A fluorescent biosensor reports the localization of CDC-42 activity in the C. elegans embryo and was used to identify regulators of CDC-42 activity, one of which is involved in a novel regulatory loop that maintains cortical PAR polarity. CDC-42 activity regulates myosin II recruitment during the maintenance phase via the kinase MRCK-1.
Abstract
The anterior–posterior axis of the Caenorhabditis elegans embryo is elaborated at the one-cell stage by the polarization of the partitioning (PAR) proteins at the cell cortex. Polarization is established under the control of the Rho GTPase RHO-1 and is maintained by the Rho GTPase CDC-42. To understand more clearly the role of the Rho family GTPases in polarization and division of the early embryo, we constructed a fluorescent biosensor to determine the localization of CDC-42 activity in the living embryo. A genetic screen using this biosensor identified one positive (putative guanine nucleotide exchange factor [GEF]) and one negative (putative GTPase activating protein [GAP]) regulator of CDC-42 activity: CGEF-1 and CHIN-1. CGEF-1 was required for robust activation, whereas CHIN-1 restricted the spatial extent of CDC-42 activity. Genetic studies placed CHIN-1 in a novel regulatory loop, parallel to loop described previously, that maintains cortical PAR polarity. We found that polarized distributions of the nonmuscle myosin NMY-2 at the cell cortex are independently produced by the actions of RHO-1, and its effector kinase LET-502, during establishment phase and CDC-42, and its effector kinase MRCK-1, during maintenance phase. CHIN-1 restricted NMY-2 recruitment to the anterior during maintenance phase, consistent with its role in polarizing CDC-42 activity during this phase.
INTRODUCTION
Many metazoan cells are polarized. Polarization usually precedes and enables asymmetric cell divisions that generate cell diversity. In the Caenorhabditis elegans embryo, cell polarization determines the pattern of cell cleavages that produce the number and diversity of cells to constitute a functional worm. Many asymmetric cleavages, including the first, occur in cells that exhibit a polarized distribution of a subset of the PAR proteins, which are necessary for cytoplasmic partitioning (Goldstein and Macara, 2007). Because the PAR proteins constitute a conserved molecular machine, insight into their segregation and mode of action may be generally applicable to many organisms.
The C. elegans embryo establishes its anterioposterior (A–P) body axis before the first embryonic cleavage: the site of sperm entry defines the posterior end of the major axis of the fertilized oocyte (Goldstein and Hird, 1996). This polarizing activity requires a functional centrosome (Schumacher et al., 1998; O'Connell et al., 2000; Hamill et al., 2002) but not the sperm nucleus (Sadler and Shakes, 2000) or many microtubules (Cowan and Hyman, 2004; Tsai and Ahringer, 2007). The cortical actomyosin cytoskeleton becomes activated during meiosis II, generating motile ruffles throughout the cortex. The polarizing cue directs cortical flows away from the sperm pronucleus–centrosome complex. These actomyosin-powered flows transport cortical components toward the anterior, accompanied by the smoothing of the posterior cortex. The flows can give rise to a transient cortical invagination, called a pseudocleavage furrow (Munro et al., 2004), which marks the boundary between distinct anterior and posterior cortical domains in wild-type embryos, although it is neither required for polarization nor is an inevitable consequence of flows (Rose et al., 1995; Maddox et al., 2005). The period from the onset of flows to the resolution of the pseudocleavage furrow comprises the establishment phase (Cuenca et al., 2003).
Two sets of cytoplasmic determinants segregated by the cortical flows transduce the polarity cue to downstream effectors. The proteins PAR-3, PAR-6, and PKC-3 accumulate to the anterior of the pseudocleavage furrow, whereas the posterior domain contains PAR-2 and PAR-1. After the resolution of the pseudocleavage furrow, these two sets of proteins are mutually exclusive for localization: the anterior PARs are generally required for the others' anterior localization, and the posterior PARs prevent the anterior PARs from expanding into the posterior domain after the cessation of cortical flows (Cuenca et al., 2003). The period in which the PAR proteins are mutually antagonistic is termed the maintenance phase (Cuenca et al., 2003). The PAR proteins are required to orient and position the first mitotic spindle, thereby defining the division plane, and to direct differential gene expression in the resulting daughter cells (Gonczy, 2008). cdc-42 is required to maintain PAR protein polarity and was the first Rho family member implicated in the polarization of the C. elegans embryo (Gotta et al., 2001; Kay and Hunter, 2001).
The Rho-family of small GTPases function as molecular switches such that their guanosine triphosphate (GTP)-bound forms activate downstream effector molecules, whereas their guanosine diphosphate (GDP)-bound forms do not. These proteins themselves are regulated by guanine nucleotide exchange factors (GEFs) that catalyze their activation, and GTPase activating proteins (GAPs) that catalyze their inactivation. In C. elegans, RHO-1 and CDC-42 are the only members of their respective subfamilies, and both are required for proper development of the one-cell embryo. RHO-1 activity is required for the cortical contractility observed during the establishment of embryonic polarization; the localization of this activity likely patterns the flows that segregate cortical determinants. Embryos depleted of RHO-1 or its putative GEF ECT-2 (Jenkins et al., 2006; Motegi and Sugimoto, 2006; Schonegg and Hyman, 2006), myosin II (Guo and Kemphues, 1996; Shelton et al., 1999), or filamentous actin (Hill and Strome, 1988) do not exhibit wild-type ruffling, cortical myosin recruitment, or cortical flows during the establishment phase. In contrast, embryos depleted either of proteins required for centrosome maturation or the paternal contribution of the RhoGAP CYK-4 (Jenkins et al., 2006) exhibit cortical ruffling and myosin recruitment but no observable cortical flows. Together, these observations suggest that RHO-1 activity initially signals myosin-mediated contractility of the entire cortex, which is reduced at the presumptive posterior in response to the centrosome-mediated polarity cue. The transduction of this cue may involve a localized high concentration of RhoGAP activity in the form of paternal CYK-4 (Jenkins et al., 2006) and a localized reduction in RhoGEF activity due to a reduction of the local ECT-2 concentration (Motegi and Sugimoto, 2006). The subsequent asymmetric contractility results in the retraction of the cortex toward the anterior half of the embryo and the creation of a noncontractile cortical domain at the posterior half. Experimental manipulation of RHO-1 activity can affect the relative sizes of the contractile and noncontractile domains during polarization but does not appreciably affect the ultimate distribution of anterior and posterior markers during first cleavage (Schmutz et al., 2007; Schonegg et al., 2007). This suggests that although the regulation of RHO-1 activity during the establishment phase is required to polarize the cell, it does not determine the extent of anterior or posterior characterization of the cell by the time of cleavage.
In contrast to RHO-1 activity, CDC-42 activity is required to maintain PAR polarity during maintenance phase and has only subtle effects during establishment phase. In maintenance phase, CDC-42–depleted embryos exhibit a cortical loss of the anterior PAR proteins (Gotta et al., 2001). PAR-6 is lost from the anterior during this phase in most cdc-42(fRNAi) embryos (Aceto et al., 2006; Motegi and Sugimoto, 2006) and may be lost well before polarization in stronger depletions (Schonegg and Hyman, 2006). This loss of cortical PAR-6, and concomitant expansion of the posterior PAR proteins into the anterior, is probably the cause of the observed defects in pronuclear rotation, asymmetric spindle and furrow positioning, and proper developmental determination of daughter cells. In the wild type, stabilization of PAR-6 at the cortex is mediated by direct interaction of PAR-6 with the active form of CDC-42 (Aceto et al., 2006), although a CDC-42–independent mechanism for PAR-6 also operates (Beers and Kemphues, 2006). This stabilization may occur by the activation of the anterior PAR complex, whose PKC-3 activity directly phosphorylates and effectively excludes the local persistence of the posterior PAR protein PAR-2 (Hao et al., 2006), which is in turn necessary to maintain PAR-1 at the posterior. Depletion of CDC-42 reduces the depth and persistence of cortical invaginations and reduces the rate of myosin flow in the establishment phase (Motegi and Sugimoto, 2006; Schonegg and Hyman, 2006). These embryos nonetheless exhibit successful anteriorward actomyosin flows. Thus, the essential polarity function of CDC-42 seems to occur in maintenance rather than establishment phase. How CDC-42 is regulated in developmental time and space to permit the maintenance of polarity is not well understood. In this study, we have characterized spatial activity patterns of CDC-42 and identified regulators that give rise to these patterns.
MATERIALS AND METHODS
Worm Stocks and Maintenance
Worm strains were maintained as described previously (Brenner, 1974). The following stains were used: Bristol N2 (wild type), DP38 (unc-119(ed3) III.), WH333 (unc-119(ed3) III; ojIs26[unc-119(+) Ppie-1::gfp::nmy-2]), KK288 (bears itIs153, which includes a Ppie-1::gfp::par-2 transgene), TH25 (expresses GFP::PAR-6 in germline), FX1909 (chin-1(tm1909 or +)/+ III.), VC506 (cgef-1(gk261) X.), and VC315 (+/eT1 III; mrck-1(ok586)/eT1 V.). FX1909 and VC506 were outcrossed six generations to N2 males. Then, gk261 animals were isolated, and tm1909 was balanced with the sC1 chromosome from strain KK747 [par-2(lw32) unc-45(e286ts)/sC1[dpy-1(e1) let-??] III.]. VC315 were outcrossed three generations to N2 males, and then ok586 animals were balanced with the nT1[qIs51] chromosome that had been outcrossed to N2 for five generations from strain VC666.
