Abstract
Autophagy, a lysosomal degradation pathway, is essential for homeostasis, development, neurological diseases, and cancer. Regulation of autophagy in human disease is not well understood. Atg9 is a transmembrane protein required for autophagy, and it has been proposed that trafficking of Atg9 may regulate autophagy. Mammalian Atg9 traffics between the TGN and endosomes in basal conditions, and newly formed autophagosomes in response to signals inducing autophagy. We identified p38IP as a new mAtg9 interactor and showed that this interaction is regulated by p38α MAPK. p38IP is required for starvation-induced mAtg9 trafficking and autophagosome formation. Manipulation of p38IP and p38α alters mAtg9 localization, suggesting p38α regulates, through p38IP, the starvation-induced mAtg9 trafficking to forming autophagosomes. Furthermore, we show that p38α is a negative regulator of both basal autophagy and starvation-induced autophagy, and suggest that this regulation may be through a direct competition with mAtg9 for binding to p38IP. Our results provide evidence for a link between the MAPK pathway and the control of autophagy through mAtg9 and p38IP.
Keywords: autophagosome, endosome, starvation, TGN, trafficking
Introduction
Autophagy is a highly regulated mechanism allowing survival during times of cellular stress or nutrient deprivation. On initiation of autophagy, an elongating membrane, termed the isolation membrane or phagophore in mammalian cells, enwraps a portion of cytoplasm. Fusion of the edges of the membrane forms a double-membrane vesicle called an autophagosome. The autophagosome then fuses with endosomes and lysosomes forming an autolysosome in which the sequestered content is degraded (for review, see Reggiori and Klionsky, 2005).
Yeast genetics have revealed a number of autophagy-related (ATG) genes that are essential for the autophagic pathway (for review, see Klionsky, 2004). Many of these ATG genes have homologues in higher eukaryotes and recent studies have identified autophagy as a contributing factor to human health and disease (Cecconi and Levine, 2008). Among these ATG genes is a conserved subset of genes required specifically for autophagosome formation. This core machinery includes the transmembrane protein Atg9 and its cycling machinery, the phosphatidylinositol (PtdIns) 3-kinase complex, and the ubiquitin-like conjugation systems that include two ubiquitin-like proteins Atg8 and Atg12.
Yeast Atg8 and its mammalian homologues LC3, GABARAP, and GATE-16, are covalently linked to phosphatidylethanolamine in a ubiquitin-like conjugation pathway on induction of autophagy. This conversion of Atg8 from its unlipidated species (form-I) to the lipidated species (form-II), leads to its recruitment to the pre-autophagosomal structure (PAS) (Kirisako et al, 2000), the site of autophagosome formation in yeast, where it has shown to be involved in the expansion of the autophagosomal membrane (Xie et al, 2008). Most Atg proteins localize to the PAS, and the correct targeting of Atg proteins to this structure is necessary for autophagosome formation (Suzuki et al, 2007; Cao et al, 2008). However, Atg9, the only identified multi-spanning membrane protein required for autophagy, localizes to multiple punctate sites, one of which corresponds to the PAS (for review, see Webber et al, 2007). Furthermore, Atg9 cycles between the PAS and these punctate structures and is retrieved from completed autophagosomes dependent upon the Atg1–Atg13 kinase complex, Atg2, Atg18, and the PtdIns 3-kinase complex (Reggiori et al, 2004).
In mammalian cells under normal growth conditions mAtg9 is found to localize to the trans-Golgi network and peripheral endosomes (Young et al, 2006). After starvation, mAtg9 redistributes to a peripheral pool that co-localizes with GFP–LC3, a marker for autophagosomes (Young et al, 2006, Yamada et al, 2005). Depletion of mAtg9 from rat hepatocytes or 293/GFP–LC3 cells results in an inhibition of the autophagic pathway (Young et al, 2006). Although starvation-induced trafficking of mAtg9 is dependent on ULK1, a homologue of Atg1, (Young et al, 2006; Chan et al, 2007) and Atg13 (Chan et al, 2009), there are no known binding partners of mAtg9.
To gain a further understanding of the function of mAtg9 in mammalian autophagy, we used a yeast two-hybrid assay to screen a human cDNA library for potential interacting proteins. We identified p38-interacting protein (p38IP) as a binding partner for the C-terminal domain of mAtg9. p38IP was first identified in a screen for gastrulation and developmental defects in mice (Zohn et al, 2006). It was shown to bind to the mitogen-activated protein kinase (MAPK) p38α and be required for its activation in vivo. MAPKs are a family of serine/threonine kinases that regulate many cellular processes in response to a variety of stimuli. p38 MAPK is typically activated after cellular stress such as osmotic shock or ionizing radiation, however, it also has an important role in a number of physiological processes, as well as being implicated in a number of different pathologies (for review, see Nebreda and Porras, 2000). Evidence is also emerging for a role of p38 in autophagy. In rat hepatocytes, activation of p38 by osmotic stress leads to an inhibition in autophagic proteolysis (Haussinger et al, 1999). Furthermore, colorectal cancer cells treated with SB203580, a p38 inhibitor, are observed to upregulate GABARAP and undergo autophagic cell death (Comes et al, 2007). However, the molecular mechanisms of how p38 may regulate autophagy have yet to be elucidated.
Here, we provide evidence that the mAtg9–p38IP interaction is important for autophagy and is regulated by the p38α MAPK. Furthermore, we propose this interaction may provide a potential mechanism to limit or control autophagy that maybe subject to stimuli-specific regulation. As the upstream signalling events leading to autophagy are still largely unknown, p38α could provide the link to nutrient-dependent signalling cascades.
Results
Identification of p38IP, a new mAtg9-interacting protein
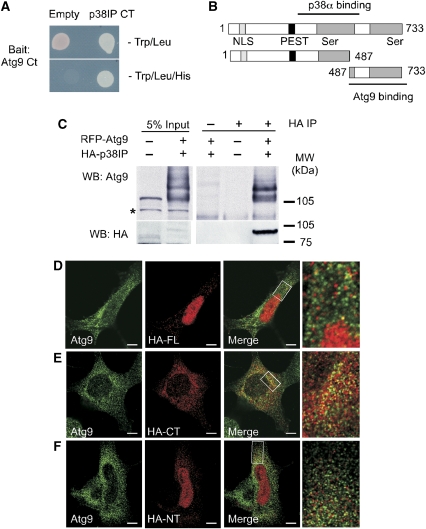
To gain additional understanding about the role of mAtg9 in mammalian autophagy, a yeast two-hybrid screen was performed to identify potential binding partners. Using the C-terminal (497–839 aa) domain of mAtg9 as bait, the initial screen of a human liver cDNA library identified a 1.2-kb fragment that encoded the C-terminal 246 amino acids of p38IP also known as FAM48A (NM_017569). This was confirmed in a directed yeast two-hybrid experiment (Figure 1A). p38IP, a 733-amino-acid protein predicted to have two serine-rich domains, a nuclear localization sequence (NLS) and PEST domain (Figure 1B), was previously identified as a binding partner for the MAPK p38α (Zohn et al, 2006). p38α was shown to bind a domain of p38IP containing the N-terminal serine-rich region (380–530 aa). To confirm the interaction of p38IP and mAtg9 in vivo, HEK293A cells were transiently transfected with RFP–mAtg9 and HA–p38IP. Co-immunoprecipitation showed efficient interaction between HA–p38IP and RFP–mAtg9 (Figure 1C). In addition, antibodies specific for endogenous mAtg9 were able to co-immunoprecipitate HA–p38IP (Figure 2D). We were unable to generate an antibody for endogenous p38IP and could not explore the interaction of endogenous p38IP with mAtg9.
Figure 1.
mAtg9 binds to p38IP in vitro and in vivo. (A) The AH109 strain was co-transformed with pGBKT7 containing the C-terminal (CT) domain of mAtg9 (bait) and pGADT7, or pACT2, containing the CT domain of p38IP (prey). The interaction was then visualized by growth of co-transformed yeast on SD-Trp/Leu/His plates for 3 days at 30°C. (B) Domain organization of p38IP, the N- (1–487 aa) and C- (487–733 aa) terminal domains. p38IP has a predicted nuclear localization signal (NLS), a PEST domain, and two serine-rich domains. The region of interaction with p38α and mAtg9 is indicated. (C) HEK293A cells were transfected with RFP–mAtg9 and HA–p38IP. HA–p38IP was immunoprecipitated with an anti-HA antibody and the co-immunoprecipitated RFP–mAtg9 was detected with an anti-mAtg9 antibody. HA–p38IP was detected with an anti-HA antibody. Asterisk indicates a non-specific band. Indirect immunofluorescence analysis of mAtg9 with (D) HA–full-length p38IP (HA–FL), (E) HA–C-terminal p38IP (HA–CT), and (F) HA–N-terminal p38IP (HA–NT). Bars=5 μm. The co-localization seen in (D) and (E) was not because of overlap of randomly distributed vesicles. Cross-correlation function (CCF) analysis (van Steensel et al, 1996) showed a peak and maximum Rp value around ΔX=0, indicating that co-localization between mAtg9 and p38IP is positively correlated and non random (Supplementary Figure S1).
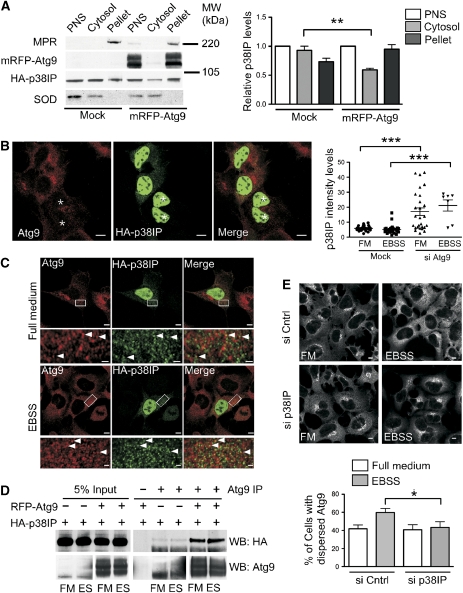
Figure 2.
p38IP is found on membranes, co-localizes with mAtg9 and is required for starvation-dependent trafficking of mAtg9. (A) HEK293A cells were transfected with HA–p38IP alone or co-transfected with HA–p38IP and RFP–mAtg9, homogenized and subjected to centrifugation, and the resulting post-nuclear supernatant (PNS) was fractionated by centrifugation at 100 000 g into membrane pellet and cytosol. Equal protein amounts were resolved by SDS–PAGE and immunoblotted with anti-mannose-6-phosphate receptor (MPR) antibody as a control for membrane-associated proteins, anti-superoxide dismutase (SOD) antibody as a control for cytosolic proteins, anti-mAtg9 antibody, and anti-HA antibody. HA–p38IP levels were quantified using ImageJ software and plotted relative to HA–p38IP in the PNS (n=4) (**P=0.0051). (B) HEK293 cells were depleted of mAtg9 using siRNA, then transfected with HA–p38IP and incubated in full medium or EBSS (data not shown). The asterisk indicates mAtg9-depleted cells. Bar is 5 μM. The intensity of the nuclear versus cytosolic fluorescence in control and cells depleted of mAtg9 under fed and starvation conditions was analysed using ImageJ software (***P=<0.0001, Students t-test). (C) HEK293A cells were transfected with HA–p38IP and processed for indirect immunofluorescence. HA–p38IP was detected with an anti-HA antibody and endogenous mAtg9 was detected with an anti-mAtg9 antibody. HA–p38IP localizes to the nucleus and cytoplasmic puncta in full medium or EBSS. HA–p38IP co-localizes with mAtg9 in the periphery of the cell (arrowheads). Bars (upper panels) 5 μM, (lower panels) 1 μM. (D) Anti-mAtg9 antibody was used to immunoprecipitate mAtg9 from HEK293 cells transfected with HA–p38IP alone or with RFP–mAtg9 and HA–p38IP. A total of 5% of lysates or immunoprecipitates were probed with anti-HA or anti-mAtg9 antibodies. The left and right hand bottom panels were obtained from the same immunoblot, but exposed for different times to allow the endogenous mAtg9 to be visualized better. (E) HEK293 cells were transfected with control or p38IP siRNA. At 72 h after transfection, cells were incubated in either full medium or EBSS for 2 h, then fixed and immunostained to detect endogenous mAtg9 localization. mAtg9 localization was quantified by visually scoring cells for either a juxta-nuclear or dispersed phenotype. Bars=5 μm (data are represented as mean±s.e.m. of 200 cells, *P=0.0413, Student's t-test).
We examined HEK293A cells transiently expressing low levels of HA–p38IP and observed that p38IP was localized to the nucleus, as well as to numerous punctate structures in the cytoplasm. Co-localization between mAtg9 and p38IP was found in peripheral punctate structures (Figure 1D). We confirmed that the C-terminal region of p38IP (487–733 aa) co-localized with mAtg9 by expressing this domain and the N-terminal domain (1–487 aa). The C-terminal domain of p38IP was found as a punctate cytosolic pool largely excluded from the nucleus (Figure 1E), and was co-localized with mAtg9 to a higher degree than the full-length p38IP (Supplementary Figure S1), whereas the N-terminal region was predominantly nuclear, exhibiting no significant co-localization with mAtg9 (Figure 1F). Thus, we have confirmed the C-terminal domain of p38IP interacts with mAtg9 in vivo.
p38IP localization to membranes through interaction with mAtg9
The punctate cytosolic co-localization of p38IP with mAtg9 suggested that a population of p38IP was membrane associated. We hypothesized that this localization may be due, in part, to its interaction with mAtg9. The requirement for mAtg9 in p38IP membrane association was tested by two approaches. We first used overexpression of mAtg9 to test whether this would increase the population of p38IP on membranes using subcellular fractionation (Figure 2A). A PNS was prepared from a lysate of HEK293A cells transiently transfected with HA–p38IP alone or co-transfected with RFP–mAtg9. After membranes were separated from cytosol by centrifugation, we observed that p38IP partitioned between the cytosol and membrane pellet. Overexpression of mAtg9 resulted in a depletion of the cytosolic pool of p38IP and an increase in the membrane pellet. Next, we used siRNA to deplete mAtg9 in HEK293 cells and analysed the distribution of p38IP between the cytosolic pool and nucleus (Figure 2B). We observed in cells depleted of mAtg9 the nucleus/cytoplasmic ratio of p38IP was significantly increased. These results support the idea that p38IP is associated to membranes through its interaction with mAtg9.
Previous studies in our laboratory have shown that mAtg9 resides in a juxta-nuclear region, corresponding to the trans-Golgi network (TGN), and in peripheral endosomes. After starvation, the juxta-nuclear pool of mAtg9 is diminished, leaving a dispersed peripheral pool that co-localizes with Rab7 and GFP–LC3, a marker for autophagosomes (Young et al, 2006). To test the sensitivity of the co-localization between p38IP and mAtg9 to starvation, HEK293 cells were incubated in either full medium or starvation medium (Earle's buffered saline solution (EBSS)). HA–p38IP and mAtg9 were found to co-localize in both full medium and EBSS (Figure 2C). In addition, the amount of p38IP in the nucleus was not affected by starvation (Figure 2B). To confirm the absence of a starvation-dependent alteration in mAtg9 and p38IP interaction, a co-immunoprecipitation experiment was performed using lysates from fed or starved cells. Immunoprecipitation of HA–p38IP by endogenous mAtg9 or RFP–mAtg9 was not affected by starvation (Figure 2D), confirming the co-localization data.
p38IP is required for starvation-induced mAtg9 trafficking
To investigate whether p38IP has a role in mAtg9 trafficking, HEK293A cells were transfected with siRNA directed against p38IP for 72 h before the induction of starvation. The level of p38IP siRNA-mediated knockdown was determined by RT–PCR (Supplementary Figure S2). As expected from our previous results, in cells treated with a control siRNA, mAtg9 resided in a juxta-nuclear position in full medium, and was dispersed on starvation (Figure 2E). However, in p38IP-depleted cells, mAtg9 did not redistribute from the juxta-nuclear pool to the peripheral pool. This result demonstrates a requirement for p38IP in starvation-induced mAtg9 trafficking and suggested that p38IP may have a role in autophagy.
p38IP is required for starvation-induced autophagy
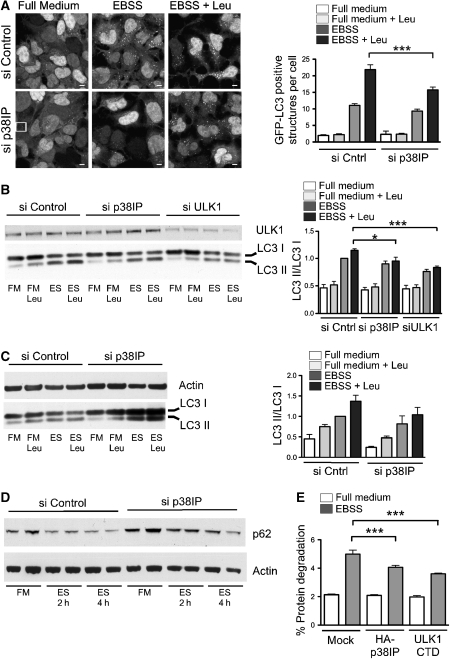
To test the hypothesis that p38IP has a role in starvation-induced autophagy, we monitored lipidation of LC3 in EBSS with and without leupeptin. LC3 or GFP–LC3 is lipidated after the induction of autophagy, resulting in a quantifiable shift in molecular weight and a translocation to punctate autophagosomal structures (Mizushima, 2004). We first used HEK293A cells stably expressing GFP–LC3 (293/GFP–LC3; Chan et al, 2007) to quantify the number of autophagosomes formed in starvation after p38IP depletion (Figure 3A). The 293/GFP–LC3 cells treated with either control or p38IP siRNA for 72 h were incubated in either full medium or EBSS for 2 h, and the localization of GFP–LC3 was analysed. In control siRNA-treated cells, numerous autophagosomes were observed after a 2-h starvation. Leupeptin was added to inhibit lysosomal proteases and accumulate autophagosomes, providing discrimination between the effects on autophagy formation and turnover. p38IP siRNA-mediated depletion had no effect in full medium, however, after starvation there was a significant reduction in the number of GFP–LC3-positive autophagosomes (Figure 3A).
