Abstract
In the biological sciences, model organisms have been used for many decades and have enabled the gathering of a large proportion of our present day knowledge of basic biological processes and their derailments in disease. Although in many of these studies using model organisms, the focus has primarily been on genetics and genomics approaches, it is important that methods become available to extend this to the relevant protein level. Mass spectrometry-based proteomics is increasingly becoming the standard to comprehensively analyze proteomes. An important transition has been made recently by moving from charting static proteomes to monitoring their dynamics by simultaneously quantifying multiple proteins obtained from differently treated samples. Especially the labeling with stable isotopes has proved an effective means to accurately determine differential expression levels of proteins. Among these, metabolic incorporation of stable isotopes in vivo in whole organisms is one of the favored strategies. In this perspective, we will focus on methodologies to stable isotope label a variety of model organisms in vivo, ranging from relatively simple organisms such as bacteria and yeast to Caenorhabditis elegans, Drosophila, and Arabidopsis up to mammals such as rats and mice. We also summarize how this has opened up ways to investigate biological processes at the protein level in health and disease, revealing conservation and variation across the evolutionary tree of life.
Well before the genomics era, the foundation for our current understanding of genetics was largely established by biological research performed using model organisms. Early genetics discoveries such as the chromosome theory of heredity and bacterial conjugation were first described in the fruit fly Drosophila melanogaster (1) and the bacterium Escherichia coli (2), respectively. Apart from these organisms, most of the current knowledge of development, evolution, and genetics originates from other classical model organisms including the bakers' yeast Saccharomyces cerevisiae, the nematode Caenorhabditis elegans, and the mouse Mus musculus. Nowadays, they hold a primary position in the analysis of biological, disease, and pharmaceutical processes in modern biology and probably claim an even more promising position in future biological research. With increasing numbers of completed genome annotations, however, the focus is also shifting somewhat from these classical model organisms toward organisms that have unique genetic properties, are economically interesting, or are more directly related to human disease such as puffer fish, rice, and Plasmodium, respectively. Consequently, the definition of a model organism has broadened over the past decade, and today model organisms are found in nearly all branches of the “tree of life,” providing extensive means to further investigate conservation or diversification of biological principles through evolution (3). This has gained momentum tremendously by the completion of genome sequencing efforts in hundreds of organisms. In relatively simple organisms (bacteria and yeast), this has allowed the systematic investigation of multiple basic biological processes conserved through evolution (e.g. apoptosis (4) and vacuolar transport (5)). Higher organisms are highly useful for the study of complex traits, which is facilitated by large collections of mutant strains (6, 7). This is of particular relevance where model systems of human physiology, either in a healthy or diseased state, are studied. Fruit flies and C. elegans, the “classical” model organisms, have been used as models for a variety of diseases (8) but also for natural processes like aging (9, 10), sleep (11, 12), and olfaction (13). Mouse and rat models have been a long-standing model for human biology (14), especially in cancer (15). Particularly the availability of strains engineered to represent human diseases has increased our understanding of pathological processes tremendously (16).
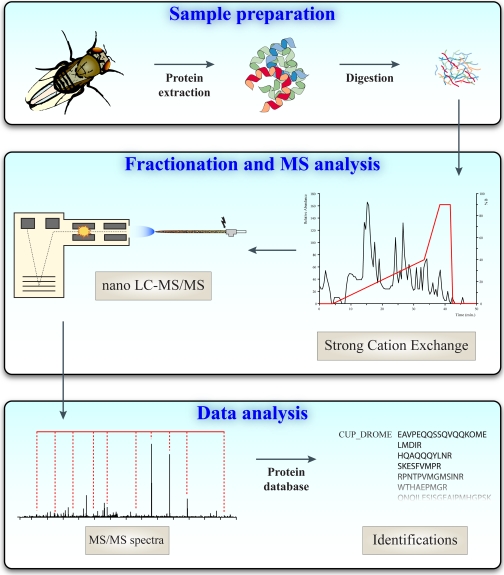
So far, the focus has primarily been on genetic and genomic aspects of these processes and disorders, but with the maturation of proteomics techniques, ways to study these at the protein level in a meaningful way are coming within reach. Over the last decade, proteomics research has experienced significant advances and has evolved into an indispensable technology to investigate the proteomic composition of biological samples. Proteomics has shifted from the analysis of small sets of proteins toward the comprehensive investigation of a much larger number of proteins expressed in a cell, tissue, or organism (17). Nowadays, a typical proteomics experiment is peptide-centric and starts with the enzymatic digestion of a protein mixture followed by fractionation using one or more chromatographic steps to reduce sample complexity (18, 19) as illustrated in Fig.1. Peptides are fragmented in the mass spectrometer as they elute, and subsequent matching of fragmentation profiles against a protein database leads to peptide and protein identification. When performed at a large scale, this can be used for the identification of thousands of proteins in cells or subcellular structures (20–23). Although such qualitative approaches are fruitful in providing information on proteins present in cells or tissues, they largely ignore the dynamics of protein expression when different conditions are to be investigated. This is highly relevant because in general only the proteins that differ between biological states (e.g. healthy/diseased) are likely to be of primary interest. Because mass spectrometry is not inherently quantitative, it is beneficial to add an internal standard as a reference for the peptide of interest. For large scale experiments, often all proteins or peptides in one sample are modified with a stable isotope-coded mass label. After mixing the labeled sample with an unmodified sample, the intensity ratio between the modified peptide and the unlabeled peptide accurately reflects the change in expression level.
Fig. 1.
Qualitative proteomics work flow. Proteins are extracted, digested, and separated by strong cation exchange. Each strong cation exchange fraction is then analyzed by nano-LC-MS/MS. Peptide fragment spectra are used in a database search to identify the peptide sequence and the corresponding protein.
Various approaches have been developed for the incorporation of stable isotopes into proteins that can be divided into in vivo and in vitro methods. In the former, isotope-enriched compounds (salts or amino acids) are added to the growth media that can be metabolized by the cell and incorporated into proteins. In vitro labeling can be established using chemical derivatization of proteins or peptides after protein extraction. The choice for either approach depends on the biological system under investigation, but there are a few considerations that should be taken into account because of their impact on the experimental work flow. In Table I, some of the strengths and weaknesses of both metabolic and chemical labeling methods are summarized.
Table I. Comparison of strengths and weaknesses of various metabolic (in vivo) and chemical (in vitro) labeling techniques.
ETD, electron transfer dissociation; ECD, electron capture dissociation; +, beneficial; −, hampering.
| Labeling method | Cost | Strengths | Weaknesses |
|---|---|---|---|
| Metabolic labeling (in vivo) | |||
| SILAC | + | Incorporation at the organism level (lowest variation). Available (free) quantitation software. | Not applicable to human samples. Arginine-to-proline conversion. Expensive and slow. Enzymes other than trypsin and/or Lys-N may produce non-quantifiable peptides. Auxotroph for the labeled amino acid(s). |
| 15N labeling | + | Incorporation at the organism level (lowest variation). All peptides can be used for quantitation regardless of the enzyme | Not applicable to human samples. Expensive and slow. Available quantitation software. Unknown mass difference prior to identification. |
| 13C labeling | + | Incorporation at the organism level (lowest variation). All peptides can be used for quantitation regardless of the enzyme. | Not applicable to human samples. Expensive and slow. Available quantitation software. Unknown mass difference prior to identification. Isotope distribution might hamper identification. |
| SMIRP | +/− | Incorporation at the organism level (lowest variation). All peptides can be used for quantitation regardless of the enzyme. | Not applicable to human samples. Slow. Available quantitation software. |
| Isotope-depleted labeling | + | Incorporation at the organism level (lowest variation). All peptides can be used for quantitation regardless of the enzyme. | Not applicable to human samples. Expensive and slow. Available quantitation software. Identification requires ECD or ETD. Quantitation at the protein level. |
| Chemical labeling (in vitro) | |||
| ICAT | +/− | Applicable to any sample. Fast. | Incorporation at the protein level (moderate variation). Only Cys-containing peptides can be used for quantitation. |
| ICPL | +/− | Applicable to any sample. Fast. | Incorporation at the protein level (moderate variation). Only Lys-containing peptides and the protein N terminus can be used for quantitation. Trypsin cleaves C-terminal to arginine residues only. |
| iTRAQ | +/− | Applicable to any sample. Fast. Simultaneous analysis of 8 labeled samples. No increase in complexity at the MS level. | Incorporation at the peptide level (high variation). Quantitation is based on 1 or a few tandem mass spectra. Requires mass spectrometers that can analyze the low m/z region. |
| 18O labeling | − | Applicable to any sample. Cheap and fast. | Incorporation at the peptide level (high variation). Difficult to reach complete labeling. Available quantitation software. |
| Dimethyl labeling | − | Applicable to any sample. Cheap and fast. Automation is possible. | Incorporation at the peptide level (high variation). Identification issues due to the number of variable modifications. |
One major consideration for labeling in vivo (metabolic) or in vitro (chemical) critically depends on whether the biological sample in question can metabolically incorporate the isotope label. Metabolic labeling requires the addition of an isotopically enriched element (e.g. 13C, 15N, or 18O in salts or amino acids) to the growth media in a form that makes it available for incorporation into the entire organism, tissue, or cell. In contrast, chemical labeling occurs after protein extraction and therefore is completely independent of the source and preparation of the sample. This has the advantage that virtually any type of biological sample can be labeled, including human tissue or body fluids. Additionally, the time needed for this type of labeling is in general much shorter than when a label is incorporated metabolically where it may take weeks to in vivo label organisms or cells depending on the growth rate. This can even increase to a few months if a secondary labeling step is required such as is the case in the 15N labeling procedure of fruit flies and worms by feeding them on labeled yeast and E. coli, respectively.
