Abstract
It is generally assumed that, in Saccharomyces cerevisiae, immature 40S ribosomal subunits are not competent for translation initiation. Here, we show by different approaches that, in wild-type conditions, a portion of pre-40S particles (pre-SSU) associate with translating ribosomal complexes. When cytoplasmic 20S pre-rRNA processing is impaired, as in Rio1p- or Nob1p-depleted cells, a large part of pre-SSUs is associated with translating ribosomes complexes. Loading of pre-40S particles onto mRNAs presumably uses the canonical pathway as translation-initiation factors interact with 20S pre-rRNA. However, translation initiation is not required for 40S ribosomal subunit maturation. We also provide evidence suggesting that cytoplasmic 20S pre-rRNAs that associate with translating complexes are turned over by the no go decay (NGD) pathway, a process known to degrade mRNAs on which ribosomes are stalled. We propose that the cytoplasmic fate of 20S pre-rRNA is determined by the balance between pre-SSU processing kinetics and sensing of ribosome-like particles loaded onto mRNAs by the NGD machinery, which acts as an ultimate ribosome quality check point.
Keywords: no go decay, ribosome biogenesis, Rio1p, translation initiation, yeast
Introduction
Ribosomes, the universal protein synthesis machinery, are ribonucleoprotein particles formed by two different subunits, which, in eukaryotic cells, are synthesized through a complex pathway taking place mostly in the nucleus and involving numerous factors. This process has been extensively studied in the yeast Saccharomyces cerevisiae and its general pattern is conserved throughout eukaryotes (Fatica and Tollervey, 2002; Fromont-Racine et al, 2003; Zemp and Kutay, 2007; Henras et al, 2008). Four different RNAs are found in eukaryotic ribosomes. Three of them, 5.8S and 25S rRNAs, which belong to the large ribosomal subunit (60S LSU), and 18S rRNA belonging to the small subunit (40S SSU), are produced by cleavage, trimming and modification of a common precursor (35S pre-rRNA in S. cerevisiae) synthesized by RNA polymerase I (Figure 1). The fourth rRNA, 5S rRNA, is produced from a distinct transcript synthesized by RNA polymerase III. Almost all ribosomal subunit maturation/assembly steps take place in the nucleus. The three rRNAs of the LSU are completely matured before its nuclear export, as opposed to 20S pre-rRNA, the immediate precursor of 18S rRNA, which is processed to 18S rRNA after pre-SSU export to the cytoplasm in which ribosomes accomplish their business: translation of the mRNAs into polypeptides. The SSU is a major player in the translation-initiation process. Once the cleavage at site D occurs, newly synthesized 40S ribosomal subunits can initiate translation by recruiting the eIF2-GTP-methionyl initiator tRNA ternary complex onto the SSU. This process is facilitated by initiation factors eIF3, eIF1 and eIF1A. The interaction between eIF3 and eIF4F, which binds the mRNA cap allows the loading of the 40S ribosomal subunit onto the mRNA. After a likely mRNA scanning by the SSU in search of the start codon, eIF5 promotes hydrolysis of GTP-eIF2 leaving the Met-tRNAi correctly positioned into the peptidyl (P) site of the SSU. At last, the 60S ribosomal subunit joins the 40S ribosomal subunit yielding an 80S ribosome capable of performing translation elongation (Hershey and Merrick, 2000; Hinnebusch, 2000; Pestova et al, 2001; Acker and Lorsch, 2008).
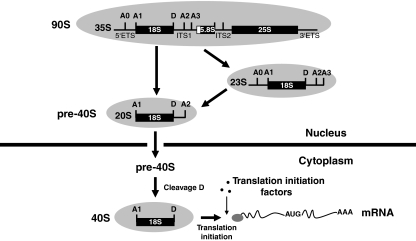
Figure 1.
Simplified scheme of 40S ribosomal subunits production in S. cerevisiae. rDNA transcription by RNA polymerase I in the nucleolus leads to 35S pre-rRNA. This transcript is processed sequentially at the A0, A1 and A2 sites yielding the 20S pre-rRNA. In a minor alternative pathway, the 35S pre-rRNA is first cleaved at A3, yielding the 23S pre-rRNA, which is then processed at A0, A1 and A2 to yield the 20S precursor (Gallagher et al, 2004). 20S pre-rRNA is exported to the cytoplasm, in which it undergoes endonucleolytic cleavage at site D, generating mature 18S rRNA contained in the 40S small subunit. Then, the small ribosomal subunit can initiate translation by binding and scanning mRNAs with the help of translation-initiation factors. The names of the pre-ribosomal particles in which these pre-rRNA intermediates are embedded are also indicated.
In their seminal work, which established that 20S pre-rRNA is processed to 18S rRNA in the cytoplasm, Udem and Warner (1972) reported also that 20S pre-rRNA is absent from polysomes. This suggests that either pre-SSU is not functional in translation initiation, or somehow excluded from it, or that processing takes place very early after pre-SSU export from the nucleus, such that all subunits are matured before having the opportunity to engage in an initiation event. However, in more recent studies, 20S pre-rRNA co-sedimentation with polysomes in sedimentation gradients analyses was observed when 20S pre-rRNA amount was increased because of processing impairment (Ford et al, 1999; Jakovljevic et al, 2004; Ferreira-Cerca et al, 2005; Granneman et al, 2005; Lacombe et al, 2009). This suggests that, at least in these particular conditions, pre-SSUs can be recruited by the translation-initiation machinery. To date, a handful of factors have been implicated in cytoplasmic processing of 20S pre-rRNA. They can be classified in two groups: (1) non-ribosomal proteins and (2) proteins stably associated with the small ribosomal subunit of translating ribosomes (i.e. rpS proteins). Factors of the first class have been identified either genetically, such as Rio1p (Vanrobays et al, 2001), by their homology with proteins involved in ribosome biogenesis, such as Rio2p (Vanrobays et al, 2003) or Tsr1p (Gelperin et al, 2001), through complex purification and analysis, such as Nob1p (Gavin et al, 2002; Fatica et al, 2003; Schafer et al, 2003), or genome-wide microarray analysis of RNA metabolism, such as Fap7p (Peng et al, 2003; Granneman et al, 2005). Besides studies focused on specific proteins of the SSU such as rpS0 (Ford et al, 1999), rpS21 (Tabb-Massey et al, 2003), rpS14 (Jakovljevic et al, 2004) and rpS31/Ubi3p (Finley et al, 1989; Lacombe et al, 2009) in which defects of cytoplasmic 20S pre-rRNA processing were observed, a systematic analysis of the function of the SSU ribosomal proteins in 40S ribosomal subunit biogenesis has shown that lack of rpS20 blocks cytoplasmic processing of 20S pre-rRNA (Ferreira-Cerca et al, 2005). Such pre-SSU defective maturation conditions commonly lead to an increase in 20S pre-rRNA amounts and a reduced production of mature 18S rRNA, but the extent of precursor accumulation is not equal to the 18S rRNA deficit indicating that cytoplasmic 20S pre-rRNA is unstable.
In this report, we analysed the fate of cytoplasmic 20S pre-rRNA. Our data strongly suggest that pre-40S subunits containing 20S pre-rRNA can complete translation initiation. We also took advantage of a hypomorphic viable allele of the essential gene RIO1 to analyse the factors determining 20S pre-rRNA stability. Finally, we suggest that the no go decay (NGD) RNA degradation pathway acts as an ultimate ribosome quality control mechanism.
Results
In our earlier work, we showed that RIO1 invalidation leads to a drastic impairment of 18S rRNA production because of a cytoplasmic 20S pre-rRNA processing defect (Vanrobays et al, 2001). We report below our analysis of the fate of the cytoplasmically accumulated 20S pre-rRNA, and bring new insights into the dynamics of its processing and of ribosome quality control.
Cells depleted of Rio1p or Nob1p accumulate cytoplasmic 20S pre-rRNA in 80S fractions
We first used the TetO7–RIO1 strain in which expression of RIO1 can be turned off using the tetracycline analogue doxycycline. The molecular phenotype associated with RIO1 depletion is the same as the one already described for a Gal–RIO1 strain (Vanrobays et al, 2001, 2003): (1) 20S pre-rRNA strongly accumulates and 18S rRNA level drops (Figure 2A), (2) in situ hybridization of an 18S rRNA-precursors specific probe (D-A2 fragment in Figure 1) leads to a strong cytoplasmic signal suggesting that pre-40S particles are exported and that cytoplasmic processing of 20S pre-rRNA is impaired (Figure 2B).
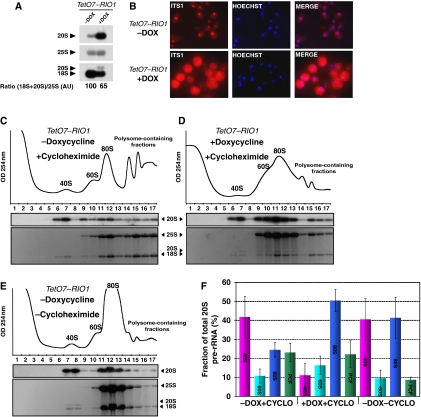
Figure 2.
Analysis of the sedimentation on a sucrose gradient of 20S pre-rRNA from a TetO7-RIO1 strain. Culture samples of a TetO7–RIO1 strain grown in YPD (−DOX) or in doxycycline containing YPD for 16 h (+DOX) were collected. From these samples, (A) total RNAs were extracted and separated in denaturing 1.2% agarose gel electrophoresis and transferred to nylon membranes. Specific RNAs were detected by hybridization with antisense oligonucleotide probes. Quantifications of the (18S+20S)/25S ratio in Rio1p-expressing cells and depleted cells were performed by phosphoimager analysis and expressed in arbitrary units (AU)—DOX condition taken as a reference. (B) Pre-18S rRNA FISH was performed with a probe complementary to the D-A2 segment of the ITS1. (C–E) Whole cell lysates were subjected to centrifugation through a sucrose gradient under polysome stabilization (+Cycloheximide) or polysomes run-off conditions (−Cycloheximide). A254 nm profiles were recorded. The peaks of free 40S and 60S subunits, 80S ribosomes and polysomes are indicated. RNA content of each fraction from the gradient was analysed by ethidium bromide staining and northern blot using a probe complementary to the D-A2 segment of the ITS1. (F) Percentage of total 20S pre-rRNA in 40S (fractions 6, 7, 8), 60S (fractions 9, 10), 80S (fractions 11, 12, 13), polysome-containing fractions (PCF, fractions 14, 15, 16, 17) in TetO7–RIO1 cells treated or not with doxycyline (DOX) or cycloheximide (CYCLO). Quantifications were performed by phosphoimager analysis. Error bars correspond to the standard deviation from three independent experiments.
To determine the distribution of 40S and 60S ribosomal subunits into free subunits, 80S ribosomes and polysomes in TetO7–RIO1 cells depleted or not depleted of Rio1p, extracts from cells treated or not treated with doxycycline were fractionated on sucrose sedimentation gradients, and gradient fractions analysed for their RNA content by northern blotting (Figure 2C). Cycloheximide, an inhibitor of translation elongation, was added shortly before cell harvest and during cell extract preparation to stabilize the polysomes complexes. Mature 18S and 25S rRNAs from the control cells were mainly distributed in the 80S and polysome-containing fractions as expected for exponentially growing cells. About half of 20S pre-rRNA was found in 40S fractions, but the other half sedimented in 80S and polysome-containing fractions (Figure 2C and F). In Rio1p-depleted cells, as expected for a condition strongly affecting the SSU maturation process, 40S and polysomes contents strongly drop, and reciprocally, the amount of free 60S subunits strongly increases, correlating with a shift of the 25S rRNA to 60S fractions (Figure 2D). In this Rio1p-depleted condition, in which few polysomes are still present, most of the 20S pre-rRNA is found in the same fractions as 80S ribosomes (Figure 2D and F). Note that the 18S/20S ratio is higher in polysome fractions than in 80S fractions (Supplementary Figure S1). Since depletion of Nob1p, the putative endonuclease involved in 20S pre-rRNA processing to 18S rRNA, also leads to a strong accumulation of cytoplasmic 20S pre-rRNA (Fatica et al, 2003, 2004), we tested whether a similar shift of 20S pre-rRNA sedimentation could be observed on Nob1p depletion. We used a Gal–HA–NOB1 strain in which NOB1 expression can be switched off by growing cells in glucose-containing medium. As observed in Rio1p-depleted cells, the 20S pre-rRNA, which accumulates in Nob1p-depleted cells mainly co-sediments with 80S complexes (Figure 3A).
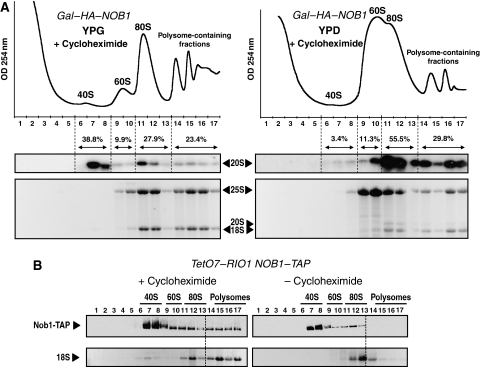
Figure 3.
Sucrose gradient sedimentation analysis of 20S pre-rRNA from a GAL–HA–NOB1 strain and of Nob1p from a TetO7–RIO1 NOB1–TAP strain. (A) Cellular extracts from cultures of the GAL–HA–NOB1 strain grown for 16 h in either galactose (YPG)- or glucose (YPD)-containing medium were subjected to sucrose gradient sedimentation analysis. RNA content of each fraction was analyzed as in Figure 2C–E. Quantifications were performed by phosphoimager analysis. (B) Whole cell lysates derived from TetO7–RIO1 NOB1–TAP cells grown in YPD were analyzed by sucrose gradient centrifugation under polysome-conserving conditions (+Cycloheximide) or polysome run-off conditions (−Cycloheximide). Fractions from the gradient were analyzed by western blotting using anti-ProtA. Sedimentation profile of the 18S rRNA is used as an internal control of polysome run off.
Altogether, these data suggest that, in Rio1p- and Nob1p-depleted cells, pre-40S particles that accumulate are incorporated into 80S complexes that may be ribosome-like particles. Moreover, even in exponentially growing cells, a fraction of 20S pre-rRNA sediments in 80S and polysome-containing fractions suggesting that this process also exists in normal cells, but is exacerbated when the last steps of 40S subunit maturation are impaired.
Pre-40S particles can be contained in polysomes
In an exponentially growing TetO7–RIO1 strain expressing Rio1p, about a quarter of 20S pre-rRNA sediments in polysome-containing fractions which suggests that pre-40S particles could be engaged in polysomes. This hypothesis was tested by omitting addition of cycloheximide before cell harvest and during cell extract preparation, thus allowing translating ribosomes to complete the ongoing elongation process. Omission of cycloheximide leads to a decrease in the amount of polysomes and to an increase in 80S run-off ribosomes (Figure 2E). Under these conditions, the fraction of 20S pre-rRNA contained in polysome fractions significantly dropped suggesting that the 20S pre-rRNA sedimenting in polysome fractions in Figure 2C is indeed associated with polysomes (Figure 2E and F). 20S pre-rRNA is generated by an early cleavage in the nucleolus (Figure 1). To confirm that the 20S pre-rRNA found in polysome-containing fractions is not nucleolar 20S pre-rRNA, we examined the distribution of nucleolar markers across the sedimentation gradients; these include 35S and 27S pre-rRNAs as well as Nop1p and Nhp2p, two core proteins of C/D and H/ACA snoRNPs, respectively (Supplementary Figure S2A). Omission of cycloheximide does not modify the sedimentation pattern of these markers suggesting that 20S pre-rRNA molecules that relocalize to 80S fractions in ribosome run-off conditions correspond to cytoplasmic pre-40S particles. This was further confirmed by fractionating a TetO7–RIO1 cellular extract into nuclear and cytoplasmic fractions and analysing their pre-rRNAs content (Supplementary Figure S2B). Assuming that 27S pre-rRNA distribution reflects the spill over of nucleoplasmic material into the cytoplasmic fraction (roughly 50% of 27S pre-rRNA is recovered in the ‘cytoplasmic' fraction), a small part of 20S pre-rRNA found in the cytoplasmic fraction could in fact originate from the nucleus. Since about 11% of total 20S pre-rRNA is found in the nuclear fraction, this means that actual nuclear 20S pre-rRNA amounts to 22% of total 20S pre-rRNA. Thus, most 20S pre-rRNA (78% of cellular 20S pre-rRNA) is indeed located in the cytoplasm. As half of total 20S pre-rRNA sediments in 80S and polysome fractions at the very least, assuming that all nuclear 20S pre-rRNA sediments in these fractions, still more than half of 20S molecules sedimenting in 80S polysomes fractions should be cytoplasmic 20S pre-rRNA. Thus, pre-40S particles can be contained in polysomes. Two models could explain the occurrence of immature 40S ribosomal subunits in polysomes. First, pre-40S particles could be engaged in translation initiation and so be associated with mRNAs upstream of translating ribosomes; second, pre-40S particles could have completed translation initiation and so associate with 60S ribosomal subunits and mRNAs upstream of translating ribosomes. The fact that, in the absence of cycloheximide, the 20S pre-rRNA contained in polysomes mainly relocalizes to 80S fractions and not to 40S fractions strongly suggests that polysomal pre-40S particles have completed translation initiation up to 60S ribosomal subunit recruitment (Figure 2E and F).
