Abstract
It is becoming increasingly clear that no single marker will have the sensitivity and specificity necessary to be used on its own for diagnosis/prognosis of tumors. Interpatient and intratumor heterogeneity provides overwhelming odds against the existence of such an ideal marker. With this in mind, our laboratory has been applying a long term systematic approach to identify multiple biomarkers that can be used for clinical purposes. As a result of these studies, we have identified and reported several candidate biomarker proteins that are deregulated in bladder cancer. Following the conceptual biomarker development phases proposed by the Early Detection Research Network, we have taken some of the most promising candidate proteins into postdiscovery validation studies, and here we report on the characterization of one such biomarker, the bladder cancer-associated protein (BLCAP), formerly termed Bc10. To characterize BLCAP protein expression and cellular localization patterns in benign bladder urothelium and urothelial carcinomas (UCs), we used two independent sets of samples from different patient cohorts: a reference set consisting of 120 bladder specimens (formalin-fixed as well as frozen biopsies) and a validation set consisting of 2,108 retrospectively collected UCs with long term clinical follow-up. We could categorize the UCs examined into four groups based on levels of expression and subcellular localization of BLCAP protein and showed that loss of BLCAP expression is associated with tumor progression. The results indicated that increased expression of this protein confers an adverse patient outcome, suggesting that categorization of staining patterns for this protein may have prognostic value. Finally, we applied a combinatorial two-marker discriminator using BLCAP and adipocyte-type fatty acid-binding protein, another UC biomarker previously reported by us, and found that the combination of the two markers correlated more closely with grade and/or stage of disease than the individual markers. The implications of these results in biomarker discovery are discussed.
Bladder cancer is the ninth most common cause of cancer worldwide for both sexes combined and the second most common malignancy of the genitourinary tract. Bladder neoplasias account for about 5% of all diagnosed cancers, affecting one in 4,000 people. An estimated 357,000 bladder cancer cases occurred in 2002 with more than 145,000 deaths reported in the same year (1). Malignancy of the urinary bladder comprises a large variety of histologically heterogeneous tumor types arising predominantly in the epithelial lining of the urinary bladder (urothelium) and the ureters. Tumor types of the urothelium include urothelial carcinomas (UCs),1 squamous cell carcinomas, and adenocarcinomas as well as other less frequent lesions (2). UCs account for more than 90% of bladder carcinomas and comprise a wide spectrum of lesions ranging in clinical severity from superficial bladder cancer to muscle-invasive and metastatic disease with the latter having a poor prognosis. Clinical management of the superficial form of the disease is currently done by endoscopic resection of the tumor supplemented with instillations of cytotoxic/cytostatic agents. Although intravesical instillations are widely used to avoid recurrences and even progression, up to 80% of patients with superficial bladder cancer lesions will recur, and of these, ∼25% will progress to invasive disease (3). Current prognostic parameters such as grade and stage, multifocality of carcinomas, and lymph node status cannot predict with certainty the long term outcome of bladder cancer, and as a result, there is a pressing need to identify markers that may be associated with bladder cancer progression and predict tumor behavior.
For the past years, our laboratory has carried out a systematic and comprehensive effort to identify protein markers that may form the basis for improved diagnosis and prognosis of patients with bladder cancer as well as identify novel potential targets for therapeutics (4). Toward this aim, we have examined the protein expression profiles of more than 1,000 samples (benign as well as tumors of various histopathological grades and stages) using gel-based proteomics, and we have identified a number of potentially useful biomarkers, such as adipocyte-type fatty acid-binding protein (A-FABP), 14-3-3 protein isoform σ, psoriasin (S100A7), GST Mu, 15-hydroxyprostaglandin dehydrogenase, tropomyosin 4, disulfide isomerase precursor (ERP60), homeobox protein (HOX-1.3), keratins 8 and 13, MRP-14, CD24, and the cytochrome oxidase III subunit among others (5–12), whose expression strongly correlates with a particular step of bladder cancer progression.
Recently, we identified bladder cancer-associated protein (BLCAP) as a novel potential biomarker based on a limited study of 30 bladder UCs (13). We showed that loss of BLCAP mRNA expression correlates with the invasive potential of UCs, and subsequent studies by others in several cancer types, such as cervical (14, 15) and renal (16) as well as human tongue carcinoma (17) and osteosarcoma (18), have all shown differential expression of this protein in cancer. Moreover, overexpression of BLCAP in the human cervical cancer HeLa cell line (14, 15) and tongue carcinoma Tca8113 cell line (17) has been shown to inhibit cell growth and to induce apoptosis, suggesting a role for this protein in the regulation of these cellular processes. BLCAP is a small (87-amino acid), evolutionary conserved protein with no homology to any know protein, and its cellular function is largely unknown. A computer-based search identified BLCAP as a target for RNA editing via adenosine to inosine (A-to-I) RNA editing catalyzed by members of the double-stranded RNA-specific adenosine deaminase acting on RNA (ADAR) family, a premise subsequently confirmed experimentally (19, 20). Selective A-to-I RNA editing is a mechanism to generate proteome variation that is carried out by the ADAR family of RNA molecules and is essential for life. Various diseases, such as epilepsy, depression, amyotrophic lateral sclerosis, and cancer, have been associated with a defective pattern of RNA editing (21–29). Although BLCAP has been shown to undergo A-to-I RNA editing (19, 20, 30–32), in bladder cancer, no correlation was found with altered BLCAP RNA editing levels and the development of transitional cell carcinoma (30). Also, the editing levels of a coding sequence target were only marginally higher in brain tumors (1.3-fold increase; p = 0.03) with lung and oral cavity samples showing no significant difference in editing levels between normal and cancer samples (26), suggesting that RNA editing of BLCAP transcript does not play a major role in cellular malignancy in these tissues either.
In the present study, we report on the detailed characterization of the BLCAP protein expression and cellular localization patterns in benign bladder urothelium and UCs in an effort to evaluate the value of BLCAP as a prospective biomarker in bladder cancer. We established novel reagents (antibodies) and characterized them extensively, and we developed and implemented a potentially useful assay, evaluated its reproducibility, and determined the appropriateness of the target for decision making in a very large number of well characterized samples (over 1,800 UCs were examined) with long term clinical follow-up. The design of the study allowed us to establish in a very large number of samples that BLCAP protein expression is lost with tumor progression. At the same time, we showed that BLCAP is overexpressed in ∼20% of the cases examined and that overexpression is linked with poor survival, suggesting that BLCAP may also have prognostic value.
MATERIALS AND METHODS
Patient Tissue Specimens
One hundred and twenty bladder specimens, consisting of random biopsies judged to be histologically normal and UCs of various grades and stages, collected over a period of 6 years at Skejby Hospital, Aarhus, Denmark were analyzed. Tumors were classified by an experienced pathologist according to Bergkvist et al. (33). All tumors were evaluated according to tumor-node-metastasis (TNM) stages and morphological grades and are presented according to standard nomenclature (e.g. pTaG1 is stage Ta, grade 1). The Scientific-Ethical Committee of Aarhus County approved the project.
Sample Preparation for Two-dimensional (2D) PAGE
Tumor samples clean of clots and contaminating tissue, as well as random biopsies diagnosed as non-malignant (clean of muscle and fat tissue) were dissected, split into small pieces with the aid of a scalpel, and subsequently labeled with [35S]methionine as previously described (6). Following labeling for 14–16h, the medium was carefully aspirated, and the pieces were dissolved in O'Farrell lysis solution (6).
Proteomics Analysis
2D PAGE was performed as described previously (6). Gels were stained with silver nitrate and subjected to autoradiography. 2D gel autoradiographs were scanned using a Molecular Imager device (Bio-Rad) and were analyzed using PDQuest 7.1 analysis software (Bio-Rad). Only gels presenting well focused spots and showing a limited amount of protein remaining at the origin were selected for further analysis.
Immunohistochemistry and Antibodies
Immunohistochemistry assays were performed using paraffin sections of mouse and human tissue samples essentially as described (34). Tissue blocks were placed in formalin fixative and subsequently paraffin-embedded for archival use. Five-micrometer sections were mounted on Super Frost Plus slides (Menzel-Gläser, Braunschweig, Germany), baked at 60 °C for 60 min, deparaffinized, and rehydrated through graded alcohol rinses. Heat-induced antigen retrieval was performed by immersing the slides in Tris/EDTA pH 9.0 buffer (10 mm Tris, 1 mm EDTA) and microwaving in a 750-watt microwave oven for 10 min. Nonspecific staining was blocked using 10% normal matched serum, 0.3% H2O2 in PBS buffer for 30 min. Antigen presence was visualized by incubation for 1 h with an appropriate primary antibody followed by detection with a suitable species-matched secondary antibody conjugated to a peroxidase complex for 30 min (Envision+ poly-HRP (horseradish peroxidase) system, Dako, Glostrup, Denmark). Color development was done using DAB+ (3,3′-diaminobenzidine) chromogen (Dako). Slides were counterstained with hematoxylin. Standardization of the incubation and development times allowed accurate comparisons in all cases. The BLCAP-specific EPO21513 (raised against a mid-protein peptide) and EPO21514 (raised against a C-terminal peptide of BLCAP) and the A-FABP-specific EPO23215 rabbit polyclonal antibodies were prepared by Eurogentec (Brussels, Belgium). Antibodies against von Willebrand factor (clone F8/86) and Ki67 (clone MIB-1) were purchased from Dako. The mouse monoclonal antibody against human FLT-4/vascular endothelial growth factor receptor 3 (VEGFR-3) (clone 9D9 F9) and the rabbit polyclonal anti-lymphatic vessel endothelial receptor 1 (LYVE-1) (SFI-16) antibody were kindly provided by Kari Alitalo. The mouse monoclonal anti-splicing factor 2/alternative splicing factor (SF2/ASF) antibody (clone AK96) was kindly provided by Adrian R. Krainer.
Tissue Microarray and Immunohistochemistry (IHC) Analysis
Construction of the bladder tumor tissue microarray (TMA) has been described in detail elsewhere (35, 36). Briefly, a total of 2,317 formalin-fixed, paraffin-embedded tissue samples of urinary bladder carcinomas representing 1,849 patients available from the archives of the Institute of Pathology at the University of Basel, the Cantonal Hospital St. Gallen, and the Triemli Hospital in Zürich, Switzerland were used. The tumors were from 1,842 patients and included 1,071 primary tumors, 207 recurrent tumors, and 1,039 biopsies with no associated information on previous history. Among the patients with multiple tumors, there were 234 with two and 92 with three or more different tumor specimens. Tumor stage and grade were defined according to UICC and WHO classifications (37, 38). There were 277 pTaG1, 567 pTaG2, 107 pTaG3, 194 pT1G2, 299 pT1G3, 142 pT2–4G2, and 423 pT2–4G3 specimens comprising a total of 2,108 UCs. Clinical data were collected retrospectively with clinical histories for 1,120 patients successfully recovered from patients' charts and contact of attending physicians. Recurrences were defined as cystoscopically visible tumors. Tumor progression was defined as the presence of muscle invasion (stage pT2 or higher) in a subsequent biopsy. Patients whose cause of death remained unknown were excluded from analyses of disease-specific survival. For TMA construction, a hematoxylin and eosin-stained section was made from each block to define representative tumor regions. Tissue cylinders with a diameter of 0.6 mm were then punched from selected tumor areas of each donor tissue block and brought into a recipient paraffin block using a custom-made precision instrument (Beecher Instruments, Silver Springs, MD).
Sections of the resulting TMA block (5 μm) were transferred to glass slides and stained essentially as described previously (11). Briefly, heat-induced antigen retrieval was performed by immersing freshly cut sections in Tris/EDTA pH 9.0 buffer (10 mm Tris, 1 mm EDTA) and microwaving in a 750-watt microwave oven for 10 min. The slides were then cooled at room temperature for 20 min and rinsed abundantly in tap water. Nonspecific staining was blocked using protein-free blocking buffer (Thermo Fisher Scientific) for 15 min, and endogenous peroxidase activity was quenched using 0.3% H2O2 in methanol for 30 min. Antigens were detected with the EPO23514 rabbit polyclonal anti-BLCAP antibody (dilution, 1:500) followed by detection of immune complexes with a horseradish peroxidase-labeled polymer (Envision+ detection kit, Dako) and 3,3′-diaminobenzidine (Dako) as a chromogen to detect bound antibody complexes. Slides were counterstained with hematoxylin.
Quantitative Assessment of IHC Staining and Data Analysis
An automated cellular imaging system, ACISTM III (DAKO), was used to digitize and quantify the IHC staining of the TMA slides. The ACIS III system was used to derive a score of staining intensity for each core in a given TMA. Briefly, the digital images from scanned TMA sections were submitted to analysis by the TMA software analysis module that is part of the ACIS III system. A mean total intensity score was generated in this manner for each core.
Indirect Immunofluorescence Analysis
Five-micrometer sections cut from paraffin blocks of bladder tissue samples were mounted on Super Frost Plus slides (Menzel-Gläser), baked at 60 °C for 60 min, deparaffinized, and rehydrated through graded alcohol rinses. Heat-induced antigen retrieval was carried out as described above. Following antigen retrieval, sections were treated with Image-iT FXTM signal enhancer (Molecular Probes, Eugene, OR) to block nonspecific staining and subsequently incubated with the relevant primary antibodies at the appropriate dilution. Detection of immune complexes was done with species-specific secondary antibodies conjugated to Alexa Fluor® 488 and Alexa Fluor 594 (Molecular Probes). Nuclear material was counterstained with TO-PRO-3TM. The sections were washed three times with cold PBS between incubations. Normal rabbit and mouse sera instead of primary antibody were used as a negative control. Sections were imaged using a Zeiss LSM510Meta confocal laser scanning microscope (Carl Zeiss MicroImaging GmbH, Göttingen, Germany).
2D Western Blot Analysis
The specificity of the BLCAP antibody was determined by 2D PAGE immunoblotting using protein extracts from COS1 cells overexpressing BLCAP according to published procedures (13, 39). Immunoblots of tissue lysates were prepared as described previously (12). Briefly, proteins were resolved by 2D gel electrophoresis, blotted onto HybondTM-C nitrocellulose membranes (Amersham Biosciences), and incubated with a BLCAP-specific rabbit polyclonal antibody (Eurogentec) followed by detection of immune complexes with a horseradish peroxidase-labeled polymer (1:200) (Envision+ detection kit, Dako).
Statistical Analysis
Contingency table analysis and χ2 tests were used to study the association between BLCAP expression/pattern of expression and histopathological classification (grade, stage, or grade/stage). Survival curves were calculated using the Kaplan-Meier method with significance evaluated using the log rank test. To assess the influence of several variables (BLCAP and A-FABP expression and patient and tumor characteristics) on the survival curves, we performed multivariate analysis using the Cox proportional hazards model. Statistical analysis of data was done with NCSS version 2007 (NCSS, Kaysville, UT).
RESULTS
BLCAP Protein Structure Analysis and Features: Generation and Characterization of Antibodies
BLCAP is a very small (87-amino acid; 10-kDa), evolutionary conserved protein (100% protein identity in Mammalia), and its cellular function is largely unknown. At present, there are no known human proteins with extended sequence similarity to BLCAP. We have shown previously, by hydropathy analysis (13), that the protein has two highly hydrophobic regions (positions 23–35 and 45–63). Although no matches to any known consensus were found, we demonstrated, by homology analysis of the individual hydrophobic and hydrophilic domains, that the second hydrophobic motif of 17 amino acid residues (positions 45–63) has high homology with transmembrane regions of various unrelated proteins (13).
Here we analyzed the primary sequence of BLCAP using protein topology prediction methods (TMHMM and TMMOD) (40, 41). These predicted BLCAP to be an integral transmembrane protein (total probability of N-in: the total probability that the N-term is on the cytoplastic side of the membrane 0.087213 for TMMOD and 0.01091 for TMHMM) with two transmembrane helices (positions 19–39 and 43–68) linked by a cytoplasmic loop (supplemental Fig. 1, a and b). Phosphorylation site analysis using NetPhos (42) and KinasePhos (43) identified three putative phosphorylation sites in the C terminus of the protein: Ser71 (Ataxia Telangiectasia Mutated phosphorylation site), Ser73 (Cdc2 phosphorylation site), and Ser78 (casein kinase II phosphorylation site). A signal peptide (region 1–19) with a single cleavage site (position 20) was predicted by SignalP (44) (shown in supplemental Fig. 1c). Based on these data, a working model of BLCAP was created (Fig. 1a) that was used to design two peptide-specific anti-BLCAP antibodies: EPO23514 raised against a unique C-terminal peptide of BLCAP (positions 74–87) and EPO23513 raised against a mid-protein peptide (positions 29–42). These antibodies, in addition to a previously described (13) rabbit polyclonal antibody raised against the full-length recombinant BLCAP protein, were used in this study to investigate the expression patterns of BLCAP in UCs.
Fig. 1.
a, structural model for BLCAP protein based on various predictive tools. According to various prediction tools, BLCAP is potentially a transmembrane protein with three putative phosphorylation sites (circled P) in the C terminus, a signal peptide (signalP) in the N-terminal part of the protein, and a cleavage site at position 20 (black arrowhead). b, determination of antibody specificity for EPO23514 by 2D PAGE Western immunoblotting (IEF) analysis of lysates from mock-transfected COS1 cells (COS) or COS1 cells overexpressing BLCAP (COS-BLCAP); a full sized immunoblot is shown for COS1 cells overexpressing BLCAP (left-hand panel) with matched relevant sections of the immunoblots from mock-transfected COS1 cells or COS1 cells overexpressing BLCAP shown in greater magnification in the right-hand panels.
Antibody specificity was determined by 2D Western blot analysis as described under “Materials and Methods,” and the results for EPO23514 are presented in Fig. 1b. Two polypeptide spots were detected in COS1 cells transiently transfected with BLCAP: a major spot corresponding to the predicted position of BLCAP (Fig. 1b, COS-BLCAP, black arrow), and one minor, more acidic spot (Fig. 1b, COS-BLCAP, white arrow). No antigen could be detected in mock-transfected COS1 cells (Fig. 1b, COS). To establish optimal IHC staining conditions and antibody performance in IHC assays, a preliminary tissue cross-reactivity screening study based on the Food and Drug Administration recommendation for antibody cross-reactivity testing was conducted using 33 different types of normal tissues. This IHC-based analysis, which will be published elsewhere, was analogous to the tissue-specific pattern of BLCAP mRNA expression (Ref. 13 and Gene Expression Omnibus GSE1133 data sets), supporting the conclusion that the antibody is specific toward the BLCAP protein in IHC-based analyses.
Expression of BLCAP in Reference Set Composed of 120 Benign Bladder Biopsies and UCs
To study changes in the pattern of BLCAP expression that might take place during progression from normal bladder urothelium to bladder cancer, we analyzed a reference set of 120 bladder specimens consisting of random biopsies judged to be histologically normal and UCs of various grades and stages (24 non-malignant biopsies, 11 pTaG1, 29 pTaG2, 10 pTaG3, 13 pT1G3, 23 pT2–4G3, and 10 pT2–4G4 tumors). This sample set consisted of formalin-fixed paraffin-embedded (FFPE) tissue samples as well as matched fresh frozen biopsies. Analysis of the specimens was performed by IHC and in many cases by gel-based proteomics.
