doi: 10.1016/s0065-2660(08)60144-3. [DOI] [PubMed] [Google Scholar]
- 3.Dynan W S, Yoo S. Nucleic Acids Res. 1998;26:1551–1559. doi: 10.1093/nar/26.7.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yaneva M, Kowalewski T, Lieber M R. EMBO J. 1997;16:5098–5112. doi: 10.1093/emboj/16.16.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hammarsten O, Chu G. Proc Natl Acad Sci USA. 1998;95:525–530. doi: 10.1073/pnas.95.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bosma G C, Custer R P, Bosma M J. Nature (London) 1983;301:527–530. doi: 10.1038/301527a0. [DOI] [PubMed] [Google Scholar]
- 7.Kirchgessner C U, Patil C K, Evans J W, Cuomo C A, Fried L M, Carter T, Oettinger M A, Brown J M. Science. 1995;267:1178–1183. doi: 10.1126/science.7855601. [DOI] [PubMed] [Google Scholar]
- 8.Peterson S R, Kurimasa A, Oshimura M, Dynan W S, Bradbury E M, Chen D J. Proc Natl Acad Sci USA. 1995;92:3171–3174. doi: 10.1073/pnas.92.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blunt T, Finnie N J, Taccioli G E, Smith G C M, Demengeot J, Gottlieb T M, Mizuta R, Varghese A J, Alt F W, Jeggo P A, Jackson S P. Cell. 1995;80:813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 10.Boubnov N V, Weaver D T. Mol Cell Biol. 1995;15:5700–5706. doi: 10.1128/mcb.15.10.5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blunt T, Gell D, Fox M, Taccioli G E, Lehmann A R, Jackson S P, Jeggo P A. Proc Natl Acad Sci USA. 1996;93:10285–10290. doi: 10.1073/pnas.93.19.10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danska J S, Holland D P, Mariathasan S, Williams K M, Guidos C J. Mol Cell Biol. 1996;16:5507–5517. doi: 10.1128/mcb.16.10.5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Araki R, Fujimori A, Hamatani K, Mita K, Saito T, Mori M, Fukumura R, Morimyo M, Muto M, Itoh M, et al. Proc Natl Acad Sci USA. 1997;94:2438–2443. doi: 10.1073/pnas.94.6.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woo R A, McLure K G, Lees-Miller S P, Rancourt D E, Lee P W K. Nature (London) 1998;394:700–704. doi: 10.1038/29343. [DOI] [PubMed] [Google Scholar]
- 15.Fulop G M, Phillips R A. Nature (London) 1990;347:479–482. doi: 10.1038/347479a0. [DOI] [PubMed] [Google Scholar]
- 16.Biedermann K A, Sun J, Giaccia A J, Tosto L M, Brown J M. Proc Natl Acad Sci USA. 1991;88:1394–1397. doi: 10.1073/pnas.88.4.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendrickson E A, Qin X-Q, Bump E A, Schatz D G, Oettinger M, Weaver D T. Proc Natl Acad Sci USA. 1991;88:4061–4065. doi: 10.1073/pnas.88.10.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schuler W, Weiler I J, Schuler A, Phillips R A, Rosenberg N, Mak T W, Kearney J F, Perry R P, Bosma M J. Cell. 1986;46:963–972. doi: 10.1016/0092-8674(86)90695-1. [DOI] [PubMed] [Google Scholar]
- 19.Lieber M R, Hesse J E, Lewis S, Bosma G C, Rosenberg N, Mizuuchi K, Bosma M J, Gellert M. Cell. 1988;55:7–16. doi: 10.1016/0092-8674(88)90004-9. [DOI] [PubMed] [Google Scholar]
- 20.Malynn B A, Blackwell T K, Fulop G M, Rathbun G A, Furley A J W, Ferrier P, Heinke L B, Phillips R A, Yancopoulos G D, Alt F W. Cell. 1988;54:453–460. doi: 10.1016/0092-8674(88)90066-9. [DOI] [PubMed] [Google Scholar]
- 21.Bosma M J, Carroll A M. Annu Rev Immunol. 1991;9:323–350. doi: 10.1146/annurev.iy.09.040191.001543. [DOI] [PubMed] [Google Scholar]
- 22.Carroll A M, Bosma M J. Genes Dev. 1991;5:1357–1366. doi: 10.1101/gad.5.8.1357. [DOI] [PubMed] [Google Scholar]
