Abstract
Several mass spectrometry-based assays have emerged for the quantitative profiling of cellular tyrosine phosphorylation. Ideally, these methods should reveal the exact sites of tyrosine phosphorylation, be quantitative, and not be cost-prohibitive. The latter is often an issue as typically several milligrams of (stable isotope-labeled) starting protein material are required to enable the detection of low abundance phosphotyrosine peptides. Here, we adopted and refined a peptidecentric immunoaffinity purification approach for the quantitative analysis of tyrosine phosphorylation by combining it with a cost-effective stable isotope dimethyl labeling method. We were able to identify by mass spectrometry, using just two LC-MS/MS runs, more than 1100 unique non-redundant phosphopeptides in HeLa cells from about 4 mg of starting material without requiring any further affinity enrichment as close to 80% of the identified peptides were tyrosine phosphorylated peptides. Stable isotope dimethyl labeling could be incorporated prior to the immunoaffinity purification, even for the large quantities (mg) of peptide material used, enabling the quantification of differences in tyrosine phosphorylation upon pervanadate treatment or epidermal growth factor stimulation. Analysis of the epidermal growth factor-stimulated HeLa cells, a frequently used model system for tyrosine phosphorylation, resulted in the quantification of 73 regulated unique phosphotyrosine peptides. The quantitative data were found to be exceptionally consistent with the literature, evidencing that such a targeted quantitative phosphoproteomics approach can provide reproducible results. In general, the combination of immunoaffinity purification of tyrosine phosphorylated peptides with large scale stable isotope dimethyl labeling provides a cost-effective approach that can alleviate variation in sample preparation and analysis as samples can be combined early on. Using this approach, a rather complete qualitative and quantitative picture of tyrosine phosphorylation signaling events can be generated.
Reversible tyrosine phosphorylation plays an important role in numerous cellular processes like growth, differentiation, and migration. Phosphotyrosine signaling is tightly controlled by the balanced action of protein-tyrosine kinases and protein-tyrosine phosphatases. Aberrant tyrosine phosphorylation has been suggested to be an underlying cause in multiple cancers (1). Therefore, the identification of tyrosine phosphorylated proteins and the investigation into their involvement in signaling pathways are important. Several groups have attempted to comprehensively study tyrosine phosphorylation by proteomics means (2–5). However, large scale identification of tyrosine phosphorylation sites by MS can be hindered by the low abundance of tyrosine phosphorylated proteins. Especially, signaling intermediates are usually low abundance proteins that show substoichiometric phosphorylation levels. In addition, the identification by mass spectrometry of phosphopeptides from a complex cellular lysate digest is often complicated by ion suppression effects due to a high background of non-phosphorylated peptides. Enrichment of tyrosine phosphorylated proteins or peptides prior to mass spectrometric detection is therefore essential. Traditionally, antibodies against phosphorylated tyrosine have been used to immunoprecipitate tyrosine phosphorylated proteins from cultured cells (2–4, 6–8). This phosphoprotein immunoaffinity purification method has for example been used to study the global dynamics of phosphotyrosine signaling events after EGF1 stimulation using stable isotope labeling by amino acids in cell culture (SILAC) (2). This approach led to the identification of known and previously unidentified signaling proteins in the EGF receptor (EGFR) pathway, including their temporal activation profile after stimulation of the EGFR, providing crucial information for modeling signaling events in the cell. However, as the identification and quantification of these phosphorylated proteins in these studies were not necessarily based on tyrosine phosphorylated peptides but largely on non-phosphorylated peptides, little information is derived on the exact site(s) of tyrosine phosphorylation. Also, binding partners of tyrosine phosphorylated proteins, which themselves are not tyrosine phosphorylated, might be co-precipitated and impair the tyrosine phosphorylation quantification. Immunoaffinity purification of phosphotyrosine peptides, rather than proteins, using anti-phosphotyrosine antibodies (5, 9–16) significantly facilitates the identification of the site(s) of phosphorylation as it greatly alleviates most of the above mentioned problems because the tyrosine phosphorylated site can be directly identified and quantified.
Accurate MS-based quantification is typically performed by stable isotope labeling. The isotopes can be incorporated metabolically during cell culture as in SILAC (17) or chemically as in an isobaric tag for relative and absolute quantitation (iTRAQ) (18) or stable isotope dimethyl labeling (19–21). Typically, the most precise quantification can be obtained by metabolic labeling as the different samples can be combined at the level of intact cells (22). However, metabolic labeling is somewhat limited to biological systems that can be grown in culture, and the medium may have an effect on the growth and development of the cells. iTRAQ has been used in conjunction with phosphotyrosine peptide immunoprecipitation (5). As the chemical labeling is performed before immunoprecipitation, the differentially labeled samples can be precipitated together, thereby neutralizing the potentially largest source of variation. However, as this phosphotyrosine peptide immunoprecipitation is typically performed on several hundreds of micrograms to milligrams of protein sample, iTRAQ provides in these cases a rather cost-prohibitive means.
Here, we present an optimized immunoaffinity purification approach for the analysis of tyrosine phosphorylation combined with stable isotope dimethyl labeling (19–21, 23). We efficiently enriched and identified by MS 1112 unique phosphopeptides derived from 4 mg of starting protein material without any further affinity chromatographic enrichment whereby up to 80% of the peptides analyzed in the final LC run were phosphotyrosine peptides. We further advanced the method by introducing triplex stable isotope dimethyl labeling prior to immunoprecipitation. We quantified differences in tyrosine phosphorylation upon pervanadate treatment or EGF stimulation to detect site-specific changes in tyrosine phosphorylation. 128 unique phosphotyrosine peptides were identified and quantified upon pervanadate treatment. By using an internal standard comprising both mock and pervanadate-treated samples, we could more confidently identify and quantify phosphorylation sites that are strongly regulated and on-off situations. Analysis of EGF-stimulated HeLa cells resulted in the quantification of 73 unique phosphotyrosine peptides. Most of the up-regulated phosphotyrosine peptides that were identified have been reported previously to be involved in the EGFR signaling pathway, validating our approach. However, for the first time, we found TFG to also become highly tyrosine phosphorylated upon EGF stimulation together with some tyrosine phosphorylation sites on for example IRS2, SgK269, and DLG3 that have not been firmly established earlier to be involved in EGFR signaling.
In general, we show that the combination of immunoaffinity purification of tyrosine phosphorylated peptides with large scale chemical stable isotope dimethyl labeling provides a cost-effective approach that can alleviate variation in immunoprecipitation and LC-MS as samples can be combined before immunoprecipitation and the necessity of performing additional enrichment is removed by an optimization of the protocol. With only a single LC-MS run, already a rather complete qualitative and quantitative picture of a signaling event can be generated.
MATERIALS AND METHODS
Cell Culture, Stimulation, and Digest Preparation
HeLa cells were grown to confluence in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum (Invitrogen) and 0.05 mg/ml penicillin/streptomycin (Invitrogen). Cells were placed in serum-free medium 16 h before EGF stimulation. Cells were stimulated with 150 ng/ml EGF for 10 or 30 min, treated with 1 mm pervanadate (prepared by incubating 1 mm orthovanadate with 1 mm hydrogen peroxide for several min) for 10 min, or left untreated. Cells were washed with cold phosphate-buffered saline and lysed. Before labeling and immunoprecipitation, cells were lysed in 8m urea, 50 mm ammonium bicarbonate, 5 mm sodium phosphate, 1 mm potassium fluoride, 1 mm sodium orthovanadate, and EDTA-free protease inhibitor mixture (Sigma). Samples were reduced with DTT at a final concentration of 10 mm at 56 °C; subsequently samples were alkylated with iodoacetamide at a final concentration of 55 mm at RT. The samples were diluted to 2 m urea, 50 mm ammonium bicarbonate, and trypsin (1:100; Promega) was added. Digestion was performed overnight at 37 °C.
Stable Isotope Labeling by Reductive Amination of Tryptic Peptides
Tryptic peptides were desalted, dried in vacuo, and resuspended in 100 μl of triethylammonium bicarbonate (100 mm). Subsequently, formaldehyde-H2 (573 μmol) was added and vortexed for 2 min followed by the addition of freshly prepared sodium cyanoborohydride (278 μmol). The resultant mixture was vortexed for 60 min at RT. A total of 60 μl of ammonia (25%) was added to consume the excess formaldehyde. Finally, 50 μl of formic acid (100%) was added to acidify the solution. For intermediate labels, formaldehyde-D2 (573 μmol) was used. For the heavy labeling, 13C-D2-formaldehyde (573 μmol) and freshly prepared cyanoborodeuteride (278 μmol) were used (20, 21). The light, intermediate, and heavy dimethyl-labeled samples were mixed in 1:1:1 ratio based on total peptide amount, which was determined by running an aliquot of the labeled samples on a regular LC-MS run and comparing overall peptide signal intensities.
Immunoprecipitation
Labeled peptides were mixed, desalted, dried down, and redissolved in immunoprecipitation (IP) buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 1% n-octyl-β-d-glucopyranoside and 1× Complete Mini (Roche Diagnostics)). Prior to IP, PY99-agarose beads (Santa Cruz Biotechnology) were washed in IP buffer. The labeled peptide mixture was added to the PY99-agarose beads, and incubation was performed overnight at 4 °C. Beads were washed three times with IP buffer and two times with water. Peptides were eluted by adding 0.15% TFA for 20 min at RT. Eluted peptides were desalted and concentrated on stop and go extraction (STAGE) tips.
