Abstract
Genetic studies in budding yeast have previously implicated SLX5 and SLX8 in the control of genome stability and sumoylation. These genes encode RING-finger domain proteins that form a complex of unknown function. Because RING-finger proteins comprise a large class of ubiquitin (Ub) ligases, Slx5 and Slx8 were tested for this activity. Here we show that the Slx5-Slx8 complex, but not its individual subunits, stimulates several human and yeast Ub conjugating enzymes, including Ubc1, 4, 5, and Ubc13-Mms2. The RING-finger domains of both subunits are genetically required for suppression of slx sgs1Δ synthetic-lethality, and point mutations that abolish Ub ligase activity in vitro also eliminate in vivo complementation. Targets of the in vitro ubiquitination reaction include the Slx5 and Slx8 subunits themselves, and the homologous recombination proteins Rad52 and Rad57. We propose that the Slx5-Slx8 complex functions as a two-component Ub ligase in vivo and that it controls genome stability and sumoylation via ubiquitination.
Keywords: genome stability, recombination, SUMO, RecQ, Sgs1 DNA helicase, Ub ligase
INTRODUCTION
As an established technique to identify genetic interactions in the yeast S. cerevisiae, synthetic-lethal screens have recently been used to characterize pathways imporant for genomic integrity.1–3 A variety of such screens identified SLX5 and SLX8 as novel genes required for viability in the absence of the SGS1 DNA helicase.4–6 The DNA helicase-topoisomerase complex encoded by SGS1, TOP3, and RMI1 has a well-documented role in maintaining genome stability,7–10 and accumulating evidence indicates that SLX5 and SLX8 define its own DNA integrity pathway.3 In a separate genetic screen, mutations in these genes were isolated together with multiple components of the SUMO pathway, as suppressors of the general transcription factor Mot1.11 Genetic and yeast two-hybrid interactions between SLX5-SLX8 and SUMO pathway genes suggest that they may regulate global sumoylation levels.11–14 At present, however, no enzymatic activity has been ascribed to Slx5 and Slx8.
On their own, null mutations in SLX5 and SLX8 produce nearly identical phenotypes. The slx5Δ and slx8Δ mutants display slow growth, a reduced plating efficiency compared to wild type (wt), and a heterogeneous colony morphology consisting of a mixture of large and small colonies with rough edges.4, 12, 14 Like sgs1Δ mutants, slx5Δ or slx8Δ cells are sensitive to the DNA synthesis inhibitor hydroxyurea (HU) and homozygous mutant cells display a reduced sporulation frequency compared to wt.4 Recent studies have shown that these mutants display elevated rates of gross chromosomal rearrangements (GCRs)15 and either elevated or reduced levels of SUMO-conjugated proteins.11, 12, 14 Where examined, these mutants display genetic epistasis such that the phenotype of the slx5Δ slx8Δ double mutant is no worse than that of either single mutant.4, 11, 15 Consistent with this epistasis, Slx5 and Slx8 proteins co-immunoprecipitate when overproduced in yeast and have been purified as a heterodimeric protein complex when expressed in recombinant form.4, 12, 16
Both Slx5 and Slx8 contain a single C-terminal “RING-finger” domain which implicates them in the process of ubiquitination.17, 18 The covalent modification of proteins with Ub is carried out through the sequential activity of a single activating (E1) enzyme, one of many conjugating (E2) enzymes, and one an ever-growing number of Ub ligases (E3).19 There are two major classes of Ub ligases and a large proportion of RING-domain proteins comprise one of these classes. RING E3s facilitate the addition of either mono-Ub, or one of several types of polyubiquitin chains, to a target protein. These chains are distinguished by their inter-Ub linkage, which can occur at one of seven lysine residues,20 and can serve an assortment of functions.21, 22 The best characterized function is the stimulation of proteosomal protein degradation by K48-linked chains. In contrast, K63-linked chains are known to promote nonproteolytic processes such as DNA damage tolerance and protein trafficking.23–25
In this study we set out to identify a biochemical activity associated with Slx5 and Slx8. In vitro assays revealed that the Slx5-Slx8 complex stimulates Ub conjugation by a subset of known Ubcs. The Slx5 and Slx8 subunits were auto-ubiquitinated in these assays whereas the individual subunits were inactive. A variety of point- and truncation-mutants confirmed that both RING-finger domains are essential for in vivo function. Moreover, using a set of purified mutant complexes we find a strong correlation between ubiquitination activity in-vitro and in vivo function. The data are consistent with the idea that Slx5-Slx8 controls genome stability by acting as a heterodimeric Ub ligase.
MATERIALS AND METHODS
Yeast Strains and Plasmids
Standard methods and media were used for the propagation, transformation and culturing of S. cerevisiae. 26 All strains are derivatives of W303-1a (MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 rad5-535).27 Mutant alleles of SLX5 and SLX8 were tested by functional complementation of strains JMY1699 (MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 rad5-535 slx5-11::hph), JMY1604 (MATa ade2-1 ura3-1 trp1-1 leu2-3,112 rad5-535 slx8-10::KAN::loxP), JMY1464 (MATa ade2 ade3::hisG ura3 his3-11,15 leu2 trp1-1 lys2 rad5-535 slx5-10::TRP1 sgs1-20::hph can1-100 + pJM500 [SGS1/URA3/ADE3]), or VCY1525 (MATa ade2-1 ade3::hisG ura3-1 his3-11,15 trp1-1 leu2-3,112 lys2 rad5-535 slx8-10::KAN sgs1-20::hph + pJM500). Yeast complementation experiments were carried out using pRS centromeric plasmids as vectors.28 Plasmids used to express recombinant proteins were constructed by amplifying the necessary protein coding regions on NdeI/BamHI fragments and ligating them into the T7 vectors pET11a or pET28a 29 as follows: pNJ6915 (His6-Mms2 + Ubc13) and pJM6996 (His6-Ubc5). The wt plasmids pJM6819 (His6-Slx5 + Slx8), pJM6813 (His6-Slx8) and pJM6511 (His6-Slx5) have been described.16
Expression and purification of recombinant proteins
His6-tagged proteins were produced in E. coli BL21-RIL cells (Stratagene) using the T7 expression system of Studier.29 Typically, a 1L culture of transformed cells were grown at 30°C until the OD600 was equal to 0.5, and protein production was induced by the addition of IPTG to 0.4 mM. Expression continued for 16 hours after which the cells were pelleted, washed, and resuspended in 40 ml Buffer N [25 mM Tris-HCl (pH 8.1), 10% glycerol, 500 mM NaCl, 0.01% NP40, 1 mM dithiothreitol (DTT) and 0.1 mM phenymethylsulfonyl floride (PMSF)] containing the following protease inhibitors: pepstatin, 10 μg/ml; leupeptin, 5 μg/ml; benzamidine, 10 mM; and bacitracin, 100 μg/ml. The cells were then incubated with 0.1 mg/ml lysozyme on ice for 15 min, sonicated three times for 1 min each, and centrifuged at 30,000 × g for 30 min. The soluble portion was collected, passed through a 0.45 mm cellulose acetate filter, made 10 mM in imidazole, and applied to a 1 ml His-Trap column on an AKTA FPLC (GE Healthcare). The affinity column was washed with 10 column volumes of Buffer N containing 10 mM imidazole and eluted with a 6 ml linear gradient of Buffer N containing 10 to 500 mM imidazole. The peak fractions were identified by SDS-PAGE, pooled, and dialyzed to buffer A [25 mM Tris-HCl (pH 7.5), 1 mM EDTA, 0.01% NP40, 1 mM DTT, 10% glycerol and 0.1 mM PMSF] containing 100 mM NaCl and stored at −80°C. Uba1, human Ubc proteins, anti-Ub, and wt and mutant Ubs were obtained from Boston Biochem. Yeast MORF proteins were partially purified from 1.5 L yeast cultures as described and were typically 50% pure.30 Purification of Slx5-Slx8 complex has been described.16
In vitro ubiquitination assay
Ubiquitination was assayed under the following conditions: 20 mM HEPES (pH 7.5), 5 mM MgCl2, 2 mM ATP, 5 μM ZnSO4, and 0.1 mM DTT. Reactions were incubated at 30°C for 10 - 60 min and contained 13 - 27 nM E1, 21 – 400 nM E2, 2 – 240 nM E3, and 1 μM Ub in a 20 μl reaction. Reaction products were immunoblotted with anti-Ub (Sigma) followed by goat anti-rabbit HRP as secondary Ab (GE Healthcare). Signals were detected using a chemiluminescent substrate (Pierce) and an LAS-3000 chemiluminescence camera (Fujifilm).