Immunohistochemistry
IHC Analysis of BLCAP Expression and Localization in Benign Specimens
IHC staining of benign bladder specimens showed that the BLCAP antigen localizes predominantly to the epithelial lining of the urinary bladder (urothelium) with weak to moderate cytoplasmic staining and interspersed moderate to strong nuclear staining of urothelial cells (Fig. 2,a and b, EPO23514, black arrow). Interestingly, nuclear staining was more frequent in the more differentiated and specialized umbrella cells that line the inner surface of the urinary bladder (Fig. 2,a and b, EPO23514, yellow arrow). Expression of BLCAP was not restricted to urothelial cells as we also detected immunostaining of vessels (Fig. 2a, EPO23514, white arrow). Moreover, sporadic nuclear staining of stromal cells was also observed. The specificity of the EPO23514 anti-BLCAP antibody for IHC analyses was further confirmed by staining consecutive bladder FFPE tissue sections with antibody preincubated with the corresponding immunizing peptide. As shown in Fig. 2c, exposure of the antibody to the immunizing peptide prior to IHC effectively blocked immunostaining of tissue sections (Fig. 2c, blocking peptide), confirming the specificity of the antibody toward the antigen. The observed expression pattern of BLCAP in non-malignant bladder tissues was further substantiated by using two additional anti-BLCAP antibodies: a second rabbit peptide-specific antibody raised against a mid-protein epitope (Fig. 2d, EPO23513) and a previously described (13) rabbit polyclonal antibody raised against the full-length recombinant BLCAP protein (Fig. 2e, antibody P1T1). The staining pattern obtained with these two additional anti-BLCAP antibodies was analogous to the pattern generated with the EPO23514 antibody, reiterating that BLCAP is expressed in the vasculature (Fig. 2,d and e, EPO23513 and P1T1, respectively, white arrows) and in bladder epithelial cells (Fig. 2, d and e, EPO23513 and P1T1, respectively, black arrows) with increased frequency of cells with nuclear expression in the umbrella cells (Fig. 2, d and e, EPO23513 and P1T1, yellow arrows).
Fig. 2.
Analysis of pattern of expression of BLCAP in non-malignant bladder urothelium. a and b, immunohistochemical staining of BLCAP protein in a non-malignant bladder section (antibody EPO23514) demonstrated the presence of the BLCAP antigen in urothelial cells with weak cytoplasmic and moderate nuclear expression. The black arrow points to a basal cell, whereas the yellow arrow points to a specialized umbrella cell with both showing nuclear expression of the BLCAP protein. In b, the white arrow indicates a capillary vessel displaying a moderate BLCAP signal. c, no immunostaining was observed in a consecutive section incubated with BLCAP antibody preincubated with immunizing peptide (blocking peptide). A pattern of stainings analogous to that of antibody EPO23514 was observed with a different peptide-specific antibody (EPO23513) (d) and an antibody against recombinant BLCAP (P1T1) (e). f, blood vessel endothelial cells stained strongly for BLCAP protein in all samples examined. The inset is a higher magnification (×400) of a capillary showing reactivity for BLCAP antigen within the vessel. g, indirect double label immunofluorescence of FFPE bladder samples using antibodies against VEGFR-3 and BLCAP. Nuclei were visualized with TO-PRO-3. Scale bar, 10 μm. h, consecutive sections stained for BLCAP (upper panel) and vWF (lower panel) showed co-localization of these antigens in endothelial cells except in lymphatic vessels (cf. upper panel black arrow with lower panel white arrow).
Examination of highly vascularized areas showed that BLCAP localization to vessels was generally limited to the endothelial cells (ECs) lining the vessels (illustrated in Fig. 2f, black arrow). In some instances, however, we also observed immunoreactivity for the BLCAP antigen within vessels (Fig. 2f, inset). To verify that BLCAP was expressed in vascular ECs, we performed double immunofluorescence analysis of bladder FFPE tissue sections using antibodies against VEGFR-3 (Fig. 2g) and LYVE-1 (data not shown), two lymphatic vessel markers (45, 46), and von Willebrand factor (vWF; Fig. 2h), an endothelial cell marker (47, 48). As illustrated in Fig. 2h, BLCAP was expressed in vascular ECs, primarily in blood vessels (Fig. 2h, compare BLCAP expression in upper panel with vWF expression in lower panel). Although most lymphatic vessel ECs did not stain for BLCAP (Fig. 2h, BLCAP upper panel, black arrow), expression of this protein was not restricted to blood vessel ECs as we were able to detect some rare VEGFR-3-positive lymphatic ECs that also expressed the protein (Fig. 2g). This observation was corroborated by double immunofluorescence staining of bladder tissue sections with BLCAP and LYVE-1 (data not shown).
IHC Analysis of BLCAP Expression in Malignant Specimens
We categorized the IHC staining patterns of all the samples analyzed, both benign and malignant lesions, as falling into one of four types termed A through D (Table I and Fig. 3). Type A was characterized by moderate/strong cytoplasmic staining with strong nuclear staining in more than 80% of the urothelial cells (illustrated in Fig. 3a). Type B was defined as weak cytoplasmic staining with interspersed (<80% of the urothelial cells) moderate/strong nuclear staining of urothelial cells (illustrated in Fig. 3b). This staining type included, with one single exception, all of the benign samples (exemplified in Fig. 2b). Type C was defined as weak cytoplasmic staining with weak/absent nuclear staining (illustrated in Fig. 3c). Type D included all samples with no detectable immunoreactivity (exemplified in Fig. 3d).
Table I.
Relation between BLCAP expression and clinicopathologic parameters in bladder specimens
| Pathology classification | BLCAP staining pattern | |||
|---|---|---|---|---|
| Type Aa | Type Bb | Type Cc | Type Dd | |
| Benign | 1 | 24 | 0 | 0 |
| CISe | 0 | 2 | 0 | 1 |
| Grade | ||||
| G1 | 2 | 3 | 3 | 5 |
| G2 | 4 | 12 | 2 | 5 |
| G3 | 12 | 9 | 14 | 23 |
| Stage | ||||
| pTa | 9 | 17 | 9 | 13 |
| pT1 | 4 | 5 | 5 | 4 |
| pT2–4 | 8 | 4 | 8 | 21 |
a Moderate/strong cytoplasmic staining with strong nuclear staining (>80% of urothelial cells).
b Weak cytoplasmic staining with moderate/strong interspersed nuclear staining (<80% of urothelial cells).
c Weak cytoplasmic staining with weak or no nuclear staining.
d No staining.
e Carcinoma in situ.
Fig. 3.
Immunohistochemistry analysis of BLCAP expression in bladder tissue specimens. Patterns of expression of BLCAP were categorized into four staining types. These are illustrated with an invasive UC (specimen MOB 154-2) showing type A immunoreactivity for BLCAP (a), a low grade UC lesion scored by IHC as type B (specimen MOB 763-2) (b), a malignant lesion showing low level homogenous expression of BLCAP classified as type C staining (c), and a lesion with no immunoreactivity for BLCAP (type D staining) (d). e and f, levels of protein expression in lysates from the same specimens assessed by 2D Western blot analysis (antibody EPO21514). For comparison purposes, in the Western blots, a section of the corresponding silver-stained gels is shown with the position for actin indicated.
A specimen was assigned to a given staining type when more than 90% of it matched the parameters established for that category. In a few cases, because of tumor heterogeneity, a specimen was judged to consist of two or more types. All slides were independently reviewed by two of the authors, and in the few discrepant cases, a consensus was reached after joint review.
Weak immunoreactivity for BLCAP with interspersed nuclear staining (type B staining) was predominant in benign lesions (24 of 25 samples), and it was more frequent in non-invasive (17 of 48 pTa tumors; 35.4%) than in invasive (8 of 41 pT2–4 tumors; 19.5%) UCs. Equally, invasive tumors tended to lose expression of BLCAP with 21 of 41 pT2–4 tumors (51.2%) showing no expression of BLCAP (type D staining) compared with 13 of 48 pTa tumors (27.1%). Increased levels of nuclear BLCAP expression (type A staining) were observed in one single benign sample and in a median of 19.2 ± 2.5% of all the examined tumor specimens, independently of histological grade and stage. Also, type C staining patterns were rather ubiquitous, accounting for 21.2 ± 6.6% of all specimens, and also without a clear correlation to grade of atypia or stage of invasion.
In all cases, we found a good correlation between immunoreactivity of samples analyzed by IHC and protein expression levels ascertained by Western immunoblotting as shown in Fig. 3 (compare a and b with e and f, respectively). In general, samples showing strong IHC reactivity for BLCAP (Fig. 3a, type A tumor) exhibited high expression levels of this protein in 2D immunoblots (Fig. 3e), whereas specimens showing low/moderate or no IHC reactivity (Fig. 3b) displayed correspondingly low levels or absence of the protein (Fig. 3f). Furthermore, the relative levels of post-translationally modified BLCAP (Fig. 3, e and f, white arrows) also increased in a proportional manner in samples showing higher immunoreactivity, indicating that the increased reactivity seen in some samples correlated with increased overall expression and not with a preferential enhancement in the expression of a modified form of BLCAP.
In addition to establishing a correlation between BLCAP expression and progression of UCs, our IHC analysis of BLCAP expression allowed us to make an interesting observation. Down-regulation of BLCAP has been demonstrated in bladder (13), cervical (14, 15), and renal cancer (16) as well as in human tongue carcinoma (17) and osteosarcoma (18). In addition, overexpression of this protein in the human cervical cancer HeLa cell line (14, 15) and tongue carcinoma Tca8113 cell line (17) inhibited cell growth and induced apoptosis, indicating a role for BLCAP in the regulation of cell proliferation and apoptosis. Together, these observations have led others to suggest a role for BLCAP in epithelial cell malignancy. However, careful analysis of the 72 urothelial carcinomas (constituting 60% of the 120 specimens that comprise the reference sample set) with no detectable expression of BLCAP, ascertained by lack of immunoreactivity in IHC analysis (type D staining), showed that in all cases loss of BLCAP expression was a collective event as both epithelial and endothelial cells lacked BLCAP immunoreactivity. This was not due to a technical artifact because in one specimen in particular (Fig. 4, a and b, MOB 1742-1) we could observe a gradient of BLCAP expression in malignant cells within the same sample (Fig. 4a) with the staining ranging from low/moderate expression to loss of expression with both vascular (Fig. 4a, compare black arrow with white arrow) and endothelial cells following this pattern (Fig. 4a, compare yellow arrow with red arrow). A consecutive section of the same specimen incubated with an antibody against A-FABP (6) is presented in Fig. 4b to illustrate that the gradual loss of expression observed for BLCAP was not displayed by other markers (Fig. 4, cf. a with b, yellow arrows). In all cases, staining of tandem sections with antibodies against cytokeratins 5, 8, and 13 was used as additional staining controls to rule out possible artifacts (data not shown).
