Abstract
The three tandem-arrayed protocadherin (Pcdh) gene clusters, namely Pcdh-α, Pcdh-β, and Pcdh-γ, play important roles in the development of the vertebrate central nervous system. To gain insight into the molecular action of PCDHs, we performed a systematic proteomics analysis of PCDH-γ-associated protein complexes. We identified a list of 154 non-redundant proteins in the PCDH-γ complexes. This list includes nearly 30 members of clustered Pcdh-α, -β, and -γ families as core components of the complexes and additionally over 120 putative PCDH-associated proteins. We validated a selected subset of PCDH-γ-associated proteins using specific antibodies. Analysis of the identities of PCDH-associated proteins showed that the majority of them overlap with the proteomic profile of postsynaptic density preparations. Further analysis of membrane protein complexes revealed that several validated PCDH-γ-associated proteins exhibit reduced levels in Pcdh-γ-deficient brain tissues. Therefore, PCDH-γs are required for the integrity of the complexes. However, the size of the overall complexes and the abundance of many other proteins remained unchanged, raising a possibility that PCDH-αs and PCDH-βs might compensate for PCDH-γ function in complex formation. As a test of this idea, RNA interference knockdown of both PCDH-αs and PCDH-γs showed that PCDHs have redundant functions in regulating neuronal survival in the chicken spinal cord. Taken together, our data provide evidence that clustered PCDHs coexist in large protein complexes and have overlapping functions during vertebrate neural development.
The development of neural circuitry involves a complex interplay of cell-cell adhesion, interneuronal signaling, and assembly of intracellular macromolecular protein complexes. A group of protocadherin genes, known as clustered protocadherins (Pcdhs),1 are among the transmembrane signaling molecules that are implicated in this developmental process (1, 2). Protocadherins are type I transmembrane proteins, which share significant sequence homology with classic cadherins in their extracellular domain but possess distinct intracellular domains (3–5).
The vertebrate genomes encode nearly 60 protocadherin genes in three gene clusters including Pcdh-α, Pcdh-β, and Pcdh-γ, which are tandem-arrayed on the same chromosome and transcribed in the same direction (6–8). Individual Pcdh-α and Pcdh-γ transcripts contain a variable exon and a cluster-specific constant exon, whereas Pcdh-βs are encoded by a single variable exon. Clustered Pcdhs are expressed predominantly in the central nervous system, and individual neurons express distinct combinations of Pcdhs through a mechanism involving multiple promoter activation and allelic exclusion (9–16). PCDH proteins are targeted partly to synapses (9, 17–19), and PCDHs have been shown to mediate both homophilic and heterophilic cell-cell adhesion (12, 20–22). Two functions of clustered PCDHs in neural development have emerged from genetic studies on Pcdh-γ and Pcdh-α clusters. First, PCDHs are essential for the survival of specific neuronal subtypes. A dramatic increase of apoptosis in spinal interneurons and retina ganglion cells has been observed in Pcdh-γ-deleted mice (19, 23, 24). Similarly, Pcdh-α knockdown by morpholino in zebrafish causes neuronal loss during neurogenesis (25). Second, PCDHs play a role in the establishment of neuronal connectivity. Abnormal axon convergence of olfactory sensory neurons has been shown in Pcdh-α mutant mice (26), and synaptic development is severely impaired in the spinal cord of Pcdh-γ-null and hypomorphic mice (27).
Despite the importance of PCDHs in vertebrate neural development, it is still unclear how PCDHs mediate their function at the molecular level. Several binding partners of clustered PCDHs have been reported. For example, the Src family tyrosine kinase FYN, neurofilament M, and fascin were found to interact with PCDH-αs (9, 28). PCDH-αs have a heterophilic, calcium-dependent cell adhesion activity with β1 integrin (20). PCDH-γ isoform B1 can interact with the microtubule-destabilizing protein SCG10 (29). PCDH-αs and PCDH-γs are also associated with metalloproteinases and γ-secretases in both extracellular and cytoplasmic domains and undergo proteolytic cleavage (30–32). Recently, we demonstrated that PCDH-αs and PCDH-γs interact with two tyrosine kinases, focal adhesion kinase and PYK2, and negatively regulate their activities (33). In addition, among clustered PCDHs, PCDH-αs and PCDH-γs appear to be in complex with each other (28, 34). Taken together, these data suggest that PCDHs might function in large protein complexes that intersect with multiple intracellular signaling pathways.
As an important step to understand molecular action of clustered protocadherins, we performed a systematic proteomics survey of PCDH-γ-associated protein complexes. Here, we show that PCDH-γs are present in large macromolecular complexes of ∼1,000 kDa using both sucrose gradient (SG) ultracentrifugation and two-dimensional blue-native (2D BN)/SDS-PAGE methods. To further define the molecular composition of the complexes, we isolated PCDH-γ-associated complexes by affinity purification and identified proteins through mass spectrometry. From this analysis, we identified 142 putative PCDH-γ-associated proteins. We validated a selected set of the mass spectrometry-identified proteins in the PCDH-γ complexes. A number of PCDH-α and PCDH-β isoforms were found in complex with PCDH-γ in the brain, suggesting that they are present in similar functional complexes. We further confirmed that PCDH-α, -β, and -γ subfamily isoforms can form complexes with each other in HEK293T cells. Moreover, simultaneous knockdown of both PCDH-αs and PCDH-γs induced apoptosis in the developing chicken spinal cord. Thus, our data provide a molecular repertoire of PCDH complexes and demonstrate overlapping functions of clustered PCDHs.
EXPERIMENTAL PROCEDURES
Mice
All experiments were carried out on 129S7/C57BL/6-Tyrc-Brd F5 hybrids. Pcdh-γfusg/+ mice were generated as described previously (9, 17–19). The experimental procedures were approved by the Northwestern University Institutional Animal Care and Use Committee.
Antibodies
The rabbit anti-pan-PCDH-γ, anti-pan-PCDH-α, anti-neural cell adhesion molecule, and anti-SAP102 antibodies were described previously (19, 33, 35). The rabbit anti-chicken caspase-3 antibody was generated by Covance using a keyhole limpet hemocyanin-conjugated peptide corresponding to cleaved chicken caspase-3 (CRGTELDSGIEAD). The chicken caspase-3 antibody was verified by using commercially available anti-human caspase-3 antibody (Cell Signaling Technology, 9661), which also weakly reacts with active chicken caspase-3 despite sequence variation between species. The other antibodies used in this study were obtained from the following sources: rat anti-GFP beads (MBL, D153-8), mouse anti-β-tubulin (Developmental Studies Hybridoma Bank, E7), rabbit anti-synapsin I (Invitrogen, A6442), mouse anti-synaptophysin (Chemicon, MAB5258), goat anti-NMDAε2 (Santa Cruz Biotechnology, sc-1469), mouse anti-pan-cadherin (Sigma, C3678), mouse anti-V5 (AbD Serotec, SV5-PK1), mouse anti-FLAG M2 affinity resin (Sigma, F3165), rabbit anti-14-3-3 (Santa Cruz Biotechnology, sc-13959), rat anti-N-cadherin (Developmental Studies Hybridoma Bank, MNCD2), rat anti-R-cadherin (Developmental Studies Hybridoma Bank, MRCD5), rabbit anti-Ca2+/calmodulin-dependent protein kinase (CAMKII)-α (Sigma, C6974), rabbit anti-CAMKII-β (Abcam, ab22131), rabbit anti-CAMKII-γ (Upstate, 07-743), rat anti-α-catenin (Developmental Studies Hybridoma Bank, NCAT2), rabbit anti-β-catenin (Santa Cruz Biotechnology, sc-7199), rabbit anti-PCDH-β22 (Santa Cruz Biotechnology, sc-68407), mouse anti-postsynaptic density (PSD)-95 (ABR Affinity BioReagents, 6G6-1C9), rabbit anti-SNAP-25-interacting protein (SNIP) (Cell Signaling Technology, 3757), rabbit anti-SRC (Cell Signaling Technology, 2109), and horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology and Invitrogen).
Sucrose Gradient Ultracentrifugation and Western Blot Analysis
Brain tissues were homogenized in a buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 10 mm NaF, and 10 mm Na3VO4) supplemented with protease inhibitor mixture (Roche Applied Science) using a Dounce tissue grinder (tight pestle, 20 strokes). Nuclei and insoluble debris were removed by a low speed centrifugation at 500 × g for 10 min. The crude membrane fraction in the supernatant was collected by centrifugation at 26,000 × g for 20 min. The pellet was washed twice with the same buffer. Membrane-bound proteins were solubilized in the same buffer supplemented with Triton X-100 (added gradually until the solution turned clear; usually 1–4% depending on the protein concentration) and clarified by centrifugation at 26,000 × g for 20 min. Equal amounts of protein extracts from Pcdh-γ+/+ and Pcdh-γdel/del were separated on a 5–50% sucrose gradient at 33,000 rpm in an SW41Ti rotor at 4 °C for 17.5 h. 750-μl fractions were collected from top to bottom. Protein complexes from individual fractions were resolved by SDS-PAGE and transferred onto a Hybond-P poly(vinylidene difluoride) membrane (GE Healthcare). Western blots were probed with specific primary antibodies and a horseradish peroxidase-conjugated secondary antibody sequentially. Probed proteins were visualized using the SuperSignal West Pico Chemiluminescent Substrate (Pierce). BSA (66 kDa, 4.4 S), thyroglobulin (669 kDa, 19.4 S), and blue dextran (2,000 kDa, 52.6 S) were used as size standards for sucrose gradient ultracentrifugation (36).
2D BN/SDS-PAGE
BN gel electrophoresis was performed based on a method by Wittig et al. (37) with modifications. Brain tissues were homogenized in a buffer containing 25 mm imidazole-HCl, pH 8.0, 500 mm ε-aminocaproic acid, 1 mm EDTA, and protease inhibitor mixture (Roche Applied Science). Tissue debris and nuclei were removed by centrifugation at 500 × g for 10 min at 4 °C. The crude membrane proteins in the supernatant were collected by centrifugation at 16,100 × g for 20 min at 4 °C. The pellet was resuspended in the same buffer supplemented with 1% (w/v) dodecylmaltoside for at least 15 min on ice and clarified by centrifugation at 16,100 × g for 20 min at 4 °C. The supernatant was transferred into a new tube. After adding 10% (v/v) glycerol (final concentration) and loading buffer (20×; 750 mm ε-aminocaproic acid and 5% (w/v) Coomassie Blue G-250), an aliquot of the sample was applied to a 2.8–12% polyacrylamide gradient gel containing 25 mm imidazole-HCl, pH 8.0 and 500 mm ε-aminocaproic acid. The gel was run in “blue” cathode buffer (50 mm Tricine, 55 mm imidazole, and 0.02% Coomassie Blue G-250) and anode buffer (20 mm imidazole-HCl, pH 8.0) at 4 °C. The NativeMark unstained protein standard (Invitrogen) was used for molecular weight estimation. For the second dimension SDS-PAGE, the first dimension gel was excised with a razor blade and incubated in the SDS sample buffer for 30 min at room temperature. The gel lane was then placed on top of an SDS-PAGE resolving gel and run at room temperature. The separated proteins were analyzed by Western blot.
Affinity Purification of PCDH-γ Complexes
The whole brain tissues from the Pcdh-γfusg/fusg mice (ages from postnatal day 0 (P0) to P5) were used as starting materials for affinity purification (19). To enrich PCDH-γ-GFP complexes and reduce the complexity of protein composition, the total water-insoluble membrane proteins were subjected to a sucrose gradient ultracentrifugation as described above. Peak PCDH-γ-GFP fractions were pooled and incubated with monoclonal anti-GFP-agarose beads (MBL) at 4 °C overnight on a rotator. Anti-GFP beads were pelleted by centrifugation at 300 × g for 1 min and washed six times with buffer containing 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 10 mm NaF, 10 mm Na3VO4, and 1% Triton X-100. The bound proteins were eluted with the SDS sample buffer without β-mercaptoethanol. After removing the anti-GFP beads by centrifugation at 300 × g for 1 min, β-mercaptoethanol was added to a final concentration of 5% (v/v), and the sample was incubated at 95 °C for 4 min. The purified immune complexes were resolved on a 5–16% gradient SDS gel. of the immune complexes was used for Western blot analysis and were used for SYPRO Ruby staining (Bio-Rad). Protein bands were visualized using a UV light, and the image was taken using a charge-coupled device camera. SYPRO Ruby-stained bands were excised and stored at −80 °C for further mass spectrometry analysis.
Mass Spectrometry Analysis
The mass spectrometry analysis was performed by the Chicago Biomedical Consortium/University of Illinois at Chicago Research Resources Center Proteomics and Informatics Services Facility. The in-gel tryptic digestion was carried out by following the protocol described by Kinter and Sherman (38). Briefly, the gel bands were cut into 1-mm3 pieces, rinsed, and dehydrated, and the protein was reduced with DTT and alkylated with iodoacetamide in the dark prior to overnight digestion with trypsin at 37 °C in 50 mm ammonium bicarbonate. The peptides were concentrated and analyzed using the Thermo LTQ mass spectrometer equipped with a Dionex 3000 nanoflow HPLC system. The RAW data file was converted to mzXML (version 2.1) using the Institute for Systems Biology's readw.exe conversion tool (version 3.5) and then submitted to a SageN Research Sorcerer appliance running the Sequest search engine (version 27, revision 11). The RAW file was also converted to the Mascot generic format (MGF) using DTASuperCharge and then submitted to a Mascot search engine (version 2.2.0). Both search engines used a custom database consisting of mouse NCBI protein sequences (June 22, 2007) plus human NCBI keratin sequences plus the GFP sequence (35,893 protein entries) and were searched using 2.0-dalton tolerance for precursor ions and 0.6-dalton tolerance for fragment ions. The maximum number of missed cleavages was set to 2. Deamidation of asparagine and glutamine, oxidation of methionine, and iodoacetamide derivative of cysteine were specified in Mascot and Sequest as variable modifications. Results of both searches were combined using Proteome Software's Scaffold 2_02_02 software (2007). Peptide identifications were accepted if they could be established at greater than 50.0% probability. Protein identifications were accepted if they could be established at greater than 95.0% probability and contained at least two identified peptides. The average false positive rate was about 3.3% at this threshold in our analysis. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins that have shared peptides were all accepted if they also passed the cutoff criteria described above.
Plasmid Construction
The expression vectors 3xFLAG-Pcdh-αC2, 3xFLAG-Pcdh-γC5, and Pcdh-γC5 were described previously (33). For Pcdh-β19-V5 construct, Pcdh-β19 single exon gene was PCR-amplified from the genomic DNA using primers 5′-GTT TCG GAT CCA TGG AGA ATC AAG AGG GAC TCT ATC TGC-3′ (forward) and 5′-GAT ACT CGA GCG ATT ACA GTC CCT AAA TAA CCC AAA ACT ATT GC-3′ (reverse) and cloned into p cDNA3.1/V5-His vector (Invitrogen) using BamHI and XhoI sites. PBase and PiggyBac (PB) shRNA vectors were described previously (39). All shRNAs against chicken PCDH-α and PCDH-γ are designed to target common exons of all chicken PCDH-α or PCDH-γ isoforms. To construct shRNAs against chicken PCDH-α or PCDH-γ, the following pairs of oligos were annealed and cloned downstream of human H1 promoter between BbsI and XhoI in PB-CAG-EGFP-H1. For PCDH-α-shRNA1, primers 2020 (5′-TTT GCA ACA GCT GGA CCT TTA AAT TAT TAA CAT TTA AAG GTC CAG CTG TTG CTT TTT C-3′) and 2021 (5′-TCG AGA AAA AGC AAC AGC TGG ACC TTT AAA TGT TAA TAA TTT AAA GGT CCA GC TGT TG-3′) were used. For PCDH-α-shRNA22, primers 2622 (5′-TTT GCA ACA GCA CCA CTG AGA ACA GAT ACT AAC TGT TCT CAG TGG TGC TGT TGC TTT TTC-3′) and 2623 (5′-TCG AGA AAA AGC AAC AGC ACC ACT GAG AAC AGT TAG TAT CTG TTC TCA GTG GTG CTG TTG-3′) were used. For PCDH-α-shRNA6, primers 2430 (5′-TTT GGA TCT CCT GCA ATC ATC TCA TTA AGA GAT GAT TGC AGG AGA TCC TTT TTC-3′) and 2431 (5′-TCG AGA AAA AGG ATC TCC TGC AAT CAT CTC TTA ATG AGA TGA TTG CAG GAG ATC-3′) were used. For PCDH-γ-shRNA1, primers 1277 (5′-TTT GCT GCT GAT GTC AAC GCA ACA CTA ATT AAT AGT GTT GCG TTG ACA TCA GCA GCT TTT TC-3′) and 1278 (5′-TCG AGA AAA AGC TGC TGA TGT CAA CGC AAC ACT ATT AAT TAG TGT TGC GTT GAC ATC AGC AG-3′) were used. For PCDH-γ-shRNA3, primers 1281 (5′-TTT GCA AGA AGG AGA AGA AGT AGA AAG GAT ATA ATC CTT TCT ACT TCT TCT CCT TCT TGC TTT TTC-3′) and 1282 (5′-TCG AGA AAA AGC AAG AAG GAG AAG AAG TAG AAA GGA TTA TAT CCT TTC TAC TTC TTC TCC TTC TTG-3′) were used. For PCDH-γ-shRNA7, primers 1708 (5′-TTT GAC TGG CGC TTC TCT CAG ATT AAT ATT CTG AGA GAA GCG CCA GTC TTT TTC-3′) and 1709 (5′-TCG AGA AAA AGA CTG GCG CTT CTC TCA GAA TAT TAA TCT GAG AGA AGC GCC AGT-3′) were used. All constructs were further verified by sequencing.
Cell Transfection and Coimmunoprecipitation Analysis
HEK293T and HeLa cells were grown in Dulbecco's modified Eagle's medium with 10% FCS. Both cells were transfected using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen). For co-immunoprecipitation analysis, transfected HEK293T cells were washed with PBS, harvested, and lysed with a buffer containing 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 10 mm NaF, 10 mm Na3VO4, 1% Triton X-100, and protease inhibitor mixture (Roche Applied Science). Lysates were spun at 16,100 × g for 20 min at 4 °C, and the supernatant was incubated with anti-FLAG M2 affinity resin (Sigma) for 2 h at 4 °C on a rotator. The resin was then washed six times with the same buffer, and the bound proteins were eluted with SDS sample buffer without β-mercaptoethanol. After removing the anti-FLAG resin by centrifugation at 300 × g for 1 min, β-mercaptoethanol was added to a final concentration of 5% (v/v) followed by heating at 95 °C for 4 min. The eluates were then subjected to SDS-PAGE and Western blot analysis.
RNAi Knockdown and Dual-Luciferase Assay
To test the relative knockdown efficiency of shRNAs against chicken PCDH-α or PCDH-γ, HeLa cells in a 24-well plate were co-transfected with CMV-Ff-Luc reporter (0.15 μg/well), CMV-Rn-Luc (0.15 μg/well), and individual effector shRNAs (0.45 μg/well) using Lipofectamine 2000. 2 days after transfection, cells were harvested in passive lysis buffer for Dual-Luciferase assays according to the manufacture's instructions (Promega).
In Ovo Electroporation of Chick Spinal Cord
In ovo electroporation was performed as described previously (33). In brief, fertilized eggs were incubated at 38 °C to stage 12 by Hamburger and Hamilton criteria. DNA mixtures (4 μg/μl) of both shRNA PB transposon and PB transposase at a ratio of 5:1 were injected into the central canal of the neural tube, and electroporation was performed at the thoracic level of the spinal cord. For immunohistochemical detection of caspase-3 and GFP, chicken embryonic tissues were dissected, fixed, cryoprotected with 30% sucrose, and embedded in Optimal Cutting Temperature Media as described previously (33). Cryostat sections were processed for indirect immunofluorescence staining after an antigen retrieval treatment.
RESULTS
PCDH-γs Are Present in Large Membrane-bound Protein Complexes
Increasing evidence suggests that PCDHs might exist in large protein complexes. To investigate the association of PCDH-γ with other proteins, we first sought to characterize PCDH-γ protein complexes using two different techniques, SG ultracentrifugation and 2D BN/SDS-PAGE, followed by Western blot analysis. Both methods are widely used in the analysis of membrane-bound protein complexes (40–42).
For SG analysis, the crude membrane fraction was first isolated from the neonatal mouse brains, and integral membrane proteins were then solubilized by a Triton X-100-containing buffer. The Triton X-100-soluble protein extracts were then separated on a 5–50% linear sucrose gradient. The sucrose gradients were fractionated into 15 fractions from top to bottom, and equal portions of individual fractions were subjected to Western blot analysis using an anti-pan-PCDH-γ antibody. The result shows that PCDH-γs peak in fraction 8 with a sedimentation coefficient close to 19.4 S (Fig. 1 A, top). To confirm that PCDH-γs are part of macromolecular complexes, we added SDS to a final concentration of 1% in the extract to disrupt non-covalent associations before SG analysis. The peak of PCDH-γs shifted to fraction 5 after SDS treatment (Fig. 1A, higher middle), suggesting that the association of PCDH-γs with the complexes is SDS-sensitive. We next asked whether the distribution and the size of PCDH-γ complexes change at different developmental stages. The SG profile of PCDH-γ complexes from P21 mice was very similar to that of P0 samples (Fig. 1A, lower middle).
Fig. 1.
Analysis of PCDH-γ macromolecular protein complexes using sucrose gradient ultracentrifugation and 2D BN/SDS-PAGE. A, SG ultracentrifugation analysis of PCDH-γ complexes. Brain membrane protein extracts from P0 or P21 mice were subjected to ultracentrifugation on a 5–50% sucrose gradient. Wild-type (WT; P0) membrane proteins were also treated with 1% SDS before SG ultracentrifugation to dissociate non-covalently linked PCDH-γ complexes. Samples from Pcdh-γdel/del mice served as a negative control to show the specificity of Western blot analysis. B, 2D BN/SDS-PAGE analysis of PCDH-γ complexes. Crude membrane protein extracts were subjected to 2D BN/SDS-PAGE analysis followed by Western blot analysis using an anti-pan-PCDH-γ antibody. The NativeMark unstained protein standard (Invitrogen) was used as molecular mass standard. The arrowhead in B indicates protein aggregation at the gel entry point (well bottom). THY, thyroglobulin; BD, blue dextran.
To confirm the results of SG analysis, we applied 2D BN/SDS-PAGE to analyze PCDH-γ-containing complexes. BN-PAGE is a non-denaturing method originally developed for separating membrane protein complexes in an enzymatically active form (43). Coomassie Blue dyes and aminocaproic acid are used in this technique to induce a charge shift and improve the solubilization of membrane proteins. To analyze PCDH-γ complexes, solubilized crude membrane fractions from either wild-type or Pcdh-γdel/del brains were subjected to BN-PAGE. After the first dimension electrophoresis, single lanes of the BN gel were excised and mounted on a denaturing SDS gel for the second dimension. Western blot analysis was used to detect PCDH-γs in the complexes. PCDH-γ complexes were observed as a relatively broad band with molecular masses ranging from ∼480 to >1236 kDa (Fig. 1B, top), suggesting that these complexes are heterogeneous. To determine whether the complexes in the SG are similar to those in BN-PAGE, we further analyzed the peak fraction (fraction 8; Fig. 1A, top) from SG ultracentrifugation using 2D BN/SDS-PAGE. A PCDH-γ band migrating at about 1,000 kDa shows that similar protein complexes were maintained in both methods (Fig. 1B, bottom). In conclusion, we showed that PCDH-γs exist as large protein complexes using two independent techniques.
Affinity Purification of Membrane-bound PCDH-γ Complexes
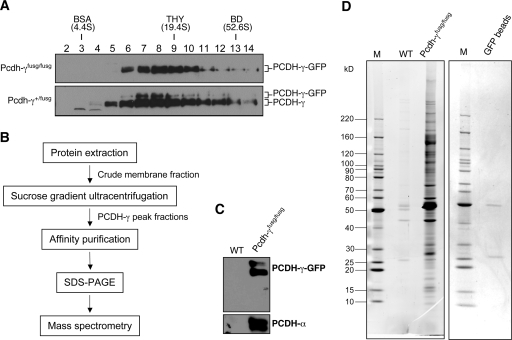
To further define the PCDH-γ complexes, we took advantage of PCDH-γ-GFP knock-in mice (Pcdh-γfusg/fusg) in which the shared C-terminal domain of PCDH-γ is fused in-frame with GFP. Pcdh-γfusg/fusg mice are normal, and PCDH-γ-GFPs exhibit similar subcellular localization as wild-type PCDH-γs (19). Therefore, we used GFP as an epitope tag for affinity purification. In this study, we first confirmed that in both homozygous (Pcdh-γfusg/fusg) and heterozygous (Pcdh-γ+/fusg) mice PCDH-γ-GFPs are cofractionated with wild-type PCDH-γs, although the expression level of PCDH-γ-GFP was lower (Fig. 2 A). Thus, PCDH-γ-GFP transgenic mice provide an optimal source for affinity purification of PCDH-γ complexes.
Fig. 2.
Affinity purification of PCDH-γ protein complexes. A, SG analysis of PCDH-γ-GFPs complexes from Pcdh-γ-GFPfusg/fusg and Pcdh-γ-GFP+/fusg mice (P0). PCDH-γ-GFPs and wild-type PCDH-γs are indicated. B, the flow chart of purification procedures. C, Western blot analysis of PCDH-γ-GFP immune complexes purified from Pcdh-γ-GFPfusg/fusg brains (P0–P5). An equal number of wild-type (WT) mouse brains served as a negative control for the purification. D, SYPRO Ruby-stained SDS-PAGE of the purified PCDH-γ-GFP immune complexes (left). To examine the possible protein contamination from anti-GFP-agarose beads, an equal amount of empty beads was eluted followed by SDS-PAGE and SYPRO Ruby staining (right). Only two faint bands, IgG heavy and light chains, were detected. M indicates the lanes loaded with marker proteins. THY, thyroglobulin; BD, blue dextran.
We have previously shown that PCDH-γs and PCDH-γ-GFPs are enriched in the “synaptosome” fraction (19). However, in the course of optimizing the experimental condition for affinity purification, we found that the synaptosome fractionation alone provided a limited amount of materials for affinity purification, whereas the crude membrane fraction had an unsatisfactory high background in the purification procedure (supplemental Fig. S1 and data not shown). To reduce purification background, we reduced the complexity of crude membrane protein extracts and enriched PCDH-γ complexes by incorporating an SG fractionation before affinity purification. A simplified purification flowchart is shown in Fig. 2B, and details are described under “Experimental Procedures”. The crude membrane fraction from 20 Pcdh-γfusg/fusg mouse brains was subjected to SG fractionation. The four most abundant PCDH-γ-GFP fractions were pooled and incubated overnight with a rat monoclonal anti-GFP antibody conjugated with agarose beads (MBL) at 4 °C. The affinity matrixes were then washed, and the bound proteins were eluted in SDS-PAGE loading buffer. As a negative control, the same procedures were applied to the wild-type sample (20 mice) in parallel. To validate our purification procedure, we used Western blot to examine both the bait PCDH-γ-GFPs and known PCDH-γ-associated proteins PCDH-αs after purification (33, 34). Both proteins were detected in the immune complex but not in the negative control (Fig. 2C). To detect PCDH-γ-GFP-associated proteins, the complexes were separated by SDS-PAGE and stained with SYPRO Ruby. Fig. 2D shows a complex pattern of the PCDH-γ-associated proteins compared with the negative control. Importantly, a vast majority of the isolated proteins in PCDH-γ complexes were specific to the PCDH-γ-GFP sample, whereas only a few faint nonspecific bands were detected in the wild-type control. The whole purification was repeated independently, and a similar result with clean background was obtained (supplemental Fig. S1D). Therefore, we concluded that we had successfully isolated PCDH-γ-associated proteins using a combination of SG fractionation and affinity purification.
Mass Spectrometry Analysis
To identify the protein composition of the purified PCDH-γ-GFP complexes, SYPRO Ruby-stained protein bands from SDS-PAGE gels were excised, destained, reduced, alkylated, and digested with trypsin in situ. All distinct bands obtained from two independent purifications were cut and pooled based on their molecular weight and relative intensity (Fig. 2D and supplemental Fig. S1). The trypsinized peptides were analyzed using LTQ-LC/MS/MS. A total of 12 independent MS analyses was performed. The MS spectra containing the information of peptide masses and sequences were searched using the Mascot and Sequest search engines against the mouse non-redundant protein database. The putative peptide results were then consolidated and filtered using the Proteome Software Scaffold. 154 non-redundant proteins including 12 PCDH-γ isoforms and GFP were identified using the criteria of ≥95% probability and ≥2 peptides matching. A subgroup of identified proteins, most of which have been verified later in this study or are previously known, is listed in Table I (for the complete list, please see supplemental Table S1). A wide range of proteins involved in different cellular functions was identified. Based on the information retrieved from Mouse Genome Informatics and Gene Ontology databases, we grouped the proteins into seven categories with regard to their functions: cytoskeleton (38 proteins), nucleic acid/protein processing (24 proteins), cell adhesion (35 proteins), trafficking/transport (18 proteins), signaling (15 proteins), metabolism (six proteins), and uncharacterized with unique or unknown functions (18 proteins) (supplemental Table S1).
