Abstract
Background
Non-coding satellite DNA (satDNA) usually has a high turn-over rate frequently leading to species specific patterns. However, some satDNA families evolve more slowly and can be found in several related species. Here, we analyzed the mode of evolution of the pDo500 satDNA family of Dolichopoda cave crickets. In addition, we discuss the potential of slowly evolving satDNAs as phylogenetic markers.
Results
We sequenced 199 genomic or PCR amplified satDNA repeats of the pDo500 family from 12 Dolichopoda species. For the 38 populations under study, 39 pDo500 consensus sequences were deduced. Phylogenetic analyses using Bayesian, Maximum Parsimony, and Maximum Likelihood approaches yielded largely congruent tree topologies. The vast majority of pDo500 sequences grouped according to species designation. Scatter plots and statistical tests revealed a significant correlation between genetic distances for satDNA and mitochondrial DNA. Sliding window analyses showed species specific patterns of variable and conserved regions. The evolutionary rate of the pDo500 satDNA was estimated to be 1.63-1.78% per lineage per million years.
Conclusions
The pDo500 satDNA evolves gradually at a rate that is only slightly faster than previously published rates of insect mitochondrial COI sequences. The pDo500 phylogeny was basically congruent with the previously published mtDNA phylogenies. Accordingly, the slowly evolving pDo500 satDNA family is indeed informative as a phylogenetic marker.
Background
Only a small fraction of a typical eukaryotic nuclear genome constitutes rDNA or protein-coding genes, whereas a large fraction of all higher eukaryotic genomes consists of repetitive sequences and heterochromatic satellite DNA (satDNA) [1,2]. Recent sequencing of various genomes have confirmed that the remarkable variability in genome size among eukaryotes is to a large extent due to different amounts of repetitive DNA, of which non-coding tandemly repeated DNA is a major part [3-7].
Typically, satDNA consists of numerous very similar repeated sequences, tandemly arranged in a head to tail orientation in large clusters up to 100 million bp in length [1]. These clusters are usually located in the heterochromatic parts of the chromosomes, mainly in the regions close to the centromeres and telomeres. Repeat size can vary largely within and between species; from only a few base pairs up to several thousand base pairs [8-11]. The biological significance of satDNA is still under discussion. No general function has been conclusively assigned to this genome component, although a number of possible functions have been put forward (summarized in [12]) out of which many challenge earlier ideas of satDNA being "junk" [13] or selfish "parasites" in the genome [14]. Most of the proposed functions of satDNA are related to heterochromatin and/or centromere formation and function. However, the enormous diversity of satDNA in nucleotide sequence, repeat length, complexity, and genomic abundance may indicate that satDNA indeed may have several different functions.
The evolutionary turnover of satDNA is usually very fast; i.e. in closely related species non-orthologous satDNAs are often found at homologous chromosomal locations [15]. However, other satDNA families evolve more slowly and are represented in several closely related species [16-18]. Some satDNAs even seem to be rather ancient and can be widely distributed among higher taxa [19,20]. Consequently, some satDNAs may be valuable taxonomic identification tools while others might be informative in phylogeny.
Most commonly, mitochondrial genes such as e.g., the ribosomal 12S and 16S, or the cytochrome oxidase subunit I (COI) and II (COII) genes [21,22], and/or nuclear ribosomal DNA (rDNA) sequences, such as the 18S and 28S genes, and the internally transcribed spacer (ITS) regions are applied as molecular markers. Nuclear protein-coding genes such as e.g., the elongation factor-1α (EF-1α), carbamyl-P synthetase/aspartate transcarbamylase/dihydroorotase (CAD), and wingless genes [23] have also been applied. Some molecular features such as e.g., an A/T bias at the third codon position of mitochondrial protein coding genes [24,25], and higher values of among-site rate variation [26] may limit the phylogenetic utility of standard markers under certain circumstances. Some studies have explored the potential of satDNAs as phylogenetic markers [18,27-33], but there is still no general agreement about the utility of satDNA in this context.
Here, we explored the mode of evolution of the specific pDo500 satDNA family in the cave cricket genus Dolichopoda Bolivar 1880 (Dolichopodinae, Rhaphidophoridae). Approximately 30 Dolichopoda species are patchily distributed throughout the North Mediterranean regions. They have colonized caves and hypogean habitats between the Pyrenees and the Caucasus. As most Dolichopoda species depend on natural caves, a high degree of geographical isolation and strictly allopatric speciation processes can be assumed [34]. This might allow insights into the short-term processes of genetic differentiation of Dolichopoda populations and species, and therefore makes the genus a suitable model for studying processes of molecular evolution of satDNA. Three specific satDNA families have previously been characterized for three geographically isolated populations of D. schiavazzii [35,36]. Two of them, the pDoP102 and pDsPv400 satDNA families, are species-specific whereas the pDo500 satDNA family occurs in the genomes of all Dolichopoda species studied so far. The pDo500 satDNA monomers have been found to include repeat motifs that resemble the structure of hammerhead ribozymes. PDo500 sequences are also transcribed to some extent and may perform self-cleavage. It has therefore been speculated that this satDNA may be under selective pressure [37].
The mitochondrial DNA phylogeny of Dolichopoda has been addressed in two recent studies [38,39]. Data on other markers, such as allozyme variability [40,41], single copy DNA-DNA hybridization [41], and RFLPs of mitochondrial DNA [42] are also available for many Dolichopoda species. These studies offer a solid background for interpreting the mode of evolution of satDNA in this genus. Here, we studied the pDo500 satDNA from twelve species of Dolichopoda in detail. We were also interested to assess whether or not a phylogenetic signal can be discerned from this satDNA family.
Results
Sequence composition and alignment
The nucleotide sequences of the pDo500 satDNA family in 12 Dolichopoda species were determined. We obtained 199 satDNA sequences with 3-9 sequences per population (Table 1). The length of the pDo500 sequences ranged from 463 bp to 505 bp. The total alignment consisted of 497 positions/characters (primer sequences excluded). There were 385 (77%) variable positions, 269 (54%) of which were parsimony informative. The average nucleotide composition was T = 35.3%, C = 24.6%, A = 21.7% and G = 18.4%. The transition/transversion rate ratios were k1 = 1.995 (purines) and k2 = 1.578 (pyrimidines). The overall transition/transversion bias was R = 0.87. The alignment of deduced population-specific consensus sequences consisted of 39 sequences and 483 positions/characters. Only for the PIL population of D. geniculata two specific consensus sequences were deduced due to too many ambiguous sites. There were 117 (24%) variable positions of which 52 (11%) were parsimony informative for the consensus sequence alignment. The average nucleotide composition of the deduced consensus sequences was almost identical to that of the total data set, i.e. T = 35.2%, C = 24.7%, A = 21.7% and G = 18.4%.
Table 1.
Dolichopoda species and populations included in this study.