On-line Nanoflow Liquid Chromatography
Nanoflow LC-MS/MS was performed by coupling an Agilent 1100 HPLC system (Agilent Technologies, Waldbronn, Germany) to an LTQ-Orbitrap mass spectrometer (Thermo Electron, Bremen, Germany) as described previously (24). Dried fractions were reconstituted in 10 μl of 0.1 m acetic acid and delivered to a trap column (AquaTM C18, 5 μm (Phenomenex, Torrance, CA); 20 mm × 100-μm inner diameter, packed in house) at 5 μl/min in 100% solvent A (0.1 m acetic acid in water). Subsequently, peptides were transferred to an analytical column (ReproSil-Pur C18-AQ, 3 μm (Dr. Maisch GmbH, Ammerbuch, Germany); 40 cm × 50-μm inner diameter, packed in house) at ∼100 nl/min in a 2- or 3-h gradient from 0 to 40% solvent B (0.1 m acetic acid in 8:2 (v/v) acetonitrile/water). The eluent was sprayed via distal coated emitter tips (New Objective) butt-connected to the analytical column. The mass spectrometer was operated in data-dependent mode, automatically switching between MS and MS/MS. Full-scan MS spectra (from m/z 300 to 1500) were acquired in the Orbitrap with a resolution of 60,000 at m/z 400 after accumulation to target value of 500,000. The three most intense ions at a threshold above 5000 were selected for collision-induced fragmentation in the linear ion trap at a normalized collision energy of 35% after accumulation to a target value of 10,000.
Data Analysis
All MS2 spectra were converted to single DTA files using Bioworks 3.3 using default settings. Runs were searched using an in-house licensed Mascot search engine (Mascot (version 2.1.0) software platform (Matrix Science, London, UK)) against the human IPI database version 3.36 (63,012 sequences) with carbamidomethylcysteine as a fixed modification. Light, intermediate, and heavy dimethylation of peptide N termini and lysine residues; oxidized methionine; and phosphorylation of tyrosine, serine, and threonine were set as variable modifications. Trypsin was specified as the proteolytic enzyme, and up to two missed cleavages were allowed. The mass tolerance of the precursor ion was set to 5 ppm, and for fragment ions the mass tolerance was set to 0.6 Da. Peptides were assigned to the first protein hit reported by Mascot. The assignment of phosphorylation sites of identified phosphopeptides was performed by the post-translational modification scoring algorithm implemented in MSQuant as described previously (25). Individual MS/MS spectra from phosphopeptides were accepted for a Mascot score ≥20. This threshold was experimentally tested (see “Results”), and the false discovery rate (FDR) at this score for phosphotyrosine peptides only was estimated to be 2% by performing a concatenated decoy database search. All identified phosphopeptides that were found to be differentially phosphorylated were manually validated.
Quantification
Quantification of peptide triplets of which at least one had a Mascot peptide score of 20 was performed using an in-house dimethyl-adapted version of MSQuant (SourceForge) as described previously (20). Briefly, peptide ratios were obtained by calculating the extracted ion chromatograms (XICs) of the “light,” “intermediate,” and “heavy” forms of the peptide using the monoisotopic peaks only. The total XIC for each of the peptide forms was obtained by summing the XICs in consecutive MS cycles for the duration of their respective LC-MS peaks in the total ion chromatogram using FT-MS scans. This total XIC was then used to compute the peptide ratio. Heavy and light labeled peptides were found to largely co-elute. Quantified proteins were normalized against the log2 of the median of all peptides quantified. StatQuant, an in-house developed program (26), was used for normalization, outlier detection, and determination of standard deviation. Ratios of phosphotyrosine levels were normalized to the ratios of (nonspecifically binding) non-phosphorylated peptides. Ratios derived from different charge states of the peptide and/or missed cleavages with the same phosphorylation sites were log-averaged. Clustering of the phosphotyrosine profiles upon EGF stimulation was performed by K-means clustering (Euclidean distance).
Data Sets
The HeLa phosphoproteome was compared with other data sets by mapping protein identifiers and phosphosite locations from Phospho.ELM (version 8.2) and Rikova et al. (10) to IPI human version 3.36. Only sites that could be mapped unambiguously to a single IPI identifier were included, resulting in 1405 and 3955 mapped phosphotyrosines, respectively. Overlap was determined by counting the number of identical sites in the different combinations of data sets.
RESULTS
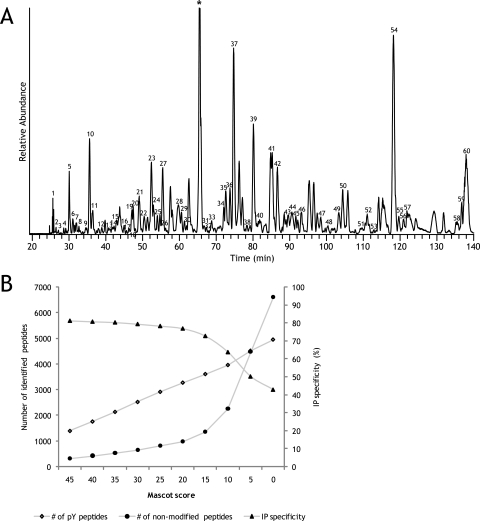
To first evaluate our protocols and to chart the potential arsenal of phosphorylation sites in HeLa cells, we performed immunoaffinity enrichment of tyrosine phosphorylated peptides from HeLa cell lysate digests in which hyperphosphorylation was induced via inhibition of phosphatases by pervanadate. To allow for a comprehensive coverage of tyrosine phosphorylation, HeLa cells that were stimulated with pervanadate or mock treated were lysed, digested by trypsin, and mixed 1:1 followed by peptide immunoaffinity purification of 4 mg of total starting protein material by an anti-phosphotyrosine antibody immobilized on agarose beads. The peptides that eluted from the immunoaffinity resin were then analyzed without any further enrichment in two separate LC-MS runs using a 2- and a 3-h LC gradient, respectively. Peptides were identified by matching the fragmentation spectra against the IPI human database (version 3.36). In Fig. 1A, a part of the base peak chromatogram of the 3-h run is shown to illustrate the efficiency and specificity of the immunoprecipitation. The resolved peaks nearly all represent tyrosine phosphorylated peptides, whereas the few non-phosphorylated peptides of abundant proteins that were detected are largely masked by these phosphopeptides. In Fig. 1B, the apparent specificity of the immunoprecipitation and the number of identified tyrosine phosphorylated peptides with decreasing Mascot threshold score are plotted. By decreasing the Mascot threshold score, the number of identified redundant tyrosine phosphorylated peptides increases considerably. However, by decreasing the threshold, the chance of including false positives is also increased substantially (27). This is reflected in the number of identified non-tyrosine phosphorylated peptides that increases exponentially below a score of 20. Consequently, the apparent immunoprecipitation specificity remains above 75% from threshold score 45 to 20 and then suddenly drops. A Mascot score of 20 was therefore taken as a threshold score as this lies above the apparent inflection point. At this score, the FDR as calculated by performing a concatenated decoy database search is seemingly large, ∼9%. However, when only tyrosine phosphorylated peptides are taken into account, the FDR is only 2%. Tandem mass spectra are available in the PRIDE (Proteomics Identifications) database under accession number 9779. To create a unique, non-redundant phosphotyrosine peptide library, we filtered the identified peptides. Phosphorylation sites that were also identified with a methionine oxidation or a miscleavage were considered redundant, but different states of additional phosphorylation on serine, threonine, and/or tyrosine were considered unique. This led to the identification of 729 unique phosphotyrosine peptides in the 2-h run and 970 unique phosphotyrosine peptides in the 3-h run. The overlap between these two runs was very large (supplemental Fig. 1A), leading to the identification of 1112 unique phosphotyrosine peptides with a total of 983 unique tyrosine phosphorylation sites. A list of these identified phosphopeptides including the sites of phosphorylation and Mascot scores is given in supplemental Table 2. This is one of the largest experimental data sets of phosphotyrosine peptides detected in a single experiment without any further TiO2 or IMAC enrichment. We compared our data set with phosphotyrosine peptides and sites reported in the recent update of the Phospho.ELM (28) database and with a recently reported large phosphotyrosine peptide data set of lung cancer cell lines consisting of the phosphotyrosine peptides cumulatively identified from 20 different cell lines and 30 different tissues samples (10). Approximately 28% of the phosphotyrosine sites that were identified in the present study overlapped with Rikova et al. (10) (supplemental Fig. 1B). The Phospho.ELM database reports whether a phosphorylation site is identified by small scale analysis (low throughput (LTP)) or large scale, typically LC-MS-based, analysis (high throughput (HTP)). The overlap of our data set with the HTP database is small with 14.3% (supplemental Fig. 1B) but still more than 3 times larger than the overlap with the LTP database (supplemental Fig. 1C).
Fig. 1.
A, LC-MS base peak chromatogram of the eluate from the phosphotyrosine-immunoprecipitated pervanadate-treated HeLa digest. The numbers indicate intense peaks representing phosphotyrosine-containing peptides. Only a few non-phosphorylated peptides were detected in this eluate. A numbered list of these abundant phosphopeptides is available as supplemental Table 1. * represents a peak of an ion that could not be identified and most likely is a non-peptide species. B, number of identified phosphotyrosine (pY) and non-phosphorylated peptides as a function of the applied Mascot threshold score revealing the apparent specificity of the immunopurification. With decreasing Mascot score threshold, the number of non-modified peptides in the data set increases exponentially below 20. The inflection point at score 20 indicates that at an even lower score relatively more false positive identifications are introduced in the data set.