Fluorescent Protein (FP) Strain Constructions
Transgenic lines used in this study were generated by bombardment of unc-119(+)-containing plasmids as described previously (Praitis et al., 2001). We subcloned the portion of the wsp-1 open reading frame (ORF) encoding amino acids 236-346 of isoform a from plasmid yk1350a08 (gift from Y. Kohara, National Institute of Genetics, Mishima, Japan) into the SpeI site of plasmid pFJ1.1 to drive expression of green fluorescent protein (GFP)-tagged GBDwsp-1 by using the pie-1 promoter and untranslated regions (UTRs). This plasmid (GenBank accession FJ602701) also contained an unc-119(+) transgene. To facilitate expression of mCherry as well as a monomeric mutant of GFP (mGFP) we constructed new plasmids from pFJ1 suitable for expressing N-terminally FP-tagged transgenes under the regulatory control of the pie-1 promoter and UTRs. In brief, this was done by removing the residual pie-1 ORF from pFJ1.1 and replacing the gfp sequence with sequences containing mgfp or mcherry. mgfp was generated by site-directed mutagenesis (using the QuikChange protocol from Stratagene, La Jolla, CA) of pFJ1.1-derived gfp sequence to induce an A206K mutation. A silent mutation was introduced into mcherry sequence (gift from A. Audhya, University of Wisconsin–Madison, Madison, WI) to remove a MluI site. These FP-encoding fragments were amplified by polymerase chain reaction (PCR) to append flanking BamHI sites and tandem SpeI and MluI sites just inside the 3′ BamHI site. These fragments were subcloned into the BamHI sites of the pie-1 ORF-deleted version of pFJ1.1. The resulting pJK3 and pJK6 vectors permit subcloning of SpeI–MluI-flanked inserts to yield plasmids suitable for expressing N-terminally tagged mGFP or mCherry under the control of pie-1 promoter and UTRs as well as an unc-119(+) rescuing fragment for transformant selection. We subcloned the ORF and introns of cdc-42 from N2 genomic DNA, full-length ORFs of cdc-42(T17N) and cdc-42(Q61L) from pJAM:yfpcdc42(T17N) and pJAM:yfpcdc42(Q61L) (gifts from D. Aceto and K. Kemphues, Cornell University, Ithaca, NY), ORF of cgef-1a from yk110c3, and the ORF and introns of chin-1 from N2 genomic DNA between the SpeI and MluI sites of pJK3 and pJK6. In this article, mgfp genes and mGFP fusions are written as “gfp” and “GFP” for simplicity.
Yeast Two-Hybrid Interaction Studies
Yeast two-hybrid studies were performed using the MatchMaker two-hybrid system (Clontech, Mountain View, CA), by using manufacturer's protocols, except as noted. The ORFs of cdc-42, cdc-42(T17N), cdc-42(Q61L), and GBDWSP-1 were subcloned into pGADT7 and pGBKT7 vectors that were modified such their respective NdeI–XhoI and NdeI–PstI fragments were replaced with SpeI–AscI and SpeI–MluI tandem cloning sites. The mutant cdc-42 sequences were gifts from D. Aceto and K. Kemphues. All two-hybrid experiments were performed in the AH109 strain, grown for 5 d at 30°C by using the His marker to test interaction.
RNA-mediated Interference (RNAi) Treatment
RNAi was performed by a previously described feeding method (Timmons and Fire, 1998). In brief, HT115(DE3) Escherichia coli were transformed with a pL4440-based vector bearing T7 promoters useful for bacterial production of double-stranded RNA (dsRNA) of the intervening sequence of interest. These bacteria were induced to transcribe dsRNA for 1 d on nematode growth media plates containing isopropyl β-d-thiogalactoside (IPTG); to deplete two genes, bacterial strains were mixed at a 1:1 ratio based on the cultures' optical densities. Worms were then fed these bacteria for 20–50 h before dissection of their embryos for imaging. Unless otherwise noted, constructs used to induce RNAi were obtained from a previously published collection produced by the Ahringer laboratory (Kamath et al., 2003). Other constructs were produced by subcloning cDNA sequences into pL4440. The cDNA clones used (gifts from Y. Kohara) include yk196b9 (for par-1), yk325e4 (par-2), yk552e12 (par-3), yk109f2 (cdc-42), yk110c3 (cgef-1), yk1083a11 (Y105E8A.24), yk1243d09 (Y95B8A.12), yk877c07 (rhgf-1), yk881c01 (Y34B4A.8), yk146f7 (rga-4), yk677g10 (2RSSE.1), and yk1707c12 (chin-1). All constructs presented were confirmed by sequencing to bear the target gene or cDNA. Embryos depleted of these gene products necessary for asymmetric cell division were imaged to the four-cell stage to demonstrate efficacy of RNAi treatment. For CHIN-1 immunolocalization, HT115(DE3) E. coli were transformed with a plasmid designed to express dsRNA corresponding to either chin-1 or the nonessential gene smd-1 and were induced to transcribe dsRNA on MYOB plates containing IPTG. N2 strain L1-stage larvae were transferred onto these plates and grown for 3 d. Adult worms were then dissected to obtain young embryos for immunostaining.
Statistical Analysis
The analysis of interactions of cgef-1 with other GEF-encoding genes was performed via logistic regression by using the statistical computing program R, version 2.7.1 (R Development Core Team, 2008), assuming a binomial outcome for survival to hatching.
Antibody Production
Affinity-purified rabbit sera produced against amino acids 137-236 of CHIN-1 and 256-375 of CGEF-1A were produced by Strategic Diagnostics (Newark, DE), by using genomic antibody technology.
Immunostaining
Embryos were fixed for 10 min at −20°C in N,N-dimethylformamide, and then briefly washed in 1× phosphate-buffered saline (PBS)/0.5% Tween/2 mM MgCl2 before blocking. The anti-α tubulin antibody DMA1 (Sigma-Aldrich, St. Louis, MO) was used at 1:1000 dilution and anti-CHIN-1 antibody at 1:4000. Isopropyl β-d-thiogalactoside was used to counterstain DNA. Staining of DNA and tubulin were used to establish embryonic staging.
Imaging
Embryos and animals for imaging were prepared either by mounting embryos on a 2% agar pad or in a hanging drop of egg buffer with subsequent sealing with Vaseline (Chesebrough-Ponds, Greenwich, CT) or by anesthetizing worms in M9 supplemented with 5 mM levamisole (Sigma-Aldrich) and mounting on a 5% agar pad with subsequent sealing with Vaseline. Multiphoton-excitation laser scanning microscopy (MPLSM) was performed with two previously described (Wokosin et al., 2003; Skala et al., 2007b), custom-built microscope systems. The MPLSM excitation sources were Ti:Sapphire lasers (Coherent Mira, Santa Clara, CA) or Spectra Physics Tsunami (Mountain View, CA) tuned to an excitation wavelength of 890 nm. The detectors were H7422P-40 GaAsP photomultiplier tubes (Hamamatsu, Hamamatsu City, Japan). Objectives used were a Super Fluor 40× 1.3 numerical aperture (NA) for imaging GFP::GBDwsp-1 and a Super Fluor 100× 1.3NA for all other probes (Nikon, Melville, NY). The MPLSMs achieve maximal resolution with the 100× objective. Images were collected at 512 × 512 pixel resolution with laboratory-developed acquisition software (WiscScan) and were analyzed with ImageJ (National Institutes of Health, Bethesda, MD). For immunolocalization of CHIN-1 and CGEF-1, confocal microscopy was performed as described previously (Song et al., 2008).
For fluorescence resonance energy transfer (FRET) studies, embryos were dissected from single mothers in egg buffer and mounted on a 2% agar pad. Mothers were the cross-progeny of mating WH363 (a line bearing an integrated Ppie-1::gfp::GBDwsp-1 transgene) males with WH423 [a line bearing nonintegrated Ppie-1::mcherry::CDC-42(Q61L) transgenes] or WH432 [a line bearing nonintegrated Ppie-1::mcherry::CDC-42(T17N) transgenes] hermaphrodites. Fluorescence lifetime measurements were made on a previously described (Skala et al., 2007a), custom-built MPLSM by using an SPC-830 time-correlated, single-photon counting (TCSPC) module (Becker & Hickl, Berlin, Germany) for fluorescence lifetime imaging microscopy (FLIM). After lifetime data collection, embryos were scored for presence or absence of mCherry expression by visual inspection via wide-field fluorescence microscopy. Data were analyzed with TCSPC analysis software (SPCImage v2.8.6.2936; Becker & Hickl) and Excel 2003 (Microsoft, Redmond, WA). For lifetime determinations, scatter was set to 0 and shift was optimized for best fit at the brightest pixel within an embryo when integration was set to 10.
Active CDC-42 Probe Construction, Validation, and Localization Pattern
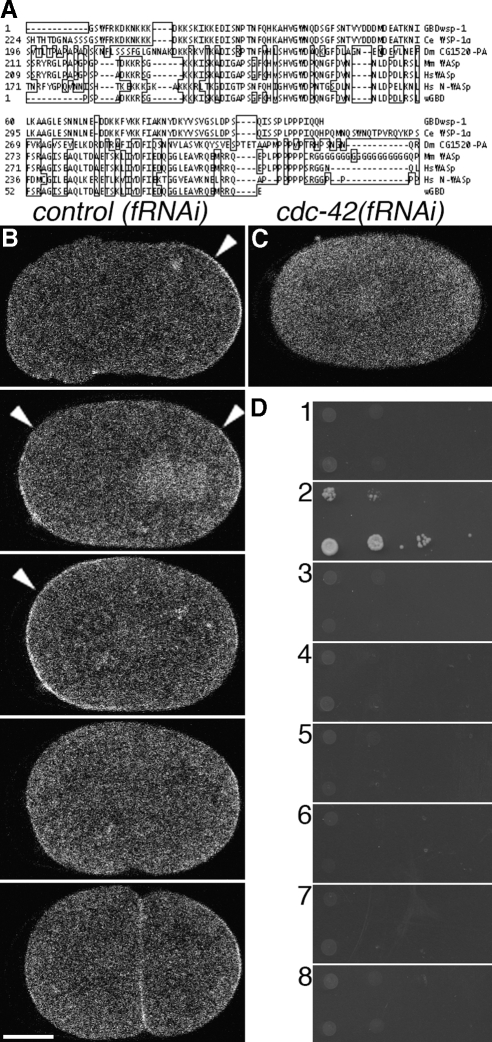
To monitor the spatiotemporal activity of CDC-42 in the early embryo, we identified a G-protein binding domain (GBD) of the C. elegans homologue of the Wiskott-Aldrich Syndrome protein WSP-1. The CRIB domain of WSP-1 binds specifically to GTP-bound (active) CDC-42. The alignment of WSP-1, vertebrate homologues of WSP-1, and a previously described biosensor that reports Cdc42 activity in Xenopus oocytes (Benink and Bement, 2005; Figure 1A) identified a fragment that includes a predicted CRIB domain, which was sufficient for specific binding to GTP-bound CDC-42. This specific binding activity was confirmed first by interaction in a yeast two-hybrid system. The putative GBD demonstrated interaction with wild-type CDC-42 and a constitutively active mutant CDC-42(Q61L) but not with a constitutively inactive mutant CDC-42(T17N) or the analogous mutations of the Rho-family GTPases CED-10/Rac and RHO-1 (Figure 1D). We confirmed the interaction in vivo using a FRET/FLIM approach. FRET was observed between GFP-tagged GBDWSP-1 and mCherry-tagged CDC-42(Q61L) but not between GFP::GBDwsp-1 and mCherry::CDC- 42(T17N). In this approach, interaction of fluorophores colocalizing on the order of 5 nm reduces the fluorescence lifetime of the donor fluorophore. Unexpectedly, the gene dose of GFP::GBDwsp-1 had a small but statistically significant effect (2569ps and 2526ps for 2 and 1 gene copies, respectively; 0.01 < p < 0.02) on observed GFP lifetime, so all comparisons were made using embryos expressing GFP::GBDwsp-1 from a single genetic copy. Embryos coexpressing one gene copy GFP::GBDwsp-1 and either mCherry::CDC-42(T17N) or mCherry::CDC- 42(Q61L) exhibited GFP lifetimes of 2523ps (p > 0.80) and 2444ps (p < 0.01), respectively (Supplemental Figure 1). Thus, FRET analysis suggests that GBDwsp-1 interacts directly with CDC-42(Q61L) but not CDC-42(T17N) probe in vivo.
Figure 1.
Construction, validation, and localization of a biosensor of CDC-42 activity. (A) Alignment of identified GBDwsp-1 (top) with WSP-1 homologues of C. elegans, Drosophila, mouse and human (middle), and a probe that reports Cdc42 activity in Xenopus (bottom). Boxed amino acids are dissimilar to those of WSP-1. (B) Localization of GFP::GBDwsp-1 in a control(fRNAi) embryo. Arrowheads indicate polarized enrichment of cortical signal. From top to bottom: phase I, transition from phase I to phase II, phase II, phase III, completion of cytokinesis. (C) Localization of GFP::GBDwsp-1 in a cdc-42(fRNAi) embryo. (D) Yeast two-hybrid showing interaction between GBDwsp-1 and (2) the active CDC-42(Q61L) mutant but not (1) the inactive CDC-42(T17N) mutant, (3) the inactive CED-10(T17N) mutant, (4) the active CED-10(Q61L) mutant, (5) the inactive RHO-1(T19N) mutant, (6) the active RHO-1(Q63L) mutant, or (7) empty-vector control. (8) There is no interaction between the paired empty-vector controls. In these (and all subsequent) micrographs of embryos, the embryo is positioned with anterior to the left and posterior to the right. Bar, 10 μm.
Transgenic worms that express a GFP-tagged GBDWSP-1 in the germline and early embryo permitted a spatiotemporal localization of CDC-42 activity by fluorescence microscopy. Fluorescence signal from GFP::GBDWSP-1 was observed at the cell cortices of the germline and young embryos (data not shown; Figure 1B and Supplemental Movie 1). RNAi-mediated depletions of CDC-42 abolished the detectible cortical enrichment (Figure 1C and Supplemental Movie 2). In these embryos, GFP::GBDWSP-1 localized throughout the cytoplasm and was enriched in the nucleoplasm. These localizations were similar to those exhibited by embryos expressing unfused GFP (data not shown). Given the demonstrations of specificity of GBDWSP-1 for the active form of CDC-42 in vitro and in vivo and the dependence upon CDC-42 for its GFP-tagged fusion protein, we interpret the cortical mobilization of the probe to reflect accessible, active CDC-42 and refer to this mobilization simply as “CDC-42 activity.”
RESULTS
Localization of CDC-42 and Its Active and Inactive Mutant Forms
Observations of the dynamic localization of polarity proteins with overexpressed, GFP-tagged transgenes revealed that CDC-42 and subsets of the PAR proteins are required for different phases of polarization in the C. elegans one-cell embryo (Cuenca et al., 2003; Motegi and Sugimoto, 2006; Schonegg and Hyman, 2006). In our transgenic lines, localizations of GFP-tagged CDC-42 and two of its mutant forms were consistent with those published previously. GFP::CDC-42 was observed to be enriched at all points on the cortex (relative to the cytoplasm) throughout the first cell cycle, but this enrichment was not uniform (Supplemental Figure 2). From polarity initiation through resolution of the pseudocleavage furrow, the cortical localization was enriched at the anterior cortex consistent with movement by bulk flow of the cortex at this time. This localization is largely consistent with those recently published (Aceto et al., 2006; Motegi and Sugimoto, 2006; Schonegg and Hyman, 2006). The anterior enrichment is retained through cytokinesis, where signal is observed most strongly at the apposition of two-cell cortices. GFP::CDC-42 was also enriched in a “cloud” near the sperm centrosome and between the sperm pronucleus and future posterior cortex (Supplemental Movie 1). This cloud flowed away from the polarization site upon initiation of cortical flows. Another novel observation is that signal also localized to the centrosomes and mitotic spindle. The significance of these noncortical localizations is unclear as is their specificity for our transgenic lines. GFP-tagged CDC-42(T17N)—which is likely constitutively inactive (Ziman et al., 1991)—also exhibited a slight cortical enrichment that was polarized at the anterior during cortical flow (Supplemental Figure 2). Between the cessation of cortical flows and onset of cytokinesis, this anterior cortical enrichment was lost, and little cortical localization was observed. By contrast, the GFP-tagged CDC-42(Q61L)—which is likely constitutively active (Ziman et al., 1991)—exhibited enhanced cortical localization and asymmetric cortical enrichment (Supplemental Figure 2). This probe was enriched at the anterior during cortical flow and remained polarized through cytokinesis. Together these observations suggest that CDC-42 activation correlates with increased cortical enrichment as would be predicted by previous descriptions where inactive Rho-family proteins are solubilized by guanosine nucleotide disassociation inhibitors, diffuse to the cytoplasm, and are able to be relocalized to the membrane upon reactivation (Hori et al., 1991; Fleming et al., 1996; Hoffman et al., 2000). Also, the bulk of cortical CDC-42 is enriched at the anterior during cortical flows. However, because the Q61L mutant cycles only very slowly from the active state (Xu et al., 1997), the constitutively active CDC-42 mutant localization probably reflects the movement of cortex-ensnared proteins rather than the localization of endogenous CDC-42 activity. Furthermore, the failure of these proteins to cycle their activity state may produce localizations that do not represent the wild-type protein. To avoid these confounding effects, we investigated the localization of endogenous CDC-42 activity by means of an effector-based biosensor approach.
GFP::GBDwsp-1 Localization
CDC-42 activity, as reported by GFP::GBDwsp-1 (see Materials and Methods for validation), localized sequentially to the posterior and anterior during polarity establishment and maintenance phases, respectively (57/57 embryos; Figure 1B and Supplemental Movie 1). The appearance of the posterior signal corresponded with the time and location of contact between the sperm pronuclear/centrosome complex and the cortex (best seen in Supplemental Movie 1). This signal then expanded toward the anterior, labeling the posterior, less contractile domain to the extent of the pseudocleavage furrow (similar to the dynamic localization of GFP::PAR-2; Cuenca et al., 2003). On entry into maintenance phase (as the anterior ruffling and pseudocleavage furrow resolve), the CDC-42 activity exhibited a change of localization from the posterior to the anterior cortex. The anterior cortical localization persisted until just before the onset of cytokinetic furrow invagination, when observable cortical enrichment was lost. We refer to the developmental phases associated with the sequential posterior, anterior and noncortical localizations of the probe as phases I, II, and III, respectively. Exogenous coexpression of a constitutively active CDC-42 mutant increases the cortical recruitment during all phases (Figure 2 and Supplemental Movie 6).
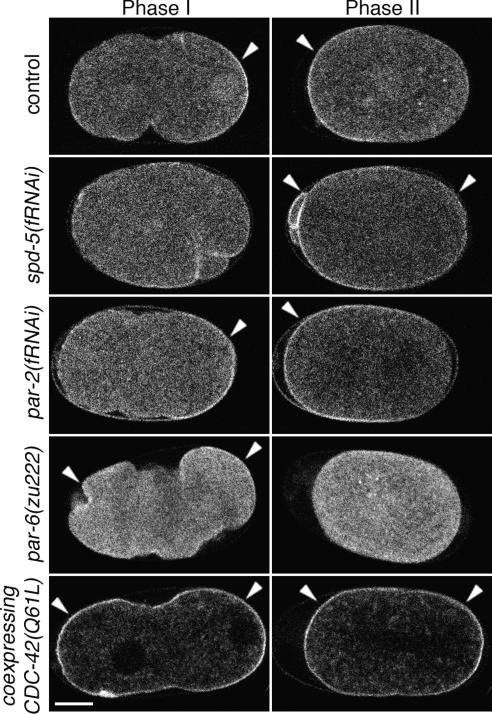
Figure 2.
Distribution of active CDC-42 depends upon identified polarity proteins. Embryos expressing GFP::GBDwsp-1 during phases I and II. Embryos from untreated mothers or those depleted of SPD-5, PAR-2 or PAR-6. Also, probe localization in an embryo coexpressing CDC-42(Q61L). Arrowheads indicate polarized enrichment of cortical signal. Bar, 10 μm.
Probe Localization Pattern Depends on Known Polarity Factors
The posterior CDC-42 activity acts at or downstream of the level of the polarity cue. To confirm that this posterior signal depends upon the sperm-provided polarization cue, we observed the probe in embryos that fail to initiate polarity as a consequence of centrosome maturation failure by depleting SPD-5 by using RNAi. These embryos did not establish a smoothed cortical domain, and the CDC-42 activity was located about the site of the previous polar body extrusion (this small, anterior patch was also transiently observed in wild-type embryos) in phase I (Figure 2 and Supplemental Movie 3). These localization patterns resemble those observed with GFP::PAR-2 in severe depletions of centrosomal function (Tsai and Ahringer, 2007). As these embryos developed, the CDC-42 activity of phase II relocalized to the cortical subdomains that were not labeled in phase I. These localization patterns recapitulate those observed with GFP::PAR-6 in severe depletions of centrosomal function (data not shown). Together, our results suggest that CDC-42 cortical activity patterns depend upon centrosome function for determination of anterior and posterior cortical domains in the same way that the polarity proteins PAR-6 and PAR-2 do.
