Abstract
At the leading edge of migrating cells, protrusive forces are developed by the assembly of actin filaments organised in a lamellipodial dendritic array at the front and a more distal lamellar linear array. Whether these two arrays are distinct or functionally linked and how they contribute to cell migration is an open issue. Tropomyosin severely inhibits lamellipodium formation and facilitates the lamellar array while enhancing migration, by a mechanism that is not understood. Here we show that the complex in vivo effects of tropomyosin are recapitulated in the reconstituted propulsion of neural Wiskott–Aldrich syndrome protein (N-WASP)-functionalised beads, which is based on the sole formation of a dendritic array of actin-related protein (Arp)2/3-branched filaments. Actin-depolymerising factor (ADF) and tropomyosin control the length of the actin tail. By competing with Arp2/3 during filament branching, tropomyosin displays opposite effects on propulsion depending on the surface density of N-WASP. Tropomyosin binding to the dendritic array is facilitated following filament debranching, causing its enrichment at the rear of the actin tail, like in vivo. These results unveil the mechanism by which tropomyosin generates two morphologically and dynamically segregated actin networks from a single one.
Keywords: actin, lamella, lamellipodium, protrusion, reconstituted motility, tropomyosin
Introduction
Cell migration is driven by site-directed assembly of actin filaments. At the leading edge of the cell, protrusive forces generate sheet-like extensions shaped by the polarised growth of a densely branched actin array defining the lamellipodium and a non-branched lamellar array visible at the rear of the flat protrusion (Svitkina and Borisy, 1999). The lamellipodium and the lamella were first identified as two morphologically distinct cellular compartments (Abercrombie et al, 1970). Measurements of actin filament turnover using fluorescence speckle microscopy later established heterogeneity in actin dynamics in these two regions at the front of the migrating cells. It was found that actin filaments turned over rapidly in the densely branched actin array and more slowly in the lamellar array (Ponti et al, 2004). The WAVE proteins (Wiskott–Aldrich syndrome protein (WASP)-family verprolin homologous), activated at the leading edge, generate the lamellipodial actin array by branching filaments with actin-related protein (Arp)2/3 complex. The origin of the lamellar array is not known: it might emerge from the time and space-dependent regulation of the branched array at the rear of the lamellipodium, or it might be initiated—either at the leading edge or at focal complexes deeper in the lamellipodium—by other nucleators like formins mDia1 and mDia2 (Gupton et al, 2007). In migrating epithelial cells, mDia2 is absent from the lamellipodium but maintains the specific dynamics of actin in the lamella (Gupton et al, 2007).
Whether these two arrays segregate or mingle in the protrusive structure and how each of them contributes to cell migration has been debated (Ponti et al, 2004; Giannone et al, 2007; Schaus et al, 2007; Koestler et al, 2008; Vallotton and Small, 2009). A cross-talk between these protrusive arrays was suggested by the observation that knock-down of WAVE2 and neural WASP (N-WASP) suppresses lamellipod formation and activates a formin-induced lamellar array (Sarmiento et al, 2008). A similar balance between the Arp2/3 complex and formin-dependent nucleation pathways exists in yeast (Gao and Bretscher, 2008).
Tropomyosin is generally absent from the lamellipodium and is present only in the lamellar array (DesMarais et al, 2002; Gupton et al, 2005; Iwasa and Mullins, 2007), although some tropomyosin isoforms have been detected close to the leading edge (Hillberg et al, 2006). A coordinating function of tropomyosin is suggested by experiments in which injection of excess skeletal tropomyosin almost abolished the formation of the lamellipodial array, leaving the lamellar array unaffected. Puzzlingly, tropomyosin microinjected cells with inhibited lamellipodia exhibited rapid migration and persistent leading edge protrusion, by a mechanism that has so far remained elusive (Gupton et al, 2005).
The standard in vitro reconstituted motility assay recapitulates the essential features of the dynamic behaviour of the lamellipodium with only actin and a minimal number of five essential proteins (bead-immobilised N-WASP, Arp2/3 complex, capping protein, actin-depolymerising factor (ADF)/cofilin and profilin) (Loisel et al, 1999). In this assay, a stationary number of growing barbed ends is maintained in close vicinity of the bead by a balance between reactions of filament branching by bead-immobilised N-WASP-Arp2/3 machinery and capping by soluble capping protein (Pantaloni et al, 2000; Carlsson, 2003). Rapid growth of the array is mediated by ADF-enhanced pointed-end depolymerisation at the rear, which builds up a high stationary amount of polymerisable monomeric actin (Loisel et al, 1999; Pantaloni et al, 2001). A constant actin:Arp2/3 and actin:capping protein molar ratio is maintained all along the actin tail initiated at the surface of a propelling N-WASP functionalised particle.
In vivo, the same basic mechanism supports lamellipodial protrusion. Membrane-immobilised WAVE protein catalyses filament branching and capping protein and ADF maintain a large pool of actin monomers that feed transient barbed-end growth at the lamellipodium tip (Kiuchi et al, 2007). However, in the lamellipodium, capped filaments have a lifetime of a few seconds and turnover within a narrow zone of 0.5 μm from the leading edge, whereas Arp2/3 complex treadmills with actin deeper within the lamellipodium (Miyoshi et al, 2006; Iwasa and Mullins, 2007; Lai et al, 2008). These data suggested that the restricted localisation of capping protein is due to rapid debranching followed by pointed-end disassembly (Bugyi et al, 2008). The dissociation of the daughter filament leading to linear filaments at the rear may be regulated by tropomyosin, coronin 1B or cortactin (Cai et al, 2008).
In conclusion, in the absence of tropomyosin, the heterogeneity seen in vivo between the highly branched array at the front and the more linear array at the rear, in the lamellar region is not observed in the actin tails in the minimum biomimetic system.
The effects of tropomyosin on the lamellipodial dendritic actin array are expected to be complex, since tropomyosin is known to interfere with two independent players in actin based motility, it binds filaments in competition with ADF (Bernstein and Bamburg, 1982; Cooper, 2002; Ono and Ono, 2002) and inhibits filament branching with N-WASP–Arp2/3 complex (Blanchoin et al, 2001).
Here we examine how tropomyosin affects the in vitro reconstituted propulsion of N-WASP functionalised beads and the morphology and dynamics of the actin array. In addition to its functional antagonism with ADF, tropomyosin has opposite effects on motility and organisation of the branched actin network, depending on N-WASP density at the surface of the bead. We demonstrate that tropomyosin binds preferentially at the rear of the actin tail with a spatial distribution that is consistent with its preferential binding to filaments following slow debranching. The results of this in vitro study shed light on the complex effects of tropomyosin on cell migration and represent a first step towards understanding the physical–chemical basis for the coordination of different actin arrays in cell protrusion.
Results
In bulk solution, tropomyosin inhibits filament branching by N-WASP in competition with Arp2/3 complex
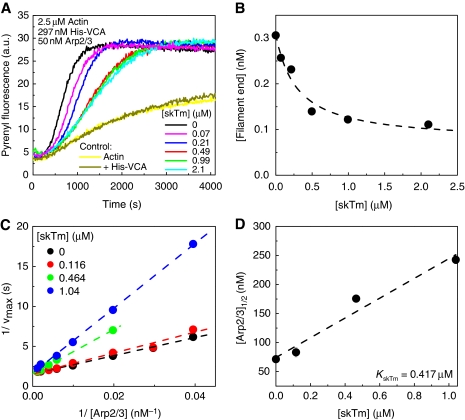
Polymerisation of actin in branched filaments with VCA (C-terminal domain of N-WASP) and Arp2/3 complex in bulk solution leads to autocatalytic polymerisation curves (Machesky et al, 1999; Pantaloni et al, 2000). In agreement with Blanchoin et al (2001), we find that tropomyosin inhibits the rate of filament branching by VCA and Arp2/3 complex in a concentration-dependent manner (Figure 1A). In contrast, in the absence of VCA and Arp2/3 complex, tropomyosin does not affect filament barbed-end assembly from either actin or profilin-actin (Supplementary Figure 1). The number of filaments created at half-polymerisation decreased by 75% upon addition of up to 2 μM tropomyosin, in the presence of 2.5 μM actin, 0.3 μM VCA and 50 nM Arp2/3 complex (Figure 1B). To understand the mechanism of inhibition of filament branching by tropomyosin, the concentration of branching complex VCA–actin–Arp2/3 was increased by increasing the concentration of Arp2/3 complex, keeping VCA and actin constant. The inhibition of filament branching by tropomyosin was relieved upon increasing the concentration of VCA–actin–Arp2/3 complex (Figure 1C). The concentration of Arp2/3 complex required to restore 50% of the maximum polymerisation rate increased linearly with the concentration of tropomyosin (Figure 1D). In other words, in the branching reaction the apparent Km of the Arp2/3 complex increases linearly with the concentration of the competitive inhibitor tropomyosin (Figure 1D). The intrinsic equilibrium dissociation constant for tropomyosin binding to F-actin, derived from the slope of the straight line, was 0.42 μM, in agreement with previous reports (Boussouf et al, 2007). Blanchoin et al (2001) had come to a different conclusion that the inhibition of branching was independent of Arp2/3 complex, but had explored a range of Arp2/3 concentrations (10–60 nM) that was too small to detect its functional competition with tropomyosin.