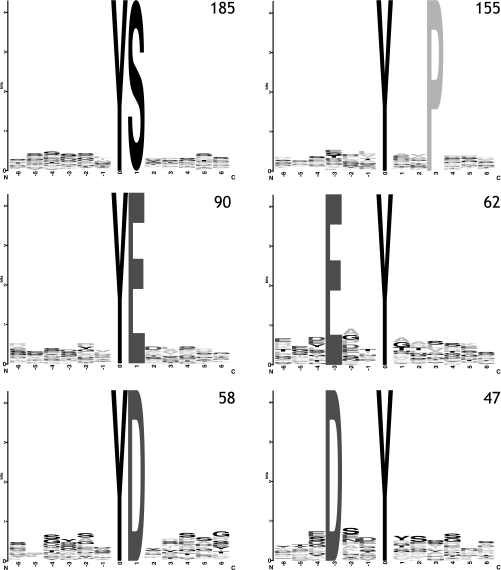
The size of the data set allowed us to do a statistically relevant motif analysis of the residues neighboring the sites of phosphorylation in our data set to identify conserved sequence motifs that can be recognized by protein-tyrosine kinases. Using motif-X (29), the occurrence of motifs from our phosphotyrosine data set was compared with the occurrence of these motifs in the total human proteome. We found six highly significantly enriched motifs that could classify two-thirds of our observed phosphotyrosine sites. These motifs are displayed as WebLogos (30) in Fig. 2. The motif with the highest occurrence has a serine residue at the P + 1 position. Surprisingly, this motif has not been reported previously, and only one rather promiscuous kinase/phosphatase motif and three also promiscuous binding motifs are known in PhosphoMotif finder (31) with a serine at that position. The second motif contains a proline at the P + 3 position. Two motifs show a negatively charged amino acid (aspartic or glutamic acid) at the P + 1 position. And two already previously identified motifs obtained from analysis in mouse brain (11) show a negatively charged amino acid at the P − 3 position. To obtain an indication of the potential kinases responsible for phosphorylation of the identified phosphotyrosine peptides, we used NetworKIN (32, 33). For 925 phosphosites (Tyr(P), Ser(P), and Thr(P)), we could identify the potential upstream kinase. Insulin receptor group kinases were shown to be the upstream kinase for more than 50% of the identified phosphotyrosine peptides that were identified in our data set whereby the insulin receptor and insulin-like growth factor 1 receptor contribute equally. Also, the EphA receptors, in particular EphA4, seem to have a high number of substrates represented in our data set (see supplemental Fig. 2).
Fig. 2.
motif-X (29) analysis of here acquired HeLa phosphotyrosine data set identified six conserved motifs, here displayed as WebLogos (30). The numbers indicate the number of peptides exhibiting this motif in the full data set.
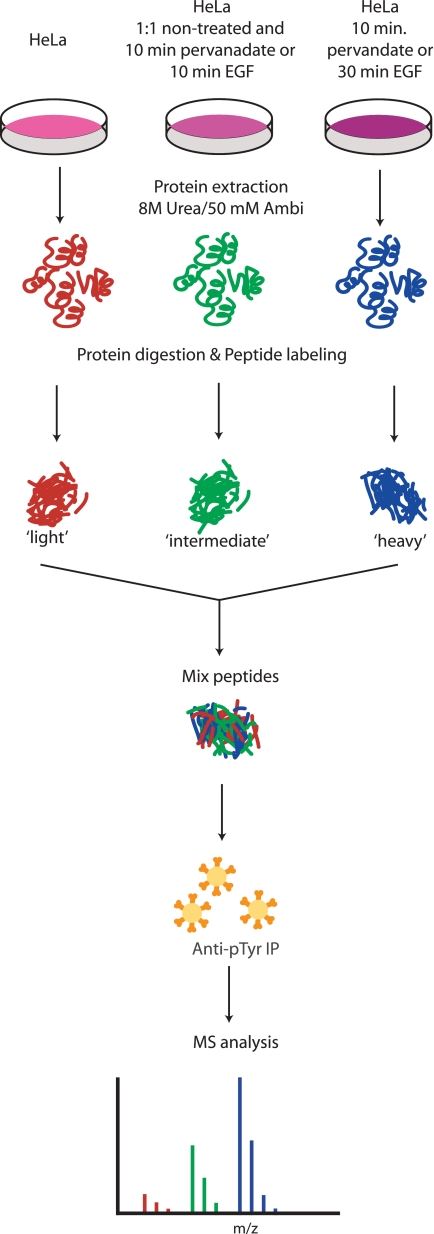
To allow for quantification of tyrosine phosphorylation, we next set out to incorporate stable isotope dimethyl labeling (19–21) into the phosphotyrosine peptide immunoprecipitation work flow. Therefore, HeLa cells were mock treated or treated with 1 mm pervanadate for 10 min. Cells were lysed, and proteins were digested. Peptides were labeled with light, intermediate, or heavy dimethyl essentially as described previously (20) but with adapted protocols to allow for the labeling of a few milligrams of sample (21). Peptides derived from the untreated cells were labeled with light dimethyl, peptides derived from a 1:1 mixture of untreated and pervanadate-treated cells were labeled with intermediate dimethyl (as an internal standard), and peptides derived from the pervanadate-treated cells were labeled with heavy dimethyl. Labeled peptides originating from 2 mg of protein sample for each of the three samples were mixed in a 1:1:1 ratio, and from this complex peptide mixture, tyrosine phosphorylated peptides were enriched by immunoprecipitation (Fig. 3). Using this approach, we were able to identify and quantify from a single LC-MS run 128 unique tyrosine phosphorylated triplet peptides originating from 99 phosphoproteins. As expected, most of the detected peptides show an abundance profile whereby the ion signal from the non-stimulated phosphopeptide is very low and the signal from the intermediate labeled phosphopeptide is equal to half of the sum of the untreated peptide signal (light) and treated phosphopeptide signal (heavy) (Fig. 4A). On average, the heavy labeled phosphopeptides were 2.22 ± 0.49 times more intense than the intermediate. A list of all quantified phosphopeptides is available as supplemental Table 3, and tandem mass spectra are available in the PRIDE database under accession number 9780. Interestingly, 15 tyrosine phosphorylated peptides seemed not to be affected by pervanadate treatment as the abundance profile did not significantly change (Table I and Fig. 5B). A gene ontology term (34) and motif-X (29) analysis was performed to find within this set of unaffected phosphopeptides and their corresponding proteins an enrichment of certain biological processes, molecular function, or conserved sequences, but the set was probably too small to detect any significant enrichment. However, an enrichment was found of phosphotyrosine sites that fall within a protein family (Pfam) domain (35) (p = 2.7e−6, Fisher’s exact test). 12 of the 15 unaffected phosphotyrosine sites (80%) fall within a Pfam domain, whereas only 18% of the remainder of phosphotyrosine sites in the quantified database fall within such globular domains, which is close to a previously reported 15–17% (36). Therefore, the exact cause of the apparent resistance to phosphatases at these particular phosphotyrosine sites remains elusive.
Fig. 3.
Experimental scheme for quantitative phosphotyrosine proteome studies. Cell cultures were mock treated or stimulated with pervanadate or EGF. After lysis and enzymatic digestion, peptides were differentially stable isotope dimethyl-labeled and combined before immunoprecipitation with a phosphotyrosine-specific antibody. The precipitate was analyzed by LC-MS followed by quantification using the triplet peaks originating from the different isotopes. AmBi, ammonium bicarbonate.
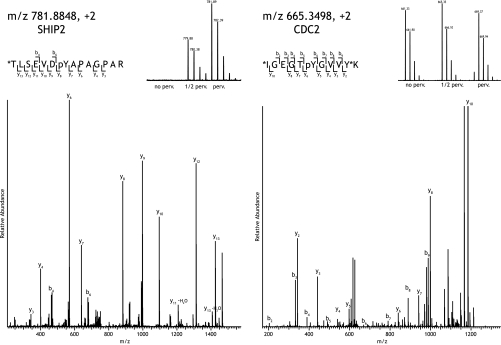
Fig. 4.
Representative examples of mass spectra and fragmentation spectra of peptides identified and quantified after pervanadate treatment. A, the abundance of SHIP-2 peptide TLSEVDpYAPAGPAR (where pY is phosphotyrosine) (fragmentation spectrum shown of m/z 781.8848, +2, heavy dimethyl-labeled) is dramatically increased by pervanadate treatment. B, the abundance of peptide IGEGTpYGVVYK (fragmentation spectrum shown of m/z 665.3498, +2, intermediate dimethyl-labeled) is not affected by pervanadate treatment. * indicates the site of stable isotope dimethyl labeling. perv., pervanadate.
Table I. Phosphotyrosine peptides whose abundance did not change significantly upon pervanadate treatment of HeLa cells.