Fig. 4.
a, immunohistochemistry analysis of BLCAP expression demonstrating that loss of BLCAP expression in urothelial cells (yellow arrow) is accompanied by loss of expression in endothelial cells (black arrow) and that in areas of the lesion that still show expression of BLCAP in epithelial cells (red arrow) the endothelial cells also express BLCAP (white arrow). b, a section of the same specimen shown in a was incubated with an antibody against A-FABP and displayed strong expression of this marker even in the areas of the lesion that have lost BLCAP expression (cf. yellow arrows).
Gel-based Proteomics
In an effort to validate the IHC studies, we carried out a systematic 2D PAGE analysis of most of the bladder biopsies of both non-malignant and tumor origin that reacted positively with the antibodies in IHC. Silver staining of the 2D gels (Fig. 5, a, c, and d) as well as in some cases metabolic labeling with [35S]methionine for increased sensitivity (Fig. 5b) revealed that the BLCAP protein in all cases was expressed at levels that were below the detection limits of the assays and that could only be detected by 2D PAGE Western immunoblotting (cf. Figs. 4f and 5b). This was true both for whole lysates from samples showing normal-like (IHC staining type B) patterns of expression (Fig. 5, a and b) and for samples showing overexpression of BLCAP (type A IHC staining; Fig 5c). In addition, we could not detect BLCAP in 2D gels of the interstitial fluid that bathes the bladder tumor microenvironment (illustrated in Fig. 5d), indicating that our inability to detect BLCAP in whole tissue lysates was not due to externalization of this protein to the extracellular space but rather to the overall low levels of expression of this protein.
Fig. 5.
Proteomics analysis of fresh bladder specimens. Relevant areas from 2D PAGE (IEF) of representative primary bladder tumors are shown. The positions of A-FABP and 14-3-3 proteins as well as the position of the BLCAP protein, inferred from the Western blot analyses, are indicated. BLCAP protein expression was below detection levels in type B specimens either silver-stained (illustrated with MOB 1753-3) (a) or in [35S]methionine metabolically labeled fresh bladder specimens (illustrated with MOB 763-2) visualized by autoradiography (b). The same was true of A type specimens in cellular lysates (c) as well as interstitial fluid (d).
Analysis of BLCAP Expression Patterns in Sample Set Consisting of 2,108 UCs: Correlation with Clinicopathologic Status of Bladder Tumors
To ensure good statistical validity and considering that having an independent test sample set is an important feature in the validation of biomarkers, we proceeded to evaluate the potential relevance of BLCAP as a tumor marker by IHC analysis using a large TMA containing 2,317 samples from 1,849 bladder cancer patients (36). In 1,730 of the 2,108 UC cases (82%), the IHC yielded interpretable results (illustrated in Fig. 6a), and these are summarized in Table II. As in the case of the reference sample set, we found that the BLCAP protein expression patterns correlated with grade of atypia (p < 0.0001) and stage of invasion (p < 0.0001). Type B staining of BLCAP antigen, which was predominant in benign lesions as shown above, tended to be more prevalent in G1 (18.9%) and pTa non-invasive tumors (10.8%) than in G3 (2.6%) and pT2–4 (2.3%) invasive tumors, respectively. Loss of BLCAP expression (type D staining) was also associated with progression with 14.2% of G1 and 19.6% of pTa tumors showing no expression of BLCAP as opposed to 26.7% of G3 and 34.5% of pT2–4 tumors, respectively. Types C and A staining were ubiquitously present, accounting for about 45 and 25% of all tumors, respectively, independently of grade of atypia or stage of disease.
Fig. 6.
a, representative BLCAP immunostaining images of one of the tissue microarrays (TMA2; left-hand panel) and tissue cores (right-hand panels) showing high and low ACIS score values (core TMA2C7f and TMA3B9d, respectively). b, Tukey outlier box and whisker plot of the correlation of ACIS intensity scores for BLCAP immunostaining with tumor grade.
Table II.
Immunohistochemical analysis of BLCAP expression in a large set of UCs
| Pathological classificationa | Statistical analysisb | Unclassified cores | BLCAP staining type | ||||
|---|---|---|---|---|---|---|---|
| A | B | C | D | Total | |||
| Grade | |||||||
| G1 | χ2 = 84.222 (p < 0.0001) | 66 | 52 (24.5%) | 40 (18.9%) | 90 (42.4%) | 30 (14.2%) | 212 |
| G2 | 153 | 171 (22%) | 58 (7.6%) | 346 (44.6%) | 200 (25.8%) | 775 | |
| G3 | 159 | 213 (28.7%) | 19 (2.6%) | 313 (42%) | 198 (26.7%) | 743 | |
| Stage | |||||||
| pTa | χ2 = 73.966 (p < 0.0001) | 191 | 180 (23.7%) | 82 (10.8%) | 349 (45.9%) | 149 (19.6%) | 760 |
| pT1 | 98 | 104 (25%) | 22 (5.3%) | 201 (48.3%) | 89 (21.4%) | 416 | |
| pT2–4 | 87 | 133 (27.8%) | 11 (2.3%) | 169 (35.4%) | 165 (34.5%) | 478 | |
| Stage/grade | |||||||
| pTaG1 | χ2 = 126.908 (p < 0.0001) | 65 | 52 (24.5%) | 40 (18.9%) | 90 (42.5%) | 30 (14.1%) | 212 |
| pTaG2 | 107 | 97 (21.1%) | 39 (8.5%) | 217 (47.1%) | 107 (23.3%) | 460 | |
| pTaG3 | 19 | 31 (35.3%) | 3 (3.4%) | 42 (47.7%) | 12 (13.6%) | 88 | |
| pT1G2 | 20 | 35 (20.1%) | 15 (8.6%) | 81 (46.6%) | 43 (24.7%) | 174 | |
| pT1G3 | 57 | 69 (28.5%) | 7 (2.9%) | 120 (49.6%) | 46 (19.0%) | 242 | |
| pT2–4G2 | 21 | 34 (28.1%) | 4 (3.3%) | 42 (34.7%) | 41 (33.9%) | 121 | |
| pT2–4G3 | 66 | 99 (27.7%) | 7 (2.0%) | 127 (35.6%) | 124 (34.7%) | 357 | |
a Tumor stage and grade were defined according to UICC and WHO classifications.
b Contingency table analysis.
The data also showed that loss of expression of BLCAP is significantly correlated with grade and stage of disease (Table II; p < 0.0001). Thus, whereas only 14.1% of pTaG1 specimens showed no immunoreactivity for BLCAP, this number was 34.7% in pT2–4G3 tumors. Concomitantly, the number of specimens showing a type B staining pattern decreased from 18.9% in pTaG1 tumors to 2.0% in pT2–4G3 UCs.
Absolute Cellular Levels of BLCAP Staining as Means of Categorizing Tumors
Although statistically, and presumably clinically, significant, our categorization of samples according to patterns of staining is an intrinsically subjective approach requiring estimation of variable protein staining intensities as well as of the distribution pattern of the staining. Because we found a good correlation between protein expression as determined by Western immunoblotting and IHC staining, we evaluated the expression of BLCAP in the specimens present in the validation TMA using a computer-assisted image analysis system, ACIS III, with a TMA-specific application (49, 50). This application overlays a user-defined grid onto an image of the TMA, thus identifying the cores to be quantified and generating quantitative data regarding intensity of brown staining for each identified core with a linear relationship (illustrated in Fig. 5a, right-hand panels). It should be noted that this approach is not able to discriminate between the relative contribution of cancer cells and endothelial cells. It is conceivable that samples containing highly vascularized areas can present skewed score values representing a possible confusion factor in any analysis based on averaging methodologies.
In agreement with the studies described in the previous section, we found that the overall expression of BLCAP decreased with increasing grade of atypia (Fig. 6b; p < 0.0001). Thus, not only did the number of specimens showing no immunoreactivity for BLCAP increase with progression, but also a progressive down-expression of this protein was found with increasing grade of atypia (Fig. 6b; G1 versus G2 versus G3, Kruskal-Wallis analysis of variance test, p < 0.0001). Correlation of BLCAP staining scores with tumor invasion (pT1 and pT2–4) showed that expression of BLCAP followed a similar behavior; overall expression of this protein decreased with invasion (pT1 versus pT2–4, Mann-Whitney analysis of variance test, p < 0.0001).
Prognostic Relevance of BLCAP Expression in UCs
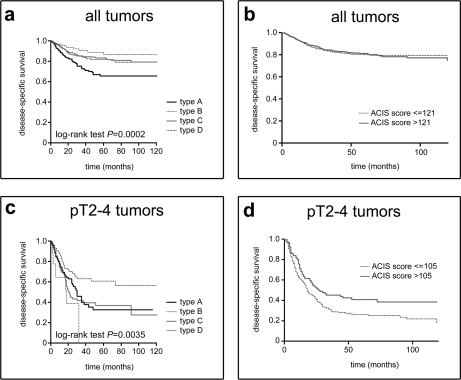
Next, we examined BLCAP expression for its prognostic relevance using the definition of prognostic factor proposed by G. M. Clark (51): “a prognostic factor is a measurement that is associated with clinical outcome in the absence of therapy or with the application of a standard therapy that patients are likely to receive.” We identified a total of 1,081 patients, with specimens present in the bladder TMA used in this study, for whom clinical data and interpretable BLCAP stainings were concurrently available. We found that the pattern of BLCAP expression, as determined by our IHC classification, had significant prognostic value with survival being correlated to the type of staining (p = 0.0002; Fig. 7a, all tumors). Thus, patients with samples with type A staining (n = 243) had a significantly shorter disease-specific survival than patients with tumors with type B (n = 80), type D (n = 276), or type C staining (n = 482).
