Metaphase spindles are steady-state ensembles of microtubules. We used single molecule imaging to measure tubulin dynamics in spindles and non-spindle assemblies in Xenopus egg extract. Our results argue that the high density of microtubules in spindles is caused by local enhancement of nucleation, and not by local stabilization.
Abstract
Metaphase spindles are steady-state ensembles of microtubules that turn over rapidly and slide poleward in some systems. Since the discovery of dynamic instability in the mid-1980s, models for spindle morphogenesis have proposed that microtubules are stabilized by the spindle environment. We used single molecule imaging to measure tubulin turnover in spindles, and nonspindle assemblies, in Xenopus laevis egg extracts. We observed many events where tubulin molecules spend only a few seconds in polymer and thus are difficult to reconcile with standard models of polymerization dynamics. Our data can be quantitatively explained by a simple, phenomenological model—with only one adjustable parameter—in which the growing and shrinking of microtubule ends is approximated as a biased random walk. Microtubule turnover kinetics did not vary with position in the spindle and were the same in spindles and nonspindle ensembles nucleated by Tetrahymena pellicles. These results argue that the high density of microtubules in spindles compared with bulk cytoplasm is caused by local enhancement of nucleation and not by local stabilization. It follows that the key to understanding spindle morphogenesis will be to elucidate how nucleation is spatially controlled.
INTRODUCTION
Mitotic and meiotic spindles are highly dynamic, self-organizing ensembles of microtubules and interacting proteins. A fundamental aspect of spindle assembly is that the concentration of microtubules is higher within the spindle than elsewhere in the mitotic cytoplasm. This is particularly evident for meiotic spindles that largely lack astral microtubules, including spindles assembled in Xenopus laevis egg extracts that we use in this study. From biochemistry (Chang et al., 2004) and electron microscopy (EM) (Coughlin and Mitchison, unpublished), we estimate the concentration of polymerized tubulin inside extract spindles is ∼60 μM, whereas outside spindles it is too small to measure, and certainly <1 μM. A similar concentration gradient is evident in vivo from images of unfertilized eggs (Wuhr et al., 2007). It is currently unclear what accounts for this strong concentration gradient. In principle, it could have several causes, including 1) local nucleation of microtubules in the spindle, 2) global nucleation of microtubules and local stabilization in the spindle, 3) global nucleation of microtubules followed by their translocation into the spindle by motors, and 4) local concentration of soluble tubulin dimers within the spindle followed by their polymerization. In the egg extract system, possibilities 3 and 4 have been largely ruled out by live imaging of spindle assembly reactions (Groen et al., 2009), although we cannot exclude the possibility that some modified form of tubulin dimer concentrates inside spindles. Local nucleation within spindles is perhaps the most intuitive model, but the mechanism and rate of nucleation in anastral meiotic spindles is currently unclear. Local stabilization is also plausible. Since the discovery of dynamic instability in the mid-1980s, models for spindle morphogenesis have proposed that microtubules are stabilized by the spindle environment (Kirschner and Mitchison, 1986; Karsenti and Vernos, 2001).
The issue of microtubule stability in spindles is complicated by the fact that they contain multiple microtubule populations whose nucleation and stabilization mechanisms may differ. Kinetochore, astral, and nonkinetochore microtubules population have been distinguished by structural criteria, and it is possible that other subpopulations exist. Kinetochore microtubules are known to be strongly stabilized in most systems, presumably because kinetochore proteins prevent rapid depolymerization of their plus ends (reviewed in Inoue and Salmon, 1995). Egg meiotic spindles contain few astral microtubules, and we do not discuss them further. Nonkinetochore microtubules are by far the most abundant class in egg meiotic spindles (e.g., see EM views in Mitchison et al., 2004) and also the most abundant class in mammalian somatic spindles in anaphase (Mastronarde et al., 1993). They have been much less studied than other microtubule classes, yet they are responsible for morphology, bipolarity, and size in egg extract spindles and probably in anastral spindles more generally. We know this because spindles with normal morphology, but lacking kinetochores and centrosomes, assemble around DNA-coated beads in egg extracts (Heald et al., 1996). Thus, if we understood how the nonkinetochore population is nucleated, spatially organized, and (perhaps) stabilized, we would understand the essence of meiotic spindle morphogenesis, and probably important aspects of the mitotic problem.
The organization and dynamics of nonkinetochore microtubules are poorly understood. In the extract system, their assembly is promoted by Ran-guanosine triphosphate (GTP) and Aurora-B kinase activity that have been proposed to instruct spindle morphogenesis (Karsenti and Vernos, 2001; Sampath et al., 2004; Maresca et al., 2009). These factors have the potential to promote both nucleation and stabilization of microtubules, by desequestering spindle assembly factors in the case of Ran-GTP (Caudron et al., 2005), and by inhibiting microtubule-destabilizing proteins in the case of Aurora-B (Ohi et al., 2004). However, the extent to which these gradients are truly responsible for morphogenesis, and the extent to which they promote nucleation versus stabilization, is unclear. Evidence that soluble gradients from chromatin promote stabilization on nonkinetochore microtubules has come from experiments where the growth of single microtubules toward versus away from chromatin masses was imaged (Athale et al., 2008). Although these reconstitution experiments allow single microtubule imaging, they may not accurately model the environment in the center of the spindle. Thus, there is a need to directly measure the stability of nonkinetochore microtubules inside spindles and to compare them with microtubules with free plus ends assembled outside spindles.
Direct observation of single microtubules in spindles is currently not possible because they are too close together to optically resolve, so a variety of bulk methods have been used to infer their dynamic behavior. Response to perturbation (Inoue and Sato, 1967) and fluorescence recovery after photobleaching (FRAP) (Inoue and Sato, 1967; Salmon et al., 1984) revealed that most spindle microtubules exchange with the soluble monomer pool on a time scale of tens of seconds. In addition, photomarking and speckle microscopy revealed that both kinetochore and nonkinetochore microtubules slide continuously toward the poles (Sawin and Mitchison, 1991; Valloton et al., 2003). All these data refer primarily to nonkinetochore microtubules, because they comprise the bulk population inside the spindles used in these studies. The data left unclear the detailed mechanism by which nonkinetochore microtubules turnover. Dynamic instability of plus ends has long been a plausible candidate (Sawin and Mitchison, 1991; Inoue and Salmon, 1995). However, many published models implicate depolymerization from minus ends as the major turnover mechanism, often proposing depolymerization by a combination of motor-driven sliding toward pole with depolymerase activity at minus ends, a class of model we refer to as “feeder-chipper” models (Gadde and Heald, 2004). One reason our understanding of turnover mechanism is limited is that bulk methods—perturbation, FRAP and photoactivation—are problematic for accurately measuring dynamics, particularly at short time scales in which monomer diffusion and polymerization dynamics both contribute to recovery kinetics. Thus, recovery kinetics in a FRAP experiment are sensitive to experimental details that are difficult to quantitatively account for, such as the exact three-dimensional shape of the bleached region (Axelrod et al., 1976), the method of normalization, and the specifics of the bleaching protocol (Weiss, 2004). Factors such as local reincorporation of bleached monomer can results in estimates of recovery times that can deviate from their true value by a factor of ≥3. Even if these systematic errors are accurately corrected, FRAP recovery time scales of binding may not directly relate to reaction rates (Beaudouin et al., 2006), implying that FRAP of fluorescently labeled tubulin in spindles might not unambiguously determine microtubule lifetimes. For example, the rate of turnover of a marked region can depend on its distance from microtubule plus ends and the extent to which released tubulin becomes reincorporated into other microtubules in the spindle. These considerations complicate interpretation of FRAP data that have suggested different turnover rates at the middle of spindles versus near poles (Pearson et al., 2006; Cheerambathur et al., 2007). Speckle microcopy provides a different approach to measuring turnover. Labeling approximately one tubulin molecule in 1000 generates a stochastic, inhomogeneous distribution of fluorescence, whose brighter regions are called “speckles.” Speckle dynamics has provided important insight into motion and turnover in cytoskeletal ensembles (Danuser and Waterman-Storer, 2006). However, one speckle corresponds to multiple flurophores, presumably incorporated into multiple microtubules, so it is difficult to directly relate speckle turnover to individual filament turnover. Taken together, published dynamics measurements have not fully answered the questions of whether the spindle environment stabilizes microtubules and whether microtubule turnover is spatially regulated in spindles nor have they elucidated the kinetic mechanism of microtubule turnover.
