Abstract
TDP-43 is an RNA/DNA-binding protein implicated in transcriptional repression and mRNA processing. Inclusions of TDP-43 are hallmarks of frontotemporal dementia and amyotrophic lateral sclerosis. Besides aggregation of TDP-43, loss of nuclear localization is observed in disease. To identify relevant targets of TDP-43, we performed expression profiling. Thereby, histone deacetylase 6 (HDAC6) downregulation was discovered on TDP-43 silencing and confirmed at the mRNA and protein level in human embryonic kidney HEK293E and neuronal SH-SY5Y cells. This was accompanied by accumulation of the major HDAC6 substrate, acetyl-tubulin. HDAC6 levels were restored by re-expression of TDP-43, dependent on RNA binding and the C-terminal protein interaction domains. Moreover, TDP-43 bound specifically to HDAC6 mRNA arguing for a direct functional interaction. Importantly, in vivo validation in TDP-43 knockout Drosophila melanogaster confirmed the specific downregulation of HDAC6. HDAC6 is necessary for protein aggregate formation and degradation. Indeed, HDAC6-dependent reduction of cellular aggregate formation and increased cytotoxicity of polyQ-expanded ataxin-3 were found in TDP-43 silenced cells. In conclusion, loss of functional TDP-43 causes HDAC6 downregulation and might thereby contribute to pathogenesis.
Keywords: frontotemporal dementia, HDAC6, microarray, motoneuron disease, TDP-43
Introduction
Transactive response DNA-binding protein (TDP-43) is a 43 kDa protein abundant in most tissues and well conserved among mammals and invertebrates (Wang et al, 2004; Ayala et al, 2005). TDP-43 contains two RNA-recognition motifs (RRMs) and a C-terminal glycine-rich domain (GRD) and was originally cloned as a novel protein binding to transactivator responsive DNA sequences within human immunodeficiency virus type 1 and acting as a strong transcriptional repressor (Ou et al, 1995). In the mouse, TDP-43 mediates epigenetic transcriptional repression of the spermatid-specific acrosomal protein SP-10 in somatic tissue (Abhyankar et al, 2007). In addition to its strong DNA binding and transcriptional shut-off activity, TDP-43 binds to RNA and mediates exon skipping of the cystic fibrosis transmembrane conductance regulator (CFTR) and apolipoprotein A2 (APOA2), in cooperation with heterogenous nuclear ribonucleoprotein (hnRNP) A/B (Buratti and Baralle, 2001; Buratti et al, 2001, 2005; Mercado et al, 2005). TDP-43 not only mediates exon skipping but also exon 7 inclusion of the survival motor neuron protein 2 (SMN2) (Bose et al, 2008).
The reported functions of TDP-43 are mainly nuclear, where most of the protein resides. Within the nucleus, mouse TDP-43 isoforms were localized in a diffuse and a punctuated pattern (Wang et al, 2002). The punctuate structures were designated T-bodies and suggested to serve as a scaffold connecting nuclear bodies (NBs) (Wang et al, 2002). Consistently, we find co-localization of human TDP-43 with markers of RNA processing bodies suggesting an involvement in pre-mRNA splicing, and perhaps in other RNA processing events.
The identification of TDP-43 as the major protein in the neuropathological hallmark lesions of patients with frontotemporal lobar degeneration with ubiquitinated inclusions (FTLD-U, meanwhile renamed to FTLD-TDP (Mackenzie et al, 2009)) and in amyotrophic lateral sclerosis (ALS) emphasizes the importance of TDP-43 (Neumann et al, 2006). Distinct neuropathological profiles distinguish particular subtypes of sporadic and familial FTLD-U and ALS, with TDP-43 immunoreactivity being present in dystrophic neurites and neuronal cytoplasmic inclusions as well as in neuronal intranuclear inclusions (Kwong et al, 2007). Moreover, TDP-43-positive cytosolic aggregates in muscle cells have been described in familiar or sporadic inclusion body myopathies (IBM) (Weihl et al, 2008; Olivé et al, 2009).
Pathological modifications of TDP-43 include ubiquitination, phosphorylation, and protein fragmentation (Neumann et al, 2006). The association of TDP-43 with disease is further substantiated genetically. Meanwhile, 30 different TDP-43 mutations, which cluster in the C-terminal GRD, have been associated with motoneuron disease, ALS, and FTD (AD&FTD mutation database http://www.molgen.ua.ac.be/FTDmutations).
Most remarkably, neurons (Neumann et al, 2006, 2007) and muscle fibres (Olivé et al, 2009; Salajegheh et al, 2009) with cytosolic inclusions showed dramatic loss of nuclear TDP-43 staining. It has been suggested that cytosolic redistribution of TDP-43 is an early event in sporadic ALS, preceding the formation of insoluble aggregates (Giordana et al, 2009). Accordingly, a common neuropathological finding in FTLD-TDP are ‘preinclusions'—defined as a cell body lacking nuclear TDP-43 but with diffuse/granular cytoplasmic TDP-43 staining (Brandmeir et al, 2008). Thus, a loss of nuclear TDP-43 protein and thereby loss of nuclear function could lead to RNA processing defects in pathologic conditions. Interestingly, only recently mutations in another DNA/RNA-binding protein, fused in sarcoma/translocation in liposarcoma, have been linked to familial ALS (Kwiatkowski et al, 2009; Vance et al, 2009), further supporting the hypothesis that defects in RNA processing may have a central function in FTLD/ALS. However, the currently known TDP-43 target mRNAs have not resolved mechanistic effects of TDP-43 misfunction in neurodegenerative diseases.
To gain further insight into the (patho)physiologically relevant functions of TDP-43, we depleted cells of TDP-43 by RNA interference. To identify TDP-43 responsive genes, we performed differential microarray analysis of human embryonic kidney HEK293E cells treated with siRNATDP-43 versus scrambled siRNA. Among the consistently altered genes, we identified histone deacetylase 6 (HDAC6). Specific downregulation of HDAC6 on TDP-43 silencing was confirmed at the mRNA and protein level in transiently as well as stably silenced non-neuronal and neuronal cell lines. Concomitant with the downregulation of HDAC6, TDP-43 silencing led to the accumulation of a major HDAC6 substrate, acetyl-tubulin (Hubbert et al, 2002). HDAC6 downregulation could be reverted after transfection with wild-type (wt) TDP-43, but not mutants lacking the RRMs, the GRD, or the nuclear localization signal (NLS). In pull-down and immunoprecipitation experiments we found that TDP-43 specifically bound to HDAC6 mRNA. Specific reduction of HDAC6 mRNA and protein was confirmed in vivo in Drosophila melanogaster lacking the TDP-43 ortholog gene, TBPH. The pathological consequences of reduced HDAC6 expression on TDP-43 silencing were assessed using ataxin-3 (Atx3) as experimental proteotoxic protein. In an HDAC6-dependent manner, TDP-43 knockdown in HEK293E cells reduced the formation of aggregates and increased cell death on transfection with Atx3 containing an expanded polyQ tract. Therefore, HDAC6 reduction and thus lowered cytosolic deacetylase activity might contribute to pathogenesis due to TDP-43 impairment.
Results
Endogenous TDP-43 in human cell lines is localized mostly in the nucleus
To characterize the distribution of endogenous TDP-43, we initially performed biochemical fractionation experiments in HEK293E cells. TDP-43 was detected mostly in the nuclear fraction and very little in the cytosol (Supplementary Figure S1A). Immunostaining further confirmed the predominant nuclear localization of TDP-43 in non-neuronal HEK293E cells (Supplementary Figure S1B) as well as in neuroblastoma SH-SY5Y cells (Supplementary Figure S1C). In addition, weak but specific granular staining of endogenous TDP-43 in the cytosol was observed. Thus, TDP-43 is a mostly nuclear protein with a small fraction in the cytosol (Wang et al, 2008).
Within the nucleus, endogenous TDP-43 is distributed in a punctuate pattern throughout the chromatin but excluded from nucleoli. Most cells display a discrete number of NBs with enriched TDP-43 signal. To characterize these T-bodies further, we performed dual-label confocal fluorescence microscopy of endogenous TDP-43 and marker proteins for known NBs. Those NBs have functions in small nuclear ribonucleoprotein biogenesis as the Cajal bodies and the closely associated gemini of Cajal bodies (gems), act as sites of splicing factor storage like SC35 speckles or have a role in transcriptional regulation as the promyelocytic leukaemia (PML) NBs (reviewed in Bernardi and Pandolfi, 2007; reviewed in Collins and Penny, 2009). Most overlap of T-body signal was observed with gems and Cajal bodies (Supplementary Figure 2A–F). Note that TDP-43 was completely buried within Cajal bodies (Supplementary Movie S1). Furthermore, T-bodies were closely associated with SC35 speckles, yet much SC35 signal was found without TDP-43 immunoreactivity (Supplementary Figure 2G–I). Finally, there was little overlap with PML NBs (Supplementary Figure 2J–L). Thus, in general agreement with a previous description of transfected mouse TDP-43 (Wang et al, 2002), endogenous human TDP-43 is diffusely distributed within the nucleus and enriched in NBs showing considerable co-localization with gems and Cajal bodies. Such distribution of TDP-43 is consistent with its broad functions reported on transcription and RNA processing and possibly as nuclear body scaffolding protein.
Identification of TDP-43 target genes
To gain insight into the physiological functions of TDP-43, we performed RNA silencing experiments in HEK293E cells. TDP-43 knockdown of up to 90% was not overtly cytotoxic and did not alter the number or morphology of TDP-43 co-localizing NBs (results not shown). As knockdown experiments did not support a general structural role of TDP-43, we screened for specific mRNA targets by microarray expression profiling. Four independent cultures of HEK293E cells were treated with siRNA against TDP-43 or control scrambled siRNA. Total RNA was extracted and each hybridized on a Human Genome U133+ 2.0 array. Knockdown efficiency was simultaneously assessed by western blot (WB) analysis of the same samples used for RNA extraction and hybridization (Supplementary Figure S3).
We first examined genes associated with the spectrum of FTDs (Kumar-Singh and Van Broeckhoven, 2007) and OMIM-annotated ALS genes (http://www.ncbi.nlm.nih.gov/sites/entrez?db=omim). As expected, TDP-43 mRNA levels were reduced more than three-fold after silencing (Table I). In contrast, expressions of the known FTD/ALS-associated genes were only minimally altered on TDP-43 knockdown. Thus, TDP-43 does not directly modulate the expression of known FTD and ALS genes in siRNATDP-43-treated HEK293E cells.
Table 1.
Influence of TDP-43 knockdown in HEK293E cells on FTD and ALS candidate genes
| Candidate genes | Fold change |
|---|---|
| FTD-associated genes | |
| MAPT | +1.25 |
| PGRN | +1.16 |
| VCP | −1.15 |
| CHMP2B | +1.08 |
| ALS-associated genes | |
| SOD1 | +1.04 |
| ALS2 | +1.16 |
| SETX | −1.05 |
| FUS/TLS | −1.27 |
| VAPB | −1.31 |
| ANG | +1.05 |
| TARDBP (TDP-43) | −3.26 |
| FIG4 | +1.08 |
| Novel TDP-43 target gene | |
| HDAC6 | −2.04 |
| Genes associated with FTD and ALS are listed together with their signal change of the GeneChip Human Genome U133+ 2.0 microarray. | |
The known TDP-43 splice targets, CFTR and APOA2 (Buratti and Baralle, 2001; Mercado et al, 2005), were expressed below meaningful detection threshold levels in HEK293E cells, precluding the analysis of TDP-43 knockdown effects. Expression of the TDP-43 splice target SMN2 was only minimally changed after TDP-43 knockdown, consistent with the original report (Bose et al, 2008). Alterations of neurofilament light chain expression (Strong et al, 2007) were not confirmed in siRNATDP-43-treated HEK293E cells. Likewise, the reported targets of TDP-43 from another microarray study performed in HeLa cells (Ayala et al, 2008) could not be confirmed in our experimental system (data not shown).
We identified a total of 402 differentially expressed genes in cells treated with siRNATDP-43 compared with scrambled siRNA controls (Supplementary Table SI). We further validated 15 altered transcripts by RT–PCR using four different TDP-43-specific siRNAs (Supplementary Figure S4). However, 60% of the tested transcripts did not pass the employed high stringency validation criteria and were thus considered as possible off-target effects. Other transcripts were regulated by each of the four TDP-43-directed siRNAs (see Supplementary Figure S4 for details). An appealing candidate among those was HDAC6.
HDAC6 is an unusual deacetylase characterized by cytoplasmic localization, ubiquitin-binding capacity, and impact on tubulin and actin cytoskeleton (Boyault et al, 2007a). Furthermore, HDAC6 has been implicated in autophagic degradation of aggregated proteins and was shown to rescue neurodegeneration in a fly model of spinal muscular atrophy (Pandey et al, 2007). Together with valosin-containing protein (VCP), another gene associated with a particular form of FTD (Watts et al, 2004), HDAC6 decides the fate of polyubiquitinated proteins (Boyault et al, 2006) and therefore putatively has an important function in proteinopathies. Owing to its functions regulating the turnover of aggregation-prone proteins associated with neurodegenerative diseases, we selected HDAC6 for further validation.
Validation of HDAC6 downregulation by TDP-43 knockdown
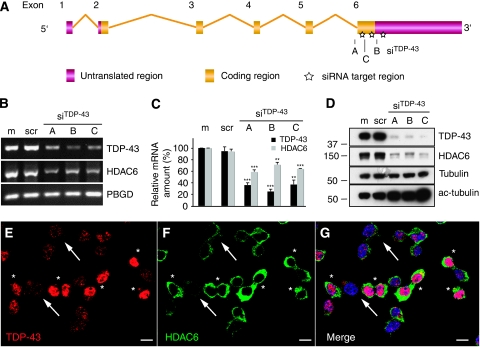
To validate the HDAC6 downregulation by TDP-43 knockdown, we treated HEK293E cells with the same siRNATDP-43 and scrambled control as for the microarray hybridizations and additionally used two more siRNAs directed against TDP-43 (Figure 1A). All siRNAs reduced TDP-43 expression and significantly downregulated HDAC6 mRNA, as visualized by semi-quantitative qRT–PCR (Figure 1B; Supplementary Figure S4) and quantitated by real-time RT–PCR (Figure 1C). This effect was confirmed at the protein level: strong knockdown of TDP-43 protein was consistently accompanied by reduced HDAC6 protein on WBs (Figure 1D). Functional depletion of the HDAC6 enzyme was shown by the concomitant increase of acetyl-tubulin (Figure 1D), a major cellular HDAC6 substrate (Hubbert et al, 2002). In addition, downregulation of HDAC6 by TDP-43 silencing was confirmed at the single cell level by dual-label confocal microscopy. Effectively silenced cells expressing low levels of TDP-43 also showed low levels of HDAC6, in contrast to adjacent non-transfected cells showing robust expression of both TDP-43 and HDAC6 (Figure 1E–G).
Figure 1.
Validation of HDAC6 downregulation in TDP-43 silenced HEK293E cells. (A) Positions of siRNA sequences within the TDP-43 locus. Boxes represent exons (gold, coding; pink, non-coding). (B–D) HEK293E cells were mock transfected (m), treated with scrambled (scr) siRNA, or the indicated TDP-43-directed siRNAs. Total RNA was extracted and subjected to semi-quantitative RT–PCR (B) using primer pairs amplifying TDP-43, HDAC6 and the housekeeping gene porphobilinogen deaminase (PBGD), as indicated. Relative qRT–PCR quantification (C) of TDP-43 (black bars) and HDAC6 (grey bars) mRNA was normalized to PBGD. Values represent the mean of three independent experiments. Compared with mock transfection, TDP-43 knockdown significantly reduced TDP-43 and HDAC6 qRT–PCR signals (**P<0.002; ***P<0.0001). (D) Cell lysates were western blotted and probed with antibodies against TDP-43, HDAC6, total α-tubulin, and acetyl-tubulin, as indicated. (E–G) HEK293E cells were transfected with siRNATDP-43 B and stained with mouse anti-TDP-43 (E, red) and rabbit anti-HDAC6 (F, green). Arrows point to transfected cells with strong TDP-43 knockdown, asterisks label cells with little knockdown. Nuclei were counterstained with Hoechst 33342 (blue); merged image is shown to the right (G). Size bars=10 μm.
Validation of HDAC6 decrease after stable lentiviral knockdown of TDP-43 in non-neuronal and neuronal cell lines
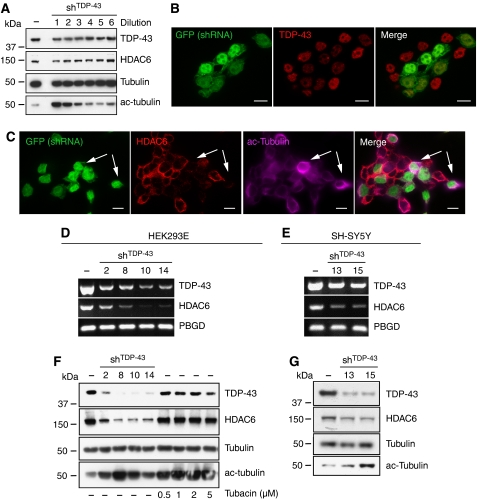
To rule out artefacts because of acute cellular stress caused by transient transfection, we validated HDAC6 downregulation in HEK293E cells with stably integrated shRNATDP-43 after lentiviral transduction. In a dose-dependent manner TDP-43 knockdown was accompanied by HDAC6 protein downregulation (Figure 2A). Even partial transduction of HEK293E cells (vector dilution 1:1024) resulted in TDP-43 knockdown (Figure 2B), strong HDAC6 decrease, and consequent acetyl-tubulin increase on single cell level as seen by dual-label confocal microscopy (Figure 2C). Stable clones of shRNATDP-43-treated HEK293E cells showed robust TDP-43 knockdown as well as strong HDAC6 mRNA (Figure 2D) and protein reduction (Figure 2F). The concomitant accumulation of the HDAC6 substrate acetyl-tubulin was comparable to the effects achieved with the specific HDAC6 inhibitor tubacin (Haggarty et al, 2003) (Figure 2F).
Figure 2.
HDAC6 downregulation in stably TDP-43 silenced cells. (A) HEK293E cells were treated with serial dilutions of lenti-shRNATDP-43 (lane 1, 1:4; lane 2, 1:16; lane 3, 1:64; lane 4, 1:256; lane 5, 1:1024; lane 6, 1:4096) or left untreated (−). WBs were prepared from cell lysates and probed with antibodies against TDP-43, HDAC6, α-tubulin, and acetyl-tubulin, as indicated. (B, C) HEK293E cells transduced with the 5th dilution (1:1024 of vector particles) were stained with anti-TDP-43 (B) or with anti-HDAC6 and anti-acetyl-tubulin (C). Epifluorescence shows the expression of green fluorescent protein (GFP) by the viral vector cassette. Note that even after low-dose transduction, successfully transduced HEK293E cells (GFP positive) have considerably strong downregulation of TDP-43 (B) and show also decrease in HDAC6 and increase in acetyl-tubulin (arrows in C). Scale bars=10 μm. (D–G) Single cell clones of shRNATDP-43-treated HEK293E (D, F) or SH-SY5Y neuroblastoma cells (E, G) were analysed for RNA and protein levels. (D, E) RNA was extracted from parental cell lines (−) and single clones and subjected to semi-quantitative RT–PCR using primer pairs against TDP-43, HDAC6, and the housekeeping gene PBGD. (F, G) WBs were prepared from protein lysates and probed with antibodies against TDP-43, HDAC6, total α-tubulin, and acetyl-tubulin, as indicated. Some cultures were treated for 5 h with the indicated concentrations of tubacin.
We furthermore used SH-SY5Y cells for validation in a neuronal cell type. Stable silencing of TDP-43 on lentiviral transduction of shRNATDP-43 in SH-SY5Y cell clones also consistently caused downregulation of HDAC6 mRNA (Figure 2E) and protein (Figure 2G). Thus, TDP-43 knockdown downregulates HDAC6 mRNA and protein expression, and reduces HDAC6 enzyme activity in non-neuronal as well as in neuronal cells.
wt TDP-43 retransfection restores HDAC6 expression
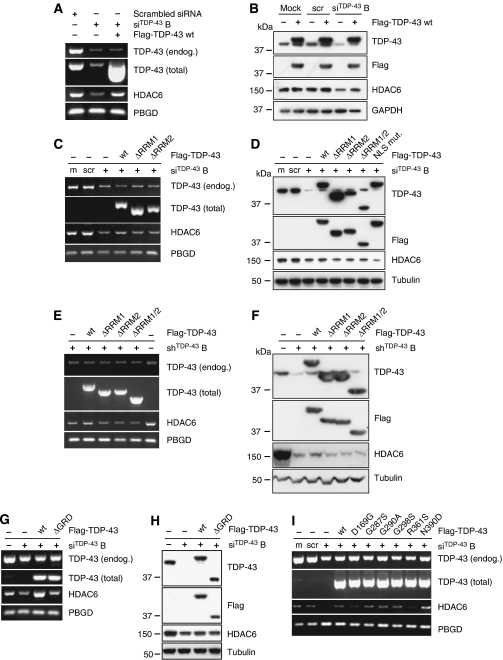
To show that HDAC6 decrease on TDP-43 silencing is a specific effect, we made use of one of the siRNAs, which targets the 3′UTR of TDP-43 (siRNATDP-43 B; see Figure 1A), and thus allows efficient retransfection with TDP-43 cDNA. HDAC6 mRNA (Figure 3A) and protein (Figure 3B) could be restored after retransfection with wt TDP-43, showing the specific dependence of HDAC6 levels on TDP-43. However, overexpression of TDP-43 wt alone did not cause further HDAC6 increase, suggesting that endogenous TDP-43 amounts are already saturating for HDAC6 expression.
Figure 3.
HDAC6 expression is specifically restored by TDP-43 retransfection. (A–D) HEK293E cells were mock transfected (m), treated with scrambled (scr) siRNA or siRNATDP-43 B. After 24 h cells were transfected with empty vector (−), or with mutant or wt Flag-TDP-43, as indicated. (A, C). Total RNA was extracted and subjected to semi-quantitative RT–PCR amplifying endogenous TDP-43, total TDP-43, HDAC6, and the housekeeping gene PBGD, as indicated. (B, D). WBs were prepared from cell lysates and probed with antibodies against TDP-43, HDAC6, total α-tubulin, or GAPDH, as indicated. (E, F). Stably silenced shRNATDP-43 HEK293E cells were retransfected with empty vector (−) or the indicated Flag-TDP-43 constructs. Extracted RNA was used for semi-quantitatve RT–PCR (E), protein lysates for WBs (F) as described above. (G, H) HEK293E cells were mock transfected (−) or transfected with siRNATDP-43 B. After 24 h cells were transfected with empty vector (−), or with wt or C-terminally truncated (ΔGRD) Flag-TDP-43. Extracted RNA was used for semi-quantitatve RT–PCR (G), protein lysates for WBs (H), as described above. (I) HEK293E cells were mock transfected (m), treated with scrambled (scr) or siRNATDP-43 B. After 24 h, TDP-43 silenced cells were transfected with empty vector (−), Flag-TDP-43 wt, or the indicated point mutants. Total RNA was extracted and subjected to semi-quantitative RT–PCR as described above.
TDP-43 contains two RRM domains that are involved in specific RNA processing functions. We have generated deletion constructs lacking RRM1 (ΔRRM1), RRM2 (ΔRRM2), and both RRMs (ΔRRM1/2). These deletion constructs failed to restore HDAC6 mRNA and protein levels on TDP-43 knockdown in transiently and stably silenced HEK293E cells (Figure 3C–F). Moreover, TDP-43 mutant variants with either impaired nuclear localization (NLSmut) (Winton et al, 2008) (Figure 3D) or lacking the C-terminal GRD (ΔGRD) (Figure 3G and H) both failed to restore HDAC6 levels. The GRD has been shown to be responsible for the interaction with hnRNPs and necessary for the exon skipping function towards CFTR (Buratti and Baralle, 2001). Thus, nuclear localization of TDP-43 as well as its nucleic acid and protein-binding capacities are important determinants of HDAC6 regulation.
To assess the effects of clinical mutations in the TDP-43 gene, we performed rescue experiments transfecting a number of full-length TDP-43 missense mutants into silenced cells. In these experiments, most disease-associated TDP-43 mutants behaved like wt, consistent with the failure to detect significant CFTR reporter splicing alterations of clinical TDP-43 mutations in the context of the full-length protein (Nonaka et al, 2009). However, [D169G]TDP-43 consistently showed a trend of reduced rescue capacity, and the R361S mutation (Kabashi et al, 2008) completely impaired the ability of TDP-43 to restore HDAC6 mRNA expression (Figure 3I). Interestingly, the R361S mutation is located within the minimal hnRNP A2 interaction domain of TDP-43 (D'Ambrogio et al, 2009).
TDP-43 specifically binds to HDAC6 mRNA
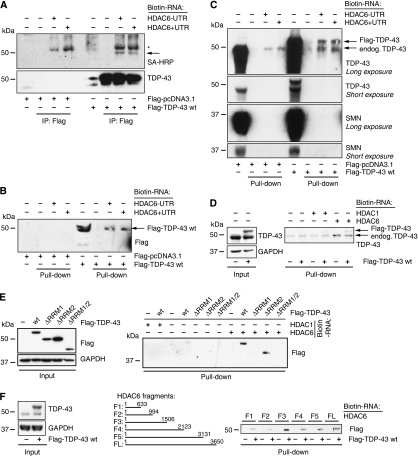
The observed effects of TDP-43 on HDAC6 expression could be due to transcriptional and/or mRNA processing/stabilizing events. To show direct effects of TDP-43 on HDAC6 mRNA, we tested whether TDP-43 binds directly to HDAC6 mRNA. For this purpose, we generated cDNA constructs containing the HDAC6 coding sequence (CDS) with and without the 3′UTR for in vitro transcription. Biotin-labelled RNA was incubated with lysates from HEK293E cells transfected with Flag-TDP-43 or vector control, crosslinked with ultraviolet light followed by immunoprecipitation and WB analysis. Flag-TDP-43 immunoprecipitates, but not control immunoprecipitates, showed the presence of biotinylated HDAC6 mRNA remnants as evidenced by probing with streptavidin conjugated to horseradish peroxidase (HRP) (Figure 4A). Reverse pull-down experiments confirmed the interaction between TDP-43 and HDAC6 mRNA. Biotinylated HDAC6 mRNA was pulled down with streptavidin-conjugated beads. Both, transfected Flag-TDP-43 and importantly endogenous TDP-43 were specifically detected in the biotinylated HDAC6 mRNA precipitates (Figure 4B and C) independently of the presence of HDAC6 3′UTR. To show specificity of this interaction, the DNA- and RNA-binding protein SMN (Lorson and Androphy, 1998) served as a control protein, which was not pulled down with HDAC6 mRNA (Figure 4C). As a complementary control for RNA specificity, we used HDAC1, another member of the HDAC family. In contrast to HDAC6, biotinylated HDAC1 did not pull down TDP-43 (Figure 4D and E). Next, we tested the ability of RRM deletion mutants to bind to HDAC6 RNA. TDP-43 ΔRRM1 as well as TDP-43 ΔRRM1/2 was not able to bind to HDAC6 RNA (Figure 4E). Interestingly, TDP-43 ΔRRM2 was capable of binding to HDAC6 mRNA similar to TDP-43 wt. Thus, direct HDAC6 RNA binding is dependent on the RRM1 of TDP-43 but not on RRM2. However, the latter might be required for correct complex formation and function as shown earlier for CFTR (Buratti and Baralle, 2001; Ayala et al, 2005). This would also explain the failure of TDP-43 ΔRRM2 to restore HDAC6 levels (Figure 3C–F).
Figure 4.
TDP-43 specifically binds to HDAC6 mRNA. HDAC6 cDNAs (comprising either the CDS alone (−UTR), or plus the 3′UTR (+UTR), or the indicated fragments) were in vitro transcribed/biotinylated and mixed with lysates from HEK293E cells transiently transfected with vector control, wt, or mutant Flag-TDP-43, as indicated. Samples were UV-crosslinked, briefly treated with RNase, and precipitated with anti-Flag beads (A) or streptavidin–agarose (B–F). Negative control precipitates were from cell lysates without added HDAC6 RNA (−), or with biotinylated HDAC1 RNA, as indicated. (A) WBs of Flag-immunoprecipitates were probed with streptavidin–HRP to visualize biotinylated HDAC6 mRNA remnants and with anti-TDP-43, as indicated. Asterisk denotes non-specific bands; arrows point to TDP-43 bands. (B–F) WBs of streptavidin pull-downs were probed with anti-Flag (B, E, F), anti-TDP-43 (C, D), or anti-SMN (C), as indicated. Biotinylated HDAC6 RNA pulled down both Flag-tagged TDP-43 wt (B) and endogenous TDP-43 but not SMN (C). In contrast, biotinylated HDAC1 RNA did not pull down TDP-43 efficiently (D, E, left lanes). HDAC6 RNA did not bind to TDP-43 lacking the RRM1 domain (E, right lanes). HDAC6 RNA fragments with progressively shortened 3′ ends were pulled down and probed for the presence of TDP-43 by anti-Flag (F). WB analyses of TDP-43 protein inputs of untransfected and Flag-TDP-43-transfected HEK293E lysates confirmed even loading for all experiments. Each experiment was performed several times with similar results.
In addition, we have roughly mapped the TDP-43-binding site on the CDS of HDAC6 mRNA. We generated progressively 3′ truncated HDAC6 RNA for in vitro crosslinking experiments. Fragments beyond ∼1000 nucleotides (nt) efficiently bound Flag-TDP-43, whereas fragments comprising only the first 663 or 994 nt did not pull down Flag-TDP-43 efficiently (Figure 4F). Hence, TDP-43 appears to bind preferentially to the 3′ CDS of HDAC6 RNA, probably within nt ∼1000–1500. This suggests that TDP-43 effects involve, but may not be limited to binding of those 500 nt within the HDAC6 mRNA.
TDP-43 knockout reduces HDAC6 mRNA in D. melanogaster
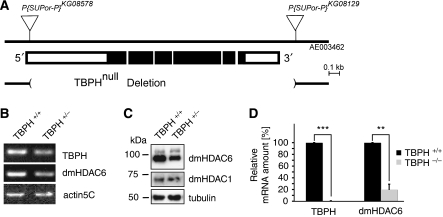
To validate the effects of TDP-43 knockout in vivo, we deleted the entire CDS of the TBPH gene, the TDP-43 ortholog of D. melanogaster (Figure 5A). In contrast to the recently described partial deletion of the TBPH gene (Feiguin et al, 2009), we were not able to obtain adult flies, because TBPH−/− individuals die as second instar larvae. To verify the deletion and the effect of reduced TBPH on HDAC6, we measured TBPH and HDAC6 mRNA levels in adult TBPH+/− flies (Figure 5B). As expected, TBPH+/− flies showed reduced mRNA levels of TBPH compared with controls. Strikingly, levels of HDAC6 were also reduced in TBPH+/− fly heads, providing in vivo evidence that TDP-43 regulates HDAC6 mRNA levels. To confirm the specific downregulation of HDAC6 protein, lysates of TBPH+/− flies and controls were subjected to WB analysis using antibodies against D. melanogaster HDAC proteins (Pandey et al, 2007). Levels of HDAC6, but not HDAC1, were reduced in TBPH+/− fly heads compared with controls (Figure 5C).
Figure 5.
Validation of HDAC6 mRNA downregulation in TBPH-knockout flies. (A) Schematic representation of D. melanogaster TBPH locus (white boxes, non-coding; black boxes, coding) and size of the TBPHnull deletion. The P-element insertions P{SUPor}KG08578 and P{SUPor}KG08129 in region 60A of the second chromosome are indicated (triangles). The TBPHnull deletion, which removes the entire TBPH coding sequence, was generated by combined P-element excision/recombination events involving P{SUPor}KG08578 and P{SUPor}KG08129. (B) Total RNA was prepared from adult flies (10 females and 10 males each of controls and TBPH+/−) and subjected to RT–PCR using primer pairs amplifying D. melanogaster TDP-43 (TBPH), HDAC6, and the housekeeping gene actin5C, as indicated. (C) Total protein lysates were prepared from adult fly heads (10 females and 10 males each of controls and TBPH+/−) and subjected to WB analysis using antibodies against D. melanogaster HDAC6, HDAC1, and α-tubulin, as indicated. (D) Brains were dissected from first instar larvae (black bars, control n=4; grey bars, TBPH−/− n=4) and transferred onto AmpliGrids. After RNA extraction and restriction digest of residual contaminating genomic DNA, qRT–PCR was performed with primer pairs amplifying TBPH, dmHDAC6, and the housekeeping gene actin5C. Expression of TBPH was not detectable in TBPH−/− larval brains within 45 amplification cycles. RNA levels of dmHDAC6 were significantly reduced in TBPH−/− compared with controls. Expression of TBPH and dmHDAC6 were normalized to actin5C levels (**P<0.002; ***P<0.000001).
To obtain measurements from tissue of D. melanogaster completely lacking TBPH we have established qRT–PCR from brains of first instar larvae. Individual brains of TBPH−/− and control larvae were transferred onto single reaction centres on AmpliGrid slides. Controls showed qRT–PCR signals for TBPH and HDAC6 mRNA (Figure 5D), showing expression of both genes in the brains of first instar larvae. As expected, TBPH mRNA was not detectable in TBPH−/− larval brains. Moreover, HDAC6 mRNA levels were dramatically reduced in TBPH−/− larval brains (Figure 5D). Thus, HDAC6 expression is reduced in D. melanogaster lacking the TDP-43 ortholog.
TDP-43 knockdown-mediated HDAC6 downregulation confers proteotoxicity
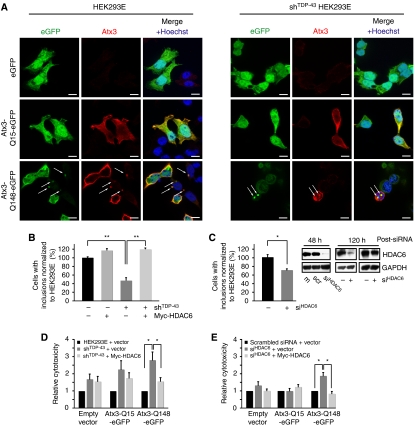
One of the potential cellular consequences of HDAC6 reduction on TDP-43 knockdown is impaired turnover of aggregating proteins. HDAC6 is known to bind misfolded, ubiquitinated proteins and connect them to the retrograde microtubular transport machinery, eventually delivering aggregating proteins to autophagic degradation (Kawaguchi et al, 2003). HDAC6 deficiency would reduce such aggresome formation, instead increasing the cellular concentration of proteotoxic species. To test the hypothesis that TDP-43-mediated downregulation of HDAC6 reduces the turnover of proteotoxic aggregates, we used very long polyQ-expanded Atx3 as an experimental model protein (Bichelmeier et al, 2007). Control and stably silenced shRNATDP-43 HEK293E cells were transiently transfected with enhanced green fluorescent protein (eGFP)-tagged Atx3 containing either 15 (Atx3-Q15–eGFP) or 148 (Atx3-Q148–eGFP) long poly-glutamine tracts or the empty vector control. Neither eGFP alone nor the non-expanded Atx3-Q15–eGFP formed distinct aggregates whereas the very long polyQ-expanded Atx3-Q148–eGFP did form large aggregates (Figure 6A). In stably silenced shRNATDP-43 cells, the percentage of aggregate-bearing cells was reduced significantly to 50% compared with parental cells (Figure 6B). Expression of Myc–HDAC6 led to a small increase in the percentage of aggregates in control HEK293E cells and importantly fully restored the aggregate formation in stably silenced shRNATDP-43 cells (Figure 6B), Conversely, aggregate formation was reduced by transient transfection of parental HEK293E with an HDAC6-specific siRNA (siRNAHDAC6) (Figure 6C). Control WBs showed very strong acute knockdown efficiency of the siRNAHDAC6 during the critical period of the Atx3-Q148–eGFP aggregation experiments, and HDAC6 protein levels remained suppressed even though there was some exhaustion of the siRNA at late time points (Figure 6C). These results indicate that TDP-43 deficiency impairs aggresome formation in an HDAC6-dependent manner.
Figure 6.
TDP-43-knockdown-mediated HDAC6 downregulation reduces protein aggregation and increases proteotoxicity. (A) Parental HEK293E cells (left panels) and stably silenced shRNATDP-43 cells (right panels) were transiently transfected with eGFP, Atx3-Q15–eGFP, or Atx3-Q148–eGFP for 96 h. Epifluorescence (green) shows the presence of eGFP (fusion proteins) in transfected cells, confirmed by immunostaining with anti-Atx3 (red). Nuclei were counterstained with Hoechst 33342. Merged images are shown to the right. Arrows point to Atx3-Q148–eGFP inclusions. Scale bars=10 μm. (B) Cells were transfected as above together with Myc–HDAC6 or empty vector control (−). After 96 h, cells with visible aggregates were counted in three independent experiments. Compared with control HEK293E cells, significantly fewer shRNATDP-43 cells contained aggregates, and aggregate formation was significantly restored by HDAC6 transfection, up to control levels (**P<0.003). (C) HEK293E cells were mock transfected (m) or transiently transfected with scrambled (scr) or HDAC6-directed siRNA. After 24 h, cells were transfected with Atx3-Q148–eGFP. After another 96 h, cells with visible Atx3 aggregates were counted and normalized to the number of aggregate-bearing cells of untransfected HEK293E cells. Compared to those, significantly less siRNAHDAC6-treated cells contained aggregates (*P<0.03). Knockdown efficiency was assessed after 48 h treatment with siRNAHDAC6, and after the end of the 120 h experimental procedure by WB analysis of protein extracts using anti-HDAC6 and anti-GAPDH. Stably silenced shRNATDP-43 cells (D) or transiently transfected with siRNAHDAC6 (E) were transfected with eGFP, Atx3-Q15–eGFP, or Atx3-Q148–eGFP together with Myc–HDAC6 (light grey bars) or empty vector control (dark grey bars). LDH release values were measured 96 h after DNA transfection and normalized to parallel non-silenced HEK293E transfections (black bars). Values represent the mean of six (D) or seven (E) independent experiments (*P<0.04).
Aggresome formation is believed to be a cellular defense mechanism sequestering toxic, misfolded proteins. Thus, the reduced formation of aggregates with the Atx3-Q148–eGFP model protein in TDP-43 silenced cells might be accompanied by increased amounts of the proteotoxic species. The aggregate preceding species of Atx3 are invisible microaggregates precluding microscopic analysis (Chai et al, 2001). However, we noted that stably silenced shRNATDP-43 HEK293E cells ‘suffered' after Atx3-Q148–eGFP transfection. Thus, we quantified cell death by lactate dehydrogenase (LDH) release of stably silenced shRNATDP-43 HEK293E cells transiently transfected with eGFP, Atx3-Q15–eGFP, or Atx3-Q148–eGFP. Compared with parental HEK293E cells, stably TDP-43 silenced cells were slightly more vulnerable to transient transfection of eGFP alone or the non-expanded Atx3-Q15–eGFP, perhaps reflecting some general viability reduction by TDP-43 knockdown. Importantly, transfection with the expanded Atx3-Q148–eGFP caused a significant three-fold increase in cell death in stably silenced shRNATDP-43 cells compared with control HEK293E cells (Figure 6D). A similar result was obtained by transient transfection of parental HEK293E cells with siRNAHDAC6 (Figure 6E). Co-transfection of Myc–HDAC6 significantly improved cellular viability in shRNATDP-43 (Figure 6D) and in siRNAHDAC6-treated (Figure 6E) HEK293E cells. Thus, HDAC6 downregulation because of TDP-43 deficiency seems to reduce the cellular turnover of toxic, aggregating proteins, thereby possibly contributing to pathogenesis.
Discussion
TDP-43 is involved in the pathogenesis of FTLD-TDP, ALS, and IBM as TDP-43 protein is a key component of inclusions in all of these disorders. In neurodegenerative disease, TDP-43 is pathologically processed to C-terminal fragment(s) and phosphorylated (Neumann et al, 2006), arguing for a scenario in which aberrant phosphorylation, cleavage, and aggregation lead to a gain-of-toxic-misfunction of TDP-43. Noteworthy, neurons and muscle cells bearing cytosolic TDP-43 inclusions lack the usual nuclear TDP-43 signal (Neumann et al, 2006; Olivé et al, 2009). Moreover, a frequent finding in diverse brain regions of FTLD-U is characterized by cytoplasmically localized granular TDP-43 staining together with lack of nuclear TDP-43 (Brandmeir et al, 2008). These findings point towards a loss of nuclear function mechanism rather than a gain of toxic misfunction. Assuming that the mostly nuclear protein TDP-43 is involved in mRNA processing events, as seen by its localization overlapping with NBs involved in splicing and mRNA metabolism, we screened for relevant TDP-43 target mRNAs by expression profiling of transiently silenced HEK293E cells.
Several genetically disease-associated mRNAs were found not significantly altered (Table I). Among the 402 transcripts altered more than two-fold by siRNATDP-43 treatment of HEK293E cells, we discovered that HDAC6 expression was downregulated on TDP-43 knockdown. This was consistently confirmed with multiple siRNA and lentiviral shRNA silencings at the mRNA and protein level, both in non-neuronal HEK293E as well as neuronal SH-SY5Y cells. Diminished HDAC6 activity on TDP-43 knockdown was evidenced by the increase in acetyl-tubulin levels. Moreover, decreased HDAC6 levels could be restored to normal levels by retransfection of silenced cells with wt TDP-43 cDNA. In contrast, RRM and NLS mutants failed to restore HDAC6 levels, showing a direct effect of nuclear and RNA-binding TDP-43 on HDAC6 mRNA levels. In addition, other (hnRNP) proteins may be necessary for HDAC6 mRNA regulation, as the truncated TDP-43 lacking the C-terminal GRD was not able to restore HDAC6 mRNA and protein levels. Interestingly, most of pathogenic TDP-43 point mutants were able to restore HDAC6 levels similar to wt protein, consistent with a previous report on CFTR splicing (Nonaka et al, 2009). We noted no obvious changes in TDP-43 protein steady-state levels, aggregation or cellular distribution for all missense mutants tested. Nevertheless, two mutations showed consistently reduced (D169G) or diminished (R361S) capacity to restore HDAC6 levels, suggesting that HDAC6 decrease might occur in individuals with specific genetic alterations.
The specific interaction of wt TDP-43 with HDAC6 RNA was shown by crosslinking experiments. Deletion of RRM1 but not of RRM2 abolished HDAC6 RNA binding, indicating that RRM2 might not be required for binding but for activity of TDP-43 on HDAC6 RNA, similar to as described for CFTR (Buratti and Baralle, 2001; Ayala et al, 2005). By rough mapping we could show that TDP-43 bind preferentially to 3′ CDS sequences of the HDAC6 RNA. Thus, the effects of TDP-43 on HDAC6 are mediated, at least in part, through direct binding of the mRNA within the coding region.
Strikingly, we provide evidence that HDAC6 regulation by TDP-43 occurs also in vivo. Reduced dmHDAC6 mRNA and protein levels were detected in adult TBPH+/− fly heads, whereas dmHDAC1 protein remained unchanged. Strong reduction of dmHDAC6 mRNA was measured in first instar larval brains of flies completely lacking TBPH. In summary, TDP-43 is necessary to maintain physiological levels of HDAC6 mRNA in vivo.
HDAC6 has multiple cellular functions, most through actions on the actin and the tubulin cytoskeleton controlling intracellular trafficking and cell motility (reviewed in Valenzuela-Fernandez et al, 2008). Moreover, HDAC6 is a key player in autophagy-mediated degradation of misfolded proteins by mediating their transport along microtubules, their aggresome formation, and recruitment of the autophagic machinery (Kawaguchi et al, 2003; Boyault et al, 2007b). In this process, HDAC6 acts in a fine-tuned balance together with VCP. While VCP promotes proteasomal degradation of aggregating proteins, HDAC6 promotes aggresome formation and autophagy (Boyault et al, 2006). Remarkably, VCP is genetically associated with IBMPFD (Watts et al, 2004), a form of FTD that is characterized by TDP-43 pathology in brain (Neumann et al, 2007) and muscle (Weihl et al, 2008). Strikingly, pathological mutated dominant-negative VCP results in dysfunction of aggresome formation of misfolded proteins, which can be rescued by HDAC6 overexpression (Ju et al, 2008). Conversely, HDAC6 decrease may lead to a similar impairment of protein degradation (Song et al, 2008). Using expanded polyQ Atx3-Q148–eGFP as model protein, we could show that TDP-43 knockdown reduces the formation of large aggregates but enhances proteotoxicity, both of which were specifically dependent on HDAC6. Thus, TDP-43 loss of nuclear function in disease may lead to HDAC6 downregulation and subsequent impairment in toxic protein turnover, contributing to the disease process of TDP-43 proteinopathies.
Materials and methods
Cloning and mutagenesis
Full-length TDP-43, HDAC1, and HDAC6 were amplified from a human brain cDNA library (Clontech). HDAC1 and HDAC6 were cloned through BglII/NotI into pCMV–Myc (Clontech). HDAC6 3′UTR was amplified from a reverse transcriptase (RT) reaction and fused to HDAC6 CDS through internal EcoRV site (EcoRV/NotI). HDAC1 and HDAC6 (without or with 3′UTR) were subcloned into pPD129.36 (BglII/NotI) under control of a T7 promotor generating pPD129.36 HDAC1, HDAC6−UTR, and HDAC6+UTR, respectively. TDP-43 was and cloned in frame (BamHI/HindIII) into pcDNA3.1(−) (Invitrogen) with a 5′-Flag tag (NotI/EcoRI). Mutant TDP-43 cDNAs were generated by site-directed mutagenesis. TDP-43 with mutated NLS (aa 82–84 changed to Ala-Ala-Ala) was cloned as described (Winton et al, 2008) and subcloned into pcDNA3.1-Flag.
Atx3 containing 15 or 148 CAG repeats (Bichelmeier et al, 2007) was subcloned through SacI/ApaI into pEGFP-N2 (Clontech) resulting in Atx3–eGFP fusion. All overexpression as well as silencing constructs (see below) were sequence verified using BigDye Terminator v.3.1 and an ABI 3100 Genetic Analyzer (Applied Biosystems). Primer sequences can be obtained on request.
Cell culture and RNA silencing
HEK293E cells were grown in Dulbecco's modified Eagle medium (DMEM), SH-SY5Y cells in DMEM-F12 medium (both Biochrom) supplemented with 10% fetal bovine serum (PAA) at 37°C under humidified 5% CO2/air. DNA transfections were performed with FuGene6 (Roche) according to manufacturer's instruction. Transient siRNA transfections were performed using HiPerfect with 5 nmol scrambled (both Qiagen) or TDP-43/HDAC6-specific siRNAs: siRNATDP-43 A, 5′-CAAUAGCAAUAGACAGUUA-3′; siRNATDP-43 B, 5′-CACUACAAUUGAUAUCAAA-3′ (both Qiagen); siRNATDP-43 C, 5′-GAATCAGGGTGGATTTGGT-3′; siRNATDP-43 D, 5′-GAAACAAUCAAGGUAGUAA-3′ (both Sigma); siRNAHDAC6, 5′-CCGUCAACGUGGCAUGGAA-3′ (Qiagen).
Stably silenced HEK293E and SH-SY5Y cells were generated by treatment with different amounts of lentiviral vector particles harbouring shRNATDP-43 for 24 h followed by isolation of single cell clones. Some cultures were treated for 5 h with the HDAC6 inhibitor tubacin.
SDS–PAGE and WB analysis
Cells were collected and lysed in lysis buffer (50 mM Tris (pH 7.4), 50 mM NaCl, 1% NP-40, 0.1% deoxycholate, and 0.1% SDS, 1 × Complete proteinase inhibitor (Roche)). Protein concentration was determined by use of bicinchoninic acid (Pierce Biotechnology). Protein was subjected to SDS–PAGE using 10 or 12% polyacrylamide gels or 4–12% Bis–Tris NuPAGE gradient gels (Invitrogen) and transferred onto nitrocellulose. Membranes were incubated with primary antibodies overnight at 4°C followed by HRP-conjugated secondary antibodies (1:15 000; Jackson ImmunoResearch Laboratories). Bands were visualized with ImmobilonWestern Chemiluminescent HRP Substrate (Millipore) on Hyperfilm ECL high performance chemiluminescence (GE Healthcare). For densitometric analysis NIH Image software version 1.63 was used.
Immunostaining
Cells were plated onto glass coverslips coated with PDL (Sigma) and collagen (Cohesion), fixed with 4% (w/v) paraformaldehyde, and permeabilized with 1% Triton-X-100. Cells were incubated with primary antibodies followed by incubation with secondary antibodies anti-mouse IgG Alexa Fluor-488 or -568 and anti-rabbit Alexa Fluor-488, -568, or -647 (Molecular Probes) diluted 1:2000. Nuclei were stained with Hoechst 33342 (Molecular Probes) diluted 1:5000. Coverslips were mounted onto microscope slides using fluorescent mounting medium (Dako). Confocal fluorescent images were taken with an AxioImager microscope equipped with an ApoTome Imaging System (Zeiss).
Antibodies
Following antibodies were used for WB analysis or immunofluorescence (IF): rabbit anti-TDP-43 (WB/IF, 1:2000; ProteinTech Group), mouse anti-TDP-43 (WB/IF, 1:2000; Abnova, clone 2E2-D3), rabbit anti-HDAC6 (WB, 1:2000; IF, 1:500; Santa Cruz (H-300)), mouse anti-acetyl-tubulin (WB/IF, 1:4000; Sigma, clone 6-11-B1), mouse anti-α-tubulin (WB, 1:5000; Sigma, clone B-5-1-2), mouse anti-Flag (WB, 1:100 000; Sigma, clone M2), mouse anti-GAPDH (WB, 1:2000; BioTrend), rabbit anti-histone H3 (WB, 1:1000; Cell Signaling), rabbit anti-HSP90 (WB, 1:35 000; Stressgen), mouse anti-SMN (WB/IF, 1:5000; BD Transduction Laboratories), mouse anti-coilin (IF, 1:5000; BD Transduction Laboratories), mouse anti-PML (IF, 1:100; Abcam), mouse anti-SC35 (IF, 1:5000; Sigma), rabbit anti-Atx3 (IF, 1:500; Schmidt et al, 1998), rabbit anti-dmHDAC1 (WB, 1:2000; Pandey et al, 2007), anti-dmHDAC6 (WB: 1:2000; Pandey et al, 2007).
Generation of lentiviral vectors encoding shRNATDP-43
TDP-43-specific shRNA was designed on the basis of siRNATDP-43 B with a loop structure of the miRNA23 (5′-CTTCCTGTCA-3′). shRNATDP-43 was cloned by ligating annealed oligos into the XbaI/BsiWI sites of the mU6-[XbaI/BsiWI]-CMV-eGFP-IRES-Hygro modified transfer plasmid (pHR'COMBI) (Baekelandt et al, 2002). Lentiviral vectors were produced by a triple transient transfection 6 × 106 of 293T cells with 20 μg of transfer plasmid, 10 μg of packaging plasmid (pCMV ΔR8.91), and 5 μg of envelope plasmid (pLP/VSVG) using polyethyleimine as transfection reagent (Lauwers et al, 2007). After overnight incubation the medium was replaced. Medium from 48 to 72 h post transfection was collected and vector particles in the medium were concentrated up to 100-fold using 50 kDa vivaspin-15 concentrators (Sartorius). Vector aliquots were frozen and kept at −80°C until use.
RNA crosslinking procedures
UV-crosslinking of in vitro transcribed biotinylated RNA to protein lysate was performed according to Vumbaca et al (2008) with modifications and subsequent immunoprecipitation according to Kashima and Manley (2003). pPD129.36 encoding HDAC6−UTR, HDAC6+UTR, or HDAC1 was linearized with NotI. HDAC6 fragments were generated by linearizing pPD129.36 HDAC6−UTR with ScaI followed by digest within HDAC6 coding region: fragment 1 (bp 1–633), StuI; fragment 2 (bp 1–994): XhoI; fragment 3 (bp 1–1506), KpnI; fragment 4 (bp 1–2123), ApaI; fragment 5 (bp 1–3131; EcoRV). After agarose gel purification 1 μg DNA was in vitro transcribed/biotinylated using biotin-16-UTP (Biotin RNA Labeling Mix) and T7 RNA Polymerase (both Roche) according to manufacturer's instructions. DNase digest was performed by adding 2 μl of RNase-free DNase (Promega) for 15 min at 37°C and the reaction stopped by addition of 1 μl of 0.5 M EDTA. RNA concentration was determined by measuring absorbance at 260 nm, and RNA size and purity were checked by agarose gel electrophoresis; 1 pmol biotin–RNA was incubated with 250 μl RNA-binding buffer (20 mM Hepes (pH 7.5), 5 mM MgCl2, 50 mM KCl, 150 mM NaCl, 0.5 mM EGTA, 0.5 mM dithiothreitol, 10% glycerol) and 500 μg HEK293E lysate for 20 min at 30°C. HEK293E lysates were generated from HEK293E cells transfected with vector alone or Flag-TDP-43 for 48 h and lysed in RNA-binding buffer +0.5% Triton-X-100. UV-irradiation was performed on ice for 10 min with 0.120 J/cm2 in a Bio-Link BLX device (Vilber-Lourmat). After brief RNase A digest (10 μg/ml, Sigma) for 15 min Flag- or Streptavidin-beads (Sigma) were added and precipitations were carried out overnight at 4°C. Beads were washed with RNA-binding buffer+0.5% Triton-X-100 and elution performed at 95°C with 1 × Lämmli buffer. Eluate and input were separated by 10% SDS–PAGE, blotted onto nitrocellulose, and probed with antibodies and streptavidin (SA)-HRP (WB, 1:5000; Amersham), respectively.
RNA extraction and RT–PCR
Cellular RNA was extracted using the RNeasy Mini kit (Qiagen) following manufacturer's instruction. RNA from D. melanogaster was isolated with Trizol (Invitrogen). For semi-quantitative RT–PCR experiments, 600 ng total RNA was reverse transcribed using Transcriptor High Fidelity cDNA Synthesis kit (Roche) and anchored oligo-dT primer. RT reaction (2 μl) was used as template for transcript amplification in a 25 μl reaction with 5 μl 5 × GoTaq Buffer, 0.1 μl GoTaq Polymerase (Promega), and 2 μM primer. Amplified PCR products were resolved by electrophoresis using 2% agarose gels and stained with ethidium bromide.
For cellular qRT–PCR, 1000 ng total RNA was reverse transcribed using the same conditions as above; 1/10 dilutions were used in triplicates with 0.2 μM primer and 5 μl LightCycler 480 SYBR Green I Master in a 10 μl reaction and qPCR executed in a 384-well block on a LightCycler 480 system (Roche). Absolute transcript levels for TDP-43, HDAC6, and PBGD were obtained by second derivative method. Relative transcript levels were calculated as TDP-43/PBGD or HDAC6/PBGD ratio and normalized to the relative expression level of the mock-transfected control. Primer sequences are available on request.
For qRT–PCR measurements of first instar larval brain, the brain and ventral ganglion of control white larvae and TBPH−/− larvae were dissected in water containing RNase inhibitor (Fermentas). Individual tissue samples were transferred onto AmpliGrid slides (Advalytix), air-dried, and frozen before analysis. The translucence of the slides allowed precise visual monitoring of optimal transfer of the tiny brain tissue. In addition, the AmpliGrid platform allows low volume PCR reactions, ideal for amplification of little template amounts.
Larval brain was covered with 0.5 μl protein lysis mix (Cell Extraction kit; Advalytix) and with 5 μl of a sealing solution to prevent evaporation for 30 min at 60°C. Possible residual genomic DNA was chopped with 0.5 μl of Alu I enzyme (0.66 U/μl) (NEB BioLabs) added to the lysis mix for 3 h at 37°C, followed by 2 min at 95°C. The RT (one step RT–PCR, Qiagen) was performed directly on the AmpliGrid reaction sites by adding 1 μl of the master mix (10 μl: 2 μl 5 × RT buffer, 0.4 μl dNTP mix (10 mM), 0.4 μl enzyme mix, 0.2 μl RNase inhibitor, 1.6 μl 5 × Q-solution, 1 μl of 4-plex RT-primer mix (3 μM) (quantitative reverse primer, Advalytix) mixed with 1:10 diluted ETSSB (NEB BioLabs) and H2O (4.4 μl)). The RT reaction was performed on the AmpliSpeed Cycler at 58°C for 30 min. For qPCR, four different master mixes were prepared adding the TBPH, HDAC6, or actin5C primer pairs (http://www.advalytix.com/primer_design_1519.htm) (20 μl each of 6 μM stocks) to 100 μl 2 × Brilliant II SYBR Green (Stratagene) and 60 μl H2O. From each master mix, 9 μl was added to 1 μl of a pre-diluted (1:3) RT mixture. qPCR was performed in quadruplicates in a Stratagene thermocycler. Ct values were calculated by the Stratagene software. Relative quantifications were obtained by the 2(−Delta Delta C(T)) method (Livak and Schmittgen, 2001).
Generation of TBPH knockout flies
The TBPHnull deletion was generated by P-element mobilization as described earlier (Voigt et al, 2002). In brief, a transposase source was introduced to transheterozygous w;P{SUPor}KG08578/P{SUPor}KG08129 flies. Excision events in the progeny were monitored by loss of the white+ eye colour marker of the P-elements. The corresponding flies were balanced with CyO to establish stable stocks. Genomic DNA was prepared and sequenced (GenterPrise GmbH) to determine the breakpoints of the deletion. All fly stocks used have been obtained from the Bloomington Stock Centre.
Atx3 aggregation and toxicity assays
For toxicity measurements, eight wells per condition were transfected with Atx3-Q15–eGFP, Atx3-Q148–eGFP, or empty vector in combination with Myc–HDAC6 or empty vector control and analysed 96 h post transfection. Four of eight wells served as a positive control for 100% LDH release/cell death and were treated with Triton-X-100 in a final concentration of 1% 1 h before LDH determination by use of CytoTox96 non-radioactive cytotoxicity assay (Promega) following manufacturer's instructions. Each condition was measured in quadruplicates and normalized to the respective Triton-X-100 controls. Resulting quotients were normalized to non-silenced HEK293E cells for each condition. Aggregation of Atx3 was analysed 96 h post transfection. Cells were fixed, permeabilized, and stained with anti-Atx3. Atx3-transfected cells (>200) were counted for inclusion formation in three independent experiments. WB analysis verified equal overexpression of Atx3 and HDAC6 in parental and silenced HEK293E cells.
Statistical analysis
Statistical analysis was performed with paired, two-sided Student's t-test. Error bars indicate s.e.m.
Public database access of microarray data
Array data is available in the GEO depository under accession number GSE18632. The microarray procedure and additional Supplementary methods are available at The EMBO Journal Online (http://www.embojournal.org).
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Movie S1
Supplementary Figure S3
Supplementary Figure S4
Supplementary Table S1
Supplementary Information
Review Process File
Acknowledgments
We thank Elena Hausherr and Susann Klein for initial HDAC6 cloning, Stuart Schreiber for the donation of tubacin, Marc Hild for the donation of antibodies against Drosophila HDAC proteins, Petra Füger for help with fly brain preparations, and Claudia Schulte for helpful suggestions. This work was supported by the German Competence Net Degenerative Dementias, the Helmholtz Alliance for Mental Health in an Aging Society, and the Hertie Foundation. Author contributions are the following: FCF conducted most experiments; technical assistance and help was provided by SSW and MLA; MW performed microarray hybridization and analysis; CVH generated lentiviruses; AV created null flies; AV, AW, KG, JVK, and PJK manipulated and measured flies; RK designed quantitative fly primer; TS contributed tools and advice for Atx3 experiments; FCF and PJK conceived and designed experiments; WS, MN, MB, JBS, MA-F, TMR, and VB provided intellectual and/or financial support; FCF and PJK wrote the paper; PJK is the principal investigator.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abhyankar MM, Urekar C, Reddi PP (2007) A novel CpG-free vertebrate insulator silences the testis-specific SP-10 gene in somatic tissues: role for TDP-43 in insulator function. J Biol Chem 282: 36143–36154 [DOI] [PubMed] [Google Scholar]
- Ayala YM, Misteli T, Baralle FE (2008) TDP-43 regulates retinoblastoma protein phosphorylation through the repression of cyclin-dependent kinase 6 expression. Proc Natl Acad Sci USA 105: 3785–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala YM, Pantano S, D'Ambrogio A, Buratti E, Brindisi A, Marchetti C, Romano M, Baralle FE (2005) Human, Drosophila, and C. elegans TDP43: nucleic acid binding properties and splicing regulatory function. J Mol Biol 348: 575–588 [DOI] [PubMed] [Google Scholar]
- Baekelandt V, Claeys A, Eggermont K, Lauwers E, De Strooper B, Nuttin B, Debyser Z (2002) Characterization of lentiviral vector-mediated gene transfer in adult mouse brain. Hum Gene Ther 13: 841–853 [DOI] [PubMed] [Google Scholar]
- Bernardi R, Pandolfi PP (2007) Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol 8: 1006–1016 [DOI] [PubMed] [Google Scholar]
- Bichelmeier U, Schmidt T, Hubener J, Boy J, Ruttiger L, Habig K, Poths S, Bonin M, Knipper M, Schmidt WJ, Wilbertz J, Wolburg H, Laccone F, Riess O (2007) Nuclear localization of ataxin-3 is required for the manifestation of symptoms in SCA3: in vivo evidence. J Neurosci 27: 7418–7428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose JK, Wang I-F, Hung L, Tarn W-Y, Shen C-KJ (2008) TDP-43 overexpression enhances exon 7 inclusion during the survival of motor neuron pre-mRNA splicing. J Biol Chem 283: 28852–28859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyault C, Gilquin B, Zhang Y, Rybin V, Garman E, Meyer-Klaucke W, Matthias P, Muller CW, Khochbin S (2006) HDAC6-p97/VCP controlled polyubiquitin chain turnover. EMBO J 25: 3357–3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyault C, