Abstract
It has been proposed that mitochondrial dysfunction is involved in the pathogenesis of type 2 diabetes (T2D). To dissect the underlying mechanisms, we performed a multiplexed proteomics study on liver mitochondria isolated from a spontaneous diabetic rat model before/after they were rendered diabetic. Altogether, we identified 1091 mitochondrial proteins, 228 phosphoproteins, and 355 hydroxyproteins. Mitochondrial proteins were found to undergo expression changes in a highly correlated fashion during T2D development. For example, proteins involved in β-oxidation, the tricarboxylic acid cycle, oxidative phosphorylation, and other bioenergetic processes were coordinately up-regulated, indicating that liver cells confronted T2D by increasing energy expenditure and activating pathways that rid themselves of the constitutively increased flux of glucose and lipid. Notably, activation of oxidative phosphorylation was immediately related to the overproduction of reactive oxygen species, which caused oxidative stress within the cells. Increased oxidative stress was also evidenced by our post-translational modification profiles such that mitochondrial proteins were more heavily hydroxylated during T2D development. Moreover, we observed a distinct depression of antiapoptosis and antioxidative stress proteins that might reflect a higher apoptotic index under the diabetic stage. We suggest that such changes in systematic metabolism were causally linked to the development of T2D. Comparing proteomics data against microarray data, we demonstrated that many T2D-related alterations were unidentifiable by either proteomics or genomics approaches alone, underscoring the importance of integrating different approaches. Our compendium could help to unveil pathogenic events in mitochondria leading to T2D and be useful for the discovery of diagnosis biomarker and therapeutic targets of T2D.
Type 2 diabetes (T2D)1 has become a global health epidemic. It is recognized as a strong risk factor for cardiovascular diseases and for associated complications that result in substantial morbidity and mortality (1). Now it is recognized that T2D is a heterogeneous disease in which almost every aspect of the body’s metabolism goes awry. The onset of T2D results from complex interactions between genetic and environmental factors. Several cellular dysfunctions and molecular defects are proposed to be associated with T2D, such as β-cell malfunction, impaired insulin secretion and function, chronic hyperglycemia, and other disturbances in systematic metabolism (1).
Besides these findings, recently the topic of mitochondrial dysfunction has also gained a lot of attention in the T2D field (2–4). Numerous studies have revealed that various mitochondrial factors are of paramount importance in the pathogenesis of diabetes. For example, high fat diet-induced alterations in the mitochondrial compartment are associated with the development of insulin resistance and ectopic fat storage in liver (5). The dysfunction of mitochondria damages the pancreatic β-cells and contributes to the development of a hyperglycemic status (6). Mutations in the mtDNA, decreased mitochondrial size, and reduced oxidative phosphorylation are observed in T2D patients (7, 8). Both mitochondrial density and copy number are dramatically decreased in insulin-resistant individuals (2), and in vivo measurements have demonstrated multiple defects in mitochondrial functions, e.g. ATP production (9). Mitochondrial biogenesis is suppressed in the diabetic animal model (10). There is also evidence of a more global effect of mitochondrial dysfunction on glucose transport (11). These clues all suggest that various mitochondrial factors are causally linked to the pathogenesis of T2D. At the same time, these results obtained so far teach us that the relation between mitochondrial dysfunction and T2D development is very complicated.
During the transition to T2D, mitochondria undergo extensive changes in gene expression and arrive at a new highly regulated, although pathological, steady state. A good approach to understand the pathophysiology of T2D is to examine changes in mitochondrial gene expression that occur during this period. By identifying groups of coordinately changed proteins, we can gain insight into potential pathways and regulatory networks that might contribute to the compensation and development of T2D. Proteomics profiling of mitochondria provides a powerful tool to search for such clues. However, to date, no study has ever comprehensively investigated T2D-derived changes in protein expression of this organelle. To this end, we performed a multiplex proteomics study on liver mitochondria isolated from a spontaneous diabetic model, the Goto-Kakizaki (GK) rat, before/after they were rendered diabetic (12). Adult GK rats exhibit a spontaneous non-insulin-dependent diabetes characterized by impaired glucose-induced insulin secretion, decreased β-cell mass, hepatic glucose overproduction, and decreased insulin sensitivity in the liver in parallel to moderate insulin resistance in extrahepatic tissue such as muscle and adipose (13). Prior studies suggested that impaired insulin secretion and excessive hepatic glucose production were both early events in the GK rat (13). Based on our study, GK rats did not exhibit basal hyperglycemia by 5 weeks of age (8.312 ± 0.482 mm, largely within normal scale) and therefore could be considered prediabetic. However, by 8 weeks of age basal hyperglycemia emerged (13.371 ± 2.494 mm) together with impaired tolerance to intravenous glucose. Hence, we suggest that the GK rat was early diabetic by 8 weeks of age. Molecular changes in this transition were more causally linked to the pathogenesis of T2D and were more informative to the early diagnosis of the disease.
Liver mitochondrial protein digests were fractionated and enriched through strong cation exchange chromatography (SCX) and strong anion exchange chromatography (SAX) followed by analysis using a linear ion trap LTQ-Orbitrap mass spectrometer. After stringent data filtration, the mitochondrial proteomes of prediabetic (5-week-old) and early diabetic (8-week-old) GK rats were identified together with two main post-translational modifications (PTMs), phosphorylation and hydroxylation. We also used a label-free quantitative approach (localized statistics of protein abundance distribution (LSPAD)) (14) to assess the T2D-derived changes in mitochondrial protein expression. We identified changes in systemic metabolism and many aberrations that might be causally related to the development of T2D. Furthermore, we complemented our proteomics data with microarray data to provide more comprehensive information unveiling the underlying biological mechanism coupled with the pathogenesis of T2D. Notably, many genes showed changes in the proteome but not transcriptome or vice versa, suggesting post-transcriptional regulation and underscoring the importance of integrating mRNA and protein expression data. We produced lists of candidate T2D genes that might be highly informative for future research efforts. Such a compendium would facilitate unveiling the disease etiology and also the discovery of diagnosis biomarkers and therapeutic targets for T2D.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
Goto-Kakizaki rats were purchased from Shanghai Laboratory Animal Center (Jiu-Ting, Shanghai, China). Analytical reagent grade chemicals were used throughout unless otherwise stated. Water was prepared using a Milli-Q system (Millipore, Bedford, MA). Nycodenz was purchased from Axis-Shield. Magnesium chloride (MgCl2), sodium orthovanadate (Na3VO4), and sodium fluoride (NaF) were obtained from Sigma. Sucrose, DTT, iodoacetamide, CHAPS, and Tris were purchased from Bio-Rad. Mannitol, EDTA, and EGTA were purchased from Amresco (Solon, OH). Sequencing grade trypsin was purchased from Promega (Madison, WI). Complete protease inhibitor mixture tablets were purchased from Roche Applied Science. Antibodies against the following proteins were used: heat shock protein 90 (Hsp90), lamin B, and fibrillarin from Santa Cruz Biotechnology (Santa Cruz, CA); porin from Merck; cytochrome c from NeoMarkers (Lab Vision and Neomarkers); thioredoxin from Abcam; pyruvate dehydrogenase E1 component α subunit (Pdha1) from MitoSciences; and pyruvate dehydrogenase phosphatase 2 (Pdp2) from Cosmo (Tokyo, Japan).
Mitochondria Preparation
For the diabetic stage, livers from six Goto-Kakizaki rats were mixed on equal weight. Mitochondria were isolated by differential centrifugation and density gradient centrifugation according to previous work (15) with a little modification. Briefly, the liver mixture was rinsed with ice-cold homogenization buffer containing 250 mm sucrose, 5 mm MgCl2, 0.2 mm Na3VO4, 1 mm NaF, and 50 mm Tris at pH 7.4 and homogenized in a Potter-Elvehjem homogenizer with a Teflon piston. After filtration through a 100 mesh filter, the homogenate was centrifuged two times at 770 × g for 10 min to remove unbroken cells and nuclei and then centrifuged at 15,000 × g for 15 min to obtain crude mitochondrial pellets. The pellets was washed three times in ice-cold washing buffer containing 200 mm mannitol, 50 mm sucrose, 1 mm EDTA, 0.5 mm EGTA, 0.2 mm Na3VO4, 1 mm NaF, and 10 mm Tris-HCl at pH 7.4 and then applied to Nycodenz density gradient centrifugation.
Mitochondrial pellets from the Nycodenz density gradient centrifugation were suspended in lysis buffer containing 8 m urea, 4% CHAPS, 65 mm DTT, and 40 mm Tris. The suspension was sonicated at 100 watts for 30 s and centrifuged at 25,000 × g for 1 h. The supernatant was collected as the mitochondrial protein lysate. The protein concentration was determined by the Bradford assay.
Trypsin Digestion
Protein concentration was adjusted to about 5 μg/μl, and 1 m DTT was added to a final concentration of 18 mm. The mixture was incubated at 37 °C for 2.5 h, and then 1 m iodoacetamide was added to a final concentration of 36 mm. The mixture was incubated for an additional 40 min at room temperature in darkness. Then 5 volumes of precooled precipitation solution containing 50% acetone, 50% ethanol, and 0.1% acetic acid were added to the protein mixture and kept at −20 °C overnight. The suspension was centrifuged at 20,400 × g for 40 min. The pellets were washed with 70% ethanol and centrifuged at 20,400 × g for 40 min. The final protein pellets were lyophilized and redissolved in 100 mm ammonium bicarbonate buffer, pH 8.5 and incubated with trypsin (50:1) at 37 °C overnight.