RESULTS
Slx5-Slx8 is a heterodimeric Ub ligase
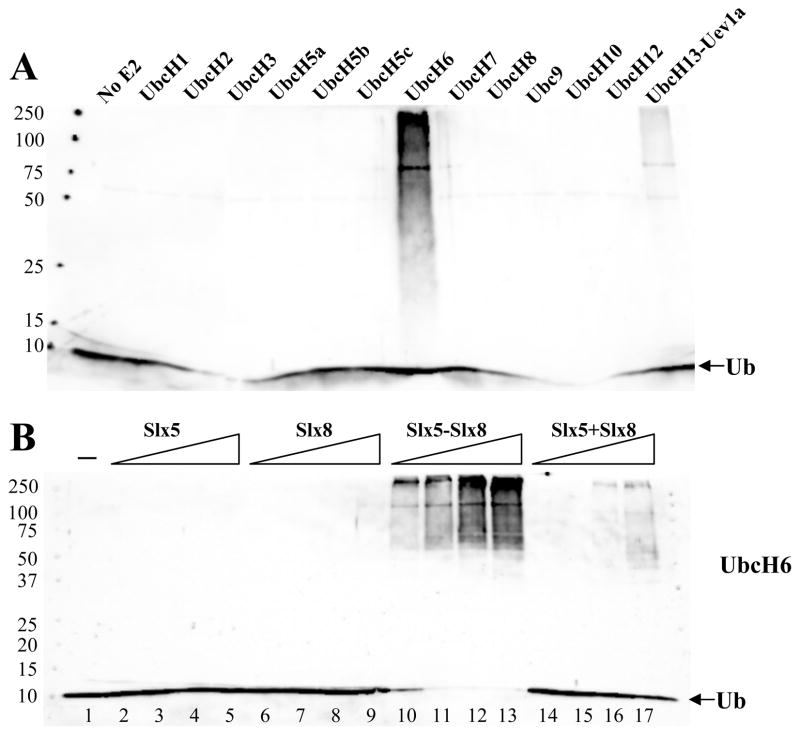
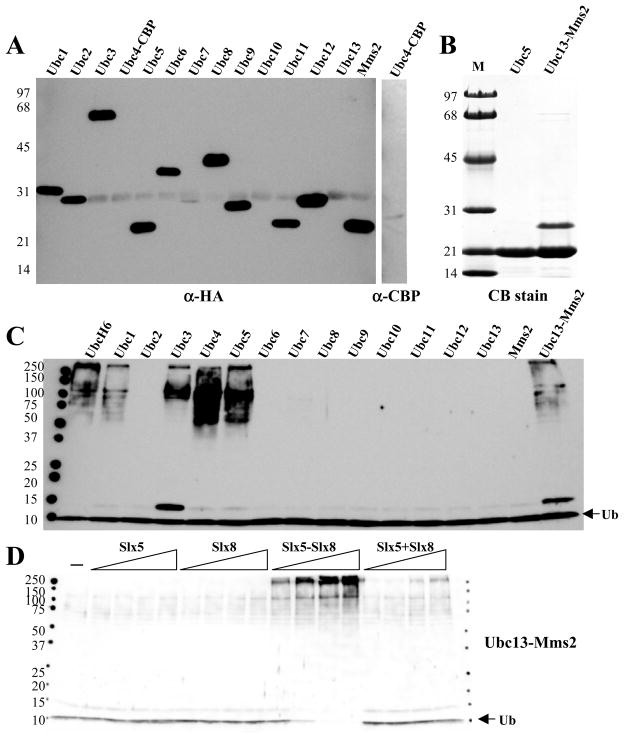
To determine their biochemical function we purified Slx5, Slx8, and a complex of the two subunits.16 These proteins were then assayed for the ability to stimulate Ub chain formation in the presence of the E1, E2, and ATP. Ub chain formation was detected by immunoblotting the reaction products with an antibody against Ub. In order to detect ubiquitin ligase activity, we first screened a collection of ubiquitin conjugating enzymes (Ubc’s) for a functional interaction with the Slx5-Slx8 complex. When the Slx5-Slx8 complex was incubated with a variety of human Ubc proteins we detected a strong signal with UbcH6 and a weaker signal with UbcH13-Uev1a (Fig. 1A). This signal consisted of a diffuse ladder of bands from about 60 kDa to the top of the resolving gel. In addition, there was an intense band at about 105 kD and a weaker band at 75 kD.
Figure 1.
The Slx5-Slx8 complex is a Ub E3 ligase. (A) Standard ubiquitination reactions were carried out as described in the Materials and Methods. Reactions contained 27 nM E1, 140 nM Slx5-Slx8, and 6 μM ubiquitin, in addition to the following E2s: Mock (lane 1), UbcH1 (40 nM), UbcH2 (20 nM), UbcH3 (30 nM), UbcH5a (20 nM), UbcH5b (20 nM), UbcH5c (20 nM), UbcH6 (48 nM), UbcH7 (55 nM), UbcH8 (55 nM), Ubc9 (40 nM), UbcH10 (50 nM), UbcH12 (50 nM), or UbcH13-Uev1a (24 nM). Following incubation (30°C, 10 min), the reaction products were analyzed by SDS-PAGE and immunoblotting using antibody against ubiquitin. (B) The Slx5-Slx8 subunit requirements were tested in ubiquitination reactions that contained 13 nM E1, 140 nM UbcH6 as E2 and various amounts of either Slx5 (3.4, 10, 34, 69 nM), Slx8 (7.6, 23, 76, 150 nM), Slx5-Slx8 complex (4.8, 14, 48, 96 nM), or Slx5 (3.4, 10, 34, 69 nM) plus Slx8 (7.6, 23, 76, or 150 nM) as indicated. Lane 1 contained no E3. Following incubation (30°C, 60 min), the reaction products were analyzed as above.
To confirm that this signal depended on Slx5-Slx8, we repeated the UbcH6 reaction in the presence of increasing levels of Slx5-Slx8, or its individual subunits. Titrating the individual subunits into reactions containing UbcH6 revealed that they were unable to stimulate Ub conjugation on their own. However, the complex (Fig. 1B, lanes 10–13), and to a lesser extent a combination of Slx5 plus Slx8 (Fig. 1B, lanes 14–17), produced Ub chains that extended to the top of the gel. At maximum input of Slx5-Slx8 there was substantial signal at the top of the gel and consumption of all free Ub. The requirement for both subunits in this assay is consistent with their genetic epistasis 4, 11, 15 and indicates that Slx5-Slx8 functions as a heterodimer.
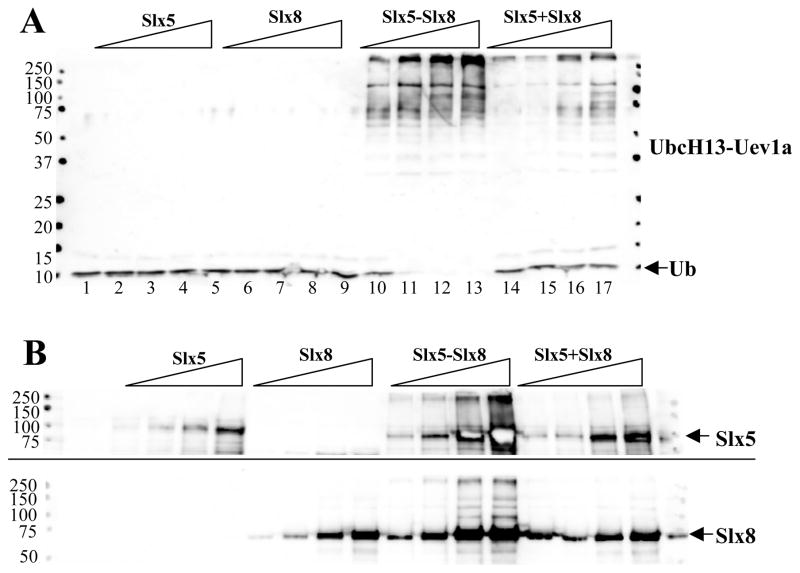
Slx5-Slx8 also stimulated Ub conjugation by UbcH13-Uev1a (Fig. 2A, lanes 10–13). The signal obtained with UbcH13-Uev1a was similar to that of UbcH6 in that it consumed all free Ub and smeared to the top of the gel, but more individual bands were distinct. As with UbcH6, the stimulation of UbcH13-Uev1a by Slx5-Slx8 was dependent on the presence of both subunits.
Figure 2.
Slx5-Slx8 stimulates UbcH13-Uev1a to ubiquitinate both E3 subunits. (A) The Slx5-Slx8 subunit requirements were tested as in Figure 1B except that ubiquitination reactions contained 90 nM UbcH13-Uev1a as E2. (B) Ubiquitination reactions were performed exactly as in (A) above, but following incubation the reaction products were analyzed by SDS-PAGE and immunoblotting using antibody against Slx5 (top) or Slx8 (bottom). Arrows indicate the position of unmodified Slx5 (top) or Slx8 (bottom).
The above reactions lack an obvious ubiquitination target, although Ub ligases are often efficiently ubiquitinated in vitro.17 To test whether these subunits were modified by ubiquitination we probed identical immunoblots with antisera that had been raised against Slx5 or Slx8.16 Slx5 (Fig. 2B, upper panel) and Slx8 (Fig. 2B, lower panel) were unmodified when assayed alone. However, when present as a dimer anti-Slx5 signal appeared as a smear to the well while anti-Slx8 signal appeared as a ladder of bands that ascended to the top of the gel. The reconstituted activity showed weak anti-Slx5 signal at the top of the gel while only a few bands larger than Slx8 were observed at the highest concentrations. Identical results were obtained using UbcH6 as the E2 enzyme (data not shown). Thus, ubiquitination of both Slx5 and Slx8 can occur but only when they are present as a complex. We note that Slx5 appears to be a better substrate for ubiquitination, perhaps because of its larger size. We conclude that Slx5-Slx8 functions as a heterodimeric Ub ligase in vitro and that it stimulates its own ubiquitination by UbcH6 and UbcH13-Uev1a.
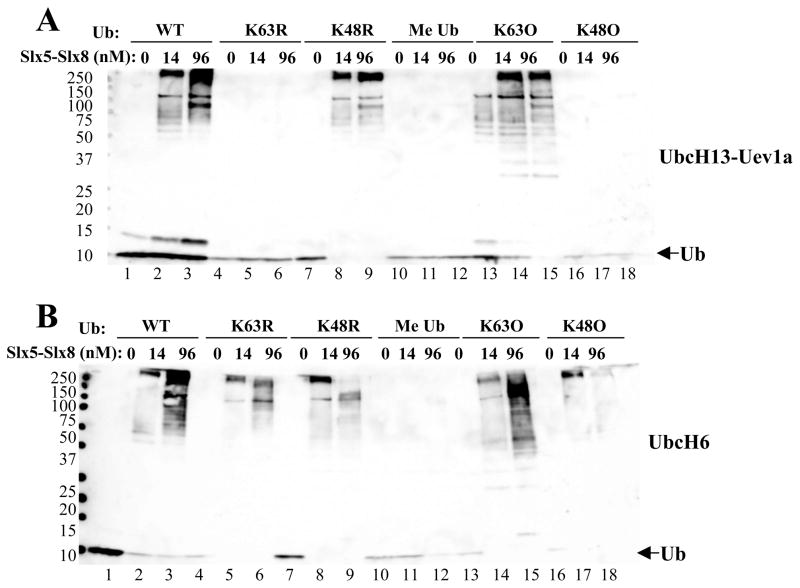
We next tested the specificity of Ub chain linkages stimulated by Slx5-Slx8. In contrast to K48-linked Ub chains that promote proteosomal degradation, UbcH13-Uev1a is known to conjugate K63-linked Ub chains like its yeast ortholog.31 Consistent with this, the UbcH13-Uev1a-conjugated chains that were stimulated by Slx5-Slx8 were dependent on Ub containing K63 and they formed even in the presence of a K48R mutant version of Ub (Fig. 3A, lanes 7–9). Moreover, a mutant Ub containing only one lysine at residue 63 (K63O) was efficiently conjugated by this E2 while the K48O variant was inactive (Fig. 3A, lanes 13–18). As expected, blocking all lysine side chains via chemical methylation of Ub (Me Ub) completely inhibited these reactions. We conclude that Slx5-Slx8 stimulates the known Ub-linkage specificity for UbcH13-Uev1a.
Figure 3.
Analysis of Ub chain linkage by two human Ubc’s. (A) Ubiquitination reactions contained 13 nM E1, 71 nM UbcH13-Uev1a as E2, the indicated concentration of Slx5-Slx8, and 1 μM of the indicated wt or mutant ubiquitin. (B) Ubiquitination reactions were performed as above but contained 140 nM UbcH6 as E2. Me Ub represents methylated ubiquitin; K63O represents Ub in which all lysines except for K63 are mutated to arginine; and K48O represents Ub in which all lysines except K48 are mutated to arginine. Following incubation (30°C, 60 min), the reaction products were immunoblotted with anti-Ub antibody.
A similar analysis was conducted with UbcH6 which is known to form K48-linked chains. Consistent with this, Slx5-Slx8 stimulated UbcH6 to conjugate Ub-K63R chains (Fig. 3B, lanes 4–6) and high molecular weight (MW) chain formation was partially inhibited when Ub-K48R was provided as substrate (Fig. 3B, lanes 7–9). At high Slx5-Slx8 concentrations, Ub-K48R polymerization was inhibited in favor of mono-ubiquitination of Slx5. Interestingly, Ub chains formed using either Ub-K63O and Ub-K48O substrates. Thus, we conclude that the linkage specificity of the ubiquitin conjugating enzymes is mostly retained, while UbcH6 can be stimulated by Slx5-Slx8 to form chains with both K48 or K63 linkages. The ability of an E3 ligase to alter the specificity of certain Ubcs or to direct the synthesis of chains with mixed linkages has previously been demonstrated. 20, 32, 33
Role of RING-finger domains in Ub E3 ligase activity
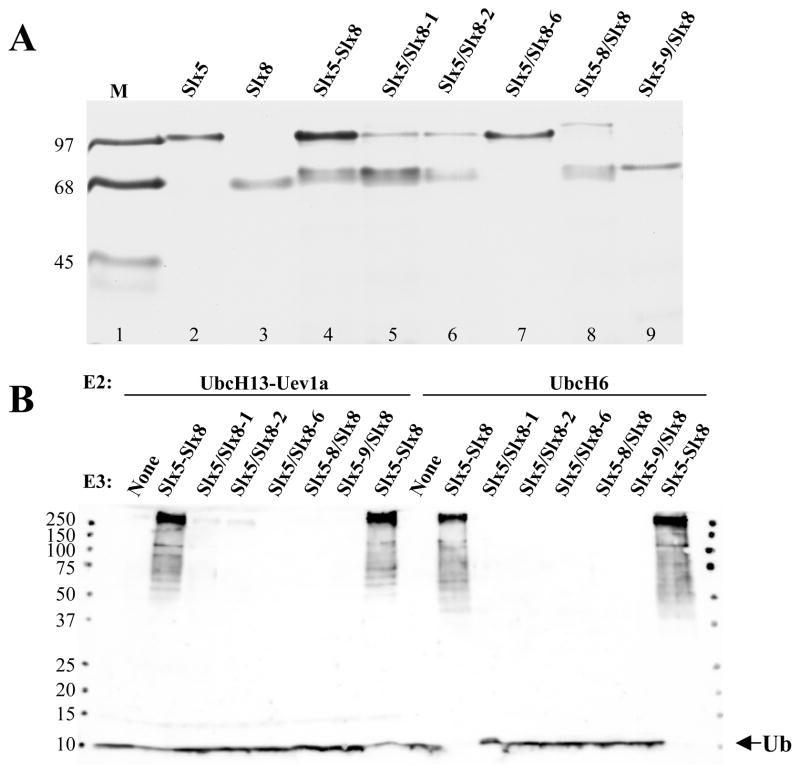
RING-domain Ub ligases are sensitive to mutation of zinc-coordinating residues.17 To test the role of these conserved cysteines and histidines in Slx5-Slx8, we expressed heterodimeric complexes in which one of the two subunits contained point mutations in these residues. Three complexes were purified that contained point mutations in Slx5 (Slx5-8/Slx8) or Slx8 (Slx5/Slx8-1, Slx5/Slx8-2) (Fig. 4A and Table 1). In all cases the proteins were well-expressed in E. coli and heterodimeric complexes were purified via the N-terminal hexa-histidine tag on Slx5 (Fig. 4A).