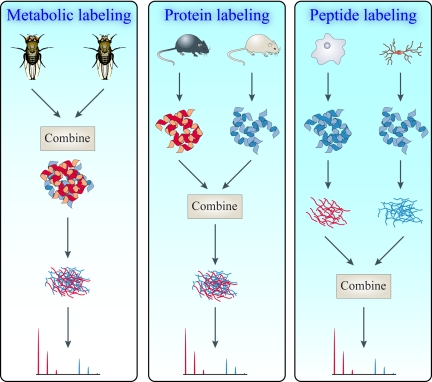
The great advantage of metabolic labeling becomes clear when the proteomics work flow is considered. Fig.2 gives an overview of the different positions in the experimental work flow where the internal standard can be introduced. Clearly, the best place to introduce an internal standard is by metabolically incorporating the stable isotope into living organisms or cells, thereby producing the lowest variation before any sample processing occurs (Fig. 2, left). When the internal standard is introduced further downstream in the work flow, higher levels of variation can be expected due to parallel sample processing as is the case with chemical derivatization of intact proteins (e.g. with ICAT and isotope-coded protein labeling (ICPL)1 (24, 25)) (Fig. 2, middle) or with chemical labeling of peptides such as isobaric tag for relative and absolute quantitation (iTRAQ) and stable isotope dimethyl labeling procedures (26–28) or proteolytic digestion in 18O-labeled water (29, 30) (Fig. 2, right).
Fig. 2.
Strategies for quantitative proteomics. Stable isotopes can be incorporated at different stages of the quantitative work flow and are indicated in black. The methods are metabolic labeling (left), protein labeling (middle), and peptide labeling (right). Relative expression levels are obtained by mass spectrometry where the signal of the unlabeled peptide is compared with that of the labeled peptide.
CHEMICAL AND LABEL-FREE APPROACHES FOR QUANTITATIVE PROTEOMICS
The advantage of using iTRAQ labeling over other (chemical) labeling strategies is the possibility to simultaneously analyze up to eight biological samples in one experiment by labeling peptides (primary amino groups) with isobaric tags that differ in reporter and balancer groups (26, 31). Quantitative information is obtained by comparing the unique reporter groups in the fragmentation spectrum, and therefore this labeling strategy is fully MS/MS-dependent, and quantitative information is only obtained from peptides that were subjected to fragmentation. The only other quantitative method that exclusively relies on fragmentation data is the label-free method of spectral counting. The abundance of a protein can be estimated by counting the number of sampling events of a peptide from this protein. It has been observed that the number of assigned MS/MS spectra directly correlates with protein abundance. In this approach, samples are analyzed and processed separately, and protein lists are compared in terms of sampling events per protein. Although spectral counting has proven to be very reliable at measuring large changes between proteins, reliability drops dramatically when smaller changes are estimated (32). In contrast, the other (chemical) labeling strategies are based on chromatographic intensities of intact peptide signals and do not require fragmentation for quantitation. However, not necessarily all peptides can be used for quantitation depending on the labeling strategy. For instance, in ICAT labeling only cysteine-containing peptides can be used for quantitation, and only lysine-containing peptides and protein N termini can be used in ICPL. Additionally, when trypsin is used as the proteolytic enzyme in ICPL, possibly longer peptides may be expected due to the modification of lysine residues. Trypsin will only cut after arginine residues, and the resulting longer peptides may be more difficult to identify. This is not the case for labeling strategies based on peptide labeling or label-free quantitation based on the ion intensity (MS mode) of peptides. Similarly to spectral counting, samples are analyzed and processed separately, but this strategy relies on extracting mass spectrometric peak areas for all peptides that are subsequently integrated for their respective retention times (33). A more detailed description of chemical and label-free approaches in quantitative proteomics and a comparison between them are not the focus of this perspective, especially as a number of excellent reviews have been published in the last few years covering these subjects (17, 34–37). Instead, we will focus specifically on strategies using metabolic incorporation of mass labels in vivo in whole organisms.
METABOLIC LABELING FOR QUANTITATIVE PROTEOMICS
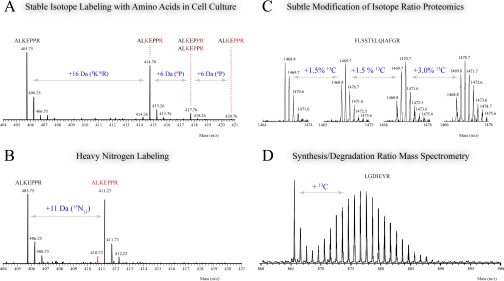
Recently, the development of strategies to label cells in culture with stable isotopes (stable isotope labeling with amino acids in cell culture (SILAC)) has found broad application and continues to expand to a wide range of cell lines. A number of first rate studies have been published (38, 39) and reviewed elsewhere (35, 40) that show the power of SILAC. Nevertheless, some limitations of this labeling procedure should be considered. For instance, the cells or organism should be auxotroph for the labeled amino acid(s), either by mutations or by using essential amino acid(s), to ensure that the labeled amino acid is the only source for protein synthesis. The widespread metabolic conversion of arginine to proline is an undesired side effect when arginine is used in SILAC labeling (41, 42). This effect is illustrated in Fig.3A where a mass spectrum shows the desired unlabeled and double labeled peptide (+16 Da, heavy lysine and arginine) but also additional unwanted peaks at positions that correspond to incorporation of single (+6 Da) and double (+12 Da) labeled proline residues in the peptide. A significant amount of signal can reside in these peaks and can negatively affect quantitation. This phenomenon is not apparent in the case of metabolic 15N labeling using salts and can be visualized by comparing the mass spectrum in Fig. 3A with that in Fig. 3B where the same peptide was 15N-labeled (11 nitrogen atoms, +11 Da). Although the arginine-to-proline conversion is most likely still taking place, the effect is eliminated because in this type of labeling the mass of the proline-converted peptide is similar to the mass of the precursor peptide. However, the mass difference between an unlabeled and 15N-labeled peptide is a priori unknown during mass spectrometric analysis and becomes apparent only after identification. This might hamper sophisticated acquisition software that only selects regulated peptides for fragmentation and/or automated quantification prior to identification. When using 15N, suboptimal labeling (exemplified by the red colored isotope at m/z 410.73 in Fig. 3B) is a recurrent issue that should be accounted for to obtain correct abundance ratios (43). Nevertheless, the procedure of 13C or 15N substitution in living organisms is very attractive because little to no side effects (cytologically or morphologically) have been reported (44, 45). Moreover, it is not essential to achieve complete labeling. The incorporation of for instance 1.5% 13C, a strategy termed subtle modification of isotope ratio proteomics (SMIRP) (Fig. 3C), is already enough to obtain good quantitative data, but automated quantitation is more difficult because of the lack of available software. Another application of incomplete labeling is to determine protein synthesis and degradation by monitoring the incorporation of labeled 13C in a time-dependent manner (Fig.3D). Also, in these cases, the lack of suitable software hampers data processing so far.
Fig. 3.
Overview of typical mass spectra that are obtained from different metabolic labeling strategies. In SILAC (A), labeled arginine can be converted to proline, indicated in red in the peptide sequence, resulting in peaks that are 6 Da higher than the labeled peptide. If such peaks are formed in heavy nitrogen labeling (B), they appear at the same position as the 15N-labeled peptide because they are isobaric. The red colored isotope at m/z 410.73 is a product of suboptimal 15N labeling. Minor enrichment of 13C in SMIRP leads to quantifiable peptide signals (C), whereas higher incorporation is used to determine protein synthesis and degradation (D).
The availability of software solutions for the analysis of quantitative proteomics data generated by LC-MS is extensive but still limited. This is caused by the fact that only a few solutions might be available for a specific type of labeling analyzed on a particular mass spectrometer. Software tools to analyze quantitative LC-MS data are usually developed for specific types of mass spectrometers, thereby restricting these tools to analyze data from only those instruments. Some labeling strategies such as SMIRP and isotope-depleted labeling even lack software solutions to automatically perform quantitation, whereas others (15N and 13C labeling) seem restricted to a few available software tools. An overview of software solutions has been published (46).
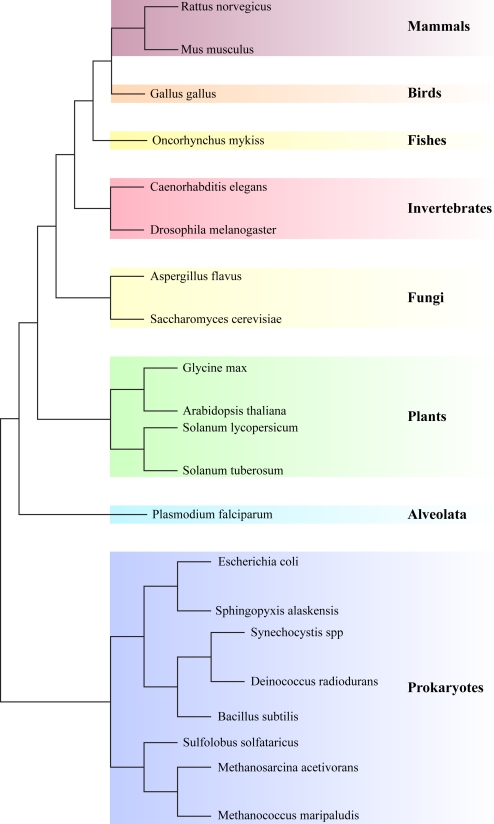
In recent years, increasing numbers of model organisms are being used for comparative proteomics, and various methodologies have been developed to metabolically label them. Shown in Fig.4 is a compendium of the current “tree of metabolically labeled life,” and it can be seen that species in almost any branch can be and have been labeled. Table II gives a more detailed overview of these species together with the type of metabolic labeling applied. Quantitative proteomics in (model) organisms, either by metabolic or chemical labeling, can be extremely powerful to elucidate biological processes. Most of these studies focus on the effect of an isolated treatment (e.g. stress condition or growth factor) of cells grown in a Petri dish, which should be regarded as a somewhat artificial system because cells have often been maintained for thousands of cell doublings over many years. Moreover, any cell system neglects the differences in responses that would normally occur in an organism (because of the nature of the cells or organs) or communication between them. If the aim is to understand cells in their natural environment, one should aim to perform studies in the intact organism. There is no doubt that quantitative approaches will be essential in uncovering relevant players, and consequently it is important to develop quantitative proteomics methods in model organisms. Therefore, here we present an overview of intact model organisms that have been metabolically labeled for the purpose of quantitative proteomics. Per species we provide the current status of the methodology as well as examples of their application to biological problems.
Fig. 4.
Tree of metabolically labeled life. Branch lengths are not proportional to evolutionary distance.
Table II. Overview of studies using metabolically labeled organisms depicting labeling strategy and corresponding references.
| Class and species | SILAC | 15N | 13C | Refs. |
|---|---|---|---|---|
| Prokaryotes | ||||
| E. coli | ✓ | ✓a | ✓a | 47, 56, 60–64 |
| D. radiodurans | ✓ | 49 | ||
| Synechocystis spp. | ✓ | ✓b | 51 | |
| S. solfataricus | ✓ | ✓ | 52–54 | |
| M. maripaludis | ✓ | 55 | ||
| M. acetivorans | ✓ | 58 | ||
| B. subtilis | ✓ | ✓ | 59 | |
| S. alaskensis | ✓ | 57 | ||
| Alveolata | ||||
| Plasmodium falciparum | ✓ | 146 | ||
| Plants | ||||
| G. max | ✓ | 101 | ||
| S. tuberosum | ✓ | 102 | ||
| A. thaliana | ✓ | ✓ | 105– | |
| S. lycopersicum | ✓ | 114 | ||
| Fungi | ||||
| S. cerevisiae | ✓ | ✓ | 23, 71–83, 85, 87–90 | |
| A. flavus | ✓ | 95, 97 | ||
| Drosophila and C. elegans | ||||
| D. melanogaster | ✓ | 43, 126, 128, 129 | ||
| C. elegans | ✓ | 43, 126, 127 | ||
| Fishes | ||||
| Oncorhynchus mykiss | ✓ | 144 | ||
| Birds and mammals | ||||
| G. gallus | ✓ | 130 | ||
| Rattus norvegicus | ✓ | 131–133 | ||
| M. musculus | ✓ | 134 |
OVERVIEW OF METABOLICALLY LABELED SPECIES
Prokaryotes
The prokaryotes E. coli, Deinococcus radiodurans, and Synechocystis spp. were initially used to establish different 15N metabolic labeling methodologies. In one of the first studies reported to accomplish this, unstressed and Cd2+-stressed E. coli were grown on normal and heavy isotope (13C, 15N, and 2H)-depleted media, respectively (47). After mixing aliquots from both cultures, relative expression levels of intact proteins were determined by fitting the experimental results with the predicted shapes of the calculated isotopic envelopes. Although this procedure readily lends itself for the determination of relative expression levels, identifying these proteins is somewhat more complicated and requires more sophisticated fragmentation methods such as electron capture dissociation and electron transfer dissociation to obtain sequence information for identification purposes (48). To overcome this identification issue, proteins can be digested into peptides for both quantitative and qualitative information. This procedure was developed by growing D. radiodurans on fully 15N-enriched (>98%) media after which proteins were digested into peptides to facilitate protein identification and quantitation (49, 50). A third method to determine protein expression levels uses the labeling of proteins with a subtle enrichment (∼1–2%) of a stable isotope instead of using fully enriched (>8%) labeled media. This approach, referred to as SMIRP, has been used to label Synechocystis spp. with various 13C/12C isotope ratios to determine the feasibility of this approach. Optimal results were obtained with enrichments as low as ∼1–2% 13C that have no effect on either data-dependent acquisition or database searching algorithms but yield a measurable effect on peptide isotopic distribution of which an isotope ratio can be inferred (51). A method similar to the labeling procedure of D. radiodurans (i.e. using fully enriched heavy nitrogen) included the incorporation of stable isotope-labeled 13C atoms in the hyperthermophilic crenarchaeon Sulfolobus solfataricus. In this approach, three different versions of the same protein are produced (unlabeled, 15N-, and 13C-labeled), allowing for the analysis of three different samples in a single experiment (52). The same group also investigated the applications and limitations of 15N labeling and showed that stable isotope labeling with heavy nitrogen in combination with mass spectrometric detection provides an excellent tool for studying protein dynamics (53, 54). This was further corroborated in another study where the archaeon Methanococcus maripaludis was 15N-labeled, and protein expression levels were validated using real time PCR (55). An additional benefit of 15N labeling is its use to increase the number and confidence of protein identifications. In 15N labeling, the number of nitrogen atoms in a peptide can easily be determined from the mass difference between labeled and unlabeled peptide pairs, which can be used as an additional criterion to accept or refute peptide identification (56). Recently, Sphingopyxis alaskensis has been 15N-labeled to investigate normalization and statistical analysis in quantitative proteomics-generated data (57). Two of the first applications utilizing metabolic labeling of a prokaryote included 15N labeling and SILAC or a combination thereof. In one study, Methanosarcina acetivorans was 15N-labeled to investigate acetate versus methanol growth conditions (58). In the other study, changes in the membrane proteome during stationary phase adaptation of Bacillus subtilis were monitored, and both techniques showed similar valuable data for quantification of bacterial membrane proteins (59). The SILAC procedure has also extensively been investigated and applied to E. coli cells. Several different labeled amino acids were used to metabolically label E. coli, including lysine (60, 61), leucine (61–63), glycine (61), and methionine (61, 64).
Fungi
S. cerevisiae
The yeast S. cerevisiae fulfills an important dual role, being an industrially applied organism in itself while serving as one of the prime model organisms for higher eukaryotes. First, this microorganism is extensively used in industry as it has the ability to produce for instance alcohol and carbon dioxide. In addition, it can be used as a host for the production of proteins and small molecules exploited by pharmaceutical companies for the production of insulin or penicillin (65). Second, yeast has been used for a long time as a model organism in the field of molecular biology. Properties such as the rapid adaptation and easy growth under numerous conditions, its lack of pathogenicity, and the availability of powerful genetic manipulation techniques make this organism perfectly suited for laboratory experiments. In addition, it was the first eukaryote to have its genome sequenced in 1996, resulting in the prediction of ∼6000 protein-coding genes (66). Since then, large amounts of data based on genome, transcriptome, proteome, and metabolome studies have been generated. A great deal of this information is curated, stored, and managed in the on-line database Saccharomyces Genome Database (67). In the postgenomics era, information from studies at different genomic levels is combined in a systems biology approach, resulting in a comprehensive understanding of eukaryotic cell biology. In this context, it is expected that yeast will continue to provide new knowledge and insights (68–70).
Metabolic labeling for quantitative proteomics is extensively applied in yeast and can be accomplished by two different approaches. Stable isotope-labeled atoms such as 15N can be incorporated by growing yeast in media containing labeled ammonium sulfate as the sole nitrogen source (71). The other method involves the incorporation of one or more stable isotope-labeled amino acids into the proteome of yeast. The former methodology was used in 1999 by Oda et al. (72) to uniformly 15N label yeast, which they used to investigate protein expression as well as site-specific changes in phosphorylation levels. They reported that relatively small changes in phosphorylation (>20%) can be reliably detected from small amounts of gel-separated proteins (72). Yates and co-workers (73–78) extensively used 15N-labeled yeast to establish methodologies for the analysis of quantitative proteomic samples as well as to address biological questions. For instance, by combining metabolic labeling with multidimensional protein identification technology, they described a system useful for detailed quantitative proteomics analysis (73) that they used to investigate the correlation between mRNA and protein expression levels (74). This approach was complemented with a correlation algorithm called RelEx for the automated analysis of quantitative data (75) and an algorithm called The Atomizer to determine isotope enrichment levels (76). Both approaches were validated using complex mixtures with known enrichment levels of 15N-labeled yeast, and moreover, the RelEx algorithm was applied to the study of NaCl osmotic stress on the protein level in yeast. An alternative data acquisition method that was developed using 15N-labeled yeast by Yates and co-workers (77) relies ondata-independent analysis where quantitative results are determined directly from tandem mass spectra (MS2) using a modified version of RelEx. In addition, this group also evaluated the use of an LTQ-Orbitrap hybrid mass spectrometer for quantitative analysis using labeled yeast (78). Other groups that focused on more fundamental research using 15N-labeled yeast investigated top-down quantitative proteomics approaches (79) and the correlation between spectral counting and metabolic labeling (80, 81). Besides this important basic research, biological effects like response to nutrient limitations (82), adaptation to anaerobiosis (83, 84), or phosphorylation level changes related to glucose activation (85) were studied using 15N-labeled yeast.
The other metabolic labeling strategy established in yeast involves the incorporation of one or more labeled amino acids. The first studies describing such an approach used deuterium-labeled amino acids such as triple deuterated methionine (Met-d3) and serine (Ser-d3) and double deuterated tyrosine (Tyr-d2) (86). Deuterated leucine was also used, but it differed in the number of deuterium atoms in the amino acid. In one case, triple deuterated leucine (Leu-d3) was used (87), whereas in the other case, decadeuterated leucine (Leu-d10) was used (88). In both cases, a yeast strain was used that is auxotrophic for leucine, ensuring that all leucine was replaced by labeled leucine. Once established, the Leu-d10 metabolic labeling approach was used to investigate the response of the yeast proteome to H2O2 (89). Besides labeled leucine, also stable isotope-labeled arginine and lysine were used to label yeast. A double auxotroph strain was used to uniformly label yeast to study the changes in pheromone-induced phosphorylation (90). However, a disadvantage of using labeled arginine in eukaryotes is that the accuracy of peptide ratio calculation is compromised by the metabolic conversion of arginine to proline, which seems to be quite efficient in yeast (41, 42). Several solutions have been published (91, 92) to correct for this effect to ensure accurate quantitation. A study that used labeled lysine only focused on determining factors that prevent complete proteome analysis (23). The sequencing speed of the mass spectrometer was found as one of these factors. The sequencing speed was determined by comparing the number of identified “light” and “heavy” peptides. If the sequencing speed is sufficiently high, both forms of a SILAC pair should be sequenced and identified, but this was shown to be true only for the more abundant peptides. The other factor that limited complete proteome coverage was the restricted dynamic range of the mass spectrometer compared with the dynamic range of the yeast proteome (i.e. 100 versus 10,000) (93). The dynamic range of the mass spectrometer was determined by comparing the intensity of the most abundant SILAC pair with the least abundant pair in the same mass spectrum (23). Recently, the same group investigated and quantified the proteomes of haploid and diploid yeast. By using the same labeling strategy in combination with several different fractionation techniques, they increased the number of identified proteins to a level of 4,399 proteins. In addition, they showed that mass spectrometry is as sensitive as other protein detection techniques such as green fluorescent protein and tandem affinity purification tagging (94).
Aspergillus flavus
Recently, two studies were published that described the SILAC labeling of the fungus A. flavus by growing this multicellular prototroph on media containing heavy arginine. This amino acid was chosen over lysine because it occurs at a frequency of about 6% in the proteome (95). By determining the incorporation efficiency of 50 proteins, they found an average enrichment level of 78%, which is similar to the enrichment level of 81% reported by the SILAC labeling of Arabidopsis suspension cells (96) (see above). This suboptimal enrichment level was sufficient to quantify protein changes in response to environmental stimuli regulating biosynthesis of the carcinogen aflatoxin (95). Another study by the same group, however, showed that this suboptimal enrichment hampered the quantification of intact proteins that have a large number of arginine residues. Here, they established a quantitative top-down proteomics approach using this labeled fungus and showed that proteins with few arginines can readily be identified and quantified (97). However, they suggest to reduce endogenous amino acid incorporation by using an arginine auxotroph strain, thereby facilitating straightforward quantification of proteins that contain larger number of arginines (97).
Plants
Metabolic labeling of plants dates back to the mid-sixties of the last century where labeled nitrogen (in the form of ammonium sulfate) was added in tracer amounts to a fertilizer and used to evaluate the efficiency of fertilizer applications in rice (Oryza sativa) (98). Metabolic incorporation of labeled nitrogen was accomplished by growing rice hydroponically (i.e. using mineral nutrient solutions instead of soil) and used to monitor how the heavy nitrogen accumulated into specific parts of the plant. Although only ∼30% enriched labeled nitrogen was used ((15NH4)2SO4 and K15NO3) to investigate the uptake and distribution of nitrogen, it was sufficient to spectroscopically determine the amount of heavy nitrogen in different organs of the rice plant (99). The same method was used to label the potato plant (Solanum tuberosum) (100). Likely the first report of a highly enriched, uniformly labeled plant was published in 1994 where soybean plants (Glycine max) were labeled with ∼98% 15N to investigate nutrient absorption and metabolism in human or animal studies (101). Next, potato plants were uniformly 15N-labeled (>98%) for the purpose of structural proteomics (102). Intact potato plants were obtained by growing aseptic seed potato tubers using a hydroponics setup for a period of 93 days in a climate-controlled greenhouse on a nutrient solution containing 99% potassium nitrate (K15NO3) as the sole nitrogen source. Although S. tuberosum was the first plant to be fully 15N-labeled, Arabidopsis thaliana is considered the prime model species in plant biology, and the genome of this plant was the first plant genome to be sequenced (103). Complemented with methods to incorporate stable isotopes in vivo, this model has facilitated mass spectrometry-based quantitative proteomics applied to plant biology.
The genome of the flowering plant Arabidopsis is one of the smallest among plants and was sequenced in 2000. It was concluded that this plant has 11,000–15,000 protein families, a number that is similar to other multicellular eukaryotes like C. elegans and Drosophila (103). The current estimate is that the Arabidopsis genome contains ∼27,000 genes encoding 35,000 proteins, but these numbers still change continuously due to combined efforts by The Arabidopsis Information Resource that updates the Arabidopsis genome annotation annually (104). Arabidopsis is an autotrophic species synthesizing all amino acids from inorganic nitrogen. This makes it particularly difficult to metabolically incorporate labeled amino acids into the proteome of the plant at high efficiency. However, labeling enrichments of 70–80% have been reported when suspension cells were grown in the presence of exogenously supplied heavy arginine for 7 days (96). In this experiment, SILAC was used to identify regulated glutathione S-transferases and 14-3-3 proteins in response to treatment with salicylic acid. Disadvantages of this approach include incomplete labeling; the use of only one labeled amino acid, reducing the number of quantifiable peptides; and expensive labeled amino acids that are needed to reach efficient enrichments. This method is therefore limited to plant cell culture. Alternatively, these drawbacks can be circumvented by growing cell cultures on media containing highly enriched 15N as the sole nitrogen source (105–108). Typically, cells were grown for around 21 days in modified liquid media containing >98% 15N, and complete uniform incorporation was achieved. No detrimental isotopic effects were observed because the morphology and growth rate of the 15N-labeled cells were indistinguishable from their 14N counterparts (105). In some cases inverse labeling was performed to confirm results (107, 108). Besides proteomics, alsometabolomics can be studied using this type of labeling, but this application is evidently limited to metabolites that contain nitrogen (105, 106). Labeling with heavy carbon (13C) is therefore considered to be more efficient for this purpose.
As for the labeling of cell cultures, heavy nitrogen has been used as the prime method to metabolically label intact plants in vivo with high levels of enrichment. The first publication of labeling of intact Arabidopsis for proteomics experiments was in 2007 (109). Since then, several different 15N labeling techniques have been investigated that include the comparison of partial versus full labeling (110), the automated analysis of uniformly labeled proteins using Mascot peptide identification in conjunction with the trans-proteomic pipeline (111), and a procedure referred to as hydroponic isotope labeling of entire plants (112). A notable aspect in the method to metabolically incorporate heavy nitrogen in intact plants is that almost all proteomics studies on Arabidopsis grow plants in liquid media. It can be argued that biologically meaningful results can only be obtained when plants are grown on solid medium, thereby simulating growth conditions as natural as possible. Leaf senescence was investigated in Arabidopsis by growing the plants on solid labeled and unlabeled media (113). Recently, the approach known as stable isotope labeling in planta was published where the tomato plant Solanum lycopersicum was 15N-labeled by growing it for 2 months on solid media (114). Besides metabolic labeling, most other quantitative proteomics methods such as iTRAQ, ICAT, and 18O labeling have been established in plants as well. For an overview of these techniques and their application in plants see Thelen and Peck (115).
D. melanogaster and C. elegans
Since the introduction of the nematode C. elegans and the fruit fly D. melanogaster as model organisms in biological research (1, 116, 117), many fundamental biological principles have been disclosed using these species. They were the first multicellular organisms to have their genomes sequenced in 1998 and 1999, respectively (118, 119), and nowadays, truly outstanding resources are available for both species (120, 121) such as the on-line databases Wormbase (122) and Flybase (123). The cellular complexity and the conservation of biological pathways between these invertebrates and higher organisms, including humans, opened the door for using them as tools to study comparative human genetics (7, 8, 124, 125). Indeed, many biomedical discoveries were fueled by C. elegans and D. melanogaster research. Of note, the processes underlying diabetes and Alzheimer disease were first discovered in C. elegans (for a review, see Ref. 8).
Although quantitative genetics and functional genomics have been established in these model organisms for some time now, quantitative proteomics based on metabolic labeling is only beginning to be introduced. Flies and worms can be metabolically 15N-labeled by feeding them on uniformly 15N-labeled S. cerevisiae and E. coli, respectively (126). Whereas flies are completely labeled after one generation, worms need to be grown on labeled media for at least one more generation after which second generation worms are completely labeled. However, due to different protein turnover rates in different tissues, it is necessary to analyze the enrichment of different proteins to ensure complete labeling. Labeled worms have been used to identify targets of insulin signaling (127), and labeled flies have been used to identify novel seminal fluid proteins (128) and proteins involved in the maternal-to-zygotic transition (129). In addition, both species have been used to optimize the identification and quantitation of 15N-labeled proteins in comparative proteomics (43). The method for metabolic labeling of both Drosophila and C. elegans should be easily adoptable by many fly and worm laboratories because it requires only minor adaptations compared with routine protocols for growing flies and worms. Therefore, we expect this approach to find broad application in fly and worm developmental biology and beyond.
Birds and Mammals
One of the first reports of stable isotope labeling in birds involved the partial SILAC labeling of the chicken Gallus gallus. A diet that consisted of 50% labeled valine was fed to 6-day-old chickens to monitor the incorporation of the heavy amino acid and thereby facilitate the determination of protein turnover rates. Although these chickens are not fully labeled, such an approach allows for the accurate detection of protein turnover rates in vivo (130). Wu et al. (131) even extended the metabolic 15N labeling approach to mammals by labeling rats using a diet source that was supplemented with 15N-enriched (>99%) algal cells. After feeding the rats for 44 days, enrichment levels ranged from 74 to 92% depending on the type of tissue (131). Using an improved labeling strategy, they increased the enrichment to 94% throughout all tissues of the rat, and these tissues can serve as internal standards to facilitate the quantitative proteomics analyses of complex mammalian tissue samples (132). For instance, labeled rat brain was used to investigate the synaptosomal proteome of the rat cerebellum during postnatal development (133). In addition to labeled rats, other mammals such as mice can be metabolically labeled by using a SILAC diet where second generation mice showing complete labeling without obvious effects on growth, behavior, or fertility were observed (134). In these experiments, 13C-labeled lysine was used; it is an essential amino acid, thus preventing synthesis from other (unlabeled) sources of amino acids. As a result, it is not straightforward to use trypsin as the preferred protease because this enzyme produces both C-terminal arginine and lysine peptides of which only the lysine-containing peptides can be used for quantitation. The endoprotease Lys-C, in contrast, produces only peptides that have a C-terminal lysine, making Lys-C the preferred choice as the proteolytic enzyme. For such studies, the recently introduced protease Lys-N may provide a very elegant alternative in the future (135, 136).
A limitation to the use of labeled rats or mice could be the investment that is required to produce isotope-enriched offspring. However, the amount of protein that can be collected from basically every tissue, even from a single animal, is sufficient for hundreds of proteomics experiments. Furthermore, if this material is produced from a wild type, commonly used strain it could be distributed and used as an internal standard even in facilities without resources to label these animals themselves.
OUTLOOK
Although stable isotope labeling has been achieved across many species in almost all branches of life (Fig. 4), there are still some model organisms where the introduction of a labeling strategy would be very useful. For example, Danio rerio (zebrafish) has been established as an important model organism to study vertebrate biology (137, 138), especially in immunology (139), cancer (140), neurological disorders (141), and toxicology (142). This research is facilitated by the development of excellent genetics techniques in zebrafish such as the targeted knockdown of genes by morpholino antisense technology (143). There are several reasons accounting for the fact that this organism has not been metabolically labeled yet. Notably, the extensive developmental period and the absence of a labeled food source limit straightforward metabolic labeling. However, metabolic labeling of fish is not unprecedented with the incorporation of 15N into rainbow trout to study protein turnover (144).
The last but certainly not least important species that has not been metabolically labeled so far is humans. Up to now, SILAC approaches to metabolically label human cell lines have been firmly established (40), but the next logical step would be to extend this to human tissue. Tissue or organs grown in the laboratory from stem cells may become a relevant source for such experiments (145). However, the holy grail of quantitative proteomics is to metabolically incorporate stable isotopes into intact humans. Heavy nitrogen would be an excellent candidate because there are delightful products for everybody including vegetarians. The menu consists of plants, rice, potatoes, insects, and rodents, but would we find volunteers?
Footnotes
* This work was supported by the Netherlands Proteomics Centre.
1 The abbreviations used are:
- ICPL
- isotope-coded protein labeling
- iTRAQ
- isobaric tag for relative and absolute quantitation
- SILAC
- stable isotope labeling with amino acids in cell culture
- SMIRP
- subtle modification of isotope ratio proteomics
REFERENCES
- 1.Bridges C. B. (1914) Direct proof through non-disjunction that the sex-linked genes of Drosophila are borne by the X-chromosome. Science 40, 107–109 [DOI] [PubMed] [Google Scholar]
- 2.Lederberg J., Tatum E. L. (1946) Gene recombination in Escherichia coli. Nature 158, 558–558 [DOI] [PubMed] [Google Scholar]
- 3.Hedges S. B. (2002) The origin and evolution of model organisms. Nat. Rev. Genet. 3, 838–849 [DOI] [PubMed] [Google Scholar]
- 4.Hamann A., Brust D., Osiewacz H. D. (2008) Apoptosis pathways in fungal growth, development and ageing. Trends Microbiol. 16, 276–283 [DOI] [PubMed] [Google Scholar]
- 5.Bowers K., Stevens T. H. (2005) Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1744, 438–454 [DOI] [PubMed] [Google Scholar]
- 6.Hirschhorn J. N., Daly M. J. (2005) Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 6, 95–108 [DOI] [PubMed] [Google Scholar]
- 7.Mackay T. F., Anholt R. R. (2006) Of flies and man: Drosophila as a model for human complex traits. Annu. Rev. Genomics Hum. Genet. 7, 339–367 [DOI] [PubMed] [Google Scholar]
- 8.Kaletta T., Hengartner M. O. (2006) Finding function in novel targets: C. elegans as a model organism. Nat. Rev. Drug Discov. 5, 387–398 [DOI] [PubMed] [Google Scholar]
- 9.Schaffitzel E., Hertweck M. (2006) Recent aging research in Caenorhabditis elegans. Exp. Gerontol. 41, 557–563 [DOI] [PubMed] [Google Scholar]
- 10.Grotewiel M. S., Martin I., Bhandari P., Cook-Wiens E. (2005) Functional senescence in Drosophila melanogaster. Ageing Res. Rev. 4, 372–397 [DOI] [PubMed] [Google Scholar]
- 11.Cirelli C., Bushey D. (2008) Sleep and wakefulness in Drosophila melanogaster. Ann. N.Y. Acad. Sci. 1129, 323–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaw P. J., Cirelli C., Greenspan R. J., Tononi G. (2000) Correlates of sleep and waking in Drosophila melanogaster. Science 287, 1834–1837 [DOI] [PubMed] [Google Scholar]
- 13.Vosshall L. B., Stocker R. F. (2007) Molecular architecture of smell and taste in Drosophila. Annu. Rev. Neurosci. 30, 505–533 [DOI] [PubMed] [Google Scholar]
- 14.Peters L. L., Robledo R. F., Bult C. J., Churchill G. A., Paigen B. J., Svenson K. L. (2007) The mouse as a model for human biology: a resource guide for complex trait analysis. Nat. Rev. Genet. 8, 58–69 [DOI] [PubMed] [Google Scholar]
- 15.Frese K. K., Tuveson D. A. (2007) Maximizing mouse cancer models. Nat. Rev. Cancer 7, 645–658 [DOI] [PubMed] [Google Scholar]
- 16.Pearson T., Greiner D. L., Shultz L. D. (2008) Humanized SCID mouse models for biomedical research. Curr. Top. Microbiol. Immunol. 324, 25–51 [DOI] [PubMed] [Google Scholar]
- 17.Aebersold R., Mann M. (2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 18.Han X., Aslanian A., Yates J. R., 3rd (2008) Mass spectrometry for proteomics. Curr. Opin. Chem. Biol. 12, 483–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Motoyama A., Yates J. R., 3rd (2008) Multidimensional LC separations in shotgun proteomics. Anal. Chem. 80, 7187–7193 [DOI] [PubMed] [Google Scholar]
- 20.Van Hoof D., Passier R., Ward-Van Oostwaard D., Pinkse M. W., Heck A. J., Mummery C. L., Krijgsveld J. (2006) A quest for human and mouse embryonic stem cell-specific proteins. Mol. Cell. Proteomics 5, 1261–1273 [DOI] [PubMed] [Google Scholar]
- 21.Brunner E., Ahrens C. H., Mohanty S., Baetschmann H., Loevenich S., Potthast F., Deutsch E. W., Panse C., de Lichtenberg U., Rinner O., Lee H., Pedrioli P. G., Malmstrom J., Koehler K., Schrimpf S., Krijgsveld J., Kregenow F., Heck A. J., Hafen E., Schlapbach R., Aebersold R. (2007) A high-quality catalog of the Drosophila melanogaster proteome. Nat. Biotechnol. 25, 576–583 [DOI] [PubMed] [Google Scholar]
- 22.Zhai B., Villén J., Beausoleil S. A., Mintseris J., Gygi S. P. (2008) Phosphoproteome analysis of Drosophila melanogaster embryos. J. Proteome Res. 7, 1675–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Godoy L. M., Olsen J. V., de Souza G. A., Li G., Mortensen P., Mann M. (2006) Status of complete proteome analysis by mass spectrometry: SILAC labeled yeast as a model system. Genome Biol. 7R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gygi S. P., Rist B., Gerber S. A., Turecek F., Gelb M. H., Aebersold R. (1999) Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 17, 994–999 [DOI] [PubMed] [Google Scholar]
- 25.Schmidt A., Kellermann J., Lottspeich F. (2005) A novel strategy for quantitative proteomics using isotope-coded protein labels. Proteomics 5, 4–15 [DOI] [PubMed] [Google Scholar]
- 26.Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 27.Hsu J. L., Huang S. Y., Chow N. H., Chen S. H. (2003) Stable-isotope dimethyl labeling for quantitative proteomics. Anal. Chem. 75, 6843–6852 [DOI] [PubMed] [Google Scholar]
- 28.Lemeer S., Jopling C., Gouw J., Mohammed S., Heck A. J., Slijper M., den Hertog J. (2008) Comparative phosphoproteomics of zebrafish Fyn/Yes morpholino knockdown embryos. Mol. Cell. Proteomics 7, 2176–2187 [DOI] [PubMed] [Google Scholar]
- 29.Mirgorodskaya O. A., Kozmin Y. P., Titov M. I., Körner R., Sönksen C. P., Roepstorff P. (2000) Quantitation of peptides and proteins by matrix-assisted laser desorption/ionization mass spectrometry using (18)O-labeled internal standards. Rapid Commun. Mass Spectrom. 14, 1226–1232 [DOI] [PubMed] [Google Scholar]
- 30.Schnölzer M., Jedrzejewski P., Lehmann W. D. (1996) Protease-catalyzed incorporation of 18O into peptide fragments and its application for protein sequencing by electrospray and matrix-assisted laser desorption/ionization mass spectrometry. Electrophoresis 17, 945–953 [DOI] [PubMed] [Google Scholar]
- 31.Choe L., D'Ascenzo M., Relkin N. R., Pappin D., Ross P., Williamson B., Guertin S., Pribil P., Lee K. H. (2007) 8-plex quantitation of changes in cerebrospinal fluid protein expression in subjects undergoing intravenous immunoglobulin treatment for Alzheimer's disease. Proteomics 7, 3651–3660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Old W. M., Meyer-Arendt K., Aveline-Wolf L., Pierce K. G., Mendoza A., Sevinsky J. R., Resing K. A., Ahn N. G. (2005) Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics 4, 1487–1502 [DOI] [PubMed] [Google Scholar]
- 33.Ono M., Shitashige M., Honda K., Isobe T., Kuwabara H., Matsuzuki H., Hirohashi S., Yamada T. (2006) Label-free quantitative proteomics using large peptide data sets generated by nanoflow liquid chromatography and mass spectrometry. Mol. Cell. Proteomics 5, 1338–1347 [DOI] [PubMed] [Google Scholar]
- 34.Gevaert K., Impens F., Ghesquière B., Van Damme P., Lambrechts A., Vandekerckhove J. (2008) Stable isotopic labeling in proteomics. Proteomics 8, 4873–4885 [DOI] [PubMed] [Google Scholar]
- 35.Ong S. E., Mann M. (2005) Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 1, 252–262 [DOI] [PubMed] [Google Scholar]
- 36.Bantscheff M., Schirle M., Sweetman G., Rick J., Kuster B. (2007) Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 389, 1017–1031 [DOI] [PubMed] [Google Scholar]
- 37.Bachi A., Bonaldi T. (2008) Quantitative proteomics as a new piece of the systems biology puzzle. J. Proteomics 71, 357–367 [DOI] [PubMed] [Google Scholar]
- 38.Andersen J. S., Lam Y. W., Leung A. K., Ong S. E., Lyon C. E., Lamond A. I., Mann M. (2005) Nucleolar proteome dynamics. Nature 433, 77–83 [DOI] [PubMed] [Google Scholar]
- 39.Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 40.Ong S. E., Mann M. (2006) A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat. Protoc. 1, 2650–2660 [DOI] [PubMed] [Google Scholar]
- 41.Everley P. A., Bakalarski C. E., Elias J. E., Waghorne C. G., Beausoleil S. A., Gerber S. A., Faherty B. K., Zetter B. R., Gygi S. P. (2006) Enhanced analysis of metastatic prostate cancer using stable isotopes and high mass accuracy instrumentation. J. Proteome Res. 5, 1224–1231 [DOI] [PubMed] [Google Scholar]
- 42.Ong S. E., Kratchmarova I., Mann M. (2003) Properties of 13C-substituted arginine in stable isotope labeling by amino acids in cell culture (SILAC). J. Proteome Res. 2, 173–181 [DOI] [PubMed] [Google Scholar]