Mass Spectrometry
On-line continuous pH and RP gradients were used for mass spectrometry analysis as described in a previous study (16) with some modifications. For SCX-RP-MS/MS analysis, 300 μg of mitochondrial protein digests were loaded onto the SCX column (320 μm × 100 mm; Column Technology) in pH 2.0 buffer and eluted by a continuous pH gradient (from pH 2.0 to pH 8.5). Finally, 10 fractions of peptides were eluted to two C18 RP trap columns (320 μm × 20 mm, C18, 5 mm; Column Technology) followed by one C18 RP capillary column (75 μm × 150 mm, C18, 5 mm; Column Technology). The RP solvents were 0.1% formic acid in either water (A) or acetonitrile (B). A linear ion trap-orbitrap hybrid mass spectrometer (LTQ-Orbitrap) (Thermo Fisher Scientific, San Jose, CA) was equipped with an ESI nanospray source. The mass spectrometer was set so that each full MS scan in the orbitrap was followed by 10 MS/MS scans in the LTQ on the 10 most intense ions from the MS spectrum. For SAX-RP-MS/MS analysis, 600 μg of protein digests were loaded onto the SAX column (320 μm × 100 mm; Column Technology) in pH 8.5 buffer. The continuous pH gradient was from pH 8.5 to pH 2.0, and 10 fractions of peptides were obtained. Other steps were the same as those for SCX-RP-MS/MS. For each sample, three replications were acquired for both SCX and SAX experiments.
Data Analysis
The BioWorksTM3.2 software suite was used to generate the peak lists of all acquired MS/MS spectra (default parameters), and then they were automatically searched against the rat International Protein Index (IPI) protein sequence database (version 3.18) (containing 38,873 protein entries) combined with target protein and reverse sequences using the SEQUEST version 2.7 (University of Washington, licensed to Thermo Finnigan) searching program. In all database searches, trypsin was designated as the protease, and up to five missed cleavages were allowed. A maximal mass deviation of 500 ppm was allowed for the precursor ion, and 1.0 Da was allowed for fragment ions. In all database searches, carbamidomethylation (+57.0125 Da) was searched as a fixed modification on cysteine, and oxidation (+15.99492 Da) was set as a variable modification on methionine. In modification database searches, phosphorylation of serine/threonine/tyrosine residues (+79.96633 Da) and hydroxylation of leucine, proline, and valine (+15.99492 Da) were allowed as variable modifications.
For protein identification, we used the IPI database, which offers complete nonredundant data sets built from the Swiss-Prot, TrEMBL, Ensembl, and RefSeq databases. A protein group was removed if all identified peptides assigned to this protein group were also assigned to another protein group. To sort out a single protein member from a protein group, we chose the protein from the Swiss-Prot database and with the highest sequence coverage. If the peptides were assigned to proteins from other databases, we chose the protein with the highest sequence coverage. In addition, to evaluate the rate of incorrect identifications (false discovery rate (FDR)), all the filtered spectra were subjected to database searching against a composite database containing rat protein sequences in both the forward (correct) and reverse (incorrect) orientations. The reverse database was positioned after the forward database. For total protein identification, all peptides matches were filtered by a set FDR index lower than 0.01, and those with a precursor ion mass tolerance less than 10 ppm and a ΔCn larger than 0.1 were kept for protein identification. Proteins identified with only one unique peptides and those with an identification coverage less than 13% were all excluded from the final result. After filtration, the FDR of peptide identification was calculated to be 0.34%, and the FDR of protein identification was 0.73% (the FDR was calculated according to the algorithm developed by Elias and Gygi (17)). For modified protein identification, first, a stricter peptide identification criteria (FDR ≤ 0.005) was set; second, a modified site(s) was considered to be unique only when the corresponding modified peptides had a ΔCn larger than 0.1 as a ΔCn larger than 0.1 is significant for discriminating the first (top) candidate peptide from the second candidate peptide (18); and third, all modified peptides were verified manually with strict criteria that an MS/MS spectrum of good quality must have its fragment ion peaks clearly above base-line noise, show sequential members of the b- or y-ion series with modified site included, and show intense proline-directed fragment ions. In addition, the phosphoric acid neutral loss peaks were checked for phosphopeptide identification. When using a label-free approach to identify differentially expressed protein and calculating the coefficient of variance, the number of spectra of each protein was logarithm-transformed.
Gene Ontology Annotation
Gene functional categorization and pathway analysis were performed with DAVID Bioinformatics Resources 2008. This provides a gene module level annotation of proteins rather than the specific function of every protein. The annotated proteins are clustered according to the biological process branch of the Gene Ontology (GO) annotation using. The statistical significance of over-representation or under-representation of proteins in each GO category is assessed using Fisher’s exact test, and the significance is indicated by the p values for each GO category. The parent general terms of these specific terms are obtained as well.
RESULTS
High Purity and Integrity of Mitochondrial Fractions from Rat Liver
To investigate the molecular consequences of T2D development in mitochondria, we conducted a comprehensive proteomics analysis on freshly isolated liver mitochondria from prediabetic (5-week-old) and early diabetic (8-week-old) male GK rats. Aberrations observed at this stage were more causally related to the pathogenesis of the disease and could be used as early diagnosis markers. Examining the protein expression pattern of the early diabetic stage compared with the prediabetic stage may provide important clues to the molecular events linked to the dynamic transition from dysfunction to failure.
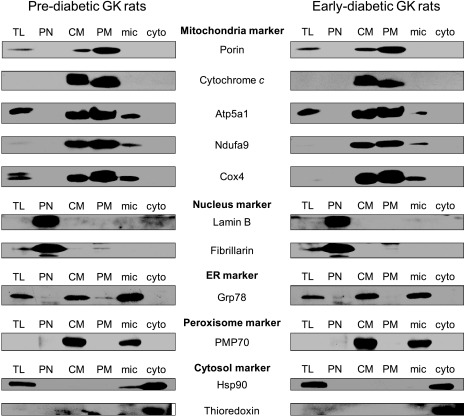
The purity and integrity of isolated mitochondria were validated by Western blot (Fig.1). The purity of mitochondria was excellent as visualized by organelle-specific markers across the different subcellular fractions. In the purified mitochondria (PM) fractions, mitochondrial markers such as porin, cytochrome c, Atp5a1, Ndufa9, and Cox4 were significantly enriched, and markers of other subcellular structures, like nucleus proteins (lamin B and fibrillarin), cytosol proteins (Hsp90 and thioredoxin), an endoplasmic reticulum protein (Grp78), and a peroxisome protein (PMP70), were distinctly absent, demonstrating the purity of isolated mitochondria. Moreover, cytochrome c could only be detected in mitochondrial fractions (crude mitochondria and PM) but not in the cytosol fraction, indicating the integrity of the mitochondria we isolated.
Fig. 1.
Purity and integrity of mitochondria fraction. Mitochondrial and other subcellular fractions isolated from rat liver were applied to Western blot. The PM fraction shows distinct enrichment of mitochondria-specific markers and the absence of markers of other subcellular fractions, indicating the purity and intactness of our mitochondrial extract. Mitochondrial proteins, porin, cytochrome c, Atp5a1, Ndufa9, and Cox4; nucleus proteins, lamin B and fibrillarin; endoplasmic reticulum (ER) protein, Grp78; peroxisome protein, PMP70; cytosol proteins, Hsp90 and thioredoxin. TL, total liver; PN, purified nucleus; CM, crude mitochondria; mic, microsome; cyto, cytosol.
Survey of GK Rat Liver Mitochondrial Proteome and Post-translational Modification Profiles
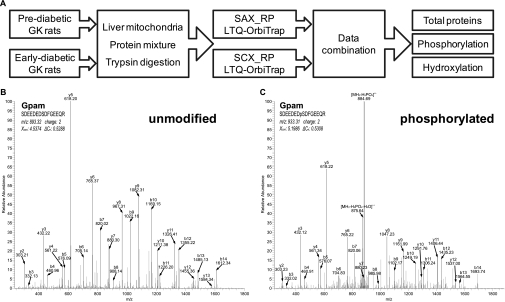
A schematic flow of our proteomics survey is illustrated in Fig.2A. To maximize the coverage of protein identification, peptide fractionation and enrichment were performed separately by SCX and SAX followed by analysis using a linear ion trap LTQ-Orbitrap mass spectrometer. Experiments were done in triplicate, and peptides were identified by stringent criteria (see “Experimental Procedures”). After removing proteins identified by only one unique peptides and those with identification coverage less than 13%, a total of 9554 unique peptides (FDR = 0.34%) corresponding to 1091 distinct proteins (FDR = 0.73%) was confidently identified in GK rat liver mitochondria. Moreover, we identified 228 phosphorylated proteins (including 447 phosphosites and 314 phosphopeptides) and 355 hydroxylated proteins (including 678 hydroxysites and 469 hydroxypeptides) after strict data processing and careful manual checking (supplemental Figs. 1 and 2). Representative MS spectra identifying an unmodified form and a phosphorylated form of the same peptide are shown in Fig. 2, B and C. All mitochondrial proteins identified in this study together with their PTM information are listed in supplemental Tables 1 and 2.
Fig. 2.
A, a schematic flow of our proteomics survey. Representative MS spectra for identification of an unmodified form (B) and phosphorylated form (C) of the same peptide of glycerol-3-phosphate acyltransferase (Gpam) are shown.
To access the reproducibility of our approach, a Venn diagram illustrating the number of proteins identified in each technical replicate and the overlap between these triplicates is shown (supplemental Fig. 3A). Altogether, 1064 different proteins were identified in prediabetic replicates of which 854 (80.26%) were common to all three replicates. Controlling for the redundancy of identifications with the approach used by Hattan et al. (19) and Zegels et al. (20), the three technical replicates therefore yielded 2906 (964 + 952 + 990) identifications of which 2562 (854 × 3) were found in all three replicates. The percentage of shared versus the total number of identifications was 88.16% (2562 of 2906); the percentage of identifications by only one replicate versus the total number of identifications was 2.62% ((23 + 22 + 31)/2906). 1049 different proteins were identified in early diabetic samples of which 815 (77.69%) were common to all three replicates. Similarly, the percentage of shared versus the total number of identifications was 87.32% (2445 of 2800); the percentage of identifications by only one replicate versus the total number of identifications was 4.03% (113 of 2800).
We semiquantitatively estimated the abundance of mitochondrial proteins using the total number of MS/MS spectra detected for each protein. To determine the technical variation, we calculated the coefficient of variance (CV = S.D./mean) of protein expression for the three technical replicates. A distribution histogram of such CV (supplemental Fig. 3B) revealed that, for both samples, the median CV was around 11%, whereas CV values of more than 75% of the proteins were below 0.3. Notably, the distribution histograms of the prediabetic and the early diabetic samples were well consistent, indicating that both samples were subject to the same technical noise.