L, light labeled peptide (mock treated); I, intermediate labeled peptide (1:1 mock and pervanadate-treated); H, heavy labeled (pervanadate-treated); pY, phosphotyrosine; pT, phosphothreonine; pS, phosphoserine; (ox)M, oxidized methionine.
| IPI accession no. | Gene | Peptide | Site 1 | Site 2 | I/L |
H/L |
||
|---|---|---|---|---|---|---|---|---|
| Ratio | S.D. | Ratio | S.D. | |||||
| IPI00016932 | SHIP-2 | NSFNNPApYpYVLEGVPHQLLPPEPPSPAR | Tyr-986 | Tyr-987 | 1.04 | 2.38 | ||
| IPI00008530 | RPLP0 | IIQLLDDpYPK | Tyr-24 | 0.81 | 0.65 | |||
| IPI00000352 | DYRK1B | IYQpYIQSR | Tyr-273 | 0.92 | 1.03 | |||
| IPI00291175 | VCL | SFLDSGpYR | Tyr-822 | 0.93 | 0.73 | |||
| IPI00396485 | EEF1A1 | EHALLApYTLGVK | Tyr-141 | 1.02 | 0.93 | |||
| IPI00026689 | CDC2 | IEKIGEGTpYGVVYK | Tyr-15 | 1.06 | 0.05 | 1.12 | 0.10 | |
| IPI00028570 | GSK3B | QLVRGEPNVSpYICSR | Tyr-216 | 1.10 | 0.01 | 1.68 | 0.15 | |
| IPI00654623 | TNS3 | KLSLGQpYDNDAGGQLPFSK | Tyr-540 | 1.15 | 1.19 | |||
| IPI00023503 | CDK3 | VEKIGEGTpYGVVYK | Tyr-43 | 1.16 | 1.41 | |||
| IPI00026689 | CDC2 | IGEGpTpYGVVYK | Thr-14 | Tyr-15 | 1.27 | 0.04 | 1.49 | 0.08 |
| IPI00013721 | PRPF4B | LCDFGSASHVADNDITPpYLVSR | Tyr-849 | 1.29 | 0.08 | 1.33 | 0.08 | |
| IPI00012885 | PTK2 | Y(ox)MEDSTpYYKASK | Tyr-576 | 1.30 | 1.42 | |||
| IPI00008438 | RPS10 | IAIpYELLFK | Tyr-12 | 1.41 | 1.70 | |||
| IPI00013981 | YES1 | LIEDNEpYTAR | Tyr-426 | 1.47 | 2.22 | |||
| IPI00166578 | PRAGMIN | EATQPEPIpYAESTKR | Tyr-411 | 1.66 | 0.03 | 2.44 | 0.23 | |
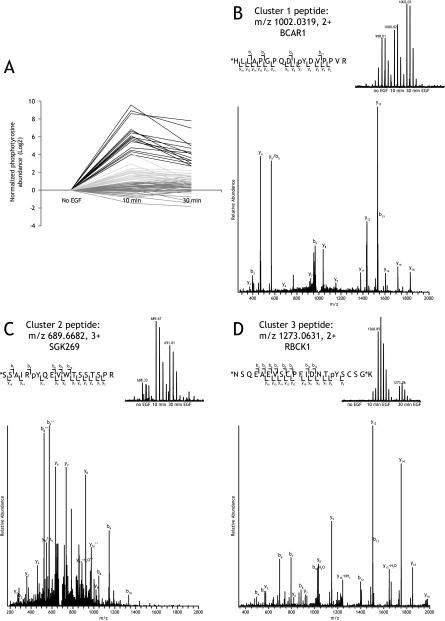
Fig. 5.
A, clustering of tyrosine phosphorylation profiles. Cluster 1 (dark gray), no or only a small change in phosphorylation levels; cluster 2 (light gray), an immediate up-regulation that remains after 30 min; cluster 3 (black), strong and quick increase in phosphorylation. B, representative examples of mass spectra and fragmentation spectra of peptides identified and quantified after EGF stimulation from each of the clusters. B, BCAR1 peptide HLLAPGPQDIpYDVPPVR (where pY is phosphotyrosine) (fragmentation spectrum shown of m/z 1002.0319, +2, heavy dimethyl-labeled) is from cluster 1 with only a slight increase in phosphorylation upon EGF stimulation. C, SGK269 peptide SSAIRpYQEVWTSSTSPR (fragmentation spectrum shown of m/z 689.6682, +3, intermediate dimethyl-labeled) is from cluster 2 with a larger increase after EGF stimulation. D, RBCK1 peptide NSQEAEVSCPFIDNTpYSCSGK (fragmentation spectrum shown of m/z 1273.0631, +2, heavy dimethyl-labeled) is from cluster 3 showing an extensive increase in abundance after EGF stimulation. * indicates the site of stable isotope dimethyl labeling.
With the quantitative method established, we next set out to investigate tyrosine phosphorylation-mediated EGFR signaling pathways using stable isotope dimethyl labeling followed by phosphotyrosine immunoprecipitation. In our study, HeLa cells were stimulated with EGF for 0, 10, or 30 min to study the temporal phosphotyrosine signaling pathways after stimulation of the EGF receptor. We used methods essentially similar to those described above for the pervanadate-treated cells (Fig. 3). Briefly, HeLa cells were stimulated with 150 ng/ml EGF for 0, 10, or 30 min. Cells were lysed, and proteins were digested. The derived peptide mixtures were stable isotope-labeled with light, intermediate, or heavy dimethyl labels and mixed in a 1:1:1 ratio after which tyrosine phosphorylated peptides were immunoprecipitated using the anti-phosphotyrosine antibody. 73 unique phosphotyrosine peptides, originating from 52 phosphoproteins, could be differentially quantified over all three time points after EGF stimulation in a single LC-MS analysis without any additional affinity enrichment. These associated phosphoproteins, including their sites of phosphorylation, are listed in Table II (for a full table with Mascot and post-translational modification scores, see supplemental Fig. 4), and tandem mass spectra are available in the PRIDE database under accession number 9777. All except one of the phosphotyrosine peptides that were identified have been reported before and can be found in the PhosphoSitePlus database (37). The observed temporal ratio profiles largely clustered into three groups (Fig. 5A). Cluster 1 consists of sites that showed no or only a small change in phosphorylation levels. Cluster 2 consists of tyrosine phosphorylation sites that showed an immediate up-regulation upon EGF stimulation that remains after 30 min. Cluster 3 comprises phosphorylation sites that showed a large and quick (10 min) increase in tyrosine phosphorylation that already diminishes at 30 min. In the latter cluster, autophosphorylation sites of EGFR are found along with established direct EGFR interactors such as Gab1, Cbl, and SHC1. In cluster 2 are downstream targets like p38a, Stat3, and GSK3-β. Some phosphotyrosine sites were found to be regulated that have not been conclusively established to be involved in EGFR signaling such as sites on SgK269, IRS2, HNRNPA1, and ATP1A1. The tyrosine phosphorylation on TFG has not been reported before. Finally, in cluster 1, phosphotyrosine peptides were detected from CTTN, ENO1, and N-WASP that showed an apparent down-regulation upon EGF stimulation. Representative examples of tyrosine phosphorylated peptides from the three clusters and their temporal profiles are shown in Fig. 5B.
Table II. Identified and quantified tyrosine phosphopeptides after EGF stimulation.
pY, phosphotyrosine; pT, phosphothreonine; pS, phosphoserine.
| IPI accession no. | Gene | Peptide | Site 1 | Site 2 | log2 ratio, 10/0 min |
S.D. | log2 ratio, 30/0 min |
S.D. |
|---|---|---|---|---|---|---|---|---|
| IPI00011676 | N-WASP | VIpYDFIEK | Tyr-256 | −1.49 | 0.66 | −1.86 | 0.74 | |
| IPI00465248 | ENO1 | AAVPSGASTGIpYEALELR | Tyr-44 | −0.90 | 0.25 | −0.08 | 0.25 | |
| IPI00029601 | CTTN | TQTPPVSPAPQPTEERLPSSPVpYEDAASFK | Tyr-421 | −0.73 | −1.47 | |||
| IPI00220644 | PKM2 | TATESFASDPILpYRPVAVALDTK | Tyr-105 | −0.68 | −0.13 | |||
| IPI00012885 | PTK2 | GSIDREDGSLQGPIGNQHIpYQPVGKPDPAAPPK | Tyr-861 | −0.52 | 0.45 | 0.43 | 0.09 | |
| IPI00216423 | ITSN2 | REEPEALpYAAVNK | Tyr-967 | −0.41 | −0.79 | |||
| IPI00021439 | ACTB | IWHHTFpYNELR | Tyr-91 | −0.32 | 0.57 | |||
| IPI00008530 | RPLP0 | IIQLLDDpYPK | Tyr-24 | −0.14 | −0.16 | |||
| IPI00215949 | HIPK2 | AVCSTpYLQSR | Tyr-361 | −0.13 | −0.34 | |||
| IPI00013721 | PRPF4B | LCDFGSASHVADNDITPpYLVSR | Tyr-849 | −0.10 | 0.09 | |||
| IPI00000352 | DYRK1B | IYQpYIQSR | Tyr-273 | −0.06 | −0.04 | |||
| IPI00217966 | LDHA | QVVESApYEVIK | Tyr-239 | 0.02 | 0.30 | |||
| IPI00641339 | BCAR1 | GLPPSNHHAVpYDVPPSVSK | Tyr-306 | 0.05 | 0.58 | |||
| IPI00552750 | TNK2 | KPTpYDPVSEDQDPLSSDFKR | Tyr-574 | 0.08 | 0.12 | −1.30 | 0.25 | |
| IPI00641339 | BCAR1 | AQQGLpYQVPGPSPQFQSPPAK | Tyr-128 | 0.16 | 0.77 | |||
| IPI00641339 | BCAR1 | HLLAPGPQDIpYDVPPVR | Tyr-249 | 0.20 | 0.15 | 0.73 | 0.22 | |
| IPI00301058 | VASP | VQIpYHNPTANSFR | Tyr-39 | 0.21 | 0.12 | 0.26 | 0.06 | |
| IPI00654623 | TNS3 | LSLGQpYDNDAGGQLPFSK | Tyr-540 | 0.24 | 0.40 | |||
| IPI00021076 | PKP4 | NNYALNTTATpYAEPYRPIQYR | Tyr-478 | 0.32 | 0.77 | |||
| IPI00306959 | KRT7 | LSSARPGGLGSSSLpYGLGASRPR | Tyr-40 | 0.32 | 0.10 | 0.82 | 0.01 | |
| IPI00641339 | BCAR1 | RPGPGTLpYDVPRER | Tyr-387 | 0.33 | 0.15 | 1.00 | 0.07 | |
| IPI00021267 | EPHA2 | TYVDPHTpYEDPNQAVLK | Tyr-594 | 0.35 | 0.69 | |||