The dynamic localization of CDC-42 activity depends upon the anterior, but not posterior, PAR proteins. Given that active CDC-42 has been demonstrated to interact with PAR-6, we determined whether the localization of CDC-42 activity depended upon the PAR proteins. Depletion of the posterior PAR proteins (PAR-1 or PAR-2) produced little alteration in the dynamic localization pattern [Figure 2 and Supplemental Movie 4 for par-2(fRNAi)]. In these embryos, the probe's phase I localization was essentially unchanged, whereas the phase II localization differed only in that it extended further toward the posterior than in the wild type. In contrast, complete embryonic depletion of the anterior PAR complex protein PAR-6 altered both the phase I and II localizations (Figure 2 and Supplemental Movie 5). In these embryos, the phase I signal localized weakly and relatively uniformly to the cortex, whereas the phase II signal showed no cortical enrichment. None of these depletions altered the phase III signal (i.e., little cortical signal observed). Together, these results indicate that CDC-42 activity is enriched at the cortical regions that are free of PAR-6 during phase I, then is enriched only at the cortical domains that contain PAR-6 during phase II, and then is not enriched at the cortex during phase III. So, our data suggest that in maintenance phase PAR-6 requires CDC-42 for localization, and CDC-42 activity requires PAR-6 for its localization.
Determination of Regulators of CDC-42 Activity
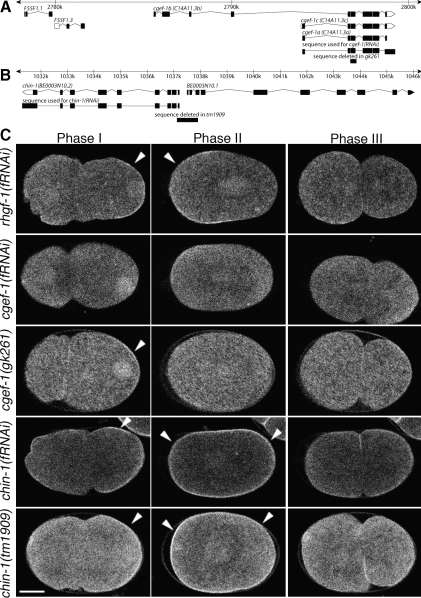
The dynamic localization pattern of the probe allowed us to identify regulators of CDC-42 activity from a candidate RNAi screen as depletions altering this pattern. Initially, we searched the genomic sequence and identified 18 candidate genes predicted to encode a RhoGEF domain (specifically, tandem DH-PH domains) containing proteins and 22 genes predicted to encode RhoGAP domain-containing proteins (Supplemental Table 1). Gene products of 39 of these 40 were individually depleted: two of these depletions were found to alter probe localization patterns. tag-150(fRNAi) embryos (depleted of a putative GEF) exhibited reduced cortical localization of the probe during all developmental time points observed (11/11 embryos; Figure 3). BE0003N10.2(fRNAi) embryos (depleted of a putative GAP) exhibited an unaltered probe localization in phase I; however, the phase II localization included an enrichment at both the anterior and poster cortical domains (6/6 embryos; Figure 3). The phase III probe localization in these embryos retained a cortical enrichment, whereas the cortical enrichment is lost at this time in the wild type. The observed loss and gain of apparent CDC-42 activity in depletions of tag-150 and BE0003N10.2 are consistent with the sequence predictions that these genes encode a GEF and a GAP, respectively. Deletion mutations predicted to disrupt these genes, gk261 and tm1909, phenocopy the RNAi phenotypes (12/12 for gk261 and 3/3 for tm1909; Figure 3 and Supplemental Movies 7 and 8). tag-150(gk261) is predicted to disrupt only tag-150. This deletion is expected to disrupt encoding of the isoform-conserved RhoGEF domain: it produces a loss of the fourth-to-final exon, encoding part of the DH domain, and a frameshift preventing the expression of the C-terminal PH domain. BE0003N10.2(tm1909) is predicted to disrupt the promoters of both that gene and the neighboring BE0003N10.1 gene, which is predicted to encode a RNA exonuclease. That both BE0003N10.2(RNAi) and tm1909 animals exhibit mislocalization of CDC-42 activity and increased embryonic lethality argues that the cause of the defects in tm1909 animals is due to the disruption of BE0003N10.2. Because loss of tag-150 affected our probe for CDC-42 activity in ways consistent with its functioning as a GEF for CDC-42, we refer to this gene as cgef-1 (for CDC-42 guanine nucleotide exchange factor). Similarly, because loss of BE0003N10.2 function affected our probe in ways consistent with its functioning as a GAP for CDC-42 and its sequence homology to vertebrate chimaerins, we refer to this gene as chin-1 (for Chimaerin-related).
Figure 3.
Wild-type CDC-42 activity requires the putative GEF CGEF-1 and the putative GAP CHIN-1. (A) Physical map of the portion of the C. elegans genome including predicted cgef-1–related ORFs, region deleted in the gk261 mutation, and the target sequence used for cgef-1(RNAi). (B) Physical map of the portion of the C. elegans genome including the predicted chin-1 and BE0003N10.1 ORFs, region deleted in the tm1909 mutation, and the target sequence used for chin-1(RNAi). (C) GFP::GBDwsp-1 localization in embryos during phases I, II, and III. Embryos from mothers treated with RNAi directed toward the RhoGEF-encoding rhgf-1, cgef-1, or chin-1. Specificity of RNAi target was tested by comparing the RNAi phenotype to the phenotype of embryos from cgef-1(gk261) or chin-1(tm1909) mothers. Note the persistence of nuclear signal in cgef-1(gk261) embryos. Arrowheads indicate polarized enrichment of cortical signal. Bar, 10 μm.
RNAi depletions of CGEF-1 and CHIN-1 did not uncover a polarity defect. Worm strains cultured under conditions of multigenerational cgef-1(fRNAi) or chin-1(fRNAi) were viable and overtly wild type. In addition, GFP::PAR-2 and GFP::PAR-6 localizations were not grossly altered in the embryos these worms produced (data not shown). These results were surprising given cgef-1 and chin-1 were identified as regulators of the activity of CDC-42, which is essential for embryonic and postembryonic development. To test whether the RNAi-mediated depletions were simply insufficient to remove function, eri-1(mg366) mutants, which are hypersensitive to exogenous RNAi (Kennedy et al., 2004), were depleted of cgef-1 or chin-1 by RNAi. In this sensitized strain, the chin-1(fRNAi) animals exhibited sterility within one generation, whereas the cgef-1(fRNAi) animals continued to exhibit generally wild-type fecundity, animal morphology, and embryonic cleavage patterns.
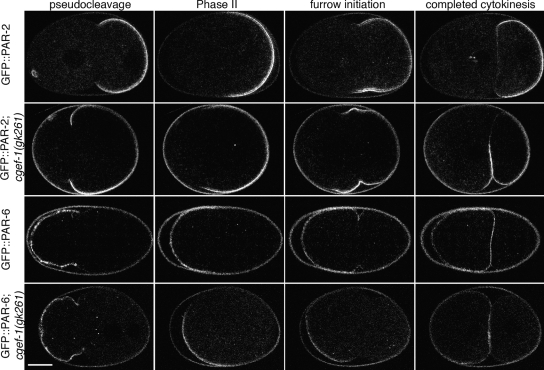
Some cgef-1 function is required for wild-type PAR protein localization dynamics. In the potentially more deleterious cgef-1(gk261) background, GFP::PAR-6 retracted further along the A-P axis during phase I than in the wild type and the GFP::PAR-6–labeled domains in these embryos remained abnormally small through phase II (8/8 embryos; Figure 4 and Supplemental Movies 9 and 10). At or about the onset of furrowing, the GFP::PAR-6 signal occupied the entire presumptive AB cell cortex, as it does in the wild type. GFP::PAR-2 dynamics were altered in ways that were roughly complementary to those of GFP::PAR-6. Namely, during phase I, the GFP::PAR-2 domain extended abnormally far toward the anterior in phases I and II (9/9 embryos; Figure 4 and Supplemental Movies 11 and 12). On furrowing, these embryos exhibited GFP::PAR-2 signal on the presumptive AB cortex that was not seen in cgef-1(+) embryos. After cleavage, the somatic cells continued to exhibit GFP::PAR-2 signal, which was not observed in the wild type.
Figure 4.
The dynamic localizations of PAR proteins are altered in cgef-1(gk261) embryos. GFP::PAR-2 or GFP::PAR-6 localization in embryos from wild-type or cgef-1(gk261) mothers. Both PAR proteins are anteriorly displaced through furrow initiation in cgef-1(gk261) embryos; however, blastomere-specific segregation is grossly intact by the two-cell stage. Some fluorescence (probably autofluorescence) can be seen. Bar, 10 μm.
Putative null mutants of the newly identified CDC-42 regulators exhibited other defects. Like cgef-1(fRNAi) embryos, cgef-1(gk261) embryos exhibited anteriorly displaced pseudocleavage furrows that persisted longer than in the wild-type. Surprisingly, no other defects in cgef-1(gk261) mutant animals or embryos were observed. The brood sizes were wild-type, and embryonic lethality was not statistically significantly elevated (Table 1). In contrast, chin-1(tm1909) animals exhibited a number of defects. chin-1(tm1909) animals exhibited both a recessive reduction of brood size and embryonic lethality (Table 1), which were cold-sensitive effects. tm1909 animals also exhibited an abnormal gonad morphology such that the distal gonad arms were enlarged and contained refractive granules in the rachis that were not observed in the wild type (data not shown).
Table 1.