In this study, we analyze the statistics of immobilization of single molecules of tubulin in spindles, which we show reports on assembly and disassembly of subunits into microtubules. This approach depends on the fact that a tubulin molecule in a microtubule moves slowly and thus appears as a point on the camera, whereas a soluble subunit diffuses rapidly compared with our image acquisition speed and therefore blurs out. Our method is a form of speckle microscopy, although a key difference is that we image the dynamics of individual tubulin molecules, so we can more directly infer the dynamics of individual microtubules. Single molecule methods allow time distributions of individual molecular events to be measured in situ, which, when combined with mathematical modeling, can be used to extract quantitative information that is difficult to obtain with bulk techniques (Sako, 2006). We find this is indeed the case for single tubulin imaging in spindles; our results answer the question of whether microtubules are stabilized in spindles and reveal fundamental aspects of the turnover mechanism. We focus on tubulin turnover and its implications. Our results are thus complementary to a recent study that analyzed movements of single tubulin molecules in X. laevus egg extract spindles but did not address turnover (Yang et al., 2007).
EXPERIMENTAL PROCEDURES
Preparation of X. laevis Extract Samples
Spindles were assembled in X. laevis egg extracts, after DNA replication, as described previously (Sawin and Mitchison, 1991). Typically, 4–6 μl of spindle reaction was squashed under 18-mm coverslips. Only one spindle was imaged per coverslip, and spindles that changed morphology or underwent substantial motion during imaging were discarded. For each condition, single molecule experiments were performed on 3 to 10 spindles per extract preparation, which was repeated on three to five different extracts prepared on different days. In total, thousands to tens of thousands of molecules were analyzed for each condition.
Reagents
2-(1-(4-Fluorophenyl)cyclopropyl)-4-(pyridin-4-yl)thiazole (FCPT) was synthesized as described previously (Groen et al., 2008) and stored as a 200 mM stock in dimethyl sulfoxide. The FCPT stock was further diluted to 20 mM in extract and added to preformed spindles to a final concentration of 200 μM. Movies were acquired rapidly after drug addition. Mitotic centromere-associated kinesin (MCAK) was immune depleted from extract, and purified baculovirus-expressed MCAK was added back to various levels as described previously (Ohi et al., 2007). Tetrahymena pellicles were purified following the protocol of Coue et al. (1991).
Single Molecule Imaging
Tubulin was directly labeled with Alexa-647, by using the protocol of Hyman et al. (1991), and added to a final concentration of ∼150 pM, such that of order one in 100,000 tubulin molecules were labeled. Using an infrared dye resulted in enhanced signal to noise compared with fluorophores that emit visible light, due to minimized image degradation by background autofluorescence (data not shown). Data were acquired on a microscope (model E800; Nikon, Tokyo, Japan), with 60× or 100× objectives (1.4 numerical aperture Plan Apo differential interference contrast; Nikon), a spinning disk confocal (CSU-10; Yokogawa, Newnan, GA), and a backilluminated electron multiplying (EM) charge-coupled device (CCD) camera (iXonEM + 897 or iXonEM 860; Andor Technology, Belfast, Northern Ireland), with maximal EM gain. Movies were taken with streaming acquisition using the SOLIS software package (Andor Technology). Each image consisted of averaging 16 separate 100-ms exposures, which reduces the noise resulting from the stochastic gain process. Illumination was provided by the 647 line of an argon-krypton mixed gas laser, with a total power of ∼3 mW after the microscope objective, corresponding to only 0.1–0.5 kW/cm2. Images were acquired 3–10 μm from the coverslip as a balance between minimize potentially perturbing effects of the boundary while maintaining high signal to noise, which rapidly degrades with depth of focus.
Particle Tracking
Images of single molecules were of high enough quality that the position of each molecule could be tracked using freely available MATLAB code developed for colloidal physics (Crocker and Grier, 1996) (http://www.seas.harvard.edu/projects/weitzlab/matlab/). In this particle tracking method (Crocker and Grier, 1996; Figure 1), raw images are background corrected with a boxcar average, convolved with a Gaussian kernel to suppress high-frequency noise, and local maxima of the filtered image above a threshold brightness are selected as candidate particles. For tracking displacements, the subpixel location of molecules was determined by fitting the intensity around each particle to a two-dimensional Gaussian (Thompson et al., 2002; Figure 1A), but the pixel location of particles is sufficient for the measurement of tubulin lifetimes, and subpixel refinement was not used in that analysis. Particle locations were linked to trajectories by a global minimization of the sum of the square of particle displacements between frames (Crocker and Grier, 1996). Additional analysis and data fitting were performed using custom written MATLAB code.
Figure 1.
(A) Raw images were filtered and smoothed, and local maxima above a threshold brightness were selected as candidate particles (Crocker and Grier, 1996). For tracking displacements, the subpixel location of molecules were determined by fitting the intensity around each particle to a two-dimensional Gaussian, but the pixel location of particles was sufficient for the measurement of tubulin lifetimes and subpixel refinement was not used in that analysis. (B, left) Particle locations were linked to form trajectories by a global optimization procedure (Crocker and Grier, 1996). (B, right) Individual particles appeared and disappeared in a step-like manner. A particle's lifetime is defined as the difference between the times of disappearance and appearance. (C) Raw image of a spindle with single molecule level labeling.
RESULTS
Single Molecule Imaging Can Be Used to Infer Polymerization Dynamics
Meiotic spindles with replicated chromatin were assembled in X. laevis egg extracts by standard methods (Desai et al., 1999). Spindles were labeled by assembly in the presence of ∼150 pM Alexa-647–labeled tubulin, such that approximately one in 100,000 molecules were labeled. In some cases, the probe was added after assembly and allowed to incorporate for >10 min with similar results. This wavelength was chosen because the probe has high molecular brightness and very slow bleaching when deoxygenated (see below), and the autofluorescence background is lower in the infrared, which greatly facilitates single molecule imaging. Spindles were identified by Hoechst staining of DNA and then imaged in the 647 nm channel with a spinning disk confocal microscope and an EM-CCD camera. Images were collected continuously every ∼2 s with no shuttering, by averaging ∼16 consecutive exposure that were collected from the camera every ∼100 ms. Images were collected from focal planes closer to the coverslip than most kinetochores, which selected for signal from nonkinetochore microtubules and improved image quality. These image sequences revealed well-defined, discrete points of light emission scattered throughout the spindle (Figure 1, A and C, and Supplemental Movie 1), which had Gaussian intensity profiles, and signal-to-noise values between 4 and 10 (Figure 1). We established that these particles are single tubulin molecules by using several criteria: They appeared and disappeared in a step-like manner during regular observation (Figure 1B and Supplemental Data); bleaching was negligible under our standard imaging conditions (see below), but when we promoted it by poisoning mitochondrial metabolism with adenosine 5′-(β,γ-imido)triphosphate) (AMPPNP), each particles bleached in a single step-like manner and the population bleached with single exponential kinetics (Supplemental Data). It has also been argued previously, using intensity measurements, that with this level of labeling in extract spindles, >98% of particles imaged by widefield microscopy were single fluorophores (Yang et al., 2007).
Particles in the image report on the location of tubulin molecules that are incorporated into spindles. Each frame in the movies took ∼2 s to acquire, in which time freely diffusing tubulin would move micrometers, so fluorescence from soluble tubulin blurs over multiple pixels (Watanabe and Mitchison, 2002; Valloton et al., 2003; Kim et al., 2006). This is the same principle that underlies speckle microscopy (Waterman-Storer and Salmon, 1998), except that in our case the signal comes from a single tubulin molecule and therefore reports on the dynamics of a single microtubule. Standard software from colloidal physics (Crocker and Grier, 1996) was used to identify and track the particles (Figure 1). We tracked particles that existed for at least two frames to eliminate spurious identification, which was a highly conservative approach as such misidentification events were not manually observed, so our effective temporal resolution was ∼4 s. All the molecules underwent extensive motion, similar to that reported in a previous single molecule study (Yang et al., 2007). Motion was directed primarily toward the nearest spindle pole (Figure 2, left), as expected from previous photoactivation (Sawin and Mitchison, 1991) and speckle microscopy (Valloton et al., 2003) observations. This motion is thought to reflect motor-driven sliding of individual microtubules within the spindle. Individual molecules displayed a wide range of velocities, and frequent velocity changes, that are not evident with averaging methods. Movement was generally slower at the poles, consistent with previous reports (Burbank et al., 2007; Yang et al., 2008). Detailed analysis of microtubule movements will be reported elsewhere.