Table I . A selected list of proteins identified by mass spectrometry analysis.
| No. | Protein | Gene name | Accession number | Molecular mass | Unique peptides | Coverage | Protein ID probability |
|---|---|---|---|---|---|---|---|
| kDa | % | % | |||||
| 1 | 14-3-3γ | Ywhaγ | gi|21464101 | 28 | 6 | 23 | 100 |
| 2 | 14-3-3 θ | Ywhaθ | gi|6756039 | 28 | 4 | 13 | 100 |
| 3 | 14-3-3 ζ | Ywhaζ | gi|6756041 | 28 | 7 | 30 | 100 |
| 4 | Cadherin 4 (R-cadherin) | Cdh4 | gi|6753376 | 100 | 5 | 6 | 100 |
| 5 | Calcium/calmodulin-dependent protein kinase II, γ | Camk2γ | gi|75991700 | 60 | 5 | 9 | 100 |
| 6 | Calcium/calmodulin-dependent protein kinase II, β | Camk2β | gi|31982483 | 60 | 8 | 14 | 100 |
| 7 | Catenin, α1 | Ctnnα1 | gi|6753294 | 100 | 2 | 3 | 100 |
| 8 | Catenin, α2 isoform 2 | Ctnnα2 | gi|6753296 | 105 | 7 | 8 | 100 |
| 9 | Catenin, β1 | Ctnnβ1 | gi|6671684 | 85 | 7 | 10 | 100 |
| 10 | Green fluorescent protein | Gfp | gi|115291372 | 29 | 9 | 31 | 100 |
| 11 | Postsynaptic density protein 95 | Dlg4 | gi|6681195 | 80 | 3 | 5 | 99 |
| 12 | Protocadherin α2 | Pcdhα2 | gi|51092277 | 102 | 2 | 2 | 99 |
| 13 | Protocadherin α3 | Pcdhα3 | gi|21426881 | 102 | 2 | 2 | 98 |
| 14 | Protocadherin α5 | Pcdhα5 | gi|20137002 | 102 | 2 | 2 | 99 |
| 15 | Protocadherin α7 | Pcdhα7 | gi|23956046 | 101 | 2 | 2 | 100 |
| 16 | Protocadherin α9 | Pcdhα9 | gi|21426885 | 107 | 2 | 2 | 100 |
| 17 | Protocadherin α11 | Pcdhα11 | gi|23956048 | 103 | 3 | 3 | 100 |
| 18 | Protocadherin α12 | Pcdhα12 | gi|21426883 | 103 | 4 | 5 | 100 |
| 19 | Protocadherin α subfamily C, 2 | Pcdhαc2 | gi|51092283 | 109 | 2 | 3 | 98 |
| 20 | Protocadherin β5 | Pcdhβ5 | gi|16716429 | 87 | 2 | 3 | 100 |
| 21 | Protocadherin β8 | Pcdhβ8 | gi|16716433 | 85 | 2 | 2 | 98 |
| 22 | Protocadherin β14 | Pcdhβ14 | gi|32189405 | 87 | 2 | 3 | 100 |
| 23 | Protocadherin β17 | Pcdhβ17 | gi|32308225 | 88 | 3 | 5 | 100 |
| 24 | Protocadherin β18 | Pcdhβ18 | gi|18087797 | 87 | 3 | 4 | 100 |
| 25 | Protocadherin β19 | Pcdhβ19 | gi|89363034 | 88 | 3 | 5 | 100 |
| 26 | Protocadherin β20 | Pcdhβ20 | gi|89994747 | 88 | 3 | 5 | 100 |
| 27 | Protocadherin β22 | Pcdhβ22 | gi|32189396 | 87 | 3 | 4 | 100 |
| 28 | Protocadherin γ subfamily A, 1 | Pcdhγa1 | gi|18087753 | 101 | 4 | 6 | 100 |
| 29 | Protocadherin γ subfamily A, 3 | Pcdhγa3 | gi|18087757 | 100 | 3 | 4 | 100 |
| 30 | Protocadherin γ subfamily A, 4 | Pcdhγa4 | gi|31982598 | 100 | 5 | 8 | 100 |
| 31 | Protocadherin γ subfamily A, 8 | Pcdhγa8 | gi|18087767 | 101 | 2 | 3 | 100 |
| 32 | Protocadherin γ subfamily A, 9 | Pcdhγa9 | gi|18087769 | 101 | 3 | 4 | 100 |
| 33 | Protocadherin γ subfamily A, 11 | Pcdhγa11 | gi|18087773 | 101 | 4 | 7 | 100 |
| 34 | Protocadherin γ subfamily A, 12 | Pcdhγa12 | gi|18087775 | 101 | 4 | 6 | 100 |
| 35 | Protocadherin γ subfamily B, 4 | Pcdhγb4 | gi|18087737 | 99 | 2 | 4 | 100 |
| 36 | Protocadherin γ subfamily B, 6 | Pcdhγb6 | gi|18087741 | 101 | 3 | 4 | 100 |
| 37 | Protocadherin γ subfamily B, 8 | Pcdhγb8 | gi|18087745 | 101 | 3 | 4 | 100 |
| 38 | Protocadherin γ subfamily C, 3 | Pcdhγc3 | gi|18087747 | 101 | 9 | 11 | 100 |
| 39 | Protocadherin γ subfamily C, 4 | Pcdhγc4 | gi|18087749 | 101 | 4 | 6 | 100 |
| 40 | SNAP-25-interacting protein | P140 | gi|116089329 | 131 | 3 | 3 | 99 |
| 41 | Tubulin, β | Tubb2β | gi|21746161 | 50 | 26 | 68 | 100 |
Previous subcellular localization studies have demonstrated that PCDH proteins are localized at synapses (9, 17–19). Therefore, we compared the list of identified PCDH-associated proteins with existing proteomic profiles of rodent PSD proteins (44–50) (see supplemental Table S2). The analysis showed that 91 of 114 identified proteins (excluding PCDHs and hypothetical proteins) in PCDH-γ complexes overlap with previously identified components in PSD preparations. Importantly, PCDH-α and PCDH-γ members were also identified in the MS analysis as part of PSD complexes (45). Therefore, our data strengthen the observation from subcellular localization studies and provide the direct evidence that PCDHs interact with synaptic components at the molecular level.
Individual neurons express distinct subsets of PCDH-γ isoforms in the brain (9–13); therefore, PCDH protein complexes purified from the whole brain tissues are heterogeneous for their protein composition. The large set of identified PCDH-γ-associated proteins reflects the heterogeneity and complexity of different PCDH complexes in the brain.
Validation of Selected Group of PCDH-γ-associated Proteins
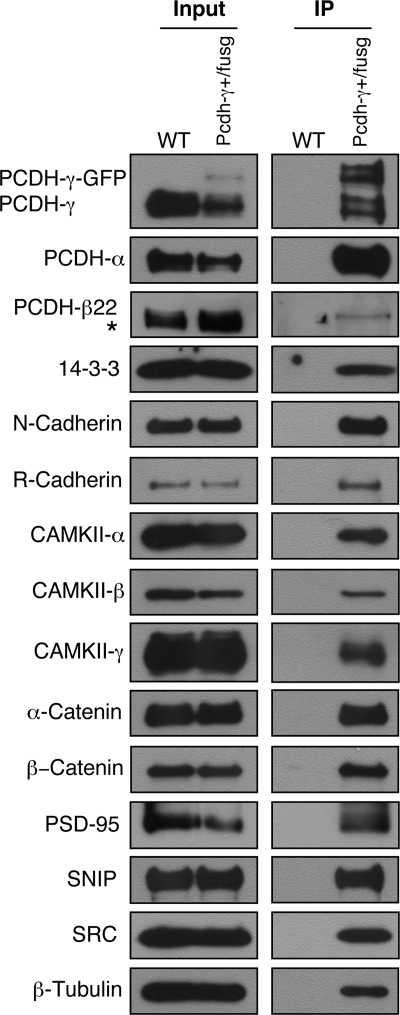
To verify the result from mass spectrometry analysis, we chose to confirm a subset of identified proteins using a co-IP and Western blot analysis. Our choice of target validation was based mostly on the availability of working antibodies. For these tests, the crude membrane extracts from Pcdh-γ+/fusg brains were subjected to co-IP with anti-GFP-agarose beads followed by Western blots with antibodies against selected proteins. Instead of Pcdh-γfusg/fusg homozygous mice, P21 Pcdh-γ+/fusg heterozygous mice were chosen as the starting material for two reasons. First, PCDH-γ-GFPs are expressed at a lower level compared with wild-type PCDH-γ. Second, PCDH-γ-GFPs are complexed with PCDH-γs. Therefore, wild-type PCDH-γs and their associated proteins can be co-purified with PCDH-γ-GFPs. Among the mass spectrometry-identified proteins, co-IP experiments confirmed 14-3-3 proteins, R-cadherin, CAMKII-β, CAMKII-γ, α-catenin, β-catenin, PCDH-αs, PCDH-β22, PSD-95, p140CAP/SNIP, and β-tubulin (Fig. 3). So far, we have been able to confirm all the candidate proteins for which working antibodies are available to us. Therefore, the target validation demonstrates that the proteomic profile of PCDH-γ complexes provides valuable information to further understand their function in neuronal signaling.