| Species | Pop. | Locality | pDO500 sequences | Mean p-distance within populations | Mean p-distances between populations |
|---|---|---|---|---|---|
| D. schiavazzii | VET | Necropoli di Vetulonia, Grosseto, Toscana, Italy | 5 | 0.024 | 0.019-0.059 |
| ORS | Grotta dei Pipistrelli, Montorsaio, Grosseto, Toscana, Italy | 3 | 0.028 | ||
| CPS | Monastero dei Fratelli Passionisti, Orbetello, Grosseto, Toscana, Italy | 6 | 0.037 | ||
| CIS | Acquedotto di Cisternino, Livorno, Toscana, Italy | 6 | 0.056 | ||
| BDO | Grotta di Buca dell'oro, Isola d'Elba, Grosseto, Toscana, Italy | 7 | 0.015 | ||
| POP | Populonia, Grosseto, Toscana, Italy | 6 | 0.019 | ||
| MRC | Marciana, Isola d'Elba, Grosseto, Toscana, Italy | 6 | 0.030 | ||
| FIC | Caverna di Fichino, Cascianna Terme, Pistoia, Toscana, Italy | 9 | 0.065 | ||
| BSC | Buca sopra cimitero, Orbetello, Grosseto, Toscana, Italy | 5 | 0.016 | ||
| D. aegilion | CAM | Miniera di Campese, Isola del Giglio, Grosseto, Toscana, Italy | 9 | 0.059 | |
| D. linderi | SIR | Sirach Cave, Eastern Pyrenees, France | 3 | 0.058 | 0.046-0.075 |
| MTB | Grotte de Montbolo, Montbolo, Eastern Pyrenees, France | 4 | 0.031 | ||
| BNP | Grotte de Bon Repaux, Bon Repaux, Ariege, Pyrenees, France | 5 | 0.061 | ||
| VMY | Grotte de Valmanya, Vinca, Eastern Pyrenees, France | 6 | 0.055 | ||
| CRQ | Mas de Crouanques, Pyrenees, France | 8 | 0.030 | ||
| D. bolivari | FRN | Forat negre cueva, Serradel, Llerida, Pyrenees, Spain | 5 | 0.030 | |
| D. cyrnensis | VAT | Grotta di Valletto, Corsica, France | 3 | 0.088 | |
| D. bormansi | BRA | Grotta di Brando, Bastia, Corsica, France | 3 | 0.075 | |
| D. baccetti | PST | Grotta di Punta degli Stretti, Orbetello, Grosseto, Toscana, Italy | 6 | 0.017 | |
| D. laetitia | FOR | Ruderi di Villa Chigi, Formello, Roma, Lazio, Italy | 4 | 0.015 | 0.015-0.019 |
| PSC | Grotta della Poscola, Monte di Malo, Priabona, Vicenza, Veneto, Italy | 4 | 0.017 | ||
| DIA | Grotta del diavolo, Semproniano, Grosseto, Toscana, Italy | 4 | 0.011 | ||
| D. palpata | TRE | Grotta di Tremusa, Scilla, Reggio di Calabria, Calabria, Italy | 4 | 0.072 | |
| D. capreensis | CPR | Grotta San Michele, Isola di Capri, Napoli, Campania, Italy | 5 | 0.056 | |
| D. geniculata | PIL | Grotta la Pila, Poggio Moiano, Rieti, Lazio, Italy | 6 | 0.039 | 0.030-0.058 |
| PAS | Grotta di Pastena, Pastena, Frosinone, Lazio, Italy | 5 | 0.027 | ||
| CLP | Grotta Regina Margherita, Collepardo, Frosinone, Lazio, Italy | 6 | 0.041 | ||
| TUS | Cunicolo dell'acquedotto, Frascati, Roma, Lazio, Italy | 4 | 0.038 | ||
| ISC | Fontana cunicoli, Isola di Ischia, Napoli, Campania, Italy | 8 | 0.030 | ||
| PRA | Grotta delle Praie, Lettomanoppello, Perugia, Umbria, Italy | 5 | 0.044 | ||
| VAL | Grotta Valmarino, Monte S. Biagio, Latina, Lazio, Italy | 4 | 0.031 | ||
| AUS | Grotta degli ausi, Prossedi, Latina, Lazio, Italy | 5 | 0.030 | ||
| PNZ | Le Forme, Isola di Ponza, Latina, Lazio, Italy | 6 | 0.043 | ||
| ZAN | Isola di Zannone, Latina, Lazio, Italy | 5 | 0.027 | ||
| D. ligustica | COR | Buco del Corno, Valle Cavallina, Zandobbio, Bergamo, Lombardia, Italy | 5 | 0.030 | 0.020-0.032 |
| PUG | Grotta del Pugnetto, Val di Lanzo, Torino, Piemonte, Italy | 6 | 0.015 | ||
| SFL | Grotta Selva, Zandobbio, Bergamo, Lombardia, Italy | 4 | 0.026 | ||
| BOS | Grotta di Bossea, Frabosa Soprana, Cuneo, Piemonte, Italy | 5 | 0.029 |
The number of pDo500 sat DNA sequences per population and the mean genetic distance within and between populations (uncorrected p-distances) are listed.
Sliding window analyses
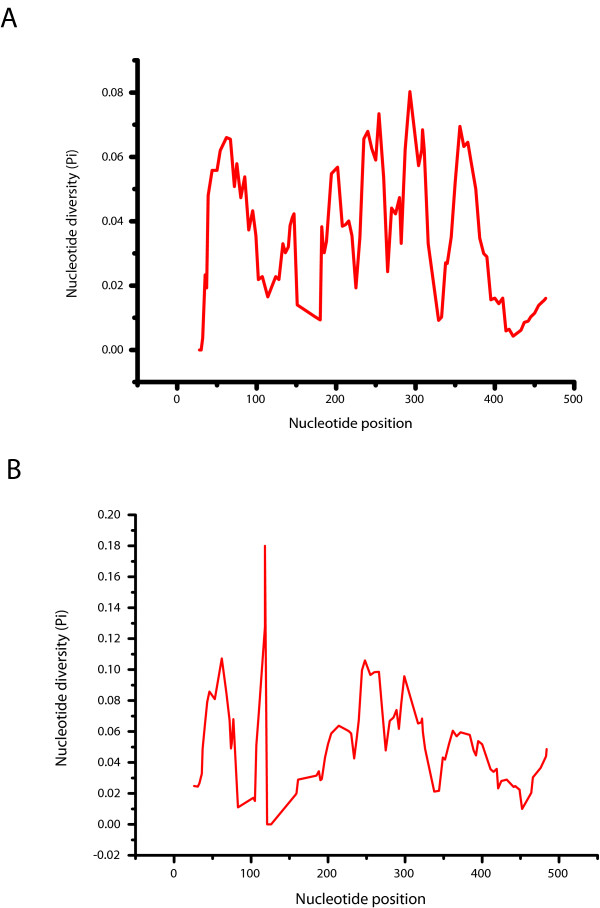
In order to identify regions within the pDo500 satDNA family with high levels of intra- and interspecific sequence conservation, sliding window analyses were performed. As shown in Figure 1A, nucleotide diversity (π) ranged between 0.009 and 0.093 for the consensus alignment. Under the applied settings, there were three regions with relatively low local minima of π (0.017, 0.009, and 0.014) at window midpoint positions 180, 329, and 424, respectively. Conversely, there were four peak areas with relatively high π values (0.075, 0.076, 0.080 and 0.093) with local maxima at window midpoint positions 202, 292, 357 and 239, respectively. When analyzing the complete alignment that also takes into account the intrapopulational variation, there were three regions with relatively low local minima of π (0.011, 0.00, and 0.010) at window midpoint positions 83, 121, and 452, respectively (Figure 1B). The peaks indicating regions with high local maxima were detected at positions 62 (π = 0.107), 118 (π = 0.180), 248 (π = 0.106), and 299 (π = 0.096). At first glance the extremely low π value around window midpoint position 121 in the total alignment may be surprising because this low local variation is not reflected in the consensus alignment to that extreme. However, the "discrepancy" can be assigned to different arrangements of indels in the two alignments and some ambiguous sites in the consensus sequences. It is also worthwhile mentioning that the area in the sliding window analysis that corresponds to the region that can form a hammerhead-like structure [37] does not show higher levels of conservation than the rest of the sequence.
Figure 1.
Sliding window analyses of the pDo500 satellite DNA of all Dolichopoda species included in this study. The results are shown for both the consensus alignment (A) and the complete alignment (B). The graphs show the value of nucleotide diversity (π) in a sliding window size of 30 with step size 5. Each value is depicted at its mid-point.
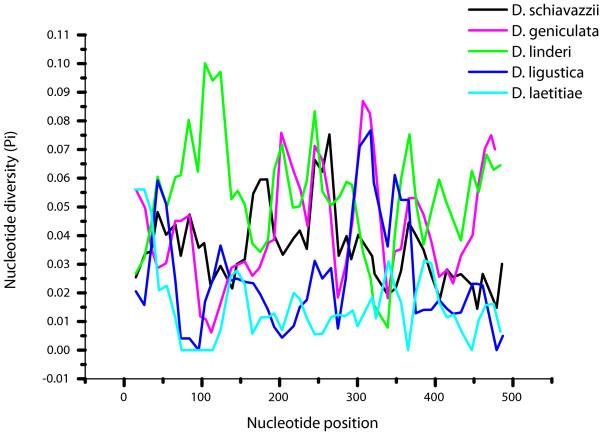
The sliding window analyses for selected species that are represented by a reasonable number of populations and individual pDo500 sequences are shown in Figure 2. Five areas with high interspecific variability were identified: i) For positions 66-124 D. laetitiae was significantly different from the other species (p < 0.05); ii) For positions 200-250 D. linderi and D. geniculata differed significantly from the other species (p < 0.05) but not from each other; iii) For positions 300-320 D. linderi and D. laetitiae were significantly different from the other species and from each other (p < 0.05); iv) For positions 350-380 D. linderi and D. laetitiae are significantly different from the other species and from each other (p < 0.05); and v) For positions 447-490 D. geniculata and D. linderi are significantly different from the other species (p < 0.05) but not from each other.
Figure 2.
Sliding window analyses of the pDo500 satellite DNA family of five Dolichopoda species separately. The graphs show the value of the nucleotide diversity (π) in a sliding window size 30 with step size 10. Each value is depicted at its mid-point.