Figure 4.
Slx5-Slx8 Ub ligase activity requires intact RING domains. (A) Recombinant heterodimers of the indicated mutant versions of Slx5-Slx8 were purifed by affinity chromatography, resolved by 15% SDS-PAGE, and detected by Coomassie blue staining. Complexes containing a mutant subunit are indicated by the hyphenated allele name and a slash between the subunits. Note that all hetero-dimers contain an Slx5 breakdown product that migrates just behind the Slx8 subunit by SDS-PAGE. (B) Ubiquitination reactions contained 13 nM E1, either UbcH13-Uev1a (71 nM; lanes 1–8) or UbcH6 (140 nM; lanes 9–16) as E2, and 48 nM of the indicated wt or mutant Slx5-Slx8 heterodimer as E3. Following incubation (30°C, 60 min), the reaction products were immunoblotted with anti-Ub antibody.
Table 1.
Summary of SLX5-SLX8 RING mutationsa
| Allele Name | RING finger Position | In vivo complementationb | In vitro activityc | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| C | C | C | H | C | C | C | C | sgs1Δ slxΔ | HU | Ub | |
| SLX5 | 494 | 497 | 556 | 558 | 561 | 564 | 599 | 604 | + | + | + |
| slx5-6 | A | A | − | − | − | ||||||
| slx5-7 | S | + | + | + | |||||||
| slx5-8 | S | A | S | − | − | − | |||||
| slx5-9 | (ΔC126) | − | − | − | |||||||
| SLX8 | 206 | 209 | 221 | 223 226 | 229 | 246 | 249 | + | + | + | |
| slx8-1 | Y | − | − | − | |||||||
| slx8-2 | S | +/− | + | +/− | |||||||
| slx8-3 | S | A | S | − | − | − | |||||
| slx8-6 | (ΔC74) | − | − | − | |||||||
The positions of the residues corresponding to the yeast Slx5 and Slx8 RING fingers are presented for the wt proteins. Where appropriate, the amino acid changes found in mutant alleles are shown relative to the RING finger. The wt Slx5 and Slx8 proteins are 619 and 274 aa, respectively.
Shown is an indication of each allele’s ability to complement slxΔ sgs1Δ synthetic lethality or sensitivity to hydroxyurea; complementation of slx8Δ sgs1Δ synthetic lethality is taken from Yang et al. (2006).
In vitro assay refers to the auto-ubiquitylation activity of an Slx5-Slx8 dimer consisting of the indicated wt or mutant subunit together with its wt counterpart in the presence of yeast Ubc5.
Symbols: + = wt activity; +/− = less than wt activity; − = no activity; ND = Not Done.
In order to completely eliminate RING function, we also expressed heterodimers in which the C-terminal RING domains of either Slx5 (Slx5-9/Slx8) or Slx8 (Slx5/Slx8-6) had been deleted. However, after purifying the N-terminally tagged Slx5 protein, we observed that Slx8-6 (ΔRING) protein failed to bind Slx5-wt (Fig. 4A, lane 7). Similarly, Slx5-9 (ΔRING) protein failed to interact stably with Slx8-wt (Fig. 4A, lane 9). Immunoblotting for these proteins revealed that all were well-expressed and that the untagged Slx8 proteins were present in the Ni-column flow-through fractions. Thus, the RING domains of Slx5 and Slx8 are required for these subunits to interact.
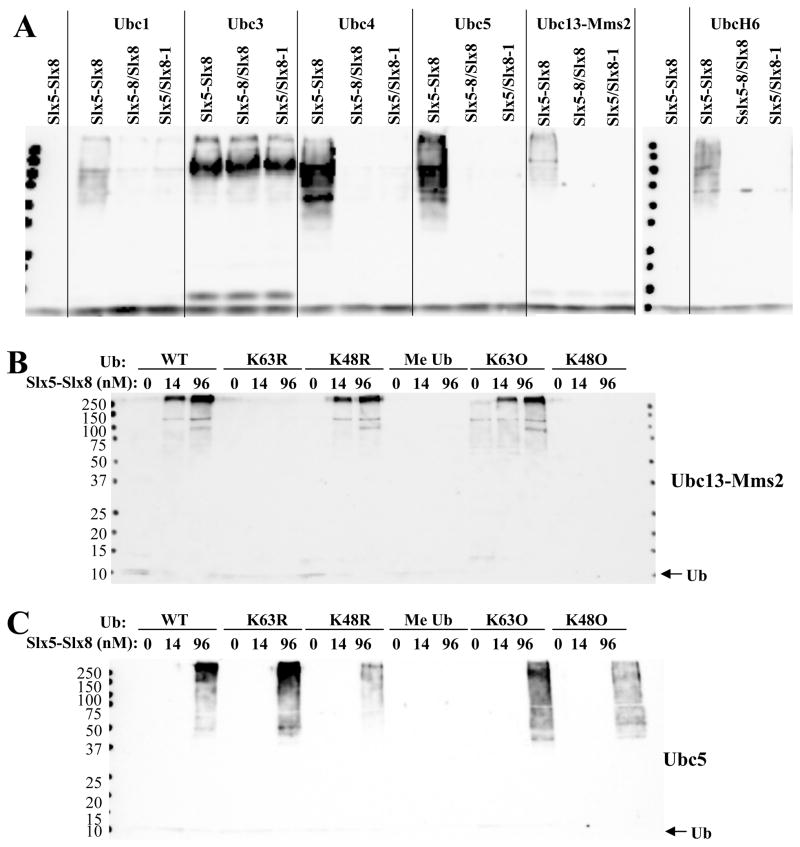
We next assayed these mutant proteins in ubiquitination reactions. As shown in Fig. 4B, complexes of Slx5-wt and an Slx8 subunit bearing a single point mutation in its RING domain (Slx5/Slx8-1, Slx5/Slx8-2) were severely compromised in their ability to stimulate conjugation by either UbcH6 or UbcH13-Uev1a. A complex containing the triple point mutation in Slx5 (Slx5-8/Slx8) was essentially non-functional. The inability of the Slx5-9/Slx8 or Slx5/Slx8-6 preparations to stimulate the reaction is likely due to the absence of the Slx8 subunit. We conclude that Slx5-Slx8 complex formation requires the RING domains of both subunits and that stimulation of Ub conjugation by the complex requires the zinc-coordinating residues in both subunits.
The RING-domains of both Slx5 and Slx8 are required for in vivo function
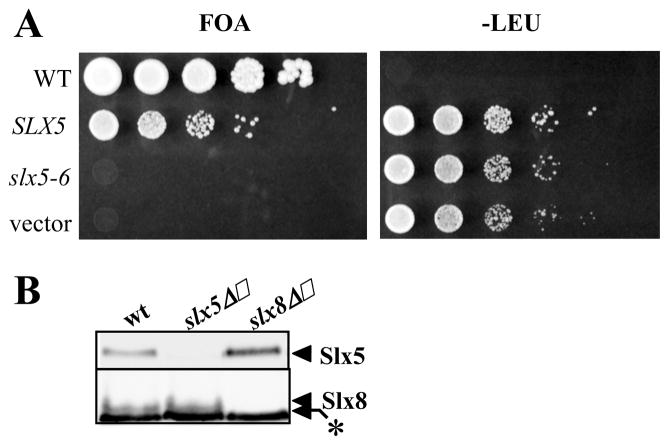
To determine whether the RING-domain of Slx5 is important in vivo, a structure-function study was performed. As shown in Table 1, conserved cysteine and histidine residues within the Slx5 RING domain were altered to produce single, double, and triple point mutants. These alleles were then assayed for their ability to complement sgs1Δ slx5Δ synthetic-lethality and slx5Δ HU sensitivity. The slx5-6 allele, encoding a double point mutant (C494A C497A), was unable to complement either of these defects (Fig. 5A and Table 1). This null phenotype was shared with slx5-9, which deletes the entire RING domain, and the triple point mutant slx5-8 (Table 1). However, the single point mutant slx5-7 showed complementation indicating that the loss of one zinc-coordinating residue can be tolerated in vivo. A structure-function analysis of SLX8 yielded similar results. SLX8 alleles that deleted the RING domain (slx8-6) or carried multiple RING domain mutations were unable to complement slx8 HUΔ sensitivity or sgs1Δ slx8Δ synthetic-lethality. In contrast, the single point mutation encoded by slx8-2 showed partial complementation in both assays.16 The ability of slx8-2 to complement HU sensitivity is probably due to its partial activity and the low resolution of this assay. Taken together, we conclude that both RING domains of Slx5 and Slx8 are critical for in vivo function.
Figure 5.
Slx5 and Slx8 require intact RING domains for in vivo function. (A) The slx5-6 allele was tested for in vivo function by complementation of sgs1Δ slx5Δ synthetic lethality. JMY1464 cells were transformed with pTI6537 (slx5-6), pJM6512 (SLX5-wt) or pRS405 (vector) that had been linearized with BstEII. Transformed JMY1464 cells or W303-1a (WT, leu2) cells were collected, adjusted to a constant cell density, and serially diluted in 10-fold steps before spotting on the indicated media. (B) Extracts from the indicated yeast strains were resolved by 12.5% SDS-PAGE and immunoblotted using antibodies against either Slx5 (upper) or Slx8 (lower). A cross-reacting band (*) migrates just below Slx8.
A trivial explanation for the above results, and for the similarity of slx5Δ and slx8Δ phenotypes, is that inactivation of one subunit leads to the degradation of its partner protein. To test this possibility, we immunoblotted for Slx5 and Slx8 in extracts of the two null strains. As shown in Figure 5B, both subunits appeared to be stable as monomers as their abundance was not significantly different than in wt extracts. Thus, Slx5 and Slx8 are important for an activity other than simply stabilizing the other subunit.
Table 1 compares these RING-domain phenotypes to the ability of corresponding mutant complexes to stimulate in vitro ubiquitination by yeast Ubc5 (Fig. S1). The data indicate that the behavior of a given allele is fairly consistent across the three assays. In cases where the in vivo complementing activity was reduced or eliminated, in vitro ubiquitination was similarly affected. Taken together, these data suggest that the RING domains of Slx5 and Slx8 are essential for their in vivo function because they are needed to stimulate ubiquitination.
Slx5-Slx8 stimulates multiple yeast Ubc enzymes
To identify yeast Ubcs that function together with Slx5-Slx8, an affinity purification technique was used to isolate enriched fractions of each of the known Ubcs in yeast. We isolated 13 epitope-tagged Ubc proteins from yeast using the MORF expression system,30 together with a TAP-tagged Ubc4 protein.34 Following proteolytic cleavage to liberate the Ubcs from the IgG affinity beads, they were immunoblotted with anti-HA (or anti-CBP) to identify the epitope-tagged proteins (Fig. 6A). These blots indicated that in all cases proteins of the expected size were obtained, although yields of Ubc7, 10, and 13 were low. We separately purified recombinant Ubc5 and Ubc13/Mms2 (Fig 6B).
Figure 6.
Slx5-Slx8 stimulates specific yeast Ub conjugating enzymes. (A) The indicated Ubc proteins were partially purified from yeast cells expressing HA (MORF) or CBP (TAP) epitope tagged fusion proteins using IgG affinity resin. Proteins were released from the beads by proteolytic cleavage, and samples (180 ng total protein) were immunoblotted with antibodies against HA or CBP. (B) Recombinant (His6)Ubc5 (2 μg) and (His6)Ubc13-Mms2 (4 μg) was purified by Ni affinity chromatography, resolved by 17% SDS-PAGE, and detected by Coomassie blue (CB) staining. (C) Stimulation of yeast Ubcs by Slx5-Slx8 was tested in ubiquitination reactions that contained 13 nM Uba1, 140 nM Slx5-Slx8, 5 μM Ub, and either UbcH6 (100 nM), Ubc13-Mms2 (400 nM), or 400 ng of the indicated partially-purified yeast Ubc. Following incubation (30°C, 10 min), the reaction products were immunoblotted with anti-Ub antibody. (D) The Slx5-Slx8 subunit requirements were tested as in Figure 1B except that ubiquitination reactions contained 100 nM Ubc13-Mms2 as E2.
Using the assay described above, each Ubc was assayed for Ub chain formation in the presence of Slx5-Slx8 dimer. As shown in Fig. 6C, Ub chain formation was observed using Ubc1, 3, 4, 5, and Ubc13-Mms2. As with the human Ubc’s, stimulation of Ubc13-Mms2 or Ubc5 (data not shown) was dependent on the presence of the Slx5-Slx8 dimer since no Ub chain formation was obtained in the presence of the individual subunits alone (Fig. 6D).
To determine whether formation of Ub chains depended on the RING domains of Slx5-Slx8, these five yeast Ubcs were assayed in the presence of either wt Slx5-Slx8 complex or complexes bearing point mutations in either Slx5 or Slx8. As shown in Fig. 7A, intact RING domains were required for the formation of Ub chains by Ubc1, 4, 5, and Ubc13-Mms2. Although Ubc3 formed chains in the presence of Slx5-Slx8, they were not dependent on intact RING domains. It is likely that Ubc3/Cdc34 behaves anomalously in this assay because it normally functions as part of larger SCF complexes. We suggest that Ubc3 uses Slx5-Slx8 as a target for ubiquitination in vitro, but not as a stimulatory factor. In contrast, the Ubcs1, 4, 5, and Ubc13-Mms2 appear to be specifically stimulated by Slx5-Slx8.
Figure 7.
Characterization of Slx5-Slx8 Ub ligase with yeast Ubcs. (A) UbcH6 (100 nM), Ubc13-Mms2 (400 nM), or 400 ng of the indicated partially-purified Ubcs from yeast were tested for sensitivity to mutant Slx5-Slx8 complex. Two control reactions lacking E2 are also shown. Ubiquitination reactions contained 13 nM E1, 5 μM Ub, and 140 nM of the indicated wt or mutant Slx5-Slx8 complex as E3. Following incubation (30°C, 10 min), the reaction products were immunoblotted with anti-Ub antibody. (B) The specificity of Ub chain conjugation by Ubc13-Mms2 was determined using mutant Ub proteins. Ubiquitination reactions contained 13 nM E1, 140 nM Ubc13-Mms2 as E2, and the indicated concentration of Slx5-Slx8 as E3, in addition to 1 μM of the indicated ubiquitin mutants. Following incubation (30°C, 60 min), the reaction products were immunoblotted with anti-Ub antibody. (C) Ubiquitination reactions were performed as in (B) except that Ubc5 (50 nM) was used as E2.