Observed brood size and embryonic lethality of cgef-1 and chin-1 mutant strains
| n | Brood size | Embryoniclethality (%) | |
|---|---|---|---|
| N2 (+/+) | 5 | 210.8 ± 13.1 | 0.4 ± 0.4 |
| cgef-1(gk261) (−/−) | 5 | 225.6 ± 9.0 | 1.2 ± 0.8 |
| N2 (+/+) | 4 | 337.5 ± 15.4 | 0.7 ± 0.6 |
| chin-1(tm1909)/+ | 5 | 339.8 ± 43.8 | 1.0 ± 0.6 |
| chin-1(tm1909) | 20 | 7.7 ± 9.5* | 98.7 ± 4.1* |
| chin-1(tm1909) (16°C) | 24 | 0.04 ± 0.2 | 100 ± 0 |
| chin-1(tm1909) (24°C) | 22 | 50.7 ± 48.5 | 75.7 ± 20.5 |
| chin-1(tm1909);cgef-1(gk261) (24°C) | 11 | 12.4 ± 13.2† | 79.4 ± 25.0 |
chin-1 and cgef-1 mutant strains were analyzed with experimentally paired observations of the same wild-type strain. Experiments were conducted at 20°C unless otherwise noted. Statistical comparisons noted are for experiments at the same temperature.
* p < 0.05, statistically significant difference from the wild type.
†p < 0.05, statistically significant difference from chin-1(tm1909) at 24°C.
cgef-1(gk261) mutants exhibited unexpectedly mild phenotypes, especially compared with depletions or deletions of cdc-42, so we tested for residual CGEF-1 function and possible redundant CDC-42 GEFs in these mutants. First, we determined that the gk261 phenotypes of slightly increased embryonic lethality and reduction of cortical recruitment of GFP::GBDwsp-1 were not more severe in cgef-1(gk261 + RNAi) strains (data not shown). Therefore, we suspect that gk261 is a null allele of cgef-1. Functional redundancy with other RhoGEFs was tested by scoring for enhancement of sterility or embryonic lethality by gk261 in worms that were depleted of the 17 other predicted RhoGEF gene products (Supplemental Table 2). The gk261 alleles enhanced the embryonic lethality observed in several of the GEF-depleted lines and reduced the brood size of a few others. Relative to gk261 animals, two of these RNAi treatments—directed against putative GEFs tag-127 and R02F2.2—markedly increased the embryonic lethality. These two depletions were repeated in gk261 worms expressing GFP::GBDwsp-1, and the observed signal localization was indistinguishable from the undepleted controls (data not shown). In addition, logistic regression analysis suggests that cgef-1 and C11D9.1 may serve overlapping essential functions and that cgef-1 and rhgf-1 may serve antagonistic functions in some essential process (Supplemental Table 2). Unfortunately, the brood reductions observed in cgef-1(gk261) animals depleted of R02F2.2 or TAG-127 were sufficiently severe to limit the statistical power of demonstrating interaction of cgef-1 with either of those two genes.
Localization of the Putative CDC-42 GAP and GEF
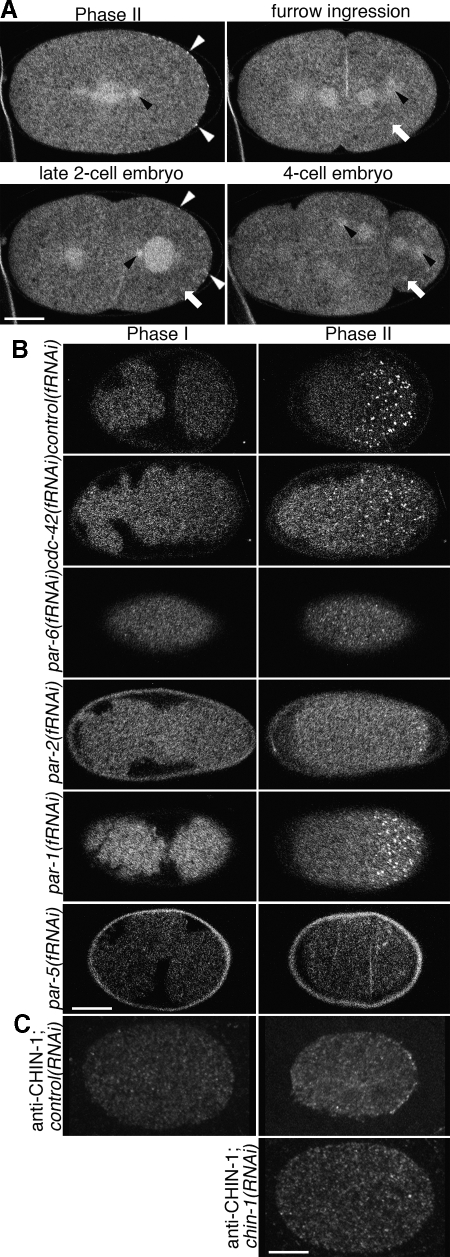
Because CHIN-1 depletions affected the spatial patterning of the CDC-42 activity biosensor, we speculated that CHIN-1 itself might exhibit a polarized cortical distribution. GFP::CHIN-1, expressed under germline regulatory control, localized to the centrosomes, pronuclei, central spindle and cell cortex in the early embryo (Figure 5A). This reporter strain exhibited no cortical enrichment during phase I but was recruited to the posterior cortex both in a diffuse pattern and in discrete puncta phase II (23/23 embryos). These puncta flowed into the ingressing cleavage furrow during phase III (Supplemental Movie 13). In older embryos, signal was observed at cell borders, nuclei and centrosomes (data not shown). cdc-42(fRNAi) (3/3 embryos), par-3(fRNAi) (3/3 embryos), pkc-3(fRNAi) (3/3 embryos), and par-6(fRNAi) (7/7 embryos) embryos exhibited a similar distribution pattern of GFP::CHIN-1 as the wild type, except that the phase II cortical localization in these embryos was not restricted to the posterior (Figure 5B and Supplemental Movies 14 and 15; par-3 and pkc-3 depletions not shown). par-1(fRNAi) embryos showed the same localizations as the wild type with the exception that the posterior, cortical localization during phase II was restricted to a smaller domain (3/3 embryos; Figure 5B and Supplemental Movie 16). Interestingly, par-2(fRNAi) embryos exhibited a different phase II localization than did par-1(fRNAi) embryos. In par-2(fRNAi) embryos, GFP::CHIN-1 localized only to the extreme posterior cortex throughout phase II (7/7 embryos; Figure 5B and Supplemental Movie 17). This observation suggests that PAR-2 is required to recruit CHIN-1 to the cortex via a mechanism that is independent of PAR-1. Unexpectedly, GFP::CHIN-1 signal is dramatically reduced in the germlines of par-5(fRNAi) animals and their embryos (3/3 animals; Figure 5B). Together, these observations suggest that PAR-2 stabilizes CHIN-1 at the posterior cortex during phase II.
Figure 5.
Localization of GFP::CHIN-1 depends on CDC-42, PAR-6 and PAR-2 but not PAR-1. Embryos expressing GFP::CHIN-1 in the early embryo. (A) Optical sections through the middle of the cell. GFP::CHIN-1 localizes to the cytoplasm, centrosomes (black arrowheads), central spindle, cortical puncta (white arrowheads) and some structures suggestive of P-granules (white arrows). The cortical puncta are only observed in the early embryo at the posterior during phase II and at the posterior during the late two-cell stage. (B) Optical sections through the cortex of the cell. Cortical localization depends upon some polarity proteins. Embryos depleted of CDC-42 or PAR-6 exhibit puncta all about the cortex in phase II. Embryos depleted of PAR-2 exhibit cortical puncta only at the extreme posterior, whereas those depleted of PAR-1 exhibit only a slight reduction of the posterior domain that contains puncta. No depletion altered the absence of puncta in phase I. Embryos depleted of PAR-5 exhibited a dramatic reduction in GFP::CHIN-1 signal and an absence of cortical puncta. (C) Optical sections through the middle of the cell. Anti-CHIN-1 antibody localizes as posterior cortical puncta during phase II. This localization is lost in chin-1(RNAi) embryos. Bar, 10 μm.
Because the GFP::CHIN-1 transgene is driven by pie-1 regulatory elements, it is possible that it reports a spurious localization. However, the endogenous protein is probably expressed in the germline (and most other cell types) as a chin-1 promoter drives expression of GFP in the germline and most cells of adults and larvae (Hunt-Newbury et al., 2007). The localization of endogenous CHIN-1 by immunolocalization recapitulates the cytoplasmic and cortical localization of the transgene with no cortical localization during phase I (0/4 embryos), posterior cortical localization in phase II (6/7 embryos) and posterior cortical localization of P1 cells in late prophase and anaphase (5/5 embryos; Figure 5C). chin-1(fRNAi) embryos exhibited no unequivocal cortical puncta in phase II (0/13 embryos; Figure 5C) or the late two-cell stage (0/8 embryos).
CGEF-1 depletions affected the cortical enrichment but not the spatial patterning of the CDC-42 biosensor. Nevertheless, the intracellular localization of CGEF-1, the a and c isoforms of which are probably expressed in the gonad (Ziel et al., 2009), might clarify mechanisms of CDC-42 activation. We found that GFP::CGEF-1A expressed under germline regulatory control localized to the cytoplasm and was enriched in the nucleoplasm and cell cortex (Supplemental Figure 3; best seen in Supplemental Movie 2). Unfortunately, the two independent transformant strains we obtained did not express at sufficient levels for unequivocal localizations. CGEF-1 immunolocalization with an antibody raised against the C-terminal portion encoded by all cgef-1 isoforms was unsuccessful (data not shown).
CDC-42 and RHO-1 Signaling Differentially Direct Cortical Recruitment of Nonmuscle Myosin II
Wild-type embryos, before the onset of cortical polarity, localize the nonmuscle myosin II NMY-2 and associated proteins to cortical foci, where they ruffle the cell surface. This process requires RHO-1, its activating GEF ECT-2 and the myosin subunit MLC-4 (Cowan and Hyman, 2007). On transmission of the polarizing signal from the sperm centrosome, we observe that NMY-2 foci were lost locally, and the remaining asymmetric NMY-2 apparently retracted the cortex toward the anterior, producing a pseudocleavage furrow at about half of the embryo's length (Figure 6, Supplemental Figure 5, and Supplemental Movie 18 [39/39 embryos; Munro et al., 2004]). This pseudocleavage furrow and anterior ruffling resolved as the NMY-2 foci dissipated, concomitant with entry into phase II. During phase II, NMY-2 localized as smaller puncta that were asymmetrically localized at the anterior cortex (also reported by Schonegg and Hyman, 2006). These puncta exhibited some cortical flow toward the anterior pole and produced a local enrichment at the posterior edge of this domain in a manner reminiscent of the local enrichment at the pseudocleavage furrow in phase I. This anterior cortical NMY-2 localization was lost before the recruitment of NMY-2 into phase I-like foci at the equatorial cortex during phase III. These foci condensed into the contractile ring that cleaved the cell.