Figure 1.
The inhibition of filament branching by tropomyosin is relieved by increasing the concentration of actin-related protein (Arp)2/3 complex. Actin (2.5 μM, 2% pyrenyl-labelled) was polymerised in the presence of 297 nM His-VCA, Arp2/3 complex and skeletal muscle tropomyosin (skTm), as indicated. (A) Time courses of actin polymerisation monitored by the change in pyrene fluorescence in the absence (black line) and presence of increasing concentration of skTm, as indicated, [Arp2/3]=50 nM. Bright and dark yellow curves show kinetics of actin polymerisation (2.5 μM) in the absence and presence of 297 nM His-VCA, respectively. (B) Concentration of filament barbed ends at half-maximum polymerisation as a function of skTm concentration. Data are derived from the polymerisation curves shown in panel (A) as described in Materials and methods. Dashed line shows the hyperbola fit to the data. (C) Double reciprocal plots of the maximum polymerisation rate as a function of Arp2/3 complex concentration at different concentrations of skTm. The linear fits of the data (shown by dashed lines in the corresponding colours) are consistent with competitive binding of Arp2/3 complex and skTm in the branching reaction. (D) Arp2/3 complex concentration required for half-maximum polymerisation ([Arp2/3]1/2), derived from data in panel (C) increases linearly with skTm concentration, consistent with a competitive binding scheme. Dashed line shows the linear fit to the data. A value of 0.42 μM of the equilibrium dissociation constant of skTm for binding to actin filaments is derived from the slope (equation, see Materials and methods). Error bars: s.d.
In the motility assay, tropomyosin inhibits or enhances bead propulsion depending on the surface density of N-WASP
Microinjection of skeletal tropomyosin into epithelial cells inhibited lamellipodium extension, yet cells exhibited persistent migration and leading edge protrusion (Gupton et al, 2005). The reconstituted motility assay of N-WASP-coated beads provides a simple fully controllable system to test the effect of tropomyosin on the sole dendritic array. Although WAVE is mainly responsible for lamellipodium protrusion, a reconstituted motility assay based on the propulsion of N-WASP-coated beads is a valid substitute to assay the effects of tropomyosin, as this protein inhibits the filament-branching activity of the constitutively active VCA domain, which is conserved among WASP family members.
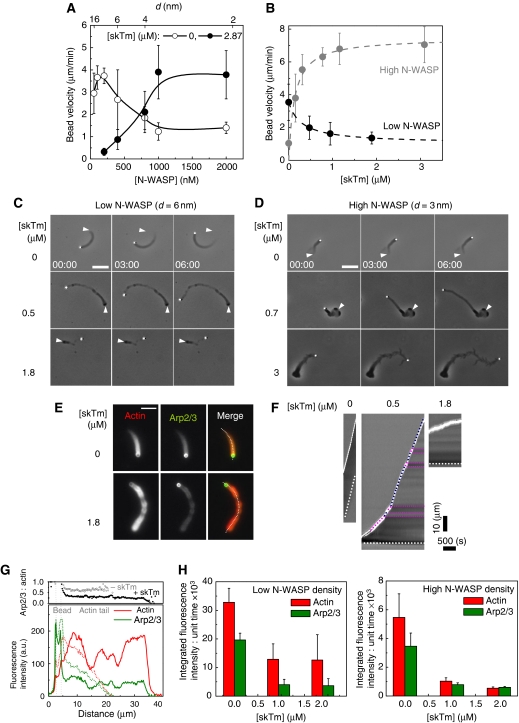
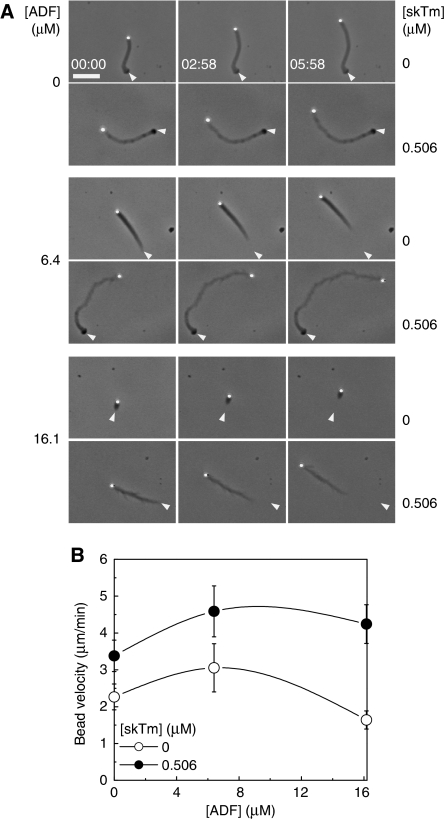
In the absence of tropomyosin, the velocity of N-WASP functionalised beads follows a bell-shape dependence on the surface density of N-WASP (Wiesner et al, 2003) and Figure 2A. A threshold N-WASP surface density (corresponding to an average distance, d, between N-WASP molecules of approximately 15 nm) is required for the establishment of a cohesive network, velocity is maximal at optimum density (d∼6 nm), whereas at surface density corresponding to close packing (d∼2 nm) movement is about four-fold slower than at the optimum density. The addition of tropomyosin to motility assay caused irregular movement and resulted in opposite phenotypes of bead propulsion depending on the surface density of N-WASP (Supplementary movies 1 and 2). Tropomyosin slowed down the movement from 3.5±1.1 μm/min to 1.3±0.3 μm/min when the beads were coated with the optimal density of N-WASP (d=6 nm) (Figure 2B), eventually at very high tropomyosin concentration (>3 μM), no tail was formed and beads did not move. In contrast, tropomyosin enhanced the velocity of highly coated beads by four-fold (d=3 nm) (Figure 2B). Both the increase in velocity of highly coated and the decrease in velocity of poorly coated beads displayed a saturation behaviour as a function of tropomyosin concentration with the same half-effect value of 0.13–0.29 μM (Figure 2B). The maximum velocity (measured at the optimum N-WASP density in the absence of tropomyosin) was unchanged, but was shifted towards higher N-WASP surface density in the presence of tropomyosin (Figure 2A).
Figure 2.