Figure 2.
Trajectories of particles in spindles are illustrated by displaying the dynamics of particles located in a particular frame. A particle's future trajectory is colored red, its past trajectory is blue, and particles that were born in the selected frame are indicated by a green square. The shorter trajectories in the FCPT-treated spindle illustrate the reduced tubulin motion in this structure. Bar, 10 μm.
In addition to moving, particles appeared and disappeared in a step-like manner (Figure 1B and Supplemental Movie 1). Because soluble tubulin cannot be visualized in these experiments, and incorporation into microtubules is the only known way that tubulin molecules become immobilized in spindles, it is likely that particle appearance reflects a tubulin molecule becoming incorporated into a growing microtubule, and disappearance is caused by the release of that molecule during microtubule depolymerization. However, disappearance of fluorescent molecules could also be caused by photobleaching or translocation out of the focal plane, and appearance could be caused by movement into the focal plane. The movement of the molecule relative to the focal plane, if that occurs, could be artifactual (due to focus drift) or biological (due to microtubule movement). We therefore systematically investigated the extent which these nonturnover processes might contribute to particle dynamics, and also the effect of perturbations that are known to change microtubule polymerization dynamics. Summarizing in advance, all out data point to the majority of particle appearance/disappearance faithfully reporting on microtubule assembly/disassembly events, and not other processes.
Characterization of Photobleaching
We first measured the number of observed tubulin molecules per spindle as a function of imaging time under our standard conditions and found they did not decrease over minutes of continuous observation (Supplemental Figure 1). This is consistent with a lack of bleaching, but the total number of fluorescent tubulin molecules in the spindle could also remain constant in the presence of photobleaching if the depletion of fluorophores is compensated by the diffusion and incorporation of new labeled tubulin into the spindle. To test the second possibility, we added two drugs that together froze all microtubule dynamics in spindles. Taxol was added at high concentration to block polymerization dynamics, and the kinesin-5 inhibitor FCPT was added to freeze microtubule motions. FCPT binds to kinesin-5 competitively with ATP, with high affinity and high specificity over a panel of eight kinesins and 38 kinases (Rickert et al., 2008). In the spindle, FCPT blocks kinesin-5 in a state with high affinity for the microtubule lattice, converting it into an immobile cross-linker (Groen et al., 2008). In the presence of FCPT, essentially all measurable tubulin motion is blocked in spindles (Figure 2, right). More commonly used allosteric inhibitors of kinesin-5, such as monastrol and S-trityl-cysteine, prevent coherent microtubule sliding to the poles (Miyamoto et al., 2004) but do not stop all motions of tubulin molecules (data not shown). Presumably, FCPT blocks motion more completely than the allosteric kinesin-5 inhibitors because FCPT not only prevents kinesin-5 motor activity but also induces cross-linking. Even with microtubule polymerization dynamics and sliding movement both frozen, the number of observed fluorescent tubulin molecules per spindle did not decrease over several minutes of continuous imaging (Supplemental Figure 2), demonstrating that photobleaching is truly negligible under our standard imaging conditions. Furthermore, under these conditions, many particles could be tracked for >100 s, much longer than was possible in untreated spindles, arguing that instrument drift in the z-axis is not responsible for particle appearance and disappearance. Z-drift would have been unlikely even from particle behavior in standard image sequences, because individual particles had a large range of lifetimes and appeared and disappeared in a step-like manner (see below), whereas Z-drift would cause gain and loss of signal in a gradual, synchronous manner. We believe that the observed extraordinary photostability is due in part to efficient depletion of oxygen by the abundant mitochondria in egg extracts (Niethammer et al., 2008). Poisoning extract metabolism with a nonhydrolysable ATP analogue AMPPNP, allowed rapid photobleaching, which we used to argue that the particles are single molecules (discussed above and in Supplemental Figure 2).
Tubulin Turnover Dynamics in Spindles in the Absence of Translocation
Because photobleaching is not significant under our standard imaging conditions, particle appearance and disappearance could be caused by assembly/disassembly or by movement through the focal plane due to microtubule translocation. To test which process dominates, and to simplify particle dynamics for initial analysis, we blocked microtubule motion in spindles by using FCPT, now in the absence of Taxol. This inhibitor blocked all detectable particle movement in the spindle (Figure 2, right), with only slight residual motion caused by small movements of the entire spindle. Despite this lack of motion, individual particles still appeared and disappeared in a step-like manner (Supplemental Movie 2). A detailed analysis of the change in intensities of individual molecules showed that they appeared and disappeared in a truly step-like manner, and not by the gradual change of intensity expected for a molecule moving through the focal plane (Supplemental Data). Thus, in these highly perturbed spindles, neither photobleaching nor movements into and out of the focal plane are significant, so particle appearance and disappearance can unambiguously be interpreted in terms of tubulin polymerization and depolymerization. Therefore, we were in a position to accurately quantify tubulin turnover in FCPT-treated spindles.
Observation of Fast Dynamics and Parameterization of Microtubule Turnover
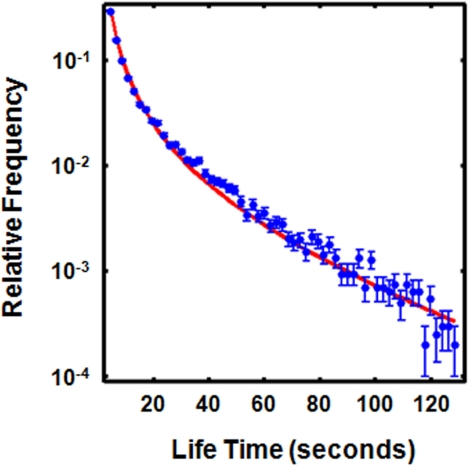
We refer to the total amount of time a tubulin molecule is immobilized—the time between its appearance and disappearance in the image sequence—as its “lifetime” (Figure 1B). The lifetime accounts for the times that tubulin is added and removed from a microtubule; both events presumably occurring at microtubule ends. Thus, this data provides information that is somewhat different from a classic FRAP experiment that measures the time of turnover of a marked region inside microtubules, typically an unknown distance from the microtubule ends. We computed histograms of lifetimes from 13 different FCPT-treated spindles and found that they were similar, so we pooled the data (20,239 events) to construct a master curve that represents a histogram of all measured lifetimes (Figure 3). Almost 25% of the lifetimes were in the shortest time bin of the histogram, corresponding to lifetimes of ∼4 s. These events correspond to very fast polymer turnover that would be difficult to distinguish from diffusion of soluble dimer into the bleach zone by FRAP. The significance of these very fast events is discussed below.
Figure 3.
Distribution of tubulin lifetimes in spindles with FCPT (blue circles). A best fit to Equation 1 is included (red line, τ = 71 ± 3 s). The data and best fit were normalized to have an area of 1.
We next sought to parameterize the entire lifetime distribution. It was manifestly nonexponential, as it is not a straight line on a linear-log plot (Figure 3). Thus, it cannot be characterized solely by its half-time, and we sought alternative, simple parameterizations that would allow quantitative comparison of different conditions. In general, there is no reason to suspect that the distribution of lifetimes for subunits in a dynamic polymer should be an exponential, or a sum of exponentials, because the dissociation process is not a simple first-order reaction if exchange is limited to the polymer ends. Rather, the distribution of subunit lifetimes is the distribution of first passage times of the polymer's ends, i.e., the time it takes the polymer end to return to the point where the molecule was incorporated. In the simplest models of polymer growth, the polymer length undergoes a biased random walk (Philips et al., 2009). The bias is a measure of the tendency of the polymer to grow or shrink on average, whereas the randomness is caused by stochasticity in the polymerization process (see Supplemental Data). A variety of more complex models of microtubule growth can be approximated as a biased random walk in the appropriate limit (Hill, 1987; Bicout, 1997). Dynamics of single microtubules in interphase cells (Vorobjev et al., 1997) and FRAP measurements of tubulin turnover (Maly, 2002) can be well described by a biased random walk model, indicating that this approximation can apply to microtubules in vivo.
