Abstract
Adenoviruses replicate primarily in the host cell nucleus, and it is well established that adenovirus infection affects the structure and function of host cell nucleoli in addition to coding for a number of nucleolar targeted viral proteins. Here we used unbiased proteomics methods, including high throughput mass spectrometry coupled with stable isotope labeling by amino acids in cell culture (SILAC) and traditional two-dimensional gel electrophoresis, to identify quantitative changes in the protein composition of the nucleolus during adenovirus infection. Two-dimensional gel analysis revealed changes in six proteins. By contrast, SILAC-based approaches identified 351 proteins with 24 proteins showing at least a 2-fold change after infection. Of those, four were previously reported to have aberrant localization and/or functional relevance during adenovirus infection. In total, 15 proteins identified as changing in amount by proteomics methods were examined in infected cells using confocal microscopy. Eleven of these proteins showed altered patterns of localization in adenovirus-infected cells. Comparing our data with the effects of actinomycin D on the nucleolar proteome revealed that adenovirus infection apparently specifically targets a relatively small subset of nucleolar antigens at the time point examined.
Human adenoviruses comprise a non-enveloped icosahedral particle of 90 nm in diameter containing a linear double-stranded DNA genome of ∼36 kbp. After virus attachment and entry to the host cell, the genome is delivered to the host cell nucleus, initiating a cascade of viral gene expression that results in viral DNA replication and accumulation of viral proteins that eventually form new infectious particles. During viral replication, host cell rRNA processing and export from the nucleus are inhibited, and a number of viral nucleolar antigens accumulate in the nucleoli of infected cells (1–10). Moreover, two cellular nucleolar antigens, UBF1 and B23.1, have been shown to be sequestered into viral DNA replication centers where they affect viral DNA replication (11–13). Thus, there is clear evidence that adenovirus infection has a substantial impact on the composition and function of the nucleolus. What is not clear is whether the sequestration of cellular nucleolar antigens is limited to a small number of proteins or widespread. In addition, it is unclear whether adenovirus primarily modulates the nucleolus for replicative advantage or whether the effects on the nucleolus are a side effect of cellular proteins being sequestered into virally induced structures.
Many other plant and mammalian viruses have also been shown to interact with the host cell nucleolus from human immunodeficiency virus to Kaposi sarcoma herpesvirus (14–21). We therefore wanted to use a systematic and unbiased approach to examine the fate of the nucleolus during a viral infection using an established model system.
The use of modern proteomics approaches to investigate viral infections is still relatively new, but recently several notable studies have been reported that illustrate the power and utility of these techniques (22–26). Most of these have used 2D gel electrophoresis-based approaches with some success. For example, 2D electrophoresis was used to examine the total proteome of West Nile virus-infected cells, identifying about 100 different proteins, many of which were clearly up- or down-regulated (27). Much less common is the use of the more powerful quantitative MS coupled with stable isotope labeling by amino acids in cell culture (SILAC) technique to compare uninfected and infected cells. However, one study has investigated the composition of lipid rafts in hepatitis C virus-infected cells, identifying almost 1000 proteins (28). Furthermore, quantitative MS/SILAC has been extensively used for proteomics analysis of the mammalian nucleolus under a range of conditions (29–32). Indeed, a key study examined the changes in composition of the nucleolus after inhibition of rRNA synthesis with actinomycin D (ActD), quantitating the effects on almost 500 proteins in the nucleolus (29).
We wanted to further explore the potential of both 2D gel electrophoresis and quantitative MS/SILAC to examine the effects of adenoviral infection on the host cell. As stated, we have previously shown that nucleolar antigens UBF and B23 are visibly depleted from the nucleolus during adenovirus infection and play functional roles in viral DNA replication (11–13). Thus, we would expect a proteomics approach to confirm these well established changes in nucleolar composition that are of functional relevance to the infection. This study not only identified proteins known to be affected by adenovirus infection, but it also identified a number of novel alterations to the nucleolar proteome that were subsequently examined by confocal microscopy for independent verification.
EXPERIMENTAL PROCEDURES
Cells and Viruses
HeLa cells were grown in Dulbecco's modified Eagle's medium + Glutamax (Invitrogen) supplemented with 10% fetal calf serum, 100 μg/ml streptomycin, and 100 IU/ml penicillin. Human adenovirus type 5 (Ad5) was propagated as described previously (33) and purified using a ViraBind adenovirus purification kit (Cellbiolabs). The HeLa cell line stably expressing GFP-tagged ribosomal protein RPL27 has been reported elsewhere (34). For microscopy, HeLa cells were grown on glass coverslips in 6-well dishes.
SILAC Labeling of HeLa Cells and Nucleolar Isolation
Isolation of purified nucleoli from uninfected and infected cells was performed as described previously (29). For both SILAC and standard 2D gel analysis, 108 HeLa cells were infected at a multiplicity of infection (m.o.i.) of 5 with Ad5 and harvested at 18 h postinfection.
For SILAC analysis, prior to infection cells were grown for five rounds of cell division in Dulbecco's modified Eagle's medium containing l-[13C6,15N4]arginine and l-[13C6,15N2]lysine (Cambridge Isotope Laboratories) supplemented with 10% dialyzed fetal calf serum (Biowest) to ensure all the cellular proteins were labeled to saturation. Prior to nucleolar isolation, equal numbers of uninfected, unlabeled cells and Ad5-infected, labeled cells were mixed. Nucleoli were then isolated from the mixed cell populations using a standard protocol (35). Nucleoli were lysed by heating in lithium dodecyl sulfate sample buffer (Invitrogen), and nucleolar proteins were separated by SDS-PAGE on a 4–12% precast gradient gel (Invitrogen). The gel was fixed, stained with colloidal Coomassie Blue (Invitrogen), and cut into six slices.
LC-MSMS Analysis
Peptides were isolated from each gel slice after in-gel digestion, desalted, and concentrated as described previously (32); separated by HPLC (Agilent) on a C18 reverse phase column; and analyzed by a QSTAR XL hybrid quadrupole TOF mass spectrometer (Applied Biosystems). The peak lists were generated by the Analyst QS software, version 1.1 (Applied Biosystems). The MS data were searched against the NCBInr database (February 19, 2006 release, 3,230,559 sequences searched) for Homo sapiens using the MASCOT search engine, version 1.9 (Matrix Science). Variable modifications used were carboxymethyl (Cys), oxidation (Met) and phospho (Ser, Thr, and Tyr) as well as the appropriate SILAC modifications. Trypsin specificity was used, two missed cleavages were allowed, and a mass tolerance of 0.5 Da was used for both precursor and fragment ions. Peptide charges of +1, +2, and +3 were selected. Individual ions with MASCOT scores higher than 20 were used, making sure the “average peptide scores” of all identified proteins exceeded 20, a threshold commonly used for confident protein identification from tandem MS data (36). Only bold red peptides were also considered, effectively removed duplicate homologous proteins from the results. Under these conditions, the estimated false positive rate is less than 5%, according to a previous analysis (37). SILAC quantitation was done by the MSQuant software, which measures the averaged MS peak areas of the isotopic pairs. Only proteins with bold red peptides and combined scores higher than 50 were quantitated. Proteins with heavy/light isotopic ratios lower than 0.01 were discarded; they mostly represented environmental contaminants. The S.D. of a protein ratio represented the variations among the measured peptide ratios for the same protein. Functional classification of the identified proteins was performed using Proteincenter (Proxeon). The biological reproducibility was addressed by a parallel 2D gel-based approach and by microscopy analysis (see below).