Tropomyosin inhibits or enhances bead propulsion depending on the surface density of neural Wiskott–Aldrich syndrome protein (N-WASP). (A) N-WASP concentration dependence of the bead velocity in the absence and presence of skeletal muscle tropomyosin (skTm), as indicated. Conditions: 7 μM F-actin, 2.6 μM profilin, 6.5 μM actin-depolymerising factor (ADF), 175 nM gelsolin and 50 nM actin-related protein (Arp)2/3 complex. Results expressed as mean±s.d., n=10–30 beads from 2–5 different fields for each condition. The top scale shows the average distance between immobilised N-WASP molecules on the bead surface (d) calculated on the basis of data from Wiesner et al (2003). (B) skTm concentration dependence of the average propulsion velocity of beads at low and high N-WASP surface density. Conditions: low N-WASP surface density: 7 μM F-actin, 2.6 μM profilin, 6.5 μM ADF, 100 nM gelsolin, 100 nM Arp2/3 complex, d=6 nm; high N-WASP surface density: 7 μM F-actin, 2.6 μM profilin, 5.2 μM ADF, 100 nM gelsolin, 50 nM Arp2/3 complex, d=3 nm. Results expressed as mean±s.d., n=11–28 beads from 2–6 different fields for each condition. (C, D) Time-lapse phase contrast images of poorly or highly coated N-WASP functionalised beads propelling in the biomimetic motility assay in the absence or presence of different concentrations of skTm, as indicated. Elapsed time in min: s, scale bar 20 μm. The initial position of the end of the actin tail is highlighted by white arrowheads on the subsequent images. Conditions as in panel (B). (E) Typical images of actin tails visualised by the fluorescence of rhodamine actin (red) and Alexa488–Arp2/3 complex (green) assembled in the absence and presence of skTm, as indicated. Conditions: 7 μM F-actin (5% rhodamine-labelled), 2.6 μM profilin, 6.5 μM ADF, 175 nM gelsolin, 208 nM Alexa488–Arp2/3 complex, d=6 nm. Scale bar 10 μm. (F) Kymographs generated using the trajectory of each bead shown in Figure 2E. White continuous and dotted lines indicate the position of the bead and of the end of the actin tail, respectively. The slope of the continuous line represents the velocity of the bead. At intermediate skTm concentration (here 0.5 μM) the actin tails frequently exhibit periodic pattern shown by the alternating darker and brighter intensity regions on phase contrast images corresponding to a lower and higher velocities, respectively (highlighted by pink and blue dotted lines, respectively). (G) Quantification of actin and Arp2/3 complex in the actin tails assembled in the absence or presence of 1.8 μM skTm. Lower panel: fluorescence intensity profiles of rhodamine actin and Alexa488–Arp2/3 complex were measured in the actin tail along a line selection shown by dotted white line in panel (E), in the absence (dotted lines) and presence (continuous lines) of 1.8 μM skTm. Upper panel: the Arp2/3:actin ratio remains constant along the actin tail both in the absence and presence of skTm. Grey dotted lines indicate the position of the bead. Conditions are as in panel (E). (H) Diagram showing the amounts of actin and Arp2/3 complex in the actin tail formed per unit time in the absence and presence of different concentrations of skTm at low (left panel) and high N-WASP surface density (right panel). Conditions: low N-WASP density: 7 μM F-actin (5% rhodamine-labelled), 2.6 μM profilin, 175 nM gelsolin, 6.5 μM ADF, 208 nM Alexa488-Arp2/3 complex, d=6 nm; high N-WASP density: 7 μM F-actin (5% rhodamine-labelled), 2.6 μM profilin, 105 nM gelsolin 0 μM ADF, 45 nM Alexa488-Arp2/3 complex, d=3 nm. Results expressed as mean±s.d., n=8–19 beads for each condition.
The actin tails assembled in the presence of tropomyosin displayed longer length and altered morphology (Figure 2C, D and Supplementary movies 1 and 2). At low N-WASP density, actin tails formed in the presence of tropomyosin often displayed alternating low- and high-density regions both in phase contrast and fluorescence (Figure 2C and E) that correlated with high and low rates of propulsion, respectively (Figure 2F). Similar irregular periodic propulsion of N-WASP-functionalised giant unilamellar vesicles (GUVs) has been observed when filament branching was reduced at low Arp2/3-complex concentration (Delatour et al, 2008). At high N-WASP density a loose, less cohesive meshwork assembled upon tropomyosin addition (Figure 2D).
Previous assays (Pantaloni et al, 2000; Wiesner et al, 2003; Disanza et al, 2004; Helfer et al, 2006; Akin and Mullins, 2008) have shown that gelsolin or any other capping protein can support actin-based motility. We have verified that the effects of tropomyosin remain qualitatively and quantitatively identical when gelsolin is replaced by the capping protein α1β2, at both low and high N-WASP density (Supplementary Figure 2). These results exclude the possibility that tropomyosin acts via a direct interaction with gelsolin (Nyakern-Meazza et al, 2002). It should be noted that although gelsolin severs filaments when it is readily added to a solution of F-actin, in the steady-state assays used here, gelsolin is either bound to barbed ends or in complex with two G-actin molecules, with such a high affinity that practically no gelsolin remains in the free state, the only state in which it can sever filaments. Incidentally, for the same reason ADF does not sever filaments on a steady-state basis.
Tropomyosin controls motility by inhibiting filament branching at the bead surface, but does not promote the dissociation of Arp2/3 complex from branched filaments
The balance between N-WASP–Arp2/3 complex and tropomyosin was analysed in deeper detail by monitoring the relative amounts of actin and of Arp2/3 complex in the actin tail, in a double fluorescence experiment. Periodic changes in Arp2/3 density paralleled the actin density variations, however, the Arp2/3:actin ratio remained constant along the tails whereas the density of the meshwork varied (Figure 2E and G). The amounts of actin and Arp2/3 complex in the tail formed per unit time upon tropomyosin addition were lowered at both low and high N-WASP density, as a consequence of the lowered filament-branching frequency. At low N-WASP density the actin and Arp2/3 complex content of the comet were decreased to 40 and 20%, respectively, of the values measured in the absence of tropomyosin (Figure 2H). Similarly, at high N-WASP density, tails formed in the presence of tropomyosin contained 20 and 10% of the amounts of actin and Arp2/3 complex found in absence of tropomyosin (Figure 2H).
The observed decrease in the amount of Arp2/3 complex in the tail might result from two different effects of tropomyosin: tropomyosin might promote debranching and dissociation of Arp2/3 complex from the actin meshwork and/or only inhibit the branching reaction at the bead surface.
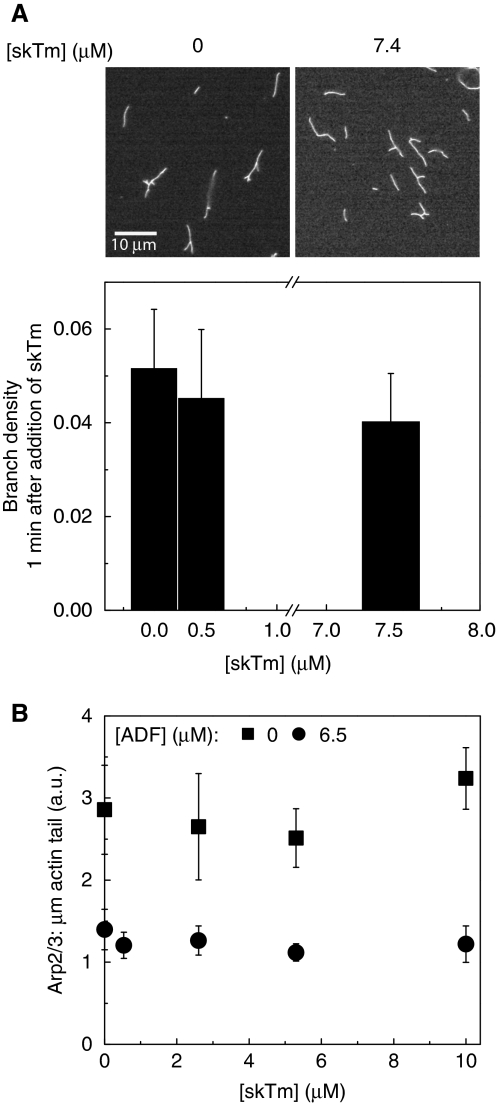
To address this issue, in a first assay, we measured that in bulk solution tropomyosin does not enhance debranching of filaments assembled with VCA–Arp2/3 complex, in agreement with Blanchoin et al (2001) (Figure 3A).
Figure 3.
Tropomyosin does not cause debranching nor dissociation of actin-related protein (Arp)2/3 complex from the actin tail. (A) Actin (4 μM) was rapidly polymerised in the presence of 100 nM Arp2/3 complex and 100 nM His-VCA in bulk solution. Then skeletal muscle tropomyosin (skTm) was added at the indicated concentrations, followed by phalloidin 1 min later and observed in fluorescence microscopy. Typical images are shown in the upper panel, skTm as indicated. The branch density was evaluated as the ratio of the number of branches to actin filament length in μm at each skTm concentrations. Results expressed as mean±s.d. (B) Neural Wiskott–Aldrich syndrome protein (N-WASP)-coated beads (d=6 nm) were incubated with the motility medium to let initiate actin tails. Then skTm was added to the preformed actin tails at the indicated concentrations. The amount of Arp2/3 complex per μm length of actin tail in the absence and presence of actin-depolymerising factor (ADF) was calculated at different concentrations of skTm as described in Materials and methods. Conditions: 7 μM F-actin, 2.6 μM profilin, 175 nM gelsolin, 50 nM Alexa488-Arp2/3 complex and either 0 or 6.5 μM ADF. Results are expressed as mean±s.d., n=0–24 beads for each condition. Note: the amount of Arp2/3 complex is lowered because of the depolymerising effect ADF (like the amount of actin, see Figure 2G).
Next, increasing amounts of tropomyosin were added to pre-assembled actin tails in the motility assay and the fluorescence of Arp2/3 complex was measured along the tail. The amount of Arp2/3 per tail unit length was unaffected by the de novo addition of tropomyosin, both in absence and presence of ADF, thus indicating that tropomyosin does not cause dissociation of Arp2/3 complex from branched filaments (Figure 3B). The immediate arrest of depolymerisation of actin in the tail following tropomyosin addition testified that tropomyosin was rapidly binding to the comets (Supplementary Figure 3). In conclusion, tropomyosin does not cause debranching nor dissociation of Arp2/3 complex from the actin meshwork. It is only by inhibiting the branching reaction at the bead surface that tropomyosin lowers the amount of Arp2/3 complex incorporated in the actin tail.
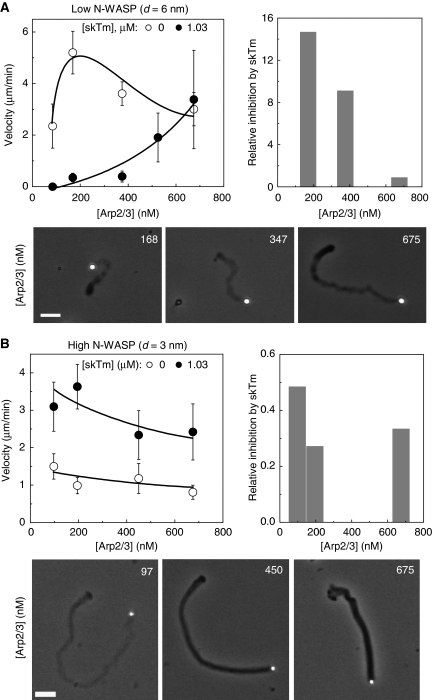
The interplay between tropomyosin and the surface density of the branching enzyme N-WASP was examined by monitoring the motility of poorly coated and highly coated beads (d=6 and 3 nm, respectively) at different Arp2/3 complex concentrations in the presence of a constant amount of tropomyosin (1.03 μM) (Figure 4). At low N-WASP density (d=6 nm), increasing Arp2/3 complex concentration relieved the inhibition of propulsion due to tropomyosin (Figure 4A). Conversely, at high N-WASP surface density (d=3 nm), increasing the concentration of Arp2/3 complex counteracted the enhancement of propulsion due to tropomyosin and slowed down propulsion (Figure 4B). In addition, the loose structure of the actin tail observed in the presence of tropomyosin reverted to a compact structure upon the addition of Arp2/3 complex (Figure 4A and B lower panels). In other words, consistent with solution studies described in Figure 1, tropomyosin and Arp2/3 complex display antagonistic competitive regulatory effects on actin-based propulsion.
Figure 4.
Antagonistic regulation of actin-based motility by actin-related protein (Arp)2/3 complex and tropomyosin. (A): low neural Wiskott–Aldrich syndrome protein (N-WASP) surface density (d=6 nm); (B): high N-WASP surface density (d=3 nm). Upper left panels: bead velocity varies in opposite directions with Arp2/3 concentration in the absence (empty circles) or in the presence of skeletal muscle tropomyosin (skTm) (filled circles). Upper right panels: histograms of the ratio of the velocity measured in the absence and in the presence of skTm. Bottom panels: phase contrast images of N-WASP functionalised beads and their actin tails in the presence of skTm (1.03 μM) and Arp2/3 complex as indicated. Scale bar 20 μm. Conditions: 7 μM F-actin, 2.6 μM profilin, 6.1 μM (A) or 5.2 μM (B) actin-depolymerising factor (ADF), 175 nM gelsolin. Results expressed as mean±s.d., n=13–32 beads from 2–5 different fields in each condition.
ADF and tropomyosin control the length of actin tails in an opposite manner
In the standard motility assay, barbed-end cappers and ADF synergise to enhance propulsion by increasing the steady-state concentration of polymerisable ATP–G-actin that fosters barbed-end growth of newly branched filaments at the bead surface (Carlier et al, 1997). The same mechanism accounts for the effect of cappers and ADF in lamellipodium motility (Karpova et al, 1995; Laine et al, 1998; Kiuchi et al, 2007). As tropomyosin competes with ADF for binding to F-actin and inhibits pointed-end depolymerisation (Broschat et al, 1989; Ono and Ono, 2002) and Supplementary Figure 4), it lowers the stationary amount of G-actin and slows down propulsion.
When added to a solution identical to the motility assay consisting of gelsolin-capped filaments, ADF and profilin, tropomyosin competes with ADF for binding to F-actin in a sedimentation assay (Supplementary Figure 5), as reported (Carlier et al, 1997; Ono and Ono, 2002); however, the change in the amount of monomeric actin in supernatants upon addition of tropomyosin in the range of 0–3 μM was too small to be detected (Supplementary Figure 5B). The lengthening of actin tails, observed upon addition of tropomyosin to motility assays, at either high or low surface density of N-WASP (Figure 2C and D) is consistent with the competition between ADF and tropomyosin in the regulation of pointed-end disassembly. Accordingly, tropomyosin abolished the exponential decrease in actin fluorescence observed in the absence of tropomyosin (Figure 2E and G). In agreement with solution studies, the comet stabilising effect of tropomyosin was competed out by ADF (Figure 5A). It is important to note that ADF affected bead velocity similarly in the absence and presence of tropomyosin (Figure 5B).
Figure 5.
Tropomyosin and actin-depolymerising factor (ADF) control the length of the actin tail in opposite manner. (A) Time-lapse phase contrast images of neural Wiskott–Aldrich syndrome protein (N-WASP) functionalised beads propelling in the biomimetic motility assay in the absence or presence of skeletal muscle tropomyosin (skTm) and ADF, as indicated. Conditions: 7 μM F-actin, 2.6 μM profilin, 175 nM gelsolin, 90 nM actin-related protein (Arp)2/3 complex, d=4 nm. Elapsed time in min: s, scale bar 20 μm. The initial position of the end of the actin tail is highlighted by white arrowheads on the subsequent images. The length of the actin tail is determined by the competition between ADF and skTm. (B) ADF concentration dependence of the bead velocity in the absence or in the presence of 0.506 μM skTm. Conditions as in panel (A). Results expressed as mean±s.d., n=9–20 beads from 2–4 different fields for each condition.
In conclusion, the effects of tropomyosin on bead movement result from its competition with ADF and Arp2/3 complex. These two effects are independent of each other and additive. However, the effect of tropomyosin on filament branching dominates on bead velocity, and overcomes its effect on ADF at high N-WASP density. On the other hand, the competition between ADF and tropomyosin regulates the length of the actin array.
The preferential localisation of tropomyosin at the rear of the actin tail is because of its facilitated binding following filament debranching
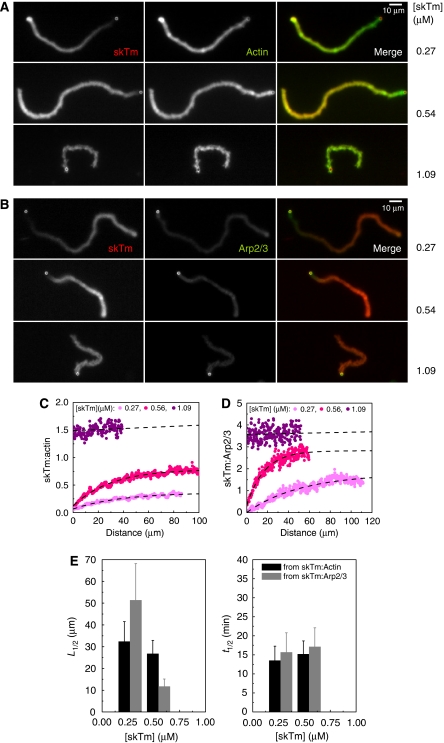
The effects of tropomyosin on bead propulsion and actin tail dynamics suggest that tropomyosin is bound to F-actin all along the tail during propulsive movement. However, we found that the steady-state distribution of fluorescently labelled tropomyosin along the actin tail is not homogeneous. Tropomyosin was enriched in old actin filaments at the rear of the actin tail and was present at a lower amount in the newly assembled actin meshwork in the close vicinity of the bead (Figure 6A and B). The tropomyosin:actin ratio increased from approximately 0.2 to 1 (arbitrary units) from the bead surface to the end of the comet (Figure 6C). Similarly, the tropomyosin:Arp2/3 ratio increased as well (Figure 6B and D). The characteristic pattern of tropomyosin fluorescence was independent of ADF concentration (data not shown). It remained stationary during propulsion, thus demonstrating that branched filament assembly and tropomyosin binding are linked by a reaction that follows network assembly and kinetically limits the binding of tropomyosin. This reaction is not the release of Pi following ATP hydrolysis on actin, as in co-sedimentation assays tropomyosin bound identically to ADP- and ADP–Pi–actin filaments, with Kd values of 0.60±0.22 and 0.64±0.10 μM, respectively (Supplementary Figure 6).
Figure 6.
Tropomyosin distribution along the actin tail correlates with filament debranching. (A, B) Typical images of actin tails assembled in the presence of different skeletal muscle tropomyosin (skTm) concentrations, as indicated, visualised by Alexa488-actin (green) and Alexa568-skTm (red) fluorescence (A) or Alexa568-skTm (red) and Alexa488-actin-related protein (Arp)2/3 (green) fluorescence (B). Conditions: 7 μM F-actin, 2.6 μM profilin, 175 nM (A) or 105 nM (B) gelsolin, 84 nM (A) or 60 nM (B) Arp2/3 complex and skTm as indicated, d=6 nm. Scale bar 10 μm. (C, D) The skTm:actin ratio (C) or the skTm:Arp2/3 ratio (D) along the actin tail at the indicated concentrations of skTm was calculated as described in Materials and methods. Representative data are shown. Black dashed lines show the exponential fit to the data. Conditions as in panel (A) for (C) or (B) for (D). (E) The distance from the bead at half saturation of skTm (L1/2) decreases with increasing skTm concentration while the corresponding half-time (t1/2) is independent of skTm concentration. Results are expressed as mean±s.d. n=14–19 beads for each condition.
The distance along the tail at which the tropomyosin:actin ratio was 50% of the maximum binding decreased from 32.4±9.2 μm at 0.27 μM tropomyosin to 26.7±6.2 μm at 0.54 μM tropomyosin, and was too small to be measured accurately at 1 μM tropomyosin (Figure 6C and E). Considering the velocity, the half-time corresponding to the half-distance was 15.3±1.5 min, independent of the tropomyosin concentration (Figure 6E). The delayed binding of tropomyosin, therefore, cannot be due to a slow association rate to the tail. This result is consistent with the known fast binding of tropomyosin to F-actin in solution (Weigt et al, 1991). On the other hand, the half-time for tropomyosin binding is identical to the half-time of filament debranching measured in solution (13.3±1.6 min (Le Clainche et al, 2003)). The results therefore suggest that tropomyosin binds preferentially to filaments following debranching.
In co-sedimentation assays no difference was detected in the binding of tropomyosin to branched or debranched filaments (Supplementary Figure 7). However, the maximum average branch spacing in this bulk solution assay was approximately 2.5 μm (calculated from data presented in Supplementary Figure 7A, and in agreement with (Pantaloni et al, 2000)), whereas in motility assay it is close to the in vivo value which is at least one order of magnitude lower (Svitkina and Borisy, 1999; Iwasa and Mullins, 2007). A sufficiently short distance between branch junctions may be required to stearically inhibit the cooperative binding of tropomyosin to filaments.
Discussion
In this paper, we showed that the addition of tropomyosin to a reconstituted motility assay based on the sole formation of a dendritic filament array causes major changes in the propulsion of N-WASP-coated beads and in the morphology of the Arp2/3-branched meshwork. These effects are explained by the interference of tropomyosin with two independent processes that control actin-based motility. First, the antagonism between tropomyosin and ADF is dominant in determining the morphology and dynamics of the actin tails. Second, the inhibition of filament branching by tropomyosin mainly controls velocity in a complex manner, which explains the puzzling effects of tropomyosin on cell migration. Tropomyosin, by decreasing branching frequency, causes slower propulsion at low N-WASP surface density, when the density of active N-WASP falls below the threshold required for building a cohesive dendritic network (Wiesner et al, 2003). Eventually at high concentrations of tropomyosin we observe the complete abolishment of comet tail formation. This total inhibition level was not reached by injecting tropomyosin in live cells, hence the presence of Arp2/3 complex was still detectable in the greatly inhibited lamellipodium (Figure 2 in (Gupton et al, 2005)). In contrast, tropomyosin accelerates propulsion at high N-WASP surface density, by preventing the frequent filament branching that generates a large fraction of transiently attached filaments acting as a brake (Mogilner and Oster, 1996). Thus, tropomyosin reveals that actin-based motility is controlled by filament branching, which corroborates previous views (Wiesner et al, 2003; Delatour et al, 2008).
Remarkably, by competing with Arp2/3 complex in the branching reaction, tropomyosin lowers the amount of Arp2/3 complex incorporated in the actin tail, but it does not cause debranching nor dissociation of Arp2/3 complex from the actin meshwork, even after debranching has been induced by hydrolysis of ATP on Arp2 (Le Clainche et al, 2003). This important result demonstrates that some irreversible reaction is associated with the branching process, which precludes subsequent displacement of Arp2/3 complex from the branch junction by tropomyosin. This conclusion is consistent with previous observations that following branch initiation Arp2/3 complex remains tightly bound to mother filaments and is recycled only as filaments depolymerise from their pointed ends (Wiesner et al, 2003; Lai et al, 2008).
Another important characteristic feature of tropomyosin interaction with the dendritic array in the biomimetic motility system is its preferred binding to filaments following debranching, demonstrated by the identical half-times of debranching and tropomyosin binding. Newly generated daughter filaments have a low probability to dissociate from the mother filament in the region proximal to the bead, however, once they dissociate, their rate of depolymerisation will be higher than at the rear of the tail because of the lower extent of tropomyosin binding in this region. In summary, the addition of tropomyosin to the motility assay based on the sole formation of a dendritic array introduces heterogeneity in the filament dynamics within the array.
The preferred localisation of tropomyosin at the rear of the lamellipodial array, and its absence in a zone of about 0.5 μm from the leading edge is also observed in vivo (DesMarais et al, 2002; Gupton et al, 2005). This zone is narrower than in the reconstituted motility assay and corresponds to a lower half-time for tropomyosin binding. The rapid turnover of capping proteins is also restricted to this narrow zone, which suggested that within a distance of 0.5 μm from the leading edge, filaments branch, grow for a few seconds, are capped and then debranching occurs, leading to a fast pointed-end depolymerisation (Miyoshi et al, 2006; Iwasa and Mullins, 2007; Bugyi et al, 2008). Hence, in vivo as well as in vitro tropomyosin binding appears facilitated by filament debranching. Our conclusions are consistent with the increased level of tropomyosin at the leading edge upon knock-down of N-WASP/WAVE branching machineries (Sarmiento et al, 2008).
Are the effects of tropomyosin on the dynamics and morphology of the sole dendritic array sufficient to account for all its effects on cell migration?
The main in vivo effects of tropomyosin on lamellipodial protrusion, including the inhibition of the dendritic array, its evolution toward a more linear lamellar-like network extending towards the leading edge and the decrease in Arp2/3 complex, ADF and barbed-end concentration near the cell edge (Gupton et al, 2005) are recapitulated in the reconstituted assay. Tropomyosin also increased the speed of migration and persistence of cell protrusions, which is mimicked by the increase in velocity of highly coated N-WASP beads induced by tropomyosin.
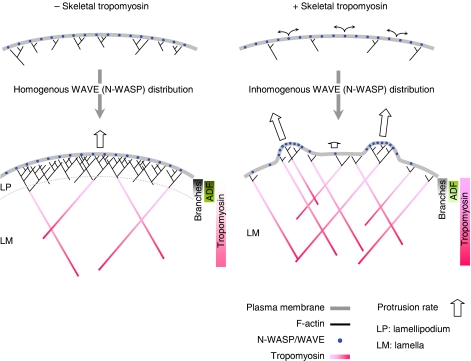
In tropomyosin-injected cells, the outline of the leading edge displayed a jagged appearance (Figure 6 in Gupton et al (2005)), suggesting the segregation of actin nucleating regions that produce force. In vivo as well as in reconstituted motility assays with functionalised GUVs, the diffusion of N-WASP in the membrane is constrained by transient Arp2/3-dependent links between N-WASP and the filaments, which results in the co-segregation of actin and N-WASP (Delatour et al, 2008; Weisswange et al, 2009). We propose that the activities of tropomyosin combine with both the mobility of N-WASP at the membrane and the autocatalytic nature of the branching reaction to induce non-homogeneous distribution of N-WASP and of force production. Within this view, distinct regions of low and high density of N-WASP would exhibit the behaviour of poorly and highly coated beads, respectively, in the presence of tropomyosin. In zones of high N-WASP density, tropomyosin would positively regulate and optimise branched filament assembly pushing the membrane forward, whereas no force would be produced in N-WASP-depleted regions wherein low frequency of branching, exacerbated by tropomyosin, leads to the arrest of the movement (Figure 7). We propose that these two opposite effects may promote the formation of the jagged leading edge.