These results suggest that pre-40S particles have the ability to complete translation initiation before their final maturation. If so, non-ribosomal factors required for cytoplasmic pre-40S particle maturation should be found co-sedimenting with polysomes. We assessed the sedimentation profile of Nob1p, the putative endonuclease required for 20S pre-rRNA processing (Fatica et al, 2003, 2004). We used a strain expressing a TAP-tagged version of Nob1p in a TetO7–RIO1 genetic background. Under conditions in which polysomes are freezed by addition of cycloheximide, Nob1p co-sediments mainly with 40S complexes, but a fraction also co-sediments with polysome-containing fractions as already observed (Fatica et al, 2003) (Figure 3B). This fraction is almost completely lost in polysome run-off conditions. Thus, the putative endonuclease Nob1p, which is specific to late pre-40S particles can interact with polysomes.
Pre-40S particles associate with 60S subunits in mRNA containing ribosome-like particles
Occurrence of 20S pre-rRNA in polysomes and persistence of 20S pre-rRNA sedimenting in ribosome fractions under run-off conditions suggest that pre-40S particles themselves interact with 60S ribosomal subunits. Moreover, in cells accumulating cytoplasmic 20S pre-rRNA, pre-40S particles, which sediment in 80S fractions, may form ribosome-like particles containing a pre-40S particle and a 60S ribosomal subunit. Thus, we tested whether immunoprecipitation (IP) of 60S subunit ribosomal proteins (rpL) would result in co-IP of some 20S pre-rRNA. TAP-tagged versions of rpL25 and rpL24a were used as baits in a TetO7–RIO1 genetic background. As expected, rpL25–TAP and rpL24a–TAP sediment in 60S, 80S and polysome-containing fractions suggesting that these proteins are fully functional (Supplementary Figure S3). The rpL25, an early ribosomal protein, associates with pre-60S particles in the nucleus, whereas rpL24a is proposed to join pre-60S particles in the cytoplasm (Kruiswijk et al, 1978; van Beekvelt et al, 2000; Saveanu et al, 2003). The rpL25–TAP and rpL24a–TAP were precipitated from total cellular extracts using IgG sepharose, and co-immunoprecipitated RNAs were extracted and analysed by northern blotting (Figure 4A). As expected for ribosomal proteins, we found that mature 25S and 18S rRNAs were co-immunoprecipitated with rpL25–TAP and rpL24a–TAP. Moreover the relatively abundant actin encoding ACT1 mRNA was immunoprecipitated as well, with an efficiency above the background levels observed in the untagged strain and similar to the IP efficiency of mature rRNAs suggesting that ribosomes engaged in translation are immunoprecipitated (Supplementary Figure S4). The near absence of tRNA in the immunoprecipitate (data not shown), and IP efficiencies of U2, an snRNA involved in splicing, and of 23S rRNA, a 5′ and 3′ extended nuclear precursor of 18S (Figure 1), which are clearly much lower with both baits, indicate that IPs of mature rRNAs and ACT1 mRNA were specific. We found that 20S pre-rRNA co-immunoprecipitates with both rpL–TAP proteins with efficiencies similar to those of mature rRNAs and ACT1 mRNA. Moreover, the amount of immunoprecipitated 20S pre-rRNA increases when pre-40S particles accumulate in the cytoplasm in Rio1p-depleted cells. Altogether, these results suggest that in exponentially growing cells, part of pre-40S particles interact with 60S subunits in ribosome-like particles and that, under conditions leading to cytoplasmic 20S pre-rRNA accumulation, most pre-40S particles are incorporated into ribosome-like particles.
Figure 4.
Pre-40S particles can associate with 60S ribosomal subunits and mRNAs. (A) IP experiments were carried out using IgG-sepharose and total cellular extracts from the parental strain lacking a tagged protein (TetO7–RIO1, lanes 1, 2, 3 and 4) or from strains expressing either rpl25–TAP (TetO7–RIO1 RPL25–TAP, lanes 5, 6, 7, 8), rpl24a–TAP (TetO7–RIO1 RPL24a–TAP, lanes 9, 10, 11, 12) or Pab1–TAP (TetO7–RIO1 PAB1–TAP, lanes 13, 14, 15, 16) treated or not with doxycycline (DOX) for 16 h. RNAs were extracted from the pellets obtained after precipitation (IP) or from an amount of cellular extract corresponding to 1/50 of that used for IP (T), and analyzed by northern blotting as described in Figure 2A. (B) IP of Nob1–TAP performed with a TetO7–RIO1 NOB1–TAP cell extract using only sepharose beads (beads) or IgG coupled to sepharose beads (IgG-beads). Co-immunoprecipitated RNAs were reverse transcribed and then rpS14, Pgk1 and Act1 mRNAs and U2 snRNA were PCR amplified (RT+ beads and RT+IgG-beads). Genomic DNA contamination was evaluated by direct PCR amplification on the material extracted from the pellet, the reverse transcription step being omitted (RT − beads and RT − IgG-beads). Co-IP of 20S pre-rRNA was assayed by northern blotting. (C) Cell extracts of a TetO7–RIO1 strain treated or not with doxycycline for 16 h were prepared using high salt containing buffer and subjected to high-salt sucrose gradient sedimentation. RNA content of each fraction was analyzed as in Figure 2C–E. Quantifications were performed by phosphoimager analysis. (D) Percentage of total 20S pre-rRNA in 80S and polysome-containing fractions (PCF) in TetO7–RIO1 cells treated or not with doxycyline (DOX) in a high-salt sedimentation assay. Percentages of 20S pre-rRNA in 80S and PCF fractions from low-salt gradients were taken from Figure 2F.
As in the TetO7–RIO1 strain expressing Rio1p, about half of 20S pre-rRNA molecules are contained in ribosomes or polysomes, pre-40S particles should interact with mRNAs. Moreover, in Rio1p-depleted conditions, pre-40S particles mainly accumulate in cytoplasmic 80S ribosome-like particles that may contain mRNAs. To assess more directly a possible interaction between mRNAs and pre-40S particles, we tested whether IP of TAP-tagged Pab1p, the poly-A-binding protein interacting with polyadenylated mRNAs, could co-immunoprecipitate 20S pre-rRNA. As shown in Figure 4A, 20S pre-rRNA is co-immunoprecipitated with Pab1p–TAP just as mature 25S and 18S rRNAs and ACT1 mRNA are (Supplementary Figure S4). On the contrary, U2 snRNA and 23S pre-rRNA do not co-immunoprecipitate with Pab1p–TAP. These results suggest that pre-40S particles and mRNAs can be contained in the same complex. Association of pre-40S particles with mRNAs was also investigated in late pre-40S particles IP experiments (Figure 4B). As mentioned above, Nob1p is a factor contained in late pre-40S subunits and the putative endonuclease processing 20S pre-rRNA to 18S rRNA. Nob1p–TAP was immunoprecipitated using IgG-sepharose beads, and RNA pull down analysed for the presence of mRNAs. Three relatively abundant mRNAs were studied: rpS14, Act1 and Pgk1. As Nob1p is a poorly abundant protein, contrarily to Pab1p, enrichment for these mRNAs in the immunoprecipitates was assayed through RT–PCR rather than using northern blotting. Clear enrichment in mRNAs content is observed in the IgG-sepharose beads immunoprecipitate compared with the control corresponding to incubation of the same extracts with sepharose beads. No U2 snRNA enrichment is observed in these conditions, validating the specificity of the interaction between Nob1p and mRNAs. Moreover, 20S pre-rRNA is clearly enriched in the immunoprecipitate, whereas 18S is not suggesting that Nob1p–TAP immunoprecipitated mRNAs co-immunoprecipitate with 20S pre-rRNA molecules.