2D Gel Electrophoresis
Nucleolar pellets were resuspended in 450 μl of 7 m urea, 2 m thiourea, 4% CHAPS, 0.002% bromphenol blue, 0.5% (v/v) IPG buffer, pH 3–11 non-linear (GE Healthcare), and 1.2% (v/v) Destreak reagent (GE Healthcare) and loaded onto 24-cm Immobiline DryStrip gels (pH 3–11 non-linear) by passive rehydration for a minimum of 12 h. Following rehydration, the DryStrip gels were transferred to an Ettan IPGPhor 3 system (GE Healthcare), and isoelectric focusing was performed by applying 500 V for 1 h, 1000 V for 1 h, and 8000 V for 10.5 h until a total of 64,000 V-h had been achieved. Following isoelectric focusing, strips were equilibrated in SDS equilibration buffer (50 mm Tris-HCl, pH 8.8, 6 m urea, 30% (v/v) glycerol, 2% (w/v) SDS, and 0.002% (w/v) bromphenol blue) containing 1% (w/v) DTT for 15 min at room temperature followed by a further incubation in fresh SDS equilibration buffer containing 2.5% (w/v) iodoacetamide for 15 min at room temperature. Strips were then applied to 12.5% (w/v) SDS-PAGE gels and run at 5 mA/gel for 1 h, 8 mA/gel for an additional hour, and then at 20 watts/gel until completion using an Ettan DALT-6 separation unit (GE Healthcare). Gels were fixed for 1 h in 50% methanol and 10% acetic acid and stained overnight using SYPRO Ruby total protein stain (Invitrogen). Following destaining in 10% methanol and 7% acetic acid, the gels were imaged using a Typhoon 9400 Variable Mode Imager (GE Healthcare).
Spot Picking, Protein Processing, and MS
Selected protein spots were cut from the gel using the Investigator ProPic automated 2D spot picking robot and digested with trypsin using the ProGest automated digestion unit (both from PerkinElmer Life Sciences).
Mass spectra were recorded in positive ion reflector mode on an Applied Biosystems 4700 MALDI mass spectrometer. The instrument was calibrated before each run using the 4700 Calibration Mix 1 (Applied Biosystems), and in addition, individual MS spectra were internally calibrated using tryptic autolytic peaks (if present). MSMS analysis was calibrated on the fragmentation of Glu-fibrinopeptide. For MSMS analysis, the top five most intense, non-tryptic precursors were selected for fragmentation by collision-induced dissociation. Neither base-line subtraction nor smoothing was applied to recorded spectra.
MS and MSMS data were analyzed, and peak lists generated using GPS Explorer 3.5 (Applied Biosystems). MS peaks were selected between 800 and 4000 Da and filtered with a signal to noise ratio greater than 15 and to exclude masses derived from trypsin autolysis (842.51, 1006.48, 1045.56, 2211.1, 2283.18, and 2299.18). MSMS peaks were selected on the basis of a signal to noise ratio greater than 10 over a mass range of 50 to 20 Da below the precursor mass. A peak density filter was used with no more than 30 peaks per 200 Da and a maximum number of peaks of 65.
Data were analyzed using the MASCOT 1.9 search engine (Matrix Science) to search against the human (148,148 sequences) MSDB protein database (Release 20063108). Search parameters allowed for one missed tryptic cleavage site, the carbamidomethylation of cysteine, and the possible oxidation of methionine; precursor ion mass tolerance was 150 ppm, and fragment ion mass tolerance was 0.25 Da. All identified proteins have a MASCOT score greater than 64 (the default MASCOT threshold for such searches), corresponding to a statistically significant (p < 0.05) confident identification.
Antibodies and Plasmids
Antisera used were anti-adenovirus protein V (5), anti-DBP (a kind gift from W. C. Russell), anti-UBF (Santa Cruz Biotechnology), anti-FLAG tag (Sigma), anti-Myc tag (Santa Cruz Biotechnology) anti-PSF/SPFQ (Sigma), anti-hnRNPU (a kind gift from P. Percipalle; Ref. 38), anti-Nopp140 (a kind gift from Tom Meir; Ref. 39), anti-U2AF65 (Santa Cruz Biotechnology), anti-hnRNPA2/B1 (Abcam), anti-S15a (a kind gift from M. Feitelson; Ref. 40), and anti-eIF6 (a kind gift from B. Stefano; Ref. 41). The expression plasmids encoding tagged plasmids were vesicular stomatitis virus-tagged hPOP1 (a kind gift from G. Pruijn; Ref. 42), FLAG-tagged RBM4 (a kind gift from W. Tarn; Ref.43), enhanced GFP-tagged Histone H1.2 (a kind gift from L. Schang; Ref. 44), Myc-tagged exportin 5 (Addgene (45)), GFP-tagged PP2C Epsilon (a kind gift from T. Kobayashi; Ref. 46), cyan fluorescent protein-tagged p87 PIKAP (a kind gift from M. Schaefer; Ref. 47), and FLAG-tagged Nop132 (a kind gift from T. Sekiguchi; Ref. 48). Transfection of tagged plasmid constructs was done using Lipofectamine 2000 (Invitrogen) according to the manufacturers' instructions. Appropriate secondary antibodies were labeled with Alexa Fluor 488 or Alexa Fluor 594 (Invitrogen).
Confocal Microscopy
HeLa cells grown on coverslips were either infected, mock infected, and/or transfected with expression plasmids encoding tagged proteins as indicated. At 18 h postinfection, cells were fixed for 5 min with 4% formaldehyde in PBS followed by washing in PBS before permeabilizing in 1% Triton X-100 in PBS for 5 min. After permeabilization, the cells were washed briefly with PBS before blocking for 30 min at room temperature in 10% FCS in PBS. The cells were then incubated with primary antibodies for 60 min, washed, and incubated with secondary antibodies. Cells were mounted with Vectashield plus DAPI to visualize the nuclear DNA. Images were taken using a Leica confocal microscope (TCS-SP2).
RESULTS
Isolation of Nucleoli from Cells Infected with Adenovirus
The rate of adenovirus replication depends on the serotype of virus, the cell type, and the m.o.i. For the prototype strain Ad5, infection of HeLa cells with a modest m.o.i. of about five infectious particles per cell results in detectable viral early gene expression by 4 h postinfection, detectable DNA replication by 10 h postinfection, production of structural proteins that will form progeny virions by about 18 h postinfection, and release of infectious particles from about 30 h onward. To ensure the isolation of intact nucleoli, we chose to examine the composition of the nucleoli at ∼18 h postinfection when late structural gene expression would be just underway in some cells but when active viral replication would be well underway in virtually all the cells examined (for a review, see Ref. 49).
In this study, we isolated nucleoli from infected and mock infected HeLa cells and compared the differences in the isolated proteins by quantitative proteomics. To test the suitability of this approach, we infected a HeLa cell line stably expressing a GFP-tagged ribosomal protein RPL27 (34) with Ad5. RPL27-GFP was used as a nucleolar marker in live cells and in isolated nucleoli. At 18 h postinfection, the cells were harvested, and nucleoli were isolated using a standard protocol (29, 35). As shown in supplemental Fig. 1, live cell imaging revealed that the RPL27-GFP signal in the nucleolus of infected cells (supplemental Fig. 1C) was similar to that in untreated cells (supplemental Fig. 1A). The GFP signals in nucleoli isolated from untreated (supplemental Fig. 1B and infected (supplemental Fig. 1D) cells were also similar, although the nucleoli from infected cells appeared slightly larger. Interestingly, adenovirus infection induced the appearance of RPL27-GFP in threadlike structures in the nucleoplasm. The significance of the accumulation of this ribosomal protein in these structures or the apparent slight enlargement of nucleoli is unknown. The threadlike GFP-containing structures were not observed in the isolated nucleolar preparation, suggesting that the preparation was not contaminated with nucleoplasmic materials and was suitable for future proteomics analysis.
SILAC Analysis of Virus-infected Cell Nucleoli
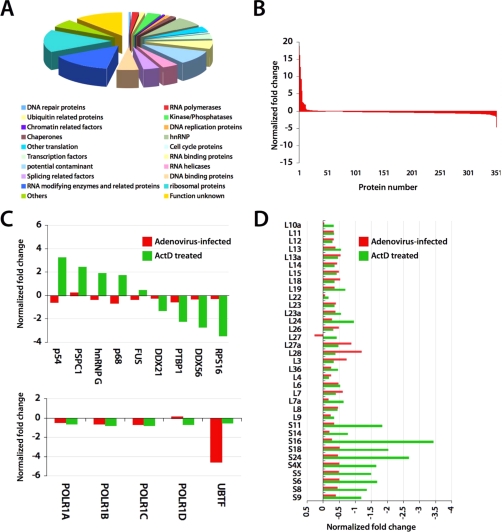
A total of 351 proteins were quantified by SILAC analysis comparing virally infected host cell nucleoli to uninfected nucleoli that were ranked according to whether there appeared to be depletion or enrichment in the infected cell nucleolus (Fig. 1 and supplemental Tables 1 and 2). Functional classification (Fig. 1A) revealed that the distribution of the functional roles of the detected proteins was similar to that of the previously reported human nucleolar proteome (29), indicating that the nucleoli isolated in this study were of purity comparable to the nucleolar preparations used in our previous proteomics studies. Interestingly, no viral proteins were detected in the nucleoli of the virally infected cells using adenoviral genome data in MASCOT searches, potentially reflecting a lack of abundance at that time point. Fig. 1B shows the changes in abundance of the detected proteins in isolated nucleoli after viral infection. Remarkably, a majority of the nucleolar proteins remained relatively unaffected by viral infection; very few proteins exhibited significant changes in levels (more than ±1-fold). A closer comparison of the nucleolar responses to viral infection and with previously reported data (29) on the effects of transcription inhibition by ActD is shown in Fig. 1C. The SILAC -fold changes of the nucleolar levels of selected proteins after viral inhibition and ActD treatment were very different, indicating that the human nucleolus reacts to these two perturbations in highly distinct manners. In particular, the effects of viral infection on components of the rRNA transcription machinery were distinct from the effects of ActD treatment. The SILAC ratios of ribosomal proteins showed a mild decrease after viral infection (Fig. 1D), whereas ActD treatment had a significant impact on a number of ribosomal proteins.
Fig. 1.
Detected nucleolar proteome. (A) Functional classification of 351 proteins identified and quantitated in untreated and adenovirus infected HeLa cells. (B) Dynamics of nucleolar proteins 18 hours after adenovirus infection. Proteins with positive normalized fold changes indicate increases in abundance in the nucleolus after viral infection. Those with negative fold changes indicate decreased abundance. (C) and (D) Comparison of responses in nucleolar levels of selected proteins after ActD treatment (green) and adenovirus infection (red). UBTF, upstream binding transcription factor.
We next investigated the adenovirus-induced changes on nucleolar composition. Of the proteins shown to be depleted, UBF was the most affected in accordance with previous reports (11). Protein B23.1 has also been shown to be sequestered into viral replication centers (12), and this depletion was also detected by our high throughput proteomics analysis as an almost 2-fold depletion compared with uninfected cells. Accordingly, we concentrated on proteins that, like B23.1 and UBF, showed a 2-fold depletion or enrichment in the nucleoli of infected cells (Table I).
Table I. List of proteins identified by SILAC as having a 2-fold quantitative change in the nucleolus after infection.
This table lists all the proteins identified as having a 2-fold enrichment or depletion in the infected cell nucleoli compared with the uninfected cell nucleoli. Each protein is listed with the gi number, the number of peptides identified, percent coverage a brief description of its function (if known), and a ratio of depletion (shown as a negative ratio) or enrichment (shown as a positive ratio). Pol, polymerase; FSCB, fibrous sheath CABYR binding protein.
| Protein | Accession number | Ratio | Peptides identified | Function |
|---|---|---|---|---|
| Proteins depleted from the nucleolus on infection | ||||
| UBF | gi|7657671 | −4.6 | 1 | Recruits RNA Pol I to rDNA promoter |
| U2AF65 | gi|228543 | −1.5 | 1 | Splicing factor |
| hPOP1 | gi|13124451 | −1.5 | 2 | pre-rRNA processing |
| Nopp140 | gi|434765 | −1.3 | 1 | Nucleolar assembly scaffold protein |
| hnRNPU isoform a | gi|74136883 | −1.2 | 1 | Member of hnRNP family |
| L29 | gi|4506629 | −1.2 | 1 | Component of the ribosome |
| SFPQ | gi|4826998 | −1.1 | 1 | Splicing factor |
| Histone H1c | gi|4885375 | −1.1 | 4 | Histone-related protein |
| hnRNPA2/B1 | gi|4504447 | −1.1 | 7 | Member of hnRNP family |
| WD repeat domain 18 | gi|3025445 | −1.0 | 3 | Unknown |
| Exportin 5 | gi|12407633 | −1.0 | 2 | Nuclear export factor |
| B23.1 | gi|10835063 | −1.0 | 4 | Ribosomal protein assembly, centrosome duplication control |
| Proteins enriched in the nucleolus on infection | ||||
| MFHAS1 | gi|4239895 | 18.8 | 1 | Unknown/potential oncogene |
| LRRC24 | gi|67003570 | 16.2 | 1 | Unknown |
| FSCB | gi|57999430 | 12.8 | 1 | Unknown |
| PIK3R6 (p87 PIKAP) | gi|58082081 | 9.4 | 1 | Kinase |
| CLPTM1 | gi|4502897 | 9.2 | 2 | Unknown |
| NUP210 | gi|27477134 | 5.8 | 2 | Nuclear pore complex component |
| Putative protein C21orf56 | gi|18202930 | 2.6 | 1 | Unknown |
| Predicted similar to hnRNPA1 | gi|42658495 | 2.4 | 2 | Unknown |
| PPM1L (PP2Cϵ) | gi|63003905 | 2.3 | 2 | PP2C group of phosphatases, regulates stress-activated kinases |
| DUSP11 | gi|4503415 | 2 | 1 | Phosphatase, binds RNA and splicing factors |
| C-NAP1 | gi|2984657 | 1.9 | 2 | Centrosome-located protein |
| Ribosomal protein S15a | gi|36142 | 1.9 | 1 | Ribosomal protein up-regulated in hepatitis B-infected liver cells |
2D Gel Analysis of Virally Infected Nucleoli
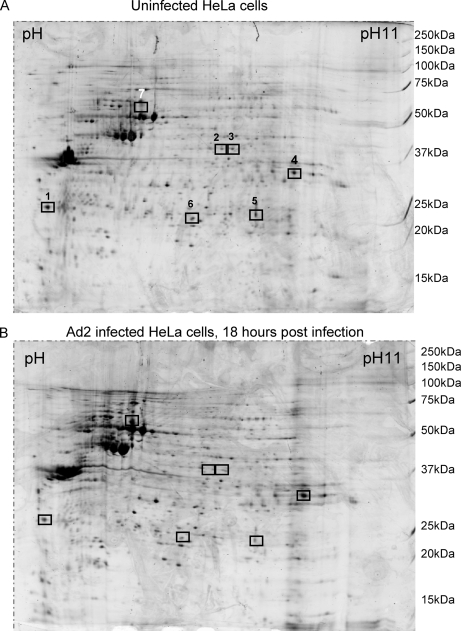
We repeated the isolation of nucleoli from uninfected and infected cells a further three times at 18 h postinfection. Western blotting confirmed the purity of the nucleolar fractions (data not shown). The nucleoli from these experiments were examined using 2D gel analysis, which showed that only a small number of proteins had clearly altered amounts at this time point. Fig. 2 shows a typical pair of gels, and seven proteins showing a change in quantity are marked. These spots were picked and sequenced as described under “Experimental Procedures.” Their identities are listed in Table II.