Figure 7.
Model scheme of the main effects of tropomyosin on the protrusive actin arrays. Left panel: in standard (non tropomyosin-injected cells) neural Wiskott–Aldrich syndrome protein (N-WASP)/WAVE molecules are maintained in a homogeneous distribution at the membrane at the leading edge where a dendritic lamellipodial actin array (LP) is formed. This actin array is highly branched and exhibits fast turnover that is regulated by actin-depolymerising factor (ADF) and fades away within a few μm from the plasma membrane. Endogenous tropomyosin is excluded from the proximal zone of lamellipodium because of the high density of branches. Debranching facilitates the association of tropomyosin to the older, more distal part of the network. Tropomyosin antagonises the depolymerising effect of ADF leading to slower turnover and extension of the non-branched lamellar actin array (LM). Right panel: In tropomyosin-injected cells, branching is inhibited, hence the fraction of non-branching, freely diffusing N-WASP/WAVE increases (curved black arrows). Diffusing N-WASP/WAVE molecules get in close proximity of a branching structure, where the availability of newly created filament barbed-ends facilitates autocatalytic branching. This process leads to the segregation of active N-WASP/WAVE and formation of discrete dendritic structures. The lower frequency of branching elicited by tropomyosin favours both alignment of these filaments and fast migration. At the low N-WASP/WAVE density regions the reduced branching by tropomyosin results in the formation of an actin array that displays the characteristics of lamella. The lack of branches allows extension of the tropomyosin-rich region up to the leading edge. Tropomyosin by antagonising to ADF establishes the slower turnover of this network. Bars on the right of each panel show the distribution of each protein throughout the network, bold and dim colours reflect the higher and lower protein concentrations, respectively.
In conclusion, the effect of tropomyosin on the lamellipodial array in itself appears sufficient to account for many aspects of the observed protrusive behaviour of cells in the presence of high levels of tropomyosin. The existence of a distinct underlying lamellar meshwork, initially proposed by Ponti et al (2004), but recently questioned (Vallotton and Small, 2009), may not be needed to explain the in vivo observations. However, additional effects of tropomyosin on formin-mediated filament bundles, evoked in recent reports (Wawro et al, 2007; Sarmiento et al, 2008) may synergise with its effects on the dendritic array described here. A more detailed understanding of how these diverse actin modules contribute to motile processes requires further experimentation using more elaborated motility assays combining the two machineries. The existence of several isoforms of tropomyosin may introduce further refinement in the regulation. Finally, long-distance effects of tropomyosin binding to other actin structures like stress fibres may have bearings on the overall steady-state level of G-actin in the cytoplasm, thus affecting lamellipodial array indirectly. Although the full complexity of the cell context is not reconstituted yet, in showing that a minimal set of proteins is sufficient to generate two self-organised networks from a single one, our work nevertheless brings primary insight into the spatio-temporal interplay between lamellipodium and lamella, and its regulation by tropomyosin.
Recently, using stochastic two-dimensional computer simulations, Huber et al (2008) found that the spatial separation of ADF and tropomyosin rich zones could simply emerge from a significant difference in the dissociation rates of ADF and tropomyosin from actin filaments. This theoretical study emphasises how complex morphological properties may also result from the functional antagonism between ADF and tropomyosin, combined with specified binding kinetics.
Materials and methods
Protein preparations and modifications
Actin was purified from rabbit skeletal muscle acetone powder (Carlier et al, 1986). Skeletal muscle tropomyosin (skTm) was purified from rabbit skeletal muscle according to Smillie (1982). Arp2/3 complex from bovine brain, human His-tagged N-WASP, human His-tagged VCA, profilin from bovine spleen, recombinant human ADF and gelsolin were purified as described (Le Clainche and Carlier, 2004). Actin was labelled with N-(1-pyrenyl)iodoacetamide (pyrene), 5-(and-6)-carboxy-tetramethylrhodamine succinimidyl ester (Molecular Probes, rhodamine) or Alexa Fluor488 carboxylic acid, succinimidyl ester (Molecular Probes, Alexa488) and Arp2/3 complex was labelled with Alexa Fluor488 C5 maleimide (Molecular Probes, Alexa568), as described previously (Kouyama and Mihashi, 1981; Isambert et al, 1995; Wiesner et al, 2003). skTm was fluorescently labelled with Alexa Fluor 568 C5 maleimide (Molecular Probes, Alexa568) as follows. 5.5 mg/ml skTm was reacted with 10-fold molar excess dye in 20 mM HEPES pH 7.5, 5 M GuHCl for 3 h at RT. The reaction was stopped by addition of 10 mM DTT. skTm was dialysed against 20 mM HEPES pH 7.5, 500 mM NaCl, 0.5 mM DTT and gel filtered using a PD-10 column (GE Healthcare). The sample was further dialysed against 20 mM HEPES pH 7.5, 50 mM NaCl, 0.5 mM DTT to renature tropomyosin and finally against 5 mM Tris–HCl pH 7.8 and 0.5 mM DTT. The final protein and probe concentrations were determined spectrophotometrically. The molar ratio of the bound probe to the skTm concentration was 0.29. Co-sedimentation assay showed that the actin-binding activity of tropomyosin was not altered by the labelling procedure (data are not shown).
Pyrene-actin polymerisation assay
Actin polymerisation was monitored at 20°C by the increase in pyrene fluorescence of a 2% labelled pyrenyl-actin solution (V=160 μl, λexc=366 nm and λem=407 nm). Experiments were performed using a Safas Xenius FLX spectrofluorimeter equipped with a 10-sample device. Polymerisation in branched filaments was initiated by the addition of 100 mM KCl, 1 mM MgCl2 and 0.2 mM EGTA to a solution of 2.5 μM Ca-G-actin containing His-VCA, Arp2/3 complex and skTm (for exact protein concentrations see figure legends). The final conditions were 5 mM Tris–HCl pH 7.8, 0.2 mM ATP, 0.1 mM CaCl2, 1 mM DTT, 100 mM KCl, 1 mM MgCl2 and 0.2 mM EGTA (F-buffer). When concentration dependence was measured the total volume of the protein-of-interest and its storing buffer was always kept constant and represented 12–18% of the total volume of the sample. The polymerisation rate at half-maximum polymerisation was derived from the pyrene fluorescence curves as described previously (Pantaloni et al, 2000). Time course of pyrenyl fluorescence (F(t)) was fitted using the following equation: ln[(F(t)−Fmin)/(Fmax−F(t))], where Fmin is the initial fluorescence intensity and Fmax is the final fluorescence intensity. The maximum polymerisation rate was determined as the maximum value of the first derivative of the fitted curve. The concentration of filament barbed end at half-maximum polymerisation (F1/2) was calculated from the maximum polymerisation rate (v1/2) using the equation v1/2=k+ × F1/2 × (c−cc)/2, where k+=10 μM−1/s (Pollard, 1986), c is the total actin concentration (c=2.5 μM) and cc is the critical concentration (cc=0.1 μM). The skTm concentration dependence of F1/2 (F1/2 (skTm)) was fit using the following equation F1/2 (skTm)=(F1/2max/(1+(x/K1/2)))+F1/2min, where F1/2max is the value of F1/2 measured in the absence of skTm, F1/2min is the value of F1/2 measured in the presence of saturating amount of skTm and K1/2 is the concentration of skTm required for half-maximum inhibition. The skTm concentration ([skTm]) dependence of the Arp2/3 complex concentration required for half-maximum polymerisation (Kmapp) was derived from the linear fit of the double reciprocal plots of the maximum polymerisation rate as a function of Arp2/3 complex concentration. Data were fit within the competitive binding model using the equation Kmapp=K0 × (1+[skTm]/KskTm), where K0 is the concentration of Arp2/3 complex required for half-maximum polymerisation in the absence of skTm and KskTm is the equilibrium dissociation constant of skTm for binding to actin.