- 23.Bogue M, Zhu C, Aguilar-Cordova E, Donehower L A, Roth D B. Genes Dev. 1996;10:553–565. doi: 10.1101/gad.10.5.553. [DOI] [PubMed] [Google Scholar]
- 24.Roth D B, Nakajima P B, Menetski J P, Bosma M J, Gellert M. Cell. 1992;69:41–53. doi: 10.1016/0092-8674(92)90117-u. [DOI] [PubMed] [Google Scholar]
- 25.Carroll A M, Slack J K, Mu X. J Immunol. 1993;150:2222–2230. [PubMed] [Google Scholar]
- 26.Carroll A M, Slack J K, Chang W-T. Mol Cell Biol. 1993;13:3632–3640. doi: 10.1128/mcb.13.6.3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pergola F, Zdzienicka M Z, Lieber M R. Mol Cell Biol. 1993;13:3464–3471. doi: 10.1128/mcb.13.6.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taccioli G E, Rathbun G, Oltz E, Stamato T, Jeggo P A, Alt F W. Science. 1993;260:207–210. doi: 10.1126/science.8469973. [DOI] [PubMed] [Google Scholar]
- 29.Nussenzweig A, Chen C, da Costa Soares V, Sanchez M, Sokol K, Nussenzweig M C, Li G C. Nature (London) 1996;382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- 30.Zhu C, Bogue M A, Lim D-S, Hasty P, Roth D B. Cell. 1996;86:379–389. doi: 10.1016/s0092-8674(00)80111-7. [DOI] [PubMed] [Google Scholar]
- 31.Gu Y, Seidl K J, Rathbun G A, Zhu C, Manis J P, van der Stoep N, Davidson L, Cheng H-L, Sekiguchi J M, Frank K, et al. Immunity. 1997;7:653–665. doi: 10.1016/s1074-7613(00)80386-6. [DOI] [PubMed] [Google Scholar]
- 32.Ouyang H, Nussenzweig A, Kurimasa A, da Costa Soares V, Li X, Cordon-Cardo C, Li W-h, Cheong N, Nussenzweig M, Iliakis G, Chen D J, Li G C. J Exp Med. 1997;186:921–929. doi: 10.1084/jem.186.6.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li G C, Ouyang H, Li X, Nagasawa H, Little J B, Chen D J, Ling C C, Fuks Z, Cordon-Cardo C. Mol Cell. 1998;2:1–8. doi: 10.1016/s1097-2765(00)80108-2. [DOI] [PubMed] [Google Scholar]
- 34.Gu Y, Jin S, Gao Y, Weaver D T, Alt F W. Proc Natl Acad Sci USA. 1997;94:8076–8081. doi: 10.1073/pnas.94.15.8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kulesza P, Lieber M R. Nucleic Acids Res. 1998;26:3944–3948. doi: 10.1093/nar/26.17.3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bogue M A, Wang C, Zhu C, Roth D B. Immunity. 1997;7:37–47. doi: 10.1016/s1074-7613(00)80508-7. [DOI] [PubMed] [Google Scholar]
- 37.Jhappan C, Morse H C, Fleischmann R D, Gottesman M M, Merlino G. Nat Genet. 1997;17:483–486. doi: 10.1038/ng1297-483. [DOI] [PubMed] [Google Scholar]
- 38.Roth D B, Menetski J P, Nakajima P B, Bosma M J, Gellert M. Cell. 1992;70:983–991. doi: 10.1016/0092-8674(92)90248-b. [DOI] [PubMed] [Google Scholar]
- 39.Han J-O, Steen S B, Roth D B. Mol Cell Biol. 1997;17:2226–2234. doi: 10.1128/mcb.17.4.2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao Y, Chaudhuri J, Zhu C, Davidson L, Weaver D T, Alt F W. Immunity. 1998;9:367–376. doi: 10.1016/s1074-7613(00)80619-6. [DOI] [PubMed] [Google Scholar]
- 41.Han J-O, Erskine L A, Purugganan M M, Stamato T D, Roth D B. Nucleic Acids Res. 1998;26:3769–3775. doi: 10.1093/nar/26.16.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melek M, Gellert M, van Gent D C. Science. 1998;280:301–303. doi: 10.1126/science.280.5361.301. [DOI] [PubMed] [Google Scholar]
- 43.Zhu C, Roth D B. Immunity. 1995;2:101–112. doi: 10.1016/1074-7613(95)90082-9. [DOI] [PubMed] [Google Scholar]
- 44.Hempel W M, Stanhope-Baker P, Mathieu N, Huang F, Schlissel M S, Ferrier P. Genes Dev. 1998;12:2305–2317. doi: 10.1101/gad.12.15.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fukumura R, Araki R, Fujimori A, Mori M, Saito T, Watanabe F, Sarashi M, Itsukaichi H, Eguchi-Kasai K, Sato K, et al. J Biol Chem. 1998;273:13058–13064. doi: 10.1074/jbc.273.21.13058. [DOI] [PubMed] [Google Scholar]