Fig. 2.
Typical 2D gel analysis of uninfected cell nucleoli compared with adenovirus-infected cell nucleoli. Isolated nucleoli were subjected to separation in two dimensions prior to staining and visualization as described under “Experimental Procedures.” Boxed proteins were consistently seen to vary in intensity in multiple experiments. Those spots were picked for sequencing. Note that spots 4 and 7 were apparently enriched in the nucleolar preparations after viral infection.
Table II. List of proteins identified by 2D analysis as having a consistent visual change in abundance in nucleoli during infection.
This table lists all the proteins identified as having a visual enrichment or depletion in the infected cell nucleoli compared with the uninfected cell nucleoli. Each protein is listed with the spot number as identified on the 2D gels in Fig. 2, the accession number, the number of peptides identified, the percent coverage, and a brief description of its function. IRES, internal ribosome entry site.
| Spot no. | Protein | Accession number | Unique peptides detected | Percent coverage | Function |
|---|---|---|---|---|---|
| % | |||||
| 1 | eIF6 (p27BBP) | gi|74717354 | 3 | 14 | Involved in ribosome biogenesis in the nucleolus and ribosome assembly in the cytoplasm |
| 2 | RBM4 | gi|74753808 | 2 | 7 | Control of splicing, promotes IRES-mediated translation |
| 3 | RBM4 | gi|74753808 | 4 | 16 | |
| 4 | hnRNPA2/B1 | gi|133257 | 2 | 7 | Member of hnRNP family |
| 5 | Cytokeratin 18 | gi|30311 | 8 | 18 | Intermediate filaments |
| 6 | KRT 8 | gi|74749889 | 3 | 12 | Intermediate filaments |
| 7 | Keratin 8 | gi|4504919 | 5 | 11 | Intermediate filaments |
Confocal Microscopy of Candidate Proteins Identified by SILAC
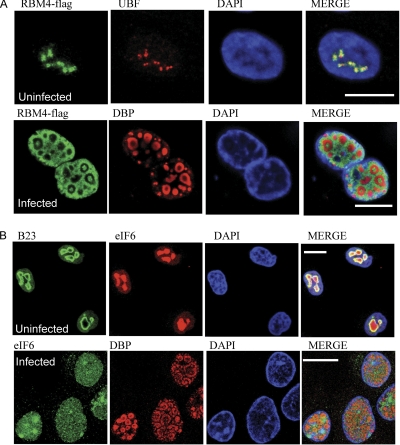
Because adenovirus induces significant changes in nuclear structure, it is possible that the nucleolar isolation protocol no longer reliably enriches intact nucleoli. Moreover, although many of the proteins identified could conceivably play a role in adenovirus infection, we wanted to independently verify any potential changes in the nucleolar proteome. This would have a further key benefit of prioritizing future work relating these changes to function. We therefore used a range of antisera and tagged expression constructs to examine the fate of a number of proteins identified as being enriched or depleted in the nucleolus. We concentrated on those proteins for which reagents and published data on subcellular location were already available so we could be confident of their locations in uninfected cells. We compared the location of target antigens relative to adenovirus DBP, a key component of the adenovirus DNA replication machinery that is expressed to high levels in the nucleoplasm but not the nucleolus in virus-infected cells. DBP has a distinct pattern of fluorescence and well defined localization relative to viral mRNA transcription and de novo viral DNA synthesis (50). Fig. 3 shows the effects of infection on those proteins apparently depleted from the nucleolus during viral infection. Two nucleolar antigens, hPOP1 and Nopp140, were clearly depleted from the nucleolus (Fig. 3, A and B). hPOP1 was sequestered from the nucleolus into a mottled pattern within the nucleoplasm and was excluded from the DBP-rich centers (Fig. 3A). Nopp140 protein showed some co-localization with viral DBP but also significant enrichment in centers adjacent to DBP, reminiscent of our previously published findings that UBF is sequestered into regions adjacent to DBP (11). We therefore compared UBF with Nopp140 to confirm that some Nopp140 is apparently sequestered in a manner similar to that of UBF (Fig. 3B, bottom row). The next group of proteins, hnRNPU isoform a, SFPQ, U2AF65, hnRNPA2/B1, histone H1c, and exportin 5 were not detectable in the nucleolus by immunofluorescence (Fig. 1, C, D, E, and F, top rows). However, they were consistently detected in the nucleolus using proteomics methods and are considered components of the nucleolar proteome (51). In adenovirus-infected cells, they all showed a pattern of exclusion from the DBP-rich replication centers apart from hnRNPA2/B1, which did not show any striking change in its distribution. However, we noted that hnRNPU, SFPQ, and U2AF65 were localized adjacent to DBP-rich centers, whereas histone H1c was clearly distinct and separated from the DBP-rich centers (Fig. 3G). Exportin 5 was also readily detectable in the cytoplasm of infected cells but not in uninfected cells (Fig. 3H).
Fig. 3.
Distribution of proteins identified by SILAC as being depleted from nucleolus during viral infection. All the images are of a fixed focal plane ∼0.3 μm in depth, the DAPI stain is in blue in all cases, and the bar represents 10 μm. A–H, in each case the top row of images is representative of the location of the indicated endogenous protein or expressed tagged fusion protein in >80% of cells examined. The second row of images shows the same indicated endogenous or expressed tagged protein in cells infected with adenovirus for 18 h. Viral infection was confirmed by anti-DBP serum. In B, a third row is shown in which cells were infected with adenovirus and the location of endogenous UBF was compared directly with that of endogenous Nopp140. A duplicate slide (not shown) confirmed that >99% of cells were infected in this experiment. In some cases, where compatible serum was available, a control nucleolar antigen, B23.1 or UBF, is also shown alongside the uninfected cell images. VSV, vesicular stomatitis virus; EGFP, enhanced GFP.
We also examined the localization of a number of proteins apparently enriched in the nucleolar fraction during infection (Fig. 4). PP2Cε is typical of proteins for which we were unable to detect any consistent change in distribution in adenovirus-infected cells (Fig. 4A). In a similar manner, we were unable to detect significant consistent changes in the subcellular location of C-NAP1 and nucleoporin 210 (data not shown). However, for PIK3-p87 (Fig. 4B) and ribosomal protein S15a (Fig. 4C), we did see some enrichment in the nuclear compartment that could account for the apparent increase in detection in the nucleolar fractions analyzed.
Fig. 4.
Distribution of proteins identified by SILAC as being enriched in nucleolus during viral infection. All the images are of a fixed focal plane ∼0.3 μm in depth, the DAPI stain is in blue in all cases, and the bar represents 10 μm. A–C, in each case the top row of images is representative of the location of the indicated endogenous protein or expressed tagged fusion protein in >80% of cells examined. The second row of images shows the same indicated endogenous or expressed tagged protein in cells infected with adenovirus for 18 h. Viral infection was confirmed by anti-DBP serum. In B, the use of a cyan fluorescent protein (CFP) fusion protein precluded the use of DAPI as a nuclear marker, and so antiserum to U2AF65 was used instead. In A, UBF is also shown alongside the uninfected cell images as a marker for the nucleolus.
Confocal Microscopy of Candidate Proteins Identified by 2D Gel Analysis
As with the SILAC data, we examined using immunofluorescence a number of candidate proteins identified by 2D gel analysis that were altered in abundance following infection. Fig. 5 shows that proteins RBM4 (Fig. 5A) and eIF6 (Fig. 5B) were both nucleolar antigens in uninfected cells. On the other hand, RBM4 was apparently recruited from the nucleolus into the nucleoplasm but with exclusion from the DBP-rich centers. However, eIF6 became more diffuse with reduced nucleolar location but, notably, without specific exclusion from DBP-rich centers.