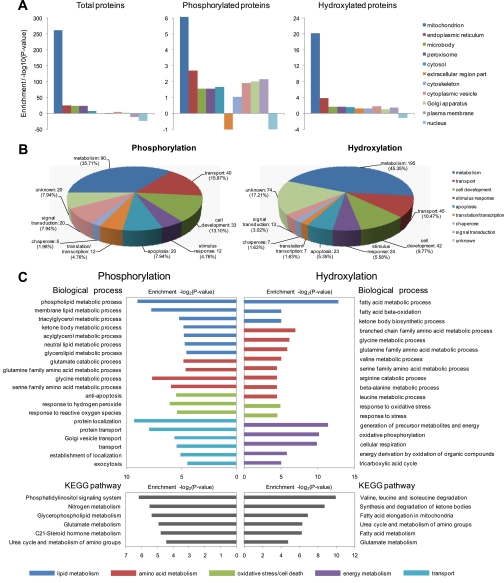
Benchmarking our results with the annotation of “cellular component” by DAVID 2008 (see “Experimental Procedures”), we demonstrated that the majority of the proteins (and modified proteins) we identified were resident in mitochondria (Fig. 3A). The proportions of phosphorylation and hydroxylation proteins identified in this study were about the same as that in prior studies (21, 22). In comparison with combined data from a variety of previous studies and databases (e.g. TargetP, MitoP, and the Gene Ontology database) (23–30), more than 70% of proteins identified in this study have been documented as mitochondrial components, and our data had a broader coverage than those of most prior individual studies (Table I).
Fig. 3.
A, enrichment of total proteins and different post-translationally modified proteins in different organelles. B, categorical analysis based on molecular function of phosphorylated proteins and hydroxylated proteins. C, GO contents based on biological process and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway for which differently modified proteins were significantly enriched.
Table I. Comparison of our data set with previous mitochondrial proteomics studies and mitochondrial protein databases.
Categorical analysis based on molecular function revealed that the majority of proteins with PTMs were involved in metabolism, transport, development, and signaling transduction (Fig. 3B). Based on GO, with regard to biological process, phosphorylated proteins were significantly enriched in biological processes such as lipid metabolism, amino acid metabolism, oxidative stress/cell death, and transport; hydroxylated proteins were enriched in lipid metabolism, amino acid metabolism, and energy metabolism (Fig. 3C). In regard to the Kyoto Encyclopedia of Genes and Genomes pathway, post-translationally modified proteins were significantly enriched in many physiological pathways that are proposed to be linked to T2D (Fig. 3C), such as glycerophospholipid metabolism, fatty acid metabolism, and synthesis and degradation of ketone bodies (31). PTMs can largely expand the variability and complicity of protein function. Accordingly, our PTM profiles could generate indispensable insight into the underlying mechanism and regulatory significance of these physiological pathways. This could largely expand the candidate proteins for mapping complex mitochondrial disorders in T2D.
Alterations of Mitochondrial Protein Expression Linked to T2D Development
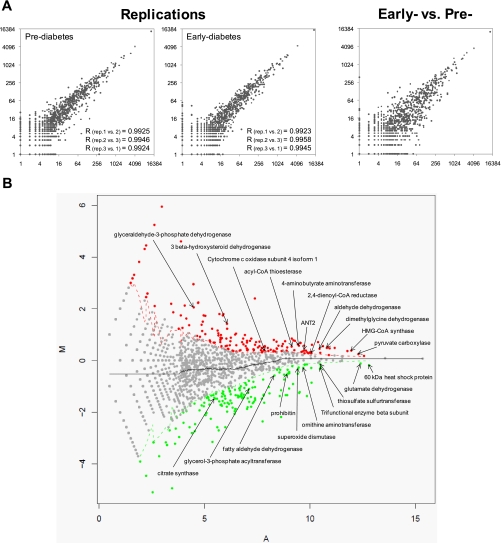
Understanding molecular consequences of T2D development has immediate relevance to the dissection of its underlying mechanisms. To this end, we investigated changes of mitochondrial protein expression between pre- and early diabetic stages using a label-free method called LSPAD (14). Accuracy and reproducibility of our proteomics approach were assessed by comparing the spectral counts of proteins between replicates and between different samples. As shown in Fig. 4A, measurements were consistent within replicates (correlation coefficient >0.99) and were distinctly more diverse between pre- and early diabetic samples. With LSPAD, for each protein, its peptide spectral count Xpre in the prediabetic stage was transformed into Ypre with formula Ypre = log2(Xpre + 1), whereas the Xearly of the early diabetic stage was transformed into Yearly with the same formula. M = Yearly − Ypre was defined as the differential protein abundance between prediabetic and early diabetic samples, and A = (Yearly + Ypre)/2 was defined as the average protein abundance. Based on these formulas, proteins were plotted as a scatter chart (Fig. 4B; M is distributed on the y axis, and A is distributed on the x axis). For a particular protein, spectral counts of neighboring proteins with an A value located within a window covering 25% of the entire length of the A axis (with the focal protein right in the middle) were summed separately for the two samples. The significance of the expression change of the focal protein between the two samples was evaluated by Fisher’s exact test with reference to the whole abundance of neighboring proteins.
Fig. 4.
A, spectral counts of different mitochondrial proteins between replicates (rep.) and between different samples. B, changes in protein expression level in the early developmental stage of T2D identified by LSPAD. Significantly up-regulated proteins (p value <0.01) are in red dots, and down-regulated proteins (p value <0.01) are in green. Genes that have already been reported to be associated with T2D are marked.
Traditional spectral counting-based quantitative approaches always use -fold change to identify altered proteins. However, such approaches are confounded by biases in protein abundance. For highly abundant proteins, a 1.5-fold increase might be too stringent, and for low abundance proteins, a 1.5-fold change might reflect nothing but technical noise (i.e. an increase in abundance from 2 to 3 is less meaningful than a change from 2000 to 3000). Contrary to such prior -fold change approaches, LSPAD is an abundance-corrected approach that uncovers many alterations that were previously indistinguishable, especially for high abundance proteins (14).
Altogether, with a threshold of p value <0.01 (Fisher’s exact test), we identified 204 up-regulated proteins and 183 down-regulated proteins during T2D development. Many differentially expressed proteins we identified were consistent with prior studies, including up-regulated proteins such as hydroxymethylglutaryl (HMG)-CoA synthase (32–34), pyruvate carboxylase (35), dimethylglycine dehydrogenase (36), aldehyde dehydrogenase (36), 2,4-dienoyl-CoA reductase (37), 4-aminobutyrate aminotransferase (36), acyl-CoA thioesterase (33), cytochrome c oxidase subunit 4 isoform 1 (38), ADP/ATP translocase 2 (ANT2) (38), 3β-hydroxysteroid dehydrogenase (33), and glyceraldehyde-3-phosphate dehydrogenase (33). Down-regulated proteins included superoxide dismutase (33, 36, 39, 40), 60-kDa heat shock protein (41), glutamate dehydrogenase (33), thiosulfate sulfurtransferase (36), trifunctional enzyme β subunit (33), ornithine aminotransferase (36), prohibitin (33), fatty aldehyde dehydrogenase (42), glycerol-3-phosphate acyltransferase (36, 43), and citrate synthase (8) (Fig. 4B).
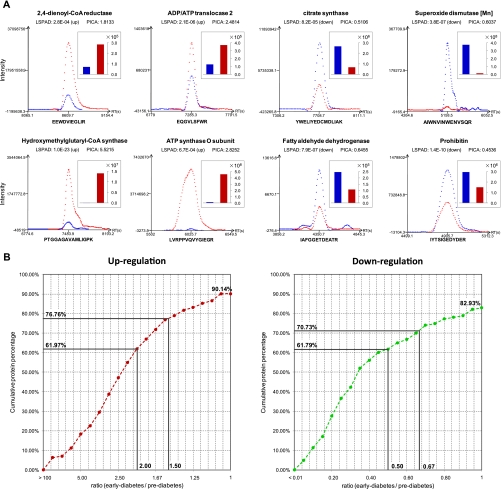
We also compared LSPAD with another label-free quantitative approach based on direct measurement of peptide ion current areas (PICAs) to evaluate its accuracy. We used Census (version 1.44) to calculate PICA for each identified peptide (44), and protein expression alteration was determined by the averaged ratio of peptide chromatographic peak areas between early and prediabetic stages. Indeed, the majority of the significant changes in protein expression identified by LSPAD were validated by PICA. Representative PICAs of significantly changed proteins identified by LSPAD are shown in Fig. 5A. We calculated the cumulative percentage of proteins with preserved expression changes allowing for different ratios of early/prediabetic PICA. As shown in Fig. 5B, 90.14% of the significantly up-regulated proteins and 82.93% of the significantly down-regulated proteins identified by LSPAD were also up- or down-regulated based on PICA; of them, 76.76% of the up-regulated proteins and 70.73% of the down-regulated proteins identified by LSPAD were more than 1.5-fold coordinately changed based on PICA.
Fig. 5.
Comparison between LSPAD and PICA. A, representative PICAs of significantly changed proteins identified by LSPAD validated the accuracy of our label-free approach. B, for significantly changed proteins identified by LSPAD, the cumulative percentage of proteins with consistent expression change identified by PICA allowing for different cutoffs of -fold change.
These results confirmed that the LSPAD method detected changes of protein expression at a high confidence level, and our data were well suited for proteomics expression profiling of T2D-derived alterations in mitochondria. All T2D-derived significant changes in mitochondrial protein expression based on LSPAD are listed in supplemental Table 3.
Coordinated Alterations in Expression of Metabolism-regulating Proteins Linked to T2D Development
Based on our data, mitochondrial proteins were found to undergo expression changes in a highly correlated fashion, especially metabolism-regulating proteins, indicating the underlying biological relevancy. We found that many mitochondrial metabolisms were significantly enhanced, potentially compensating for restrained glucose metabolism by insulin resistance.