Figure 6.
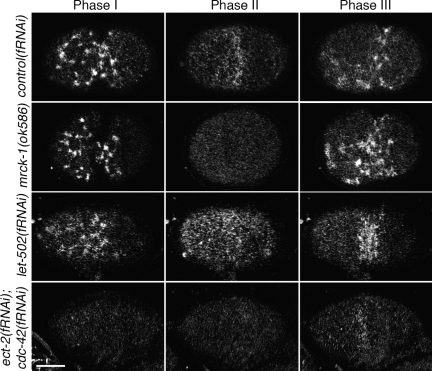
RHO-1 and CDC-42 pathway components are required to recruit GFP::NMY-2 to the cortex in distinct phases. Optical sections through the cell cortex of embryos expressing GFP::NMY-2 in the early embryo. In the wild type, GFP::NMY-2 localizes to the cortex in distinct morphological distributions in phases I and II. The robust myosin foci observed in phase I depend upon ECT-2, RHO-1, and LET-502 [represented here by let-502(fRNAi)]. The robust, diffuse, and polar distribution of smaller myosin puncta in phase II depends upon CGEF-1, CDC-42, and MRCK-1 [represented here by mrck-1(ok586)]. Disruption of either pathway does not abolish the recruitment by the other, and disruption of both pathways disrupts both recruitment patterns. Shown here are embryos from wild-type, mrck-1(ok586), let-502(fRNAi), and ect-2(fRNAi); cdc-42(fRNAi) mothers. Embryos from each of the six single disruptions are presented in Supplemental Figure 3. Bar, 10 μm.
Disrupting CDC-42 signaling did little to the phase I localization of NMY-2 but did affect its phase II localization. Although most published defects in cdc-42(RNAi) embryos occur during phase II, the posterior enrichment we observed in our CDC-42 activity biosensor suggested that CDC-42 may play a role during the phase I cortical flows. To determine the biological significance of the posterior phase I localization of the active CDC-42 probe, we used RNAi to deplete candidate CDC-42 effectors. Phase I cortical flow rates were measured as described previously (Munro et al., 2004) in the background of our gfp::nmy-2 transgene with embryonic expression driven by a pie-1 promoter. In contrast to a published finding (Motegi and Sugimoto, 2006), cdc-42(fRNAi) embryos did not exhibit a significantly reduced flow rate. Few of the CDC-42 upstream regulator or effector depletion conditions tested (Supplemental Figure 4) detectably altered the average rates of individual foci, though par-1(fRNAi) and wve-1(fRNAi) treatments increased flow rates. However, cgef-1(fRNAi), cdc-42(fRNAi) and tag-59(fRNAi) embryos exhibited a striking, reduced GFP::NMY-2 localization to the anterior cortex during phase II. This effect of cdc-42(RNAi) has been reported previously (Schonegg and Hyman, 2006). tag-59 has homology to vertebrate myotonic dystrophy-related Cdc42 binding kinase, so we refer to tag-59 as mrck-1 (for myotonic dystrophy-related Cdc42 binding kinase homologue). The phase II cortical recruitment of GFP::NMY-2 was nearly abolished in cgef-1(gk261) (8/8 embryos), cdc-42(fRNAi) (14/14 embryos) and mrck-1(fRNAi or ok586) embryos (2/19 RNAi and 7/7 ok586 embryos) (Figure 6, Supplemental Figure 5, and Supplemental Movies 19–21). These observations suggest a pathway by which CGEF-1 activity produces a recruitment of NMY-2 to the cortex via the actions of CDC-42 and the kinase MRCK-1 during phase II. Further supporting this hypothesized pathway, depletion of CHIN-1 produces an increase of NMY-2 recruitment. In chin-1(fRNAi) embryos, the phase I myosin localization seems unaltered; however, the diffuse myosin recruited to cortex in phase II is spatially expanded toward the posterior (Supplemental Figure 5).
In contrast to the effects of disrupting the CDC-42 pathway, disrupting a putative RHO-1 pathway produces defects primarily in establishment phase/phase I (Jenkins et al., 2006; Motegi and Sugimoto, 2006; Schonegg and Hyman, 2006; Schmutz et al., 2007; Schonegg et al., 2007). We reproduced the experimental result that depletions of the putative RHO-1 GEF ECT-2 (3/3 embryos) or RHO-1 (2/2 embryos) itself reduce cortical recruitment of NMY-2 to the cortex in phase I (Supplemental Figure 5 and Supplemental Movies 22–23). To expand upon these findings, we depleted candidate RHO-1 effectors and identified LET-502, a previously described Rho-binding kinase (Wissmann et al., 1997), as a likely effector linking RHO-1 signaling to NMY-2 recruitment. let-502(fRNAi) embryos were found to have reduced cortical GFP::NMY-2 recruitment in phase I (Figure 6, Supplemental Figure 5, and Supplemental Movie 24) (7/8 embryos), reduced average speeds of foci movement during cortical flow (Supplemental Figure 4), and dramatically reduced cortical retraction. Together with previously published results, this observation suggests a pathway by which ECT-2 activity produces a recruitment of NMY-2 to the cortex via the actions of RHO-1 and LET-502 during phase I. Although cyk-1(fRNAi) embryos exhibited marked increased maximum cortical flow puncta speeds (Supplemental Figure 4), this treatment did not disrupt the overall developmental pattern of cortical flows.
Embryos that failed to localize NMY-2 to the anterior in phase I because of a Rho pathway defect, nonetheless localized NMY-2 to the anterior in phase II. Embryos depleted of ECT-2, RHO-1, or LET-502 exhibited reduced cortical flows during phase I, but the production of a myosin-enriched domain at the anterior by the end of phase II occurred (Figure 6, Supplemental Figure 5, and Supplemental Movies 22–24) nonetheless. Given that the morphology of the phase II, cortical NMY-2 signal in Rho pathway-depleted embryos resembles that of the wild-type phase II morphology, we assayed cortical myosin recruitment in embryos depleted of both a Rho and Cdc42 pathway member. Simultaneous disruption of both the Rho and Cdc42 pathways produced a loss of both phase I and phase II myosin recruitment (Figure 6, Supplemental Figure 5, and Supplemental Movie 25). This suggests that the Rho pathway is active during phase I and that the Cdc42 pathway is active in phase II.
DISCUSSION
Utility of Localizing Protein Signaling Activity
The localization of CDC-42 activity that we observed does not correlate with the localization of either CDC-42 or its fixed-activity mutant forms. Previous studies demonstrated that CDC-42 is not required for the cortical recruitment of the anterior PAR complex during phase I in the C. elegans early embryo but is required for its maintenance during phase II (Gotta et al., 2001; Motegi and Sugimoto, 2006). That a GFP-tagged mutant PAR-6 defective for CDC-42-binding in cdc-42(+) exhibits the same localization patterns in phase I as wild-type PAR-6 in cdc-42(RNAi) embryos suggests that PAR-6 can be localized specifically to the anterior cortex without directly interacting with CDC-42. However, this mutant PAR-6 is lost from the cortex in phase II, just about when our probe reports increased CDC-42 activity at that location (Aceto et al., 2006). The active CDC-42 probe reports anterior cortical activity specifically during the period that PAR-6 requires interaction with active CDC-42 for cortical localization, suggesting that both PAR-6 and the probe can access the pool of active CDC-42 during phase II. The dynamics of recruitment of this probe indicates that anterior activation of CDC-42 occurs only in phase II. However, it is possible that activation occurs also in phase I, but that active CDC-42 may be masked from the probe during this time by competitively binding PAR-6. Although we cannot eliminate the second possibility, it seems unlikely that such a competition would occur in phase I but not phase II, when PAR-6 requires CDC-42 to remain enriched at the cortex. Under either scenario, our observation of the localization of endogenous CDC-42 activity suggests several interesting possibilities: between phases I and II there is an increase in the anterior cortical activity of CDC-42; there is a substantive, but as yet unnoticed reduction in anterior PAR protein concentration; or the anterior PAR complex's binding to active CDC-42 becomes less tight.
The Phase II Balance of the PAR Proteins Regulates, and Is Regulated by, CDC-42 Activity
Previous work has shown that cortical flows establishing polarity in the C. elegans one-cell embryo do not determine the position of first cleavage. Embryos depleted of RHO-1–oriented GAPs exhibit exaggerated cortical flows during phase I that produce small anterior domains (Schmutz et al., 2007; Schonegg et al., 2007). These exaggerated flows, however, alter neither the ultimate relative size of the polarized domains nor the site of first cleavage. By contrast, we found that depletion of the putative CDC-42 GAP CHIN-1 produced an enlarged anterior cortical domain.
We propose that the anterior PARs engage in a negative regulatory feedback loop with PAR-2 and the putative CDC-42 GAP CHIN-1. In this model, the anterior PAR proteins and CDC-42 are enriched at the anterior by phase I cortical flows as proposed previously (Munro et al., 2004). During maintenance phase (Figure 7), some timing cue recruits CHIN-1 to the cortex, which is destabilized at the cortex by the anterior PAR complex. Anterior PAR complexes that migrate into the posterior are destabilized by CHIN-1–mediated inactivation of CDC-42. This model is insufficient to fully describe the mutually antagonistic relationship between the two sets of PAR proteins. PAR-1 depletions result in an expanded anterior domain during phase II (Cuenca et al., 2003; Hao et al., 2006) but do not prevent the cortical recruitment of CHIN-1. Therefore, the regulation of CDC-42 activity must be in a pathway redundant to the mutual antagonism mediated by the phosphorylation of PAR-3 by PAR-1 and of PAR-1 by PKC-3 (Hurov et al., 2004; Suzuki et al., 2004; Hao et al., 2006) that causes cortical loss of the phosphorylated proteins. Interestingly, both of these proposed pathways incorporate PAR-5, which is necessary to stabilize CHIN-1 and to mediate cortical exclusion of certain phosphorylated PAR proteins. The redundant regulatory path provided by this model explains an observation that is otherwise difficult to explain, namely, that ectopic expression of CDC-42(Q61L) expands the PAR-6-labeled anterior domain (Aceto et al., 2006). This mutant form of CDC-42 would not be subject to inactivation by the posterior GAP activity and could therefore bias the regulatory balance of the contested border of the two PAR domains in favor of the anterior complex.