Fig. 5.
Distribution of proteins identified by 2D as being depleted from nucleolus during viral infection. All the images are of a fixed focal plane ∼0.3 μm in depth, the DAPI stain is in blue in all cases, and the bar represents 10 μm. A and B, in each case the top row of images is representative of the location of the indicated endogenous protein or expressed tagged fusion protein in >80% of cells examined. The second row of images shows the same indicated endogenous or expressed tagged protein in cells infected with adenovirus for 18 h. Viral infection was confirmed by anti-DBP serum. In addition, as indicated in the images, a control nucleolar antigen (UBF or B23) is also shown in the uninfected cells.
Confocal Microscopy of Proteins Identified by SILAC as Having No Change in Nucleolar Abundance at 18 h Postinfection
For completeness, we tested a protein that did not change its location at this time point postinfection. Nop132 is a nucleolar antigen that has not been previously investigated in the context of adenovirus infection. We used a FLAG-tagged construct to examine its location in uninfected and infected cells. As can be seen from Fig. 6, Nop132 maintained its apparent nucleolar location with no detectable depletion from the nucleolus to other compartments.
Fig. 6.
Distribution of proteins identified by SILAC as having no change in distribution. All the images are of a fixed focal plane ∼0.3 μm in depth, the DAPI stain is in blue in all cases, and the bar represents 10 μm. As before, the top row of images is representative of the location of FLAG-tagged Nop132 fusion protein in >80% of cells examined. The second row of images shows the location of FLAG-tagged Nop132 in cells infected with adenovirus for 18 h. Viral infection was confirmed by anti-DBP serum.
DISCUSSION
Adenovirus Infection Induces a Change in Nucleolar Proteome
We utilized both quantitative MS-based proteomics techniques and more traditional 2D gel analysis to provide the first detailed analysis of changes in the human nucleolar proteome after infection with a human pathogen. These data reveal novel nucleolar antigens that were affected by adenovirus infection and therefore identify new candidate proteins that may play either a direct or indirect role in viral infection. In addition, our data illustrate the benefits offered by confocal microscopy when assessing proteomics analysis of a subcellular fraction. We feel this combination of techniques is crucial for several reasons. First, viral infection may have unsuspected effects on the integrity of cellular compartments that could affect the fractionation process. Second, this combination enables targets identified by proteomics analysis to be prioritized for further in-depth analysis. Finally, a proportion of the proteins in our SILAC analysis were only identified by one peptide, and for these proteins further verification is important.
Changes in Nucleolar Proteome: Adenovirus Compared with Actinomycin D
Comparison with the response of the nucleolar proteome after transcriptional inhibition by ActD treatment (in which the flux of ∼500 nucleolar proteins was characterized) was very revealing (29). Comparison with our data set showed that the effect of adenovirus infection is very specific with only a limited range of proteins showing a relative change in abundance (e.g. compare our Fig. 1B with Fig. 4a of Andersen et al. (29)). For example, although transcription inhibition by ActD causes depletion of ribosomal proteins of the 40 S subunit and, to a lesser extent, the 60 S subunit (29), this pattern was not observed in viral infection (Fig. 1D). This was interesting because adenovirus has been shown to eventually affect ribosomal RNA export (2). However, our chosen time point of 18 h postinfection is the earliest time that such defects in rRNA export can be detected (2). As such, it is probable that defects in processing or export of ribosomal proteins may be only just beginning at this time point and are therefore not yet readily detectable by our methods.
Thus, adenovirus infection induces a change in the nucleolar proteome that is distinct from the classical nucleolar disruption caused either by transcription inhibition or by other general cellular stresses. Viral infection in this case induces a much more limited and targeted effect on the nucleolar proteome. Moreover, the 2D approach reinforces the observation from SILAC analysis that adenovirus infection does not induce dramatic changes in the nucleolar proteome at this 18-h postinfection time point. These observations are important because they argue against the effects of adenovirus infection on the nucleolus being a nonspecific consequence of viral damage to the host cell. Further work will establish how this targeted effect on the nucleolus relates to the adenovirus replication cycle.
Specific Effects of Adenovirus Infection on rRNA Transcription Complex
Fig. 1C (top panel) also reinforces the distinct effects of adenovirus infection compared with ActD treatment. Looking at rRNA transcription, upon ActD treatment, RNA polymerase I subunits and accessory transcription factors were similarly depleted from the nucleolus. However, in virally infected cells, nucleolar levels of the RNA polymerase subunits and transcription factor UBF were affected differently (Fig. 1C, bottom panel). This is consistent with previous observations during adenovirus infection where UBF was depleted from the nucleolus whereas RNA polymerase I remained nucleolar as evidenced by immunofluorescence using antibodies specific for RNA polymerase I, in situ visualization of ongoing RNA synthesis, and S1 nuclease protection assay of rRNA synthesis initiation (11). This suggests that adenovirus induces a specific removal of UBF from the nucleolus, independently of other components of the rDNA transcription complex. This gives us confidence that our data reflect genuine changes in the nucleolar proteome because we know of no other way of inducing this pattern of changes on the rDNA transcription complex.
Comparison with Other Established Effects of Adenovirus on Nucleolus
The SILAC data point to depletion of UBF and B23.1 during viral infection, which reflects previously published data by ourselves and others that UBF and B23.1 are functionally sequestered upon adenovirus infection into viral DNA replication centers (11–13). Another example is U2AF65, a splicing factor known to play a role in splicing of adenovirus transcripts (52, 53). Finally, exportin 5 is known to export adenovirus VA RNA, competing with cellular microRNA export and potentially modulating cellular protein expression (54). The identification of these proteins by SILAC/MS analysis further validates our approach because they are all well established functional partners in adenovirus infection. This gives us confidence that some, if not all, of the novel proteins examined here by microscopy will prove to have functional relevance to adenoviral infection.
Novel Proteins Depleted from Nucleolus on Infection
Of those proteins examined that show a clear nucleolar localization in uninfected cells, all had a dramatic redistribution in infected cells. For example, hPOP1 is normally involved in rRNA processing (42, 55), and its mislocalization may reflect data showing that adenovirus eventually inhibits rRNA processing during infection (2). Nopp140 mislocalization also fits with these observations because it is a chaperone for small nucleolar RNA (56, 57). However, its co-localization with UBF and DBP implies that it may be an accessory factor in viral DNA replication during viral infection. Notably, in interphase cells, UBF and Nopp140 are co-localized and are known to be recruited together (56). RBM4 has been reported to be either predominantly nuclear or predominantly nucleolar, depending on experimental conditions (43, 58–60). In our hands, the protein was nucleolar in >80% of cells. RBM4 has been implicated in the modulation of splice site selection, something of clear relevance to adenovirus infection, but it has also been suggested that this protein may inhibit cap-dependent translation while promoting internal ribosome entry site-mediated translation (43). This too may be significant because at later times in infection adenovirus makes use of a ribosome shunting method to promote the translation of its structural proteins at the expense of cellular mRNA translation (61). In a similar vein, ribosome assembly is regulated by eIF6, and it too may be used by adenoviruses to maximize the suitability of the infected cell for viral protein expression (41, 62).
Changes Detected in Predominantly Nuclear Antigens
Many proteins in the current nucleolar proteome were not readily visualized in the nucleolus by fluorescence microscopy. However, our data show that even in these cases quantitative changes in nucleolar content are reflected in visible changes in protein distribution. For example, U2AF65, hnRNPU, and SFPQ (also known as PSF) are all known to be involved in mRNA metabolism, and at least U2AF65 has been shown to play a role in adenovirus mRNA metabolism (38, 52, 63). As such, it seems reasonable to expect that these factors will be sequestered into regions adjacent to adenovirus DBP because these regions are known to be active in viral mRNA synthesis (50). Indeed, U2AF65, hnRNPU, and SFPQ were all apparently sequestered into a similar pattern that would be consistent with a role for these proteins in viral mRNA metabolism. However, we do not see any particular sequestration into these regions for hnRNPA2/B1, which weakens the case hnRNPA2/B1 plays a role in viral mRNA metabolism, something which can only be determined through further experimentation.