In Rio1p-depleted cells, most 20S pre-rRNA accumulates in 80S ribosome-like particles, which may be composed of two subpopulations similar to 80S ribosomes: free 80S ribosome-like particles that do not contain mRNAs and 80S ribosome-like particles that have completed translation initiation and so are bound to mRNAs. It is assumed that free 80S ribosomes are dissociated under high-salt conditions, whereas mRNA-associated 80S ribosomes are not (Martin and Hartwell, 1970; Decatur et al, 2007). To determine to what extent 80S ribosome-like particles are associated with mRNAs, cell extracts from TetO7–RIO1 cells grown in the presence or in absence of doxycycline were spun through high-salt sucrose gradients and the sedimentation properties of 20S pre-rRNA analysed. As shown in Figure 4C and D, 20S pre-rRNA sedimentation pattern is quite similar in high-salt or in low-salt conditions. Notably, the amount of pre-40S particles co-sedimenting with polysomes is not modified in high-salt conditions, suggesting that these particles indeed are associated to polysomes. Furthermore, the same results are observed whether cells are Rio1p depleted or not, indicating that in both conditions most, if not all, 20S pre-rRNA molecules contained in 80S ribosomes belong to mRNA-associated ribosome-like particles.
These data indicate that in exponentially growing cells, a fraction of pre-40S particles is contained in ribosome-like particles that are associated with mRNAs. This fraction becomes the major one when cytoplasmic pre-40S particles accumulate.
Translation initiation factors Tif11p, Rpg1p and Prt1p interact with 20S pre-rRNA
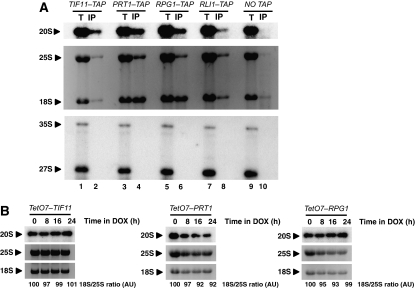
The results reported above suggest that pre-40S particles can be associated with 60S ribosomal subunits and mRNA. Such immature 40S ribosomal subunits would have completed translation initiation, indicating that pre-40S particles are able to initiate translation. If pre-40S particles initiate translation through the same pathway as mature 40S subunits do, they must interact with canonical translation-initiation factors. Thus, we assessed whether 20S pre-rRNA can be co-immunoprecipitated with Rpg1p and Prt1p, two factors belonging to the eIF3 complex, or Tif11p, the eIF1A orthologue. Rli1p, a putative ATPase implicated in translation initiation and ribosome biogenesis, was used as a positive control, as it interacts with eIF5 as well as with eIF2 and eIF3 subunits, and also with 20S pre-rRNA (Dong et al, 2004; Kispal et al, 2005; Yarunin et al, 2005). Total cellular extracts of strains expressing TAP-tagged version of Tif11p, Prt1p and Rpg1p were used in IP assays (Figure 5A). In this set of experiments, IPs were performed according to the method used in the study describing the Rli1p interaction with 20S pre-rRNA (Yarunin et al, 2005). Compared with the untagged strain, 18S rRNA and 25S rRNA were enriched in the IP material especially in Prt1p–TAP and Rpg1p–TAP IPs assays. Moreover, and as expected, as these factors mainly co-sediment with 40S particles in sedimentation assays, 18S rRNA enrichment was higher than that of 25S rRNA (Phan et al, 2001; Valásek et al, 2001a; Dong et al, 2004). Finally, clear co-IP of 20S pre-rRNA with these TAP-tagged proteins is observed, particulary in the case of Prt1–TAP and Rpg1p–TAP IPs. Nuclear rRNA precursors such as 35S and 23S were not immunoprecipitated suggesting a specific interaction between these initiation factors and 20S pre-rRNA. Thus, translation-initiation factors interact with pre-40S particles suggesting that immature 40S ribosomal subunits, which engage in translation initiation do so through the canonical initiation pathway.
Figure 5.
Initiation factors Tif11p, Rpg1p and Prt1p interact with 20S pre-rRNA, but are not required for 40S ribosomal subunit maturation. (A) IP experiments were carried out using IgG-sepharose beads. Aliquots of precipitated RNAs (IP and lanes 2, 4, 6, 8 and 10) and 1/80 of the corresponding amount of total RNAs from the input extract (T and lanes 1, 3, 5, 7 and 9) were submitted to denaturing 1.2% agarose gel electrophoresis and transferred to nylon membranes. Northern blotting was performed as in Figure 1A. (B) Total RNAs were submitted to northern blot analysis after separation by electrophoresis through a 1.2% agarose denaturing gel. Quantifications of 18S/25S were performed by analysis of the ethidium bromide staining by Quantity One Analysis software (Bio-Rad). h: hours, DOX: doxycycline.
We also investigated whether depletion of initiation factors may affect 20S pre-rRNA processing. Mature rRNA production and 20S pre-rRNA processing were followed after addition of doxycycline to the growth medium of strains expressing Tet-regulated translation-initiation factors, namely Tif11p, Prt1p and Rpg1p. As expected, depletion of these factors led to a translation-initiation defect, itself resulting in a decreased amount of polysomes and an increase in 80S ribosomes (Supplementary Figure S5): polysome-engaged ribosomes terminate translation, disengage from mRNAs and accumulate as vacant 80S couples. Clearly, depletion of these factors did not affect either 20S pre-rRNA processing or mature rRNAs synthesis (Figure 5B). Other authors have already reported that depletion for translation-initiation factors does not significantly affect pre-rRNA processing (Senger et al, 2001; Yarunin et al, 2005). However, this is not an absolute rule, as depletion or inactivation of eIF3j/Hcr1, a non-essential factor, leads to a delay in the maturation process affecting 25S and 18S rRNAs synthesis, a small reduction in SSU steady state level and ribosomal subunits nuclear export defect (Valásek et al, 2001b; Yarunin et al, 2005). These results indicate that 20S pre-rRNA processing can take place independently from translation initiation.
NGD is implicated in the degradation of ribosome-like particles
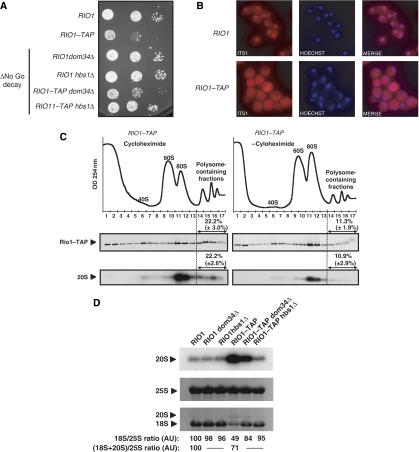
As pre-40S particles can engage in translation initiation, a fraction of Rio1p may interact with polysomes just as Nob1p does. To determine the sedimentation pattern of Rio1p, we constructed an RIO1–TAP strain. Strikingly, this RIO1–TAP strain exhibits a clear growth defect compared with the non-tagged isogenic strain (Figure 6A). This mutant phenotype is certainly due to the presence of the tag, which somehow impairs Rio1p–TAP functionality. Interestingly, in the RIO1–TAP strain, 18S rRNA production is reduced and 20S pre-rRNA levels are increased compared with its untagged counterpart (Figure 6D). Moreover, in situ hybridization of a probe complementary to the ITS1 produces a cytoplasmic signal in the RIO1–TAP strain, whereas in the RIO1 strain, a predominant nucleolar staining is observed (Figure 6B). Thus, the growth defect of the RIO1–TAP strain may be the consequence of a marked 20S pre-rRNA processing delay making that strain sick, but still viable. As shown in Figure 6C, free 60S subunits accumulate in RIO1–TAP cells in accordance with the observed 20S pre-rRNA processing defect. As expected, a fraction of Rio1p–TAP sediments in polysome-containing fractions and this localization is dependent on the presence of cycloheximide. Thus, Rio1p–TAP interacts with polysomes. As the 20S pre-rRNA accumulates in the cytoplasm, the main part of 20S pre-rRNA is expected to be found in 80S ribosome-like particles, which is indeed the case, cycloheximide being present or not (Figure 6C). Moreover, in this strain, the fraction of 20S pre-rRNA contained in polysomes is cycloheximide dependent in the same way as observed in the untagged strain. Altogether, these data indicate that the RIO1–TAP strain behaves as a viable mimic of Rio1p-depleted cells.