Genetic distance analyses
The mean K2P and uncorrected p distances of the pDo500 satDNA sequences within and between populations as well as between species are given in Additional files 1 and 2, respectively.
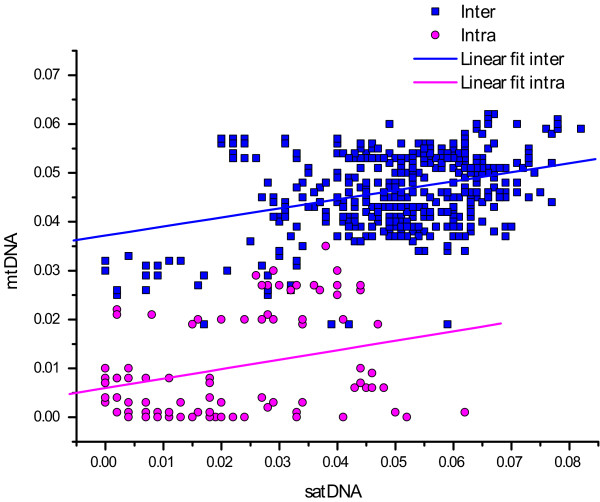
The scatter plot (Figure 3) relates the mean interpopulational uncorrected p-distances of pDo500 satDNA sequences to the average mtDNA distances calculated from partial 16S and COI sequences [38,39]. There was a highly significant correlation between genetic distances based on the two molecular markers (Mantel test: r = 0.616, p = 0.0001). The Spearman Ranks Correlation tests revealed that also the interpopulational genetic distances within and between species of the pDo500 satDNA and the mtDNA were correlated, Rs = 0.234 (p < 0.0001) and Rs = 0.231 (p = 0.03), respectively. The biological relevance of the correlations from the Spearmans Ranks test might be debated due to the low Rs values. There was also a significant correlation between the genetic distances based on satDNA and the geographic distances between the populations (Mantel test: r = 0.274, p = 0.0001). Despite the observed significant overall association, it is noteworthy that for the pDo500 satDNA, a number of intraspecific genetic distance values are higher than some of the interspecific values, e.g. between some D. linderi populations and some D. schiavazzii populations. On the other hand, some interspecific genetic distance values are quite low, e.g. D. laetitiae (DIA) vs D. geniculata. For comparison, for the mitochondrial DNA the vast majority of the intraspecific genetic distance values are lower than the interspecific values.
Figure 3.
Scatter plot of inter- and intraspecific genetic distances (uncorrected p-distances), blue squares and red circles, respectively. Trend lines from linear regression analyses are shown for visual purposes. The linear regression lines are Y = 0,005977 + 0.193514X and Y = 0,037182 + 0.184845X for the intra- and interpopulational genetic distances, respectively.
The saturation plot with uncorrected P-distances versus GTR-distances yielded a literally straight line (data not shown). Separate plots with the transition and transversion ratios showed two straight lines, transversions being more frequent than transitions (data not shown). This plot indicated that there was no substitution saturation in the pDo500 data set.
Phylogenetic analyses
The phylogenetic hypothesis depicted in Figure 4 resulted from the Bayesian analysis of the consensus alignment of the pDo500 satDNA sequences. The respective tree for the entire dataset is provided in Additional file 3. In order to allow for a comparison with the previously published mitochondrial DNA phylogenies [38,39], the satDNA tree was rooted with D. bolivari. The three different phylogenetic analyses (MP, MrBayes and ML) resulted in very similar tree topologies with only a few minor incongruencies. The MP analysis yielded 425 trees of equal length (252 steps) whereas ML analysis resulted in two trees; in both cases, the 50% majority-rule consensus trees were congruent with the phylogenetic hypothesis obtained with the Bayesian analysis.
Figure 4.
Bayesian phylogeny of the 39 derived consensus sequences the pDo500 satellite DNA family of Dolichopoda cave crickets. The Parsimony and Maximum Likelihood analyses yielded largely congruent trees. Posterior probabilities (PP) and bootstrap support > 50 from the MP and ML analyses are given for each node in the following order PP/MP/ML. Abbreviations for populations are as in Table 1.
As shown in Figure 4, the pDo500 sequences clustered in three main groups in the Bayesian tree: 1) D. bolivari and the majority of the sequences from D. linderi, 2) D. ligustica and D. schiavazzii, and 3) D. geniculata, D. laetitiae, D. aegilion, D. baccettii, D. cyrnensis, D. capreensis and D. palpata, in addition to two sequences from D. linderi (VMY and MTB). In contrast to high posterior probability values in the Bayesian analysis the bootstrap support values from the MP analysis are quite low for many nodes.
The phylogeny obtained with the complete pDo500 alignment (Additional file 3) is congruent with the phylogeny of the consensus alignment. However, there are also some sequences that do not cluster conspecifically, which reflected the intraspecific variation among pDo500 repeats. To some extent, sequences from the same population cluster together, but there is little intraspecific structure in the phylogeny of the complete alignment. This means that hardly any population-specific signatures could be attributed to the pDo500 sequences.
Evolutionary rate estimates
The pDo500 sequences evolve on average 1.48 times faster than the COI sequences from Dolichopoda. Multiplied by the previously reported substitution rate for insect COI genes of 1.1-1.2% per lineage per million years, per silent sites [43], this gives an evolutionary rate of 1.63-1.78% per lineage per million years for the pDo500 sequences.
The congruency of the satDNA dataset and the previously published mtDNA dataset were evaluated by a partition homogeneity test/ILD and Partitioned Bremer Support (PBS) values. Three different tests were performed for the ILD: i) pDo500 + COI, ii) pDo500 + 16S, and iii) pDo500 + COI + 16S. The null hypothesis of homogeneity of the phylogenetic signal among the data sets were rejected for all three tests (p = 0.001). This may indicate that the two data sets reflect different phylogenetic signals that should yield incongruent phylogenetic trees. PBS values were determined for one of the MPTs, the strict consensus and the 50% majority rule consensus tree. The strict consensus tree did not yield informative PBS values probably due to lack of structure in the tree, i.e. too many polytomies. The PBS values for the MPT and the 50% MJR consensus tree (Additional files 4 and 5) were to some extent indicating conflict between the two datasets and thus in line with the results of the ILD tests. However, the satellite DNA and the mtDNA sequences do not contribute equally to the tree. The mtDNA sequences are approximately three times as long as the pDo500 satDNA sequences and accordingly contribute more parsimony informative sites (296) than the pDo500 satDNA consensus sequences (52). Nodes with PBS values indicating strongest conflict were usually those with both low bootstrap support and posterior probabilities in the phylogenetic analyses. Not surprinsingly, for nodes with strong statistical support in the phylogenetic analyses, the PBS values indicated only little conflict. However, conflicting PBS values are certainly to some extent due to the high number of equally parsimonious trees obtained in the analysis of the combined data set as described by Lambkin et al. [44].
Discussion
In the current paper, we study the mode of evolution of the tandemly arranged satellite DNA family pDo500 in Dolichopoda cave crickets. Scatter plots and the related statistical analyses showed a significant correlation between the K2P distances calculated for mtDNA and the pDo500 satDNA. The pDo500 sequences evolve on average 1.48 times faster than the COI sequences from Dolichopoda. This is lower than previously observed for Hawaiian Tetragnatha spiders, for which a four times increased nucleotide substitution rate has been reported for satDNA compared to mtDNA [30]. The among-site rate variation of the pDo500 was higher than estimated for mtDNA from the same species, which may indicate that some regions of the pDo500 sequences are under selection. Accordingly, previous observations on preliminary data from populations of the D. laetitae-geniculata complex already indicated that this family of sequences is not informative at the intraspecific level [45]. As has been reported for most satDNA families the pDo500 family is slightly AT-rich (~57%), which is also common for mtDNA. Nevertheless, homoplasy is expected to be limited for the pDo500 satDNA family because no saturation was detected. Whether or not the pDo500 family is a typically slowly evolving satDNA with respect to among-site rate variation and saturation is difficult to assess. Among-site rate variation has hardly ever been reported for satDNA families, and saturation tests have only rarely been performed, e.g. in Pons et al. (2002) [29] who report slight saturation for the PIM357 satDNA family in Pimelia. Nevertheless, the high among-site rate variation did not obscure the phylogenetic signal in the pDo500 satDNA sequences. This conclusion may to some extent contradict the results of the sliding window analyses. For at least some species such as e.g. D. ligustica, D. schiavazzii, and D. linderi the sliding window profiles differed significantly.
pDo500 satDNA based phylogeny in Dolichopoda
The phylogeny obtained with the pDo500 satDNA (Figure 4 and Additional file 3) is largely congruent to those previously published on the basis of mtDNA markers [38,39]. The main differences between these phylogenies can be summarized as follows: i) Two well supported clusters in the mtDNA tree appear in different positions in the satDNA phylogeny. These clusters are D. geniculata + D. laetitiae and D. schiavazzii, which swap positions. ii) With satDNA as marker the two Corsican species D. cyrnensis and D. bormansi do not cluster together. iii) In two cases (D. linderi and D. geniculata) satDNA consensus sequences representing different populations do not cluster conspecifically.