In a biased random walk approximation, the distribution of first passage times for a microtubule of length x to return to length x after growing to some greater length—and hence the predicted probability, P, of observing a tubulin molecule with a lifetime of t when dissociation occurs at the same end as association—is given by:
 |
where τ is four times the expected lifetime of a microtubule of average length (Bicout, 1997; Supplemental Data). Figure 3 (red line) shows a fit of this equation to the measured lifetime distribution in FCPT-treated spindles. An excellent fit to Equation 1 is obtained with τ = 71 ± 3 s. Thus, the entire lifetime distribution can be characterized with just a single parameter, τ. The lifetime data could also be fit to a sum of two exponentials (data not shown), but such a fit involves two additional freely adjustable parameters (the amplitude and time constant of the second exponential), produces a fit of worse quality (the sum of residuals for the two exponential best fit is 3.6 × 10−3, compared with 0.53 × 10−3 for the biased random walk model), and we are unaware of any theoretical justification for suspecting that the lifetime data should be described as a sum of exponentials. We therefore used fits to the biased random walk model to parameterize our data. It is important to emphasize that this model is a phenomenological model that can at best be considered as an approximation to the true microscopic dynamics of microtubules. Mechanistic implications of the excellent fit to a random walk model are considered in the discussion section.
Tubulin Turnover Dynamics in Unperturbed Spindles
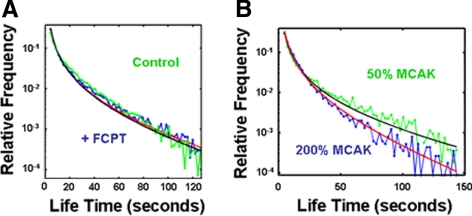
The measurements of tubulin lifetimes described above were made on spindles in the presence of FCPT, which eliminated potential artifacts caused by tubulin translocation, but is a highly perturbing treatment that might modify microtubule polymerization. We therefore performed similar measurements of tubulin lifetimes on unperturbed spindles. We pooled data from 27,478 single molecules from 13 different spindles to construct the lifetime distribution. Although tubulin molecules in control spindles undergo extensive motion, unlike in FCPT-treated spindles in which no detectable translocation is observed (Figure 2 and Supplemental Movies 1 and 2), the lifetime distribution for both sets of spindles was strikingly similar (Figure 4A). The similarities of the distribution of tubulin lifetimes in the absences of translocation (with FCPT) and in the presence of translocation (control) further argues that movement through the focal plane does not contribute to the observed lifetimes.
Figure 4.
(A)Tubulin lifetimes are the same in unperturbed spindles (control, green symbols) and when microtubule translocation is blocked with FCPT (+FCPT, blue symbols). Best fits to Equation 1 are displayed without FCPT (black line, τ = 64 ± 6 s) and with FCPT (red line, τ = 71 ± 3 s). (B) Increasing concentration of MCAK, a microtubule depolymerizer, results in decreasing tubulin lifetimes. Spindles were assembled in extract with MCAK reduced to 50% native levels (green symbols and black line best fit to Equation 1, τ = 97 ± 11 s) or 200% native levels (blue symbols and red best fit, τ = 47 ± 8 s).
A large fraction of fast turnover events were observed in unperturbed spindles, and again the entire lifetime distribution was well fit by the biased random walk model. For the unperturbed spindles, a best fit to Equation 1 yields τ = 64 ± 6 s and cannot be distinguished from the value of τ = 71 ± 3 s obtained for FCPT-treated structures (P ∼ 0.30, using a two-sample two-sided t test). Below, we show the same is true even if only lifetimes >30 s are analyzed. This result argues that kinesin-5 activity is not a major determinate of microtubule turnover dynamics in X. laevis egg extract spindles, in contrast to what has recently been suggested in yeast spindles (Gardner et al., 2008). The yeast data pertained mainly to kinetochore microtubules, and it is possible that kinesin-5 can alter kinetochore microtubule dynamics indirectly, by influencing chromosome motion. We confirmed that FCPT does not strongly alter bulk tubulin turnover by a cruder, but more familiar measurement, of the dynamics of incorporation of a pulse of fluorescently labeled tubulin into spindles. Spindles assembled in Alexa-488 tubulin, pretreated with FCPT or not, were mixed with extract containing XRhodamine tubulin and observed by live imaging. The time scale of incorporation of the red labeled tubulin was unaffected by FCPT (Supplemental Figure 4), confirming the quantitative conclusion from single molecule imaging. We conclude that neither kinesin-5 activity, nor microtubule sliding, are required for fast turnover of nonkinetochore microtubules in X. laevis egg extract spindles.
Perturbing Microtubule Stability Alters Tubulin Lifetimes in the Expected Manner
To further test whether the distribution of single molecule tubulin lifetimes measures microtubule stability, and to quantify the effects of an important dynamics regulator, we changed the concentration of MCAK (also called XKCM1), a microtubule-depolymerizing kinesin-13 that is a major negative regulator of polymerization in X. laevis egg extracts (Walczak et al., 1996). MCAK was immunodepleted from extracts; pure, expressed MCAK (Ohi et al., 2007) was added back to various concentrations; and spindles were assembled and studied with single tubulin imaging. Average lifetimes increased as MCAK concentration decreased; best fits to Equation 1 resulted in τ = 97 ± 11 s with 50% endogenous MCAK, τ = 61 ± 5 s with 100% MCAK, and τ = 47 ± 3 s with 200% MCAK (Figure 4B and Table 1). In the biased random walk model, the expected lifetime of a microtubule of average length is given by (Bicout, 1997), so we infer that changing the concentration of MCAK causes a change in microtubule stability. Spindle length also increased with decreasing concentrations of MCAK (Ohi et al., 2007), as might be expected if MCAK shortens microtubules. Thus, a molecule-specific perturbation known to effect microtubule polymerization dynamics effects tubulin lifetimes in the expected manner, supporting our conclusion that the tubulin lifetime distribution provide a quantitative measure of microtubule stability in spindles.
Table 1.
Table summarizing the measured microtubule stability, τ, determined from fits to the tubulin lifetime distribution, and typical microtubule sliding rate, obtained from a linear fit to the square root of the average mean-squared displacement of tubulin molecules, for different conditions
| Control | + FCPT | 200% MCAK | 100% MCAK | 50% MCAK | |
|---|---|---|---|---|---|
| τ (s) | 64 ± 6 | 71 ± 3(0.30) | 47 ± 3(0.014) | 61 ± 5(0.70) | 97 ± 11(0.012) |
| Sliding rate (μm/min) | 2.17 ± 0.62 | 1.65 ± 0.55(0.53) | 2.14 ± 0.28(0.96) | 2.00 ± 0.47(0.83) |
The p values from a two-sample two-sided t test, testing whether the measurements are different from those obtained from control spindles, are recorded under each entry. Values that are significantly different from control spindles (p < 0.05) are in bold. Within the error of the measurement, no motion is detected in FCPT-treated spindles (apart from small bulk movements of the entire structure).
Turnover Kinetics Are Spatially Uniform in Spindles
Many models have proposed that microtubule stability is spatially regulated in spindles. To critically test this, we divided spindles into five equal subregions (Figure 5A) and separately quantified dynamics in each region, pooling data from 13 unperturbed spindles. First, we measured the ratio of particle deaths to births in each region. This quantifies whether there is a net bias toward assembly or disassembly. There were more total events in the middle of the spindle (data not shown), presumably because there are more plus ends there (Burbank et al., 2006), but the ratio of deaths to births was nearly equal to one at all positions (Figure 5B). There is a slight tendency toward more deaths at the poles and births at the center, but it is not clear whether this is statistically significant. At steady state, there must be some preference for birth near the spindles middle and deaths near poles to balance microtubule sliding, but this effect is evidently very small. The majority of tubulin molecules disappeared in the same region they appeared in, reflecting our observation that turnover (measured as lifetimes) was fast compared with sliding velocities. Approximately 75% of tubulin molecules disappeared in <20 s, and in this time they only moved ∼0.5 μm, although there was large variation between individual molecules in both velocity and lifetime. This conclusion is consistent with results from early FRAP studies, which were incapable of measuring translocation in spindles because turnover was so fast (Salmon et al., 1984).
Figure 5.
(A) Spindles were divided into fifths; two pole regions (green), two midregions (blue), and one central region (red). Bar, 10 μm. (B) Ratio of particle deaths to births is nearly equal to one in all regions. (C) Distributions of tubulin lifetimes in each region are the same.
We measured the lifetime distribution in different subregions of spindles to investigate potential spatial variations in microtubule stability (Figure 5C). The three distributions were indistinguishable, implying uniform lifetimes, and therefore spatially uniform microtubule stability. This result argues against the proposal that bulk microtubule stability increases near chromosomes, as proposed in chromatin gradient models. Our observation that microtubule turnover rate is the same at different positions in egg extract spindles is supported by FRAP experiments (Supplemental Figure 5A; Uteng et al., 2008).
Comparison of Tubulin Turnover Inside and Outside Spindles
We next tested whether microtubules are stabilized by the spindle environment. We did this in two ways, by comparing our lifetime data to literature observations of single microtubule growing from centrosomes, far removed from spindles, in X. laevis extracts, and by directly measuring single tubulin lifetimes in a nonspindle assembly. As described above, our measured distribution of tubulin lifetimes in spindles can be accounted for by a very simple model of microtubule dynamics with just one freely adjustable parameter, τ (Figures 3 and 4). A best fit for unperturbed spindles is obtained with τ = 64 ± 6 s. In a phenomenological model of microtubule dynamics as a biased random walk, τ is 4 times the expected lifetime of a microtubule of average length (Bicout, 1997). Our estimate for microtubule lifetimes in spindles can be compared with the estimated average lifetime of single microtubules elongating from centrosomes in mitotic egg extract, which was ∼20 s in two published studies (Belmont et al., 1990; Verde et al., 1992). The similarity of this value to (∼16 s) suggests that nonkinetochore microtubules in the spindle environment are no more stable than microtubules nucleated from isolated centrosomes in mitotic egg extract. However, this argument is based on comparing two different types of experimental measurements using the biased random walk model to infer the half life of a microtubule from the single molecule tubulin lifetime, so it was also important to perform a head-to-head comparison of spindle and nonspindle arrays independent of any specific theory.
To directly compare microtubule stability inside and outside of spindles, we performed a single molecule lifetime analysis on a nonspindle assembly. Our initial attempts using microtubules nucleated from centrosomes and axonemes failed because the density of microtubules is too low in these structures to obtain sufficient data to construct a lifetime distribution. We therefore turned to Tetrahymena pellicles, which contain hundreds of basal bodies, capable of nucleating thousands of microtubules (Heidemann and McIntosh, 1980; Coue et al., 1991). Pellicles incubated in X. laevis egg extract generated a dense array of microtubules on their surface (Figure 6A) that allowed measurement of single molecule lifetimes by the same protocol that was used for spindles. A lifetime distribution of tubulin molecules was computed from 7,362 events from nine pellicles. The resulting distribution is visually indistinguishable from the one obtained from control spindles (Figure 6B), and a fit to Equation 1 yields τ = 71 ± 9 s, implying, within error (P ∼0.52, using a two-sample two-sided t test), the same microtubule stability as measured in spindles (τ = 64 ± 6 s). This result directly demonstrates that nonkinetochore microtubules are not stabilized (or destabilized) by the spindle environment.
Figure 6.
(A) Microtubules growing off of pellicles in X. laevis egg extracts. (A, top) Heavy labeling with Alexa-488 tubulin allows all microtubules to be visualized, but individual filaments cannot be resolved. (A, bottom) The same region as in A with single molecule level Alexa-647 tubulin. Bar, 10 μm. (B) Distributions of tubulin lifetimes from pellicles (pellicles, blue) and unperturbed spindles (control spindles, green) are very similar. Best fits to Equation 1 are displayed for pellicles (red, τ = 71 ± 9 s) and spindles (black, τ = 64 ± 6 s).
Analysis of Only Long-lived Tubulin Molecules
The biased random walk model provides an excellent fit to our lifetime data (Figure 3). However, this cannot be taken as proof that this model is a literal description of microtubule dynamics. Rather, it is only phenomenological model that can quantitatively explain all of our data. Even though the biased random walk model can explain all of our lifetime data, covering >3 orders of magnitude of relative frequency (Figure 3), it is still possible that distinct portions of the lifetime curve are caused by mechanistically distinct process. For example, shorter lifetime events might correspond to rapid exchange of subunits within the GTP-tubulin cap (Howard and Hyman, 2009), whereas longer lifetimes might correspond to the more global dynamics of length changes of the entire polymer. We therefore performed a further analysis restricted to molecules with lifetimes >30 s. For these long times, both the biased random walk model and a single exponential provide an excellent fit to the lifetime data (Supplemental Figure 6), with R2 values >0.999. Exponential fits to lifetimes >30 s gives decay times of 22.0 ± 0.7 s for control spindles, 26.1 ± 0.8 s for FCPT-treated spindles, and 23.3 ± 1.6 s for pellicle-nucleated arrays of microtubules. Using these measures, data from the pellicle-nucleated arrays cannot be distinguished from control spindles (P ∼ 0.45, using a two-sample two-sided t test). The FCPT-treated value is statistically significant difference from control (p < 0.001, using a two-sample two-sided t test), but the magnitude of the effect is <10%. We again conclude that the spindle environment does not stabilize microtubules compared with pellicle-nucleated arrays and that kinesin-5 and microtubule sliding makes at most a small contribution to turnover mechanism for bulk nonkinetochore microtubules.
We also tested the possibility that there is a small population of more stable tubulin molecules, with a half-life of 10 min, by fitting the lifetime data for control spindles, restricted to times >30 s, to a sum of two exponentials, one exponential with a decay time of 10 min and the other exponential with a freely adjustable decay (data not shown). Kinetochore microtubules have been suggested previously to have a half-life of ∼10 min (DeLuca et al., 2006). This two component fit gave the fraction of the fast exponential decay as 99.98 ± 0.28%, which makes the contribution of the 10-min decay indistinguishable from zero (P ∼ 0.95). Thus our data does not provide evidence for the existence of a more stable subset of microtubules, but we cannot rule out the possibility that such microtubules exist. As noted above, images were acquired close to the coverslip to improve imaging and minimize the contribution from kinetochore microtubules, so we cannot rule out the possibility that a more stable subset of microtubules exists at other locations.
DISCUSSION
In summary, we developed a new method to quantify the turnover of individual microtubules in spindles, accurately quantified spindle polymerization dynamics in the 4- to 20-s time scale for the first time and provided evidence that nonkinetochore microtubules are not measurably stabilized by proximity to chromosomes or by the spindle environment. Our measurements and conclusions apply to nonkinetochore microtubules, which comprise the majority in X. laevis egg extract spindles, and not to kinetochore microtubules, which are known to be preferentially stabilized in many systems. Because only nonkinetochore microtubules are required for extract spindle assembly (Heald et al., 1996), our data imply that anastral spindle assembly can occur without microtubule stabilization. This finding held when we analyzed all lifetimes (i.e., >4 s) and also when we restricted the analysis to lifetimes >30 s, to remove possible complication from microtubule tip dynamics that do not contribute to micrometer-scale growth and shrinkage. Mitotic spindles in mammalian somatic cells can also assemble without kinetochore microtubules, although in this case their structure is abnormal, with lower microtubule density and increased pole–pole distance (DeLuca et al., 2002), so it is not clear to what extent our conclusion holds for astral, somatic mitotic spindles. More generally, spindles from different organisms—such as yeast, flies, humans, and frogs—are known to be different and results from one system might not extrapolate to other systems.