The specificity of the Ub chain linkage catalyzed by Ubc13-Mms2 was tested using Ub mutants. As with UbcH13-Uev1a, stimulation of Ubc13-Mms2 by Slx5-Slx8 required Ub substrates containing K63, and was unaffected by the K48R mutant (Fig. 7B). We conclude that Slx5-Slx8 stimulates Ubc13-Mms2 without altering its known linkage specificity. Chain formation by yeast Ubc5 was partially inhibited by Ub-K48R but not by Ub-K63R (Fig. 7C). Although this is consistent with the known activity of Ubc5 stimulating K48-linked chains, Ubc5 could be stimulated by Slx5-Slx8 to form Ub chains containing K63 only or K48 only. This lack of Ub chain specificity is reminiscent of that observed with UbcH6 in the presence of Slx5-Slx8 (Fig. 3B). Finally, this interaction is likely to be biologically relevant as Ubc5 was associated with both Slx5 and Slx8 in extracts of yeast overexpressing these proteins (Fig. S2).
Slx5-Slx8 can ubiquitinate proteins involved in homologous recombination
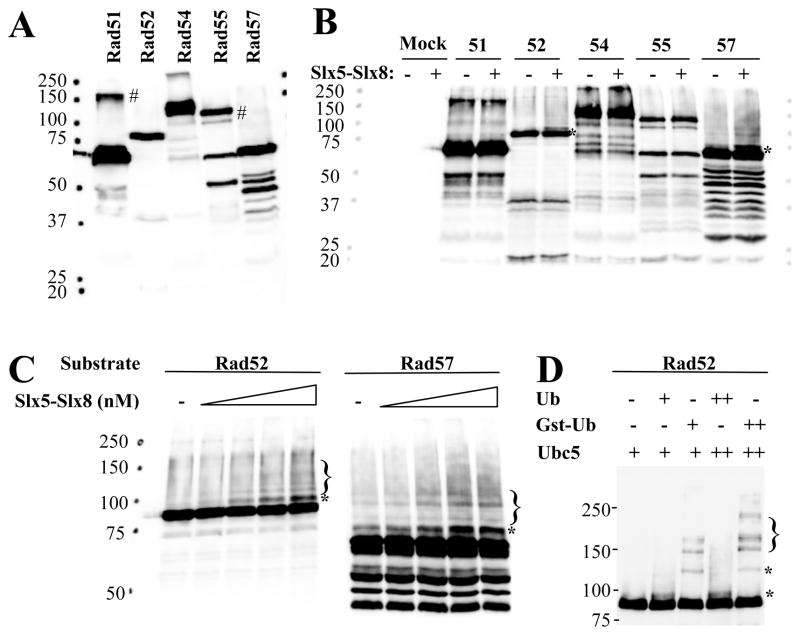
We searched for potential targets of Slx5-Slx8-stimulated ubiquitination by testing proteins that are known to play roles in the maintenance of genome stability. Five members of the RAD52-epistasis group were purified from yeast on IgG beads using the MORF expression system and subjected to ubiquitination using recombinant yeast Ubc5. These candidate proteins were first immunoblotted with peroxidase-conjugated anti-peroxidase (PAP) to detect the Protein A-tagged target proteins. As shown in Fig. 8A, bands of the expected size were obtained 30 although Rad51 and Rad55 contained unidentified bands at 160 and 120 kD, respectively. After subjecting these samples to ubiquitination reactions, the products were immunoblotted as above. Rad52 and Rad57 gave rise to novel higher molecular-weight bands that were dependent on Slx5-Slx8 (Fig. 8B). To confirm their dependence on Slx5-Slx8, titration reactions were analyzed by higher-resolution immunoblotting. In both cases, an Slx5-Slx8-dependent band was observed that migrated appromixately 10 kD larger than the original substrate (Fig. 8C). We interpret these bands to represent mono-ubiquitinated Rad52 and Rad57. At higher levels of Slx5-Slx8, a ladder of bands representing Ub-chains was apparent. To confirm this conclusion, ubiquitination reactions were performed on Rad52-beads in the presence of Ub or a larger GST-Ub fusion protein (30 kD). As shown in Figure 8D, the retarded band is Ub-dependent and supershifted when ubiquitinated with GST-Ub. We conclude that Slx5-Slx8 is capable of stimulating the ubiquitination of target proteins in vitro.
Figure 8.
Ubc5-dependent ubiquitination of Rad52 and Rad57 is stimulated by Slx5-Slx8. (A) The indicated Protein A epitope-tagged Rad proteins were expressed in yeast using the MORF expression system and partially purified using IgG affinity resin. Proteins were released from the resin with SDS sample buffer and immunoblotted with peroxidase-conjugated anti-peroxidase (PAP) to detect the Protein A tag. # indicates a non-specific cross-reacting band. (B) IgG beads containing no protein (Mock) or the indicated Protein A-tagged Rad protein were used as substrates for ubiquitination assays in the absence (−) or presence (+) of 140 nM Slx5-Slx8. Ubiquitination assays contained 27 nM E1, 230 nM recombinant yeast Ubc5 as E2, and 6 μM Ub. Following incubation (30°C, 30 min), the reaction products were solubilized in SDS sample buffer and immunoblotted with PAP. Asterisks indicate mono-ubiquitinated bands. (C) IgG beads containing Rad52 or Rad57 were used as substrates for ubiquitination assays as above, but contained 0, 2.4, 24, 120, or 240 nM Slx5-Slx8. Following incubation (30°C, 20 min), the reaction products were treated as above. Brackets indicate poly-ubiquitinated forms. (D) IgG beads containing Rad52 were treated as in (C) except that reactions contained either 0, 2, or 4 uM Ub, or GST-Ub, as indicated. Slx5-Slx8 was present at 100 nM and Ubc5 was present at 50 (+) or 100 nM (++).
DISCUSSION
SLX5 and SLX8 were first identified in yeast as related genes required for viability in the absence of the Bloom’s Syndrome homolog, Sgs1.4 An extensive analysis of such synthetic-fitness interactions led to the notion that SLX5 and SLX8 define a common pathway required for genome maintenance,3 and this idea has recently been verified by the discovery of increased recombination and gross chromosomal rearrangements in the two mutants.14, 15 In the current study we addressed the biochemical function of these proteins with a focus on identifying an activity specific to the Slx5-Slx8 complex. Our major finding is that both subunits are required for Ub ligase activity in vitro. Although there are at least 15 known RING-domain Ub ligases in yeast, none of them function as heterodimers analogous to the two-component systems in higher eukaryotes. These two-component systems comprise an important class of regulators including BRCA1-BARD117, 35, 36 and the p53 Ub ligase MDM2, which is synergistically activated by the MDMX subunit.37
So, what is the target of the Slx5-Slx8 Ub ligase? Although we have yet to identify an authentic in vivo target, Slx5-Slx8 may act in multiple pathways based on the various Ubcs that it stimulates. Ubc13-Mms2 promotes stable K63-linked Ub chains and at least one role is to act downstream of Rad6/Ubc2 to modify PCNA in response to DNA damage.38 It is intriguing to speculate that Slx5-Slx8 promotes K63-linked modfications in response to DNA damage, however, there is no evidence that SLX5-SLX8 functions in the RAD6 post-replicative repair pathway. And while it is likely that Ubc13-Mms2 acts independently of RAD6, where it could interact with Slx5-Slx8 to promote K63-linked modifications, this pathway is unlikely to be the one needed in the absence of SGS1 since ubc13Δ sgs1Δ double mutants are viable.