Figure 3.
p38IP is required for mammalian autophagy.(A) 293/GFP–LC3 cells were transfected with control or p38IP siRNA. At 72 h after transfection, cells were incubated in either full medium (FM), full medium with leupeptin (FM Leu), EBSS (ES), or EBSS with leupeptin (ES Leu) for 2 h. FM Leu (data not shown) is identical to FM alone. Quantification of GFP–LC3-positive autophagosome number was performed by counting in a blinded experiment. Bars=5 μm (data are represented as mean±s.e.m. of 60 cells, ***P=0.0002, Students t-test). (B) HEK293A cells were transfected with siRNA control, siRNA for p38IP, and siRNA for ULK1. Cell lysates were analysed by SDS–PAGE for endogenous LC3 lipidation using an anti-LC3 antibody, or immunoblotted with anti-ULK1 antibody. (C) HeLa cells were transfected with siRNA control, and siRNA for p38IP. After incubation for 2 h as in (A), samples were immunoblotted with anti-LC3 and anti-actin antibodies. Endogenous LC3II/LC3I levels were quantified and the ratio presented as arbitrary units. In (B), n=4, ***P=<0.0001 and *P=0.0324, Students t-test. In (C), data are representative of two experiments. (D) 293/GFP–LC3 cells were transfected with siRNA control or siRNA p38IP, and incubated in full medium, or for 2 and 4 h in EBSS. Samples were immunoblotted with anti-p62 antibodies, and actin as a loading control. (E) HEK293A cells were transfected with HA–p38IP or myc–ULK1 C-terminal domain (CTD). After 24 h, cells were labelled with [14C]valine as described in Material and Methods section, and incubated in either full medium or EBSS for 2 h. Cells were then collected and analysed for long-lived protein degradation (data are represented as mean±s.e.m. of triplicates, representative of two experiments, EBSS mock versus EBSS HA–p38IP (***P=0.036); EBSS mock versus ULK1 CTD (***P=<0.0001); Students t-test).
To further quantify the extent of inhibition, we analysed the appearance of GFP–LC3II in 293/GFP–LC3 cells treated with either control or p38IP siRNA. In cells treated with control siRNA, a 2-h starvation with leupeptin induced GFP–LC3II (Supplementary Figure S3), whereas in cells treated with p38IP siRNA, levels of GFP–LC3II in starvation were significantly reduced. We next analysed the effects of p38IP depletion on autophagy by following endogenous LC3 lipidation in HEK293A and HeLa cells (Figure 3B and C). ULK1, the mammalian homologue of yeast Atg1, is required for starvation-induced autophagy in these cell lines (Chan et al, 2007) and depletion of this protein was used as a positive control. In both HEK293A and HeLa cells, p38IP depletion caused an inhibition of LC3II in EBSS with leupeptin (Figure 3B and C, respectively). In HEK293 cells, this inhibition was comparable to 50% of that observed by ULK1 depletion. The siRNA-mediated depletion of p38IP could be recovered by transfection of an siRNA-resistant HA–p38IP construct (Supplementary Figure S4). An LC3-independent measure of autophagy was then used to further assess the requirement for p38IP. We monitored the levels of p62/SQSTM1, a LC3-binding protein, which is sequestered and degraded in autophagosomes (Klionsky et al, 2008). As shown in Figure 3D, siRNA-depletion of p38IP resulted in an inhibition of p62/SQSTM1 degradation during starvation. Taken together these results indicate that p38IP is required for starvation-induced autophagy.
We hypothesized that p38IP levels may be important for the regulation of autophagy through its binding to mAtg9, and that overexpression of p38IP may cause an imbalance in the autophagy flux. We assessed the impact of p38IP overexpression by long-lived protein degradation. In starved cells, as expected from previous results, we found a two- to three-fold increase in long-lived protein degradation. This starvation-dependent increase was significantly reduced after overexpression of p38IP (Figure 3E). The extent of inhibition was comparable to that observed after overexpression of the C-terminal domain of ULK1, a potent inhibitor of autophagy (Chan et al, 2009).
Activation of p38 inhibits starvation-induced autophagy
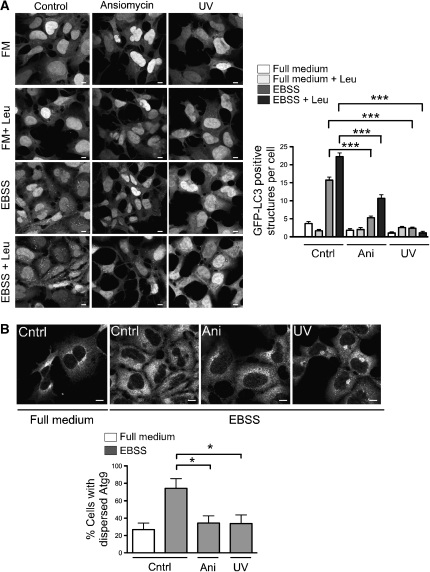
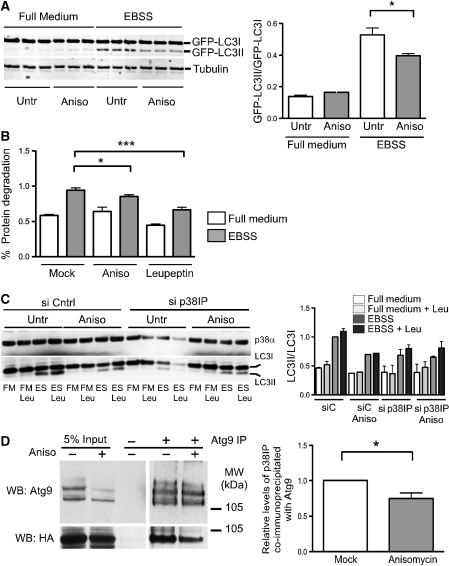
As p38IP was shown to bind p38α and is required for activation of p38α in vivo (Zohn et al, 2006), we analysed the effect of activation of p38 MAPK on the autophagic pathway. The 293/GFP–LC3 cells were pre-treated for 30 min with anisomycin, a protein synthesis inhibitor, or UV irradiated for 3 min followed by a 40-min recovery (Raingeaud et al, 1995, Hazzalin et al, 1998, Kang et al, 2006) to activate p38 MAPK. As shown in Figure 4A, both anisomycin and UV irradiation inhibited the formation of GFP–LC3-positive autophagosomes in EBSS, and in EBSS containing leupeptin in 293/GFP–LC3 cells. There was no effect of either treatment on cells in full medium. We next asked whether the activation of p38 also resulted in an alteration of mAtg9 distribution. As shown in Figure 4B, after anisomycin treatment or UV irradiation there was an inhibition of mAtg9 dispersion in EBSS. As we hypothesized that p38IP is associated with membranes through mAtg9 and that the distribution of mAtg9 is altered after p38 activation, we asked whether p38IP membrane-association was similarly altered by anisomycin. We found a decrease in the membrane-associated pool of p38IP after anisomycin treatment (Supplementary Figure S5), suggesting that p38IP interaction with mAtg9 is also decreased after p38 activation.
Figure 4.