- 43.Gouw J. W., Tops B. B., Mortensen P., Heck A. J., Krijgsveld J. (2008) Optimizing identification and quantitation of 15N-labeled proteins in comparative proteomics. Anal. Chem. 80, 7796–7803 [DOI] [PubMed] [Google Scholar]
- 44.Meselson M., Stahl F. W. (1958) The replication of DNA in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A 44, 671–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uphaus R. A., Flaumenhaft E., Katz J. J. (1967) A living organism of unusual isotopic composition. Sequential and cumulative replacement of stable isotopes in Chlorella vulgaris. Biochim. Biophys. Acta 141, 625–632 [DOI] [PubMed] [Google Scholar]
- 46.Mueller L. N., Brusniak M. Y., Mani D. R., Aebersold R. (2008) An assessment of software solutions for the analysis of mass spectrometry based quantitative proteomics data. J. Proteome Res. 7, 51–61 [DOI] [PubMed] [Google Scholar]
- 47.Pasa-Tolic L., Jensen P. K., Anderson G. A., Lipton M. S., Peden K. K., Martinovic S., Tolic N., Bruce J. E., Smith R. D. (1999) High throughput proteome-wide precision measurements of protein expression using mass spectrometry. J. Am. Chem. Soc. 121, 7949–7950 [Google Scholar]
- 48.McLafferty F. W., Breuker K., Jin M., Han X., Infusini G., Jiang H., Kong X., Begley T. P. (2007) Top-down MS, a powerful complement to the high capabilities of proteolysis proteomics. FEBS J. 274, 6256–6268 [DOI] [PubMed] [Google Scholar]
- 49.Conrads T. P., Alving K., Veenstra T. D., Belov M. E., Anderson G. A., Anderson D. J., Lipton M. S., Pasa-Toliæ L., Udseth H. R., Chrisler W. B., Thrall B. D., Smith R. D. (2001) Quantitative analysis of bacterial and mammalian proteomes using a combination of cysteine affinity tags and 15N-metabolic labeling. Anal. Chem. 73, 2132–2139 [DOI] [PubMed] [Google Scholar]
- 50.Smith R. D., Anderson G. A., Lipton M. S., Pasa-Tolic L., Shen Y., Conrads T. P., Veenstra T. D., Udseth H. R. (2002) An accurate mass tag strategy for quantitative and high-throughput proteome measurements. Proteomics 2, 513–523 [DOI] [PubMed] [Google Scholar]
- 51.Whitelegge J. P., Katz J. E., Pihakari K. A., Hale R., Aguilera R., Gómez S. M., Faull K. F., Vavilin D., Vermaas W. (2004) Subtle modification of isotope ratio proteomics; an integrated strategy for expression proteomics. Phytochemistry 65, 1507–1515 [DOI] [PubMed] [Google Scholar]
- 52.Snijders A. P., de Vos M. G., Wright P. C. (2005) Novel approach for peptide quantitation and sequencing based on 15N and 13C metabolic labeling. J. Proteome Res. 4, 578–585 [DOI] [PubMed] [Google Scholar]
- 53.Snijders A. P., de Koning B., Wright P. C. (2005) Perturbation and interpretation of nitrogen isotope distribution patterns in proteomics. J. Proteome Res. 4, 2185–2191 [DOI] [PubMed] [Google Scholar]
- 54.Snijders A. P., de Vos M. G., de Koning B., Wright P. C. (2005) A fast method for quantitative proteomics based on a combination between two-dimensional electrophoresis and 15N-metabolic labelling. Electrophoresis 26, 3191–3199 [DOI] [PubMed] [Google Scholar]
- 55.Xia Q., Hendrickson E. L., Zhang Y., Wang T., Taub F., Moore B. C., Porat I., Whitman W. B., Hackett M., Leigh J. A. (2006) Quantitative proteomics of the archaeon Methanococcus maripaludis validated by microarray analysis and real time PCR. Mol. Cell. Proteomics 5, 868–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhong H., Marcus S. L., Li L. (2004) Two-dimensional mass spectra generated from the analysis of 15N-labeled and unlabeled peptides for efficient protein identification and de novo peptide sequencing. J. Proteome Res. 3, 1155–1163 [DOI] [PubMed] [Google Scholar]
- 57.Ting L., Cowley M. J., Hoon S. L., Guilhaus M., Raftery M. J., Cavicchioli R. (2009) Normalization and statistical analysis of quantitative proteomics data generated by metabolic labeling. Mol. Cell. Proteomics 8, 2227–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li L., Li Q., Rohlin L., Kim U., Salmon K., Rejtar T., Gunsalus R. P., Karger B. L., Ferry J. G. (2007) Quantitative proteomic and microarray analysis of the archaeon Methanosarcina acetivorans grown with acetate versus methanol. J. Proteome Res. 6, 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dreisbach A., Otto A., Becher D., Hammer E., Teumer A., Gouw J. W., Hecker M., Völker U. (2008) Monitoring of changes in the membrane proteome during stationary phase adaptation of Bacillus subtilis using in vivo labeling techniques. Proteomics 8, 2062–2076 [DOI] [PubMed] [Google Scholar]
- 60.Gu S., Pan S., Bradbury E. M., Chen X. (2002) Use of deuterium-labeled lysine for efficient protein identification and peptide de novo sequencing. Anal. Chem. 74, 5774–5785 [DOI] [PubMed] [Google Scholar]
- 61.Engen J. R., Bradbury E. M., Chen X. (2002) Using stable-isotope-labeled proteins for hydrogen exchange studies in complex mixtures. Anal. Chem. 74, 1680–1686 [DOI] [PubMed] [Google Scholar]
- 62.Veenstra T. D., Martinoviæ S., Anderson G. A., Pasa-Toliæ L., Smith R. D. (2000) Proteome analysis using selective incorporation of isotopically labeled amino acids. J. Am. Soc. Mass Spectrom. 11, 78–82 [DOI] [PubMed] [Google Scholar]
- 63.Martinoviæ S., Veenstra T. D., Anderson G. A., Pasa-Toliæ L., Smith R. D. (2002) Selective incorporation of isotopically labeled amino acids for identification of intact proteins on a proteome-wide level. J. Mass Spectrom. 37, 99–107 [DOI] [PubMed] [Google Scholar]
- 64.Chen X., Smith L. M., Bradbury E. M. (2000) Site-specific mass tagging with stable isotopes in proteins for accurate and efficient protein identification. Anal. Chem. 72, 1134–1143 [DOI] [PubMed] [Google Scholar]
- 65.Kjeldsen T. (2000) Yeast secretory expression of insulin precursors. Appl. Microbiol. Biotechnol. 54, 277–286 [DOI] [PubMed] [Google Scholar]
- 66.Goffeau A., Barrell B. G., Bussey H., Davis R. W., Dujon B., Feldmann H., Galibert F., Hoheisel J. D., Jacq C., Johnston M., Louis E. J., Mewes H. W., Murakami Y., Philippsen P., Tettelin H., Oliver S. G. (1996) Life with 6000 genes. Science 274, 546, 563–567 [DOI] [PubMed] [Google Scholar]
- 67.Cherry J. M., Adler C., Ball C., Chervitz S. A., Dwight S. S., Hester E. T., Jia Y., Juvik G., Roe T., Schroeder M., Weng S., Botstein D. (1998) SGD: Saccharomyces Genome Database. Nucleic Acids Res. 26, 73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Castrillo J. I., Oliver S. G. (2004) Yeast as a touchstone in post-genomic research: strategies for integrative analysis in functional genomics. J. Biochem. Mol. Biol. 37, 93–106 [DOI] [PubMed] [Google Scholar]
- 69.Kumar A., Snyder M. (2001) Emerging technologies in yeast genomics. Nat. Rev. Genet. 2, 302–312 [DOI] [PubMed] [Google Scholar]
- 70.Perocchi F., Mancera E., Steinmetz L. M. (2008) Systematic screens for human disease genes, from yeast to human and back. Mol. Biosyst. 4, 18–29 [DOI] [PubMed] [Google Scholar]
- 71.Gao H., Shen Y., Veenstra T. D., Harkewicz R., Anderson G. A., Bruce J. E., Pasa-Tolic L., Smith R. D. (2000) Two-dimensional electrophoretic/chromatographic separations combined with electrospray ionization FTICR mass spectrometry for high throughput proteome analysis. J. Microcolumn Sep. 12, 383–390 [Google Scholar]
- 72.Oda Y., Huang K., Cross F. R., Cowburn D., Chait B. T. (1999) Accurate quantitation of protein expression and site-specific phosphorylation. Proc. Natl. Acad. Sci. U.S.A 96, 6591–6596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Washburn M. P., Ulaszek R., Deciu C., Schieltz D. M., Yates J. R., 3rd (2002) Analysis of quantitative proteomic data generated via multidimensional protein identification technology. Anal. Chem. 74, 1650–1657 [DOI] [PubMed] [Google Scholar]
- 74.Washburn M. P., Koller A., Oshiro G., Ulaszek R. R., Plouffe D., Deciu C., Winzeler E., Yates J. R., 3rd (2003) Protein pathway and complex clustering of correlated mRNA and protein expression analyses in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A 100, 3107–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.MacCoss M. J., Wu C. C., Liu H., Sadygov R., Yates J. R., 3rd (2003) A correlation algorithm for the automated quantitative analysis of shotgun proteomics data. Anal. Chem. 75, 6912–6921 [DOI] [PubMed] [Google Scholar]
- 76.MacCoss M. J., Wu C. C., Matthews D. E., Yates J. R., 3rd (2005) Measurement of the isotope enrichment of stable isotope-labeled proteins using high-resolution mass spectra of peptides. Anal. Chem. 77, 7646–7653 [DOI] [PubMed] [Google Scholar]
- 77.Venable J. D., Dong M. Q., Wohlschlegel J., Dillin A., Yates J. R. (2004) Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nat. Methods 1, 39–45 [DOI] [PubMed] [Google Scholar]