The other three E2s that were activated by Slx5-Slx8, Ubc1, Ubc4, and Ubc5, are known to form a family with redundant functions.39 These enzymes mediate the destruction of short-lived proteins and are needed in response to stress and during germination. Ubc4 and Ubc5 are nearly identical (93% identity) and loss of any two of the three genes results in synthetic fitness defects, while the triple mutant is inviable.39 Thus, Slx5-Slx8 may cooperate with these Ubcs to degrade damaged or short-lived DNA repair proteins. Interestingly, ubc4Δ slx8Δ double mutants display a synthetic fitness defect,3 consistent with the idea that Slx5-Slx8 acts in the Ubc1-Ubc4-Ubc5 pathway. Although we have no data on potential targets of these pathways in vivo, our biochemical data suggest that the Rad52 and Rad57 recombination proteins may be substrates. As a working model we propose that Slx5-Slx8 targets such DNA repair proteins for destruction via Ubc4/Ubc5 when they are damaged or irreversibly modified. Failure to degrade such proteins may lead to the observed growth defects including hyper-recombination and sensitivity to HU.
One question we have yet to answer is why mutations in SLX5 and SLX8 are synthetically lethal with sgs1Δ. Many of the mutations that are lethal in the absence of SGS1 occur in genes that play roles in DNA repair, replication, or recombination (e.g., Rad27/FEN1, Mus81-Mms4, Srs2, Slx1-4).5, 6 The need for the Slx5-Slx8 Ub ligase to suppress hyper-recombination and GCRs suggests that SGS1 may be required to resolve the increased number of recombination intermediates that occur in slx5Δ or slx8Δ cells. Alternatively, protein modification by the Slx5-Slx8 Ub ligase may become essential in the absence of the SGS1-TOP3-RMI1 pathway.
A second question that remains to be answered concerns the connection between Slx5-Slx8 and the SUMO pathway. In addition to interacting genetically with a variety of SUMO pathway mutants, slx5Δ or slx8Δ mutants display increased levels of global sumoylation11 and a reduction in damage-induced SUMO-conjugation of some proteins.14 Given the link between sumoylation and recombination,14, 40, 41 our data suggest a simple explanation for two of these phenotypes. First, the hyper-sumoylation phenotype might be explained if Slx5-Slx8 is involved in the proteolytic turnover of sumoylated proteins. For example, multiply sumoylated proteins, or proteins carrying poly-SUMO chains, might be targets for ubiquitination by Slx5-Slx8. In this case, we would expect Slx5-Slx8 to stimulate Ubc 1, 4, or 5 to promote their destruction. Second, it is possible that Slx5-Slx8 targets SUMO-modified repair proteins to remove them from chromatin after they have done their job. Failure to eliminate these SUMO-modified proteins may lead to persistent lesions that stimulate recombination. Although de-sumoylating enzymes such as Ulp1 or Ulp2 could theoretically carry out this function,42, 43 there may be cases where proteolytic destruction is preferable to simply removing the conjugated SUMO. The recent in-situ localization of Slx5-Slx8 to DNA replication centers14 and its interaction with DNA16 are consistent with this idea. In addition, we have shown that Slx5-Slx8 can bind SUMO and stimulate Ubc9-dependent SUMO chain formation in vitro.12 Although we cannot rule out the possibility that Slx5-Slx8 catalyzes both Ub and SUMO ligation in vivo, the fact that sumoylation does not depend on its RING domains or dimer formation12 suggests that this is not the primary function of Slx5-Slx8. Perhaps the stimulation of sumoylation by Slx5-Slx8 reflects its ability to bind multiple SUMO residues in a manner that promotes their conjugation into chains by Ubc9. Such an interpretation is consistent with the idea that the Slx5-Slx8 Ub ligase targets substrates that are poly-sumoylated.
While future studies should address these specific models, we are intrigued by several similarities between Slx5-Slx8 and the human BRCA1-BARD1 complex.44 Although larger and significantly more complex than Slx5 and Slx8, BRCA1 and BARD1 contain essential RING domains at their N-termini that are required for subunit interaction.45–47 Like Slx5-Slx8, BRCA1-BARD1 appears to be an obligate heterodimeric Ub ligase.32, 36, 46 And Slx5-Slx8 binds DNA in vitro which is an activity shared with BRCA1.16, 48 Finally, BRCA1 is known to associate with the Rad51 recombinase on mitotic and meiotic chromosomes49 and to control genomic stability and homologous recombination.50, 51 These findings suggest that the similar phenotypes of BRCA1−/− and slx5Δ-slx8Δ cells may be more than just coincidence. Given the importance of BRCA1 in human health, it will be interesting to determine whether these Ub ligases have common targets.
Supplementary Material
Acknowledgments
We thank Marc Gartenberg, Kiran Madura, KJ Myung, and Eric Phizicky for providing strains, plasmids, and other reagents. Thanks also to Kiran Madura for critical reading of the manuscript and to Reema Koikkara and Suzanne Shanower for invaluable technical assistance. This work was supported by grant R01GM071268 from the National Institutes of Health.
The abbreviations used are
- wt
wild type
- Ub
ubiquitin
- Ubc
ubiquitin conjugating enzyme
- Ab
antibody
- HA
hemmaglutinin
- DTT
dithiothreitol
- TAP
tandem affinity purification
- CBP
calmodulin binding protein
- MORF
moveable ORF
- GCR
gross chromosomal rearrangement
- HU
hydoxyurea
- MW
molecular weight
References
- 1.St Onge RP, Mani R, Oh J, Proctor M, Fung E, Davis RW, Nislow C, Roth FP, Giaever G. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat Genet. 2007;39:199–206. doi: 10.1038/ng1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, Ding H, Xu H, Han J, Ingvarsdottir K, Cheng B, Andrews B, Boone C, Berger SL, Hieter P, Zhang Z, Brown GW, Ingles CJ, Emili A, Allis CD, Toczyski DP, Weissman JS, Greenblatt JF, Krogan NJ. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–10. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 3.Pan X, Ye P, Yuan DS, Wang X, Bader JS, Boeke JD. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–81. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- 4.Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2001;157:103–18. doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ooi SL, Shoemaker DD, Boeke JD. DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. Nat Genet. 2003;35:277–86. doi: 10.1038/ng1258. [DOI] [PubMed] [Google Scholar]
- 6.Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, Chen Y, Cheng X, Chua G, Friesen H, Goldberg DS, Haynes J, Humphries C, He G, Hussein S, Ke L, Krogan N, Li Z, Levinson JN, Lu H, Menard P, Munyana C, Parsons AB, Ryan O, Tonikian R, Roberts T, Sdicu AM, Shapiro J, Sheikh B, Suter B, Wong SL, Zhang LV, Zhu H, Burd CG, Munro S, Sander C, Rine J, Greenblatt J, Peter M, Bretscher A, Bell G, Roth FP, Brown GW, Andrews B, Bussey H, Boone C. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–13. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 7.Watt PM, Louis EJ, Borts RH, Hickson ID. Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell. 1995;81:253–60. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]
- 8.Myung K, Datta A, Chen C, Kolodner RD. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat Genet. 2001;27:113–6. doi: 10.1038/83673. [DOI] [PubMed] [Google Scholar]
- 9.Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol Cell Biol. 1994;14:8391–8. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang M, Bellaoui M, Zhang C, Desai R, Morozov P, Delgado-Cruzata L, Rothstein R, Freyer GA, Boone C, Brown GW. RMI1/NCE4, a suppressor of genome instability, encodes a member of the RecQ helicase/Topo III complex. EMBO J. 2005;24:2024–33. doi: 10.1038/sj.emboj.7600684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z, Jones GM, Prelich G. Genetic analysis connects SLX5 and SLX8 to the SUMO pathway in Saccharomyces cerevisiae. Genetics. 2006;172:1499–509. doi: 10.1534/genetics.105.052811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ii T, Mullen JR, Slagle CE, Brill SJ. Stimulation of in-vitro sumoylation by Slx5-Slx8: evidence for a functional interaction with the SUMO pathway. DNA Repair. 2007 doi: 10.1016/j.dnarep.2007.06.004. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bialkowska A, Kurlandzka A. Proteins interacting with Lin 1p, a putative link between chromosome segregation, mRNA splicing and DNA replication in Saccharomyces cerevisiae. Yeast. 2002;19:1323–33. doi: 10.1002/yea.919. [DOI] [PubMed] [Google Scholar]