We found that proteins involved in glucose metabolism and the tricarboxylic acid cycle were distinctly up-regulated in the progression of T2D such that 12 of the 15 significantly changed proteins were up-regulated (Table II). Protein expression for important enzymes of the tricarboxylic acid cycle, such as succinate dehydrogenase flavoprotein subunit (Sdha), isocitrate dehydrogenase (NADP) (Idh1), and 2-oxoglutarate dehydrogenase E1 component (Ogdh), were all up-regulated (Fig. 6 and Table II). These proteins play crucial roles in the tricarboxylic acid cycle catalyzing reactions that generate reducing equivalents (NADH and FADH2) and GTP. This up-regulation should be expected as T2D might be forestalled through compensation with increased enzymes of the tricarboxylic acid cycle to counteract the superfluous plasma glucose.
Table II. Functional categorization of proteins differentially expressed during development of GK rats.
(+), up-regulated; (−), down-regulated. The p value was calculated through the LSPAD approach. p, phosphorylation; h, hydroxylation.
| IPI number | Protein name | Gene symbol | Change, p value | Modification |
|---|---|---|---|---|
| Energy generation | ||||
| Oxidative phosphorylation | ||||
| IPI00561513 | NADH dehydrogenase (ubiquinone) 1 α subcomplex 10 | Ndufa10 | (+) 1.0e−04 | |
| IPI00189176 | NADH dehydrogenase (ubiquinone) 1 α subcomplex subunit 11 | Ndufa11 | (+) 5.3e−04 | |
| IPI00366206 | NADH dehydrogenase (ubiquinone) 1 α subcomplex, 12 | Ndufa12 | (±) 6.5e−03 | |
| IPI00365436 | NADH dehydrogenase (ubiquinone) 1 α subcomplex, 3 | Ndufa3 | (±) 8.0e−04 | |
| IPI00213436 | NADH dehydrogenase (ubiquinone) 1 α subcomplex, 4 | Ndufa4 | (±) 2.4e−03 | |
| IPI00191312 | NADH dehydrogenase (ubiquinone) 1 α subcomplex, 7 | Ndufa7 | (±) 6.1e−05 | |
| IPI00358441 | NADH dehydrogenase (ubiquinone) 1 α subcomplex, 9 | Ndufa9 | (±) 6.9e−03 | |
| IPI00191112 | NADH dehydrogenase (ubiquinone) 1, α/β subcomplex, 1 | Ndufab1 | (±) 6.9e−03 | |
| IPI00372179 | NADH dehydrogenase (ubiquinone) 1 β subcomplex, 5 | Ndufb5 | (±) 5.8e−04 | |
| IPI00364850 | NADH dehydrogenase (ubiquinone) 1 β subcomplex, 6 | Ndufb6 | (±) 4.1e−03 | |
| IPI00358033 | NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75 kDa | Ndufs1 | (±) 2.8e−03 | h |
| IPI00200659 | Succinate dehydrogenase (ubiquinone) flavoprotein subunit | Sdha | (±) 3.8e−05 | |
| IPI00208288 | Succinate dehydrogenase Ip subunit | Sdhb | (±) 1.6e−04 | |
| IPI00371518 | Succinate dehydrogenase complex, subunit C | Sdhc | (±) 6.4e−06 | |
| IPI00395281 | Electron transfer flavoprotein-ubiquinone oxidoreductase | Etfdh | (±) 1.5e−06 | h |
| IPI00734686 | Cytochrome oxidase subunit II | Cox2 | (±) 2.6e−03 | h |
| IPI00194222 | Cytochrome c oxidase subunit 4 isoform 1 | Cox4i1 | (±) 5.3e−04 | |
| IPI00192246 | Cytochrome c oxidase polypeptide Va | Cox5a | (±) 2.9e−05 | |
| IPI00193918 | Cytochrome c oxidase polypeptide Vb | Cox5b | (±) 9.1e−03 | |
| IPI00230832 | Cytochrome c oxidase polypeptide VIc-2 | Cox6c | (±) 4.1e−03 | |
| IPI00195860 | Cytochrome c oxidase polypeptide VIIa | Cox7a2 | (±) 2.5e−03 | |
| IPI00193233 | Cytochrome b5 outer mitochondrial membrane isoform precursor | Cyb5b | (±) 7.4e−09 | |
| IPI00231864 | Cytochrome c | Cycs | (±) 3.6e−05 | |
| IPI00201307 | Ubiquinol-cytochrome c reductase-binding protein | Uqcrb | (±) 1.9e−07 | |
| IPI00396910 | ATP synthase α chain | Atp5a1 | (±) 1.8e−05 | |
| IPI00231411 | ATP synthase ϵ chain | Atp5e | (±) 6.3e−03 | |
| IPI00196107 | ATP synthase B chain | Atp5f1 | (±) 3.1e−03 | |
| IPI00231978 | ATP synthase e chain | Atp5i | (±) 7.0e−06 | |
| IPI00204316 | ATP synthase coupling factor 6 | Atp5j | (±) 9.1e−03 | |
| IPI00390086 | ATP synthase, H+-transporting, mitochondrial F0 complex, subunit F2 | Atp5j2 | (±) 2.8e−04 | |
| IPI00195123 | ATP synthase O subunit | Atp5o | (±) 6.7e−04 | |
| IPI00421711 | ATP synthase, H+-transporting, mitochondrial F0 complex, subunit G | MGC72942 | (±) 1.3e−05 | h |
| IPI00200466 | ADP/ATP translocase 2 | Slc25a5 | (±) 2.1e−06 | p |
| IPI00191913 | NADH dehydrogenase (Ubiquinone) flavoprotein 1, 51 kDa | Ndufv1 | (−) 4.0e−03 | |
| IPI00286154 | Cytochrome b5 | Cyb5 | (−) 7.7e−03 | |
| IPI00471577 | Ubiquinol-cytochrome c reductase core protein I | Uqcrc1 | (−) 3.1e−04 | |
| IPI00195772 | ATP synthase protein 8 | Atp8 | (−) 9.2e−03 | |
| Glucose metabolism and tricarboxylic acid cycle | ||||
| IPI00231714 | Dihydrolipoamide S-acetyltransferase | Dlat | (±) 6.5e−04 | |
| IPI00231745 | Fructose-1,6-bisphosphatase 1 | Fbp1 | (±) 3.1e−05 | |
| IPI00199663 | Glycerol-3-phosphate dehydrogenase | Gpd2 | (±) 3.0e−03 | |
| IPI00202658 | 3-Hydroxyisobutyrate dehydrogenase | Hibadh | (±) 5.1e−03 | |
| IPI00194045 | Isocitrate dehydrogenase (NADP) | Idh1 | (±) 8.6e−05 | |
| IPI00194047 | Isocitrate dehydrogenase (NAD) subunit γ | Idh3g | (±) 4.5e−04 | |
| IPI00215093 | 2-Oxoglutarate dehydrogenase E1 component | Ogdh | (±) 4.3e−03 | |
| IPI00554039 | Glyceraldehyde-3-phosphate dehydrogenase | RGD1565368 | (±) 5.9e−09 | |
| IPI00200659 | Succinate dehydrogenase (ubiquinone) flavoprotein subunit | Sdha | (±) 3.8e−05 | |
| IPI00208288 | Succinate dehydrogenase Ip subunit | Sdhb | (±) 1.6e−04 | |
| IPI00371518 | Succinate dehydrogenase complex, subunit C | Sdhc | (±) 6.4e−06 | |
| IPI00231767 | Triose-phosphate isomerase | Tpi1 | (±) 4.4e−05 | |
| IPI00360011 | Citrate synthase | Cs | (−) 8.2e−05 | |
| IPI00231611 | Isoform mitochondrial of fumarate hydratase | Fh1 | (−) 2.6e−06 | |
| IPI00209753 | Pyruvate dehydrogenase phosphatase 2 | Pdp2 | (−) 1.0e−02 | |
| Lipid metabolism | ||||
| β-Oxidation | ||||
| IPI00211225 | Long chain-specific acyl-CoA dehydrogenase | Acadl | (±) 6.6e−04 | h |
| IPI00212015 | Medium chain-specific acyl-CoA dehydrogenase | Acadm | (±) 4.6e−21 | h |
| IPI00231359 | Acyl-coenzyme A dehydrogenase, short chain | Acads | (±) 5.7e−03 | h |
| IPI00215574 | 3,2-trans-Enoyl-CoA isomerase | Dci | (±) 5.5e−32 | |
| IPI00213659 | 2,4-Dienoyl-CoA reductase | Decr1 | (±) 2.8e−04 | h |
| IPI00207217 | Enoyl-CoA hydratase | Echs1 | (±) 3.2e−19 | |
| IPI00395281 | Electron transfer flavoprotein-ubiquinone oxidoreductase | Etfdh | (±) 1.5e−06 | h |
| IPI00205157 | Short chain 3-hydroxyacyl-CoA dehydrogenase | Hadh | (±) 1.4e−13 | h |
| IPI00231253 | 3-Hydroxyacyl-CoA dehydrogenase type-2 | Hsd17b10 | (±) 1.4e−18 | |
| IPI00200489 | Isoform mitochondrial of malonyl-CoA decarboxylase | Mlycd | (±) 1.9e−04 | |
| IPI00213057 | Very long chain-specific acyl-CoA dehydrogenase | Acadvl | (−) 7.6e−04 | |
| IPI00195593 | Carnitine O-palmitoyltransferase 2 | Cpt2 | (−) 7.1e−03 | |
| IPI00198467 | Trifunctional enzyme β subunit | Hadhb | (−) 2.6e−09 | |
| Ketone body metabolism | ||||
| IPI00480620 | 3-Hydroxybutyrate dehydrogenase | Bdh1 | (±) 6.4e−06 | h |
| IPI00210444 | Hydroxymethylglutaryl-CoA synthase | Hmgcs2 | (±) 1.1e−23 | h |
| Phospholipid/glucolipid metabolism | ||||
| IPI00189554 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase | Cdipt | (−) 3.6e−03 | |
| IPI00205564 | Glycerol-3-phosphate acyltransferase | Gpam | (−) 2.1e−07 | p |
| IPI00191385 | Phosphoinositide phosphatase SAC1 | Sacm1l | (−) 9.8e−04 | |
| Oxidative stress/cell death | ||||
| IPI00231742 | Catalase | Cat | (±) 5.6e−03 | p |
| IPI00196118 | Isoform 1 of thioredoxin reductase 2 | Txnrd2 | (±) 5.0e−03 | h |
| IPI00562214 | Fatty aldehyde dehydrogenase | Aldh3a2 | (−) 7.9e−07 | |
| IPI00190701 | Apolipoprotein E precursor | Apoe | (−) 3.4e−03 | |
| IPI00206624 | 78-kDa glucose-regulated protein precursor | Hspa5 | (−) 5.7e−103 | p/h |
| IPI00363265 | Stress-70 protein | Hspa9 | (−) 5.2e−04 | |
| IPI00326433 | 10-kDa heat shock protein | Hspd1 | (−) 5.5e−09 | |
| IPI00339148 | 60-kDa heat shock protein | Hspd1 | (−) 2.5e−26 | h |
| IPI00198887 | Protein-disulfide isomerase precursor | P4hb | (−) 4.6e−53 | h |
| IPI00324741 | Protein-disulfide isomerase A3 precursor | Pdia3 | (−) 2.2e−22 | |
| IPI00212220 | Protein-disulfide isomerase A4 precursor | Pdia4 | (−) 1.7e−68 | |
| IPI00391087 | Protein-disulfide isomerase family A, member 4 | Pdia4 | (−) 3.1e−65 | |
| IPI00373089 | Protein-disulfide isomerase A5 precursor | Pdia5 | (−) 1.3e−03 | |
| IPI00365929 | Thioredoxin domain-containing 7 | Pdia6 | (−) 1.2e−14 | p |
| IPI00211593 | Superoxide dismutase (manganese) | Sod2 | (−) 3.8e−07 | |
| IPI00365985 | Tumor rejection antigen gp96 | Tra1 | (−) 1.7e−31 | h |
| IPI00365626 | Thioredoxin domain-containing 1 | Txndc1 | (−) 3.1e−07 | p |
| IPI00365087 | Thioredoxin domain-containing protein 12 precursor | Txndc12 | (−) 3.6e−03 |
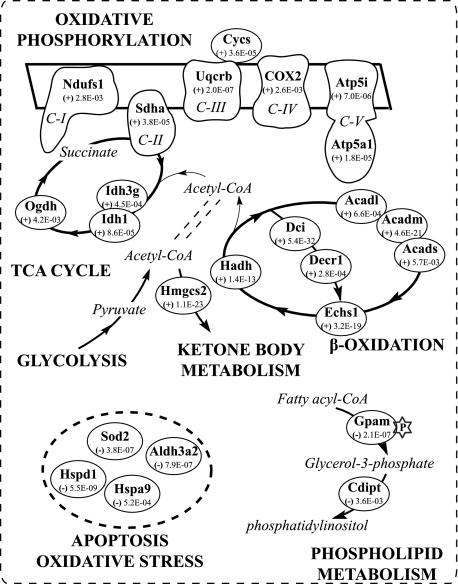
Fig. 6.