- 78.Venable J. D., Wohlschlegel J., McClatchy D. B., Park S. K., Yates J. R., 3rd (2007) Relative quantification of stable isotope labeled peptides using a linear ion trap-Orbitrap hybrid mass spectrometer. Anal. Chem. 79, 3056–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Du Y., Parks B. A., Sohn S., Kwast K. E., Kelleher N. L. (2006) Top-down approaches for measuring expression ratios of intact yeast proteins using Fourier transform mass spectrometry. Anal. Chem. 78, 686–694 [DOI] [PubMed] [Google Scholar]
- 80.Zybailov B., Coleman M. K., Florens L., Washburn M. P. (2005) Correlation of relative abundance ratios derived from peptide ion chromatograms and spectrum counting for quantitative proteomic analysis using stable isotope labeling. Anal. Chem. 77, 6218–6224 [DOI] [PubMed] [Google Scholar]
- 81.Zybailov B., Mosley A. L., Sardiu M. E., Coleman M. K., Florens L., Washburn M. P. (2006) Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 5, 2339–2347 [DOI] [PubMed] [Google Scholar]
- 82.Kolkman A., Daran-Lapujade P., Fullaondo A., Olsthoorn M. M., Pronk J. T., Slijper M., Heck A. J. (2006) Proteome analysis of yeast response to various nutrient limitations. Mol. Syst. Biol. 2, 2006.0026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.de Groot M. J., Daran-Lapujade P., van Breukelen B., Knijnenburg T. A., de Hulster E. A., Reinders M. J., Pronk J. T., Heck A. J., Slijper M. (2007) Quantitative proteomics and transcriptomics of anaerobic and aerobic yeast cultures reveals post-transcriptional regulation of key cellular processes. Microbiology 153, 3864–3878 [DOI] [PubMed] [Google Scholar]
- 84.Daran-Lapujade P., Rossell S., van Gulik W. M., Luttik M. A., de Groot M. J., Slijper M., Heck A. J., Daran J. M., de Winde J. H., Westerhoff H. V., Pronk J. T., Bakker B. M. (2007) The fluxes through glycolytic enzymes in Saccharomyces cerevisiae are predominantly regulated at posttranscriptional levels. Proc. Natl. Acad. Sci. U.S.A 104, 15753–15758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lecchi S., Nelson C. J., Allen K. E., Swaney D. L., Thompson K. L., Coon J. J., Sussman M. R., Slayman C. W. (2007) Tandem phosphorylation of Ser-911 and Thr-912 at the C terminus of yeast plasma membrane H+-ATPase leads to glucose-dependent activation. J. Biol. Chem. 282, 35471–35481 [DOI] [PubMed] [Google Scholar]
- 86.Hunter T. C., Yang L., Zhu H., Majidi V., Bradbury E. M., Chen X. (2001) Peptide mass mapping constrained with stable isotope-tagged peptides for identification of protein mixtures. Anal. Chem. 73, 4891–4902 [DOI] [PubMed] [Google Scholar]
- 87.Zhu H., Pan S., Gu S., Bradbury E. M., Chen X. (2002) Amino acid residue specific stable isotope labeling for quantitative proteomics. Rapid Commun. Mass Spectrom. 16, 2115–2123 [DOI] [PubMed] [Google Scholar]
- 88.Jiang H., English A. M. (2002) Quantitative analysis of the yeast proteome by incorporation of isotopically labeled leucine. J. Proteome Res. 1, 345–350 [DOI] [PubMed] [Google Scholar]
- 89.Jiang H., English A. M. (2006) Evaluation of D10-Leu metabolic labeling coupled with MALDI-MS analysis in studying the response of the yeast proteome to H2O2 challenge. J. Proteome Res. 5, 2539–2546 [DOI] [PubMed] [Google Scholar]
- 90.Gruhler A., Olsen J. V., Mohammed S., Mortensen P., Faergeman N. J., Mann M., Jensen O. N. (2005) Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell. Proteomics 4, 310–327 [DOI] [PubMed] [Google Scholar]
- 91.Van Hoof D., Pinkse M. W., Oostwaard D. W., Mummery C. L., Heck A. J., Krijgsveld J. (2007) An experimental correction for arginine-to-proline conversion artifacts in SILAC-based quantitative proteomics. Nat. Methods. 4, 677–678 [DOI] [PubMed] [Google Scholar]
- 92.Bendall S. C., Hughes C., Stewart M. H., Doble B., Bhatia M., Lajoie G. A. (2008) Prevention of amino acid conversion in SILAC experiments with embryonic stem cells. Mol. Cell. Proteomics 7, 1587–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Global analysis of protein expression in yeast. Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 94.de Godoy L. M., Olsen J. V., Cox J., Nielsen M. L., Hubner N. C., Fröhlich F., Walther T. C., Mann M. (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251–1254 [DOI] [PubMed] [Google Scholar]
- 95.Georgianna D. R., Hawkridge A. M., Muddiman D. C., Payne G. A. (2008) Temperature-dependent regulation of proteins in Aspergillus flavus: whole organism stable isotope labeling by amino acids. J. Proteome Res. 7, 2973–2979 [DOI] [PubMed] [Google Scholar]
- 96.Gruhler A., Schulze W. X., Matthiesen R., Mann M., Jensen O. N. (2005) Stable isotope labeling of Arabidopsis thaliana cells and quantitative proteomics by mass spectrometry. Mol. Cell. Proteomics 4, 1697–1709 [DOI] [PubMed] [Google Scholar]
- 97.Collier T. S., Hawkridge A. M., Georgianna D. R., Payne G. A., Muddiman D. C. (2008) Top-down identification and quantification of stable isotope labeled proteins from Aspergillus flavus using online nano-flow reversed-phase liquid chromatography coupled to a LTQ-FTICR mass spectrometer. Anal. Chem. 80, 4994–5001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Patnaik S., Broadbent F. E. (1967) Utilization of tracer nitrogen by rice in relation to time of application. Agron. J. 59, 287–288 [Google Scholar]
- 99.Muhammad S., Kumazawa K. (1974) The absorption, distribution, and redistribution of 15N-labelled ammonium and nitrate nitrogen administered at different growth stages of rice. Soil Sci. Plant Nutr. 20, 47–55 [Google Scholar]
- 100.Osaki M., Shirai J., Shinano T., Tadano T. (1995) 15N-Allocation of 15NH4-N and 15NO3-N to nitrogenous compounds at the vegetative growth stage of potato plants. Soil Sci. Plant Nutr. 41, 699–708 [Google Scholar]
- 101.Grusak M., Pezeshgi S. (1994) Uniformly 15N-labeled soybean seeds produced for use in human and animal nutrition studies: description of a recirculating hydroponic growth system and whole plant nutrient and environmental requirements. J. Sci. Food Agric. 64, 223–230 [Google Scholar]
- 102.Ippel J. H., Pouvreau L., Kroef T., Gruppen H., Versteeg G., van den Putten P., Struik P. C., van Mierlo C. P. (2004) In vivo uniform (15)N-isotope labelling of plants: using the greenhouse for structural proteomics. Proteomics 4, 226–234 [DOI] [PubMed] [Google Scholar]
- 103.The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815 [DOI] [PubMed] [Google Scholar]
- 104.Swarbreck D., Wilks C., Lamesch P., Berardini T. Z., Garcia-Hernandez M., Foerster H., Li D., Meyer T., Muller R., Ploetz L., Radenbaugh A., Singh S., Swing V., Tissier C., Zhang P., Huala E. (2008) The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 36, D1009–D1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim J. K., Harada K., Bamba T., Fukusaki E., Kobayashi A. (2005) Stable isotope dilution-based accurate comparative quantification of nitrogen-containing metabolites in Arabidopsis thaliana T87 cells using in vivo 15N-isotope enrichment. Biosci. Biotechnol. Biochem. 69, 1331–1340 [DOI] [PubMed] [Google Scholar]
- 106.Engelsberger W. R., Erban A., Kopka J., Schulze W. X. (2006) Metabolic labeling of plant cell cultures with K15NO3 as a tool for quantitative analysis of proteins and metabolites. Plant Methods 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Benschop J. J., Mohammed S., O'Flaherty M., Heck A. J., Slijper M., Menke F. L. (2007) Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell. Proteomics 6, 1198–1214 [DOI] [PubMed] [Google Scholar]
- 108.Lanquar V., Kuhn L., Lelièvre F., Khafif M., Espagne C., Bruley C., Barbier-Brygoo H., Garin J., Thomine S. (2007) 15N-metabolic labeling for comparative plasma membrane proteomics in Arabidopsis cells. Proteomics 7, 750–754 [DOI] [PubMed] [Google Scholar]
- 109.Nelson C. J., Huttlin E. L., Hegeman A. D., Harms A. C., Sussman M. R. (2007) Implications of 15N-metabolic labeling for automated peptide identification in Arabidopsis thaliana. Proteomics 7, 1279–1292 [DOI] [PubMed] [Google Scholar]
- 110.Huttlin E. L., Hegeman A. D., Harms A. C., Sussman M. R. (2007) Comparison of full versus partial metabolic labeling for quantitative proteomics analysis in Arabidopsis thaliana. Mol. Cell. Proteomics 6, 860–881 [DOI] [PubMed] [Google Scholar]
- 111.Palmblad M., Bindschedler L. V., Cramer R. (2007) Quantitative proteomics using uniform 15N-labeling, MASCOT, and the trans-proteomic pipeline. Proteomics 7, 3462–3469 [DOI] [PubMed] [Google Scholar]
- 112.Bindschedler L. V., Palmblad M., Cramer R. (2008) Hydroponic isotope labelling of entire plants (HILEP) for quantitative plant proteomics; an oxidative stress case study. Phytochemistry 69, 1962–1972 [DOI] [PubMed] [Google Scholar]
- 113.Hebeler R., Oeljeklaus S., Reidegeld K. A., Eisenacher M., Stephan C., Sitek B., Stühler K., Meyer H. E., Sturre M. J., Dijkwel P. P., Warscheid B. (2008) Study of early leaf senescence in Arabidopsis thaliana by quantitative proteomics using reciprocal 14N/15N labeling and difference gel electrophoresis. Mol. Cell. Proteomics 7, 108–120 [DOI] [PubMed] [Google Scholar]
- 114.Schaff J. E., Mbeunkui F., Blackburn K., Bird D. M., Goshe M. B. (2008) SILIP: a novel stable isotope labeling method for in planta quantitative proteomic analysis. Plant J. 56, 840–854 [DOI] [PubMed] [Google Scholar]