Figure 6.
Inactivation of NGD restores cell growth and 20S pre-rRNA processing in RIO1 TAP mutants. (A) RIO1, RIO1–TAP, RIO1 dom34Δ, RIO1 hbs1Δ, RIO1–TAP dom34Δ and RIO1–TAP hbs1Δ cells were spotted in 10-fold serial dilution steps onto YPD plates, which were incubated at 30°C for 2 days. (B) Pre-18S rRNA FISH was performed with a probe complementary to the D-A2 segment of the ITS1. (C) WCEs of RIO1–TAP strain were analyzed by sucrose gradient sedimentation under polysome stabilizing conditions (+Cycloheximide) or polysome run-off conditions (−Cycloheximide). A254 nm profiles were recorded. Rio1p–TAP protein content in each fraction was determined by western blotting and quantified using LAS-4000 and Multi gauge software (Fujifilm). RNA content in each fraction from the gradient was determined by northern blotting using a probe complementary to the D-A2 segment of the ITS1. Standard deviations from three independent experiments are indicated. (D) Total RNAs were extracted and separated by denaturing 1.2% agarose gel electrophoresis and transferred to nylon membranes. Specific RNAs were detected by hybridization with antisense oligonucleotide probes. Quantifications of 18S/25S ratio and (18S+20S)/25S ratios were performed as in Figure 2A and expressed in arbitrary units (AU), RIO1 strain taken as a reference.
Strikingly, in RIO1–TAP cells, as in Rio1p-depleted cells, the amount of 20S pre-rRNA accumulated does not correspond to the 18S deficit relative to 25S rRNA (Figures 2A and 6D) showing that, although in these cells part of 20S pre-rRNA is matured into 18S rRNA, a large part of it is unstable. As most 20S pre-rRNA accumulates in 80S fractions when cytoplasmic maturation of 40S ribosomal subunits is impaired, ribosome-like particles may not be competent for translation elongation, or not fully processive for it. Consequently, on a given mRNA molecule, translating ribosomes downstream from a ribosome-like particle will complete their translation round, but the presence of this stalled inactive or poorly active ribosome-like particle will prevent mature ribosomes from initiating new translation runs. Interestingly, the NGD is a mechanism in which ribosome stalling on mRNA leads to the degradation of the mRNA (Doma and Parker, 2006; Tollervey, 2006). The molecular basis for this degradation remains elusive, although two factors showing homology with translation termination factors eRF1 and eRF3, Dom34p and Hbs1p, respectively, involved in this process have been identified (Doma and Parker, 2006). Assuming that ribosome-like particles do not enter translation elongation or have a low processivity in it, which may cause them to stall on the mRNA, we investigated whether the NGD pathway influences the stability of 20S pre-rRNA contained in those ribosome-like particles. Thus, we assayed the effects of DOM34 or HBS1 invalidation on the growth properties of the RIO1–TAP strain and 20S pre-rRNA accumulation in these cells. As shown in Figure 6A, NGD inactivation suppresses at least partially the growth defect associated with the RIO1–TAP allele. At the same time, 20S pre-rRNA processing is restored to wild-type levels in RIO1–TAP hbs1Δ cells or greatly improved in the case of RIO1–TAP dom34Δ (Figure 6D). This suggests that in the RIO1–TAP context, NGD inactivation allows more 20S pre-rRNA molecules to be processed possibly because of an increase of the amount of 20S pre-rRNA available to the poorly efficient processing machinery of the RIO1–TAP strain, and thus to an increased processing speed. In an NGD inactivation context, Rio1p–TAP level is not modified (Supplementary Figure S6), showing that restoration of the 20S pre-rRNA processing is not attributable to a direct Rio1–TAP mRNA stabilization, or an increase of the amount of Rio1p–TAP in these NGD deficient conditions. However, we cannot exclude that the observed phenotype could be an indirect consequence of NGD inactivation leading to increased 20S pre-rRNA processing efficiency.
Discussion
In the yeast S. cerevisiae, final maturation of the small ribosomal subunit, which requires processing of the 3′ end of 20S pre-rRNA at site D, occurs in the cytoplasm (Fatica and Tollervey, 2002; Fromont-Racine et al, 2003; Zemp and Kutay, 2007; Henras et al, 2008). From the seminal work of Udem and Warner (1972) , it is assumed that pre-40S particles are not competent for translation initiation, or at least do not complete translation initiation, as in sedimentation gradients analyses no 20S pre-rRNA was found in polysomes. Thus, processing of 20S pre-rRNA has to take place before completion of translation initiation. It has to be mentioned that these studies did not report the rRNA composition of the ribosome fractions.
Immature small ribosomal subunits may complete translation initiation
In this work, we show that part of 20S pre-rRNA is associated with translating ribosomes and polysomes. We observed that roughly half of 20S pre-rRNA sediments in ribosome and polysome fractions. In ribosome run-off conditions, the amount of polysomes drastically decreases and the 20S pre-rRNA initially associated with the polysome fractions relocates to ribosome fractions in the same way as mature 18S and 25S rRNA do. Furthermore, the sedimentation pattern of 20S pre-rRNA is not modified under conditions that dissociate free 80S couples (Figure 4D) indicating that 20S pre-rRNA is indeed associated to translating complexes. Presence of pre-SSUs in translating complexes is further supported by the co-IP of 20S pre-rRNA with ribosomal proteins of the large subunit as well as with Pab1p. Furthermore, we show that mRNAs are co-immunoprecipitated with the late pre-SSU processing factor Nob1p. Altogether, these data show that, even in exponentially growing cells, part of the synthesized pre-SSUs complete translation initiation. On depletion of pre-SSU processing factors, namely Rio1p and Nob1p, immature SSUs accumulate in the cytoplasm, and again a large part of this material is found in ribosome and polysome fractions (70% in Rio1p-depleted cells, 85% in Nob1p-depleted cells). Altogether, these data suggest that in these conditions, a large part of immature SSU is associated with translating complexes. Moreover, the fact that in polysome run-off conditions, these immature SSUs still sediment with 80S complexes indicate that these pre-40S particles are associated with 60S ribosomal subunits. This strongly suggests that pre-40S particles can complete translation initiation.
In wild-type conditions, at least half of pre-SSUs are matured before completion of translation initiation. As mentioned above, in the pioneering work of the early days of yeast ribosome processing studies (Udem and Warner, 1972), all pre-SSUs were processed before completion of initiation, as no 20S pre-rRNA was found in polysomes. This discrepancy can be attributed to (1) the methodology, as in these studies freshly synthesized 20S pre-rRNAs were followed, whereas we analysed all 20S pre-rRNAs (although the half-life of this processing intermediate being quite short, most of this pre-rRNA should correspond to recently synthesized molecules); (2) growth conditions: spheroplasts incubated in minimal medium versus cells grown in complete medium and (3) differences in the genetic backgrounds of the strains used in these works, which implies that our strains (BY 4741/4742 background) possess a low efficiency 20S pre-rRNA processing machinery. In similar experiments realized with S288C wild-type strain, much less 20S pre-rRNA sediments in the ribosome (6%) and polysome (7%) fractions (data not shown). Such amounts would have been barely detectable using Udem's and Warner's protocols.
Presence of 20S pre-rRNA in ribosome-polysome fractions of sedimentation gradients is also observed on depletion of rpS0 or Fap7p, or when C-terminal mutants of Rps14p or truncated alleles of RPS31/UBI3 are expressed (Ford et al, 1999; Jakovljevic et al, 2004; Ferreira-Cerca et al, 2005; Granneman et al, 2005; Lacombe et al, 2009). Under all these conditions, a strong cytoplasmic accumulation of 20S pre-rRNA is observed. Thus, association of immature SSUs to translating complexes is not specific to the depletion of ‘late' processing factors such as Rio1p and Nob1p. However, as mentioned above and elsewhere (Ford et al, 1999), the higher 18S rRNA/20S pre-rRNA ratio observed in the polysomes fractions compared with 80S fractions (Supplementary Figure S1) suggests that ribosome-like particles are poorly efficient in translation elongation, if at all.