Activation of p38 inhibits autophagy and mAtg9 trafficking. (A) 293/GFP–LC3 cells were treated with 10 μM anisomycin for 30 min, or exposed to UV irradiation for 3 min followed by a 40-min recovery and incubation in full medium, full medium with leupeptin, EBSS, or EBSS with leupeptin for 2 h. Cells were then fixed and visualized by confocal microscopy. GFP–LC3-positive structures per cell were quantified as in Figure 3A. Bars=5 μm. Data are represented as mean±s.e.m. n=60 cells, mock versus anisomycin EBSS (***P=<0.0001); mock versus UV EBSS (***P=<0.0001); mock versus anisomycin EBSS with leupeptin (***P=<0.0001); mock versus EBSS with leupeptin UV (***P=<0.0001). All analysed using Student's t-test. (B) HEK293A cells were pretreated with 10 μM anisomycin for 30 min, or UV irradiation for 3 min followed by a 40-min recovery, before incubation in EBSS for 2 h. Images were analysed as in Figure 2E. Bars=5 μm. Data are represented as mean±s.e.m. of 60 cells, EBSS mock versus EBSS anisomycin (*P=0.0142); EBSS mock versus EBSS UV (*P=0.0150); Student's t-test.
To quantify the effect of p38 MAPK activation on autophagy, GFP–LC3II levels were measured after activation of p38 followed by a 2-h starvation. We found that anisomycin treatment reduced GFP–LC3II levels (Figure 5A). Similar effects on GFP–LC3II levels were also observed when p38 was activated with UV irradiation (data not shown). To determine whether p38 activation also affects the autophagic proteolytic capacity of cells, we examined the affects on long-lived protein degradation. On activation of p38 with anisomycin 30 min before and during starvation, we observed an inhibition of protein degradation that was comparable to the inhibition observed with leupeptin (Figure 5B). Taken together these results suggest that activation of p38 is able to inhibit starvation-induced dispersal of mAtg9 and autophagy.
Figure 5.
The role of p38IP in p38α-dependent inhibition of autophagy. (A) 293/GFP–LC3 cells were pre-treated with 10 μM anisomycin for 30 min before incubation in either full medium or EBSS. Cell lysates were analysed by SDS–PAGE for GFP–LC3 lipidation using an anti-LC3 antibody. The membrane was also probed with anti-β tubulin. The GFP–LC3 lipidation was quantified as the amount of GFP–LC3II/GFP–LC3I (data are represented as mean±s.e.m. of triplicates, P=0.0091, Student's t-test). (B) HEK293A cells labelled with [14C]valine, as described in Material and Methods section, were pretreated with 10 μM anisomycin for 30 min before incubation in either full medium, EBSS, or EBSS with leupeptin for 2 h. Cells were then analysed for long-lived protein degradation (data are represented as mean±s.e.m. of triplicates, representative of two experiments, EBSS untreated versus anisomycin treated (P=0.0363); EBSS untreated versus leupeptin treated (P=<0.0001), Student's t-test). (C) HEK293 cells were transfected with control siRNA or siRNA for p38IP. After 24-h incubation, control or p38IP-depleted cells were pre-treated with anisomycin, followed by a 2-h incubation with full medium, full medium with leupeptin, EBSS, or EBSS with leupeptin. Cell lysates were analysed with anti-p38α or anti-LC3 antibodies. LC3II/LC3I levels were quantified and presented normalized to control, untreated cells incubated in EBSS. Data are representative of two experiments. (D) HEK293A cells transiently transfected with HA-p38IP and RFP-mAtg9 were treated with anisomycin for 30 min and mAtg9 was immunoprecipitated. 5% of the lysates or the immunoprecipitates were probed with anti-HA and anti-mAtg9 antibodies (data is representative of 3 experiments, *P=0.0332).
We next asked whether the reduction in autophagy after p38 activation was modulated by p38IP. The loss of p38IP was previously shown to cause a decrease in phosphorylated p38α and its downstream effectors ATF2 and CREB (Zohn et al, 2006). Thus, we predicted that loss of p38IP would result in a reduction in the inhibition of autophagy by anisomycin. However, this experiment was complicated by the requirement of p38IP for autophagy. By monitoring LC3II formation, as shown in Figure 5C, we found that siRNA depletion of p38IP inhibited LC3II formation, as did anisomycin treatment. The combined effects of siRNA depletion of p38IP and anisomycin resulted in an inhibition of LC3II formation comparable with the effect of either treatment alone. The lack of an additive effect suggests that p38α and p38IP function in the same pathway.
To expand these results, we examined the effect of loss of p38IP on the phosphorylation of p38α in our cell model. Zohn et al (2006) have shown that loss of p38IP decreases p38α phosphorylation in mutant mouse embryos. To test whether phospho-p38α levels are affected by p38IP depletion in our system, we probed for phospho-p38α after anisomycin treatment after siRNA depletion of p38IP. Surprisingly, we did not observe a decreased phosphorylation of p38α in the absence of p38IP (Supplementary Figure S6A).
Furthermore, as shown in Figure 3E, overexpression of p38IP inhibited long-lived protein degradation. Therefore, to determine the effect of overexpression of p38IP on p38α phosphorylation, we analysed cells untreated or treated with anisomycin after p38IP overexpression for phospho-p38α (Supplementary Figure S6B). p38IP overexpression alone did not significantly affect phospho-p38α levels, nor did it alter phospho-p38α after anisomycin treatment. Thus, in our cell model, the anisomycin-activated pool of phospho-p38α remains after loss of p38IP or overexpression of p38IP, and overexpression of p38IP does not cause an increase in phospho-p38α.
p38α regulates localization of p38IP and binding to mAtg9
Our data suggest that p38α is a negative regulator of autophagy and implies that its inhibitory effect is not regulated by a p38IP-dependent activation of p38α phosphorylation, but perhaps by a phosphorylation-dependent interaction with p38IP. Thus, ectopic activation of p38α would be predicted to cause a change in the localization of p38IP. Anisomycin treatment generates an increased pool of phosphorylated p38α, and causes a loss of p38IP from membranes (Supplementary Figure S5), suggesting that the increased cytosolic pool of p38IP results from an increased interaction with p38α. To test this, we determined whether p38IP interaction with p38α was increased when the pool of phosphorylated p38α was increased. We treated cells with anisomycin, immunoprecipitated phosphorylated p38α with a phospho-specific antibody, and probed for p38IP (Supplementary Figure S6C). After anisomycin treatment, we immunoprecipitated an increased amount of phospho-p38α and co-immunoprecipitated a proportionally increased amount of p38IP. Furthermore, after anisomycin treatment, the amount of p38IP co-immunoprecipitated with mAtg9 was reduced (Figure 5D).
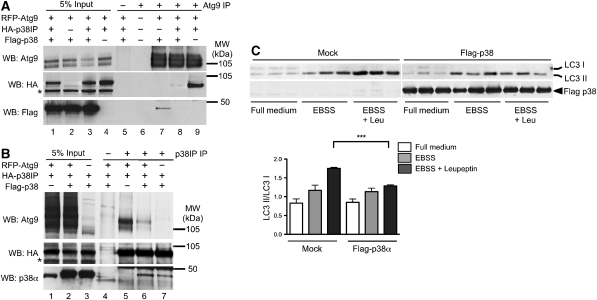
As activation of p38α results in a loss of p38IP on membranes, we asked whether p38α regulates p38IP binding to mAtg9. HEK293A cells were transfected with a combination of Flag–p38α, HA–p38IP, and RFP–mAtg9 and subjected to immunoprecipitation with an anti-mAtg9 antibody. p38IP co-immunoprecipitated with mAtg9 as expected (Figure 6A, lane 9). However, the interaction between HA–p38IP and RFP–mAtg9 was lost on overexpression of Flag–p38α (Figure 6A, lane 8 and 6B, lane 6). Interestingly, p38α was also observed to co-immunoprecipitate with mAtg9 in cells expressing RFP–mAtg9 and Flag–p38α (Figure 6A, lane 7) and this interaction was diminished on overexpression of HA–p38IP (Figure 6A, lane 8). The reciprocal experiment was performed by immunoprecipitation of p38IP (Figure 6B). On overexpression of all three proteins binding of mAtg9 to HA–p38IP was reduced, whereas binding of p38α was unaffected (Figure 6B, lanes 6 and 7). These results suggest that the affinity of p38α for p38IP is greater than that of mAtg9 for p38IP, and p38α can compete with mAtg9 for p38IP interaction. Taken together these results indicate that p38α regulates the binding of p38IP to mAtg9 and that this differential binding may be a mechanism by which p38α exerts its control on autophagy. In support of this, overexpression of p38α, leading to the recruitment of p38IP away from mAtg9, would also be predicted to inhibit autophagy. As shown in Figure 6C, overexpression of p38α in HEK293A cells significantly inhibited starvation-induced LC3II formation.
Figure 6.