Mitochondria underwent highly coordinated T2D-derived changes in systematic metabolisms. Key players involved in such metabolic pathways and the significance of their protein expression change identified by LSPAD are marked. Ndufs1, NADH dehydrogenase (ubiquinone) Fe-S protein 1; Sdha, succinate dehydrogenase (ubiquinone) flavoprotein subunit; Uqcrb, ubiquinol-cytochrome c reductase-binding protein; Cycs, cytochrome c; Cox2, cytochrome oxidase subunit II; Atp5i, ATP synthase e chain; Atp5a1, ATP synthase α chain; Ogdh, 2-oxoglutarate dehydrogenase E1 component; Idh1, isocitrate dehydrogenase (NADP); Idh3g, isocitrate dehydrogenase (NAD) subunit γ; Acadl, long chain-specific acyl-CoA dehydrogenase; Acadm, medium chain-specific acyl-CoA dehydrogenase; Acads, acyl-coenzyme A dehydrogenase, short chain; Dci, 3,2-trans-enoyl-CoA isomerase; Decr1, 2,4-dienoyl-CoA reductase; Echs1, enoyl-CoA hydratase; Hadh, short chain 3-hydroxyacyl-CoA dehydrogenase; Hmgcs2, hydroxymethylglutaryl-CoA synthase; Cdipt, CDP-diacylglycerol-inositol 3-phosphatidyltransferase; Gpam, glycerol-3-phosphate acyltransferase; Sod2, superoxide dismutase (manganese); Aldh3a2, fatty aldehyde dehydrogenase; Hspd1, 60-kDa heat shock protein; Hspa9, stress-70 protein. The numbers in the circles are p values calculated through the LSPAD approach. “(+),” up-regulation of the protein; “−,” down-regulation of the protein. TCA, tricarboxylic acid, C-I, -II, -III, -IV and -V, oxidative phosphorylation complex I, II, III, IV and V.
There was also a coordinated up-regulation of multiple proteins involved in fatty acid oxidation. Our results also revealed that 10 of the 13 significantly changed proteins of the β-oxidation pathway were significantly up-regulated (Table II). These enzymes work in a stepwise fashion to metabolize fats and convert them to energy. These included three kinds of acyl-CoA dehydrogenases, the long chain-specific acyl-CoA dehydrogenase (Acadl), the medium chain-specific acyl-CoA dehydrogenase (Acadm), and the short chain acyl-coenzyme A dehydrogenase (Acads). Other enzymes involved in the fatty acid β-oxidation spiral, such as electron transfer flavoprotein-ubiquinone oxidoreductase (Etfdh), short chain 3-hydroxyacyl-CoA dehydrogenase (Hadh), and enoyl-CoA hydratase (Echs1), were also significantly up-regulated in the process of T2D. 3,2-trans-Enoyl-CoA isomerase (Dci) and 2,4-dienoyl-CoA reductase 1 (Decr1), which are involved in the oxidation of unsaturated fatty enoyl-CoA esters, were also coordinately up-regulated.
We also found that proteins involved in OXPHOS were distinctly up-regulated in the progress of T2D (33 of the 37 significantly changed proteins were up-regulated) (Table II). Oxidative phosphorylation and ATP production occur in a system consisting of five multiprotein complexes. Notably, each of the five complexes composing the mitochondrial electron chain had at least one subunit significantly up-regulated (Fig. 6 and Table II), including NADH dehydrogenase (ubiquinone) Fe-S protein 1 (Ndufs1), succinate dehydrogenase (ubiquinone) flavoprotein subunit (Sdha), ubiquinol-cytochrome c reductase-binding protein (Uqcrb), cytochrome oxidase subunit II (Cox2), and ATP synthase α chain (Atp5a1), consistent with prior claims that genes involved in OXPHOS are up-regulated in the livers of diabetic patients (38, 45). A mitochondrial carrier, ADP/ATP translocase 2 (Slc25a5), which facilitates the exchange of ADP and ATP across the inner mitochondrial membrane, was also up-regulated, consistent with a prior observation in a human T2D patient (38).
We suggest that the changes of protein expression observed involving the tricarboxylic acid cycle, fatty acid β-oxidation, and OXPHOS were highly coordinated during the development of T2D. The β-oxidation pathway, which results in an increased intramitochondrial NADH level, was activated in the process of T2D. Through acetyl-CoA, glucose and fatty acids could be fed into the tricarboxylic acid cycle. Accordingly, at least in part, the flux through the tricarboxylic acid cycle was increased to keep up with the activated lipolysis and β-oxidation. Moreover, the major portion of the energy from the oxidation of carbohydrates and fatty acids comes from the generation of reducing equivalents in the tricarboxylic acid cycle and the subsequent generation of ATP via redox reactions of the electron transport chain. Accordingly, the coordinated activation of the OXPHOS pathway might be driven by the demands of the increased flux to reduce equivalents generated by activated β-oxidation and the tricarboxylic acid cycle.
Ketone bodies are mainly biosynthesized in the liver and delivered to other tissues as an energy source (46). Mitochondrial HMG-CoA synthase (Hmgcs2), a key player in ketogenesis, was significantly up-regulated. HMG-CoA synthase catalyzes the condensation of acetoacetyl-CoA acetyl-CoA to form HMG-CoA. Accumulated mitochondrial HMG-CoA synthase may result in increased ketone body production in T2D, and this reaction occurs at a faster rate in patients with diabetes (32, 34).
Several important enzymes involved in phospholipid metabolism were down-regulated in the development of T2D, including glycerol-3-phosphate acyltransferase (Gpam), which participates in the first committed rate-limiting step of triacylglycerol and glycerophospholipid synthesis (43), and CDP-diacylglycerol-inositol 3-phosphatidyltransferase (Cdipt), which catalyzes the production of phosphatidyl-1-d-myo-inositol. Moreover, our results revealed that multiple proteins involved in oxidative stress and apoptosis were significantly down-regulated (16 of 18 proteins significantly changed were down-regulated), including superoxide dismutase (Sod2), fatty aldehyde dehydrogenase (Aldh3a2), stress-70 protein (Hspa9), 60-kDa heat shock protein (Hspd1), and thioredoxin domain-containing proteins (Txndc1 and Txndc12). Sod2 is the key enzyme responsible for the mitochondrial detoxification of oxygen radicals that decreases superoxide and subsequently blocks caspase-3 activation (40, 47). Deficiency of Sod2 in the T2D state might result in accumulation of superoxide, activation of caspase-3, and induced injury to β-cells (40). Accordingly, deficiency of Sod2 in T2D might be a crucial alteration that accounts for many of the changes observed and be causally linked to the pathogenesis of T2D.
During the development of T2D, proteins involved in β-oxidation, the tricarboxylic acid cycle, OXPHOS, and other bioenergetic processes were coordinately up-regulated, indicating that liver cells confronted T2D by increasing energy expenditure and activating pathways that rid themselves of the constitutively increased flux of glucose and lipid. This might be immediately related to the overproduction of reactive oxygen species, which are the by-product of OXPHOS and cause oxidative stress within the cell (48). Moreover, depression of antioxidant protein might result in increased susceptibility to oxidative damage and partly account for the observed increase in the hydroxylation level during T2D development (supplemental Table 4 and also see “Discussion”).