The fact that pre-SSUs can engage in initiation of translation is further supported by our observations that (1) 20S pre-rRNA co-immunoprecipitates with tagged translation-initiation factors and (2) mRNAs co-immunoprecipitate with Nob1p. Furthermore, Prt1p, Rpg1p, Tif35p and Nip1p subunits of eIF3 translation-initiation factor, Rli1p, a substeochiometric component of eIF3, as well as Sui3p, an eIF2 component, and Tif5p, the translation-initiation factor eIF5, were identified in mass-spectrometry analysis of proteins, which co-purify with Rio1p–TAP (data not shown). These data suggest that pre-40S particles, which would engage in translation initiation would do so through the canonical mechanism. Although depletion of the translation-initiation factors Tif11p, Prt1p or Rpg1p lead to an increase of free ribosomes, it does not impair ribosomal subunit maturation, and levels of 20S pre-rRNA are not affected under such conditions, showing that pre-SSU processing can take place normally in the absence of translation initiation.
To summarize, from our data we can conclude that (1) pre-SSU processing does not require a functional translation-initiation machinery, (2) immature 40S ribosomal subunits can complete translation initiation, thus pre-SSU maturation and translation initiation are functionally independent processes.
NGD pathway acts as a ribosome quality check point
Association of rRNA precursors with polysomes has been reported in the mold Dictyostelium discoideum (Mangiarotti et al, 1997). In this organism, translation elongation is required for complete rRNA maturation, suggesting that a late quality control mechanism validates correct ribosome assembly and/or efficient translation elongation.
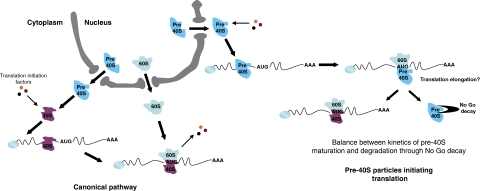
As mentioned above, in cells defective for late pre-SSU maturation, 20S pre-rRNA accumulates in the cytoplasm. Although 20S pre-rRNA originates from the same transcripts as 25S rRNA, the amount of accumulated 20S pre-rRNA is much lower than what would be expected from the deficit in 18S rRNA, as shown by the large excess of 25S rRNA over (20S+18S) RNAs observed in these cells (Ford et al, 1999; Jakovljevic et al, 2004; Ferreira-Cerca et al, 2005; Granneman et al, 2005; Lacombe et al, 2009). This implies that under these conditions, most 20S pre-rRNA is degraded by an unknown mechanism. To date, the non-functional rRNA decay (NRD) pathway is the only cytoplasmic rRNA degradation process identified in exponentially growing yeast cells (LaRiviere et al, 2006). The molecular basis of this mechanism, which eliminates 25S rRNA molecules defective in peptide bond formation as well as 18S rRNAs impaired in tRNA/mRNA base pairing is unknown. Besides this mechanism eliminating translation defective ribosomes, the NGD pathway acts on mRNAs on which translating ribosomes are stalled (Doma and Parker, 2006). Whether this process directs only the mRNAs to degradation or is also active towards stalled ribosomes has not been addressed. The RIO1–TAP allele behaves as a viable mimic of a 20S pre-rRNA processing mutant: it accumulates cytoplasmic 20S pre-rRNA, the majority of which is associated with ribosomes polysomes (Figure 6B and C). Our data show that NGD pathway inactivation effectively suppresses the growth defect of the RIO1–TAP strain, and at the same time partially (RIO1–TAP dom34Δ strain) or completely (RIO1–TAP hbs1Δ strain) restores 20S pre-rRNA processing and 18S rRNA synthesis. Thus, the poorly active Rio1p produced by the leaky RIO1–TAP allele leads to slow processing of 20S pre-rRNA, the amount of 18S rRNA produced still permitting cell growth, although at slow pace. Depletion of NGD components by extending 20S pre-rRNA half-life and diverting part of 20S pre-rRNA to the processing pathway allows an increased production of mature 18S rRNA and correlatively improves cell fitness. This suggests that the half-life of translating ribosome-associated 20S pre-rRNA is determined by a balance between processing to 18S rRNA and degradation triggered by its detection as stalled ‘ribosome' by the NGD pathway. In NGD deficient cells, not only is 18S synthesis restored, but also 20S accumulation is decreased, suggesting that 20S pre-rRNA processing kinetics is improved in these conditions. This improvement could be simply attributable to the increased amount of 20S substrate available for the processing machinery, or an indirect effect of NGD inactivation on the intrinsic activity of processing factors. As NGD acts onto stalled translation elongation complexes, these results show that (1) ribosome-polysome-associated pre-SSUs can be processed to mature ribosomal subunits, (2) pre-SSUs are poorly, or not at all, able to participate in translation elongation, (3) NRD and NGD pathways could be one and the same mechanism, or at least share some molecular components that similarly to the unknown mechanism at work in D. discoideum acts as an ultimate ribosome quality check point (Figure 7).
Figure 7.
Cytoplasmic fate of 20S pre-rRNA. Once exported from the nucleus, pre-40S particles can be matured before translation initiation, but are also competent for translation initiation using the same translation-initiation machinery as mature 40S ribosomal subunits. After completing translation initiation, pre-40S particles are either processed to mature 40S ribosomal subunits or sensed as stalled ribosomal-like particles and degraded through the NGD pathway.
In HeLa cells, just as in S. cerevisiae, the last step of SSU maturation occurs in the cytoplasm by cleavage of the 18S-E pre-rRNA, the 18S rRNA direct precursor. This cleavage involves RioK2, the human orthologue of Rio2p (Vanrobays et al, 2003; Rouquette et al, 2005). Interestingly, when RioK2 is depleted through RNA interference, 18S-E pre-rRNA can be detected in 80S and polysome-containing fractions just as in Rio1p- or Nob1p-depleted yeast cells (Rouquette et al, 2005). Thus, it is very tempting to propose that the new pathway described in our study has been conserved throughout evolution.
While this work was under revision, Cole and collaborators (Cole et al, 2009) reported that translation inactive mutant 18S rRNAs elimination by the NRD pathway (LaRivière et al, 2006) involves factors acting in the NGD pathway. Thus, as suggested above, NRD and NGD share molecular components.
Materials and methods
Yeast strains and microbiological methods
For strains used and details of construction, see Supplementary Table S1. S. cerevisiae strains were grown either in YP medium (1% yeast extract, 1% peptone) supplemented with 2% galactose or 2% glucose as the carbon source, or in YNB medium (0.17% yeast nitrogen base, 0.5% (NH4)2SO4) supplemented with 2% galactose or 2% glucose and the required amino acids and bases. When required, G418 and Doxycycline were added at 0.2 mg/ml and 30 μg/ml final concentrations, respectively.
Total RNA extractions and northern hybridizations
RNA extractions were performed by vortexing cells with glass beads in the presence of Trizol (Invitrogen). Electrophoreses of glyoxylated RNAs through agarose gels were performed as reported (Sambrook and Russell, 2001). Pre-rRNA precursors, mature rRNAs, actin mRNA and U2 snRNA analyses by northern hybridization were carried out by use of 32P-labelled oligodeoxynucleotide probes. Sequences of antisense oligonucleotides used to detect these RNAs have been reported earlier (Henras et al, 1998; Lebaron et al, 2005). Blots were hybridized with 5′ end-labelled oligonucleotide probes using Rapid Hyb Buffer (GE Healthcare).
Fluorescence in situ hybridization and immunofluorescence microscopy
Pre-rRNAs were localized by FISH as described (Gleizes et al, 2001) with the following oligonucleotidic probe: TT*GCACAGAAATCTCT*CACCGTTTGGAAT*AGC AAGAAAGAAACT*TACAAGCT*T (ITS1 probe), in which T* represents amino-modified deoxythymidine conjugated to Cy3. DNA was counterstained with Hoechst 33258. Images were captured with a CoolSnap colour CCD camera (Photometrics) mounted on a DMRB microscope (Leica) and processed with Metamorph software version 6 (Universal Imaging).