Figure 7.
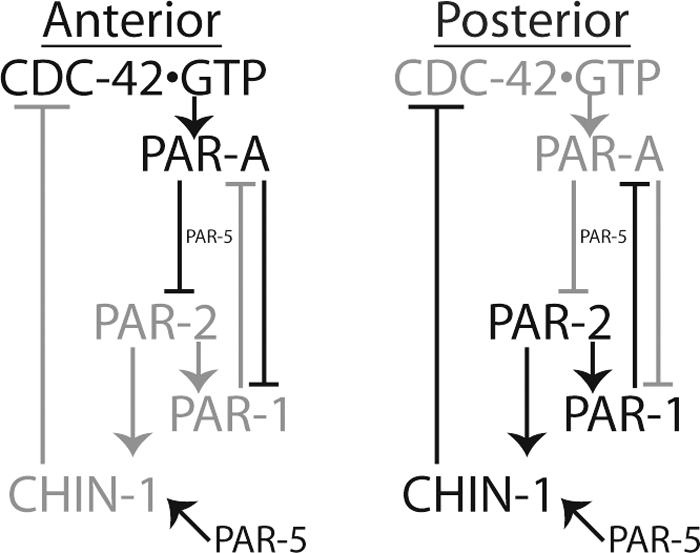
Model of CDC-42activity regulation in phase II polarity maintenance. Here, regulatory associations are positive for arrows and negative for crossed lines. Gray associations are not operative, whereas dark associations are operative. For a fuller description of the model, see Discussion.
Spatial restriction of CDC-42 activity also plays a key role in the polarization of cells in other contexts. The establishment of “inner-outer” PAR asymmetry of embryonic blastomeres later in development also requires the restriction of CDC-42 activity by the action of the GAP PAC-1 (Anderson et al., 2008). In budding yeast, the GEF Cdc24 is strictly required to recruit Cdc42 to the cortex and for initiation of budding (Park and Bi, 2007), whereas the GAP Rga1 is required to restrict this bud selection site from the previous bud scar (Tong et al., 2007). In the Drosophila neuroblast, Cdc42 activity is required to prevent the expansion of the basal determinants, whereas the constitutively active Cdc42 mutant expands the apical domain to the entire cortex (Atwood et al., 2007). Given that no identified basal component is required to restrict apical component in the way that PAR-1 and PAR-2 exclude the anterior PARs in C. elegans, this raises the prospect that an unidentified Cdc42 GAP may serve this function in the Drosophila neuroblast.
The developmental timing of CHIN-1 recruitment to cortical puncta suggests that its mobilization is under significant regulatory control at the transition from phase I to phase II. In this study, we did not identify upstream regulators of this transition. As CHIN-1 is predicted to encode a C2 domain, we tested whether depleting phospholipase C-encoding gene products might affect the localization of CHIN-1. Individual RNAi-mediated depletions of plc-1, plc-2, plc-3, plc-4, and egl-8 did not alter the localization of GFP::CHIN-1 in the embryos examined (n = 2 each condition; data not shown). We speculate that identifying cell regulators of CHIN-1 cortical recruitment may mechanistically link the phase I-to-phase II timing cue and the spatial regulation of PAR protein polarity.
RHO-1 and CDC-42-mediated Pathways Independently Direct Anteriorward Cortical Flows
In the early embryo, the polarization initiated by the sperm centrosome is spatially elaborated by a myosin II-powered cortical flow regulated by RHO-1 signaling (Cowan and Hyman, 2007). Although this flow is essential for establishing the PAR protein polarity, it does not determine the size of the PAR-labeled domains at cleavage (Cuenca et al., 2003; Schmutz et al., 2007; Schonegg et al., 2007). The refinement of this boundary between PAR domains occurs at the same time that actin and myosin exhibit anteriorly restricted cortical localizations (Schonegg and Hyman, 2006; Velarde et al., 2007). That this second, polarized myosin distribution requires CDC-42 (Schonegg and Hyman, 2006) suggests that these might represent accumulations of myosin in response to distinct G protein-mediated signals. Indeed, we observed that these two phases of asymmetric myosin enrichment occur independently as consequences of signals mediated by ECT-2, RHO-1, and LET-502 in phase I and CGEF-1, CDC-42, and MRCK-1 in phase II (Figure 6). This second, anteriorly localized accumulation of the actomyosin cortex may provide a mechanism, perhaps redundantly with unequal strength of mutual PAR antagonism, to refine the boundary of polarized domains during phase II that ultimately determine the cleavage plane and blastomere size. How the PAR-dependent spatial information is propagated to spindle positioning proteins such as LET-99, GPR-1/2 and LIN-5 is not understood at a mechanistic level. Our observation that regulation of CDC-42 signaling can alter the size of the myosin-enriched anterior domain provides another potential mechanism by which this positioning information can be transduced. It should be interesting to determine to what extent this polarized recruitment of NMY-2 affects the observed refinement of the boundary defined by segregation of the PAR proteins. The identification of MRCK-1 as a mediator of CDC-42–dependent myosin recruitment might allow a dissection of a specific role for NMY-2 in the refinements of PAR polarity observed in phase II. The speculation that CDC-42 might play a conserved role in regulating cortical determinant segregation through actinomyosin regulation is consistent with the observation that Cdc42p organizes the assembly of actin cables by which polarized transport of a spindle pole and cell fate determinants occur in Saccharomyces cerevisiae (reviewed in Pruyne and Bretscher, 2000). Also, in the Drosophila neuroblast model system, Cdc42 localizes to an apical cap where the enrichment of myosin II is required to restrict basal determinants to the basal cortex (Barros et al., 2003; Atwood et al., 2007). Further work will define the role of the CDC-42-dependent, phase II actin and myosin enrichments in the refinement of polarity during this phase of polarization in the early C. elegans embryo.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kenneth Kemphues, Ellen Batchelder, and Julie Ahringer for helpful comments and suggestions on this work. We are grateful to Drs. Yuji Kohara, Anjon Audhya, Donato Aceto, Kenneth Kemphues, and Julie Ahringer as well as the Caenorhabditis Genetics Center and National BioResource Project of Japan for reagents and constructs. We thank Long Yan, Muhammad Nazir, and Steve Trier for advice on FLIM and Jens Eickhoff for assistance with logistic regression analysis. This work was supported by National Institutes of Health grants GM-052454 (to J.G.W.) and NIBIB EB000184 (to K.W.E. and J.G.W.). K.T.K. was supported by the Biotechnology Training Program (NIH 5 T32 GM-08349) at the University of Wisconsin–Madison.
Abbreviations used:
- GAP
GTPase activating protein
- GBD
G protein binding domain
- GEF
guanine nucleotide exchange factor
- FLIM
fluorescence lifetime imaging microscopy
- FP
fluorescent protein
- FRET
Förster resonance energy transfer
- MPLSM
multiphoton laser-scanning microscopy/microscope
- ORF
open reading frame
- RNAi
RNA-mediated interference
- TCSPC
time-correlated single photon counting
- UTR
untranslated region
- WT
wild type.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-01-0060) on November 18, 2009.