Fig. 7.
Prognostic relevance of BLCAP expression. a, Kaplan-Meier survival curves by BLCAP expression pattern according to IHC analysis in all tumors. b, Kaplan-Meier survival curves by dichotomized BLCAP staining ACIS intensity score (median cutoff point). c, Kaplan-Meier survival curves by BLCAP expression pattern according to IHC analysis in pT2–4 UCs. d, Kaplan-Meier survival curves by dichotomized BLCAP staining ACIS intensity score (105 cutoff point is set to no immunoreactivity according to no-antibody control stainings) in pT2–4 UCs.
Survival analysis performed by the Cox proportional hazards model using the ACIS III intensity score as a continuous variable showed no prognostic value of the overall expression of BLCAP (hazard ratio (HR), 0.9992; 95% CI, 0.9943–1.0042). Dichotomization of intensity scores by use of the score median (intensity score, 121) and the 25th (intensity score, 112) and 75th (intensity score, 131) percentiles showed that none of these cutoff points were statistically significant (illustrated for score median in Fig. 7b; HR, 1.001; 95% CI, 0.7536–1.330), confirming that there was no significant association between overall expression of BLCAP and disease-specific survival.
Given that loss of BLCAP expression is associated with progression, itself a poor prognosis factor, we determined correlation of BLCAP staining with prognosis among patients bearing highly invasive tumors only (pT2–4). We found that expression of BLCAP correlated with disease-specific survival in these patients in an inverse fashion (p = 0.0035; Fig. 7c, pT2–4 tumors). In these cases, patients with tumors lacking BLCAP (type D; n = 77) had a better prognosis than patients bearing tumors with type A (n = 107), type C (n = 99), or type B staining (n = 7). Dichotomization of BLCAP to the cutoff point for staining intensity scores of BLCAP corresponding to lack of immunoreactivity (intensity score, 105; determined from no-antibody control stainings) was prognostic of disease-specific survival (HR, 1.455; 95% CI, 1.113–1.900) (Fig. 7d). The interesting additional implication of these results is that expression of BLCAP in tumors may worsen disease outcome for patients.
We also found that there was an increased likelihood of recurrence and shorter time to progression in samples showing type A staining (median, 13-month disease-free survival, 35 months to progression) as compared with type D staining (median, 19-month disease-free survival, 43 months to progression). However, none of these associations reached statistical significance.
Addressing Molecular Mechanisms Involved in BLCAP Deregulation
The interspersed character of the nuclear expression of BLCAP in benign samples was suggestive of an intermittent event such as cell cycle progression or a signaling event. Furthermore, the staining of serial sections with antibodies against BLCAP and cell cycle-regulated proteins (Ki67, cyclin D1, and proliferating cell nuclear antigen) showed no correlation (illustrated in Fig. 8a for Ki67 and data not shown). The one benign specimen that showed type A BLCAP staining displayed overall strong nuclear expression (data not shown), indicating that cellular differentiation state is not a major determinant of BLCAP overexpression and nuclear localization. Also, staining of serial sections with BLCAP and known differentiation state-specific proteins (uroplakin and 15-prostaglandin dehydrogenase) and oncoproteins (p53, c-Jun, and ErbB2) did not show any significant correlation (data not shown), suggesting that nuclear localization of BLCAP may instead occur in response to stimuli from the microenvironment.
Fig. 8.
Molecular mechanisms involved in BLCAP deregulation. a, immunohistochemistry analysis of tandem sections of a low grade tumor stained with antibodies against BLCAP and Ki67 showing the lack of correlation between the two stainings. b, indirect double label immunofluorescence analysis of tissue sections incubated with BLCAP (Alexa Fluor 488; green) and SF2/ASF (Alexa Fluor 594; red channel) antibodies and counterstained with the nuclear stain TO-PRO-3 (blue). In the merged image, co-localization of both antigens shows as yellow. Scale bar, 5 μm.
Our studies showed that about 20% of the lesions examined displayed strong nuclear expression of BLCAP (type A staining) and that this phenotype was associated with overall poor disease outcome. We addressed the possible nuclear function(s) of BLCAP by performing indirect immunofluorescence analysis of some of the FFPE tissue samples that showed type A or type B staining. We found that BLCAP was diffusely distributed throughout the nucleoplasm and showed a strong, irregular, punctate fluorescent pattern that did not include the nucleoli (Fig. 8b, BLCAP). This nuclear staining pattern is compatible with localization of this protein to a nuclear body/domain such as Cajal bodies, nuclear speckles, or promyelocytic leukemia bodies (52, 53). To address this possibility, we performed indirect double immunofluorescence analysis using antibodies against BLCAP and proteins that are archetypal markers of these different nuclear bodies (coilin, SF2/ASF, and promyelocytic leukemia protein) (53). We found that BLCAP co-localized with SF2/ASF (Fig. 8b), demonstrating its association with nuclear speckles and suggesting an involvement of BLCAP in RNA metabolism, an interesting possibility given that BLCAP mRNA undergoes ADAR-mediated selective A-to-I RNA editing (19, 20).
Multiprotein Pattern Analysis of UCs
The old paradigm of one protein-one biomarker-one clinical decision is currently shifting with single biomarker analysis being replaced by multiparametric analysis of genes or proteins, and protein patterns are currently considered to offer better diagnostic possibilities than a single biomarker (54). Earlier we presented evidence that loss of A-FABP expression correlates with both tumor stage and tumor grade (11), and therefore it seemed relevant to ascertain whether the combination of BLCAP and A-FAPB would be advantageous as compared with a single marker setup. To address this possibility, we stained the reference set of 120 bladder specimens with an antibody specific for A-FABP (11). As illustrated in Fig. 9, staining of non-malignant specimens showed that both the cellular expression and the localization of BLCAP and A-FABP were very different (Fig. 9, a and b, respectively) with A-FABP staining being stronger in the basal and suprabasal layers of the transitional epithelia and weak, but detectable, in the intermediate and more differentiated cell layers (Fig. 9b). Nuclear expression of BLCAP was not correlated with that of A-FABP as basal cells expressing A-FABP at high levels did (Fig. 9c, white arrow) or did not (Fig. 9c, yellow arrow) display a nuclear presence of BLCAP.
Fig. 9.
Immunohistochemistry analysis of tandem sections of benign specimen stained with antibodies against BLCAP (a) and A-FABP (b). c, indirect double label immunofluorescence analysis of a benign tissue section incubated with BLCAP (Alexa Fluor 488; green) and A-FABP (Alexa Fluor 594; red channel) antibodies and counterstained with the nuclear stain TO-PRO-3 (blue). Scale bar, 10 μm. Cells expressing A-FABP did (white arrow) or did not (yellow arrow) express BLCAP.
We originally categorized UCs into six groups based on their A-FABP IHC staining pattern (types A–F) (11); these are illustrated in Fig. 10a. Combining the different groups defined by categorization of IHC patterns of the two markers, BLCAP and A-FABP, generates 24 possible outcomes, but most of these are only represented by a few cases in our TMA. To simplify the multimarker-based analysis of the UCs in the TMA while allowing for sufficient number of cases in each category for appropriate statistical analysis, we reclassified the IHC stainings of all samples in the TMA using a simpler categorization system we termed normal-like/up-regulated/down-regulated (NUD) score. Thus, A-FABP staining types B and D were classified as normal-like (NA-FABP); types C, E, and F were classified as down-regulated (DA-FABP); and type A was classified as up-regulated (UA-FABP). Similarly, BLCAP staining type B was reclassified as normal-like (NBLCAP), types C and D were classified as down-regulated (DBLCAP), and type A was classified as up-regulated (UBLCAP). Correlation of the combined classifiers with grade of atypia and/or stage of disease is shown in Table III. As can be seen, some classifiers such as UBLCAPDA-FABP, which applies to 6.2% of pTaG1 tumors and 31.2% of pT2–4G3 tumors, respectively, and classifier DBLCAPUA-FABP, which applies to 17.6% of pTaG1 tumors and 0.8% of pT2–4G3 tumors, respectively, correlate very well with grade and/or stage of disease, indicating that multimarker categorization reflects the pathological state of UCs more closely than the single marker patterns.
Fig. 10.
Retrospective multimarker-based categorization of UCs using tissue microarray analysis. a, immunohistochemistry analysis of A-FABP in UC specimens illustrating the six different staining types we defined previously. Kaplan-Meier survival curves by BLCAP expression alone (b), A-FABP expression alone (c), and combination of both parameters (d) are shown.
Table III.
Association of two marker-based classification with clinicopathological variables in UCs
| Pathological classificationa | Total cores (n) | Combined NUD scores | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NBLCAP NA-FABP | NBLCAP UA-FABP | NBLCAP DA-FABP | DBLCAP UA-FABP | UBLCAP NA-FABP | UBLCAP UA-FABP | DBLCAP NA-FABP | UBLCAP DA-FABP | DBLCAP DA-FABP | ||
| % | ||||||||||
| Grade | ||||||||||
| G1 | 210 | 3.3 | 4.8 | 11.0 | 17.6 | 3.8 | 4.3 | 6.2 | 6.2 | 42.9 |
| G2 | 766 | 2.2 | 0.7 | 4.6 | 7.8 | 4.7 | 5.1 | 8.5 | 16.3 | 50.1 |
| G3 | 734 | 0.3 | 0.3 | 2.2 | 1.8 | 2.9 | 1.6 | 4.5 | 22.5 | 64.0 |
| Stage | ||||||||||
| pTa | 712 | 2.5 | 2.2 | 6.7 | 11.8 | 4.6 | 5.1 | 4.6 | 11.2 | 51.1 |
| pT1 | 452 | 1.8 | 0 | 3.5 | 3.8 | 3.8 | 3.3 | 1.3 | 17.3 | 65.3 |
| pT2–4 | 538 | 0 | 0.2 | 1.9 | 1.7 | 2.4 | 1.7 | 13.4 | 26.6 | 52.2 |
| Stage/grade | ||||||||||
| pTaG1 | 210 | 3.3 | 4.8 | 11.0 | 17.6 | 3.8 | 4.3 | 6.2 | 6.2 | 42.9 |
| pTaG2 | 455 | 2.4 | 1.3 | 4.8 | 9.7 | 4.8 | 5.3 | 12.1 | 13.4 | 46.2 |
| pTaG3 | 86 | 0 | 0 | 3.5 | 3.5 | 3.5 | 3.5 | 4.7 | 7.0 | 74.4 |
| pT1G2 | 173 | 3.5 | 0 | 5.2 | 4.6 | 4.6 | 5.2 | 5.2 | 15.0 | 56.6 |
| pT1G3 | 239 | 0.4 | 0 | 2.5 | 2.5 | 2.5 | 1.7 | 8.4 | 15.1 | 66.9 |
| pT2–4G2 | 119 | 0 | 0 | 3.4 | 5.0 | 3.4 | 3.4 | 0.8 | 27.7 | 56.3 |
| pT2–4G3 | 353 | 0 | 0.3 | 1.7 | 0.8 | 2.5 | 1.4 | 1.4 | 31.2 | 60.6 |
a Tumor stage and grade were defined according to UICC and WHO classifications.