Increased oxidative stress and depression of antiapoptosis proteins can lead to β-cell dysfunction and apoptosis. Subsequently, T2D evolves when the β-cells fail to release appropriate amounts of insulin, causing metabolic dysregulation and hyperglycemia. Our results could explain why there is indeed a loss of β-cell mass in both T2D patients (49) and GK rats (13, 50). Accordingly, we suggest that many of the changes we observed in systematic metabolisms were causally linked to T2D development. All metabolism-regulating proteins with protein expression changes are listed in Table II. However, we caution that it is so far unclear whether these aberrations we observed are all causally related to the development of the disease. Alternatively, some changes in systemic metabolism might have no role in the generation of T2D but are simply consequences of other turbulence.
Case Study of Pdp2 and Pdha1
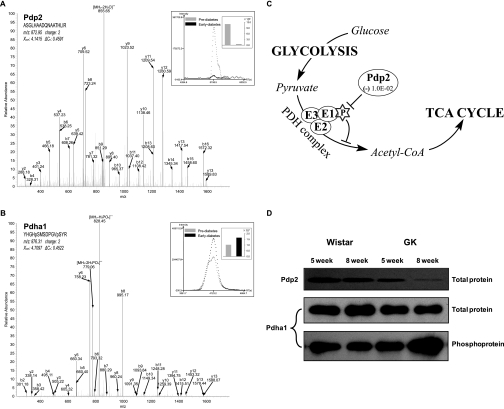
Based on our quantitative approach we have pointed out a number of factors that highlight the pathogenetic role of mitochondrial perturbations in T2D. Of them, an interesting finding is the reciprocal changes between the protein expression of Pdp2 and the phosphorylation level of its substrate, namely Pdha1. Pdha1 is a component of PDH complex, which catalyzes the irreversible oxidative decarboxylation of pyruvate to acetyl-CoA (Fig. 7C). This important process links glycolysis with the tricarboxylic acid cycle and plays a crucial role in cellular energy production. Phosphorylation deactivates Pdha1 and inhibits the activity of PDH complex. Pdp2 is the phosphatase of Pdha1 that restores the activity of PDH complex by dephosphorylating Pdha1 (51, 52). MS spectra for identification and quantification of Pdp2 and Pdha1 are shown in Fig. 7, A and B. Our result revealed that Pdp2 was significantly down-regulated in the development of T2D (Table II and Fig. 7A), which was consistent with the observation that there are more phosphopeptides in the early diabetic sample than in the prediabetic sample (Fig. 7B). The deficiency of Pdp2 and the associated hyperphosphorylation of Pdha1 in the development of T2D were confirmed by Western blot, and we also found that the total protein expression of Pdha1 remained unchanged (Fig. 7D). This result emphasizes the importance of PTM profiling in unveiling complex issues surrounding the pathogenesis of T2D. PTMs could largely expand the complexity and regulatory function of the protein, many important changes may not be at protein expression level but at the PTM level, and changes in protein abundance would in turn have prolonged functional influence especially on those with functionally important PTMs.
Fig. 7.
A, MS spectrum of a representative peptide of Pdp2. B, MS spectrum of a phosphopeptide of Pdha1. The top right panel shows both the peptide ion current areas and the corresponding values between the pre- and early diabetic samples. C, the metabolic pathway of the involved proteins that links glycolysis with the tricarboxylic acid cycle. D, Western blot validating the proteomics observation that protein expression of Pdp2 was down-regulated in the development of T2D and that the phosphorylation level of its substrate Pdha1 was coordinately up-regulated. TCA, tricarboxylic acid.
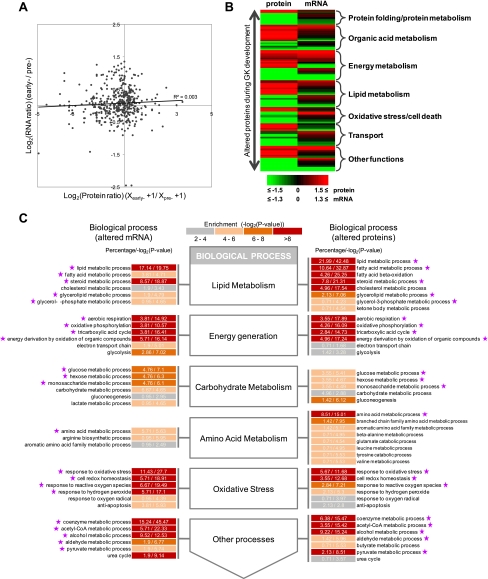
Correlation between Changes in Protein Expression and mRNA Abundance
We obtained gene expression profiles of T2D rat liver at pre- and early diabetic stages from published data sets (36) and sought to compare the changes in protein expression with those in mRNA abundance. Altogether, we obtained 437 mitochondrial genes with both data sets available. Fig. 8A shows early/prediabetic protein expression ratios versus the corresponding mRNA ratios. This result revealed that the overall expression changes in the mitochondrial proteome and transcriptome were poorly correlated (r = 0.06, p value >0.05). For the 165 significantly changed proteins identified by LSPAD (proteomics data), we classified them by biological function and compared the protein expression ratio versus the corresponding mRNA expression ratio (Fig. 8B). About 20.61% of the differentially expressed proteins showed preserved changes in mRNA (>1.3-fold), 12.72% were discordant (i.e. having higher protein expression but lower mRNA expression), and the remaining 66.67% showed changes in the proteome but not in the transcriptome. These results indicate that many gene expression changes would be ignored without integrating mRNA and proteomics data together. Incongruent variation between mRNA and protein could stem from many biological aspects, including different kinds of post-transcriptional modifications and different rates of protein synthesis and degradation. We caution, however, that such discrimination could be artificial because of diverse technical factors, such as different throughput and sensitivity of the approaches and procedures of data processing. These issues were non-trivial, and we would leave any such consideration to further analysis.
Fig. 8.
Correlation between mRNA ratios and protein levels of differentially expressed proteins. A, scatter plots of protein expression ratios from proteomics data against mRNA abundance ratios from microarray data. B, a pictorial comparison of significantly changed protein ratios against the corresponding mRNA ratio with functional clustering based on the biological process category of Gene Ontology. Red and green colors represent an increase or decrease in -fold change, respectively. C, gene module-based correlation analysis between the proteomics and microarray data. After partitioning genes with altered mRNA or protein expression separately into small function categories, or gene modules, we compared GO contents between mRNA data and protein data in response to T2D and found that the results were highly coordinate.
To unveil the potential biological relevancy behind this incompatibility, we adopted a gene module-based comparison between these two data sets. If a priori changes in protein and mRNA levels are functionally coordinated then we would expect the functional modules in which differentially expressed protein/mRNA were enriched to be highly similar. Consistent with such a hypothesis, as shown in Fig. 8C, the majority of the GO contents significantly affected were consistent between the data sets (labeled by a star). For example, changes in gene expression identified by microarray and in the proteome were all enriched in organic acid metabolism, lipid metabolism, transport, oxidative stress, and energy metabolism processes. Specific GO terms including carboxylic acid metabolism, amino acid metabolism, amine metabolism, lipid metabolism, fatty acid metabolism, intracellular transport, electron transport, response to hypoxia, and oxidative phosphorylation were all consistently and heavily affected in the development of T2D. These results indicated that T2D development affects different aspects of gene expression. Accordingly, integrating changes in protein expression, mRNA abundance, and PTM profiles could yield more comprehensive information for better understanding of its underlying regulatory mechanisms.
DISCUSSION
Despite numerous efforts made in investigating the etiology of T2D, we are still grappling with the enormous complexity of the disease process in which almost every aspect of the body’s metabolism goes awry. Dysfunction of mitochondria underlies a broad spectrum of human diseases including T2D. Although much effort had been made (2, 9, 11), identifying proteins resident in this organelle and understanding how they are functionally integrated into pathways remain a major challenge. The discovery of mitochondrial proteins affected in T2D will provide key insights into the mechanisms that actively promote T2D and will be useful for the design of targeted therapeutic approaches.
In this study, we comprehensively identified mitochondrial proteins and their PTM profiles in rat liver; we also closely investigated T2D-derived changes in their protein expression. Our proteomics approach added to the supposition that development of T2D has profound consequences in mitochondria. Additionally, our efforts also mark the first large scale survey on mitochondrial protein hydroxylation. Assuming that mitochondrial factors are closely linked to the pathogenesis of T2D, our results likely revealed many previously unrecognized components that could facilitate the research, diagnosis, and therapies of this disease in the future.
Based on our label-free quantitative approach, either directly or indirectly, we found that T2D development leads to extensive changes in mitochondrial protein expression and post-translational modification. During the transition to diabetes, mitochondria underwent extensive changes in protein expression and arrived at a new highly regulated, although pathological, steady state. These changes were functionally correlated, and the metabolism pathways were altered in a highly coordinated fashion. This coordinated program of changes in systematic metabolisms could underlie the mechanisms through which GK rats develop and adapt toT2D. In this study, a particular emphasis was placed on changes in metabolic enzymes and function. The emerging picture is that alterations in mitochondria may be a culprit in the pathogenetic processes culminating in T2D. Such processes may prove to be targets for therapeutic interventions in the disease.
Indeed, liver plays a pivotal role in the regulation of lipid metabolism, accounting for the creation and degradation of fatty acid, bile salts, cholesterols, eicosanoids, glycolipids, ketone bodies, fatty acids, phospholipids, sphingolipids, steroids, and triacylglycerols. Our results revealed that almost every important aspect of lipid metabolism had gone awry during T2D development, such as fatty acid metabolism, ketone body metabolism, and glycerolipid/phospholipid metabolism. This may be causally linked to or caused by the aberrant accumulation of lipolytic products. To dispose of such lipolytic products, the cell up-regulated a series of oxidative catabolic pathways such as β-oxidation, the tricarboxylic acid cycle, and OXPHOS. The accumulation of circulating free fatty acids and fat, possibly by impaired fatty acid metabolism (53, 54), might have deleterious influences on insulin signaling, glucose uptake, and glycogen synthesis and lead to T2D development (55).