Polysome analysis and sucrose gradient fractionation
For polysome analysis, yeast cells grown in 500 ml YPD to OD600=0.6–0.9 and treated with 100 μg/ml cycloheximide for 10 min were harvested by centrifugation, washed in 10 ml ice-cold buffer K (20 mM Tris–HCl at pH 7.5, 50 mM KCl, 10 mM MgCl2) or buffer K 800 (20 mM Tris–HCl at pH 7.5, 800 mM KCl, 10 mM MgCl2) containing 100 μg/ml cycloheximide. The cell pellet was recovered in a onefold-packed cell volume of buffer K or buffer K 800 containing the same concentration of cycloheximide plus 0.1 unit/μl RNasin (Promega), protease inhibitors (Roche) and 1 mM DTT. For the polysome run-off experiments, cycloheximide was omitted. Whole cell extracts (WCE) were prepared by homogenizing the washed cells by vortexing with glass beads. Lysates were cleared briefly at 9300 g for 5 min followed by a 10 min 9300 g centrifugation (Eppendorf 5415D) to give the final WCEs. Between 20 and 30 A260 units of WCEs were layered on 4.5–45% sucrose gradient prepared in buffer K or buffer K 800 and centrifuged for 2.5 h at 39 000 r.p.m. in a Beckman SW41 rotor. Positions of ribosomal species in the gradient were determined by A254 scanning with the ISCO UA-6 gradient fraction collector. Fractions of 500 μl were collected. A total of 150 μl of each fraction were precipitated with trichloroacetic acid, and protein content analysed by electrophoresis in 4–20 or 8% pre-cast polyacrylamide-sodium dodecyl sulphate gels (Invitrogen) and western blotting. TAP-tagged proteins and Nop1p were detected as described (Dez et al, 2002). Nhp2p was detected as described (Henras et al, 2001). For RNA analysis, 200 μl of 4 M guanidinium isothiocyanate solution, 2 μl of glycogen solution, 150 μl of a 100 mM NaAc (pH 5), 10 mM Tris–HCl (pH 8.0), 1 mM EDTA solution, 225 μl of phenol and 225 μl of chloroform were added to 150 μl of every fraction. The samples were thoroughly mixed, incubated 5 min at 65°C and centrifuged 5 min at 16 000 g at 4°C. A total of 350 μl of the aqueous phase were then precipitated with ethanol.
rpL25p–TAP, rpL24ap–TAP, Pab1p–TAP and Nob1p–TAP IPs
To preserve polysomes, IP conditions were adapted from an already described method (Inada et al, 2002). Yeast cells, grown to OD600=0.6–0.9 in 500 ml YPD and treated with 100 μg/ml cycloheximide for 10 min, were harvested by centrifugation, washed in 10 ml lysis buffer (20 mM HEPES at pH 7.4, 2 mM Mg(OAc)2, 100 mM KOAc, 100 μg/ml cycloheximide, 0.5 mM DTT) and resuspended in a onefold-packed cell volume of lysis buffer containing 0.1 unit/μl RNasin (Promega) and protease inhibitors (Roche). WCE were prepared as described above. A total of 25 A260 units of extracts were mixed with an equal volume of 2X-binding buffer (100 mM Tris–HCl at pH 7.5, 24 mM Mg(OAc)2, 1 mM DTT, 0.1 unit/μl RNasin (Promega) and protease inhibitors (Roche)) and 50 μl (bead volume) of Sepharose 6 Fast Flow (GE Healthcare) or IgG-Sepharose 6 Fast Flow (GE Healthcare). IP was performed at 4°C for 2 h on a shaking table. Beads were then washed five times with 0.8 ml of IXA-100 buffer (50 mM Tris–HCl at pH 7.5, 100 mM KCl, 12 mM Mg(OAc)2, 1 mM DTT). RNAs associated with beads were extracted as reported earlier (Lebaron et al, 2005). For Nob1–TAP IP, extracted RNAs were reverse transcribed using the Superscript II reverse transcriptase (Invitrogen) and random primers. rpS14, Act1, Pgk1 and U2 cDNAs were then amplified by PCR using TAQ polymerase (New England Biolabs) and specific oligos (see Supplementary Table S2).
IPs of translation-initiation factors
Cell extracts were prepared as reported above except that buffers used were replaced by the A200 buffer (20 mM Tris–HCl (pH 8.0), 5 mM MgAc, 0.2% Triton X-100, 150 mM NaCl, 1 mM dithiothreitol (DTT), 0.1 U/μl of RNasin (Promega) and protease inhibitors (Roche)). IPs and RNA extractions were performed as already described (Dez et al, 2004; Lebaron et al, 2005; Yarunin et al, 2005).
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Tables S1 and S2, Supplementary Figure Legends
Review Process File
Acknowledgments
We thank Célia Bensafi for constant support. We are grateful to members of the Ferrer and Henry laboratory for help and numerous discussions and especially Yves Henry for critical reading of this manuscript. We are also thankful to G Stahl, I Léger-Silvestre and A Henras for advices. We thank B Monsarrat and A Stella for Mass Spectrometry analysis, D Villa for artwork and F Moutahir for expert technical assistance. JS is a recipient of a post-graduate fellowship from the Fondation pour la Recherche Médicale. This work was supported by the CNRS, the Université Paul Sabatier and grants from La Ligue Nationale contre le Cancer (‘Equipe Labellisée') and the Agence Nationale de la Recherche.
Footnotes
The authors declare that they have no conflict of interest.
References
- Acker MG, Lorsch JR (2008) Mechanism of ribosomal subunit joining during eukaryotic translation initiation. Biochem Soc Trans 36 (Part 4): 653–657 [DOI] [PubMed] [Google Scholar]
- Cole SE, LaRiviere FJ, Merrikh CN, Moore MJ (2009) A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay. Mol Cell 34: 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decatur WA, Liang XH, Piekna-Przybylska D, Fournier MJ (2007) Identifying effects of snoRNA-guided modifications on the synthesis and function of the yeast ribosome. Methods Enzymol 425: 283–316 [DOI] [PubMed] [Google Scholar]
- Dez C, Froment C, Noaillac-Depeyre J, Monsarrat B, Caizergues-Ferrer M, Henry Y (2004) Npa1p, a component of very early pre-60S ribosomal particles, associates with a subset of small nucleolar RNPs required for peptidyl transferase center modification. Mol Cell Biol 24: 6324–6337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dez C, Noaillac-Depeyre J, Caizergues-Ferrer M, Henry Y (2002) Naf1p, an essential nucleoplasmic factor specifically required for accumulation of box H/ACA small nucleolar RNPs. Mol Cell Biol 22: 7053–7065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma MK, Parker R (2006) Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440: 561–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Lai R, Nielsen K, Fekete CA, Qiu H, Hinnebusch AG (2004) The essential ATP-binding cassette protein RLI1 functions in translation by promoting preinitiation complex assembly. J Biol Chem 279: 42157–42168 [DOI] [PubMed] [Google Scholar]
- Fatica A, Oeffinger M, Dlakic M, Tollervey D (2003) Nob1p is required for cleavage of the 3′ end of 18S rRNA. Mol Cell Biol 23: 1798–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Tollervey D (2002) Making ribosomes. Curr Opin Cell Biol 14: 313–318 [DOI] [PubMed] [Google Scholar]
- Fatica A, Tollervey D, Dlakic M (2004) PIN domain of Nob1p is required for D-site cleavage in 20S pre-rRNA. RNA 10: 1698–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira-Cerca S, Poll G, Gleizes PE, Tschochner H, Milkereit P (2005) Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol Cell 20: 263–275 [DOI] [PubMed] [Google Scholar]
- Finley D, Bartel B, Varshavsky A (1989) The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature 338: 394–401 [DOI] [PubMed] [Google Scholar]
- Ford CL, Randal-Whitis L, Ellis SR (1999) Yeast proteins related to the p40/laminin receptor precursor are required for 20S ribosomal RNA processing and the maturation of 40S ribosomal subunits. Cancer Res 59: 704–710 [PubMed] [Google Scholar]
- Fromont-Racine M, Senger B, Saveanu C, Fasiolo F (2003) Ribosome assembly in eukaryotes. Gene 313: 17–42 [DOI] [PubMed] [Google Scholar]
- Gallagher JE, Dunbar DA, Granneman S, Mitchell BM, Osheim Y, Beyer AL, Baserga SJ (2004) RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev 18: 2506–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147 [DOI] [PubMed] [Google Scholar]