Cellular Proteins Are Enriched in Nucleolus after Adenovirus Infection
That we detected cellular proteins enriched in the nucleolus was intriguing, and this was reflected in changes in distribution of p87 PIKAP and S15a by immunofluorescence (Fig. 4, B and C). That ribosomal protein S15a is enriched in the nucleus/nucleolus during infection again may reflect eventual interference by adenovirus with aspects of rRNA maturation and/or export.
The enrichment of p87 PIKAP (also known as PIK3R6) is also interesting. This protein is a regulatory subunit of phosphoinositide 3-kinase γ, which is part of the phosphoinositide 3-kinase signaling pathway. These pathways cover a very wide range of responses from cellular growth to cellular motility. However, activation of phosphoinositide 3-kinase γ is linked mainly to host-wide responses to injury or infection (64). Indeed, it is clear that adenovirus does directly modulate the phosphoinositide 3-kinase pathway (65).
A further set of proteins apparently enriched in the nucleolus were not detected by microscopy, possibly reflecting differences in sensitivity of proteomics methods compared with microscopy. Thus, a protein may be enriched in the nucleolus but still be below the level of detection by microscopic methods.
Proteins Unaffected by Adenovirus Infection
We also thought it a relevant control to examine a protein that did not apparently change in nucleolar abundance by 18 h postinfection. Nop132 is thought to help recruit dead box protein DDX47 to the nucleolus and therefore play a role in rRNA processing (48). As can be seen in Fig. 6, there is no apparent change in the pattern of distribution of Nop132 at the time examined.
Localization Patterns of Sequestered Cellular Antigens Are Heterogeneous
Interestingly, there was a diverse range of localization changes in response to infection. For example, eIF6 was diffuse throughout the infected nucleoplasm. Alternatively, hnRNPU, SFPQ, and U2AF65 were adjacent to DBP-rich centers, whereas histone H1c was cleared from the DBP-rich centers and regions adjacent to the DBP centers. This suggests that exclusion of cell proteins from DBP-rich centers is not simply a nonspecific feature of the adenovirus-infected cell nucleoplasm. Moreover, hPOP1 was excluded from the DBP-rich centers but was clearly enriched in a mottled pattern distinct from Nopp140 and U2AF65 for example. Thus, there is, as might be expected, enrichment of nucleolar antigens in distinct regions of the infected nucleoplasm. This suggests, not surprisingly, that distinct aspects of adenovirus replication benefit from different nucleolar antigens.
Comparison between SILAC and 2D Gel Analysis
Comparing 2D gel analysis and SILAC approaches revealed that the SILAC method provided more information. However, only one protein (hnRNPA2/B1) was identified by both methods, and the depletion of eIF6 and RBM4 was not detected by SILAC. This reflects two key points: first, the lower sensitivity of the 2D gel approach, and second, that for the SILAC analysis, we only divided the one-dimensional gel electrophoresis sample into six slices prior to analysis.
As with previous proteomics investigations of subcellular components, we anticipate that further refinement over time will be possible and will yield a more detailed picture of the effects of adenovirus on this important structure both at the 18-h postinfection time point analyzed here and other time points. Also of note is the lack of viral proteins identified by either method. In part, this is a consequence of the time point chosen as the expression of late adenovirus proteins that are known nucleolar antigens would have only just begun. Other explanations are inherent in the methodology; i.e. the viral proteins may dissociate from nucleoli under the conditions present in the nucleolar isolation protocol.
Several members of the cytoskeletal family were also highlighted by the 2D gel approach. This may well reflect the observation that adenovirus protease cleaves cytokeratin 18 during infection, leading to altered cytoskeletal structure (66, 67).
Concluding Remarks
Our data show that adenovirus specifically targets aspects of the nucleolar proteome in contrast to nonspecific disruption by agents like ActD, providing the first high throughput insight into viral-nucleolar interactions. Moreover, by examining the nucleolar proteome, we were able to highlight misdistribution of antigens from all compartments of the cell. We believe this demonstrates the utility of proteomics approaches to generate meaningful data sets that will help us understand the role of the nucleolus in viral infection and to uncover previously unsuspected effects on the host cell during viral infection. Indeed, subcellular fractionation of this sort coupled with confocal microscopy provides a powerful new tool for understanding viral host cell interactions in a broad context and is a positive step toward a systematic understanding of virus host cell interactions.
Acknowledgments
We thank the Wolfson Bioimaging facility at the University of Bristol.
Footnotes
 The on-line version of this article (available at http://www.mcponline.org)
contains supplemental Fig. 1 and Tables 1 and 2.
The on-line version of this article (available at http://www.mcponline.org)
contains supplemental Fig. 1 and Tables 1 and 2.
1 The abbreviations used are:
- UBF
- upstream binding factor
- 2D
- two-dimensional
- ActD
- actinomycin D
- Ad5
- adenovirus type 5
- DAPI
- 4′,6-diamidino-2-phenylindole
- DBP
- DNA-binding protein
- GFP
- green fluorescent protein
- eIF6
- eukaryotic translation initiation factor 6
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- hPOP1
- human processing of precursor 1
- m.o.i.
- multiplicity of infection
- NOLC1
- nucleolar and coiled body phosphoprotein 1
- Nop132
- nucleolar protein 132
- Nopp140
- nucleolar phosphoprotein 140
- PIKAP
- phosphoinositide 3-kinase adapter protein
- PP2C
- protein phosphatase 2C
- RBM4
- RNA binding motif protein 4
- SFPQ
- splicing factor, proline- and glutamine-rich
- SILAC
- stable isotope labeling by amino acids in cell culture
- snoRNP
- small nucleolar ribonucleoprotein
- U2AF
- U2 small nuclear ribonucleoprotein particle auxiliary factor
- PSF
- pre-mRNA splicing factor.