- 115.Thelen J. J., Peck S. C. (2007) Quantitative proteomics in plants: choices in abundance. Plant Cell 19, 3339–3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics 77, 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Castle W. E. (1906) Inbreeding, cross-breeding and sterility in Drosophila. Science 23153. [DOI] [PubMed] [Google Scholar]
- 118.Adams M. D., Celniker S. E., Holt R. A., Evans C. A., Gocayne J. D., Amanatides P. G., Scherer S. E., Li P. W., Hoskins R. A., Galle R. F., George R. A., Lewis S. E., Richards S., Ashburner M., Henderson S. N., Sutton G. G., Wortman J. R., Yandell M. D., Zhang Q., Chen L. X., Brandon R. C., Rogers Y. H., Blazej R. G., Champe M., Pfeiffer B. D., Wan K. H., Doyle C., Baxter E. G., Helt G., Nelson C. R., Gabor G. L., Abril J. F., Agbayani A., An H. J., Andrews-Pfannkoch C., Baldwin D., Ballew R. M., Basu A., Baxendale J., Bayraktaroglu L., Beasley E. M., Beeson K. Y., Benos P. V., Berman B. P., Bhandari D., Bolshakov S., Borkova D., Botchan M. R., Bouck J., Brokstein P., Brottier P., Burtis K. C., Busam D. A., Butler H., Cadieu E., Center A., Chandra I., Cherry J. M., Cawley S., Dahlke C., Davenport L. B., Davies P., de Pablos B., Delcher A., Deng Z., Mays A. D., Dew I., Dietz S. M., Dodson K., Doup L. E., Downes M., Dugan-Rocha S., Dunkov B. C., Dunn P., Durbin K. J., Evangelista C. C., Ferraz C., Ferriera S., Fleischmann W., Fosler C., Gabrielian A. E., Garg N. S., Gelbart W. M., Glasser K., Glodek A., Gong F., Gorrell J. H., Gu Z., Guan P., Harris M., Harris N. L., Harvey D., Heiman T. J., Hernandez J. R., Houck J., Hostin D., Houston K. A., Howland T. J., Wei M. H., Ibegwam C., Jalali M., Kalush F., Karpen G. H., Ke Z., Kennison J. A., Ketchum K. A., Kimmel B. E., Kodira C. D., Kraft C., Kravitz S., Kulp D., Lai Z., Lasko P., Lei Y., Levitsky A. A., Li J., Li Z., Liang Y., Lin X., Liu X., Mattei B., McIntosh T. C., McLeod M. P., McPherson D., Merkulov G., Milshina N. V., Mobarry C., Morris J., Moshrefi A., Mount S. M., Moy M., Murphy B., Murphy L., Muzny D. M., Nelson D. L., Nelson D. R., Nelson K. A., Nixon K., Nusskern D. R., Pacleb J. M., Palazzolo M., Pittman G. S., Pan S., Pollard J., Puri V., Reese M. G., Reinert K., Remington K., Saunders R. D., Scheeler F., Shen H., Shue B. C., Sidén-Kiamos I., Simpson M., Skupski M. P., Smith T., Spier E., Spradling A. C., Stapleton M., Strong R., Sun E., Svirskas R., Tector C., Turner R., Venter E., Wang A. H., Wang X., Wang Z. Y., Wassarman D. A., Weinstock G. M., Weissenbach J., Williams S. M., Woodage T., Worley K. C., Wu D., Yang S., Yao Q. A., Ye J., Yeh R. F., Zaveri J. S., Zhan M., Zhang G., Zhao Q., Zheng L., Zheng X. H., Zhong F. N., Zhong W., Zhou X., Zhu S., Zhu X., Smith H. O., Gibbs R. A., Myers E. W., Rubin G. M., Venter J. C. (2000) The genome sequence of Drosophila melanogaster. Science 287, 2185–2195 [DOI] [PubMed] [Google Scholar]
- 119.C. elegans Sequencing Consortium (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282, 2012–2018 [DOI] [PubMed] [Google Scholar]
- 120.Antoshechkin I., Sternberg P. W. (2007) The versatile worm: genetic and genomic resources for Caenorhabditis elegans research. Nat. Rev. Genet. 8, 518–532 [DOI] [PubMed] [Google Scholar]
- 121.Matthews K. A., Kaufman T. C., Gelbart W. M. (2005) Research resources for Drosophila: the expanding universe. Nat. Rev. Genet. 6, 179–193 [DOI] [PubMed] [Google Scholar]
- 122.Rogers A., Antoshechkin I., Bieri T., Blasiar D., Bastiani C., Canaran P., Chan J., Chen W. J., Davis P., Fernandes J., Fiedler T. J., Han M., Harris T. W., Kishore R., Lee R., McKay S., Müller H. M., Nakamura C., Ozersky P., Petcherski A., Schindelman G., Schwarz E. M., Spooner W., Tuli M. A., Van Auken K., Wang D., Wang X., Williams G., Yook K., Durbin R., Stein L. D., Spieth J., Sternberg P. W. (2008) WormBase 2007. Nucleic Acids Res. 36, D612–D617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wilson R. J., Goodman J. L., Strelets V. B. (2008) FlyBase: integration and improvements to query tools. Nucleic Acids Res. 36, D588–D593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bier E. (2005) Drosophila, the golden bug, emerges as a tool for human genetics. Nat. Rev. Genet. 6, 9–23 [DOI] [PubMed] [Google Scholar]
- 125.Markow T. A., O'Grady P. M. (2007) Drosophila biology in the genomic age. Genetics 177, 1269–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Krijgsveld J., Ketting R. F., Mahmoudi T., Johansen J., Artal-Sanz M., Verrijzer C. P., Plasterk R. H., Heck A. J. (2003) Metabolic labeling of C. elegans and D. melanogaster for quantitative proteomics. Nat. Biotechnol. 21, 927–931 [DOI] [PubMed] [Google Scholar]
- 127.Dong M. Q., Venable J. D., Au N., Xu T., Park S. K., Cociorva D., Johnson J. R., Dillin A., Yates J. R., 3rd (2007) Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science 317, 660–663 [DOI] [PubMed] [Google Scholar]
- 128.Findlay G. D., Yi X., Maccoss M. J., Swanson W. J. (2008) Proteomics reveals novel Drosophila seminal fluid proteins transferred at mating. PLoS Biol. 6e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gouw J. W., Pinkse M. W., Vos H. R., Moshkin Y., Verrijzer C. P., Heck A. J., Krijgsveld J. (2009) In vivo stable isotope labeling of fruit flies reveals post-transcriptional regulation in the maternal-to-zygotic transition. Mol. Cell. Proteomics 8, 1566–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Doherty M. K., Whitehead C., McCormack H., Gaskell S. J., Beynon R. J. (2005) Proteome dynamics in complex organisms: using stable isotopes to monitor individual protein turnover rates. Proteomics 5, 522–533 [DOI] [PubMed] [Google Scholar]
- 131.Wu C. C., MacCoss M. J., Howell K. E., Matthews D. E., Yates J. R., 3rd (2004) Metabolic labeling of mammalian organisms with stable isotopes for quantitative proteomic analysis. Anal. Chem. 76, 4951–4959 [DOI] [PubMed] [Google Scholar]
- 132.McClatchy D. B., Dong M. Q., Wu C. C., Venable J. D., Yates J. R., 3rd (2007) 15N metabolic labeling of mammalian tissue with slow protein turnover. J. Proteome Res. 6, 2005–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.McClatchy D. B., Liao L., Park S. K., Venable J. D., Yates J. R. (2007) Quantification of the synaptosomal proteome of the rat cerebellum during post-natal development. Genome Res. 17, 1378–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Krüger M., Moser M., Ussar S., Thievessen I., Luber C. A., Forner F., Schmidt S., Zanivan S., Fässler R., Mann M. (2008) SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell 134, 353–364 [DOI] [PubMed] [Google Scholar]
- 135.Boersema P. J., Raijmakers R., Lemeer S., Mohammed S., Heck A. J. (2009) Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 4, 484–494 [DOI] [PubMed] [Google Scholar]
- 136.Boersema P. J., Taouatas N., Altelaar A. F., Gouw J. W., Ross P. L., Pappin D. J., Heck A. J., Mohammed S. (2009) Straightforward and de novo peptide sequencing by MALDI-MS/MS using a Lys-N metalloendopeptidase. Mol. Cell. Proteomics 8, 650–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Grunwald D. J., Eisen J. S. (2002) Headwaters of the zebrafish—emergence of a new model vertebrate. Nat. Rev. Genet. 3, 717–724 [DOI] [PubMed] [Google Scholar]
- 138.Lieschke G. J., Currie P. D. (2007) Animal models of human disease: zebrafish swim into view. Nat. Rev. Genet. 8, 353–367 [DOI] [PubMed] [Google Scholar]
- 139.Langenau D. M., Zon L. I. (2005) The zebrafish: a new model of T-cell and thymic development. Nat. Rev. Immunol. 5, 307–317 [DOI] [PubMed] [Google Scholar]
- 140.Feitsma H., Cuppen E. (2008) Zebrafish as a cancer model. Mol. Cancer Res. 6, 685–694 [DOI] [PubMed] [Google Scholar]
- 141.Flinn L., Bretaud S., Lo C., Ingham P. W., Bandmann O. (2008) Zebrafish as a new animal model for movement disorders. J. Neurochem. 106, 1991–1997 [DOI] [PubMed] [Google Scholar]
- 142.McGrath P., Li C. Q. (2008) Zebrafish: a predictive model for assessing drug-induced toxicity. Drug Discov. Today 13, 394–401 [DOI] [PubMed] [Google Scholar]
- 143.Eisen J. S., Smith J. C. (2008) Controlling morpholino experiments: don't stop making antisense. Development 135, 1735–1743 [DOI] [PubMed] [Google Scholar]
- 144.Carter C., Owen S., He Z., Watt P., Scrimgeour C., Houlihan D., Rennie M. (1994) Determination of protein synthesis in rainbow trout, Oncorhynchus mykiss, using a stable isotope. J. Exp. Biol. 189, 279–284 [DOI] [PubMed] [Google Scholar]
- 145.Sato T., Vries R. G., Snippert H. J., van de Wetering M., Barker N., Stange D. E., van Es J. H., Abo A., Kujala P., Peters P. J., Clevers H. (2009) Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 [DOI] [PubMed] [Google Scholar]
- 146.Nirmalan N., Sims P. F., Hyde J. E. (2004) Quantitative proteomics of the human malaria parasite Plasmodium falciparum and its application to studies of development and inhibition. Mol. Microbiol. 52, 1187–1199 [DOI] [PubMed] [Google Scholar]