- Gelperin D, Horton L, Beckman J, Hensold J, Lemmon SK (2001) Bms1p, a novel GTP-binding protein, and the related Tsr1p are required for distinct steps of 40S ribosome biogenesis in yeast. RNA 7: 1268–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleizes PE, Noaillac-Depeyre J, Leger-Silvestre I, Teulieres F, Dauxois JY, Pommet D, Azum-Gelade MC, Gas N (2001) Ultrastructural localization of rRNA shows defective nuclear export of preribosomes in mutants of the Nup82p complex. J Cell Biol 155: 923–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S, Nandineni MR, Baserga SJ (2005) The putative NTPase Fap7 mediates cytoplasmic 20S pre-rRNA processing through a direct interaction with Rps14. Mol Cell Biol 25: 10352–10364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras A, Dez C, Noaillac-Depeyre J, Henry Y, Caizergues-Ferrer M (2001) Accumulation of H/ACA snoRNPs depends on the integrity of the conserved central domain of the RNA-binding protein Nhp2p. Nucleic Acids Res 29: 2733–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras A, Henry Y, Bousquet-Antonelli C, Noaillac-Depeyre J, Gelugne JP, Caizergues-Ferrer M (1998) Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J 17: 7078–7090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y (2008) The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci 65: 2334–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey JWB, Merrick WC (2000) In Translational Control of Gene Expression, Sonenberg N, Herskey JWB, Mathews MB (eds), pp 33–88. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Hinnebusch AG (2000) In Translational Control of Gene Expression, Sonenberg N, Herskey JWB, Mathews MB (eds), pp 185–243. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Inada T, Winstall E, Tarun SZ Jr, Yates JR III, Schieltz D, Sachs AB (2002) One-step affinity purification of the yeast ribosome and its associated proteins and mRNAs. RNA 8: 948–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakovljevic J, de Mayolo PA, Miles TD, Nguyen TM, Leger-Silvestre I, Gas N, Woolford JL Jr (2004) The carboxy-terminal extension of yeast ribosomal protein S14 is necessary for maturation of 43S preribosomes. Mol Cell 14: 331–342 [DOI] [PubMed] [Google Scholar]
- Kispal G, Sipos K, Lange H, Fekete Z, Bedekovics T, Janaky T, Bassler J, Aguilar Netz DJ, Balk J, Rotte C, Lill R (2005) Biogenesis of cytosolic ribosomes requires the essential iron-sulphur protein Rli1p and mitochondria. EMBO J 24: 589–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruiswijk T, Planta RJ, Mager WH (1978) Quantitative analysis of the protein composition of yeast ribosomes. Eur J Biochem 83: 245–252 [DOI] [PubMed] [Google Scholar]
- Lacombe T, Garcia-Gomez JJ, de la Cruz J, Roser D, Hurt E, Linder P, Kressler D (2009) Linear ubiquitin fusion to Rps31 and its subsequent cleavage are required for the efficient production and functional integrity of 40S ribosomal subunits. Mol Microbiol 72: 69–84 [DOI] [PubMed] [Google Scholar]
- LaRiviere FJ, Cole SE, Ferullo DJ, Moore MJ (2006) A late-acting quality control process for mature eukaryotic rRNAs. Mol Cell 24: 619–626 [DOI] [PubMed] [Google Scholar]
- Lebaron S, Froment C, Fromont-Racine M, Rain JC, Monsarrat B, Caizergues-Ferrer M, Henry Y (2005) The splicing ATPase prp43p is a component of multiple preribosomal particles. Mol Cell Biol 25: 9269–9282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangiarotti G, Chiaberge S, Bulfone S (1997) rRNA maturation as a ‘quality' control step in ribosomal subunit assembly in Dictyostelium discoideum. J Biol Chem 272: 27818–27822 [DOI] [PubMed] [Google Scholar]
- Martin TE, Hartwell LH (1970) Resistance of active yeast ribosomes to dissociation by KCl. J Biol Chem 245: 1504–1506 [PubMed] [Google Scholar]
- Peng WT, Robinson MD, Mnaimneh S, Krogan NJ, Cagney G, Morris Q, Davierwala AP, Grigull J, Yang X, Zhang W, Mitsakakis N, Ryan OW, Datta N, Jojic V, Pal C, Canadien V, Richards D, Beattie B, Wu LF, Altschuler SJ et al. (2003) A panoramic view of yeast noncoding RNA processing. Cell 113: 919–933 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG, Lomakin IB, Pilipenko EV, Shatsky IN, Agol VI, Hellen CU (2001) Molecular mechanisms of translation initiation in eukaryotes. Proc Natl Acad Sci USA 98: 7029–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan L, Schoenfeld LW, Valasek L, Nielsen KH, Hinnebusch AG (2001) A subcomplex of three eIF3 subunits binds eIF1 and eIF5 and stimulates ribosome binding of mRNA and tRNA(i)Met. EMBO J 20: 2954–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouquette J, Choesmel V, Gleizes PE (2005) Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells. EMBO J 24: 2862–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual, 3rd edn, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Saveanu C, Namane A, Gleizes PE, Lebreton A, Rousselle JC, Noaillac-Depeyre J, Gas N, Jacquier A, Fromont-Racine M (2003) Sequential protein association with nascent 60S ribosomal particles. Mol Cell Biol 23: 4449–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer T, Strauss D, Petfalski E, Tollervey D, Hurt E (2003) The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J 22: 1370–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger B, Lafontaine D, Graindorge J-S, Gadal O, Camasses A, Sanni A, Garnier J-M, Bteitenbach M, Hurt E, Fasiolo F (2001) The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol Cell 8: 1363–1373 [DOI] [PubMed] [Google Scholar]
- Tabb-Massey A, Caffrey JM, Logsden P, Taylor S, Trent JO, Ellis SR (2003) Ribosomal proteins Rps0 and Rps21 of Saccharomyces cerevisiae have overlapping functions in the maturation of the 3′ end of 18S rRNA. Nucleic Acids Res 31: 6798–6805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D (2006) Molecular biology: RNA lost in translation. Nature 440: 425–426 [DOI] [PubMed] [Google Scholar]
- Udem SA, Warner JR (1972) Ribosomal RNA synthesis in Saccharomyces cerevisiae. J Mol Biol 65: 227–242 [DOI] [PubMed] [Google Scholar]
- Valásek L, Hasek J, Nielsen KH, Hinnebusch AG (2001a) Dual function of eIF3j/Hcr1p in processing 20 S pre-rRNA and translation initiation. J Biol Chem 276: 43351–43360 [DOI] [PubMed] [Google Scholar]
- Valásek L, Phan L, Schoenfeld LW, Valaskova V, Hinnebusch AG (2001b) Related eIF3 subunits TIF32 and HCR1 interact with an RNA recognition motif in PRT1 required for eIF3 integrity and ribosome binding. EMBO J 20: 891–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beekvelt CA, Kooi EA, de Graaff-Vincent M, Riet J, Venema J, Raue HA (2000) Domain III of Saccharomyces cerevisiae 25 S ribosomal RNA: its role in binding of ribosomal protein L25 and 60 S subunit formation. J Mol Biol 296: 7–17 [DOI] [PubMed] [Google Scholar]
- Vanrobays E, Gelugne JP, Gleizes PE, Caizergues-Ferrer M (2003) Late cytoplasmic maturation of the small ribosomal subunit requires RIO proteins in Saccharomyces cerevisiae. Mol Cell Biol 23: 2083–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanrobays E, Gleizes PE, Bousquet-Antonelli C, Noaillac-Depeyre J, Caizergues-Ferrer M, Gelugne JP (2001) Processing of 20S pre-rRNA to 18S ribosomal RNA in yeast requires Rrp10p, an essential non-ribosomal cytoplasmic protein. EMBO J 20: 4204–4213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarunin A, Panse VG, Petfalski E, Dez C, Tollervey D, Hurt EC (2005) Functional link between ribosome formation and biogenesis of iron-sulfur proteins. EMBO J 24: 580–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemp I, Kutay U (2007) Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett 581: 2783–2793 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Tables S1 and S2, Supplementary Figure Legends
Review Process File