Fig. 3.
Confirmation of mass spectrometry-identified proteins using Western blots. The crude membrane protein extracts from P21 Pcdh-γ+/fusg mouse brains were immunoprecipitated with anti-GFP beads. The purified immune complexes were used for Western blot analyses using antibodies against PCDH-γs, PCDH-αs, PCDH-β22, 14-3-3, N-cadherin, R-cadherin, CAMKII-α, CAMKII-β, CAMKII-γ, α-catenin, β-catenin, PSD-95, SNIP, SRC family kinases, and β-tubulin. Wild-type (WT) mouse brains were used as a negative control. Inputs are equal amounts of the crude membrane protein extracts from both genotypes. “*” marks a nonspecific band partially overlapped with PCDH-β22 (also seen in Fig. 5).
We also chose to verify some additional proteins that are closely related to the proteins we identified. For instance, N-cadherin, closely related to the MS-identified R-cadherin, was also found to be associated with PCDH-γs. Similarly, CAMKII-α, as a major component of CAMKII multisubunit holoenzyme in the brain (51), was detected by Western blot with an antibody against CAMKII-α (Fig. 3). In addition, we confirmed that members of SRC family kinases were present in the immune complex with an anti-pan-SRC family kinase antibody (Fig. 3) because FYN, a SRC family tyrosine kinase, was previously shown to interact with PCDH-αs (9). Taken together, these results show that our current one-dimensional gel-based mass spectrometry analysis was not comprehensive enough to retrieve all the protein components in the immune complex. Future analysis with the multidimensional protein identification technology method or a Western blot-based screen for candidate proteins may ensure more complete identifications.
Clustered PCDH Subfamily Isoforms Are Associated with Each Other
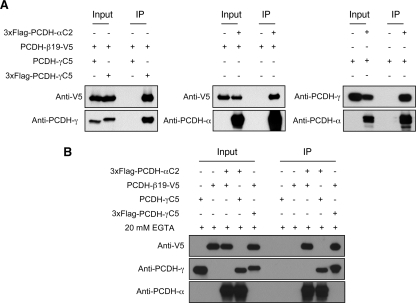
In agreement with the previous finding that that PCDH-αs and PCDH-γs interact with each other (33, 34), the mass spectrometry analysis recovered eight different PCDH-α isoforms together with 12 different PCDH-γs. Unexpectedly, eight different PCDH-β isoforms were also identified, providing the evidence, for the first time, that PCDH-βs are associated with PCDH-γ complexes (Table I). Co-IP experiments further confirmed that PCDH-γs interact with PCDH-αs and PCDH-β22 (Fig. 3, top three panels). Despite the previous finding on PCDH-αs, the surprisingly high abundance of PCDH-α and PCDH-β isoforms in the PCDH-γ complexes prompted us to examine the possibility that different subfamilies of clustered PCDHs coexist in similar macromolecular protein complexes. We first investigated whether PCDH-βs can interact with either PCDH-αs or PCDH-γs in vitro (Fig. 4). We co-transfected HEK293T cells with different pairs of expression vectors including 3xFLAG-Pcdh-γC5/Pcdh-β19-V5, 3xFLAG-Pcdh-αC2/Pcdh-β19-V5, and 3xFLAG- Pcdh-αC2/Pcdh-γC5 followed by co-IP analysis using anti-FLAG M2 affinity resins and Western blot using anti-V5, anti-pan-PCDH-α, and anti-pan-PCDH-γ antibodies. For the negative controls, single transfection of the prey construct or co-transfection of the bait without epitope tag was used. We found that PCDH-α, -β, and -γ isoforms can form complexes with each other (Fig. 4A). In addition, to test whether PCDH interactions are dependent on calcium as previously described for classic cadherins, we repeated the co-IP experiments in the presence of EGTA to deplete calcium ions (Fig. 4B). We found that these interactions are calcium-independent (Fig. 4B).
Fig. 4.
PCDH-α, PCDH-β, and PCDH-γ isoforms interact with each other in vitro. A, co-IP experiments show that PCDH-α isoform C2, PCDH-γ isoform C5, and PCDH-β isoform 22 interact with each other in transfected cells. HEK293T cells were co-transfected with different combinations of expression vectors followed by co-IP using the anti-FLAG M2 affinity resin and Western blot with anti-V5, anti-pan-PCDH-α, and anti-pan-PCDH-γ antibodies. B, interactions among different PCDH isoforms are calcium-independent. Similar experiments as in A were carried out with an addition of 20 mm EGTA in the lysis buffer to deplete calcium.
As previously demonstrated (33), wild-type PCDH-γ proteins were present in PCDH-γ-GFP immune complexes when purified from Pcdh+/fusg heterozygous mice (Fig. 3), showing that PCDH-γs can form complexes with members of the same PCDH-γ subfamily. Similar results have also been reported for PCDH-αs (28, 34). Taken together, these results suggest that clustered PCDHs are associated with each other not only between different subfamilies (forming hetero-oligomers) but also within the same subfamily (forming hetero- or homo-oligomers).
To further analyze the clustered PCDH protein complexes, we compared SG and 2D BN-PAGE profiles of PCDH-αs, PCDH-βs, and PCDH-γs. PCDH-αs and PCDH-β22 cofractionated with PCDH-γs in both techniques (Fig. 5), whereas other proteins including presynaptic, postsynaptic, and adhesion molecular markers displayed different profiles in the analyses. For example, two presynaptic markers, synapsin I and synaptophysin, were present in much smaller complexes in SG fractionation and showed no overlap with PCDH-γs and PCDH-αs in the 2D BN-PAGE system. The postsynaptic SAP102 and NMDAε2 and neural cell adhesion molecule exhibited some overlaps with PCDH-γs in both techniques, but the overall distributions and spot patterns were different (Fig. 5, A and B). Taken together, the results suggest that clustered PCDH-αs, -βs, and -γs might constitute the core components of multiprotein PCDH complexes, which are distinct from other synaptic protein complexes or other cell adhesion protein complexes.
Fig. 5.
Comparison of PCDH-α, PCDH-β, and PCDH-γ protein complexes from mouse brain using SG and 2D BN/SDS-PAGE. The SG and 2D BN/SDS-PAGE profiles of PCDH-γs, PCDH-αs, PCDH-β22, and other marker proteins are compared using P0 mouse brain samples. The SG and 2D BN/SDS-PAGE analyses were performed as in Fig. 1 with the indicated antibodies. “*” indicates nonspecific bands detected by anti-PCDH-α or anti-PCDH-β22 antibody. A, SG analysis. B, 2D BN/SDS-PAGE analysis. THY, thyroglobulin; BD, blue dextran; N-CAM, neural cell adhesion molecule.
Protein Complex Formation in Pcdh-γdel/del Mice
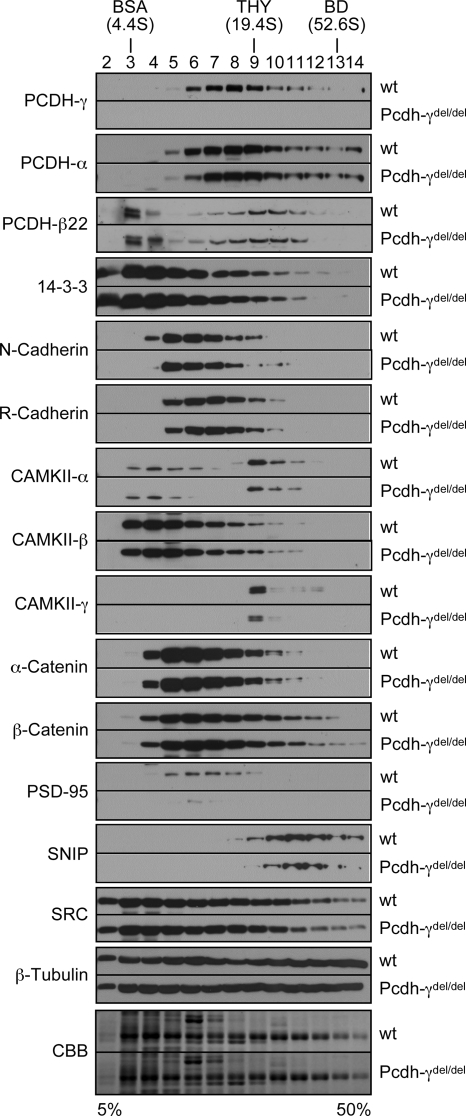
To further investigate the relationships between PCDH-γs and those verified PCDH-γ-associated proteins, we asked whether the deletion of PCDH-γ genes affects those components in vivo with regard to protein levels and complex sizes. We performed SG analysis and compared the SG profiles of the verified proteins in P0 wild-type and Pcdh-γdel/del brains. Equal amounts of wild-type and Pcdh-γdel/del membrane protein extracts were subjected to a 5–50% linear SG ultracentrifugation. Aliquots of individual fractions with equal amounts of proteins were subjected to SDS-PAGE followed by Western blot analyses. Fig. 6 shows the SG profiles of individual confirmed proteins from both wild-type and Pcdh-γdel/del mice. The SG profiles of PCDH-γ-associated proteins are partially overlapped with but different from those of PCDHs. For example, SRC family kinases are more evenly distributed in all fractions, whereas CAMKII-γ has a sharp peak at fraction 9. All the checked proteins, including PCDH-αs and PCDH-β22, have very similar SG distributions in Pcdh-γdel/del samples compared with the wild-type controls. In terms of protein levels, we found that the levels of CAMKII-α, CAMKII-γ, PSD-95, and SNIP are notably reduced in Pcdh-γdel/del mice, whereas the others had no significant changes. Western blot analysis further confirmed that decreased levels of CAMKII-α, CAMKII-γ, PSD-95, and SNIP associated with the membrane protein complexes were not due to lower expression levels of these proteins in the brain (supplemental Fig. S2). Therefore, these results show that PCDH-γs are required for the association of a subset of proteins with the PCDH complexes. Importantly, CAMKII-α, CAMKII-γ, PSD-95, and SNIP are all known components of PSD complexes. The decrease of these proteins in PSD complexes might have contributed to the synaptic deficit observed in Pcdh-γ mutant mice. However, for most PCDH-γ-associated proteins, depletion of PCDH-γs had no impact on the overall size and the abundance of complexes. Thus, it is possible that PCDH-γ complexes that contain these proteins are less abundant than other macromolecular complexes that share these proteins. The resolution of complex analysis using SG ultracentrifugation is too low to reveal subtle differences. Alternatively, other proteins such as PCDH-αs and PCDH-βs might compensate for the loss of PCDH-γs in the formation of some functional complexes. In this case, clustered PCDHs might have redundant function in complex formation.