| IPI00166578 | PRAGMIN | EATQPEPIpYAESTK | Tyr-411 | 0.39 | 0.14 | 0.20 | 0.23 | |
| IPI00291175 | VCL | SFLDSGpYR | Tyr-822 | 0.40 | 0.35 | |||
| IPI00220030 | PXN | VGEEEHVpYSFPNK | Tyr-118 | 0.43 | 0.19 | 0.50 | 0.11 | |
| IPI00013981 | Src | LIEDNEpYTAR | Tyr-426 | 0.44 | 0.72 | |||
| IPI00418471 | VIM | SLYASSPGGVpYATR | Tyr-61 | 0.45 | 0.65 | |||
| IPI00220030 | PXN | FIHQQPQSSSPVpYGSSAK | Tyr-88 | 0.47 | 0.50 | |||
| IPI00396485 | EEF1A1 | STTTGHLIpYK | Tyr-29 | 0.50 | 0.32 | |||
| IPI00298347 | PTPN11 | IQNTGDpYYDLYGGEK | Tyr-62 | 0.50 | 0.75 | |||
| IPI00026689 | CDC2 | IGEGpTpYGVVYK | Thr-14 | Tyr-15 | 0.58 | 0.00 | 1.22 | 0.00 |
| IPI00026689 | CDC2 | IGEGTYGVVpYKGR | Tyr-19 | 0.59 | 0.15 | 0.49 | 0.03 | |
| IPI00016932 | SHIP-2 | ERLpYEWISIDKDEAGAK | Tyr-886 | 0.63 | 1.55 | |||
| IPI00021267 | EPHA2 | VLEDDPEATpYTTSGGKIPIR | Tyr-772 | 0.65 | 1.04 | |||
| IPI00220030 | PXN | FIHQQPQSSpSPVpYGSSAK | Ser-85 | Tyr-88 | 0.74 | 0.00 | 0.91 | 0.00 |
| IPI00396485 | EEF1A1 | EHALLApYTLGVK | Tyr-141 | 0.80 | 0.61 | |||
| IPI00182469 | CTNND1 | SLDNNpYSTPNER | Tyr-898 | 0.90 | 0.33 | |||
| IPI00012885 | PTK2 | YMEDSTpYYK | Tyr-576 | 0.91 | 0.71 | |||
| IPI00182469 | CTNND1 | LNGPQDHSHLLpYSTIPR | Tyr-96 | 0.98 | 1.03 | |||
| IPI00737545 | SGK269 | NAIKVPIVINPNApYDNLAIYK | Tyr-635 | 1.17 | 0.98 | |||
| IPI00022353 | TYK2 | LLAQAEGEPCpYIR | Tyr-292 | 1.19 | 1.01 | |||
| IPI00215965 | HNRNPA1 | SSGPpYGGGGQYFAKPR | Tyr-341 | 1.22 | 0.45 | |||
| IPI00028570 | GSK3B | GEPNVSpYICSR | Tyr-216 | 1.34 | 0.74 | 0.95 | 0.39 | |
| IPI00176903 | PTRF | SFTPDHVVpYAR | Tyr-308 | 1.44 | 1.15 | |||
| IPI00014454 | RIN1 | EKPAQDPLpYDVPNASGGQAGGPQRPGR | Tyr-36 | 1.53 | 0.05 | 1.81 | 0.11 | |
| IPI00412752 | STAT3 | YCRPESQEHPEADPGSAAPpYLK | Tyr-705 | 1.56 | 0.96 | |||
| IPI00029601 | CTTN | GPVSGTEPEPVpYSMEAADYR | Tyr-446 | 1.57 | 1.21 | |||
| IPI00026689 | CDC2 | IEKIGEGTpYGVVYK | Tyr-15 | 1.66 | 0.71 | |||
| IPI00334715 | GRLF1 | NEEENIpYSVPHDSTQGK | Tyr-1105 | 1.69 | 0.00 | 1.64 | 0.04 | |
| IPI00552750 | TNK2 | VSSTHpYYLLPERPSYLER | Tyr-913 | 1.87 | 0.89 | |||
| IPI00216423 | ITSN2 | LIpYLVPEK | Tyr-552 | 1.93 | 1.26 | |||
| IPI00022462 | TFRC | SAFSNLFGGEPLSpYTR | Tyr-20 | 2.04 | 1.41 | |||
| IPI00737545 | SGK269 | SSAIRpYQEVWTSSTSPR | Tyr-531 | 2.04 | 1.63 | |||
| IPI00464978 | IRS2 | SYKAPYTCGGDSDQpYVLMSSPVGR | Tyr-825 | 2.16 | 1.61 | |||
| IPI00006482 | ATP1A1 | GIVVpYTGDR | Tyr-260 | 2.28 | 1.88 | |||
| IPI00031068 | GAB1 | APSASVDSSLpYNLPR | Tyr-259 | 2.42 | 2.19 | |||
| IPI00002857 | p38a | HTDDEMTGpYVATR | Tyr-182 | 2.98 | 0.36 | |||
| IPI00294619 | TFG | NRPPFGQGpYTQPGPGYR | Tyr-392 | 4.13 | 1.04 | |||
| IPI00003479 | Erk1 | VADPDHDHTGFLTEpYVATR | Tyr-187 | 3.98 | 0.29 | 2.68 | 0.25 | |
| IPI00003479 | Erk1 | VADPDHDHTGFLpTEpYVATR | Thr-185 | Tyr-187 | 4.31 | 2.98 | ||
| IPI00016932 | SHIP-2 | NSFNNPApYYVLEGVPHQLLPPEPPSPAR | Tyr-986 | 4.49 | 0.27 | 3.29 | 0.48 | |
| IPI00023343 | DLG3 | RDNEVDGQDpYHFVVSR | Tyr-673 | 4.74 | 4.15 | |||
| IPI00018274 | EGFR | GSHQISLDNPDpYQQDFFPK | Tyr-1172 | 5.49 | 4.27 | |||
| IPI00016932 | SHIP-2 | TLSEVDpYAPAGPAR | Tyr-1135 | 5.70 | 3.35 | |||
| IPI00020178 | STAM | QQSTTLSTLpYPSTSSLLTNHQHEGR | Tyr-198 | 5.84 | 0.24 | 3.64 | 0.25 | |
| IPI00513796 | SHC1 | ELFDDPSpYVNVQNLDKAR | Tyr-317 | 5.97 | 0.69 | 4.71 | 0.61 | |
| IPI00290542 | STAM2 | LVNEAPVpYSVYSK | Tyr-371 | 6.12 | 2.78 | |||
| IPI00002495 | EPN1 | NIVHNpYSEAEIK | Tyr-17 | 6.45 | 5.19 | |||
| IPI00018274 | EGFR | GSTAENAEpYLR | Tyr-1197 | 6.68 | 4.07 | |||
| IPI00027269 | CBL | IKPSSSANAIpYSLAARPLPVPK | Tyr-674 | 6.81 | 0.05 | 3.04 | 0.35 | |
| IPI00031068 | GAB1 | SSGSGSSVADERVDpYVVVDQQK | Tyr-659 | 8.59 | 7.76 | |||
| IPI00010748 | RBCK1 | NSQEAEVSCPFIDNTpYSCSGK | Tyr-288 | 8.93 | 6.95 | |||
| IPI00290542 | STAM2 | SLpYPSSEIQLNNK | Tyr-192 | 9.57 | 4.95 |
Comparing the identified phosphotyrosine peptides from the three separate experiments, a large overlap can be observed between the more comprehensive data set of 1112 phosphopeptides in the pervanadate-blocked cells and the two other quantitative experiments (supplemental Fig. 3). 86% of the phosphotyrosine peptides identified in the quantitative pervanadate study overlapped with the library, whereas 74% of the EGF study overlapped with the library set. Also, 51% of the phosphotyrosine peptides identified in the EGF experiment overlapped with those identified in the quantitative pervanadate experiment. Ratios of these phosphopeptides were plotted against each other (supplemental Fig. 4). Some of the sites that showed an increase in tyrosine phosphorylation upon phosphatase inhibition showed no increase upon EGF stimulation. However, tyrosine phosphorylation events that showed a large increase upon EGF stimulation also showed a strong increase upon phosphatase inhibition by pervanadate. Interestingly, some of the phosphotyrosine sites that were not affected by pervanadate treatment, such as CDC2 (Tyr(P)-15) and GSK3β (Tyr(P)-216), did increase in abundance upon EGF stimulation.
DISCUSSION
Analysis of tyrosine phosphorylation in cells or tissue is extremely important to understand critical signaling processes involved in processes like development and human disease. In this work, we explored and further optimized an enrichment method for phosphotyrosine peptides based on immunoaffinity purification using phosphotyrosine-specific antibodies. Our protocol allowed a very high level of enrichment efficiency, providing an eluted fraction dominated by phosphotyrosine peptides that required no further IMAC or TiO2 enrichment prior to LC-MS/MS analysis. From just two of these eluates, we were able to identify 1112 unique, non-redundant phosphotyrosine peptides derived from only 4 mg of starting material. The overlap of our HeLa cells phosphotyrosine sites with the data set reported in Rikova et al. (10) is 28%. Notably, the data set was of Rikova et al. (10) was obtained by analyzing several dozens of non-small cell lung cancer cell lines and non-small cell lung cancer tumors. The different constitutions of their and our cells, together with a different antibody used for phosphotyrosine peptide enrichment, might explain the relatively small overlap in tyrosine phosphopeptides, whereas the greater protein amount and larger number of LC-MS analyses might have allowed them an even deeper penetration into the tyrosine phosphoproteome. The overlap between LTP and HTP phosphosites in the Phospho.ELM database is surprisingly low, indicating that the tyrosine phosphoproteome has not yet been fully mapped (28). Not unexpectedly, the overlap between our data set and the HTP data set is larger than with the smaller LTP data set. Apparently, a different subset of phosphosites is detected by classical biology means than by LC-MS even though data sets derived by the latter method typically contain hundreds of phosphosites. It is estimated that of the peptides that are identified in a vertebrate cell to be phosphorylated ∼0.05–2% are phosphorylated on tyrosine residues (25, 38). If these numbers are taken as true, the number of phosphotyrosine sites identified in this study (over 1000) would suggest that there may be concurrently even up to 1,000,000 serine and threonine phosphorylation sites present as well. Therefore, even with ever improving analytical tools, it still remains a great challenge to comprehensively analyze the complete cellular phosphoproteome.