In earlier studies on the phylogeny of Dolichopoda using the mitochondrial 16S and cytochrome oxidase I genes, the northern Italian D. ligustica grouped with the Central- and Southern-Italian D.laetitiae-geniculata complex. With respect to biogeography this grouping was difficult to explain [39]. However, in the satDNA phylogeny, D. ligustica grouped with the more northern species D. schiavazzii. Similarly, the two southernmost species D. palpata and D. capreensis grouped with the Northern Italian D. schiavazzii in the mtDNA phylogeny, which apparently did not conform to the biogeographical pattern. In contrast, the satDNA phylogeny indicates that D. palpata and D. capreensis are closely related to D. laetitae-geniculata complex that is also geographically closest. Thus, we believe that at least in these instances, the tree topology obtained with the pDo500 data is a better reflection of the Dolichopoda phylogeny than those obtained with mtDNA markers.
However, D. baccettii and its sister taxon D. aegilion are geographically more close to D. schiavazzii, but in the satDNA phylogeny, as opposed to the mtDNA phylogeny, D. baccettii and D. aegilion are found closely related to D. laetitiae-geniculata. This is in line with the results of a preliminary study [46]. Interestingly, the D. laetitiae-geniculata complex is also found in the mid-Italian region as is D. baccettii, therefore, the relationships found in the satDNA tree may be plausible. However, morphological features are more in favour of the phylogeny as resolved by mtDNA sequences, since D. schiavazzii, D. bormansi and D. cyrnensis share spinulation on the femurs and are grouped in the same subgenus Chopardina. Furthermore, D. bormansi, and D. cyrnensis have been recorded from the same caves and even hybridization of the two species has been hypothesized [47]. The incongruence between the satDNA and mtDNA may have been expected at least according to the results of the sliding window analyses. These results indicated that the intra- and interspecific variation of different regions of the pDo500 repeats differ significantly. Accordingly, among-site rate variation in the dataset was high. However, the incongruencies may also be due to stochastic processes in the usually small Dolichopoda populations such as random sorting of ancestral lineages during the short internodes, homoplasy in the mtDNA data, or both. The processes of molecular drive (see [48] for a review on molecular drive) affecting the pDo500 satDNA may also lead to an accumulation of variants that do not perfectly reflect the phylogenetic relationships.
As mentioned above, the level of the among-site rate variation of the pDo500 is higher (α = 0.59) than estimated for mtDNA (α = 0.88). This could be due to selection pressure on the region where the previously reported hammerhead structure is found [37]. However, according to the sliding window analyses performed here, this region is overall not more conserved than the rest of the pDo500 repeat. Nevertheless, since it is the secondary structure that is most crucial to the hammerhead and not the sequence itself, we can not rule out that the potential hammerhead structure has a profound influence on the evolution of the pDo500 satDNA family.
The congruence of the phylogenies obtained with the pDo500 satDNA and mtDNA markers is to some extent contradicted by the results of the ILD test and the PBS values. The partition homogeneity test indicates disagreement between the phylogenetic signal in the data sets based on mtDNA and satDNA. However, several authors have questioned the validity of this test as a criterion for congruence and combinability [49,50]. It has been demonstrated that the ILD test can wrongly reject the null hypothesis of congruence if the two data sets compared have different among-site rate variation [51]. Whether this applies to our data sets is uncertain as the satDNA and the mtDNA data sets differ only slightly in among-site variation. However, the ILD test is also sensitive for homoplasy, meaning random noise that is unequally distributed between the partitions [50]. There is homoplasy in both data sets due to little intraspecific structure; for example, within D. schiavazzii the populations can change position in the tree topology without altering the tree length. In our dataset a number of species were represented by several populations all with very similar if not identical pDo500 satDNA sequences. It is thus not surprising that parsimony analyses yielded many equally parsimonious trees. However, this may lead for several nodes to PBS values indicating conflicts between the combined satDNA and mtDNA datasets. However, it should be noted that nodes with PBS values indicating strongest conflict were usually those with both low bootstrap support and low posterior probabilities in the phylogenetic analyses.
Our study has added to the evidence that there is a clear potential for using slowly evolving satDNA families as phylogenetic markers. This is in line with a number of previous studies e.g. [17,18,30,52] that also used consensus sequences of satDNA and obtained phylogenies that were largely congruent with those based on other markers. In addition, several studies have successfully used satDNA as taxonomic and phylogenetic markers to solve issues that other markers such as e.g., mtDNA markers could not resolve [53,54].
There is an extensive literature on the characteristics of molecular phylogenetic markers e.g. [22,26,55-65], and at least at first glance satDNA seems to violate all requirements. In brief, a phylogenetic molecular marker must show an appropriate level of sequence conservation for the taxa of interest, while at the same time providing a sufficient number of variable and informative sites. Ideally, all sites should vary with equal probability because high among-site rate variation might hamper obtaining the true phylogeny. Furthermore, equal base composition will keep homoplasy low. Single-copy sequences are preferred in order to avoid paralogous comparisons. Finally, methodological aspects, such as ease to amplify by PCR or the availability of universal primers, are relevant as well. No marker will satisfy all of the above criteria and systematists therefore have developed a toolbox of most commonly used markers.
In comparison to protein-coding and ribosomal genes, the evolutionary turnover of non-coding satDNA is usually very high, leading to marked sequence divergence between species. Thus, there is often little or no phylogenetic signal when comparing satDNA from closely related species e.g. [66]. Accordingly, two species-specific satDNAs have earlier been described also for Dolichopoda schiavazzii [16,36]. For the pDo500 satDNA family, we have also shown high among-site rate variation, and for tandemly repeated satDNA paralogous comparisons are certainly an issue. In addition, there are no universal satDNAs and slowly evolving satDNA families to be used as phylogenetic markers need to be identified case by case. Nevertheless, several satDNAs such as the pDo500 satDNA family have been described as gradually evolving, and are conserved over considerable evolutionary time. Such satDNA may be phylogenetically informative [16,17,29,67,68]. For the pDo500 satDNA family, once sequences were available, the alignment of repeats was straightforward because there were only few indels.
Conclusion
Most satellite DNAs described so far have high evolutionary turn-over rates leading to rapid changes between species (interspecific heterogeneity) in contrast to homogenization and fixation within species (intraspecific homogeneity). However, as discussed here, some satDNA families such as the pDo500 satDNA family of Dolichopoda cave crickets may evolve rather slowly with an evolutionary rate only slightly faster than mtDNA. In Dolichopoda, both the fast evolving and the more slowly evolving satDNA have been found. In D. schiavazzii, two species-specific satDNA families have been identified, the pDoP102 (102 bp) and pDsPv400 (400 bp) in addition to the genus-specific family of pDo500 studied here [35,36]. These three satellite DNA families differ significantly in their molecular characteristics. A comparison showed a trend of sequence variability and copy number being positively correlated, and a trend of sequence variability and length of repeat being negative correlation [36], but data sets from further species are needed in order assess if these trends reflect general patterns. Unfortunately, very few other examples are found in the literature with both fast-evolving and slowly-evolving satDNAs found within the same species e.g. [69,70].
For some satDNA families a high degree of conservation was observed for species that diverged millions of years ago [52,71-73]. Such slowly evolving satDNA can even be a useful tool for phylogenetic analyses at higher taxonomic levels. Our study illustrates that satellite DNA can be successfully used as a molecular marker in phylogenetic analyses. In a phylogeographic context we believe that the pDo500 in some cases yields even better hypotheses than mtDNA. Compared with other commonly used markers - mtDNA and nuclear protein-coding and rDNA genes - particular gradually evolving satellite DNA families may fulfil the criteria of a good phylogenetic marker satisfactorily.
Methods
Material
We included samples from 38 populations representing 12 Dolichopoda cave cricket species from Italy and Spain (Table 1). The species' distributions are given in Additional file 6. Previously reported sequences of the pDo500 satDNA family from three populations of D. schiavazzii (VET, CPS, and BDO) [36] were also included in the analyses.