Overexpression of p38α inhibits autophagy and competes with mAtg9 for binding to HA–p38IP. (A) HEK293A cells were transfected with RFP–mAtg9 and Flag–p38 (lanes 2 and 7), RFP–mAtg9, Flag–p38 and HA–p38IP (lanes 3 and 8), or RFP–mAtg9 and HA–p38IP (lanes 4 and 9). RFP–mAtg9 was immunoprecipitated with an anti-mAtg9 antibody, the co-immunoprecipitated HA–p38IP was detected with an anti-HA antibody, and co-immunoprecipitated Flag–p38 was detected with an anti-Flag antibody. RFP–mAtg9 was detected with an anti-Atg9 antibody. (B) HEK293A cells were transfected with RFP–Atg9 and HA–p38IP (lanes 1 and 5), RFP–mAtg9, Flag–p38, and HA–p38IP (lanes 2 and 6), or Flag–p38 and HA–p38IP (lanes 3 and 7). HA–p38IP was immunoprecipitated with an anti-HA antibody, the co-immunoprecipitated RFP–Atg9 was detected with an anti-Atg9 antibody, and co-immunoprecipitated Flag–p38 was detected with an anti-p38α antibody. HA–p38IP was detected with an anti-HA antibody. (C) HEK293A cells were transfected with Flag–p38α for 24 h. Cells were incubated in either full medium, EBSS, or EBSS plus leupeptin for 2 h, lysed, and analysed for endogenous LC3 lipidation using an anti-LC3 antibody, and immunoblotted with anti-Flag antibody. LC3 lipidation was quantified as the amount of LC3II/LC3I (data are represented as mean±s.e.m. of triplicates, n=2 experiments, EBSS control versus EBSS with Flag-p38α, ***P=0.0026).
p38α is a negative regulator of autophagy
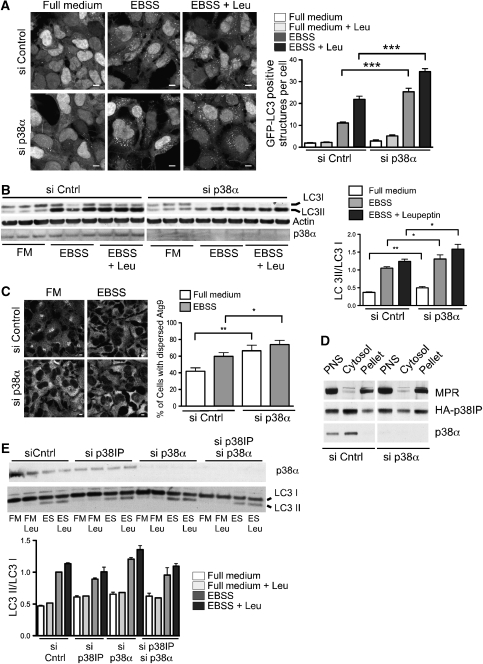
To further understand the role of p38α in autophagy, cells were depleted of p38α and autophagy was measured. We observed an increased number of GFP-positive autophagosomes after starvation in 293/GFP–LC3 cells depleted of p38α (Figure 7A). Interestingly, p38α depletion also resulted in an increased basal level of GFP–LC3-positive structures (Figure 7A, full medium). Furthermore, in Figure 7B, LC3 lipidation was analysed in HEK293A cells treated with control or p38α siRNA. In fed cells, depleted of p38α, levels of LC3II are greater than those seen in siRNA control-treated cells again suggesting an increase in basal autophagy. Furthermore, levels of LC3I decreased after starvation and conversion of LC3I to II increased (Figure 7B).
Figure 7.
p38α requires p38IP for inhibition of autophagy. (A) 293/GFP–LC3 cells were transfected with control or p38α siRNA. At 72 h after transfection, cells were incubated in either full medium, EBSS, or EBSS with leupeptin for 2 h, then fixed and visualized by confocal microscopy. Bars=5 μm (data are represented as mean±s.e.m. n=60 cells, EBSS control versus p38α siRNA (***P=<0.0001); EBSS with leupeptin control versus p38α siRNA (***P=<0.0001), Student's t-test). (B) HEK293A cells were transfected with control or p38α siRNA. At 72 h after transfection, cells were incubated in either full medium, EBSS, or EBSS containing leupeptin for 2 h. Cells lysates were analysed for LC3 lipidation using an anti-LC3 antibody. The membrane was also probed with anti-actin and anti-p38α antibodies. LC3 lipidation was quantified as the amount of LC3II/LC3I (data are represented as mean ±s.e.m. n=5, full medium control versus p38α siRNA (**P=0.0056); EBSS control versus p38α siRNA (**P=0.048); EBSS with Leu control versus p38α siRNA (**P=0.04), Student's t-test). (C) HEK293A cells were transfected with control or p38α siRNA. At 72 h after transfection, cells were incubated in either full medium or EBSS for 2 h, then fixed and immunostained to detect endogenous mAtg9 localization. The percentage of cells with dispersed mAtg9 was quantified as in Figure 2E (data are represented as mean±s.e.m. n=200 cells, full medium control versus p38α siRNA (**P=0.0027); EBSS control versus p38α siRNA (*P=0.0459); Student's t-test). Bars=5 μm (D) HEK293A cells were transfected with siRNA for p38α, homogenized and subjected to centrifugation, and the resulting post-nuclear supernatant (PNS) was fractionated by centrifugation at 100 000 g into membrane pellet and cytosol as in Figure 2A. Equal protein amounts were resolved by SDS–PAGE and immunoblotted with anti-MPR, anti-HA, and anti-p38α antibodies. Data are representative of four experiments. (E) HEK293A cells were transfected with control, p38α siRNA or both p38α siRNA, and p38IP siRNA. At 72 h after transfection, cells were incubated in either full medium, full medium with leupeptin, EBSS, or EBSS with leupeptin for 2 h. Cells lysates were analysed for LC3 lipidation using an anti-LC3 antibody. The membrane was also probed with anti-p38α. LC3 lipidation was quantified as the amount of LC3II/LC3I. Data are representative of two experiments.
We examined mAtg9 localization in fed HEK293A cells depleted of p38α and found that mAtg9 re-distributed and dispersed from the juxta-nuclear pool, in contrast to control cells in which it remained juxta-nuclear (Figure 7C). We quantified the effect of p38α depletion on mAtg9 localization and observed an increase in the dispersed mAtg9 phenotype in both full medium and after starvation compared with control siRNA. p38IP localization was also affected by the loss of p38α. As shown in Figure 7D, loss of p38α resulted in an increased amount of p38IP in the membrane pellet, suggesting an increased binding to mAtg9. These results suggest that p38α may be a negative regulator of autophagy in both basal and starvation conditions, and that this regulation is through p38IP and mAtg9 trafficking and distribution.
If the upregulation of autophagy, which occurs as a result of the loss of the negative regulator p38α, is via p38IP, then we would predict that loss of p38IP would ablate the upregulation due to the loss of the negative regulator. Thus, depletion of p38α using siRNA increased LC3II lipidation (Figure 7E), and siRNA depletion of p38IP reduced LC3II levels, whereas, as expected the combined depletion of p38α and p38IP resulted in a decrease in the p38α-mediated induction of autophagy.
Discussion
There are two central questions surrounding the autophagic process: how is autophagy initiated and how is it regulated? Our previous experiments have shown trafficking of mAtg9, a multi-spanning membrane protein, is required for autophagy, and our data presented here suggests that Atg9 trafficking is also important for the regulation of autophagy. mAtg9 redistributes to endosomes and autophagosomes from the TGN after amino-acid starvation (Young et al, 2006) dependent on ULK1 and Atg13 (Young et al, 2006; Chan et al, 2009). We have previously suggested that mAtg9 trafficking could respond to diverse cellular stimuli by using different sorting machinery for different transport steps (Webber et al, 2007). Our experiments aimed to identify new proteins that control mAtg9 trafficking, focusing on the cytosolic N- and C-terminal domains of mAtg9, which have no homology to the respective yeast domains. We identified p38IP (also known as Fam48A) through a yeast two-hybrid approach and have shown that it interacts in vivo with the C-terminal domain of mAtg9, and controls the trafficking of mAtg9 during starvation. It is noteworthy that this is, to the best of our knowledge, the first study to identify and characterize an mAtg9-interacting protein in a mammalian system.
p38IP has been shown to bind to p38α and is required for p38 MAPK activation during mouse development (Zohn et al, 2006). Our data is suggestive of a role for p38IP in the autophagic pathway, although it is not an autophagy-related protein. p38IP mutant mice (p38IPDrey) exhibit defects in neural tube closure, including exencephaly and spina bifida that are reminiscent of those observed in the Ambra1-deficient mice. Ambra1 is required for autophagy during neuronal development (Fimia et al, 2007). The defects in neural tube closure observed in the drey mutant mice may be due to a defect in autophagy, as well as specific defects in EMT observed by Zohn et al (2006). Further insight into this could be obtained by examining p38IPDrey mice to determine whether there is an autophagic defect.