Several stable isotope labeling-based quantification methods have been used in combination with phosphoproteomics approaches, including chemical labeling such as iTRAQ (4, 5, 13) and metabolic labeling such as SILAC (2, 39). The advantage of a chemical modification approach over metabolic labeling is the ability to label samples after cell lysis and digestion. This makes the approach more generically applicable as it also allows the quantitative analysis of biological samples that cannot be grown in culture, such as human body fluids or tissue. The benefit of peptide level immunoprecipitation combined with stable isotope labeling is that differently labeled samples can be combined prior to immunoprecipitation, thereby neutralizing the potentially largest source of variation. Furthermore, quantification at the peptide level allows the separate analysis of phosphorylation events on the same protein. Here, we introduced stable isotope dimethyl labeling for quantification of immunoprecipitated phosphotyrosine peptides. As the starting material for these immunoprecipitations is typically several milligrams of protein, the use of iTRAQ labeling can be cost-prohibitive, whereas stable isotope dimethyl labeling is performed with inexpensive generic reagents and thereby does not pose financial restrictions to the amount of sample to be labeled (21). Importantly, the chemical labeling does not seem to significantly affect the immunoprecipitation process itself.
We were able to identify and quantify 128 unique phosphotyrosine peptides upon pervanadate induction, all detected by their characteristic peptide triplets that can be easily distinguished in the LC-MS spectra. As expected, most of the peptide ion signals of the internal standard intensity were equal to half of the sum of the untreated and treated signal intensities, confirming that stable isotope dimethyl labeling does not alter phosphorylation states or impair immunoprecipitation. For most phosphotyrosine peptides, the detected ratios were close to the expected 0:1:2, indicating that tyrosine phosphorylation was largely enhanced upon pervanadate treatment. In Fig. 4A, an example of such a phosphotyrosine peptide can be seen. No signal is detected at the m/z where the light labeled, non-pervanadate treated phosphotyrosine peptide would reside. The intermediate labeled phosphotyrosine peptide from the internal standard is half the intensity of the heavy labeled phosphotyrosine peptide. The internal standard facilitates the detection and quantification of such an on-off situation. Without the internal standard, only a single isotope envelope would be visible. The internal standard confirms the on-off situation and validates the identified sequence by the number of lysine residues that can be easily determined based on the mass shift between the intermediate and heavy labeled peptides. Surprisingly, about 15 of 128 of the quantified peptides showed a close to 1:1:1 ratio in the three samples, indicating that these phosphorylation sites were not affected by pervanadate treatment. Pervanadate is thought to inhibit phosphatases by irreversibly oxidizing the catalytic cysteine (40). The fact that all of these phosphopeptides were selected by the mass spectrometer for sequencing indicates that these phosphopeptides are relatively abundant. These sites might therefore be constitutively phosphorylated. However, some of the sites showed an increase in phosphorylation upon EGF stimulation, and assuming that the time scale of 10 min is too low for protein synthesis, this suggests that some non-phosphorylated tyrosine residues are phosphorylated upon EGF stimulation. Explanations for the unaltered phosphorylation levels upon pervanadate treatment might involve specific phosphatases that are not inhibited by pervanadate or phosphorylation sites that are not easily accessible for these phosphates so that inhibition does not lead to increased phosphorylation under the condition used here. The here observed enrichment of phosphotyrosine sites residing in Pfam domains, which are protein domains with a known function and tertiary structure, for this class of peptides could be in agreement with the latter hypothesis. It has been suggested that proteins become typically phosphorylated on sites outside these functional domains (36, 41) as phosphorylation sites on the interspersing variable regions might be more exposed for kinases and phosphatases. We hypothesize that the unaffected phosphorylation sites might therefore be structurally essential or concealed in a non-accessible tertiary structural element.
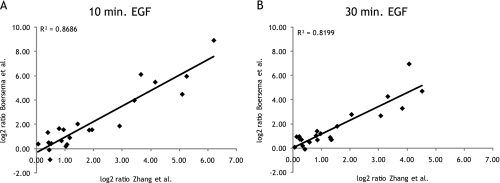
The EGF receptor plays an important role in a variety of cellular processes and has been studied intensively over the years. The EGFR signaling pathway has been investigated by a variety of (phospho)proteomics approaches (2, 4, 5, 9, 13, 25, 42, 43). Upon activation by a growth factor, the protein-tyrosine kinase activity of EGFR is enhanced (44). Our quantitative phosphotyrosine peptide immunoprecipitation, therefore, provides an excellent way to study EGFR signaling. For this reason and as the pathway is very well documented in the literature, we took it here as a model system to further test our new approach. We analyzed HeLa cells that were stimulated for 0, 10, or 30 min by EGF. 73 unique tyrosine phosphorylated triplet peptides were identified and quantified over all time points. Most of the tyrosine phosphopeptides we identified have been reported before also in the context of EGF stimulation. For example, more than 30% of the here identified phosphosites overlap with those identified in Zhang et al. (5). In that report, similar EGF stimulation conditions were used, and the effect was also measured after 0, 10, and 30 min. Reassuringly, the observed ratios for these overlapping phosphopeptides 10 and 30 min after EGF stimulation correlated very well with those found in our study (correlation coefficients, 0.87 and 0.82, respectively; see Fig. 6 and Table III). This is especially remarkable considering that different cell lines were used (HeLa versus 184A1 parental human mammary epithelial cells), suggesting a high conservation of the EGFR signaling pathway between these two cell lines. Moreover, it reveals that quantitative tyrosine phosphoproteome studies can provide highly reproducible results, even between different laboratories and different cell lines, which is a status not yet readily achieved in global phosphoproteome studies. Most of the identified phosphotyrosine peptides that are increased upon EGF stimulation are known members of the EGF signaling pathway. The EGFR autophosphorylation sites Tyr(P)-1172 and Tyr(P)-1197 showed an increase in phosphorylation after 10 min of EGF stimulation whereby this phosphorylation decreased again after 30 min, consistent with previously published data (5, 45). Furthermore, phosphopeptides of Gab1 (Tyr(P)-659), STAM2 (Tyr(P)-192 and Tyr(P)-371), RBCK1 (Tyr(P)-288), Cbl (Tyr(P)-674), SHIP-2 (Tyr(P)-986 and Tyr(P)-1135), and Epsin (Tyr(P)-17) showed an extensive increase in phosphorylation and have shown to be involved in EGF signaling and internalization (5, 12, 15, 46, 47).
Fig. 6.
Ratios of changes in tyrosine phosphorylation 10 (A) and 30 min (B) after EGF stimulation detected in present study (Boersema et al.) plotted against ratios of overlapping phosphotyrosine sites found in Zhang et al.(5).
Table III. Ratios of changes in tyrosine phosphorylation upon EGF stimulation of phosphorylation sites detected in the present study and Zhang et al. (5).
pY, phosphotyrosine; pS, phosphoserine.