Single marker association analysis with disease-specific survival showed that the NUD scores for BLCAP (p = 0.0013) and A-FABP (p = 0.001) have prognostic value with up-regulation of BLCAP (UBLCAP; Fig. 10b, U) and down-regulation of A-FABP (DA-FABP; Fig. 10c, D) being associated with poor disease outcome. The combination of the two markers showed a stronger association with disease outcome (p < 0.0001). The groups that do not contain any of the poor prognostic parameters (NBLCAPNA-FABP, NBLCAPUA-FABP, DBLCAPNA-FABP, and DBLCAPUA-FABP; Fig. 8d, NN, NU, DN, and DU, respectively) had the best prognosis followed by the groups containing a single poor prognostic parameter (NBLCAPDA-FABP and DBLCAPDA-FABP or UBLCAPNA-FABP and UBLCAPUA-FABP; Fig. 8d, ND and DD or UN and UU) with the group containing both poor prognostic parameters (UBLCAPDA-FABP) having the worse disease outcome, suggesting that the combination of these two biomarkers in multimarker analyses appears to add prognostic information (Fig. 10d).
DISCUSSION
Detecting a tumor at an early stage and predicting how it will behave are critical parameters in the clinical management of patients with malignancy of the urinary bladder. Current prognostic parameters such as grade and stage, multifocality of carcinomas, and lymph node status cannot predict with certainty the long term outcome of bladder cancer, and as a result, there is a pressing need to identify individual biomarkers or a group of biomarkers that may be associated with bladder cancer progression and that predict tumor behavior. The need to predict which superficial tumors will recur and/or progress has been a major driving force in our studies, and toward this aim, we have examined the protein expression profiles of more than 1,000 biopsies (both benign as well as of tumor origin of various histopathological grades and stages) using gel-based proteomics (2D gels). As a result of these studies, we have identified several potentially useful biomarkers (55). Differential protein expression of a few of these markers, including 14-3-3 protein isoform σ (12) and A-FABP (11), were confirmed using specific antibodies and large sample sets.
One of the main hurdles to the implementation of molecular markers into routine clinical practice is that most studies reporting the discovery of a novel cancer biomarker are largely of exploratory nature, and more often than not, promising results in the initial studies cannot be corroborated in follow-up studies. Moreover, there is often a substantial time gap between the original observation and subsequent validation studies, a crucial next step in the development of a biomarker for clinical use. The National Cancer Institute's Early Detection Research Network has set forth a series of five phases for developing biomarkers (56). Although this proposal does not solve all problems associated with biomarker development (for a broader discussion on the subject see Ref. 57), it does provide a formalized framework to guide this process and has been adopted by the Early Detection Research Network itself as well as other biomarker research projects. As a first approximation, our laboratory is currently in the process of validating the proteins we previously reported as being deregulated in bladder cancer as potentially useful biomarkers of malignancy, alone or in combination, with the aim of subsequently testing those that show some promise of clinical usefulness. Thus, as part of this ongoing effort, we have so far examined and characterized in more detail the expression of two potential biomarkers, A-FABP and 14-3-3 protein isoform σ (11, 12), and here we report on a third biomarker, BLCAP. These studies are important as they represent a required key step toward the development of a novel biomarker following its discovery.
We generated and characterized antibodies that specifically recognize BLCAP and were able to show that BLCAP localizes predominantly to the epithelial lining of the urinary bladder with weak to moderate cytoplasmic staining and interspersed moderate to strong nuclear staining of urothelial cells. We found that BLCAP IHC staining pattern types B and D, which were generally associated with benign/low grade and high grade invasive lesions, respectively, may be valuable as diagnostic indicators even if the type A and C staining patterns are not good classifiers because they appeared rather ubiquitously in all grades and stages. Interestingly, staining type A was significantly associated with poor disease-specific survival. 2D Western blot analysis of samples classified by IHC as type A or B (Fig. 3, e and f, respectively) showed that the increased immunoreactivity for BLCAP antigen observed by IHC in specimens corresponds to elevated levels of protein expression and that expression of both polypeptides (unmodified and modified forms; Fig. 3, e and f, black and white arrows, respectively) is increased. These data indicate that loss of BLCAP expression is associated with tumor progression, whereas an increased percentage of cells with high levels of nuclear BLCAP confers a poor prognosis. We showed that BLCAP is overexpressed in ∼20% of the cases examined and is linked with poor survival, suggesting that BLCAP expression may be of prognostic value. In invasive tumors, expression of BLCAP confers an adverse patient outcome with patients bearing tumors that have lost expression of BLCAP faring better than those with tumors expressing BLCAP at any levels of expression (Fig. 7c, pT2–4 tumors).
The concomitant loss of expression of BLCAP we observed in both bladder epithelial and vascular endothelial cells suggests that this phenomenon most likely reflects a change(s) in the cellular microenvironment rather than being a causal process in epithelial carcinogenesis. Several different cancer types, such as cervical cancer (14, 15), renal cancer (16), human tongue carcinoma (17), and osteosarcoma (18), have all been shown to have differential expression of BLCAP, implying that the change(s) in the microenvironment that triggers this cellular response may be of a general character rather than tissue-specific.
Clearly, the expression of BLCAP in bladder tissue is under various regulatory constraints, and Fig. 11 depicts a working model for the regulation of BLCAP expression that takes into consideration the different influences and staining types we observed. On the one hand, interspersed nuclear expression seems to be a response to cues/stimuli from the microenvironment, whereas on the other hand, expression is also dependent on cell differentiation. Another possible degree of complexity lies in the manifestation of genetic or epigenetic alterations that lead to epithelial overexpression of BLCAP in ∼20% of all tumor samples irrespective of tumor stage/grade (Table II). The latter suggest that some malignancy-associated event directly or indirectly, for example as a result of activating mutations in a transcriptional regulator, leads to increased expression of this protein. It should be noted that all examined samples showing no immunohistochemical reactivity to BLCAP had concurrent loss of expression in endothelial cells, ruling out the possibility that loss of expression can also be due to genetic or epigenetic changes. In addition, although some samples displayed augmented relative levels of BLCAP, assessed by either increased immunohistochemical reactivity or Western blot analysis, the absolute levels of this protein in all cases were never within the detection limits of 2D gel-based analyses. These data are consistent with the observation that high levels of BLCAP protein are deleterious for cells.2
Fig. 11.
Working model for regulation of BLCAP expression. The expression of BLCAP is interspersed nuclear in benign specimens and can remain so in tumors, but changes in the microenvironment associated with tumor progression lead to loss of BLCAP expression, presumably in a progressive manner, which would generate the samples showing homogeneous low level expression of BLCAP. Genetic/epigenetic alterations and/or alterations in the tumor microenvironment can lead to overexpression of BLCAP with a strong nuclear presence.
During the last two decades, a large number of molecular markers of bladder cancer with potential diagnostic and prognostic value have been identified, but when tested for their prognostic value, most markers have failed to provide any added prognostic information as compared with the clinicopathological index of the primary tumor. Presently, there is not a single marker able to predict with accuracy the biological potential of bladder cancer, and as a result, molecular prognostication is moving toward a multiplex marker setting (58–62). Multimarker phenotype analysis has been used by different groups to increase the diagnostic or prognostic value of markers. The combination of candidate markers spanning different biological pathways, in particular, has been investigated. We have explored a similar approach but using our own markers. Accordingly, we tried to determine whether the combination of two of the markers we have identified would be advantageous compared with a single marker setup. We used a two-marker discriminator that included BLCAP and A-FABP, another marker that we have previously identified (11), and found that it correlates very well with grade and/or stage of disease, suggesting that multimarker categorization using these two markers may reflect the pathological state of UCs more closely than using single markers on their own. In addition, the combination of these two markers showed a stronger association with disease outcome than single marker analysis, suggesting that combination of these markers in multimarker analyses may add prognostic information (Fig. 10d). Additional studies using more homogeneous sample cohorts (e.g. identical treatment, age-matched, and disease-matched) should establish whether the combination of these markers can contribute to a more accurate treatment of UCs. However, the acquisition of a large series of cases associated with high quality samples with clinical data is not a trivial task as it will necessitate the involvement of many laboratories in different countries in a large scale multicenter research project, a challenging prospect by all accounts (63).
In conclusion, we have been able to establish that down-regulation of BLCAP protein is associated with tumor progression presumably in response to changes in the microenvironment and that increased expression of this protein is detrimental for patients, indicating that categorization of staining patterns for this protein has prognostic value. Consequently, it is important to examine further the prognostic effects of BLCAP in particular to determine its ability to predict a differential clinical benefit from a specific treatment. The interspersed nuclear staining of BLCAP, with some cells showing the presence of this protein in the cytoplasm but not in the nucleus, suggests that localization of this protein to the nucleus is a regulated function. In this respect it is particularly interesting that protein topology prediction methods (supplemental Fig. 1, a and b) suggest BLCAP to be a transmembrane protein, whereas we observed nuclear and cytoplasmic localization. It is conceivable therefore that localization to the nucleus is a regulated process in response to stimuli from the microenvironment. Accordingly, BLCAP under certain conditions could localize transiently to the cytoplasmic membrane and in response to effectors present in the microenvironment move to the nucleus and/or cytoplasm.