Coordinated regulation of autophagy through mAtg9 and p38IP by p38α
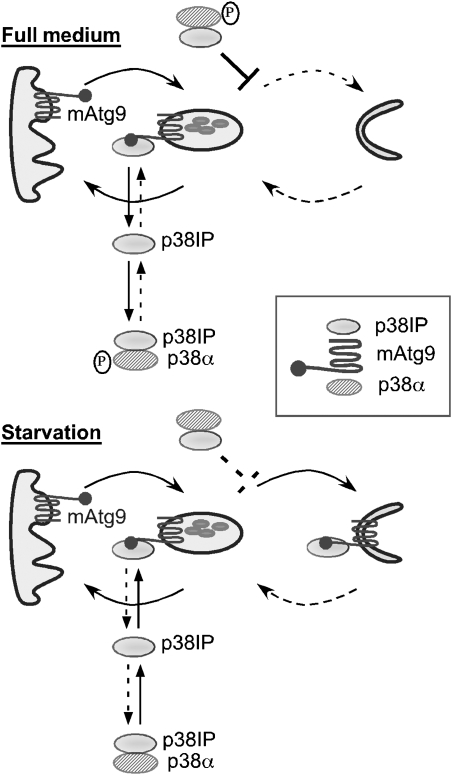
On the basis of our data, we propose a model that aims to explain our results and suggest a role for mAtg9 trafficking in the regulation of starvation-dependent autophagy (Figure 8). We suggest that in full medium a pool of p38α is phosphorylated and binds p38IP. Under these conditions, mAtg9 traffics between a juxta-nuclear pool and peripheral endosomes. p38IP is also bound to mAtg9 on peripheral endosomes, and this pool is in equilibrium with the pool bound to phospho-p38α. Autophagy occurs at a low basal rate, we hypothesize, because phospho-p38α binds p38IP with a higher affinity than mAtg9. Phospho-p38α bound to p38IP may also inhibit mAtg9 trafficking. On starvation, reduced amino-acid levels cause a decrease in phospho-p38α. The decreased affinity of p38IP for dephosphorylated p38α may favour the interaction between p38IP and mAtg9. Either the change in the equilibrium between p38IP and mAtg9, or the loss of the inhibition by phospho-p38α allows mAtg9 to traffic to forming autophagosomes, resulting in an increase in autophagy. We also speculate that recovery from starvation (data not shown in model) occurs when amino-acid pools are replenished, promoting phosphorylation of p38α and restoring the original equilibrium between p38α, p38IP, and mAtg9. During the initial recovery period, p38IP can dissociate from mAtg9 on the autophagosome enabling recycling of mAtg9 to the endosome.
Figure 8.
Model for regulation of autophagy through mAtg9, p38IP, and p38 MAPK. In full medium, mAtg9 traffics between the TGN and endosomes. Autophagy is inhibited by phosphorylated p38α. p38IP is found in the pool of mAtg9 in peripheral endosomes, and in complex with p38α. It should be noted that the two pools of p38α–p38IP are identical and shown separately for clarity. In starvation, p38α is dephosphorylated and binds p38IP with a lower affinity, and can no longer inhibit autophagy (shown as a dashed line). The pool of p38IP released from p38α would facilitate mAtg9 trafficking and autophagy.
p38α activation is modulated by nutrients and regulates p38IP binding to mAtg9
In support of our model, p38α has been shown to be activated on stimulation with amino acids (Casas-Terradellas et al, 2008), which agrees with our findings that levels of phosphorylated p38α are greater in full medium than after a 2-h starvation (Supplementary Figure S6). We have shown that levels of p38IP associated with membranes are reduced when p38α is activated, and predict that the amount of p38IP available to bind mAtg9 is less. We have also shown that activation of p38α by anisomycin results in an increase in the p38α–p38IP complex. In support of the hypothesis that under nutrient-rich conditions p38α preferentially binds p38IP, we find that when p38α, p38IP, and mAtg9 are overexpressed, the mAtg9–p38IP complex level is decreased. We believe this is due to a direct competition for binding to p38IP rather than a decrease in the interaction modulated by phosphorylation of either mAtg9 or p38IP by p38α, as neither p38IP nor mAtg9 immunoprecipitated from cell lysates was phosphorylated by recombinant activated p38α (JW and ST, unpublished observations).
In further support of this hypothesis, siRNA depletion of p38α in full medium results in the dispersion of mAtg9 from the trans-Golgi network, an increase in the membrane-associated pool of p38IP, and increased LC3II levels. Taken together, these data suggest that p38α functions to negatively regulate the p38IP–mAtg9 interaction in full medium, thereby preventing re-distribution of mAtg9 to the autophagosome thus controlling levels of autophagy in fed cells.
Induction of autophagy through p38IP binding to Atg9
We have shown that p38IP is required for mAtg9 trafficking under starvation conditions. In cells depleted of p38IP, mAtg9 remains in the trans-Golgi network and LC3 lipidation is inhibited. In addition, we have observed that p38IP and mAtg9 do not co-localize in the juxta-nuclear region under any conditions so far tested. Therefore, we hypothesize that mAtg9 and p38IP associate at the endosome and that during starvation, when the p38α-bound pool of p38IP is diminished, the mAtg9 associated with p38IP in a peripheral endosomal pool increases and instead of returning to the TGN mAtg9 is trafficked to the autophagosome. However, we did not observe an increase in HA–p38IP binding to mAtg9 during starvation (Figure 2D), as our model would predict. A plausible explanation for this result is that overexpression of HA–p38IP may alter the p38IP–mAtg9 and p38IP–p38α pools, and we speculate that overexpression of HA–p38IP may overcome the starvation-induced decrease in binding of p38IP to dephosphorylated p38α. We have shown that overexpression of p38α allows the interaction of p38IP and p38α to be restored resulting in a disruption in the mAtg9–HA–p38IP interaction (Figure 6A and B). As we were unable to develop an antibody against p38IP, we were unable to investigate the nutrient-dependent interaction of endogenous p38IP with mAtg9.
Previous data have shown that activated p38α is present on endosomes and regulates endocytosis (Cavalli et al, 2001; Delcroix et al, 2003; Pelkmans et al, 2005; Walchli et al, 2008). Although we observed in our cell lines that the majority of p38α is cytosolic, we found that activation of p38 increased the amount of p38α bound to p38IP, and decreased the amount of p38IP associated with membranes. Further study is required to determine where the activated pool of p38α, which binds p38IP, resides. However, as we have shown that when mAtg9 cycles constitutively in full medium it co-localizes with late endosomes, we speculate that mAtg9 may transiently associate with p38IP on this compartment, but may not be retained by p38IP and return to the TGN. During starvation, as p38α is dephosphorylated and the interaction of p38α–p38IP decreases, we speculate that the p38IP–mAtg9 interaction on the endosome would increase and facilitate autophagy, although this remains to be directly demonstrated. Our data rely on overexpression of p38IP that may alter the equilibrium between mAtg9, p38IP, and p38α. However, in support of this, we see that anisomycin inhibits retention of mAtg9 in the peripheral pool and inhibits LC3II formation and autophagy under starvation conditions.
Alternatively, after starvation mAtg9 could interact with proteins such as ULK1, or proteins regulated by ULK1 activity, and be retained on endosomes, allowing p38IP to be recruited to this compartment. In support of this, we find that depletion of ULK1 inhibits retention of mAtg9 at the peripheral pool. However, further experiments are required to determine how and where ULK1 exerts its influence on mAtg9 and whether this is sensitive to activation of p38α. In addition, insight into the question of how p38α regulates mAtg9 trafficking from the endosome could be obtained by studying yeast Atg9p distribution. Interestingly, it has been shown that the induction of autophagy by osmotic stress requires HOG1, the yeast p38α MAPK orthologue. However, so far no obvious p38IP orthologue has been identified in yeast, suggesting that the regulation in yeast may be different from mammalian cells.
Evidence for negative regulation by p38α
Conflicting data exist regarding the role of p38α in autophagy. In support of a role for p38 as a negative regulator, it has been shown that in rat liver when p38 is activated autophagy is inhibited (Haussinger et al, 1999). Consistently, in colorectal cancer cells, the p38 inhibitor, SB202190, was observed to induce autophagy and cell death. Furthermore, GABARAP, a homologue of LC3, was upregulated in these cells after 24-h SB202190 treatment (Comes et al, 2007). However, on accumulation of glial fibrillary acidic protein (GFAP) in astrocytes (Tang et al, 2008), p38 is activated and involved in the induction of autophagy. Addition of the diterpenoid, oridonin, in HeLa cells was previously reported to induce autophagy through p38 (Cui et al, 2007), although inhibition of JNK and not p38 was found to suppress oridonin-induced autophagy in a later study (Zhang et al, 2009). These conflicting results may suggest that the role of p38α in autophagy is dependent on the cell type or the stimulus responsible for its activation.