Our results contradict models proposing that microtubules are preferentially stabilized by the spindle environment, and this stabilization is the driving force for spindle assembly (Kirschner and Mitchison, 1986; Karsenti and Vernos, 2001). However, we view them as consistent with most direct observations of dynamics in spindles. For example, the classic finding from FRAP that the majority of spindle microtubules turn over in tens of seconds (Salmon et al., 1984) hardly suggests that the spindle is a stabilizing environment. The widespread credence given to stabilization models may reflect the fact that students of the spindle tend to focus on kinetochore microtubules, which clearly are stabilized in many systems (Inoue and Salmon, 1995). Stabilized microtubules also assemble in the midzone during cytokinesis, presumably in response to decreasing Cdk1 activity and relocalization of Aurora-B to the midzone (Eggert et al., 2006), but there is no evidence for such a stabilized midzone population in metaphase.
Can we reconcile our findings with previous arguments that chromatin stabilizes microtubules in the egg extract system? In a recent experiment, the shape of asters was measured near chromatin in TPX2-inhibited egg extracts, and these data were interpreted as predicting an approximately three- to fourfold increase in the lifetime of microtubule with free plus ends in spindles (Athale et al., 2008). Our data clearly reject this degree of stabilization for the majority of microtubules. In different experiments where dynamics were not perturbed we observed best-fit τ values that varied by <10%, which gives an approximate measure of the accuracy of our method. Even halving the amount of MCAK did not cause a three- to fourfold increase in stability. Adding constitutively active Ran to X. laevis egg extracts promotes microtubule assembly (Carazo-Salas et al., 2001) and stabilization (Caudron et al., 2005). These data were interpreted as showing that Ran-GTP causes microtubule stabilization near chromatin in spindles, but other interpretations are possible. For example, Ran-GTP may promote nucleation but not stabilization, and/or it may selectively stabilize kinetochore microtubules. It is also possible that chromatin derived factors act locally in spindles to destabilize microtubules, opposing the stabilizing effects of the Ran-GTP and Aurora-B gradients, and these influences were missed in observations made outside true spindles. For example, the dynamics of microtubules in fully formed spindles—investigated here—might be substantially different from microtubule dynamics during the initial phases of spindle assembly, perhaps corresponding to the stabilization suggested by Athale et al. (2008). Overall, our data strongly rejects the idea that all spindle microtubules are stabilized three- to fourfold near chromatin in spindles. Our methods might fail to detect a small subset of microtubules that were selectively stabilized, if this population was <1% of the total, but we see no evidence of a longer lived tail in any of our lifetime distributions. We cannot rigorously rule out the existence of a structurally or biochemically differentiated, more stable, small subset of nonkinetochore microtubules, but we know of no data that support the existence of such a subset. A special subset of more stable microtubules, called the midzone array, does assemble near the cell equator during cytokinesis (discussed above), but there is no evidence that it exists during metaphase. It is possible that the pellicle-nucleated arrays we used to model the environment outside spindles somehow induce microtubule stabilization. Arguing against a major effect of this kind, the kinetics of single microtubule assembly from centrosomes in mitotic egg extract (Belmont et al., 1990; Verde et al., 1992) is consistent with the average microtubule lifetimes we infer in spindles.
Our data also speak strongly to the mechanism of rapid turnover of nonkinetochore microtubules in spindles that has been mysterious since it was discovered by Inoue in the 1950s (Inoue and Sato, 1967). Our lifetime distribution was well fit by a model for first passage times of a biased random walk process, with an average lifetime of a microtubule of average length of 16 s. This fit is consistent with turnover occurring by a rapid, stochastic process at microtubule ends, most likely dynamic instability of free plus ends. We cannot rule out a contribution from some kind of stochastic dynamics at minus ends, or slow depolymerization from minus ends that makes little contribution to the lifetime distribution. Our data do argue strongly against a treadmilling model for turnover (Margolis et al., 1978), because this class of model is expected to show a peak in the lifetime distribution, corresponding to the average length of time it takes a subunit incorporated at one end of the microtubules to be lost at the other. Also, the lack of effect on turnover rate of blocking microtubule sliding with FCPT shows that microtubule turnover and movement are mechanistically distinct processes. This argues strongly against feeder-chipper models for turnover, because these require translocation of microtubules into static depolymerase sites (Gadde and Heald, 2004).
The biased random walk model is a phenomenological model that can quantitatively account for our lifetime data and provides a convenient means of parameterizing these data. Evaluating the validity of more detailed, microscopic models of microtubule dynamics will require additional data that this simple model cannot explain. Another widely used simple model of microtubule dynamics is the two-state model (Verde et al., 1992; Dogterom and Leibler, 1993), in which microtubules are taken to stochastically switch between phases of constant polymerization and constant depolymerization. These simple models ignore many features known to be present in real microtubules, such as nanoscale fluctuations (Schek et al., 2007) and complex structural transitions at the polymer ends (Anrnal et al., 2000). The two-state model predicts an exponential distribution of tubulin lifetimes in the absence of rescues (transitions from shrinking to growing; Bicout, 1997). Our measured lifetime distributions are clearly nonexponential, but it is possible that the large turnover at short times is caused by fluctuations of the tubulin-GTP cap (Howard and Hyman, 2009), whereas the turnover at longer times is dominated by the length changes of the microtubule. Although it would surely be possible to construct models of microtubule dynamics of that nature that are consistent with our measured lifetime distribution, because our data can already be accounted for by an even simpler model, we believe that it is more judicious to wait for additional data that demonstrate the deficiencies of the biased random walk model. As an additional precaution, we separately analyzed the lifetime data for times >30 s, which can be well fit to a single exponential, and all of the conclusion obtained from analyzing the full data set hold equally well when only this restrict subset is considered.
Our interpretation makes testable predictions about the average length and length distribution of spindle microtubules that are largely independent of the details of plus-end dynamics. If the spindle does not cause stabilization as we assert, nonkinetochore microtubules in spindles should have the same length distribution as microtubules grown from centrosomes in egg extracts, which is approximately exponential, with an average length of ∼3 μm (Verde et al., 1992). This proposed length distribution differs from published estimates using other methods. Correlated motions of individual tubulin molecules were used to infer a mean length of ∼20 μm and a bell-shaped distribution (Yang et al., 2007), but cross-linking might give rise to correlated motions of tubulin molecules in different microtubules, potentially complicating this analysis. A different length estimate based on measuring tubulin incorporation inferred an average length of ∼14 μm (Burbank et al., 2006). However, this technique used a number of unproven assumptions, such as the hypothesis that a microtubule's orientations can be definitively determined from its direction of motion, so the resulting estimate may be faulty. All of the current estimates of microtubule lengths in X. laevis egg extract spindles are indirect, including our own, and direct measurements by electron microscopy will be necessary to unambiguously resolve this issue.
If the locally high concentration of microtubules in spindles is not caused by local stabilization, what does cause it? We postulate it is solely due to an increased rate of nucleation in the spindle compared with bulk cytoplasm, and furthermore, that understanding how nucleation is spatially regulated will be the key to understanding morphogenesis of meiotic spindles. Given that microtubule turnover is fast relative to transport, nucleation has to occur throughout the spindle, as proposed by others (Mahoney et al., 2006). More specifically, during the metaphase steady state the pattern of nucleation must preserve local microtubule density and local microtubule polarity. Nucleation is globally controlled by the Ran pathway (Karsenti and Vernos, 2001), but it is still unclear how this generates spindle structure and gives rise to a sharp boundary between the inside and outside of the spindle. Mechanisms in which pre-existing microtubules locally stimulate nucleation are a possibility (Clausen and Ribbeck, 2007), as per the Arp2/3 complex for actin. One candidate mechanism might involve orientation of the γ-tubulin complex on microtubules by the augmin complex (Goshima et al., 2008). It is also possible that nucleation occurs with random orientation, and newborn microtubules are rapidly reoriented by motor proteins. Measurements of tubulin turnover, such as those presented here and elsewhere in the literature, cannot be used to directly probe microtubule nucleation because the majority of tubulin is incorporated into preexisting microtubules in the spindle. Hopefully, new experimental techniques will be developed to characterize the nature and spatial regulation of microtubule nucleation in spindles.