Activation of OXPHOS may produce deleterious oxidation products (such as reactive oxygen species) that would be likely candidates that are causally linked to T2D development in the GK rat (48). Both experimental and clinical studies have provided compelling evidence that oxidative stress plays a crucial role in the pathogenesis of diabetes, leading to insulin resistance, β-cell dysfunction, impaired glucose tolerance, and ultimately T2D (56, 57). Hydroxylation is a kind of oxidative modification that reflects oxidant damage (58). Our results demonstrated that increased oxidative stress could also be reflected by an elevated hydroxylation level. Actually, hydroxyleucine and hydroxyvaline have already been used to assess the activity of oxidative stress in atherosclerotic plaques and nuclear cataractogenesis (59, 60). In this study, we found that the hydroxylation levels of many mitochondrial proteins were up-regulated in the development of T2D (supplemental Table 4), for example the enzymes of lipid metabolism (hydroxymethylglutaryl-CoA synthase, medium chain-specific acyl-CoA dehydrogenase, 3-hydroxybutyrate dehydrogenase, Δ3,5-Δ2,4-dienoyl-CoA isomerase, and 3-ketoacyl-CoA thiolase), a component of oxidative phosphorylation (ATP synthase g subunit), and two proteins of the metabolic process (pyruvate carboxylase and methylmalonate-semialdehyde dehydrogenase). This result indicated that the oxidative damage occurs at the very early stage of T2D development. The superoxide proteins we identified would help to evaluate the extent of oxidative stress of the cell and also to elucidate the mechanisms by which increased oxidative stress accelerates the generation of diabetic complications.
Moreover, we have learned the importance of integrating different approaches to yield more comprehensive information that might otherwise be overlooked by using only one of these approaches. For example, many genes were altered at either the mRNA or protein level. In addition, many proteins may have changes at the PTM level rather than at the mRNA/protein expression level. For example, although Pdha1 protein was consistently expressed between the diabetic and normal stages, its higher phosphorylation level could still inhibit its activity and impair the biological processes in which it participates. Furthermore, our gene module-based comparison between the proteomics data and microarray data was indicative. In this way, we could discover the potential underlying biological relevancy of incompatible data sets and identify the core regulatory changes or disturbances in response to the metabolic status of concern.
In summary, mitochondria are a primary concern in the disease processes of T2D, and in this study we attempted to build a comprehensive proteome and post-translational modification profiles of liver mitochondria. We also systematically elucidated alterations in mitochondria at both protein expression and PTM levels. Variations in systematic metabolisms and their biological implications were closely discussed. We also integrated our proteomics data with microarray data to yield more comprehensive information. Our results have created a framework to explore correlated alterations in systematic metabolisms during the compensation and development of T2D. This will help to gain insight into potential pathways and regulatory networks of T2D and also facilitate the discovery of diagnostic biomarkers and therapeutic targets for T2D.
Supplementary Material
Footnotes
* This work was supported by National Natural Science Foundation Grants 30425021 and 30821065, Basic Research Foundation Grants 2006CB910700 and 2006CB503900, Chinese Academy of Sciences Project Grants KSCX2-YW-R-106 and KSCX1-YW-02, and Proteomage from the European Union.
 The on-line version of this article (available at http://www. mcponline.org) contains supplemental Figs. 1–3 and Tables 1–4.
The on-line version of this article (available at http://www. mcponline.org) contains supplemental Figs. 1–3 and Tables 1–4.
1 The abbreviations used are:
- T2D
- type 2 diabetes
- GK
- Goto-Kakizaki
- PTM
- post-translational modification
- SCX
- strong cation exchange chromatography
- SAX
- strong anion exchange chromatography
- OXPHOS
- oxidative phosphorylation
- LSPAD
- localized statistics of protein abundance distribution
- Pdha1
- pyruvate dehydrogenase E1 component α subunit
- Pdp2
- pyruvate dehydrogenase phosphatase 2
- RP
- reverse phase
- IPI
- International Protein Index
- FDR
- false discovery rate
- GO
- Gene Ontology
- PM
- purified mitochondria
- Cox
- cytochrome oxidase
- CV
- coefficient of variance
- HMG
- hydroxymethylglutaryl
- PICA
- peptide ion current area
- PDH
- pyruvate dehydrogenase.
REFERENCES
- 1.Zimmet P., Alberti K. G., Shaw J. (2001) Global and societal implications of the diabetes epidemic. Nature 414, 782–787 [DOI] [PubMed] [Google Scholar]
- 2.Abdul-Ghani M.A, DeFronzo R. A. (2008) Mitochondrial dysfunction, insulin resistance, and type 2 diabetes mellitus. Curr. Diab. Rep 8, 173–178 [DOI] [PubMed] [Google Scholar]
- 3.Leahy J. L. (2005) Pathogenesis of type 2 diabetes mellitus. Arch. Med. Res 36, 197–209 [DOI] [PubMed] [Google Scholar]
- 4.Skyler J. S. (2004) Diabetes mellitus: pathogenesis and treatment strategies. J. Med. Chem 47, 4113–4117 [DOI] [PubMed] [Google Scholar]
- 5.Raffaella C., Francesca B., Italia F., Marina P., Giovanna L., Susanna I. (2008) Alterations in hepatic mitochondrial compartment in a model of obesity and insulin resistance. Obesity 16, 958–964 [DOI] [PubMed] [Google Scholar]
- 6.Wollheim C. B. (2000) Beta-cell mitochondria in the regulation of insulin secretion: a new culprit in type II diabetes. Diabetologia 43, 265–277 [DOI] [PubMed] [Google Scholar]
- 7.Becker R., Laube H., Linn T., Damian M. S. (2002) Insulin resistance in patients with the mitochondrial tRNA(Leu(UUR)) gene mutation at position 3243. Exp. Clin. Endocrinol. Diabetes 110, 291–297 [DOI] [PubMed] [Google Scholar]
- 8.Kelley D. E., He J., Menshikova E. V., Ritov V. B. (2002) Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes 51, 2944–2950 [DOI] [PubMed] [Google Scholar]
- 9.Lowell B. B., Shulman G. I. (2005) Mitochondrial dysfunction and type 2 diabetes. Science 307, 384–387 [DOI] [PubMed] [Google Scholar]
- 10.Rong J. X., Qiu Y., Hansen M. K., Zhu L., Zhang V., Xie M., Okamoto Y., Mattie M. D., Higashiyama H., Asano S., Strum J. C., Ryan T. E. (2007) Adipose mitochondrial biogenesis is suppressed in db/db and high-fat diet-fed mice and improved by rosiglitazone. Diabetes 56, 1751–1760 [DOI] [PubMed] [Google Scholar]
- 11.Gerbitz K. D., Gempel K., Brdiczka D. (1996) Mitochondria and diabetes. Genetic, biochemical, and clinical implications of the cellular energy circuit. Diabetes 45, 113–126 [DOI] [PubMed] [Google Scholar]
- 12.Goto Y., Kakizaki M., Masaki N. (1976) Production of spontaneous diabetic rats by repetition of selective breeding. Tohoku J. Exp. Med 119, 85–90 [DOI] [PubMed] [Google Scholar]
- 13.Picarel-Blanchot F., Berthelier C., Bailbé D., Portha B. (1996) Impaired insulin secretion and excessive hepatic glucose production are both early events in the diabetic GK rat. Am. J. Physiol. Endocrinol. Metab 271, E755–E762 [DOI] [PubMed] [Google Scholar]
- 14.Li R. X., Chen H. B., Tu K., Zhao S. L., Zhou H., Li S. J., Dai J., Li Q. R., Nie S., Li Y. X., Jia W. P., Zeng R., Wu J. R. (2008) Localized-statistical quantification of human serum proteome associated with type 2 diabetes. PLoS One 3, e3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang X. S., Zhou H., Zhang L., Sheng Q.H., Li S.J., Li L., Hao P., Li Y.X., Xia Q.C., Wu J.R., Zeng R. (2004) A high-throughput approach for subcellular proteome: identification of rat liver proteins using subcellular fractionation coupled with two-dimensional liquid chromatography tandem mass spectrometry and bioinformatic analysis. Mol. Cell. Proteomics 3, 441–455 [DOI] [PubMed] [Google Scholar]
- 16.Zhou H., Dai J., Sheng Q.H., Li R.X., Shieh C.H., Guttman A., Zeng R. (2007) A fully automated 2-D LC-MS method utilizing online continuous pH and RP gradients for global proteome analysis. Electrophoresis 28, 4311–4319 [DOI] [PubMed] [Google Scholar]
- 17.Elias J. E., Gygi S.P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 18.Yates J. R., 3rd, Eng J.K., McCormack A.L., Schieltz D. (1995) Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem 67, 1426–1436 [DOI] [PubMed] [Google Scholar]
- 19.Hattan S. J., Marchese J., Khainovski N., Martin S., Juhasz P. (2005) Comparative study of [Three] LC-MALDI workflows for the analysis of complex proteomic samples. J. Proteome Res 4, 1931–1941 [DOI] [PubMed] [Google Scholar]
- 20.Zegels G., Van Raemdonck G. A., Coen E. P., Tjalma W. A., Van Ostade X. W. (2009) Comprehensive proteomic analysis of human cervical-vaginal fluid using colposcopy samples. Proteome Sci 7, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee J., Xu Y., Chen Y., Sprung R., Kim S. C., Xie S., Zhao Y. (2007) Mitochondrial phosphoproteome revealed by an improved IMAC method and MS/MS/MS. Mol. Cell. Proteomics 6, 669–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meany D. L., Xie H., Thompson L. V., Arriaga E. A., Griffin T.J. (2007) Identification of carbonylated proteins from enriched rat skeletal muscle mitochondria using affinity chromatography-stable isotope labeling and tandem mass spectrometry. Proteomics 7, 1150–1163 [DOI] [PubMed] [Google Scholar]