- 46.Priestley A, Beamish H J, Gell D, Amatucci A G, Muhlmann-Diaz M C, Singleton B K, Smith G C M, Blunt T, Schalkwyk L C, Bedford J S, et al. Nucleic Acids Res. 1998;26:1965–1973. doi: 10.1093/nar/26.8.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agrawal A, Schatz D G. Cell. 1997;89:43–53. doi: 10.1016/s0092-8674(00)80181-6. [DOI] [PubMed] [Google Scholar]
- 48.Hiom K, Gellert M. Cell. 1997;88:65–72. doi: 10.1016/s0092-8674(00)81859-0. [DOI] [PubMed] [Google Scholar]
- 49.Roth D B, Proctor G N, Stewart L K, Wilson J H. Nucleic Acids Res. 1991;19:7201–7205. doi: 10.1093/nar/19.25.7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baumann P, West S. Proc Natl Acad Sci USA. 1998;95:14066–14070. doi: 10.1073/pnas.95.24.14066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peterson S R, Stackhouse M, Waltman M J, Chen F, Sato K, Chen D J. J Biol Chem. 1997;272:10227–10231. doi: 10.1074/jbc.272.15.10227. [DOI] [PubMed] [Google Scholar]
- 52.Lin J Y-D, Muhlmann-Diaz M C, Stackhouse M A, Robinson J F, Taccioli G E, Chen D J, Bedford J S. Radiat Res. 1997;147:166–171. [PubMed] [Google Scholar]
- 53.Lees-Miller S P, Godbout R, Chan D W, Weinfeld M, Day R S, Barron G M, Allalunis-Turner J. Science. 1995;267:1183–1185. doi: 10.1126/science.7855602. [DOI] [PubMed] [Google Scholar]
- 54.Errami A, He D M, Friedl A A, Overkamp W J I, Morolli B, Hendrickson E A, Eckardt-Schupp F, Oshimura M, Lohman P H M, Jackson S P, Zdzienicka M Z. Nucleic Acids Res. 1998;26:3146–3153. doi: 10.1093/nar/26.13.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wiler R, Leber R, Moore B B, VanDyk L F, Perryman L E, Meek K. Proc Natl Acad Sci USA. 1995;92:11485–11489. doi: 10.1073/pnas.92.25.11485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shin E K, Perryman L E, Meek K. J Immunol. 1997;158:3565–3569. [PubMed] [Google Scholar]
- 57.Li Z, Otevrel T, Gao Y, Cheng H-L, Seed B, Stamato T D, Taccioli G E, Alt F W. Cell. 1995;83:1079–1089. doi: 10.1016/0092-8674(95)90135-3. [DOI] [PubMed] [Google Scholar]
- 58.Ramsden D A, Gellert M. EMBO J. 1998;17:609–614. doi: 10.1093/emboj/17.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Critchlow S E, Bowater R P, Jackson S P. Curr Biol. 1997;7:588–598. doi: 10.1016/s0960-9822(06)00258-2. [DOI] [PubMed] [Google Scholar]
- 60.Grawunder U, Wilm M, Wu X, Kulesza P, Wilson T E, Mann M, Lieber M R. Nature (London) 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 61.Boulton S J, Jackson S P. Nucleic Acids Res. 1996;24:4639–4648. doi: 10.1093/nar/24.23.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Porter S E, Greenwell P W, Ritchie K B, Petes T D. Nucleic Acids Res. 1996;24:582–585. doi: 10.1093/nar/24.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nugent C I, Bosco G, Ross L O, Evans S K, Salinger A P, Moore J K, Haber J E, Lundblad V. Curr Biol. 1998;8:657–660. doi: 10.1016/s0960-9822(98)70253-2. [DOI] [PubMed] [Google Scholar]
- 64.Herrmann G, Lindahl T, Schär P. EMBO J. 1998;17:4188–4198. doi: 10.1093/emboj/17.14.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taccioli G E, Amatucci A G, Beamish H J, Gell D, Xiang X H, Arzayus M I T, Priestley A, Jackson S P, Rothstein A M, Jeggo P A, Herrera V L M. Immunity. 1998;9:355–366. doi: 10.1016/s1074-7613(00)80618-4. [DOI] [PubMed] [Google Scholar]