| Peptide sequence/phosphosite | Tyr(P) site | Zhang et al. (5) |
Present study |
||
|---|---|---|---|---|---|
| log2 ratio, 10/0 min |
log2 ratio, 30/0 min |
log2 ratio, 10/0 min |
log2 ratio, 30/0 min |
||
| AAVPSGASTGIpYEALELR | Tyr-44 | 0.47 | 0.42 | −0.90 | −0.08 |
| LCDFGSASHVADNDITPpYLVSR | Tyr-849 | 0.44 | 0.07 | −0.10 | 0.09 |
| EATQPEPIpYAESTKR | Tyr-411 | 0.06 | 0.34 | 0.39 | 0.20 |
| VGEEEHVpYSFPNK | Tyr-118 | 0.52 | 0.59 | 0.43 | 0.50 |
| TYVDPHTpYEDPNQAVLK | Tyr-594 | 1.07 | 0.86 | 0.35 | 0.69 |
| YMEDSTpYYK | Tyr-576 | 1.16 | 1.35 | 0.91 | 0.71 |
| IGEGTpYGVVYK | Tyr-15 | 0.79 | 0.30 | 1.66 | 0.71 |
| HLLAPGPQDIpYDVPPVR | Tyr-249 | 1.03 | 1.33 | 0.20 | 0.73 |
| IQNTGDpYYDLYGGEK | Tyr-62 | 0.42 | 0.25 | 0.50 | 0.75 |
| VSSTHpYYLLPERPSYLER | Tyr-857 | 2.91 | 1.31 | 1.87 | 0.89 |
| YCRPESQEHPEADPGSAAPpYLK | Tyr-705 | 1.94 | 0.23 | 1.56 | 0.96 |
| GEPNVSpYICSR | Tyr-279 | 0.39 | 0.14 | 1.34 | 0.95 |
| VLEDDPEATpYTTSGGKIPIR | Tyr-772 | 0.87 | 0.82 | 0.65 | 1.04 |
| GPVSGTEPEPVpYSMEAADYR | Tyr-446 | 0.95 | 0.97 | 1.57 | 1.21 |
| SAFSNLFGGEPLSpYTR | Tyr-20 | 1.44 | 0.85 | 2.04 | 1.41 |
| EKPAQDPLpYDVPNASGGQAGGPQRPGR | Tyr-36 | 1.84 | 1.55 | 1.53 | 1.81 |
| VADPDHDHTGFLTEpYVATR | Tyr-204 | 3.42 | 3.07 | 3.98 | 2.68 |
| LVNEAPVYSVpYSK | Tyr-374 | 3.66 | 2.06 | 6.12 | 2.78 |
| NSFNNPApYYVLEGVPHQLLPPEPPpSPARAPVPSATK | Tyr-986/ Ser-1003 | 5.10 | 3.83 | 4.49 | 3.29 |
| GSHQISLDNPDpYQQDFFPK | Tyr-1172 | 4.16 | 3.32 | 5.49 | 4.27 |
| ELFDDPSpYVNVQNLDK | Tyr-317 | 5.26 | 4.52 | 5.97 | 4.71 |
| NSQEAEVSCPFIDNTpYSCSGK | Tyr-288 | 6.21 | 4.07 | 8.93 | 6.95 |
Some of the tyrosine phosphorylated peptides that showed an increase in abundance after EGF stimulation have not been reported before, or the phosphosite has not been described to be profoundly involved in EGFR signaling. For example, here we observed IRS2 Tyr(P)-260 to become more phosphorylated upon EGF stimulation (48, 49). Also, TFG (Tyr(P)-392) showed high tyrosine phosphorylation in response to EGF treatment, but the phosphosite has not been reported before. Recently, this protein has been described to be a substrate of Src (50) like SgK269, a pseudokinase for which we found two phosphosites (Tyr(P)-531 and Tyr(P)-635) to be up-regulated (51, 52). Up-regulation of both these phosphotyrosine sites has not been associated with EGF stimulation before, but Tyr(P)-635 was suggested to be involved in Src interaction (52) and was shown to be down-regulated upon drug-induced EGFR inhibition (9). With these two proteins, we might shed new light on a less well established Src-based part of the EGFR signaling pathway. Next, DLG3 (Tyr(P)-673), which was shown to be down-regulated upon drug-induced EGFR inhibition (9), was found here, in agreement, to be up-regulated upon EGF stimulation.
Finally, only a few tyrosine phosphorylation sites were observed to be significantly down-regulated upon EGF stimulation: N-WASP (Tyr(P)-256), ENO1 (Tyr(P)-44), and CTTN (Tyr(P)-421). Interestingly, CTTN has been shown to bind N-WASP. Tyr(P)-421 is one of three residues known to be phosphorylated by Src, whereas Erk can phosphorylate CTTN at two other serine residues (53, 54). This Erk and Src phosphorylation of CTTN was suggested to act as a switch on-switch off mechanism, respectively, for the activation of N-WASP leading to actin polymerization, which is important for example in cytokinesis (55). The exact outcome of the down-regulation of these sites regarding actin polymerization remains to be elucidated.
Our quantitative phosphotyrosine profiling in HeLa cells enables a direct comparison of the response of individual phosphorylation sites upon EGF stimulation and pervanadate treatment. Some sites showed a strong increase after pervanadate treatment but were seemingly unaffected by EGF stimulation, whereas others were increased in abundance after both EGF stimulation and pervanadate treatment. The existence of the latter category would suggest that in the pervanadate treatment conditions EGFR stimuli exist or kinases are active that target substrates that are also in the EGFR signaling pathway. This is not surprising as the fetal calf serum on which these cells are grown contains hormones and growth factors. This is reflected in the overrepresentation of insulin receptor, insulin-like growth factor receptor, and ephrin receptors in the upstream kinases as predicted by NetworKIN in the larger library phosphotyrosine data set.
In conclusion, immunoprecipitation of tyrosine phosphorylated stable isotope dimethyl-labeled peptides allows the quantitative analysis of tyrosine phosphorylation. As isotope labeling is performed after cell lysis and enzymatic digestion, the method is applicable to virtually any sample type, including human tissue. As demonstrated for the EGFR signaling pathway, by these means protein-tyrosine kinase signaling pathways can be studied in a relatively quick and inexpensive manner. Several phosphotyrosine sites were newly identified or further substantiated to be involved in EGFR signaling. By performing quantification on the peptide rather than protein levels, different phosphorylation events on the same protein can be readily monitored.
Supplementary Material
Acknowledgments
We thank Dr. Rune Linding for help with the NetworKIN analysis.
Footnotes
* This work was supported by The Netherlands Proteomics Centre, a program embedded in The Netherlands Genomics Initiative.
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental Tables 1–4 and Figs. 1–4.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental Tables 1–4 and Figs. 1–4.
1The abbreviations used are:
- EGF
- epidermal growth factor
- EGFR
- EGF receptor
- FDR
- false discovery rate
- HTP
- high throughput
- iTRAQ
- isobaric tag for relative and absolute quantitation
- LTP
- low throughput
- Pfam
- protein family
- PRIDE
- Proteomics Identifications
- SILAC
- stable isotope labeling by amino acids in cell culture
- XIC
- extracted ion chromatogram
- RT
- room temperature
- IP
- immunoprecipitation
- IPI
- International Protein Index
- Erk
- extracellular signal-regulated kinase
- TFG
- TRK-fused gene
- CTTN
- cortactin
- N-WASP
- neuronal Wiskott-Aldrich syndrome protein
REFERENCES
- 1.Blume-Jensen P., Hunter T. (2001) Oncogenic kinase signalling. Nature 411, 355–365 [DOI] [PubMed] [Google Scholar]
- 2.Blagoev B., Ong S. E., Kratchmarova I., Mann M. (2004) Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nat. Biotechnol 22, 1139–1145 [DOI] [PubMed] [Google Scholar]
- 3.Steen H., Kuster B., Fernandez M., Pandey A., Mann M. (2002) Tyrosine phosphorylation mapping of the epidermal growth factor receptor signaling pathway. J. Biol. Chem 277, 1031–1039 [DOI] [PubMed] [Google Scholar]
- 4.Thelemann A., Petti F., Griffin G., Iwata K., Hunt T., Settinari T., Fenyo D., Gibson N., Haley J. D. (2005) Phosphotyrosine signaling networks in epidermal growth factor receptor overexpressing squamous carcinoma cells. Mol. Cell. Proteomics 4, 356–376 [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y., Wolf-Yadlin A., Ross P. L., Pappin D. J., Rush J., Lauffenburger D. A., White F. M. (2005) Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol. Cell. Proteomics 4, 1240–1250 [DOI] [PubMed] [Google Scholar]
- 6.Pandey A., Podtelejnikov A. V., Blagoev B., Bustelo X. R., Mann M., Lodish H. F. (2000) Analysis of receptor signaling pathways by mass spectrometry: identification of Vav-2 as a substrate of the epidermal and platelet-derived growth factor receptors. Proc. Natl. Acad. Sci. U.S.A 97, 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amanchy R., Kalume D. E., Iwahori A., Zhong J., Pandey A. (2005) Phosphoproteome analysis of HeLa cells using stable isotope labeling with amino acids in cell culture (SILAC). J. Proteome Res 4, 1661–1671 [DOI] [PubMed] [Google Scholar]
- 8.Hinsby A. M., Olsen J. V., Bennett K. L., Mann M. (2003) Signaling initiated by overexpression of the fibroblast growth factor receptor-1 investigated by mass spectrometry. Mol. Cell. Proteomics 2, 29–36 [DOI] [PubMed] [Google Scholar]
- 9.Guo A., Villén J., Kornhauser J., Lee K. A., Stokes M. P., Rikova K., Possemato A., Nardone J., Innocenti G., Wetzel R., Wang Y., MacNeill J., Mitchell J., Gygi S. P., Rush J., Polakiewicz R. D., Comb M. J. (2008) Signaling networks assembled by oncogenic EGFR and c-Met. >Proc. Natl. Acad. Sci. U.S.A 105, 692–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., Nardone J., Lee K., Reeves C., Li Y., Hu Y., Tan Z., Stokes M., Sullivan L., Mitchell J., Wetzel R., Macneill J., Ren J. M., Yuan J., Bakalarski C. E., Villen J., Kornhauser J. M., Smith B., Li D., Zhou X., Gygi S. P., Gu T. L., Polakiewicz R. D., Rush J., Comb M. J. (2007) Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131, 1190–1203 [DOI] [PubMed] [Google Scholar]
- 11.Ballif B. A., Carey G. R., Sunyaev S. R., Gygi S. P. (2008) Large-scale identification and evolution indexing of tyrosine phosphorylation sites from murine brain. J. Proteome Res 7, 311–318 [DOI] [PubMed] [Google Scholar]