- 23.Pagliarini D. J., Calvo S. E., Chang B., Sheth S. A., Vafai S. B., Ong S. E., Walford G. A., Sugiana C., Boneh A., Chen W. K., Hill D. E., Vidal M., Evans J. G., Thorburn D. R., Carr S. A., Mootha V. K. (2008) A mitochondrial protein compendium elucidates complex I disease biology. Cell 134, 112–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calvo S., Jain M., Xie X., Sheth S. A., Chang B., Goldberger O. A., Spinazzola A., Zeviani M., Carr S. A., Mootha V. K. (2006) Systematic identification of human mitochondrial disease genes through integrative genomics. Nat. Genet 38, 576–582 [DOI] [PubMed] [Google Scholar]
- 25.Forner F., Foster L. J., Campanaro S., Valle G., Mann M. (2006) Quantitative proteomic comparison of rat mitochondria from muscle, heart, and liver. Mol. Cell. Proteomics 5, 608–619 [DOI] [PubMed] [Google Scholar]
- 26.Zhang J., Li X., Mueller M., Wang Y., Zong C., Deng N., Vondriska T. M., Liem D. A., Yang J. I., Korge P., Honda H., Weiss J. N., Apweiler R., Ping P. (2008) Systematic characterization of the murine mitochondrial proteome using functionally validated cardiac mitochondria. Proteomics 8, 1564–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mootha V. K., Bunkenborg J., Olsen J. V., Hjerrild M., Wisniewski J. R., Stahl E., Bolouri M. S., Ray H. N., Sihag S., Kamal M., Patterson N., Lander E. S., Mann M. (2003) Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell 115, 629–640 [DOI] [PubMed] [Google Scholar]
- 28.Gaucher S. P., Taylor S. W., Fahy E., Zhang B., Warnock D. E., Ghosh S. S., Gibson B. W. (2004) Expanded coverage of the human heart mitochondrial proteome using multidimensional liquid chromatography coupled with tandem mass spectrometry. J. Proteome Res 3, 495–505 [DOI] [PubMed] [Google Scholar]
- 29.McDonald T., Sheng S., Stanley B., Chen D., Ko Y., Cole R. N., Pedersen P., Van Eyk J. E. (2006) Expanding the subproteome of the inner mitochondria using protein separation technologies: one- and two-dimensional liquid chromatography and two-dimensional gel electrophoresis. Mol. Cell. Proteomics 5, 2392–2411 [DOI] [PubMed] [Google Scholar]
- 30.Sickmann A., Reinders J., Wagner Y., Joppich C., Zahedi R., Meyer H. E., Schönfisch B., Perschil I., Chacinska A., Guiard B., Rehling P., Pfanner N., Meisinger C. (2003) The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. U.S.A 100, 13207–13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prentki M., Madiraju S. R. (2008) Glycerolipid metabolism and signaling in health and disease. Endocr. Rev 29, 647–676 [DOI] [PubMed] [Google Scholar]
- 32.Quant P.A. (1994) The role of mitochondrial HMG-CoA synthase in regulation of ketogenesis. Essays Biochem 28, 13–25 [PubMed] [Google Scholar]
- 33.Lu H., Yang Y., Allister E. M., Wijesekara N., Wheeler M. B. (2008) The identification of potential factors associated with the development of type 2 diabetes: a quantitative proteomics approach. Mol. Cell. Proteomics 7, 1434–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hegardt F. G. (1998) Transcriptional regulation of mitochondrial HMG-CoA synthase in the control of ketogenesis. Biochimie 80, 803–806 [DOI] [PubMed] [Google Scholar]
- 35.Weinberg M. B., Utter M. F. (1980) Effect of streptozotocin-induced diabetes mellitus on the turnover of rat liver pyruvate carboxylase and pyruvate dehydrogenase. Biochem. J 188, 601–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suh Y. H., Kim Y., Bang J. H., Choi K. S., Lee J. W., Kim W. H., Oh T. J., An S., Jung M. H. (2005) Analysis of gene expression profiles in insulin-sensitive tissues from pre-diabetic and diabetic Zucker diabetic fatty rats. J. Mol. Endocrinol 34, 299–315 [DOI] [PubMed] [Google Scholar]
- 37.Willsky G. R., Chi L. H., Liang Y., Gaile D. P., Hu Z., Crans D. C. (2006) Diabetes-altered gene expression in rat skeletal muscle corrected by oral administration of vanadyl sulfate. Physiol. Genomics 26, 192–201 [DOI] [PubMed] [Google Scholar]
- 38.Takamura T., Misu H., Matsuzawa-Nagata N., Sakurai M., Ota T., Shimizu A., Kurita S., Takeshita Y., Ando H., Honda M., Kaneko S. (2008) Obesity upregulates genes involved in oxidative phosphorylation in livers of diabetic patients. Obesity 16, 2601–2609 [DOI] [PubMed] [Google Scholar]
- 39.Kartha G. K., Moshal K. S., Sen U., Joshua I. G., Tyagi N., Steed M. M., Tyagi S. C. (2008) Renal mitochondrial damage and protein modification in type-2 diabetes. Acta Diabetol 45, 75–81 [DOI] [PubMed] [Google Scholar]
- 40.Mizobuchi N., Nakata H., Horimi T., Takahashi I. (1993) Serum superoxide dismutase (SOD) activity in diabetes mellitus. Rinsho Byori 41, 673–678 [PubMed] [Google Scholar]
- 41.Chen H. S., Wu T. E., Juan C. C., Lin H. D. (2009) Myocardial heat shock protein 60 expression in insulin-resistant and diabetic rats. J. Endocrinol 200, 151–157 [DOI] [PubMed] [Google Scholar]
- 42.Demozay D., Rocchi S., Mas J. C., Grillo S., Pirola L., Chavey C., Van Obberghen E. (2004) Fatty aldehyde dehydrogenase: potential role in oxidative stress protection and regulation of its gene expression by insulin. J. Biol. Chem 279, 6261–6270 [DOI] [PubMed] [Google Scholar]
- 43.Farese R. V., Standaert M. L., Yamada K., Huang L. C., Zhang C., Cooper D. R., Wang Z., Yang Y., Suzuki S., Toyota T., Larner J. (1994) Insulin-induced activation of glycerol-3-phosphate acyltransferase by a chiro-inositol-containing insulin mediator is defective in adipocytes of insulin-resistant, type II diabetic, Goto-Kakizaki rats. Proc. Natl. Acad. Sci. U.S.A 91, 11040–11044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park S. K., Venable J. D., Xu T., Yates J. R., 3rd (2008) A quantitative analysis software tool for mass spectrometry-based proteomics. Nat. Methods 5, 319–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Misu H., Takamura T., Matsuzawa N., Shimizu A., Ota T., Sakurai M., Ando H., Arai K., Yamashita T., Honda M., Yamashita T., Kaneko S. (2007) Genes involved in oxidative phosphorylation are coordinately upregulated with fasting hyperglycaemia in livers of patients with type 2 diabetes. Diabetologia 50, 268–277 [DOI] [PubMed] [Google Scholar]
- 46.Hegardt F.G. (1999) Mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase: a control enzyme in ketogenesis. Biochem. J 338, 569–582 [PMC free article] [PubMed] [Google Scholar]
- 47.Ukkola O., Erkkilä P. H., Savolainen M. J., Kesäniemi Y. A. (2001) Lack of association between polymorphisms of catalase, copper-zinc superoxide dismutase (SOD), extracellular SOD and endothelial nitric oxide synthase genes and macroangiopathy in patients with type 2 diabetes mellitus. J. Intern. Med 249, 451–459 [DOI] [PubMed] [Google Scholar]
- 48.Wallace D.C. (2001) A mitochondrial paradigm for degenerative diseases and ageing. Novartis Found. Symp 235, 247–266 [DOI] [PubMed] [Google Scholar]
- 49.Butler A. E., Janson J., Bonner-Weir S., Ritzel R., Rizza R. A., Butler P. C. (2003) Beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes. Diabetes 52, 102–110 [DOI] [PubMed] [Google Scholar]
- 50.Portha B. (2005) Programmed disorders of beta-cell development and function as one cause for type 2 diabetes? The GK rat paradigm. Diabetes Metab. Res. Rev 21, 495–504 [DOI] [PubMed] [Google Scholar]
- 51.Piccinini M., Mostert M., Alberto G., Ramondetti C., Novi R. F., Dalmasso P., Rinaudo M. T. (2005) Down-regulation of pyruvate dehydrogenase phosphatase in obese subjects is a defect that signals insulin resistance. Obes. Res 13, 678–686 [DOI] [PubMed] [Google Scholar]
- 52.Wu P., Inskeep K., Bowker-Kinley M. M., Popov K. M., Harris R. A. (1999) Mechanism responsible for inactivation of skeletal muscle pyruvate dehydrogenase complex in starvation and diabetes. Diabetes 48, 1593–1599 [DOI] [PubMed] [Google Scholar]
- 53.Bennett M.J. (2007) Assays of fatty acid beta-oxidation activity. Methods Cell Biol 80, 179–197 [DOI] [PubMed] [Google Scholar]
- 54.Woldseth B., Rootwelt T. (2006) Mitochondrial beta-oxidation defects. Tidsskr. Nor. Laegeforen 126, 756–759 [PubMed] [Google Scholar]
- 55.Iizuka Y., Murase T., Iizuka Y. (1997) Abnormalities in lipid metabolism associated with diabetes mellitus. Nippon Rinsho 55, (suppl.) 603–608 [PubMed] [Google Scholar]
- 56.Santos D. L., Palmeira C. M., Seiça R., Dias J., Mesquita J., Moreno A. J., Santos M. S. (2003) Diabetes and mitochondrial oxidative stress: a study using heart mitochondria from the diabetic Goto-Kakizaki rat. Mol. Cell. Biochem 246, 163–170 [PubMed] [Google Scholar]
- 57.Forbes J. M., Coughlan M. T., Cooper M. E. (2008) Oxidative stress as a major culprit in kidney disease in diabetes. Diabetes 57, 1446–1454 [DOI] [PubMed] [Google Scholar]
- 58.Davies M. J., Fu S., Wang H., Dean R. T. (1999) Stable markers of oxidant damage to proteins and their application in the study of human disease. Free Radic. Biol. Med 27, 1151–1163 [DOI] [PubMed] [Google Scholar]
- 59.Fu S., Davies M. J., Stocker R., Dean R. T. (1998) Evidence for roles of radicals in protein oxidation in advanced human atherosclerotic plaque. Biochem. J 333, 519–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fu S., Dean R., Southan M., Truscott R. (1998) The hydroxyl radical in lens nuclear cataractogenesis. J. Biol. Chem 273, 28603–28609 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.