Fig. 6.
Sucrose gradient analyses of mass spectrometry-identified proteins in wild-type and Pcdh-γdel/del mice. Equal amounts of membrane protein extracts from neonatal wild-type (wt) and Pcdh-γdel/del mouse brains were analyzed on a 5–50% sucrose gradient by ultracentrifugation. Equal amounts of individual fractions were separated by SDS-PAGE and analyzed by Western blots using antibodies against PCDH-γs, PCDH-αs, PCDH-β22, 14-3-3, N-cadherin, R-cadherin, CAMKII-α, CAMKII-β, CAMKII-γ, α-catenin, β-catenin, PSD-95, SNIP, SRC family kinases, and β-tubulin. The Coomassie Brilliant Blue (CBB)-stained gels of both wild-type and Pcdh-γdel/del at the area of around 55 kDa were used as loading controls to show an equal amount of proteins loaded. Each pair of Western blots from two genotypes were probed and developed at the same time. THY, thyroglobulin; BD, blue dextran.
PCDH-αs and PCDH-γs Have Overlapping Function in Regulating Neuronal Survival
It is difficult to generate a genetic null of all Pcdh clusters for testing their functional redundancy because other apparently essential genes such as TATA box-binding protein (TAF55) reside within the complex Pcdh gene clusters. However, loss-of-function experiments on Pcdh-γ in mice and Pcdh-α in zebrafish have demonstrated that PCDHs are required for neuronal survival in the developing spinal cord (19, 25). To test the functional redundancy of clustered PCDHs, we examined whether PCDH-αs and PCDH-γs have overlapping functions in the regulation of neuronal survival using a PB-mediated stable RNAi approach (39). First, we constructed several pairs of shRNAs against the shared constant exons of chicken PCDH-α and PCDH-γ mRNAs. PCDH-βs were excluded from this analysis because individual PCDH-βs have no common sequences for designing effective shRNAs. To screen the most efficient shRNAs against PCDH-α and PCDH-γ, respectively, we used firefly luciferase (Ff-Luc) reporters with the shared PCDH-α or PCDH-γ constant exons as the target sequences to compare the relative knockdown efficiency (Fig. 7, A and B). To validate the knockdown in the chicken spinal cord, we microinjected and electroporated PB transposons encoding different shRNAs with PBase-expressing plasmid into one side of the neural tube at the thoracic level at Hamburger and Hamilton stage 12. 4 days after electroporation, Western blot analysis showed that different shRNAs against chicken PCDH-α or PCDH-γ reduced PCDH-α and PCDH-γ protein levels in the electroporated side of the spinal cord (Fig. 7C). Thus, we next asked whether knockdown of PCDH-α, PCDH-γ, or both can lead to an increase of apoptosis in the developing spinal cord. Apoptotic neurons were detected by a cleaved caspase-3 antibody staining 4 days after electroporation (Fig. 7D and supplemental Fig. S3). The cell death index is defined by subtracting the number of dying cells in the non-electroporated side from that of the electroporated side in the spinal cord to exclude cell death that normally occurs during development. Quantitative analysis of apoptotic cells showed that combinations of PCDH-α and PCDH-γ shRNAs induce a statistically significant increase of spinal neuronal apoptosis compared with single PCDH-α or PCDH-γ shRNAs (Fig. 7E). To exclude the possibility that the phenotype was induced by an off-target effect of a specific shRNA, we used three combinations of PCDH-α and PCDH-γ shRNAs and obtained similar results (Fig. 7E). In addition, any single shRNA against PCDH-α or PCDH-γ did not have a phenotype, further eliminating the possibility of an off-target effect in these experiments (Fig. 7E). Thus, our data demonstrate that PCDH-αs and PCDH-γs have overlapping functions in promoting neuronal survival in the chicken spinal cord.
Fig. 7.
PCDH-γs and PCDH-αs have overlapping function in regulating neuronal survival in developing chicken spinal cord. A, a diagram showing the PB transposon expressing shRNAs and the Ff-Luc reporter to test shRNA knockdown. B, relative efficiency of shRNA knockdown against the chicken PCDH-γ (left) or PCDH-α target sequence (right). HeLa cells were co-transfected with the Ff-Luc PCDH-α or PCDH-γ target reporter, individual effector shRNAs against PCDH-α or PCDH-γ, and Renilla luciferase (Rn-Luc). The relative knockdown efficiency is reflected by relative ratios of Ff/Rn-Luc activity using a GFP shRNA as the negative control. Shown are triplicates of transfection experiments. Bars show standard errors of the mean (SEM). The more effective shRNAs (γ-si1, γ-si3, α-si1, and α-si22) were identified for subsequent studies. C, to knock down endogenous chicken PCDH-αs and PCDH-γs, individual PB shRNAs were co-electroporated with a PBase expression vector into chicken neural tube at stage 12. The spinal cord was dissected into the electroporated side (E) and non-electroporated side (C) 4 days after electroporation (EP). The dissected tissues were used for detecting PCDH-αs and PCDH-γs. Both PCDH proteins are reduced in the electroporated side of the spinal cord. Note that residual levels of PCDH proteins remain possibly due to incompleteness of both electroporation and RNAi knockdown. D and E, simultaneous knockdown of PCDH-α and PCDH-γ induces neuronal apoptosis in the spinal cord. Different combinations of the indicated shRNAs were electroporated into one side of the spinal cord. Cryostat sections of the electroporated spinal cords were double stained with anti-active caspase-3 (red) and anti-GFP (green). D, representative images of active caspase-3-positive cells in the PCDH shRNA electroporated spinal cord (left in each panel). The yellow arrowheads mark the apoptotic cells. Composite images double stained with anti-GFP (green) and 4′,6-diamidino-2-phenylindole (blue) are shown for each panel on the right, demonstrating the stable transposition efficiency. E, quantitative analysis of cell death in the shRNA-expressing spinal cord. To exclude some apoptotic cells (mainly motoneurons) during normal development, Cell death index = Number of caspase-3-positive cells per section on the electroporated side − Number of caspase-3-positive cells per section on the non-electroporated side. Plotted are average cell death index numbers from individual electroporated chicken embryos. For each embryo, five randomly picked GFP-positive sections were used to generate the average cell death index. The results were subjected to a two-tail t test, and p values <0.0001 were obtained between the three combinations of PCDH-α and PCDH-γ shRNAs and other control samples. TR, translated region.
DISCUSSION
Genetic studies have demonstrated that clustered PCDHs play important roles in regulating neuronal survival and synaptic connectivity in the central nervous system. To understand the molecular mechanisms of PCDH signaling, in this study, we performed a proteomics profiling of PCDH-γ protein complexes. Our analysis led to identification of nearly 140 PCDH-γ-associated proteins including cytoskeleton components, cell adhesion molecules, vesicle trafficking/transport proteins, and signaling molecules (see supplemental Table S1). Strikingly, the vast majority of PCDH-associated proteins are previously identified by proteomics analysis as components of postsynaptic density complexes. Thus, our list provides molecular evidence that PCDHs interact with synapses. Furthermore, this list will facilitate future studies to dissect the function of clustered PCDHs at the molecular level.
Although the verification and functional analysis of each protein on our list are beyond the scope of the current study, it is worth discussing several candidate proteins and their potential links to protocadherin functions. For most of the identified proteins, we do not know whether their associations with PCDHs are direct or indirect. It is interesting to note that 14-3-3, an important signaling adaptor protein (52), is present in PCDH-γ complexes. This protein has also been identified in our yeast two-hybrid screen using the common cytoplasmic domain of PCDH-γs (data not shown). Furthermore, bacterially expressed 14-3-3 directly interacts with the PCDH-γ cytoplasmic domain in vitro (data not shown). Thus, 14-3-3 is most likely a direct binding partner for PCDH-γs. Other direct target proteins on our list also include previously identified SRC kinases for PCDH-αs (9). On the other hand, some proteins, for example α-catenin and β-catenin, appear to be indirect binding proteins for PCDHs (3). They are present in the complexes likely due to binding to classic cadherins, R-cadherin and N-cadherin. The identification of R-cadherins and N-cadherin associated with PCDH-γs suggests an intriguing possibility of cross-talking between the classic cadherin and clustered protocadherin pathways. Future studies may improve our understanding in this regard. A group of identified proteins including PSD-95 and components of CAMKII kinase complexes has known functions in postsynaptic differentiation (53, 54). Noticeably, both PSD-95 and CAMKII-γ exhibited decreased levels in the membrane protein complexes from Pcdh-γ-deficient mice. This observation is consistent with the synaptic deficit observed in Pcdh-γ mutant mice (27). Another set of PCDH-γ-associated proteins includes adaptor protein AP2A, AP2B, SV2, clathrin, kinesin 2A, and RAB-interacting proteins, which are clearly important for vesicle transport and trafficking. It has been reported that PCDHs are enriched in intracellular vesicles (17, 55), and our findings might identify a molecular anchor for PCDHs on these vesicles. Nuclear proteins such as histone were also identified in our MS analysis. It is currently unclear whether this is biologically relevant or simply a purification artifact. Interestingly, the proteolytically processed cytoplasmic domains of PCDH-γs and PCDH-αs have been proposed to have a nuclear function like the NOTCH intracellular domain (30–32, 56).