DNA extraction, PCR, cloning and sequencing
Genomic DNA was extracted from femurs and heads of Dolichopoda using the QIAamp DNA purification kit (Qiagen) following the manufacturer's instructions, or by standard procedures as described in Sambrook et al. [74].
Sequences of the pDo500 satDNA family used in this study were obtained by two different approaches: i) Genomic DNA was digested with the restriction enzyme PstI and subsequently electrophoresed on 5% polyacrylamide gels. The ladder-like strongly stained bands were cloned into standard plasmid vectors such as e.g., pUC19 [74]. Plasmid preparation was done according to the protocol of the QIAprep Spin Miniprep Kit (Qiagen). This approach can only discover pDo500 copies containing the PstI cleavage site. ii) Copies of the pDo500 satDNA family were amplified from genomic template DNA using the primers 5'-GTTTTACACGTTCACTGCAG-3' and 5'-GACACATTGATGAGACTGCAG-3' [36]. The obtained PCR products were cloned using the Zero Blunt® TOPO® Cloning Kit (Invitrogen). Positive clones were selected through PCR amplification using the M13 forward and M13 reverse primers. Sequencing was performed on an ABI 3100 automatic sequencer using BigDye chemistry (Applied Biosystems). This approach may be biased by preferential annealing of the primers to certain pDo500 variants.
All sequences have been deposited in GenBank, accession numbers: GU322143-GU322341
Sequence Analyses
The sequences were aligned using the software BioEdit [75]. Due to the high level of sequence similarity aligning all sequences was straightforward. The primer sequences of the PCR derived pDo500 repeats were excluded from the subsequent analyses.
Following the concept of concerted evolution [76], population-specific consensus sequences were deduced manually by evaluating each position in the alignment according to the six classes of transitional stages in tandem repetitive DNA described by Strachan et al. [77]. The different stages represent various steps in the homogenization and fixation process amongst repeats of tandemly repeated DNA families between pairs of species and populations. In essence this means that we used a 50% majority rule to deduce the consensus sequence. In some cases we either used the standard IUPAC symbols for ambiguous sites or deduced more than one consensus sequence for the respective populations.
Nucleotide composition, number of variable and parsimony informative sites, the transition/transversion bias, and genetic distance values were calculated using Mega version 4.0.1 [78]. All positions containing alignment gaps and missing data were excluded in pair-wise sequence comparisons.
Phylogenetic reconstruction and statistics
In order to assess whether nucleotide substitutions reach saturation, both transitions and transversions, and uncorrected genetic distances (p-distance), were plotted in Microsoft Excel against distances based on the general-times reversible model (GTR). The GTR distances were calculated in PAUP through the Bioportal at the University of Oslo, Norway http://www.bioportal.uio.no/.
Phylogenetic analyses were conducted using Bayesian, Maximum Likelihood (ML), and Maximum Parsimony (MP) approaches. The phylogenetic analyses were performed on both the complete alignment and the consensus sequences. The appropriate substitution models were determined using MrModeltest [79]. For both alignments, the general time reversible substitution model with gamma distribution for the among-site rate variation (GTR+Γ) obtained highest score according to the Akaike information criterion. The shape parameter of the gamma distribution (α) is inversely related to the rate of variation, and low values (α < 0.5) suggest extreme rate heterogeneity [80]. Accordingly, for the pDo500 sequences α = 0.59 suggests a relatively high rate heterogeneity.
The Bayesian phylogenetic analysis was conducted using MrBayes [81]. Each analysis was run with 6 million generations, 4 chains (one cold, three heated) and a sampling frequency of 100. A 50% majority rule consensus tree was made from each analysis with the first 12,000 trees ignored as burn-in. The ML using PAUP [82] was conducted through the Bioportal at the University of Oslo, Norway http://www.bioportal.uio.no/. MP was done using the program TNT (Tree analysis using New Technology) [83] made available online with the sponsorship of the Willi Hennig Society http://www.cladistics.org/tnt.html. The TNT searches were conducted with gaps being treated as fifth character state. Bootstrap support values [84] were estimated for both the ML analysis and the MP analyses, with 500 and 1000 repetitions respectively.
The evolutionary rate of the pDo500 satDNA family was estimated in relation to a previously reported substitution rate (1.1-1.2% per lineage, per million years, per silent sites) for insect mitochondrial COI sequences [43]. Interspecific K2P distances [85] for the pDo500 satDNA data set were related to interspecific genetic distances for the COI data set from the same species in order to estimate the relative evolutionary rate. The species specific average multiplied by the 1.1-1.2% rate estimate yielded the evolutionary rate of pDo500. These estimates include all pDo500 sequences obtained in this study.
Sliding window analyses were performed in DnaSP [86] in order to detect regions of high sequence conservation. In DnaSP, gaps are not treated as a fifth character state when analyzing the data, but in the sliding window analysis it is possible to consider sites with gaps in the length of the windows. We conducted the analyses with both options. The window sizes were set as 50, 30 and 20, respectively, with three different step sizes, 1, 5 and 10, for all three window sizes. The analysis was performed on both the consensus alignment and the complete alignment. Since the sliding window analysis in DnaSP can not handle more than 181 sequences, our complete alignment needed to be slightly reduced. Sequences to be excluded from the analysis were randomly chosen from those populations with the highest number of sequences. We also analyzed the sequences of D. schiavazzii, D. geniculata, D. linderi, and D. ligustica separately. Since DnaSP does not handle standard IUPAC symbols for ambiguous sites, such positions in the consensus alignment were replaced by N's.
From the sliding window analyses of D. schiavazzii, D. geniculata, D. linderi, D. laetitiae and D. ligustica, regions in the pDo500 satDNA sequences with high variability between the species were identified. Uncorrected p-distances within each species were calculated for these regions [78], and subjected to an Analysis of Variance (Kruskal-Wallis ANOVA) with subsequent Post-hoc tests - Tukey honest significant difference test for unequal sample size [87]. The tests were performed in Statistica [88].
The congruence of the satDNA and the mitochondrial DNA data sets was addressed with a partition homogeneity test/incongruence-length difference test (ILD) and by Partitioned Bremer Support (PBS). The ILD test was performed in PAUP [82] and TNT; the Partitioned Bremer Support was obtained using TreeRot [89] in combination with PAUP (following the instructions of the TreeRot manual). In PAUP, the number of ILD replicates was set to 1,000, with 10 random addition sequence replicates, holding one tree per replicate. A time limit for each replicate was set to 200 seconds. In TNT, the analysis was done with a script provided by Pablo Goloboff with some modifications [90]. The number of replicates was set to 10,000, with 10 random addition sequences holding 10 trees per replicate, and branch swapping with TBR (tree bisection-reconnection). For calculating the PBS parsimony analysis was conducted in PAUP on the satDNA consensus sequences combined with the mtDNA sequences. The analysis yielded 14,232 most parsimonious trees (MPTs). For the analysis in TreeRot, we used both of the consensus trees and one of the equally parsimonious MPTs (the first one to appear in the tree file from PAUP).
Scatter plots showing interpopulational and interspecific distances from pDo500 sequences versus mitochondrial DNA sequences were made in Microsoft Excel. Two different plots at each taxonomic level were made: one using all the pDo500 sequences, and one using only the consensus sequences. The results were similar and we only present the plot with the consensus sequences.
Two Mantel tests were performed with 10,000 random iterations using Mantel2 [91]: i) to test the correlation between genetic distances of satDNA and genetic distances of mitochondrial DNA; and ii) to test the correlation between genetic distances of satDNA and geographic distances between populations. P-distances between populations [78], were used as input data for the genetic distances. The program ArcView Gis 3.3 was used to obtain geographic distances.
Spearman Rank Correlation tests were performed in order to assess the correlation between genetic distances based on satDNA and mtDNA in within- and between species comparison, respectively. The tests were performed using the Free Statistics and Forecasting Software at http://www.wessa.net[92].
Authors' contributions
LB, FV and VS planned the project, conducted labwork, and participated in writing and editing the manuscript. LM conducted the labwork, analyzed the data and participated in writing and editing the manuscript. AJ participated in data analyses and writing and editing of the manuscript. All authors approved the final version of the manuscript.
Supplementary Material
Interspecific genetic distances of the pDo500 satDNA sequences. Kimura (1980) two-parameter distances (above diagonal) and uncorrected p-distance (below diagonal) of 199 genomic or PCR amplified pDo500 satDNA sequences from Dolichopoda.