Finally, our observation that the tubulin lifetime distribution in the complex environment of the spindle can be well described by a simple biased random walk model with a single freely adjustable parameter is encouraging for efforts to provide a quantitative basis for cytoskeleton biology. Similar methods may be useful in investigating different microtubule populations and the dynamics of other biological polymers.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Woods Hole MBL Cell Division Group, especially Jay Gatlin, for helpful discussions. We thank Alex Lobkvosky and Bulbul Chakraborty for insightful conversations regarding models of dynamic instability. This work was supported by National Institutes of Health grant GM-39565 (to T.J.M.) and a fellowship (to D.J.N.) from the Life Science Research Foundation, sponsored by Novartis.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-09-0816) on November 25, 2009.
REFERENCES
- Anrnal I., Karsenti E., Hyman A. A. Structural transitions at microtubule ends correlate with their dynamic properties in Xenopus egg extracts. J. Cell Biol. 2000;149:767–774. doi: 10.1083/jcb.149.4.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athale C. A., Dinarina A., Mora-Coral M., Pugieux C., Nedelec F., Karsenti E. Regulation of microtubule dynamics by reaction cascades around chromosomes. Science. 2008;322:1243–1247. doi: 10.1126/science.1161820. [DOI] [PubMed] [Google Scholar]
- Axelrod D., Koppel D. E., Schlessinger J., Elson E., Webb W. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys. J. 1976;16:1055–1069. doi: 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudouin J., Mora-Bermudez F., Klee T., Daigle N., Ellenberg J. Dissecting the contribution of diffusion and interactions to the mobility of nuclear proteins. Biophys. J. 2006;90:1878–1894. doi: 10.1529/biophysj.105.071241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont L. D., Hyman A. A., Sawin K. E., Mitchison T. J. Real-time visualization of cell cycle-dependent changes in microtubule dynamics in cytoplasmic extracts. Cell. 1990;62:579–589. doi: 10.1016/0092-8674(90)90022-7. [DOI] [PubMed] [Google Scholar]
- Bicout D. J. Green's function and first passage time distribution for dynamic instability of microtubules. Phys. Rev. E. 1997;56:6656. [Google Scholar]
- Burbank K. S., Groen A. C., Perlman Z. E., Fisher D. S., Mitchison T. J. A new method reveals microtubule minus ends throughout the meiotic spindle. J. Cell Biol. 2006;175:369–375. doi: 10.1083/jcb.200511112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbank K. S., Mitchison T. J., Fisher D. S. Slide-and-cluster models for spindle assembly. Curr. Biol. 2007;17:1373–1383. doi: 10.1016/j.cub.2007.07.058. [DOI] [PubMed] [Google Scholar]
- Carazo-Salas R. E., Gruss O. J., Mattaj I. W., Karsenti E. Ran-GTP coordinated regulation of microtubule nucleation and dynamics during mitotic-spindle assembly. Nat. Cell Biol. 2001;3:228–234. doi: 10.1038/35060009. [DOI] [PubMed] [Google Scholar]
- Caudron M., Bunt G., Bastiaens P., Karsenti E. Spatial coordination of spindle assembly by chromosome-mediated signaling gradients. Science. 2005;309:1373–1376. doi: 10.1126/science.1115964. [DOI] [PubMed] [Google Scholar]
- Chang P., Jacobson M. K., Mitchison T. J. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 2004;432:645–649. doi: 10.1038/nature03061. [DOI] [PubMed] [Google Scholar]
- Cheerambathur D. K., Civelekoglu-Scholey G., Brust-Mascher I., Sommi P., Mogilner A., Scholey J. M. Quantitative analysis of an anaphase B switch: predicted role for a microtubule catastrophe gradient. J. Cell Biol. 2007;177:995–1004. doi: 10.1083/jcb.200611113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen T., Ribbeck K. Self-organization of anastral spindles by synergy of dynamic instability, autocatalytic microtubule production, and a spatial signaling gradient. PLoS One. 2007:e244. doi: 10.1371/journal.pone.0000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coue M., Lombillo V. A., McIntosh J. R. Microtubule depolymerization promotes particle and chromosome movement in vitro. J. Cell Biol. 1991;112:1165–1175. doi: 10.1083/jcb.112.6.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker J. C., Grier D. G. Methods of digital video microscopy for colloidal studies. J. Colloidal Interface Sci. 1996;179:298–310. [Google Scholar]
- Danuser G., Waterman-Storer C. M. Quantitative fluorescent speckle microscopy of cytoskeleton dynamics. Ann. Rev. Biophys. Biomol. Struct. 2006;35:361–387. doi: 10.1146/annurev.biophys.35.040405.102114. [DOI] [PubMed] [Google Scholar]
- DeLuca J., Gall W. E., Ciferri C., Cimini D., Muszcchio A., Salmon E. D. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- DeLuca J. G., Moree B., Hickey J. M., LKilmartin J. V., Salmon E. D. hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J. Cell Biol. 2002;159:549–555. doi: 10.1083/jcb.200208159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Murray A., Mitchison T. J., Walczak C. E. The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 1999;61:385–412. doi: 10.1016/s0091-679x(08)61991-3. [DOI] [PubMed] [Google Scholar]
- Dogterom M., Leibler S. Physical aspects of the growth and regulation of microtubule structures. Phys. Rev. Lett. 1993;70:1347. doi: 10.1103/PhysRevLett.70.1347. [DOI] [PubMed] [Google Scholar]
- Eggert U. S., Mitchison T. J., Field C. M. Animal cytokinesis: from parts list to mechanisms. Annu. Rev. Biochem. 2006;75:543–566. doi: 10.1146/annurev.biochem.74.082803.133425. [DOI] [PubMed] [Google Scholar]
- Gadde S., Heald R. Mechanisms and molecules of the mitotic spindle. Curr. Biol. 2004;14:R797–R805. doi: 10.1016/j.cub.2004.09.021. [DOI] [PubMed] [Google Scholar]
- Gardner M. K., et al. Chromosome congression by kinesin-5 motor-mediated disassembly of longer kinetochore microtubules. Cell. 2008;135:894–906. doi: 10.1016/j.cell.2008.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Mayer M., Zhang N., Stuurman N., Vale R. D. Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J. Cell Biol. 2008;181:421–429. doi: 10.1083/jcb.200711053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen A. C., Maresca T. J., Gatlin J. C., Salmon E. D., Mitchison T. J. Functional overlap of microtubule assembly factors in chromatin-promoted spindle assembly. Mol. Biol. Cell. 2009;20:2766–2773. doi: 10.1091/mbc.E09-01-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen A. C., Needleman D. J., Brangwynne C., Gradinaru C., Fowler B., Mazitschek R., Mitchison T. J. A novel small-molecule inhibitor reveals a possible role of kinesin-5 in anastral spindle-pole assembly. J. Cell Sci. 2008;121:2293–2300. doi: 10.1242/jcs.024018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R., Tournebize R., Blank T., Sandaltzopoulos R., Becker P., Hyman A. A., Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Heidemann S. R., McIntosh J. R. Visualization of the structural polarity of microtubules. Nature. 1980;286:517–519. doi: 10.1038/286517a0. [DOI] [PubMed] [Google Scholar]
- Hill T. L. Linear Aggregation Theory in Cell Biology. New York: Springer-Verlag; 1987. [Google Scholar]
- Howard J., Hyman A. A. Growth, fluctuation and switching at microtubule plus ends. Nat. Rev. Mol. Cell Biol. 2009;10:569–574. doi: 10.1038/nrm2713. [DOI] [PubMed] [Google Scholar]
- Hyman A. A., Drechsel D., Kellogg D., Salser S., Sawin K. E., Steffen P., Wordeman L., Mitchison T. J. Preparation of modified tubulins. Methods Enzymol. 1991;196:478–485. doi: 10.1016/0076-6879(91)96041-o. [DOI] [PubMed] [Google Scholar]