- 12.Wolf-Yadlin A., Hautaniemi S., Lauffenburger D. A., White F. M. (2007) Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc. Natl. Acad. Sci. U.S.A 104, 5860–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolf-Yadlin A., Kumar N., Zhang Y., Hautaniemi S., Zaman M., Kim H. D., Grantcharova V., Lauffenburger D. A., White F. M. (2006) Effects of HER2 overexpression on cell signaling networks governing proliferation and migration. Mol. Syst. Biol 2, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng H., Hu P., Quinn D. F., Wang Y. K. (2005) Phosphotyrosine proteomic study of interferon alpha signaling pathway using a combination of immunoprecipitation and immobilized metal affinity chromatography. Mol. Cell. Proteomics 4, 721–730 [DOI] [PubMed] [Google Scholar]
- 15.Tong J., Taylor P., Jovceva E., St-Germain J. R., Jin L. L., Nikolic A., Gu X., Li Z. H., Trudel S., Moran M. F. (2008) Tandem immunoprecipitation of phosphotyrosine-mass spectrometry (TIPY-MS) indicates C19ORF19 becomes tyrosine-phosphorylated and associated with activated epidermal growth factor receptor. J. Proteome Res 7, 1067–1077 [DOI] [PubMed] [Google Scholar]
- 16.Rush J., Moritz A., Lee K. A., Guo A., Goss V. L., Spek E. J., Zhang H., Zha X. M., Polakiewicz R. D., Comb M. J. (2005) Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol 23, 94–101 [DOI] [PubMed] [Google Scholar]
- 17.Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 18.Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 19.Hsu J. L., Huang S. Y., Chow N. H., Chen S. H. (2003) Stable-isotope dimethyl labeling for quantitative proteomics. Anal. Chem 75, 6843–6852 [DOI] [PubMed] [Google Scholar]
- 20.Boersema P. J., Aye T. T., van Veen T. A., Heck A. J., Mohammed S. (2008) Triplex protein quantification based on stable isotope labeling by peptide dimethylation applied to cell and tissue lysates. Proteomics 8, 4624–4632 [DOI] [PubMed] [Google Scholar]
- 21.Boersema P. J., Raijmakers R., Lemeer S., Mohammed S., Heck A. J. (2009) Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc 4, 484–494 [DOI] [PubMed] [Google Scholar]
- 22.Bantscheff M., Schirle M., Sweetman G., Rick J., Kuster B. (2007) Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem 389, 1017–1031 [DOI] [PubMed] [Google Scholar]
- 23.Lemeer S., Jopling C., Gouw J., Mohammed S., Heck A. J., Slijper M., den Hertog J. (2008) Comparative phosphoproteomics of zebrafish Fyn/Yes morpholino knockdown embryos. Mol. Cell. Proteomics 7, 2176–2187 [DOI] [PubMed] [Google Scholar]
- 24.Raijmakers R., Berkers C. R., de Jong A., Ovaa H., Heck A. J., Mohammed S. (2008) Automated online sequential isotope labeling for protein quantitation applied to proteasome tissue-specific diversity. Mol. Cell. Proteomics 7, 1755–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 26.van Breukelen B., van den Toorn H. W., Drugan M. M., Heck A. J. (2009) StatQuant: A post quantification analysis toolbox for improving quantitative mass spectrometry. Bioinformatics 25, 1472–1473 [DOI] [PubMed] [Google Scholar]
- 27.Käll L., Storey J. D., MacCoss M. J., Noble W. S. (2008) Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J. Proteome Res 7, 29–34 [DOI] [PubMed] [Google Scholar]
- 28.Diella F., Gould C. M., Chica C., Via A., Gibson T. J. (2008) Phospho.ELM: a database of phosphorylation sites update 2008. Nucleic Acids Res 36, D240–D244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwartz D., Gygi S. P. (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol 23, 1391–1398 [DOI] [PubMed] [Google Scholar]
- 30.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amanchy R., Periaswamy B., Mathivanan S., Reddy R., Tattikota S. G., Pandey A. (2007) A curated compendium of phosphorylation motifs. Nat. Biotechnol 25, 285–286 [DOI] [PubMed] [Google Scholar]
- 32.Linding R., Jensen L. J., Ostheimer G. J., van Vugt M. A., Jørgensen C., Miron I. M., Diella F., Colwill K., Taylor L., Elder K., Metalnikov P., Nguyen V., Pasculescu A., Jin J., Park J. G., Samson L. D., Woodgett J. R., Russell R.B., Bork P., Yaffe M. B., Pawson T. (2007) Systematic discovery of in vivo phosphorylation networks. Cell 129, 1415–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Linding R., Jensen L. J., Pasculescu A., Olhovsky M., Colwill K., Bork P., Yaffe M. B., Pawson T. (2008) NetworKIN: a resource for exploring cellular phosphorylation networks. Nucleic Acids Res 36, D695–D699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. (2000) Gene Ontology: tool for the unification of biology. Nat. Genet 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sonnhammer E. L., Eddy S. R., Durbin R. (1997) Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins 28, 405–420 [DOI] [PubMed] [Google Scholar]
- 36.Boekhorst J., van Breukelen B., Heck A. J., Snel B. (2008) Comparative phosphoproteomics reveals evolutionary and functional conservation of phosphorylation across eukaryotes. Genome Biol 9, R144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hornbeck P. V., Chabra I., Kornhauser J. M., Skrzypek E., Zhang B. (2004) Phosphosite: a bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics 4, 1551–1561 [DOI] [PubMed] [Google Scholar]
- 38.Mann M., Ong S. E., Grønborg M., Steen H., Jensen O. N., Pandey A. (2002) Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol 20, 261–268 [DOI] [PubMed] [Google Scholar]
- 39.Kratchmarova I., Blagoev B., Haack-Sorensen M., Kassem M., Mann M. (2005) Mechanism of divergent growth factor effects in mesenchymal stem cell differentiation. Science 308, 1472–1477 [DOI] [PubMed] [Google Scholar]
- 40.Huyer G., Liu S., Kelly J., Moffat J., Payette P., Kennedy B., Tsaprailis G., Gresser M. J., Ramachandran C. (1997) Mechanism of inhibition of protein-tyrosine phosphatases by vanadate and pervanadate. J. Biol. Chem 272, 843–851 [DOI] [PubMed] [Google Scholar]
- 41.Nühse T. S., Stensballe A., Jensen O. N., Peck S. C. (2004) Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell 16, 2394–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morandell S., Stasyk T., Skvortsov S., Ascher S., Huber L. A. (2008) Quantitative proteomics and phosphoproteomics reveal novel insights into complexity and dynamics of the EGFR signaling network. Proteomics 8, 4383–4401 [DOI] [PubMed] [Google Scholar]
- 43.Huang P. H., Mukasa A., Bonavia R., Flynn R. A., Brewer Z. E., Cavenee W. K., Furnari F. B., White F. M. (2007) Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc. Natl. Acad. Sci. U.S.A 104, 12867–12872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schlessinger J. (2002) Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell 110, 669–672 [DOI] [PubMed] [Google Scholar]
- 45.Schulze W. X.., Deng L., Mann M. (2005) Phosphotyrosine interactome of the ErbB-receptor kinase family. Mol. Syst. Biol 1, 2005 0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pesesse X., Dewaste V., De Smedt F., Laffargue M., Giuriato S., Moreau C., Payrastre B., Erneux C. (2001) The Src homology 2 domain containing inositol 5-phosphatase SHIP2 is recruited to the epidermal growth factor (EGF) receptor and dephosphorylates phosphatidylinositol 3,4,5-trisphosphate in EGF-stimulated COS-7 cells. J. Biol. Chem 276, 28348–28355 [DOI] [PubMed] [Google Scholar]
- 47.Prasad N. K., Decker S. J. (2005) SH2-containing 5′-inositol phosphatase, SHIP2, regulates cytoskeleton organization and ligand-dependent down-regulation of the epidermal growth factor receptor. J. Biol. Chem 280, 13129–13136 [DOI] [PubMed] [Google Scholar]
- 48.Yamauchi T., Ueki K., Tobe K., Tamemoto H., Sekine N., Wada M., Honjo M., Takahashi M., Takahashi T., Hirai H., Tushima T., Akanuma Y., Fujita T., Komuro I., Yazaki Y., Kadowaki T. (1997) Tyrosine phosphorylation of the EGF receptor by the kinase Jak2 is induced by growth hormone. Nature 390, 91–96 [DOI] [PubMed] [Google Scholar]
- 49.Gogg S., Smith U. (2002) Epidermal growth factor and transforming growth factor alpha mimic the effects of insulin in human fat cells and augment downstream signaling in insulin resistance. J. Biol. Chem 277, 36045–36051 [DOI] [PubMed] [Google Scholar]
- 50.Amanchy R., Zhong J., Molina H., Chaerkady R., Iwahori A., Kalume D. E., Grønborg M., Joore J., Cope L., Pandey A. (2008) Identification of c-Src tyrosine kinase substrates using mass spectrometry and peptide microarrays. J. Proteome Res 7, 3900–3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leroy C., Fialin C., Sirvent A., Simon V., Urbach S., Poncet J., Robert B., Jouin P., Roche S. (2009) Quantitative phosphoproteomics reveals a cluster of tyrosine kinases that mediates Src invasive activity in advanced colon carcinoma cells. Cancer Res 69, 2279–2286 [DOI] [PubMed] [Google Scholar]
- 52.Luo W., Slebos R. J., Hill S., Li M., Brábek J., Amanchy R., Chaerkady R., Pandey A., Ham A. J., Hanks S. K. (2008) Global impact of oncogenic Src on a phosphotyrosine proteome. J. Proteome Res 7, 3447–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Head J. A., Jiang D., Li M., Zorn L. J., Schaefer E. M., Parsons J. T., Weed S. A. (2003) Cortactin tyrosine phosphorylation requires Rac1 activity and association with the cortical actin cytoskeleton. Mol. Biol. Cell 14, 3216–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nieto-Pelegrin E., Martinez-Quiles N. (2009) Distinct phosphorylation requirements regulate cortactin activation by TirEPEC and its binding to N-WASP. Cell Commun. Signal 7, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martinez-Quiles N., Ho H. Y., Kirschner M. W., Ramesh N., Geha R. S. (2004) Erk/Src phosphorylation of cortactin acts as a switch on-switch off mechanism that controls its ability to activate N-WASP. Mol. Cell. Biol 24, 5269–5280 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.