Perhaps the most revealing finding from our proteomics survey of PCDH-γ complexes is the surprisingly high abundance of PCDH-α and PCDH-β isoforms in the PCDH-γ complexes. Although the interaction between PCDH-α and PCDH-γ was reported previously (28, 34), the significance of this interaction was not well appreciated because the relative abundance of the complexes was not evaluated. Using SG and particularly 2D BN-PAGE techniques, we further demonstrated that clustered PCDHs form stable macromolecular complexes distinct from other synaptic proteins and cell adhesion molecules. To support the notion that different clusters of PCDHs coexist in similar functional complexes and might have overlapping functions, we analyzed complex formation in Pcdh-γ-deficient mice and found that all verified PCDH-γ-associated proteins are present in similar protein complexes with a minority of these proteins showing lower abundance. Therefore, it is possible that other PCDHs compensate for PCDH-γ function in complex formation. RNAi experiments in the chicken spinal cord further strengthen this interpretation by showing that combinations of shRNAs against both PCDH-α and PCDH-γ induced apoptosis, whereas PCDH-α or PCDH-γ shRNAs alone did not. Clustered Pcdh genes are ubiquitously expressed in all neuronal populations. A very attractive hypothesis has been proposed based on the diversity and combinatorial expression patterns of Pcdhs (6, 57–59). In this model, cell-specific combinatorial PCDH molecules might provide a surface barcode for establishing synaptic specificity. However, mouse knock-out experiments did not provide conclusive evidence to either affirm or disprove this hypothesis. Although regional and subpopulation-specific phenotypes have been described (19, 23, 26, 27), no pleiotropic effects are observed for either Pcdh-γ- or Pcdh-α-null mice. A plausible explanation for the lack of pleiotropic neuronal phenotype in the Pcdh-α- or Pcdh-γ-deficient mice is the genetic redundancy and dosage compensation among different clustered Pcdhs. Our proteomics data presented in this study provide direct evidence that clustered PCDHs have functional redundancy both biochemically and biologically. Thus, deletion of all clustered Pcdhs in model systems might be necessary to reveal perhaps even more striking neuronal phenotypes during development.
Supplementary Material
Acknowledgments
We thank Dr. L. Helseth for mass spectrometry data analysis, Dr. Y. Lu for advice on electroporation, Dr. C. C. Garner for anti-SAP102 antibody, and Dr. R. Holmgren for critical reading of the manuscript. Proteomics and informatics services were provided by the Chicago Biomedical Consortium/University of Illinois at Chicago Research Resources Center Proteomics and Informatics Services Facility, which was established by a grant from The Searle Funds at the Chicago Community Trust to the Chicago Biomedical Consortium.
* This work was supported, in whole or in part, by National Institutes of Health Grant 5R01NS051253 to (X. W.).
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental Figs. S1–S3 and Tables S1 and S2.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental Figs. S1–S3 and Tables S1 and S2.
1 The abbreviations used are:
- PCDH
- protocadherin
- BN
- blue native
- PB
- PiggyBac
- PSD
- postsynaptic density
- RNAi
- RNA interference
- SG
- sucrose gradient
- SNIP
- SNAP-25-interacting-protein
- 2D
- two-dimensional
- GFP
- green fluorescent protein
- CAMK
- Ca2+/calmodulin-dependent protein kinase
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- LTQ
- linear trap quadrupole
- shRNA
- short hairpin RNA
- IP
- immunoprecipitation
- Ff
- firefly
- Luc
- luciferase
- Rn
- Renilla
- PBase
- PiggyBac transposase
- P
- postnatal day
- CMV
- cytomegalovirus.
REFERENCES
- 1.Benson D. L., Colman D. R., Huntley G. W. (2001) Molecules, maps and synapse specificity. Nat. Rev. Neurosci. 2, 899–909 [DOI] [PubMed] [Google Scholar]
- 2.Takeichi M. (2007) The cadherin superfamily in neuronal connections and interactions. Nat. Rev. Neurosci. 8, 11–20 [DOI] [PubMed] [Google Scholar]
- 3.Sano K., Tanihara H., Heimark R. L, Obata S., Davidson M., St John T., Taketani S., Suzuki S. (1993) Protocadherins: a large family of cadherin-related molecules in central nervous system. EMBO J. 12, 2249–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morishita H., Yagi T. (2007) Protocadherin family: diversity, structure, and function. Curr. Opin. Cell Biol. 19, 584–592 [DOI] [PubMed] [Google Scholar]
- 5.Halbleib J. M., Nelson W. J. (2006) Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 20, 3199–3214 [DOI] [PubMed] [Google Scholar]
- 6.Wu Q., Maniatis T. (1999) A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell 97, 779–790 [DOI] [PubMed] [Google Scholar]
- 7.Noonan J. P., Grimwood J., Schmutz J., Dickson M., Myers R. M. (2004) Gene conversion and the evolution of protocadherin gene cluster diversity. Genome Res. 14, 354–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Q. (2005) Comparative genomics and diversifying selection of the clustered vertebrate protocadherin genes. Genetics 169, 2179–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kohmura N., Senzaki K., Hamada S., Kai N., Yasuda R., Watanabe M., Ishii H., Yasuda M., Mishina M., Yagi T. (1998) Diversity revealed by a novel family of cadherins expressed in neurons at a synaptic complex. Neuron 20, 1137–1151 [DOI] [PubMed] [Google Scholar]
- 10.Wang X., Su H., Bradley A. (2002) Molecular mechanisms governing Pcdh-gamma gene expression: evidence for a multiple promoter and cis-alternative splicing model. Genes Dev. 16, 1890–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tasic B., Nabholz C. E., Baldwin K. K., Kim Y., Rueckert E. H., Ribich S. A., Cramer P., Wu Q., Axel R., Maniatis T. (2002) Promoter choice determines splice site selection in protocadherin alpha and gamma pre-mRNA splicing. Mol. Cell 10, 21–33 [DOI] [PubMed] [Google Scholar]
- 12.Frank M., Ebert M., Shan W., Phillips G. R., Arndt K., Colman D. R., Kemler R. (2005) Differential expression of individual gamma-protocadherins during mouse brain development. Mol. Cell. Neurosci. 29, 603–616 [DOI] [PubMed] [Google Scholar]
- 13.Zou C., Huang W., Ying G., Wu Q. (2007) Sequence analysis and expression mapping of the rat clustered protocadherin gene repertoires. Neuroscience 144, 579–603 [DOI] [PubMed] [Google Scholar]
- 14.Esumi S., Kakazu N., Taguchi Y., Hirayama T., Sasaki A., Hirabayashi T., Koide T., Kitsukawa T., Hamada S., Yagi T. (2005) Monoallelic yet combinatorial expression of variable exons of the protocadherin-alpha gene cluster in single neurons. Nat. Genet. 37, 171–176 [DOI] [PubMed] [Google Scholar]
- 15.Kaneko R., Kato H., Kawamura Y., Esumi S., Hirayama T., Hirabayashi T., Yagi T. (2006) Allelic gene regulation of Pcdh-alpha and Pcdh-gamma clusters involving both monoallelic and biallelic expression in single Purkinje cells. J. Biol. Chem. 281, 30551–30560 [DOI] [PubMed] [Google Scholar]
- 16.Ribich S., Tasic B., Maniatis T. (2006) Identification of long-range regulatory elements in the protocadherin-alpha gene cluster. Proc. Natl. Acad. Sci. U.S.A. 103, 19719–19724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phillips G. R., Tanaka H., Frank M., Elste A., Fidler L., Benson D. L., Colman D. R. (2003) Gamma-protocadherins are targeted to subsets of synapses and intracellular organelles in neurons. J. Neurosci. 23, 5096–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Junghans D., Heidenreich M., Hack I., Taylor V., Frotscher M., Kemler R. (2008) Postsynaptic and differential localization to neuronal subtypes of protocadherin beta16 in the mammalian central nervous system. Eur. J. Neurosci. 27, 559–571 [DOI] [PubMed] [Google Scholar]
- 19.Wang X., Weiner J. A., Levi S., Craig A. M., Bradley A., Sanes J. R. (2002) Gamma protocadherins are required for survival of spinal interneurons. Neuron 36, 843–854 [DOI] [PubMed] [Google Scholar]
- 20.Mutoh T., Hamada S., Senzaki K., Murata Y., Yagi T. (2004) Cadherin-related neuronal receptor 1 (CNR1) has cell adhesion activity with beta1 integrin mediated through the RGD site of CNR1. Exp. Cell Res. 294, 494–508 [DOI] [PubMed] [Google Scholar]
- 21.Obata S., Sago H., Mori N., Rochelle J. M., Seldin M. F., Davidson M., St John T., Taketani S., Suzuki S. T. (1995) Protocadherin Pcdh2 shows properties similar to, but distinct from, those of classical cadherins. J. Cell Sci. 108, 3765–3773 [DOI] [PubMed] [Google Scholar]
- 22.Fernández-Monreal M., Kang S., Phillips G. R. (2009) Gamma-protocadherin homophilic interaction and intracellular trafficking is controlled by the cytoplasmic domain in neurons. Mol. Cell. Neurosci. 40, 344–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lefebvre J. L., Zhang Y., Meister M., Wang X., Sanes J. R. (2008) {gamma}-Protocadherins regulate neuronal survival but are dispensable for circuit formation in retina. Development 135, 4141–4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prasad T., Wang X., Gray P. A., Weiner J. A. (2008) A differential developmental pattern of spinal interneuron apoptosis during synaptogenesis: insights from genetic analyses of the protocadherin-{gamma} gene cluster. Development 135, 4153–4164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emond M. R., Jontes J. D. (2008) Inhibition of protocadherin-alpha function results in neuronal death in the developing zebrafish. Dev. Biol. 321, 175–187 [DOI] [PubMed] [Google Scholar]
- 26.Hasegawa S., Hamada S., Kumode Y., Esumi S., Katori S., Fukuda E., Uchiyama Y., Hirabayashi T., Mombaerts P., Yagi T. (2008) The protocadherin-alpha family is involved in axonal coalescence of olfactory sensory neurons into glomeruli of the olfactory bulb in mouse. Mol. Cell. Neurosci. 38, 66–79 [DOI] [PubMed] [Google Scholar]
- 27.Weiner J. A., Wang X., Tapia J. C., Sanes J. R. (2005) Gamma protocadherins are required for synaptic development in the spinal cord. Proc. Natl. Acad. Sci. U.S.A. 102, 8–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Triana-Baltzer G. B., Blank M. (2006) Cytoplasmic domain of protocadherin-alpha enhances homophilic interactions and recognizes cytoskeletal elements. J. Neurobiol. 66, 393–407 [DOI] [PubMed] [Google Scholar]
- 29.Gayet O., Labella V., Henderson C. E., Kallenbach S. (2004) The b1 isoform of protocadherin-gamma (Pcdhgamma) interacts with the microtubule-destabilizing protein SCG10. FEBS Lett. 578, 175–179 [DOI] [PubMed] [Google Scholar]
- 30.Haas I. G., Frank M., Véron N., Kemler R. (2005) Presenilin-dependent processing and nuclear function of gamma-protocadherins. J. Biol. Chem. 280, 9313–9319 [DOI] [PubMed] [Google Scholar]
- 31.Hambsch B., Grinevich V., Seeburg P. H., Schwarz M. K. (2005) {gamma}-Protocadherins, presenilin-mediated release of C-terminal fragment promotes locus expression. J. Biol. Chem. 280, 15888–15897 [DOI] [PubMed] [Google Scholar]
- 32.Bonn S., Seeburg P. H., Schwarz M. K. (2007) Combinatorial expression of alpha- and gamma-protocadherins alters their presenilin-dependent processing. Mol. Cell. Biol. 27, 4121–4132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J., Lu Y., Meng S., Han M. H., Lin C., Wang X. (2009) alpha- and gamma-Protocadherins negatively regulate PYK2. J. Biol. Chem. 284, 2880–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murata Y., Hamada S., Morishita H., Mutoh T., Yagi T. (2004) Interaction with protocadherin-gamma regulates the cell surface expression of protocadherin-alpha. J. Biol. Chem. 279, 49508–49516 [DOI] [PubMed] [Google Scholar]
- 35.Müller B. M., Kistner U., Kindler S., Chung W. J., Kuhlendahl S., Fenster S. D., Lau L. F., Veh R. W., Huganir R. L., Gundelfinger E. D., Garner C. C. (1996) SAP102, a novel postsynaptic protein that interacts with NMDA receptor complexes in vivo. Neuron 17, 255–265 [DOI] [PubMed] [Google Scholar]
- 36.Cole D. G., Diener D. R., Himelblau A. L., Beech P. L., Fuster J. C., Rosenbaum J. L. (1998) Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 141, 993–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wittig I., Braun H. P., Schägger H. (2006) Blue native PAGE. Nat. Protoc. 1, 418–428 [DOI] [PubMed] [Google Scholar]
- 37.Kinter M., Sherman N. E. (2000) Protein Sequencing and Identification Using Tandem Mass Spectrometry, pp. 152–160, Wiley-Interscience, Inc., New York [Google Scholar]
- 39.Lu Y., Lin C., Wang X. (2009) PiggyBac transgenic strategies in the developing chicken spinal cord. Nucleic Acids Res., Sept. 15 Epub ahead of print [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hansson M., Dupuis T., Strömquist R., Andersson B., Vener A. V., Carlberg I. (2007) The mobile thylakoid phosphoprotein TSP9 interacts with the light-harvesting complex II and the peripheries of both photosystems. J. Biol. Chem. 282, 16214–16222 [DOI] [PubMed] [Google Scholar]
- 41.Nicchia G. P., Cogotzi L., Rossi A., Basco D., Brancaccio A., Svelto M., Frigeri A. (2008) Expression of multiple AQP4 pools in the plasma membrane and their association with the dystrophin complex. J. Neurochem. 105, 2156–2165 [DOI] [PubMed] [Google Scholar]
- 42.Schägger H., Cramer W. A., von Jagow G. (1994) Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217, 220–230 [DOI] [PubMed] [Google Scholar]
- 43.Schägger H., von Jagow G. (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199, 223–231 [DOI] [PubMed] [Google Scholar]
- 44.Witzmann F. A., Arnold R. J., Bai F., Hrncirova P., Kimpel M. W., Mechref Y. S., McBride W. J., Novotny M. V., Pedrick N. M., Ringham H. N., Simon J. R. (2005) A proteomic survey of rat cerebral cortical synaptosomes. Proteomics 5, 2177–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trinidad J. C., Thalhammer A., Specht C. G., Lynn A. J., Baker P. R., Schoepfer R., Burlingame A. L. (2008) Quantitative analysis of synaptic phosphorylation and protein expression. Mol. Cell. Proteomics 7, 684–696 [DOI] [PubMed] [Google Scholar]
- 46.Li K. W., Hornshaw M. P., Van Der Schors R. C., Watson R., Tate S., Casetta B., Jimenez C. R., Gouwenberg Y., Gundelfinger E. D., Smalla K. H., Smit A. B. (2004) Proteomics analysis of rat brain postsynaptic density. Implications of the diverse protein functional groups for the integration of synaptic physiology. J. Biol. Chem. 279, 987–1002 [DOI] [PubMed] [Google Scholar]
- 47.Jordan B. A., Fernholz B. D., Boussac M., Xu C., Grigorean G., Ziff E. B., Neubert T. A. (2004) Identification and verification of novel rodent postsynaptic density proteins. Mol. Cell. Proteomics 3, 857–871 [DOI] [PubMed] [Google Scholar]
- 48.Dosemeci A., Makusky A. J., Jankowska-Stephens E., Yang X., Slotta D. J., Markey S. P. (2007) Composition of the synaptic PSD-95 complex. Mol. Cell. Proteomics 6, 1749–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collins M. O., Husi H., Yu L., Brandon J. M., Anderson C. N., Blackstock W. P., Choudhary J. S., Grant S. G. (2006) Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. J. Neurochem. 97, Suppl. 1, 16–23 [DOI] [PubMed] [Google Scholar]
- 50.Cheng D., Hoogenraad C. C., Rush J., Ramm E., Schlager M. A., Duong D. M., Xu P., Wijayawardana S. R., Hanfelt J., Nakagawa T., Sheng M., Peng J. (2006) Relative and absolute quantification of postsynaptic density proteome isolated from rat forebrain and cerebellum. Mol. Cell. Proteomics 5, 1158–1170 [DOI] [PubMed] [Google Scholar]
- 51.Hudmon A., Schulman H. (2002) Structure-function of the multifunctional Ca2+/calmodulin-dependent protein kinase II. Biochem. J. 364, 593–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jin J., Smith F. D., Stark C., Wells C. D., Fawcett J. P., Kulkarni S., Metalnikov P., O'Donnell P., Taylor P., Taylor L., Zougman A., Woodgett J. R., Langeberg L. K., Scott J. D., Pawson T. (2004) Proteomic, functional, and domain-based analysis of in vivo 14–3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr. Biol. 14, 1436–1450 [DOI] [PubMed] [Google Scholar]
- 53.Kim E., Sheng M. (2004) PDZ domain proteins of synapses. Nat. Rev. Neurosci. 5, 771–781 [DOI] [PubMed] [Google Scholar]
- 54.Colbran R. J., Brown A. M. (2004) Calcium/calmodulin-dependent protein kinase II and synaptic plasticity. Curr. Opin. Neurobiol. 14, 318–327 [DOI] [PubMed] [Google Scholar]
- 55.Phillips G. R., Huang J. K., Wang Y., Tanaka H., Shapiro L., Zhang W., Shan W. S., Arndt K., Frank M., Gordon R. E., Gawinowicz M. A., Zhao Y., Colman D. R. (2001) The presynaptic particle web: ultrastructure, composition, dissolution, and reconstitution. Neuron 32, 63–77 [DOI] [PubMed] [Google Scholar]
- 56.Reiss K., Maretzky T., Haas I. G., Schulte M., Ludwig A., Frank M., Saftig P. (2006) Regulated ADAM10-dependent ectodomain shedding of gamma-protocadherin C3 modulates cell-cell adhesion. J. Biol. Chem. 281, 21735–21744 [DOI] [PubMed] [Google Scholar]
- 57.Serafini T. (1999) Finding a partner in a crowd: neuronal diversity and synaptogenesis. Cell 98, 133–136 [DOI] [PubMed] [Google Scholar]
- 58.Shapiro L., Colman D. R. (1999) The diversity of cadherins and implications for a synaptic adhesive code in the CNS. Neuron 23, 427–430 [DOI] [PubMed] [Google Scholar]
- 59.Yagi T., Takeichi M. (2000) Cadherin superfamily genes: functions, genomic organization, and neurologic diversity. Genes Dev. 14, 1169–1180 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.