REFERENCES
- Aceto D., Beers M., Kemphues K. J. Interaction of PAR-6 with CDC-42 is required for maintenance but not establishment of PAR asymmetry in C. elegans. Dev. Biol. 2006;299:386–397. doi: 10.1016/j.ydbio.2006.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. C., Gill J. S., Cinalli R. M., Nance J. Polarization of the C. elegans embryo by RhoGAP-mediated exclusion of PAR-6 from cell contacts. Science. 2008;320:1771–1774. doi: 10.1126/science.1156063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood S. X., Chabu C., Penkert R. R., Doe C. Q., Prehoda K. E. Cdc42 acts downstream of Bazooka to regulate neuroblast polarity through Par-6 aPKC. J. Cell Sci. 2007;120:3200–3206. doi: 10.1242/jcs.014902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros C. S., Phelps C. B., Brand A. H. Drosophila nonmuscle myosin II promotes the asymmetric segregation of cell fate determinants by cortical exclusion rather than active transport. Dev. Cell. 2003;5:829–840. doi: 10.1016/s1534-5807(03)00359-9. [DOI] [PubMed] [Google Scholar]
- Beers M., Kemphues K. Depletion of the co-chaperone CDC-37 reveals two modes of PAR-6 cortical association in C. elegans embryos. Development. 2006;133:3745–3754. doi: 10.1242/dev.02544. [DOI] [PubMed] [Google Scholar]
- Benink H. A., Bement W. M. Concentric zones of active RhoA and Cdc42 around single cell wounds. J. Cell Biol. 2005;168:429–439. doi: 10.1083/jcb.200411109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan C. R., Hyman A. A. Centrosomes direct cell polarity independently of microtubule assembly in C. elegans embryos. Nature. 2004;431:92–96. doi: 10.1038/nature02825. [DOI] [PubMed] [Google Scholar]
- Cowan C. R., Hyman A. A. Acto-myosin reorganization and PAR polarity in. C. elegans. Development. 2007;134:1035–1043. doi: 10.1242/dev.000513. [DOI] [PubMed] [Google Scholar]
- Cuenca A. A., Schetter A., Aceto D., Kemphues K., Seydoux G. Polarization of the C. elegans zygote proceeds via distinct establishment and maintenance phases. Development. 2003;130:1255–1265. doi: 10.1242/dev.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming I. N., Elliott C. M., Exton J. H. Differential translocation of rho family GTPases by lysophosphatidic acid, endothelin-1, and platelet-derived growth factor. J. Biol. Chem. 1996;271:33067–33073. doi: 10.1074/jbc.271.51.33067. [DOI] [PubMed] [Google Scholar]
- Goldstein B., Hird S. N. Specification of the anteroposterior axis in Caenorhabditis elegans. Development. 1996;122:1467–1474. doi: 10.1242/dev.122.5.1467. [DOI] [PubMed] [Google Scholar]
- Goldstein B., Macara I. G. The PAR proteins: fundamental players in animal cell polarization. Dev. Cell. 2007;13:609–622. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy P. Mechanisms of asymmetric cell division: flies and worms pave the way. Nat. Rev. Mol. Cell Biol. 2008;9:355–366. doi: 10.1038/nrm2388. [DOI] [PubMed] [Google Scholar]
- Gotta M., Abraham M. C., Ahringer J. CDC-42 controls early cell polarity and spindle orientation in C. elegans. Curr. Biol. 2001;11:482–488. doi: 10.1016/s0960-9822(01)00142-7. [DOI] [PubMed] [Google Scholar]
- Guo S., Kemphues K. J. A non-muscle myosin required for embryonic polarity in Caenorhabditis elegans. Nature. 1996;382:455–458. doi: 10.1038/382455a0. [DOI] [PubMed] [Google Scholar]
- Hamill D. R., Severson A. F., Carter J. C., Bowerman B. Centrosome maturation and mitotic spindle assembly in C. elegans require SPD-5, a protein with multiple coiled-coil domains. Dev. Cell. 2002;3:673–684. doi: 10.1016/s1534-5807(02)00327-1. [DOI] [PubMed] [Google Scholar]
- Hao Y., Boyd L., Seydoux G. Stabilization of cell polarity by the C. elegans RING protein PAR-2. Dev. Cell. 2006;10:199–208. doi: 10.1016/j.devcel.2005.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill D. P., Strome S. An analysis of the role of microfilaments in the establishment and maintenance of asymmetry in Caenorhabditis elegans zygotes. Dev. Biol. 1988;125:75–84. doi: 10.1016/0012-1606(88)90060-7. [DOI] [PubMed] [Google Scholar]
- Hoffman G. R., Nassar N., Cerione R. A. Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell. 2000;100:345–356. doi: 10.1016/s0092-8674(00)80670-4. [DOI] [PubMed] [Google Scholar]
- Hori Y., Kikuchi A., Isomura M., Katayama M., Miura Y., Fujioka H., Kaibuchi K., Takai Y. Post-translational modifications of the C-terminal region of the rho protein are important for its interaction with membranes and the stimulatory and inhibitory GDP/GTP exchange proteins. Oncogene. 1991;6:515–522. [PubMed] [Google Scholar]
- Hunt-Newbury R., et al. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 2007;5:e237. doi: 10.1371/journal.pbio.0050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurov J. B., Watkins J. L., Piwnica-Worms H. Atypical PKC phosphorylates PAR-1 kinases to regulate localization and activity. Curr. Biol. 2004;14:736–741. doi: 10.1016/j.cub.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Jenkins N., Saam J. R., Mango S. E. CYK-4/GAP provides a localized cue to initiate anteroposterior polarity upon fertilization. Science. 2006;313:1298–1301. doi: 10.1126/science.1130291. [DOI] [PubMed] [Google Scholar]
- Kamath R. S., et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kay A. J., Hunter C. P. CDC-42 regulates PAR protein localization and function to control cellular and embryonic polarity in C. elegans. Curr. Biol. 2001;11:474–481. doi: 10.1016/s0960-9822(01)00141-5. [DOI] [PubMed] [Google Scholar]
- Kennedy S., Wang D., Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- Maddox A. S., Habermann B., Desai A., Oegema K. Distinct roles for two C. elegans anillins in the gonad and early embryo. Development. 2005;132:2837–2848. doi: 10.1242/dev.01828. [DOI] [PubMed] [Google Scholar]
- Motegi F., Sugimoto A. Sequential functioning of the ECT-2 RhoGEF, RHO-1 and CDC-42 establishes cell polarity in Caenorhabditis elegans embryos. Nat. Cell Biol. 2006;8:978–985. doi: 10.1038/ncb1459. [DOI] [PubMed] [Google Scholar]
- Munro E., Nance J., Priess J. R. Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev. Cell. 2004;7:413–424. doi: 10.1016/j.devcel.2004.08.001. [DOI] [PubMed] [Google Scholar]
- O'Connell K. F., Maxwell K. N., White J. G. The spd-2 gene is required for polarization of the anteroposterior axis and formation of the sperm asters in the Caenorhabditis elegans zygote. Dev. Biol. 2000;222:55–70. doi: 10.1006/dbio.2000.9714. [DOI] [PubMed] [Google Scholar]
- Park H. O., Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol. Mol. Biol. Rev. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V., Casey E., Collar D., Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D., Bretscher A. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J. Cell Sci. 2000;113:365–375. doi: 10.1242/jcs.113.3.365. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. Vienna, Austria: R Development Core Team; 2008. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Rose L. S., Lamb M. L., Hird S. N., Kemphues K. J. Pseudocleavage is dispensable for polarity and development in C. elegans embryos. Dev. Biol. 1995;168:479–489. doi: 10.1006/dbio.1995.1096. [DOI] [PubMed] [Google Scholar]
- Sadler P. L., Shakes D. C. Anucleate Caenorhabditis elegans sperm can crawl, fertilize oocytes and direct anterior-posterior polarization of the 1-cell embryo. Development. 2000;127:355–366. doi: 10.1242/dev.127.2.355. [DOI] [PubMed] [Google Scholar]
- Schmutz C., Stevens J., Spang A. Functions of the novel RhoGAP proteins RGA-3 and RGA-4 in the germ line and in the early embryo of C. elegans. Development. 2007;134:3495–3505. doi: 10.1242/dev.000802. [DOI] [PubMed] [Google Scholar]
- Schonegg S., Constantinescu A. T., Hoege C., Hyman A. A. The Rho GTPase-activating proteins RGA-3 and RGA-4 are required to set the initial size of PAR domains in Caenorhabditis elegans one-cell embryos. Proc. Natl. Acad. Sci. USA. 2007;104:14976–14981. doi: 10.1073/pnas.0706941104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonegg S., Hyman A. A. CDC-42 and RHO-1 coordinate acto-myosin contractility and PAR protein localization during polarity establishment in C. elegans embryos. Development. 2006;133:3507–3516. doi: 10.1242/dev.02527. [DOI] [PubMed] [Google Scholar]
- Schumacher J. M., Ashcroft N., Donovan P. J., Golden A. A highly conserved centrosomal kinase, AIR-1, is required for accurate cell cycle progression and segregation of developmental factors in Caenorhabditis elegans embryos. Development. 1998;125:4391–4402. doi: 10.1242/dev.125.22.4391. [DOI] [PubMed] [Google Scholar]
- Shelton C. A., Carter J. C., Ellis G. C., Bowerman B. The nonmuscle myosin regulatory light chain gene mlc-4 is required for cytokinesis, anterior-posterior polarity, and body morphology during Caenorhabditis elegans embryogenesis. J. Cell Biol. 1999;146:439–451. doi: 10.1083/jcb.146.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skala M. C., Riching K. M., Bird D. K., Gendron-Fitzpatrick A., Eickhoff J., Eliceiri K. W., Keely P. J., Ramanujam N. In vivo multiphoton fluorescence lifetime imaging of protein-bound and free nicotinamide adenine dinucleotide in normal and precancerous epithelia. J. Biomed. Opt. 2007a;12 doi: 10.1117/1.2717503. 024014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skala M. C., Riching K. M., Gendron-Fitzpatrick A., Eickhoff J., Eliceiri K. W., White J. G., Ramanujam N. In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc. Natl. Acad. Sci. USA. 2007b;104:19494–19499. doi: 10.1073/pnas.0708425104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M. H., Aravind L., Muller-Reichert T., O'Connell K. F. The conserved protein SZY-20 opposes the Plk4-related kinase ZYG-1 to limit centrosome size. Dev. Cell. 2008;15:901–912. doi: 10.1016/j.devcel.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A., et al. aPKC acts upstream of PAR-1b in both the establishment and maintenance of mammalian epithelial polarity. Curr. Biol. 2004;14:1425–1435. doi: 10.1016/j.cub.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Timmons L., Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Tong Z., Gao X. D., Howell A. S., Bose I., Lew D. J., Bi E. Adjacent positioning of cellular structures enabled by a Cdc42 GTPase-activating protein-mediated zone of inhibition. J. Cell Biol. 2007;179:1375–1384. doi: 10.1083/jcb.200705160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M. C., Ahringer J. Microtubules are involved in anterior-posterior axis formation in C. elegans embryos. J. Cell Biol. 2007;179:397–402. doi: 10.1083/jcb.200708101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velarde N., Gunsalus K. C., Piano F. Diverse roles of actin in C. elegans early embryogenesis. BMC Dev. Biol. 2007;7:142. doi: 10.1186/1471-213X-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissmann A., Ingles J., McGhee J. D., Mains P. E. Caenorhabditis elegans LET-502 is related to Rho-binding kinases and human myotonic dystrophy kinase and interacts genetically with a homolog of the regulatory subunit of smooth muscle myosin phosphatase to affect cell shape. Genes Dev. 1997;11:409–422. doi: 10.1101/gad.11.4.409. [DOI] [PubMed] [Google Scholar]
- Wokosin D. L., Squirrell J. M., Eliceiri K. W., White J. G. Optical workstation with concurrent, independent multiphoton imaging and experimental laser microbeam capabilities. Rev. Sci. Instrum. 2003;74:193–201. doi: 10.1063/1.1524716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Wang Y., Barry D. C., Chanock S. J., Bokoch G. M. Guanine nucleotide binding properties of Rac2 mutant proteins and analysis of the responsiveness to guanine nucleotide dissociation stimulator. Biochemistry. 1997;36:626–632. doi: 10.1021/bi962059h. [DOI] [PubMed] [Google Scholar]
- Ziel J. W., Matus D. Q., Sherwood D. R. An expression screen for RhoGEF genes involved in C. elegans gonadogenesis. Gene Expr. Patterns. 2009;9:397–403. doi: 10.1016/j.gep.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman M., O'Brien J. M., Ouellette L. A., Church W. R., Johnson D. I. Mutational analysis of CDC42Sc, a Saccharomyces cerevisiae gene that encodes a putative GTP-binding protein involved in the control of cell polarity. Mol. Cell. Biol. 1991;11:3537–3544. doi: 10.1128/mcb.11.7.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.