Our data suggest that in HEK293 cells, p38α MAPK is an important negative regulator of autophagy. Furthermore, we have shown that this negative regulation is diminished in the absence of p38IP, suggesting that the regulation of autophagy by p38α MAPK is through p38IP. Surprisingly, we did not observe any effect on phospho-levels of p38α after either overexpression or knockdown of p38IP. This is consistent with a model in which the phosphorylated form of p38α binds p38IP. We did, however, observe a decrease in levels of phospho-CREB and ATF2 (Supplementary Figure S6), suggesting that p38IP has a role in downstream events of the p38α MAPK pathway.
Conclusions
In this study, we have shown that p38IP is a new interacting protein of mAtg9 and that this interaction is regulated by the MAPK p38α. The effects observed on the autophagic pathway on manipulation of p38α are likely to be due to disruption of the starvation-induced trafficking of mAtg9 to forming autophagosomes. Yeast Atg9p is involved in the localization of Atg8p to the PAS (Suzuki et al, 2007), and we have shown that LC3 lipidation is inhibited after depletion of mAtg9 (Young et al, 2006). Geng and Klionsky (2008) have shown that recruitment of Atg8p and Atg9p to the PAS is increased during autophagy. Atg9 trafficking, therefore, is a key step to allow the progression of autophagosome formation.
In summary, our data support a role for mAtg9 in regulation of the autophagic pathway through interaction with p38IP, providing a mechanism by which p38α is able to exert its control on the autophagic pathway and potentially limit the extent of autophagy. This regulation may prevent autophagic cell death, thereby linking cellular signalling to autophagosome formation and protein degradation. Previous data has shown that inhibition of p38 leads to an induction of autophagy initially, but that prolonged p38 blockade results in cell death in colorectal cancer cells (Simone, 2007). It would, therefore, be beneficial to the cell to have a negative feedback mechanism by which p38α is re-activated after sufficient rounds of mAtg9 trafficking and protein degradation, preventing unwanted autophagic cell death. As autophagy is emerging as an important pathway in health and disease, further understanding the mechanism by which p38 is able to exert its influence on the autophagic pathway and mAtg9 trafficking in different cell types will be important for understanding potential points at which we can intervene and either inhibit or upregulate autophagy.
Materials and methods
Cell culture
HEK293A, HeLa, and 293/GFP–LC3 cells (previously described by Kochl et al (2006)) were maintained in DMEM supplemented with 10% FBS, which also served as full medium in analytical experiments. Starvation medium consisted of EBSS plus 0.5 mM leupeptin where indicated.
Antibodies and fluorescent reagents
Monoclonal anti-mAtg9 antibody was generated in hamsters injected with the C-terminal 20 aa. Polyclonal anti-mAtg9 has been described previously by Young et al (2006). Monoclonal anti-p38 and polyclonal anti phospho-p38 antibodies were obtained from NEB (UK). Polyclonal anti-Stx6 antibody was purchased from Synaptic Systems. Monoclonal anti-Flag M2 and monoclonal anti-actin antibodies were purchased from Sigma Aldrich (USA). Monoclonal anti-HA antibody was obtained from Covance (UK). Polyclonal anti-SOD antibody was purchased from Abcam. Polyclonal anti-MPR antibody was obtained from CRUK and monoclonal anti-p62 antibody was purchased from Santa Cruz Biotechnology. Polyclonal anti-beta-tubulin antibody was from Abcam. Fluorescent secondary antibodies were purchased from Molecular Probes (Invitrogen).
DNA constructs
Plasmids for HA–p38IP and Flag–p38 were kind gifts from Jiahuai Han (Scripps Research Institute, CA) and have been described previously by Zohn et al (2006). CT and NT-p38IP constructs were amplified by PCR using primers having an HA tag into a pcDNA 3.1 vector. Monomeric RFP–mAtg9 has been described previously by Young et al (2006).
Yeast two-hybrid screen
The yeast 2-hybrid screen was carried out by the Yeast Two-hybrid Core facilities at the University of Helsinki. The mAtg9 C-terminal domain (residues 1489–2520) was cloned into pGBKT7. A human liver library cloned into pACT2 was used for screening. The directed yeast two-hybrid analysis was performed using the Matchmaker 2-Hybrid System 3 (Clonetech). Transformants were selected on SD-trp/leu plates. The interaction was visualized by growth on SD-trp/leu/his plates after 3 days at 30°C.
siRNA knockdown
siRNA duplexes (HP GenomeWide siRNA) were obtained from Qiagen and Dharmacon. The duplex specific for p38α with most efficient level of knockdown by RT–PCR corresponds to: Hs_MAPK14_3_HP 5′-CAGTCCATCATTCATGCGAAA-3′. The siRNA targeting p38IP with the most efficient level of knockdown by RT–PCR corresponds to: Hs_FAM48A_10_HP 5′-TGCGATTCTTGCAGCATCAAA-3′ (Qiagen) and 5′-GCAACAAGCTTTAGAACTATT-3′ (Dharmacon). Experimental and control siRNAs were transfected into HEK293 and 293/GFP–LC38 cells using oligofectamine as described in the manufacturer's protocol. The siRNA-resistant plasmid was made with the following primer using multi-site-directed mutagenesis kit (Qiagen): 5′-ACTGAAACCGTGTCAGTTCAGTCTTCGGTATTGGGGAA-3′.
Confirmation of siRNA knockdown by RT–PCR was carried out in cell samples after 72 h of knockdown, using SYBR green RT–PCR as described by Chan et al (2007). Primer sets were obtained for detection of the p38IP and p38 transcripts, and the β-actin control.
Immunoprecipitation
Cells were lysed in ice-cold TNTE buffer (20 mM Tris (pH 7.5), 150 mM NaCl, 1% w/v Chaps, 5 mM EDTA) containing the Complete EDTA-free protease inhibitor cocktail (Roche). Lysates cleared by centrifugation were incubated with protein-G sepharose beads coupled to anti-HA monoclonal 12CA5 antibody (CRUK) or with protein-A Sepharose beads coupled to anti-Atg9 rabbit polyclonal antibody for 3 h and then washed thrice with TNTE. Proteins were eluted using 5 × SDS sample buffer (with heating to 65°C for 5 min) and analysed by SDS–PAGE.
Immunofluorescence microscopy
Immunofluorescence was performed as described previously by Young et al (2006). mAtg9 was analysed using a hamster anti-Atg9 monoclonal antibody generated using a C-terminal peptide described previously by Young et al (2006). Cells were imaged with a LSM 510 laser scanning confocal microscope equipped with a × 63, 1.4 NA, Plan Apochromat oil-immersion objective (both Carl Zeiss MicroImaging). Images were processed using LSM 510 software. p38IP intensity was measured in the nucleus and cytoplasm using Image J software and plotted as a ratio of p38IP intensity in the nucleus/p38IP intensity in the cytoplasm.
Immunoblotting
To detect GFP–LC3 lipidation, 293/GFP–LC3 cells were lysed, after various treatments, in 1 × SDS sample buffer. Lysates were then heated to 37°C for 5 min and passed through a 27G needle five times to reduce viscosity before analysis on 10% Laemmli SDS–PAGE. To detect lipidation of endogenous LC3, HEK293A cells were lysed after various treatments in ice-cold TNTE (20 mM Tris (pH 7.5), 150 mM NaCl, 0.3% v/v Triton X-100, 5 mM EDTA) containing Complete EDTA-free protease inhibitor cocktail (Roche). Lysates cleared by centrifugation were mixed with 5 × SDS sample buffer, heated to 37°C for 5 min, and then analyzed on 4–12% bis-Tris NuPAGE gels (MES SDS running conditions) (Invitrogen). Proteins were transferred to PVDF membranes. GFP–LC3 and endogenous LC3 were detected using 5F10 anti-LC3 monoclonal antibody (Nanotools, Teningen, Germany).
After incubation with primary antibodies, GFP–LC3 signals were detected and quantified using secondary antibodies coupled to infra-red chromophores and a 2-channel scanning method (Licor Odyssey) or chemilumensecence as described previously (Chan et al, 2007). Statistical analyses for various pairwise comparisons were performed using a Student's two-tailed t-test on sample sets with equal variances.
Long-lived protein degradation
Autophagy-dependent degradation of [14C]valine-labelled cellular proteins was measured in transfected HEK293A cells as previously described (Chan et al, 2007). The extent of long-lived protein degradation was expressed as the percentage of the TCA-soluble counts in the medium compared with the total TCA-soluble counts in the medium and TCA-insoluble counts from the cells.