- Inoue S., Salmon E. D. Force generation by microtubule assembly disassembly in mitosis and related movements. Mol. Biol. Cell. 1995;6:1619–1640. doi: 10.1091/mbc.6.12.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S., Sato H. Cell motility by labile association of molecules. The nature of mitotic spindle fibers and their role in chromosome movement. J. Gen. Physiol. 1967;50:259–292. [PMC free article] [PubMed] [Google Scholar]
- Karsenti E., Vernos I. The mitotic spindle: a self-made machine. Science. 2001;294:543–547. doi: 10.1126/science.1063488. [DOI] [PubMed] [Google Scholar]
- Kim S. Y., Gitai Z., Kinkhabwala A., Shapiro L., Moerner W. E. Singe molecules of the bacterial actin MreB undergo directed treadmilling motion in Caulobacter crescentus. Proc. Natl. Acad. Sci. USA. 2006;103:10929–10934. doi: 10.1073/pnas.0604503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner M. W., Mitchison T. J. Beyond self-assembly: from microtubules to morphogenesis. Cell. 1986;45:329–342. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- Mahoney N. M., Goshima G., Douglass A. D., Vale R. D. Making microtubule and mitotic spindles in cells without functional centrosomes. Curr. Biol. 2006;16:564–569. doi: 10.1016/j.cub.2006.01.053. [DOI] [PubMed] [Google Scholar]
- Maly I. V. Diffusion approximation of the stochastic process of microtubule assembly. Bull. Math. Biol. 2002;64:213–238. doi: 10.1006/bulm.2001.0265. [DOI] [PubMed] [Google Scholar]
- Maresca T. J., Groen A. C., Gatlin J. C., Ohi R., Mitchison T. J., Salmon E. D. Spindle assembly in the absence of a RanGTP gradient requires localized CPC activity. Curr. Biol. 2009;19:1210–1215. doi: 10.1016/j.cub.2009.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis R. L., Wilson L., Keifer B. I. Mitotic mechanism based on intrinsic microtubule behavior. Nature. 1978;272:450–452. doi: 10.1038/272450a0. [DOI] [PubMed] [Google Scholar]
- Mastronarde D. N., McDonald K. L., Ding R., McIntosh J. R. Interpolar spindle microtubules in PTK cells. J. Cell Biol. 1993;123:1475–1489. doi: 10.1083/jcb.123.6.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T. J., Maddox P. S., Groen A. C., Cameron L. A., Perlman Z. E., Ohi R., Desai A., Salmon E. D., Kapoor T. M. Bipolarization and poleward flux correlate during Xenopus extract spindle assembly. Mol. Biol. Cell. 2004;15:5603–5615. doi: 10.1091/mbc.E04-05-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto D. T., Perlman Z. E., Burbank K. S., Groen A. C., Mitchison T. J. The kinesin Eg5 drives poleward microtubule flux in xenopus laevis egg extract spindles. J. Cell Biol. 2004;167:813–818. doi: 10.1083/jcb.200407126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niethammer P., Kueh H. Y., Mitchison T. J. Spatial patterning of metabolism by mitochondria, oxygen, and energy sinks in a model cytoplasm. Curr. Biol. 2008;18:586–591. doi: 10.1016/j.cub.2008.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi R., Burbank K. S., Liu Q., Mitchison T. J. Non-redundant functions of kinesin-13s during meiotic spindle assembly. Curr. Biol. 2007;17:953–959. doi: 10.1016/j.cub.2007.04.057. [DOI] [PubMed] [Google Scholar]
- Ohi R., Sapra T., Howard J., Mitchison T. J. Differentiation of cytoplasmic and meiotic spindle assembly MCAK functions by Aurora B-dependent phosphorylation. Mol. Biol. Cell. 2004;15:2895–2906. doi: 10.1091/mbc.E04-02-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C. G., Gardner M. K., Paliulis L. V., Salmon E. D., Odde D. J., Bloom K. Measuring nanometer scale gradients in spindle microtubule dynamics using model convolution microscopy. Mol. Biol. Cell. 2006;17:4069–4079. doi: 10.1091/mbc.E06-04-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips R., Kondev J., Theriot J. Physical Biology of the Cell. New York: Garland Science; 2009. [Google Scholar]
- Rickert K., et al. Discovery and biochemical characterization of selective ATP competitive inhibitors of the human mitotic kinesin KSP. Arch. Biochem. Biophys. 2008;469:220–231. doi: 10.1016/j.abb.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Sako Y. Imaging single molecules in living cells for systems biology. Mol. Syst. Biol. 2006;56:1–6. doi: 10.1038/msb4100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon E. D., Leslie R. J., Saxton W. M., Karow M. L., McIntosh J. R. Spindle microtubule dynamics in sea urchin embryos: analysis using a fluorescein-labeled tubulin and measurements of fluorescence redistribution after laser photobleaching. J. Cell Biol. 1984;99:2165–2174. doi: 10.1083/jcb.99.6.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath S. C., Ohi R., Leismann O., Salic A., Pozniakovsky A., Funabiki H. The chromosome passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell. 2004;118:187–202. doi: 10.1016/j.cell.2004.06.026. [DOI] [PubMed] [Google Scholar]
- Sawin K. E., Mitchison T. J. Poleward microtubule flux in mitotic spindles assembled in vitro. J. Cell Biol. 1991;112:941–954. doi: 10.1083/jcb.112.5.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schek H. T., Gardner M. K., Cheng J., Odde D. J., Hunt A. J. Microtubule assembly dynamics at the nanoscale. Curr. Biol. 2007;17:1–11. doi: 10.1016/j.cub.2007.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R., Larson D., Webb W. Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uteng M., Hentrich C., Miura K., Bieling P., Surrey T. Poleward transport of Eg5 by dynein-dynactin in Xenopus laevis egg extract spindles. J. Cell Biol. 2008;182:715–726. doi: 10.1083/jcb.200801125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valloton P., Ponti A., Waterman-Storer C. M., Salmon E. D., Danuser G. Recovery, visualization, and analysis of actin and tubulin polymer flow in live cells: a fluorescent speckle microscopy study. Biophys. J. 2003;85:1289–1306. doi: 10.1016/S0006-3495(03)74564-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde F., Dogterom M., Stelzer E., Karsenti E., Leibler S. Control of microtubule dynamics and length by cyclin a- and cyclin b-dependent kinases in Xenopus egg extracts. J. Cell Biol. 1992;118:1097–1108. doi: 10.1083/jcb.118.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev I. A., Svitkina T. M., Borisy G. G. Cytoplasmic assembly of microtubules in cultured cells. J. Cell Sci. 1997;110:2635–2645. doi: 10.1242/jcs.110.21.2635. [DOI] [PubMed] [Google Scholar]
- Walczak C. E., Mitchison T. J., Desai A. XKCM1, a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- Watanabe N., Mitchison T. J. Single-molecule speckle analysis of actin filament turnover in lamellipodia. Science. 2002;295:1083–1086. doi: 10.1126/science.1067470. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer C. M., Salmon E. D. How microtubules get fluorescent speckles. Biophys. J. 1998;75:2059–2069. doi: 10.1016/S0006-3495(98)77648-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss M. Challenges and artifacts in quantitative photobleaching experiments. Traffic. 2004;5:662–671. doi: 10.1111/j.1600-0854.2004.00215.x. [DOI] [PubMed] [Google Scholar]
- Wuhr M., Chen Y., Dumont S., Groen A. C., Needleman D. J., Salic A., Mitchison T. J. Evidence for an upper limit to mitotic spindle length. Curr. Biol. 2007;18:1256–1261. doi: 10.1016/j.cub.2008.07.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Cameron L. A., Maddox P. S., Salmon E. D., Danuser G. Regional variation of microtubule flux reveals microtubule organization in the metaphase meiotic spindles. J. Cell Biol. 2008;182:631–639. doi: 10.1083/jcb.200801105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Houghtaling B. R., Gaetz J., Liu J. Z., Danuser G., Kapoor T. M. Architectural dynamics of the meiotic spindle revealed by single-fluorophore imaging. Nat. Cell Biol. 2007;9:1233–1242. doi: 10.1038/ncb1643. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.