Our results also suggested a relation between BLCAP nuclear expression levels and disease outcome, but the mechanism(s) underlying this phenomenon is at present unknown. We determined that BLCAP co-localizes to nuclear speckles with spliceosome factor SF2/ASF, a proto-oncogene that is up-regulated in various human tumors (64). Thus, the association we identified between nuclear patterns of staining for BLCAP and prognosis could reflect a potential role of this protein in RNA splicing. Given the nuclear localization of BLCAP, our laboratory is now investigating the possibility of BLCAP being functionally involved in RNA metabolism.
These data together with the results presented in this report underline the need for further research into a possible dual role of BLCAP in cancer cells. Dissecting the various pathways controlling BLCAP expression and localization and determining its function might provide a clearer understanding of the regulatory networks controlling expression and localization of this protein, facts that will enhance its usefulness as a bladder cancer biomarker alone or in combination with others and may provide additional insight into the molecular mechanisms underlying pathogenesis of the urinary bladder.
Supplementary Material
Acknowledgments
We thank Teresa Cabezón and Gary Clark for critical reading and comments on our paper that greatly improved it. We also thank Pamela Celis, Hanne Nors, Michael Radich Johansen, Ahmad Abdul-Al, Sofia Svensson, and Tanja Poulsen for expert technical assistance. We thank K. Alitalo (Biomedicum Helsinki) and A. Krainer (Cold Spring Harbor Laboratory) for providing reagents.
Footnotes
* This work was supported by grants from the Danish Cancer Society (to J. M. A. M. and J. E. C.) and from the Novo Nordisk Foundation (to I. G.).
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental Fig. 1.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental Fig. 1.
2 J. M. A. Moreira, unpublished observation.
1 The abbreviations used are:
- UC
- urothelial carcinoma
- BLCAP
- bladder cancer-associated protein
- IHC
- immunohistochemistry
- A-FABP
- adipocyte-type fatty acid-binding protein
- A-to-I
- adenosine to inosine
- ADAR
- adenosine deaminase acting on RNA
- 2D
- two-dimensional
- TMA
- tumor tissue microarray
- ACIS
- Automated Cellular Imaging System
- FFPE
- formalin-fixed paraffin-embedded
- VEGFR-3
- vascular endothelial growth factor receptor 3
- UICC
- International Union against Cancer
- WHO
- World Health Organization
- LYVE-1
- lymphatic vessel endothelial receptor 1
- SF2/ASF
- splicing factor 2/alternative splicing factor
- NUD
- normal-like/up-regulated/down-regulated
- EC
- endothelial cell
- vWF
- von Willebrand factor
- HR
- hazard ratio
- CI
- confidence interval
- TMHMM
- transmembrane hidden Markov model.
REFERENCES
- 1.Parkin D. M. (2008) The global burden of urinary bladder cancer. Scand. J. Urol. Nephrol. Suppl 12–20 [DOI] [PubMed] [Google Scholar]
- 2.Pauli B. U., Alroy J., Weinstein R. S. (1983) in The Pathology of Bladder Cancer (Bryan G. T., Cohen S. M. eds) Vol. II, pp. 41–140, CRC Press, Boca Raton, FL [Google Scholar]
- 3.Heney N. M. (1992) Natural history of superficial bladder cancer. Prognostic features and long-term disease course. Urol. Clin. North Am 19, 429–433 [PubMed] [Google Scholar]
- 4.Celis J. E., Gromov P. (2003) Proteomics in translational cancer research: toward an integrated approach. Cancer Cell 3, 9–15 [DOI] [PubMed] [Google Scholar]
- 5.Gromova I., Gromov P., Celis J. E. (1999) Identification of true differentially expressed mRNAs in a pair of human bladder transitional cell carcinomas using an improved differential display procedure. Electrophoresis 20, 241–248 [DOI] [PubMed] [Google Scholar]
- 6.Celis J. E., Ostergaard M., Basse B., Celis A., Lauridsen J. B., Ratz G. P., Andersen I., Hein B., Wolf H., Orntoft T. F., Rasmussen H. H. (1996) Loss of adipocyte-type fatty acid binding protein and other protein biomarkers is associated with progression of human bladder transitional cell carcinomas. Cancer Res 56, 4782–4790 [PubMed] [Google Scholar]
- 7.Ostergaard M., Wolf H., Orntoft T. F., Celis J. E. (1999) Psoriasin (S100A7): a putative urinary marker for the follow-up of patients with bladder squamous cell carcinomas. Electrophoresis 20, 349–354 [DOI] [PubMed] [Google Scholar]
- 8.Gromova I., Gromov P., Wolf H., Celis J. E. (1998) Protein abundancy and mRNA levels of the adipocyte type fatty acid binding protein correlate in non-invasive and invasive bladder transitional cell carcinomas. Int. J. Oncol 13, 379–383 [DOI] [PubMed] [Google Scholar]
- 9.Celis J. E., Celis P., Palsdottir H., Østergaard M., Gromov P., Primdahl H., Ørntoft T. F., Wolf H., Celis A., Gromova I. (2002) Proteomic strategies to reveal tumor heterogeneity among urothelial papillomas. Mol. Cell. Proteomics 1, 269–279 [DOI] [PubMed] [Google Scholar]
- 10.Ostergaard M., Rasmussen H. H., Nielsen H. V., Vorum H., Orntoft T. F., Wolf H., Celis J. E. (1997) Proteome profiling of bladder squamous cell carcinomas: identification of markers that define their degree of differentiation. Cancer Res 57, 4111–4117 [PubMed] [Google Scholar]
- 11.Ohlsson G., Moreira J. M., Gromov P., Sauter G., Celis J. E. (2005) Loss of expression of the adipocyte-type fatty acid-binding protein (A-FABP) is associated with progression of human urothelial carcinomas. Mol. Cell. Proteomics 4, 570–581 [DOI] [PubMed] [Google Scholar]
- 12.Moreira J. M., Gromov P., Celis J. E. (2004) Expression of the tumor suppressor protein 14-3-3σ is down-regulated in invasive transitional cell carcinomas of the urinary bladder undergoing epithelial-to-mesenchymal transition. Mol. Cell. Proteomics 3, 410–419 [DOI] [PubMed] [Google Scholar]
- 13.Gromova I., Gromov P., Celis J. E. (2002) Bc10: a novel human bladder cancer-associated protein with a conserved genomic structure downregulated in invasive cancer. Int. J. Cancer 98, 539–546 [DOI] [PubMed] [Google Scholar]
- 14.Zuo Z.H., Zhao M., Liu J., Wei Y., Wu X. X. (2006) Inhibitory effect of bladder cancer related protein gene on HeLa cell proliferation. Ai Zheng 25, 811–817 [PubMed] [Google Scholar]
- 15.Zuo Z., Zhao M., Liu J., Gao G., Wu X. (2006) Functional analysis of bladder cancer-related protein gene: a putative cervical cancer tumor suppressor gene in cervical carcinoma. Tumour Biol 27, 221–226 [DOI] [PubMed] [Google Scholar]
- 16.Rae F. K., Stephenson S. A., Nicol D. L., Clements J. A. (2000) Novel association of a diverse range of genes with renal cell carcinoma as identified by differential display. Int. J. Cancer 88, 726–732 [DOI] [PubMed] [Google Scholar]
- 17.Yao J., Duan L., Fan M., Yuan J., Wu X. (2007) Overexpression of BLCAP induces S phase arrest and apoptosis independent of p53 and NF-kappaB in human tongue carcinoma: BLCAP overexpression induces S phase arrest and apoptosis. Mol. Cell. Biochem 297, 81–92 [DOI] [PubMed] [Google Scholar]
- 18.Su H. C., Zhao Y. H., Fan D. G., Fan Q. Y., Zhang P., Wen Y. H., Liu Y. Y. (2003) Relationship between expression of BLCAP protein and malignancy of osteosarcoma. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 19, 465–466 [PubMed] [Google Scholar]
- 19.Levanon E. Y., Hallegger M., Kinar Y., Shemesh R., Djinovic-Carugo K., Rechavi G., Jantsch M. F., Eisenberg E. (2005) Evolutionarily conserved human targets of adenosine to inosine RNA editing. Nucleic Acids Res 33, 1162–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riedmann E. M., Schopoff S., Hartner J. C., Jantsch M. F. (2008) Specificity of ADAR-mediated RNA editing in newly identified targets. RNA 14, 1110–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brusa R., Zimmermann F., Koh D. S., Feldmeyer D., Gass P., Seeburg P. H., Sprengel R. (1995) Early-onset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science 270, 1677–1680 [DOI] [PubMed] [Google Scholar]
- 22.Gurevich I., Tamir H., Arango V., Dwork A. J., Mann J. J., Schmauss C. (2002) Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron 34, 349–356 [DOI] [PubMed] [Google Scholar]
- 23.Kawahara Y., Ito K., Sun H., Aizawa H., Kanazawa I., Kwak S. (2004) Glutamate receptors: RNA editing and death of motor neurons. Nature 427, 801. [DOI] [PubMed] [Google Scholar]
- 24.Maas S., Patt S., Schrey M., Rich A. (2001) Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc. Natl. Acad. Sci. U.S.A 98, 14687–14692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cenci C., Barzotti R., Galeano F., Corbelli S., Rota R., Massimi L., Di Rocco C., O'Connell M. A., Gallo A. (2008) Downregulation of RNA editing in pediatric astrocytomas: ADAR2 editing activity inhibits cell migration and proliferation. J. Biol. Chem 283, 7251–7260 [DOI] [PubMed] [Google Scholar]