Interspecific genetic distances based on pDo500 satDNA consensus sequences. Interspecific Kimura (1980) two-parameter distances (above diagonal) and uncorrected p-distance (below diagonal) of 32 derived population specific pDo500 satDNA consensus sequences from Dolichopoda
Unrooted Bayesian phylogeny of the 199 repeats of the pDo500 satDNA family of Dolichopoda cave crickets. Posterior probabilities (PP) > 50% are given. Abbreviations for populations are as in Table 1. The Maximum Parsimony and the Maximum Likelihood analyses were congruent with the Bayesian analysis.
Partition Bremer Support values for one of the most parsimonious trees from the pDo500 satDNA consensus and the mtDNA sequences. The PBS values for the two data partitions are given as satDNA/mtDNA.
Partition Bremer Support values for the 50% majority-rule consensus tree derived for the pDo500 satDNA consensus and the mtDNA sequences. The 50% majority-rule consensus tree is based on the MPTs from the combined parsimony analysis of the satDNA consensus and the mtDNA sequences. The PBS values for the two data partitions are given as satDNA/mtDNA.
Map of the geographical distributions of Dolichopoda species included in this study. The map is showing the distribution areas for the Dolichopoda species in the West Mediterranean region.
Contributor Information
Lene Martinsen, Email: lene.martinsen@nhm.uio.no.
Federica Venanzetti, Email: f.venanzetti@inail.it.
Arild Johnsen, Email: arild.johnsen@nhm.uio.no.
Valerio Sbordoni, Email: valerio.sbordoni@uniroma2.it.
Lutz Bachmann, Email: lutz.bachmann@nhm.uio.no.
Acknowledgements
The manuscript is dedicated to Prof. Dr. Diether Sperlich on the occasion of his 80th birthday.
The project was supported by the 'National Centre for Biosystematics' (Project no. 146515/420), co-funded by the Norwegian Research Council and the Natural History Museum, University of Oslo, Norway.
References
- Charlesworth B, Sniegowski P, Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature London. 1994;371(6494):215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- Lewin B. Genes VIII. Upper Saddle River, NJ 07458: Pearson Prentice Hall; 2004. [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA. The Sequence of the Human Genome. Science. 2001;291(5507):1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Sharma S, Raina SN. Organization and evolution of highly repeated satellite DNA sequences in plant chromosomes. Cytogenet Genome Res. 2005;109(1-3):15–26. doi: 10.1159/000082377. [DOI] [PubMed] [Google Scholar]
- Bosco G, Campbell P, Leiva-Neto JT, Markow TA. Analysis of Drosophila species genome size and satellite DNA content reveals significant differences among strains as well as between species. Genetics. 2007;177(3):1277–1290. doi: 10.1534/genetics.107.075069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TGS Consortium. The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452:949. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- Wang S, Lorenzen M, Beeman R, Brown S. Analysis of repetitive DNA distribution patterns in the Tribolium castaneum genome. Genome Biol. 2008;9(3):R61. doi: 10.1186/gb-2008-9-3-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgtorf C, Bünemann H. A telomere-like satellite (GGGTCAT)n comprises 4% of genomic DNA of Drosophila hydei and is located mainly in centromeric heterochromatin of all large acrocentric autosomes. Gene. 1993;137(2):287–291. doi: 10.1016/0378-1119(93)90022-U. [DOI] [PubMed] [Google Scholar]
- King LM, Cummings MP. Satellite DNA repeat sequence variation is low in three species of burying beetles in the genus Nicrophorus (Coleoptera: Silphidae) Mol Biol Evol. 1997;14(11):1088–1095. doi: 10.1093/oxfordjournals.molbev.a025718. [DOI] [PubMed] [Google Scholar]
- Lorite P, Carrillo JA, Tinaut A, Palomeque T. Evolutionary dynamics of satellite DNA in species of the Genus Formica (Hymenoptera, Formicidae) Gene. 2004;332:159–168. doi: 10.1016/j.gene.2004.02.049. [DOI] [PubMed] [Google Scholar]
- Palomeque T, Lorite P. Satellite DNA in insects: a review. Heredity. 2008;100(6):564–573. doi: 10.1038/hdy.2008.24. [DOI] [PubMed] [Google Scholar]
- Plohl M, Luchetti A, Mestrovic N, Mantovani B. Satellite DNAs between selfishness and functionality: Structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin. Gene. 2008;409(1-2):72–82. doi: 10.1016/j.gene.2007.11.013. [DOI] [PubMed] [Google Scholar]
- Ohno S. So much junk in our genome. Brookhaven Sym Biol. 1972;23:366–370. [PubMed] [Google Scholar]
- Orgel LE, Crick FHC. Selfish DNA - The ultimate parasite. Nature. 1980;284(5757):604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- Csink AK, Henikoff S. Something from nothing: the evolution and utility of satellite repeats. Trends Genet. 1998;14(5):200–204. doi: 10.1016/S0168-9525(98)01444-9. [DOI] [PubMed] [Google Scholar]
- Bachmann L, Sperlich D. Gradual evolution of a specific satellite DNA family in Drosophila ambigua, D. tristis, and D. obscura. Mol Biol Evol. 1993;10(3):647–659. doi: 10.1093/oxfordjournals.molbev.a040029. [DOI] [PubMed] [Google Scholar]
- Mantovani B, Tinti F, Bachmann L, Scali V. The Bag320 satellite DNA family in Bacillus stick insects (Phasmatodea): different rates of molecular evolution of highly repetitive DNA in bisexual and parthenogenic taxa. Mol Biol Evol. 1997;14(12):1197–1205. doi: 10.1093/oxfordjournals.molbev.a025729. [DOI] [PubMed] [Google Scholar]
- Watabe H, Bachmann L, Haring E, Sperlich D. Taxonomic and molecular studies on Drosophila sinobscura and D. hubeiensis, two sibling species of the D. obscura group. J Zool Syst Evol Research. 1997;35:81–94. [Google Scholar]
- Blake RD, Wang JZ, Beauregard L. Repetitive sequence families in Alces alces americana. J Mol Evol. 1997;44(5):509–520. doi: 10.1007/PL00006175. [DOI] [PubMed] [Google Scholar]
- Modi WS, Ivanov S, Gallagher DS. Concerted evolution and higher-order repeat structure of the 1.709 (satellite IV) family in bovids. J Mol Evol. 2004;58(4):460–465. doi: 10.1007/s00239-003-2567-6. [DOI] [PubMed] [Google Scholar]
- Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. Evolution, weighting, and phylogenetic utility of mitochondrial gene-sequences and a compilation of conserved polymerase chain-reaction primers. Ann Entomol Soc Am. 1994;87(6):651–701. [Google Scholar]
- Caterino MS, Cho S, Sperling FAH. The current state of insect molecular systematics: A thriving tower of babel. Annu Rev Entomol. 2000;45(1):1–54. doi: 10.1146/annurev.ento.45.1.1. [DOI] [PubMed] [Google Scholar]