Membrane association
After transfection and treatments as indicated, HEK293A cells were scraped into ice-cold Homogenization Buffer (HB) (20 mM HEPES (pH 7.2), 1 mM EGTA, 55 mM MgCl2, 150 mM KCl) containing the Complete EDTA-free protease (Roche). Cells were disrupted by passing 20 times through a 27G needle and then cleared using a low-speed (1000 g) centrifugation. These cleared lysates were then centrifuged at 100 000 g for 1 h (4°C) to obtain a crude membrane pellet and resulting supernatant fraction. Membrane pellets were finally re-suspended in ¼ volume of HB containing 1% Triton X-100. Membrane and supernatant fractions were mixed with 5 × SDS sample buffer, heated to 65°C for 5 min, and resolved by SDS–PAGE for immunoblotting.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure Legends
Acknowledgments
We thank Irene Zohn for advice, the SPW lab for all of their help, in particular, Joëlle Morvan and Minoo Razi for help with yeast two-hybrid screen and confocal imaging respectively, and Alex Massey for continuing support. We would also like to thank Joëlle Morvan, Minoo Razi, Harold Jefferies and Nicole McKnight for critical reading of the paper. Finally, we would like to thank Cancer Research UK for support.
References
- Cao Y, Cheong H, Song H, Klionsky DJ (2008) In vivo reconstitution of autophagy in Saccharomyces cerevisiae. J Cell Biol 182: 703–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas-Terradellas E, Tato I, Bartrons R, Ventura F, Rosa JL (2008) ERK and p38 pathways regulate amino acid signalling. Biochim Biophys Acta 1783: 2241–2254 [DOI] [PubMed] [Google Scholar]
- Cavalli V, Vilbois F, Corti M, Marcote MJ, Tamura K, Karin M, Arkinstall S, Gruenberg J (2001) The stress-induced MAP kinase p38 regulates endocytic trafficking via the GDI:Rab5 complex. Mol Cell 7: 421–432 [DOI] [PubMed] [Google Scholar]
- Cecconi F, Levine B (2008) The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell 15: 344–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EY, Kir S, Tooze SA (2007) siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J Biol Chem 282: 25464–25474 [DOI] [PubMed] [Google Scholar]
- Chan EY, Longatti A, McKnight NC, Tooze SA (2009) Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domain using an Atg13-independent mechanism. Mol Cell Biol 29: 157–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comes F, Matrone A, Lastella P, Nico B, Susca FC, Bagnulo R, Ingravallo G, Modica S, Lo Sasso G, Moschetta A, Guanti G, Simone C (2007) A novel cell type-specific role of p38alpha in the control of autophagy and cell death in colorectal cancer cells. Cell Death Differ 14: 693–702 [DOI] [PubMed] [Google Scholar]
- Cui Q, Tashiro S, Onodera S, Minami M, Ikejima T (2007) Oridonin induced autophagy in human cervical carcinoma HeLa cells through Ras, JNK, and P38 regulation. J Pharmacol Sci 105: 317–325 [DOI] [PubMed] [Google Scholar]
- Delcroix JD, Valletta JS, Wu C, Hunt SJ, Kowal AS, Mobley WC (2003) NGF signaling in sensory neurons: evidence that early endosomes carry NGF retrograde signals. Neuron 39: 69–84 [DOI] [PubMed] [Google Scholar]
- Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, Gruss P, Piacentini M, Chowdhury K, Cecconi F (2007) Ambra1 regulates autophagy and development of the nervous system. Nature 447: 1121–1125 [DOI] [PubMed] [Google Scholar]
- Geng J, Klionsky DJ (2008) Quantitative regulation of vesicle formation in yeast nonspecific autophagy. Autophagy 4: 955–957 [DOI] [PubMed] [Google Scholar]
- Haussinger D, Schliess F, Dombrowski F, Vom Dahl S (1999) Involvement of p38MAPK in the regulation of proteolysis by liver cell hydration. Gastroenterology 116: 921–935 [DOI] [PubMed] [Google Scholar]
- Hazzalin CA, Le Panse R, Cano E, Mahadevan LC (1998) Anisomycin selectively desensitizes signalling components involved in stress kinase activation and fos and jun induction. Mol Cell Biol 18: 1844–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang YJ, Seit-Nebi A, Davis RJ, Han J (2006) Multiple activation mechanisms of p38α mitogen-activated protein kinase. J Biol Chem 281: 26225–26234 [DOI] [PubMed] [Google Scholar]
- Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y (2000) The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol 151: 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ (2004) Cell biology: regulated self-cannibalism. Nature 431: 31–32 [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, Bamber BA, Bassham DC, Bergamini E, Bi X, Biard-Piechaczyk M, Blum JS, Bredesen DE, Brodsky JL, Brumell JH, Brunk UT et al. (2008) Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4: 151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochl R, Hu XW, Chan EY, Tooze SA (2006) Microtubules facilitate autophagosome formation and fusion of autophagosomes with endosomes. Traffic 7: 129–145 [DOI] [PubMed] [Google Scholar]
- Mizushima N (2004) Methods for monitoring autophagy. Int J Biochem Cell Biol 36: 2491–2502 [DOI] [PubMed] [Google Scholar]
- Nebreda AR, Porras A (2000) p38 MAP kinases: beyond the stress response. Trends Biochem Sci 25: 257–260 [DOI] [PubMed] [Google Scholar]
- Pelkmans L, Fava E, Grabner H, Hannus M, Habermann B, Krausz E, Zerial M (2005) Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature 436: 78–86 [DOI] [PubMed] [Google Scholar]
- Raingeaud J, Gupta S, Rogers JS, Dickens M, Han J, Ulevitch RJ, Davis RJ (1995) Pro-inflammatory cytokines and environemntal stress cause p38 mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine. J Biol Chem 270: 7420–7426 [DOI] [PubMed] [Google Scholar]
- Reggiori F, Klionsky DJ (2005) Autophagosomes: biogenesis from scratch? Curr Opin Cell Biol 17: 415–422 [DOI] [PubMed] [Google Scholar]
- Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ (2004) The Atg1–Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell 6: 79–90 [DOI] [PubMed] [Google Scholar]
- Simone C (2007) Signal-dependent control of autophagy and cell death in colorectal cancer cell: the role of the p38 pathway. Autophagy 3: 468–471 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Kubota Y, Sekito T, Ohsumi Y (2007) Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 12: 209–218 [DOI] [PubMed] [Google Scholar]
- Tang G, Yue Z, Talloczy Z, Hagemann T, Cho W, Messing A, Sulzer DL, Goldman JE (2008) Autophagy induced by Alexander disease-mutant GFAP accumulation is regulated by p38/MAPK and mTOR signaling pathways. Hum Mol Genet 17: 1540–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, van Binnendijk EP, Hornsby CD, van der Voort HT, Krozowski ZS, de Kloet ER, van Driel R (1996) Partial colocalization of glucocorticoid and mineralocorticoid receptors in discrete compartments in nuclei of rat hippocampus neurons. J Cell Sci 109 (Part 4): 787–792 [DOI] [PubMed] [Google Scholar]
- Walchli S, Skanland SS, Gregers TF, Lauvrak SU, Torgersen ML, Ying M, Kuroda S, Maturana A, Sandvig K (2008) The mitogen-activated protein kinase p38 links Shiga toxin-dependent signaling and trafficking. Mol Biol Cell 19: 95–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber JL, Young AR, Tooze SA (2007) Atg9 trafficking in Mammalian cells. Autophagy 3: 54–56 [DOI] [PubMed] [Google Scholar]
- Xie Z, Nair U, Klionsky DJ (2008) Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell 19: 3290–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Carson AR, Caniggia I, Umebayashi K, Yoshimori T, Nakabayashi K, Scherer SW 2005. Endothelial nitric-oxide synthase antisense (NOS3AS) gene encodes an autophagy-related protein (APG9-like2) highly expressed in trophoblast. J Biol Chem 280: 18283–18290 [DOI] [PubMed] [Google Scholar]
- Young AR, Chan EY, Hu XW, Kochl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA (2006) Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci 119 (Part 18): 3888–3900 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wu Y, Tashiro S, Onodera S, Ikejima T (2009) Involvement of PKC signal pathways in oridonin-induced autophagy in HeLa cells: a protective mechanism against apoptosis. Biochem Biophys Res Commun 378: 273–278 [DOI] [PubMed] [Google Scholar]
- Zohn IE, Li Y, Skolnik EY, Anderson KV, Han J, Niswander L (2006) p38 and a p38-interacting protein are critical for downregulation of E-cadherin during mouse gastrulation. Cell 125: 957–969 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure Legends