- 26.Paz N., Levanon E. Y., Amariglio N., Heimberger A. B., Ram Z., Constantini S., Barbash Z. S., Adamsky K., Safran M., Hirschberg A., Krupsky M., Ben-Dov I., Cazacu S., Mikkelsen T., Brodie C., Eisenberg E., Rechavi G. (2007) Altered adenosine-to-inosine RNA editing in human cancer. Genome Res 17, 1586–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sommer B., Köhler M., Sprengel R., Seeburg P. H. (1991) RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 67, 11–19 [DOI] [PubMed] [Google Scholar]
- 28.Burns C. M., Chu H., Rueter S. M., Hutchinson L. K., Canton H., Sanders-Bush E., Emeson R. B. (1997) Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387, 303–308 [DOI] [PubMed] [Google Scholar]
- 29.Hoopengardner B., Bhalla T., Staber C., Reenan R. (2003) Nervous system targets of RNA editing identified by comparative genomics. Science 301, 832–836 [DOI] [PubMed] [Google Scholar]
- 30.Zilberman D. E., Safran M., Paz N., Amariglio N., Simon A., Fridman E., Kleinmann N., Ramon J., Rechavi G. (2009) Does RNA editing play a role in the development of urinary bladder cancer? Urol Oncol, in press [DOI] [PubMed] [Google Scholar]
- 31.Clutterbuck D. R., Leroy A., O'Connell M. A., Semple C. A. (2005) A bioinformatic screen for novel A-I RNA editing sites reveals recoding editing in BC10. Bioinformatics 21, 2590–2595 [DOI] [PubMed] [Google Scholar]
- 32.Kwak S., Nishimoto Y., Yamashita T. (2008) Newly identified ADAR-mediated A-to-I editing positions as a tool for ALS research. RNA Biol 5, 193–197 [DOI] [PubMed] [Google Scholar]
- 33.Bergkvist A., Ljungqvist A., Moberger G. (1965) Classification of bladder tumours based on the cellular pattern. Preliminary report of a clinical-pathological study of 300 cases with a minimum follow-up of eight years. Acta Chir. Scand 130, 371–378 [PubMed] [Google Scholar]
- 34.Celis J. E., Gromova I., Cabezón T., Gromov P., Shen T., Timmermans-Wielenga V., Rank F., Moreira J. M. (2007) Identification of a subset of breast carcinomas characterized by expression of cytokeratin 15: relationship between CK15+ progenitor/amplified cells and pre-malignant lesions and invasive disease. Mol. Oncol 1, 321–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richter J., Wagner U., Kononen J., Fijan A., Bruderer J., Schmid U., Ackermann D., Maurer R., Alund G., Knönagel H., Rist M., Wilber K., Anabitarte M., Hering F., Hardmeier T., Schönenberger A., Flury R., Jäger P., Fehr J. L., Schraml P., Moch H., Mihatsch M. J., Gasser T., Kallioniemi O. P., Sauter G. (2000) High-throughput tissue microarray analysis of cyclin E gene amplification and overexpression in urinary bladder cancer. Am. J. Pathol 157, 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nocito A., Bubendorf L., Tinner E. M., Süess K., Wagner U., Forster T., Kononen J., Fijan A., Bruderer J., Schmid U., Ackermann D., Maurer R., Alund G., Knönagel H., Rist M., Anabitarte M., Hering F., Hardmeier T., Schoenenberger A. J., Flury R., Jäger P., Fehr J. L., Schraml P., Moch H., Mihatsch M. J., Gasser T., Sauter G. (2001) Microarrays of bladder cancer tissue are highly representative of proliferation index and histological grade. J. Pathol 194, 349–357 [DOI] [PubMed] [Google Scholar]
- 37.International Union against Cancer (2002) in TNM Classification of Malignant Tumours (Sobin L. H., Wittekind C. eds) 6th Ed., John Wiley and Sons, New York [Google Scholar]
- 38.Mostofi F. (1973) in Histological Typing of Urinary Bladder Tumors, World Health Organization, Geneva [Google Scholar]
- 39.Celis J. E., Gromov P. (2000) High-resolution two-dimensional gel electrophoresis and protein identification using western blotting and ECL detection. EXS 88, 55–67 [DOI] [PubMed] [Google Scholar]
- 40.Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 41.Kahsay R. Y., Gao G., Liao L. (2005) An improved hidden Markov model for transmembrane protein detection and topology prediction and its applications to complete genomes. Bioinformatics 21, 1853–1858 [DOI] [PubMed] [Google Scholar]
- 42.Blom N., Gammeltoft S., Brunak S. (1999) Sequence- and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol 294, 1351–1362 [DOI] [PubMed] [Google Scholar]
- 43.Huang H. D., Lee T. Y., Tzeng S. W., Horng J. T. (2005) KPhos: a web tool for identifying protein kinase-specific phosphorylation sites. Nucleic Acids Res 33, W226–W229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. (2004) Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol 340, 783–795 [DOI] [PubMed] [Google Scholar]
- 45.Petrova T. V., Bono P., Holnthoner W., Chesnes J., Pytowski B., Sihto H., Laakkonen P., Heikkilä P., Joensuu H., Alitalo K. (2008) VEGFR-3 expression is restricted to blood and lymphatic vessels in solid tumors. Cancer Cell 13, 554–556 [DOI] [PubMed] [Google Scholar]
- 46.Baluk P., McDonald D. M. (2008) Markers for microscopic imaging of lymphangiogenesis and angiogenesis. Ann. N.Y. Acad. Sci 1131, 1–12 [DOI] [PubMed] [Google Scholar]
- 47.Matsumoto K., Soh S., Satoh T., Iwamura M., Ishikawa Y., Ishii T., Baba S. (2008) Distribution of lymphatic vessel network in normal urinary bladder. Urology 72, 706–710 [DOI] [PubMed] [Google Scholar]
- 48.Sehested M., Hou-Jensen K. (1981) Factor VII related antigen as an endothelial cell marker in benign and malignant diseases. Virchows Arch. A Pathol. Anat. Histol 391, 217–225 [DOI] [PubMed] [Google Scholar]
- 49.Wang S., Saboorian M. H., Frenkel E. P., Haley B. B., Siddiqui M. T., Gokaslan S., Wians F. H., Jr., Hynan L., Ashfaq R. (2001) Cellular Imaging System (ACIS)-assisted quantitation of immunohistochemical assay achieves high accuracy in comparison with fluorescence in situ hybridization assay as the standard. Am. J. Clin. Pathol 116, 495–503 [DOI] [PubMed] [Google Scholar]
- 50.Faith D. A., Isaacs W. B., Morgan J. D., Fedor H. L., Hicks J. L., Mangold L. A., Walsh P. C., Partin A. W., Platz E. A., Luo J., De Marzo A. M. (2004) Trefoil factor 3 overexpression in prostatic carcinoma: prognostic importance using tissue microarrays. Prostate 61, 215–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clark G. M. (2008) Prognostic factors versus predictive factors: examples from a clinical trial of erlotinib. Mol. Oncol 1, 406–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spector D. L. (2001) Nuclear domains. J. Cell Sci 114, 2891–2893 [DOI] [PubMed] [Google Scholar]
- 53.Spector D. L. (2006) SnapShot: cellular bodies. Cell 127, 1071. [DOI] [PubMed] [Google Scholar]
- 54.Rosenblatt R., Jonmarker S., Lewensohn R., Egevad L., Sherif A., Kälkner K. M., Nilsson S., Valdman A., Ullén A. (2008) Current status of prognostic immunohistochemical markers for urothelial bladder cancer. Tumour Biol 29, 311–322 [DOI] [PubMed] [Google Scholar]
- 55.Celis J. E., Gromova I., Moreira J. M., Cabezon T., Gromov P. (2004) Impact of proteomics on bladder cancer research. Pharmacogenomics 5, 381–394 [DOI] [PubMed] [Google Scholar]
- 56.Pepe M. S., Etzioni R., Feng Z., Potter J. D., Thompson M. L., Thornquist M., Winget M., Yasui Y. (2001) Phases of biomarker development for early detection of cancer. J. Natl. Cancer Inst 93, 1054–1061 [DOI] [PubMed] [Google Scholar]
- 57.Ransohoff D. F. (2007) How to improve reliability and efficiency of research about molecular markers: roles of phases, guidelines, and study design. J. Clin. Epidemiol 60, 1205–1219 [DOI] [PubMed] [Google Scholar]
- 58.Syrigos K. N., Karapanagiotou E., Harrington K. J. (2004) The clinical significance of molecular markers to bladder cancer. Hybrid Hybridomics 23, 335–342 [DOI] [PubMed] [Google Scholar]
- 59.Stavropoulos N. E., Filiadis I., Ioachim E., Hastazeris K., Tsimaris I., Kalogeras D., Stefanaki S., Agnantis N. J. (2002) Prognostic significance of p53, bcl-2 and Ki-67 in high risk superficial bladder cancer. Anticancer Res 22, 3759–3764 [PubMed] [Google Scholar]
- 60.Pfister C., Moore L., Allard P., Larue H., Lacombe L., Têtu B., Meyer F., Fradet Y. (1999) Predictive value of cell cycle markers p53, MDM2, p21, and Ki-67 in superficial bladder tumor recurrence. Clin. Cancer Res 5, 4079–4084 [PubMed] [Google Scholar]
- 61.Jain K. K. (2008) Innovations, challenges and future prospects of oncoproteomics. Mol. Oncol 2, 153–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zieger K. (2008) High throughput molecular diagnostics in bladder cancer—on the brink of clinical utility. Mol. Oncol 1, 384–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Riegman P. H., Morente M. M., Betsou F., de Blasio P., Geary P.; Marble Arch International Working Group on Biobanking for Biomedical Research (2008) Biobanking for better healthcare. Mol. Oncol 2, 213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karni R., deStanchina E., Lowe S. W., Sinha R., Mu D., Krainer A. R. (2007) The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol 14, 185–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.