- 14.Burgess RC, Rahman S, Lisby M, Rothstein R, Zhao X. The Slx5/8 complex affects sumoylation of DNA repair proteins and negatively regulates recombination. Mol Cell Biol. 2007 doi: 10.1128/MCB.00787-07. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang C, Roberts TM, Yang J, Desai R, Brown GW. Suppression of genomic instability by SLX5 and SLX8 in Saccharomyces cerevisiae. DNA Repair (Amst) 2006;5:336–46. doi: 10.1016/j.dnarep.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 16.Yang L, Mullen JR, Brill SJ. Purification of the yeast Slx5-Slx8 protein complex and characterization of its DNA-binding activity. Nucleic Acids Res. 2006;34:5541–51. doi: 10.1093/nar/gkl685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA. 1999;96:11364–9. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joazeiro CA, Weissman AM. RING finger proteins: mediators of ubiquitin ligase activity. Cell. 2000;102:549–52. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- 19.Ardley HC, Robinson PA. E3 ubiquitin ligases. Essays Biochem. 2005;41:15–30. doi: 10.1042/EB0410015. [DOI] [PubMed] [Google Scholar]
- 20.Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, King RW, Gygi SP. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8:700–10. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- 21.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–6. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 22.Sun L, Chen ZJ. The novel functions of ubiquitination in signaling. Curr Opin Cell Biol. 2004;16:119–26. doi: 10.1016/j.ceb.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 23.Spence J, Sadis S, Haas AL, Finley D. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol Cell Biol. 1995;15:1265–73. doi: 10.1128/mcb.15.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ulrich HD. How to activate a damage-tolerant polymerase: consequences of PCNA modifications by ubiquitin and SUMO. Cell Cycle. 2004;3:15–8. [PubMed] [Google Scholar]
- 25.Galan JM, Haguenauer-Tsapis R. Ubiquitin lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J. 1997;16:5847–54. doi: 10.1093/emboj/16.19.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rose MD, Winston F, Hieter P. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 27.Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–30. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 28.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;12:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Meth Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 30.Gelperin DM, White MA, Wilkinson ML, Kon Y, Kung LA, Wise KJ, Lopez-Hoyo N, Jiang L, Piccirillo S, Yu H, Gerstein M, Dumont ME, Phizicky EM, Snyder M, Grayhack EJ. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 2005;19:2816–26. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hofmann RM, Pickart CM. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96:645–53. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- 32.Xia Y, Pao GM, Chen HW, Verma IM, Hunter T. Enhancement of BRCA1 E3 ubiquitin ligase activity through direct interaction with the BARD1 protein. J Biol Chem. 2003;278:5255–63. doi: 10.1074/jbc.M204591200. [DOI] [PubMed] [Google Scholar]
- 33.Wu-Baer F, Lagrazon K, Yuan W, Baer R. The BRCA1/BARD1 heterodimer assembles polyubiquitin chains through an unconventional linkage involving lysine residue K6 of ubiquitin. J Biol Chem. 2003;278:34743–6. doi: 10.1074/jbc.C300249200. [DOI] [PubMed] [Google Scholar]
- 34.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–41. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 35.Ruffner H, Joazeiro CA, Hemmati D, Hunter T, Verma IM. Cancer-predisposing mutations within the RING domain of BRCA1: loss of ubiquitin protein ligase activity and protection from radiation hypersensitivity. Proc Natl Acad Sci USA. 2001;98:5134–9. doi: 10.1073/pnas.081068398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hashizume R, Fukuda M, Maeda I, Nishikawa H, Oyake D, Yabuki Y, Ogata H, Ohta T. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J Biol Chem. 2001;276:14537–40. doi: 10.1074/jbc.C000881200. [DOI] [PubMed] [Google Scholar]
- 37.Badciong JC, Haas AL. MdmX is a RING finger ubiquitin ligase capable of synergistically enhancing Mdm2 ubiquitination. J Biol Chem. 2002;277:49668–75. doi: 10.1074/jbc.M208593200. [DOI] [PubMed] [Google Scholar]
- 38.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–41. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 39.Seufert W, McGrath JP, Jentsch S. UBC1 encodes a novel member of an essential subfamily of yeast ubiquitin-conjugating enzymes involved in protein degradation. EMBO J. 1990;9:4535–41. doi: 10.1002/j.1460-2075.1990.tb07905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Branzei D, Sollier J, Liberi G, Zhao X, Maeda D, Seki M, Enomoto T, Ohta K, Foiani M. Ubc9- and Mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell. 2006;127:509–22. doi: 10.1016/j.cell.2006.08.050. [DOI] [PubMed] [Google Scholar]
- 41.Soustelle C, Vernis L, Freon K, Reynaud-Angelin A, Chanet R, Fabre F, Heude M. A new Saccharomyces cerevisiae strain with a mutant Smt3-deconjugating Ulp1 protein is affected in DNA replication and requires Srs2 and homologous recombination for its viability. Mol Cell Biol. 2004;24:5130–43. doi: 10.1128/MCB.24.12.5130-5143.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bylebyl GR, Belichenko I, Johnson ES. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J Biol Chem. 2003;278:44113–20. doi: 10.1074/jbc.M308357200. [DOI] [PubMed] [Google Scholar]
- 43.Li SJ, Hochstrasser M. A new protease required for cell-cycle progression in yeast. Nature. 1999;398:246–51. doi: 10.1038/18457. [DOI] [PubMed] [Google Scholar]
- 44.Parvin JD. Overview of history and progress in BRCA1 research: the first BRCA1 decade. Cancer Biol Ther. 2004;3:505–8. doi: 10.4161/cbt.3.6.839. [DOI] [PubMed] [Google Scholar]
- 45.Meza JE, Brzovic PS, King MC, Klevit RE. Mapping the functional domains of BRCA1. Interaction of the ring finger domains of BRCA1 and BARD1. J Biol Chem. 1999;274:5659–65. doi: 10.1074/jbc.274.9.5659. [DOI] [PubMed] [Google Scholar]
- 46.Wu LC, Wang ZW, Tsan JT, Spillman MA, Phung A, Xu XL, Yang MC, Hwang LY, Bowcock AM, Baer R. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat Genet. 1996;14:430–40. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- 47.Brzovic PS, Rajagopal P, Hoyt DW, King MC, Klevit RE. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat Struct Biol. 2001;8:833–7. doi: 10.1038/nsb1001-833. [DOI] [PubMed] [Google Scholar]
- 48.Simons AM, Horwitz AA, Starita LM, Griffin K, Williams RS, Glover JN, Parvin JD. BRCA1 DNA-binding activity is stimulated by BARD1. Cancer Res. 2006;66:2012–8. doi: 10.1158/0008-5472.CAN-05-3296. [DOI] [PubMed] [Google Scholar]
- 49.Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston DM. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997;88:265–75. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 50.Shen SX, Weaver Z, Xu X, Li C, Weinstein M, Chen L, Guan XY, Ried T, Deng CX. A targeted disruption of the murine BRCA1 gene causes gamma-irradiation hypersensitivity and genetic instability. Oncogene. 1998;17:3115–24. doi: 10.1038/sj.onc.1202243. [DOI] [PubMed] [Google Scholar]
- 51.Moynahan ME, Chiu JW, Koller BH, Jasin M. BRCA1 controls homology-directed DNA repair. Mol Cell. 1999;4:511–8. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.