REFERENCES
- 1.Matthews D. A. (2001) Adenovirus protein V induces redistribution of nucleolin and B23 from nucleolus to cytoplasm. J. Virol. 75, 1031–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castiglia C. L., Flint S. J. (1983) Effects of adenovirus infection on rRNA synthesis and maturation in HeLa cells. Mol. Cell. Biol. 3, 662–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee T. W., Lawrence F. J., Dauksaite V., Akusjärvi G., Blair G. E., Matthews D. A. (2004) Precursor of human adenovirus core polypeptide Mu targets the nucleolus and modulates the expression of E2 proteins. J. Gen. Virol. 85, 185–196 [DOI] [PubMed] [Google Scholar]
- 4.Lee T. W., Blair G. E., Matthews D. A. (2003) Adenovirus core protein VII contains distinct sequences that mediate targeting to the nucleus and nucleolus, and colocalization with human chromosomes. J. Gen. Virol. 84, 3423–3428 [DOI] [PubMed] [Google Scholar]
- 5.Matthews D. A., Russell W. C. (1998) Adenovirus core protein V is delivered by the invading virus to the nucleus of the infected cell and later in infection is associated with nucleoli. J. Gen. Virol. 79, 1671–1675 [DOI] [PubMed] [Google Scholar]
- 6.Puvion-Dutilleul F., Christensen M. E. (1993) Alterations of fibrillarin distribution and nucleolar ultrastructure induced by adenovirus infection. Eur. J. Cell Biol. 61, 168–176 [PubMed] [Google Scholar]
- 7.Rodrigues S. H., Silva N. P., Delício L. R., Granato C., Andrade L. E. (1996) The behavior of the coiled body in cells infected with adenovirus in vitro. Mol. Biol. Rep. 23, 183–189 [DOI] [PubMed] [Google Scholar]
- 8.Lutz P., Puvion-Dutilleul F., Lutz Y., Kedinger C. (1996) Nucleoplasmic and nucleolar distribution of the adenovirus IVa2 gene product. J. Virol. 70, 3449–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miron M. J., Gallouzi I. E., Lavoie J. N., Branton P. E. (2004) Nuclear localization of the adenovirus E4orf4 protein is mediated through an arginine-rich motif and correlates with cell death. Oncogene 23, 7458–7468 [DOI] [PubMed] [Google Scholar]
- 10.Martinez-Palomo A., Le Buis J., Bernhard W. (1967) Electron microscopy of adenovirus 12 replication. 1. Fine structural changes in the nucleus of infected KB cells. J. Virol. 1, 817–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawrence F. J., McStay B., Matthews D. A. (2006) Nucleolar protein upstream binding factor is sequestered into adenovirus DNA replication centres during infection without affecting RNA polymerase I location or ablating rRNA synthesis. J. Cell Sci. 119, 2621–2631 [DOI] [PubMed] [Google Scholar]
- 12.Hindley C. E., Davidson A. D., Matthews D. A. (2007) The relationship between adenovirus DNA replication proteins and nucleolar proteins B23.1 and B23.2. J. Gen. Virol. 88, 3244–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okuwaki M., Iwamatsu A., Tsujimoto M., Nagata K. (2001) Identification of nucleophosmin/B23, an acidic nucleolar protein, as a stimulatory factor for in vitro replication of adenovirus DNA complexed with viral basic core proteins. J. Mol. Biol. 311, 41–55 [DOI] [PubMed] [Google Scholar]
- 14.Hiscox J. A. (2002) The nucleolus—a gateway to viral infection? Arch. Virol. 147, 1077–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hiscox J. A. (2007) RNA viruses: hijacking the dynamic nucleolus. Nat. Rev. Microbiol. 5, 119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyne J. R., Whitehouse A. (2006) Nucleolar trafficking is essential for nuclear export of intronless herpesvirus mRNA. Proc. Natl. Acad. Sci. U.S.A. 103, 15190–15195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michienzi A., Cagnon L., Bahner I., Rossi J. J. (2000) Ribozyme-mediated inhibition of HIV 1 suggests nucleolar trafficking of HIV-1 RNA. Proc. Natl. Acad. Sci. U.S.A. 97, 8955–8960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stow N. D., Evans V. C., Matthews D. A. (2009) Upstream-binding factor is sequestered into herpes simplex virus type 1 replication compartments. J. Gen. Virol. 90, 69–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiscox J. A., Wurm T., Wilson L., Britton P., Cavanagh D., Brooks G. (2001) The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J. Virol. 75, 506–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S. H., Macfarlane S., Kalinina N. O., Rakitina D. V., Ryabov E. V., Gillespie T., Haupt S., Brown J. W., Taliansky M. (2007) Interaction of a plant virus-encoded protein with the major nucleolar protein fibrillarin is required for systemic virus infection. Proc. Natl. Acad. Sci. U.S.A. 104, 11115–11120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim S. H., Ryabov E. V., Kalinina N. O., Rakitina D. V., Gillespie T., MacFarlane S., Haupt S., Brown J. W., Taliansky M. (2007) Cajal bodies and the nucleolus are required for a plant virus systemic infection. EMBO J. 26, 2169–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saphire A. C., Gallay P. A., Bark S. J. (2006) Proteomic analysis of human immunodeficiency virus using liquid chromatography/tandem mass spectrometry effectively distinguishes specific incorporated host proteins. J. Proteome Res. 5, 530–538 [DOI] [PubMed] [Google Scholar]
- 23.Fang C., Yi Z., Liu F., Lan S., Wang J., Lu H., Yang P., Yuan Z. (2006) Proteome analysis of human liver carcinoma Huh7 cells harboring hepatitis C virus subgenomic replicon. Proteomics 6, 519–527 [DOI] [PubMed] [Google Scholar]
- 24.Lai C. C., Jou M. J., Huang S. Y., Li S. W., Wan L., Tsai F. J., Lin C. W. (2007) Proteomic analysis of up-regulated proteins in human promonocyte cells expressing severe acute respiratory syndrome coronavirus 3C-like protease. Proteomics 7, 1446–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang X. S., Tang L. Y., Dai J., Zhou H., Li S. J., Xia Q. C., Wu J. R., Zeng R. (2005) Quantitative analysis of severe acute respiratory syndrome (SARS)-associated coronavirus-infected cells using proteomic approaches: implications for cellular responses to virus infection. Mol. Cell. Proteomics 4, 902–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brasier A. R., Spratt H., Wu Z., Boldogh I., Zhang Y., Garofalo R. P., Casola A., Pashmi J., Haag A., Luxon B., Kurosky A. (2004) Nuclear heat shock response and novel nuclear domain 10 reorganization in respiratory syncytial virus-infected a549 cells identified by high-resolution two-dimensional gel electrophoresis. J. Virol. 78, 11461–11476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pastorino B., Boucomont-Chapeaublanc E., Peyrefitte C. N., Belghazi M., Fusaï T., Rogier C., Tolou H. J., Almeras L. (2009) Identification of cellular proteome modifications in response to West Nile virus infection. Mol. Cell. Proteomics 8, 1623–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mannová P., Fang R., Wang H., Deng B., McIntosh M. W., Hanash S. M., Beretta L. (2006) Modification of host lipid raft proteome upon hepatitis C virus replication. Mol. Cell. Proteomics 5, 2319–2325 [DOI] [PubMed] [Google Scholar]
- 29.Andersen J. S., Lam Y. W., Leung A. K., Ong S. E., Lyon C. E., Lamond A. I., Mann M. (2005) Nucleolar proteome dynamics. Nature 433, 77–83 [DOI] [PubMed] [Google Scholar]
- 30.Boisvert F. M., van Koningsbruggen S., Navascués J., Lamond A. I. (2007) The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 8, 574–585 [DOI] [PubMed] [Google Scholar]
- 31.Lam Y. W., Trinkle-Mulcahy L., Lamond A. I. (2005) The nucleolus. J. Cell Sci. 118, 1335–1337 [DOI] [PubMed] [Google Scholar]
- 32.Andersen J. S., Lyon C. E., Fox A. H., Leung A. K., Lam Y. W., Steen H., Mann M., Lamond A. I. (2002) Directed proteomic analysis of the human nucleolus. Curr. Biol. 12, 1–11 [DOI] [PubMed] [Google Scholar]
- 33.Matthews D. A., Russell W. C. (1994) Adenovirus protein-protein interactions: hexon and protein VI. J. Gen. Virol. 75, 3365–3374 [DOI] [PubMed] [Google Scholar]
- 34.Lam Y. W., Lamond A. I., Mann M., Andersen J. S. (2007) Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr. Biol. 17, 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lam Y. W., Lamond A. I. (2006) Isolation of nucleoli, in Cell Biology: a Laboratory Handbook (Celis J. E. ed) pp. 103–108, Elsevier, Burlington, MA [Google Scholar]
- 36.Chepanoske C. L., Richardson B. E., von Rechenberg M., Peltier J. M. (2005) Average peptide score: a useful parameter for identification of proteins derived from database searches of liquid chromatography/tandem mass spectrometry data. Rapid Commun. Mass Spectrom. 19, 9–14 [DOI] [PubMed] [Google Scholar]
- 37.Rudnick P. A., Wang Y., Evans E., Lee C. S., Balgley B. M. (2005) Large scale analysis of MASCOT results using a Mass Accuracy-based THreshold (MATH) effectively improves data interpretation. J. Proteome Res. 4, 1353–1360 [DOI] [PubMed] [Google Scholar]
- 38.Obrdlik A., Kukalev A., Louvet E., Farrants A. K., Caputo L., Percipalle P. (2008) The histone acetyltransferase PCAF associates with actin and hnRNP U for RNA polymerase II transcription. Mol. Cell. Biol. 28, 6342–6357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kittur N., Zapantis G., Aubuchon M., Santoro N., Bazett-Jones D. P., Meier U. T. (2007) The nucleolar channel system of human endometrium is related to endoplasmic reticulum and R-rings. Mol. Biol. Cell 18, 2296–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lian Z., Liu J., Li L., Li X., Tufan N. L., Wu M. C., Wang H. Y., Arbuthnot P., Kew M., Feitelson M. A. (2004) Human S15a expression is upregulated by hepatitis B virus X protein. Mol. Carcinog. 40, 34–46 [DOI] [PubMed] [Google Scholar]
- 41.Gandin V., Miluzio A., Barbieri A. M., Beugnet A., Kiyokawa H., Marchisio P. C., Biffo S. (2008) Eukaryotic initiation factor 6 is rate-limiting in translation, growth and transformation. Nature 455, 684–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Welting T. J., Kikkert B. J., van Venrooij W. J., Pruijn G. J. (2006) Differential association of protein subunits with the human RNase MRP and RNase P complexes. RNA 12, 1373–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin J. C., Hsu M., Tarn W. Y. (2007) Cell stress modulates the function of splicing regulatory protein RBM4 in translation control. Proc. Natl. Acad. Sci. U.S.A. 104, 2235–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conn K. L., Hendzel M. J., Schang L. M. (2008) Linker histones are mobilized during infection with herpes simplex virus type 1. J. Virol. 82, 8629–8646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yi R., Doehle B. P., Qin Y., Macara I. G., Cullen B. R. (2005) Overexpression of exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. RNA 11, 220–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saito S., Matsui H., Kawano M., Kumagai K., Tomishige N., Hanada K., Echigo S., Tamura S., Kobayashi T. (2008) Protein phosphatase 2Cepsilon is an endoplasmic reticulum integral membrane protein that dephosphorylates the ceramide transport protein CERT to enhance its association with organelle membranes. J. Biol. Chem. 283, 6584–6593 [DOI] [PubMed] [Google Scholar]
- 47.Voigt P., Dorner M. B., Schaefer M. (2006) Characterization of p87PIKAP, a novel regulatory subunit of phosphoinositide 3-kinase gamma that is highly expressed in heart and interacts with PDE3B. J. Biol. Chem. 281, 9977–9986 [DOI] [PubMed] [Google Scholar]
- 48.Sekiguchi T., Hayano T., Yanagida M., Takahashi N., Nishimoto T. (2006) NOP132 is required for proper nucleolus localization of DEAD-box RNA helicase DDX47. Nucleic Acids Res. 34, 4593–4608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shenk T. (2001) Adenoviridae: the viruses and their replication, in Fields Virology (Knipe D. ed) 4th Ed., Vol. 2, pp. 2265–2299, Lippincott-Raven, Philadelphia [Google Scholar]
- 50.Pombo A., Ferreira J., Bridge E., Carmo-Fonseca M. (1994) Adenovirus replication and transcription sites are spatially separated in the nucleus of infected cells. EMBO J. 13, 5075–5085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ahmad Y., Boisvert F. M., Gregor P., Cobley A., Lamond A. I. (2009) NOPdb: Nucleolar Proteome Database—2008 update. Nucleic Acids Res. 37, D181–D184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gama-Carvalho M., Krauss R. D., Chiang L., Valcárcel J., Green M. R., Carmo-Fonseca M. (1997) Targeting of U2AF65 to sites of active splicing in the nucleus. J. Cell Biol. 137, 975–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lützelberger M., Backström E., Akusjärvi G. (2005) Substrate-dependent differences in U2AF requirement for splicing in adenovirus-infected cell extracts. J. Biol. Chem. 280, 25478–25484 [DOI] [PubMed] [Google Scholar]
- 54.Aparicio O., Razquin N., Zaratiegui M., Narvaiza I., Fortes P. (2006) Adenovirus virus-associated RNA is processed to functional interfering RNAs involved in virus production. J. Virol. 80, 1376–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Welting T. J., van Venrooij W. J., Pruijn G. J. (2004) Mutual interactions between subunits of the human RNase MRP ribonucleoprotein complex. Nucleic Acids Res. 32, 2138–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prieto J. L., McStay B. (2007) Recruitment of factors linking transcription and processing of pre-rRNA to NOR chromatin is UBF-dependent and occurs independent of transcription in human cells. Genes Dev. 21, 2041–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang C., Query C. C., Meier U. T. (2002) Immunopurified small nucleolar ribonucleoprotein particles pseudouridylate rRNA independently of their association with phosphorylated Nopp140. Mol. Cell. Biol. 22, 8457–8466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Markus M. A., Heinrich B., Raitskin O., Adams D. J., Mangs H., Goy C., Ladomery M., Sperling R., Stamm S., Morris B. J. (2006) WT1 interacts with the splicing protein RBM4 and regulates its ability to modulate alternative splicing in vivo. Exp. Cell Res. 312, 3379–3388 [DOI] [PubMed] [Google Scholar]
- 59.Markus M. A., Morris B. J. (2006) Lark is the splicing factor RBM4 and exhibits unique subnuclear localization properties. DNA Cell Biol. 25, 457–464 [DOI] [PubMed] [Google Scholar]
- 60.Markus M. A., Morris B. J. (2009) RBM4: a multifunctional RNA-binding protein. Int. J. Biochem. Cell Biol. 41, 740–743 [DOI] [PubMed] [Google Scholar]
- 61.Yueh A., Schneider R. J. (1996) Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev. 10, 1557–1567 [DOI] [PubMed] [Google Scholar]
- 62.Ceci M., Gaviraghi C., Gorrini C., Sala L. A., Offenhäuser N., Marchisio P. C., Biffo S. (2003) Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature 426, 579–584 [DOI] [PubMed] [Google Scholar]
- 63.Patton J. G., Porro E. B., Galceran J., Tempst P., Nadal-Ginard B. (1993) Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 7, 393–406 [DOI] [PubMed] [Google Scholar]
- 64.Hawkins P. T., Stephens L. R. (2007) PI3Kgamma is a key regulator of inflammatory responses and cardiovascular homeostasis. Science 318, 64–66 [DOI] [PubMed] [Google Scholar]
- 65.Shimwell N. J., Martin A., Bruton R. K., Blackford A. N., Sedgwick G. G., Gallimore P. H., Turnell A. S., Grand R. J. (2009) Adenovirus 5 E1A is responsible for increased expression of insulin receptor substrate 4 in established adenovirus 5-transformed cell lines and interacts with IRS components activating the PI3 kinase/Akt signalling pathway. Oncogene 28, 686–697 [DOI] [PubMed] [Google Scholar]
- 66.Brown M. T., McBride K. M., Baniecki M. L., Reich N. C., Marriott G., Mangel W. F. (2002) Actin can act as a cofactor for a viral proteinase in the cleavage of the cytoskeleton. J. Biol. Chem. 277, 46298–46303 [DOI] [PubMed] [Google Scholar]
- 67.Chen P. H., Ornelles D. A., Shenk T. (1993) The adenovirus L3 23-kilodalton proteinase cleaves the amino-terminal head domain from cytokeratin 18 and disrupts the cytokeratin network of HeLa cells. J. Virol. 67, 3507–3514 [DOI] [PMC free article] [PubMed] [Google Scholar]