Motility assay
Motility assays using N-WASP-coated beads were performed as described (Wiesner et al, 2003; Le Clainche and Carlier, 2004). Carboxylated polystyrene microspheres (2 μm diameter, Polysciences, 2.5% solid) were functionalised by incubating with N-WASP in Xb buffer (10 mM HEPES, pH7.8, 100 mM KCl, 1 mM MgCl2, 0.1 mM CaCl2 and 1 mM ATP) for 1 h on ice during continuous stirring. The reaction was stopped with the addition of BSA (100 mg/ml, for 15 min), the beads were washed twice by centrifugation (13 000 r.p.m., 4°C, 5 min) and stored on ice in Xb buffer supplemented with 1 mg/ml BSA and used within 2 days. The motility medium was prepared by mixing 7 μM F-actin, ADF, profilin, gelsolin, Arp2/3 complex and skTm at the indicated concentrations, 5 mg/ml BSA, 0.2% methylcellulose (w/v) (M-0512, Sigma-Aldrich), 6.6 mM DTT, 0.15 mM DABCO (1.4-diazabicyclo-[2.2.2]octane, D2522, Sigma-Aldrich), 2 mM ATP and 4 mM MgCl2 in buffer Xb. The samples were incubated for 30 min before addition of the beads (final concentration 0.01% (v/v)). Samples of 4 μl were placed between a slide and coverslip sealed with VALAP. The beads were observed either in phase contrast or fluorescence microscopy (AX70, Olympus) using × 20 or × 60 objectives and a CCD camera (OrcaII ERG Hamamatsu).
Measurement of bead propulsion velocity
Time-lapse series of freely moving beads were acquired and analysed using the template recognition based tracking option of Metamorph 6.0 (Universal Imaging) The average velocities were calculated for sets of 8–30 beads selected from 3–5 different fields in each condition.
Quantification of actin, Arp2/3 complex and skTm in the actin tails
Fluorescent images of comets assembled in the presence of fluorescently labelled proteins were recorded at steady state, and the average emission intensities of rhodamine, Alexa488 or Alexa568 were measured in the actin comet excluding at least 1-bead diameter immediately behind the bead and at the end of the comet (Wiesner et al, 2003) using ImageJ (http://rsbweb.nih.gov/ij/index.html). The intensity values were corrected for background fluorescence. Control experiments were performed to make sure that no emission overlap existed between the two fluorescence channels, if it was necessary. The bound label:protein ratios were 1 for Arp2/3 complex, 0.05 for actin and 0.29 for skTm.
The Arp2/3 complex:actin ratio was calculated as the ratio of the integrated fluorescence intensities of Arp2/3 complex and actin measured in a square selection of the actin comet at least 1-bead diameter behind the bead. The same results were obtained when the corresponding fluorescence intensities were measured along a line selection along the axis of the comet (data are not shown).
The skTm:actin or skTm:Arp2/3 complex ratio was calculated as the ratio of the integrated fluorescence intensities of skTm and actin or Arp2/3 complex as described for Arp2/3:actin, measured along a line selection along the axis of the comet. The data were fit with an exponential function y(L)=y0+A × (1−exp(−L/L1/2)), where L is the distance measured from the bead, y0 is the skTm:actin ratio at L=0, y0+A is the maximum value of the skTm:actin or skTm:Arp2/3 complex ratio and L1/2 is the distance at half maximum skTm:actin or skTm:Arp2/3 complex ratio. To obtain the half-time for skTm binding (t1/2) to the comet the values of L1/2 were normalised with the actual bead velocities.
To study the effect of skTm on the dissociation of Arp2/3 complex from actin tails, N-WASP-coated beads were incubated with the motility medium in an eppendorf tube until steady state (1.5–2 h) (for exact protein concentrations see figure legend). Then skTm was added to the preformed actin tails at different concentrations and samples were further incubated for 2 min. For quantification aliquots of 2 μl were fixed by gently mixing with 2 μl 1% glutaraldehyde and processed for microscopic observation. The amount of Arp2/3 complex was determined as the integrated fluorescence intensity measured along a line selection along the axis of the comet excluding at least one bead diameter immediately behind the bead and two bead diameters at the end of the comet (Wiesner et al, 2003) using ImageJ.
Quantification of branch density
Branch density at different concentrations of skTm was measured by light microscopy (Le Clainche et al, 2003). Actin (4 μM) was rapidly polymerised in the presence of 100 nM Arp2/3 complex and 100 nM His-VCA. The reaction was followed by pyrenyl-actin fluorescence. When 90% completion of the polymerisation was reached (approximately 200 s) the sample was split into several aliquots that were supplemented with different amounts of skTm. After 1 min incubation Alexa Fluor488-phalloidin (Molecular Probes) was added in a 1:1 molar ratio to actin. Samples were diluted 200-fold and processed for microscopy observation. The number of branches per μm actin filament was measured using ImageJ software. In each condition a total filament length of 5000–10 000 μm and about 250–500 branches were counted.
Supplementary Material
Supplementary Movie 1
Supplementary Movie 2
Supplementary Material
Review Process File
Acknowledgments
La Ligue Contre le Cancer (MFC: équipe labellisée); ANR-PCV 2006; STREP ‘Biomics' (MFC); EMBO Long-Term Fellowship ALTF 626-2006 (BB); La Ligue Contre le Cancer, Allocation postdoctorale pour jeune chercheur confirmé, 2009 (BB).
Authors' contributions: BB and MFC conceived the experiments and wrote the paper, BB conducted the experiments and analysed the results, BB and DD purified the proteins.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abercrombie M, Heaysman JE, Pegrum SM (1970) The locomotion of fibroblasts in culture. 3. Movements of particles on the dorsal surface of the leading lamella. Exp Cell Res 62: 389–398 [DOI] [PubMed] [Google Scholar]
- Akin O, Mullins RD (2008) Capping protein increases the rate of actin-based motility by promoting filament nucleation by the Arp2/3 complex. Cell 133: 841–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BW, Bamburg JR (1982) Tropomyosin binding to F-actin protects the F-actin from disassembly by brain actin-depolymerizing factor (ADF). Cell Motil 2: 1–8 [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD, Hitchcock-DeGregori SE (2001) Inhibition of the Arp2/3 complex-nucleated actin polymerization and branch formation by tropomyosin. Curr Biol 11: 1300–1304 [DOI] [PubMed] [Google Scholar]
- Boussouf SE, Maytum R, Jaquet K, Geeves MA (2007) Role of tropomyosin isoforms in the calcium sensitivity of striated muscle thin filaments. J Muscle Res Cell Motil 28: 49–58 [DOI] [PubMed] [Google Scholar]
- Broschat KO, Weber A, Burgess DR (1989) Tropomyosin stabilizes the pointed end of actin filaments by slowing depolymerization. Biochemistry 28: 8501–8506 [DOI] [PubMed] [Google Scholar]
- Bugyi B, Le Clainche C, Romet-Lemonne G, Carlier MF (2008) How do in vitro reconstituted actin-based motility assays provide insight into in vivo behavior? FEBS Lett 582: 2086–2092 [DOI] [PubMed] [Google Scholar]
- Cai L, Makhov AM, Schafer DA, Bear JE (2008) Coronin 1B antagonizes cortactin and remodels Arp2/3-containing actin branches in lamellipodia. Cell 134: 828–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Laurent V, Santolini J, Melki R, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D (1997) Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J Cell Biol 136: 1307–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Pantaloni D, Korn ED (1986) Fluorescence measurements of the binding of cations to high-affinity and low-affinity sites on ATP-G-actin. J Biol Chem 261: 10778–10784 [PubMed] [Google Scholar]
- Carlsson AE (2003) Growth velocities of branched actin networks. Biophys J 84: 2907–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JA (2002) Actin dynamics: tropomyosin provides stability. Curr Biol 12: R523–R525 [DOI] [PubMed] [Google Scholar]
- Delatour V, Helfer E, Didry D, Le KH, Gaucher JF, Carlier MF, Romet-Lemonne G (2008) Arp2/3 controls the motile behavior of N-WASP-functionalized GUVs and modulates N-WASP surface distribution by mediating transient links with actin filaments. Biophys J 94: 4890–4905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesMarais V, Ichetovkin I, Condeelis J, Hitchcock-DeGregori SE (2002) Spatial regulation of actin dynamics: a tropomyosin-free, actin-rich compartment at the leading edge. J Cell Sci 115: 4649–4660 [DOI] [PubMed] [Google Scholar]
- Disanza A, Carlier MF, Stradal TE, Didry D, Frittoli E, Confalonieri S, Croce A, Wehland J, Di Fiore PP, Scita G (2004) Eps8 controls actin-based motility by capping the barbed ends of actin filaments. Nat Cell Biol 6: 1180–1188 [DOI] [PubMed] [Google Scholar]
- Gao L, Bretscher A (2008) Analysis of unregulated formin activity reveals how yeast can balance F-actin assembly between different microfilament-based organizations. Mol Biol Cell 19: 1474–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannone G, Dubin-Thaler BJ, Rossier O, Cai Y, Chaga O, Jiang G, Beaver W, Dobereiner HG, Freund Y, Borisy G, Sheetz MP (2007) Lamellipodial actin mechanically links myosin activity with adhesion-site formation. Cell 128: 561–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupton SL, Anderson KL, Kole TP, Fischer RS, Ponti A, Hitchcock-DeGregori SE, Danuser G, Fowler VM, Wirtz D, Hanein D, Waterman-Storer CM (2005) Cell migration without a lamellipodium: translation of actin dynamics into cell movement mediated by tropomyosin. J Cell Biol 168: 619–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupton SL, Eisenmann K, Alberts AS, Waterman-Storer CM (2007) mDia2 regulates actin and focal adhesion dynamics and organization in the lamella for efficient epithelial cell migration. J Cell Sci 120: 3475–3487 [DOI] [PubMed] [Google Scholar]
- Helfer E, Nevalainen EM, Naumanen P, Romero S, Didry D, Pantaloni D, Lappalainen P, Carlier MF (2006) Mammalian twinfilin sequesters ADP-G-actin and caps filament barbed ends: implications in motility. EMBO J 25: 1184–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillberg L, Zhao Rathje LS, Nyakern-Meazza M, Helfand B, Goldman RD, Schutt CE, Lindberg U (2006) Tropomyosins are present in lamellipodia of motile cells. Eur J Cell Biol 85: 399–409 [DOI] [PubMed] [Google Scholar]
- Huber F, Kas J, Stuhrmann B (2008) Growing actin networks form lamellipodium and lamellum by self-assembly. Biophys J 95: 5508–5523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isambert H, Venier P, Maggs AC, Fattoum A, Kassab R, Pantaloni D, Carlier MF (1995) Flexibility of actin filaments derived from thermal fluctuations. Effect of bound nucleotide, phalloidin, and muscle regulatory proteins. J Biol Chem 270: 11437–11444 [DOI] [PubMed] [Google Scholar]
- Iwasa JH, Mullins RD (2007) Spatial and temporal relationships between actin-filament nucleation, capping, and disassembly. Curr Biol 17: 395–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpova TS, Tatchell K, Cooper JA (1995) Actin filaments in yeast are unstable in the absence of capping protein or fimbrin. J Cell Biol 131: 1483–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiuchi T, Ohashi K, Kurita S, Mizuno K (2007) Cofilin promotes stimulus-induced lamellipodium formation by generating an abundant supply of actin monomers. J Cell Biol 177: 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koestler SA, Auinger S, Vinzenz M, Rottner K, Small JV (2008) Differentially oriented populations of actin filaments generated in lamellipodia collaborate in pushing and pausing at the cell front. Nat Cell Biol 10: 306–313 [DOI] [PubMed] [Google Scholar]
- Kouyama T, Mihashi K (1981) Fluorimetry study of N-(1-pyrenyl)iodoacetamide-labelled F-actin. Local structural change of actin protomer both on polymerization and on binding of heavy meromyosin. Eur J Biochem 114: 33–38 [PubMed] [Google Scholar]
- Lai FP, Szczodrak M, Block J, Faix J, Breitsprecher D, Mannherz HG, Stradal TE, Dunn GA, Small JV, Rottner K (2008) Arp2/3 complex interactions and actin network turnover in lamellipodia. EMBO J 27: 982–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine RO, Phaneuf KL, Cunningham CC, Kwiatkowski D, Azuma T, Southwick FS (1998) Gelsolin, a protein that caps the barbed ends and severs actin filaments, enhances the actin-based motility of Listeria monocytogenes in host cells. Infect Immun 66: 3775–3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Clainche C, Carlier MF (2004) Actin-based motility assay. Curr Protoc Cell Biol Chapter 12, unit 12.7.1–12.7.20 [DOI] [PubMed] [Google Scholar]
- Le Clainche C, Pantaloni D, Carlier MF (2003) ATP hydrolysis on actin-related protein 2/3 complex causes debranching of dendritic actin arrays. Proc Natl Acad Sci USA 100: 6337–6342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loisel TP, Boujemaa R, Pantaloni D, Carlier MF (1999) Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature 401: 613–616 [DOI] [PubMed] [Google Scholar]
- Machesky LM, Mullins RD, Higgs HN, Kaiser DA, Blanchoin L, May RC, Hall ME, Pollard TD (1999) Scar, a WASP-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc Natl Acad Sci USA 96: 3739–3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Tsuji T, Higashida C, Hertzog M, Fujita A, Narumiya S, Scita G, Watanabe N (2006) Actin turnover-dependent fast dissociation of capping protein in the dendritic nucleation actin network: evidence of frequent filament severing. J Cell Biol 175: 947–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilner A, Oster G (1996) Cell motility driven by actin polymerization. Biophys J 71: 3030–3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyakern-Meazza M, Narayan K, Schutt CE, Lindberg U (2002) Tropomyosin and gelsolin cooperate in controlling the microfilament system. J Biol Chem 277: 28774–28779 [DOI] [PubMed] [Google Scholar]
- Ono S, Ono K (2002) Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J Cell Biol 156: 1065–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaloni D, Boujemaa R, Didry D, Gounon P, Carlier MF (2000) The Arp2/3 complex branches filament barbed ends: functional antagonism with capping proteins. Nat Cell Biol 2: 385–391 [DOI] [PubMed] [Google Scholar]
- Pantaloni D, Le Clainche C, Carlier MF (2001) Mechanism of actin-based motility. Science 292: 1502–1506 [DOI] [PubMed] [Google Scholar]
- Pollard TD (1986) Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J Cell Biol 103: 2747–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponti A, Machacek M, Gupton SL, Waterman-Storer CM, Danuser G (2004) Two distinct actin networks drive the protrusion of migrating cells. Science 305: 1782–1786 [DOI] [PubMed] [Google Scholar]
- Sarmiento C, Wang W, Dovas A, Yamaguchi H, Sidani M, El-Sibai M, Desmarais V, Holman HA, Kitchen S, Backer JM, Alberts A, Condeelis J (2008) WASP family members and formin proteins coordinate regulation of cell protrusions in carcinoma cells. J Cell Biol 180: 1245–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaus TE, Taylor EW, Borisy GG (2007) Self-organization of actin filament orientation in the dendritic-nucleation/array-treadmilling model. Proc Natl Acad Sci USA 104: 7086–7091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie LB (1982) Preparation and identification of alpha- and beta-tropomyosins. Methods Enzymol 85 (Pt B): 234–241 [DOI] [PubMed] [Google Scholar]
- Svitkina TM, Borisy GG (1999) Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol 145: 1009–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallotton P, Small JV (2009) Shifting views on the leading role of the lamellipodium in cell migration: speckle tracking revisited. J Cell Sci 122: 1955–1958 [DOI] [PubMed] [Google Scholar]
- Wawro B, Greenfield NJ, Wear MA, Cooper JA, Higgs HN, Hitchcock-DeGregori SE (2007) Tropomyosin regulates elongation by formin at the fast-growing end of the actin filament. Biochemistry 46: 8146–8155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigt C, Wegner A, Koch MH (1991) Rate and mechanism of the assembly of tropomyosin with actin filaments. Biochemistry 30: 10700–10707 [DOI] [PubMed] [Google Scholar]
- Weisswange I, Newsome TP, Schleich S, Way M (2009) The rate of N-WASP exchange limits the extent of Arp2/3-complex-dependent actin-based motility. Nature 458: 87–91 [DOI] [PubMed] [Google Scholar]
- Wiesner S, Helfer E, Didry D, Ducouret G, Lafuma F, Carlier MF, Pantaloni D (2003) A biomimetic motility assay provides insight into the mechanism of actin-based motility. J Cell Biol 160: 387–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movie 1
Supplementary Movie 2
Supplementary Material
Review Process File