- Pauls SU, Graf W, Haase P, Lumbsch HT, Waringer J. Grazers, shredders and filtering carnivores -The evolution of feeding ecology in Drusinae (Trichoptera: Limnephilidae): Insights from a molecular phylogeny. Mol Phylogenet Evol. 2008;46(2):776–791. doi: 10.1016/j.ympev.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooers AØ, Holmes EC. The evolution of base composition and phylogenetic inference. Trends Ecol Evol. 2000;15(9):365–369. doi: 10.1016/S0169-5347(00)01934-0. [DOI] [PubMed] [Google Scholar]
- Kim I, Cha SY, Yoon MH, Hwang JS, Lee SM, Sohn HD, Jin BR. The complete nucleotide sequence and gene organization of the mitochondrial genome of the oriental mole cricket, Gryllotalpa orientalis (Orthoptera: Gryllotalpidae) Gene. 2005;353(2):155–168. doi: 10.1016/j.gene.2005.04.019. [DOI] [PubMed] [Google Scholar]
- Yang Z. On the Best Evolutionary Rate for Phylogenetic Analysis. Syst Biol. 1998;47(1):125–133. doi: 10.1080/106351598261067. [DOI] [PubMed] [Google Scholar]
- Garrido-Ramos MA, de la Herran R, Jamilena M, Lozano R, Ruiz Rejon C, Ruiz Rejon M. Evolution of centromeric satellite DNA and its use in phylogenetic studies of the Sparidae family (Pisces, Perciformes) Mol Phylogenet Evol. 1999;12(2):200–204. doi: 10.1006/mpev.1998.0609. [DOI] [PubMed] [Google Scholar]
- Picariello O, Feliciello I, Bellinello R, Chinali G. S1 satellite DNA as a taxonomic marker in brown frogs: molecular evidence that Rana graeca graeca and Rana graeca italica are different species. Genome. 2002;45(1):63. doi: 10.1139/g01-125. [DOI] [PubMed] [Google Scholar]
- Pons J, Petitpierre E, Juan C. Evolutionary dynamics of satellite DNA family PIM357 in species of the genus Pimelia (Tenebrionidae, Coleoptera) Mol Biol Evol. 2002;19(8):1329–1340. doi: 10.1093/oxfordjournals.molbev.a004194. [DOI] [PubMed] [Google Scholar]
- Pons J, Gillespie RG. Evolution of satellite DNAs in a radiation of endemic Hawaiian spiders: Does concerted evolution of highly repetitive sequences reflect evolutionary history? J Mol Evol. 2004;59(5):632–641. doi: 10.1007/s00239-004-2655-2. [DOI] [PubMed] [Google Scholar]
- Grechko VV, Ciobanu DG, Darevsky IS, Kosushkin SA, Kramerov DA. Molecular evolution of satellite DNA repeats and speciation of lizards of the genus Darevskia (Sauria: Lacertidae) Genome. 2006;49(10):1297–1307. doi: 10.1139/G06-089. [DOI] [PubMed] [Google Scholar]
- Pathak D, Srivastava J, Premi S, Tiwari M, Garg LC, Kumar S, Ali S. Chromosomal localization, copy number assessment, and transcriptional status of BamHI repeat fractions in water buffalo Bubalus bubalis. Dna Cell Biol. 2006;25(4):206–214. doi: 10.1089/dna.2006.25.206. [DOI] [PubMed] [Google Scholar]
- Suárez-Santiago VN, Blanca G, Ruiz-Rejón M, Garrido-Ramos MA. Satellite-DNA evolutionary patterns under a complex evolutionary scenario: The case of Acrolophus subgroup (Centaurea L., Compositae) from the western Mediterranean. Gene. 2007;404(1-2):80. doi: 10.1016/j.gene.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Sbordoni MC, Allegrucci G, Caccone A, Carchini G, Cesaroni D. In: Evolutionary Biology of Orthopteroid Insects. Baccetti B, editor. Vol. 1. Chichester: Ellis Horwood Limited; 1987. Microevolutionary studies in Dolichopodinae cave crickets; pp. 514–540. [Bowman C (Series Editor): Ellis Horwood Series in Entomology and Acarology] [Google Scholar]
- Bachmann L, Venanzetti F, Sbordoni V. Characterization of a species-specific satellite DNA family of Dolichopoda schiavazzii (Orthoptera, Rhaphidophoridae) cave crickets. J Mol Evol. 1994;39(3):274–281. doi: 10.1007/BF00160151. [DOI] [PubMed] [Google Scholar]
- Bachmann L, Venanzetti F, Sbordoni V. Tandemly repeated satellite DNA of Dolichopoda schiavazzii: a test for models on the evolution of highly repetitive DNA. J Mol Evol. 1996;43(2):135–144. doi: 10.1007/BF02337358. [DOI] [PubMed] [Google Scholar]
- Rojas AA, Vazquez-Tello A, Ferbeyre G, Venanzetti F, Bachmann L, Paquin B, Sbordoni V, Cedergren R. Hammerhead-mediated processing of satellite pDo500 family transcripts from Dolichopoda cave crickets. Nucl Acids Res. 2000;28(20):4037–4043. doi: 10.1093/nar/28.20.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allegrucci G, Todisco V, Sbordoni V. Molecular phylogeography of Dolichopoda cave crickets (Orthoptera, Rhaphidophoridae): A scenario suggested by mitochondrial DNA. Mol Phylogenet Evol. 2005;37(1):153–164. doi: 10.1016/j.ympev.2005.04.022. [DOI] [PubMed] [Google Scholar]
- Martinsen L, Venanzetti F, Johnsen A, Bachmann L. Phylogeography and mitochondrial DNA divergence in Dolichopoda cave crickets (Orthoptera, Rhaphidophoridae) Hereditas. 2008;146(2):33–45. doi: 10.1111/j.1601-5223.2008.02068.x. [DOI] [PubMed] [Google Scholar]
- Allegrucci G, Cesaroni D, Sbordoni V. Adaptation and Speciation of Dolichopoda Cave Crickets (Orthoptera, Rhaphidophoridae) - Geographic-Variation of Morphometric Indexes and Allozyme Frequencies. Biol J Linn Soc. 1987;31(2):151–160. doi: 10.1111/j.1095-8312.1987.tb01986.x. [DOI] [Google Scholar]
- Allegrucci G, Caccone A, Cesaroni D, Sbordoni V. Evolutionary divergence in Dolichopoda cave crickets: a comparison of single copy DNA hybridization data with allozymes and morphometric distances. J Evol Biol. 1992;5(1):121–148. doi: 10.1046/j.1420-9101.1992.5010121.x. [DOI] [Google Scholar]
- Venanzetti F, Cesaroni D, Mariottini P, Sbordoni V. Molecular phylogenies in Dolichopoda cave crickets and mtDNA rate calibration. Mol Phylogenet Evol. 1993;2(4):275–280. doi: 10.1006/mpev.1993.1026. [DOI] [PubMed] [Google Scholar]
- Brower AVZ. Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial DNA evolution. Proc Natl Acad Sci USA. 1994;91(14):6491–6495. doi: 10.1073/pnas.91.14.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambkin CL, Lee MSY, Winterton SL, Yeates DK. Partitioned Bremer support and multiple trees. Cladistics-Int J Willi Hennig Soc. 2002;18(4):436–444. doi: 10.1111/j.1096-0031.2002.tb00159.x. [DOI] [PubMed] [Google Scholar]
- Livi S. Caratterizzazione, variazione e significato filogenetico della famiglia di DNA satellite pDo500 nelle Dolichopoda del gruppo laetitiae-geniculata (Orthoptera, Rhaphidophoridae) Tesi di Laurea, Corso di Laurea in Scienze Biologiche, Facoltà di Scienze MFN, Università degli Studi di Roma "Tor Vergata". 1996.
- Miggiano E. Significato filogenetico della famiglia di DNA satellite pDo500 in Dolichopoda (Orthoptera, Rhaphidophoridae) Tesi di Laurea, Corso di Laurea in Scienze Biologiche, Facoltà di Scienze MFN, Università degli Studi di Roma "Tor Vergata". 1997.
- Saltet P. Données actuelles sur les Dolichopodes de Corse (Orth. Raphidophoridae) Extrait du bulletin de la société d'histoire naturelle de Toulouse. 1962;T.97:413–416. [Google Scholar]
- Dover G. Molecular drive. Trends Genet. 2002;18(11):587–589. doi: 10.1016/S0168-9525(02)02789-0. [DOI] [PubMed] [Google Scholar]
- Barker FK, Lutzoni FM. The utility of the incongruence length difference test. Syst Biol. 2002;51(4):625–637. doi: 10.1080/10635150290102302. [DOI] [PubMed] [Google Scholar]
- Quicke DLJ, Jones OR, Epstein DR. Correcting the problem of false incongruence due to noise imbalance in the incongruence length difference (ILD) test. Syst Biol. 2007;56(3):496–503. doi: 10.1080/10635150701429974. [DOI] [PubMed] [Google Scholar]
- Sullivan J. Combining data with different distributions of among-site rate variation. Syst Biol. 1996;45(3):375–380. [Google Scholar]
- Robles F, de la Herran R, Ludwig A, Ruiz Rejon C, Ruiz Rejon M, Garrido-Ramos MA. Evolution of ancient satellite DNAs in sturgeon genomes. Gene. 2004;338(1):133–142. doi: 10.1016/j.gene.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Lopez-Flores I, de la Herran R, Garrido-Ramos MA, Boudry P, Ruiz-Rejon C, Ruiz-Rejon M. The molecular phylogeny of oysters based on a satellite DNA related to transposons. Gene. 2004;339:181–188. doi: 10.1016/j.gene.2004.06.049. [DOI] [PubMed] [Google Scholar]
- Mahendran B, Acharya C, Dash R, Ghosh SK, Kundu SC. Repetitive DNA in tropical tasar silkworm Antheraea mylitta. Gene. 2006;370:51–57. doi: 10.1016/j.gene.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Sullivan J, Holsinger KE, Simon C. Among-site rate variation and phylogenetic analysis of 12S rRNA in sigmodontine rodents. Mol Biol Evol. 1995;12(6):988–1001. doi: 10.1093/oxfordjournals.molbev.a040292. [DOI] [PubMed] [Google Scholar]
- Yang ZH. Among-site rate variation and its impact on phylogenetic analyses. Trends in Ecology & Evolution. 1996;11(9):367–372. doi: 10.1016/0169-5347(96)10041-0. [DOI] [PubMed] [Google Scholar]
- Nei M, Kumar S. Molecular evolution and phylogenetics. New York Oxford University Press; 2000. [Google Scholar]
- Krzywinski J, Wilkerson RC, Besansky NJ. Toward Understanding Anophelinae (Diptera, Culicidae) Phylogeny: Insights from Nuclear Single-Copy Genes and the Weight of Evidence. Syst Biol. 2001;50(4):540. doi: 10.1080/106351501750435095. [DOI] [PubMed] [Google Scholar]
- Cruickshank RH. Molecular markers for the phylogenetics of mites and ticks. Syst Appl Acarol. 2002;7:3–14. [Google Scholar]
- Avise JC. Molecular markers, Natural history, and Evolution. Second. Sunderland: Sinauer Associates, Inc. Publisher; 2004. [Google Scholar]
- Lin C-P, Danforth BN. How do insect nuclear and mitochondrial gene substitution patterns differ? Insights from Bayesian analyses of combined datasets. Mol Phylogenet Evol. 2004;30(3):686–702. doi: 10.1016/S1055-7903(03)00241-0. [DOI] [PubMed] [Google Scholar]
- Moulton JK, Wiegmann BM. Evolution and phylogenetic utility of CAD (rudimentary) among Mesozoic-aged Eremoneuran Diptera (Insecta) Mol Phylogenet Evol. 2004;31(1):363. doi: 10.1016/S1055-7903(03)00284-7. [DOI] [PubMed] [Google Scholar]
- Rokas A, Carroll SB. Bushes in the tree of life. PLoS Biology. 2006;4(11):1899–1904. doi: 10.1371/journal.pbio.0040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A. Genomics and the Tree of Life. Science. 2006;313(5795):1897–1899. doi: 10.1126/science.1134490. [DOI] [PubMed] [Google Scholar]
- Whitfield JB, Kjer KM. Ancient Rapid Radiations of Insects: Challenges for Phylogenetic Analysis. Annu Rev Entomol. 2008;53(1):449–472. doi: 10.1146/annurev.ento.53.103106.093304. [DOI] [PubMed] [Google Scholar]
- Ugarkovic T, Plohl M. New EMBO Member's Review: Variation in satellite DNA profiles--causes and effects. EMBO J. 2002;21(22):5955–5959. doi: 10.1093/emboj/cdf612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta J, Mandal RK. A Conserved Tandemly Repeated DNA Sequence In Cruciferae. J Genet. 1990;69(3):169–177. doi: 10.1007/BF02927977. [DOI] [Google Scholar]
- Garrido-Ramos MA, Jamilena M, Lozano R, Rejon CR, Rejon MR. The EcoRI centromeric satellite DNA of the Sparidae family (Pisces, Perciformes) contains a sequence motive common to other vertebrate centromeric satellite DNAs. Cytogenet Cell Genet. 1995;71(4):345–351. doi: 10.1159/000134137. [DOI] [PubMed] [Google Scholar]
- Yoshimura A, Nakata A, Mito T, Noji S. The characteristics of karyotype and telomeric satellite DNA sequences in the cricket, Gryllus bimaculatus (Orthoptera, Gryllidae) Cytogenet Genome Res. 2006;112(3-4):329–336. doi: 10.1159/000089889. [DOI] [PubMed] [Google Scholar]
- Kuhn GCS, Sene FM, Moreira-Filho O, Schwarzacher T, Heslop-Harrison JS. Sequence analysis, chromosomal distribution and long-range organization show that rapid turnover of new and old pBuM satellite DNA repeats leads to different patterns of variation in seven species of the Drosophila buzzatii cluster. Chromosome Res. 2008;16(2):307–324. doi: 10.1007/s10577-007-1195-1. [DOI] [PubMed] [Google Scholar]
- Cafasso D, Cozzolino S, De Luca P, Chinali G. An unusual satellite DNA from Zamia paucijuga (Cycadales) characterised by two different organisations of the repetitive unit in the plant genome. Gene. 2003;311:71–79. doi: 10.1016/S0378-1119(03)00555-9. [DOI] [PubMed] [Google Scholar]
- Mravinac B, Plohl M, Mestrovic N, Ugarkovic D. Sequence of PRAT Satellite DNA "Frozen" in some Coleopteran Species. J Mol Evol. 2002;54:774–783. doi: 10.1007/s0023901-0079-9. [DOI] [PubMed] [Google Scholar]
- Li YX, Kirby ML. Coordinated and conserved expression of alphoid repeat and alphoid repeat-tagged coding sequences. Dev Dyn. 2003;228(1):72–81. doi: 10.1002/dvdy.10355. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Dover G, Brown S, Coen E, Dallas J, Strachan T, Trick M. In: Genome evolution. Dover GA, Flavell RB, editor. Vol. 20. London; New York: Published for the Systematics Association Academic Press; 1982. The dynamics of genome evolution and species differentiation; pp. 343–372. [Google Scholar]
- Strachan T, Webb D, Dover GA. Transition stages of molecular drive in multiple-copy DNA families in Drosophila. Embo Journal. 1985;4(7):1701–1708. doi: 10.1002/j.1460-2075.1985.tb03839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Nylander JAA. Program distributed by the author. Evolutionary Biology Centre, Uppsala University, Sweden; 2004. MrModeltest v2. [Google Scholar]
- Tateno Y, Takezaki N, Nei M. Relative Efficiencies of the Maximum-Likelihood, Neighbor-Joining, and Maximum-Parsimony Methods when Substition Rate Varies with Site. Mol Biol Evol. 1994;11(2):261–277. doi: 10.1093/oxfordjournals.molbev.a040108. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Swofford DL Phylogenetic Analysis Using Parsimony (* and Other Methods). Version 4 2002Sinauer Associates, Sunderland, Massachusetts; 12504223 [Google Scholar]
- Goloboff PA, Farris JS, Nixon KC. TNT, a free program for phylogenetic analysis. Cladistics. 2008;24:774–786. doi: 10.1111/j.1096-0031.2008.00217.x. [DOI] [Google Scholar]
- Felsenstein J. Conficence-limits on phylogenies - an approach using the bootstrap. Evolution. 1985;39(4):783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19(18):2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Spjotvoll E, Stoline M. An extension of the T-method of multiple comparison to include the cases with unequal sample sizes. J Am Stat Assoc. 1973;68:976–978. doi: 10.2307/2284534. [DOI] [Google Scholar]
- StatSoft I. STATISTICA (data analysis software system) version 7.1. edition. 2005. http://www.statsoft.com
- Sorenson MD, Franzosa EA. TreeRot. version 3. Boston University, Boston, MA; 2007. [Google Scholar]
- Ramirez MJ. Further problems with the incongruence length difference test: "hypercongruence" effect and multiple comparisons. Cladistics. 2006;22(3):289–295. [Google Scholar]
- Liedloff AC. Mantel Nonparametric Test Calculator. Version 2.0. School of Natural Resource Sciences, Queensland University of Technology, Australia; 1999. [Google Scholar]
- Free Statistics Software, Office for Research Development and Education, version 1.1.23-r3. http://www.wessa.net/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Interspecific genetic distances of the pDo500 satDNA sequences. Kimura (1980) two-parameter distances (above diagonal) and uncorrected p-distance (below diagonal) of 199 genomic or PCR amplified pDo500 satDNA sequences from Dolichopoda.
Interspecific genetic distances based on pDo500 satDNA consensus sequences. Interspecific Kimura (1980) two-parameter distances (above diagonal) and uncorrected p-distance (below diagonal) of 32 derived population specific pDo500 satDNA consensus sequences from Dolichopoda
Unrooted Bayesian phylogeny of the 199 repeats of the pDo500 satDNA family of Dolichopoda cave crickets. Posterior probabilities (PP) > 50% are given. Abbreviations for populations are as in Table 1. The Maximum Parsimony and the Maximum Likelihood analyses were congruent with the Bayesian analysis.
Partition Bremer Support values for one of the most parsimonious trees from the pDo500 satDNA consensus and the mtDNA sequences. The PBS values for the two data partitions are given as satDNA/mtDNA.
Partition Bremer Support values for the 50% majority-rule consensus tree derived for the pDo500 satDNA consensus and the mtDNA sequences. The 50% majority-rule consensus tree is based on the MPTs from the combined parsimony analysis of the satDNA consensus and the mtDNA sequences. The PBS values for the two data partitions are given as satDNA/mtDNA.
Map of the geographical distributions of Dolichopoda species included in this study. The map is showing the distribution areas for the Dolichopoda species in the West Mediterranean region.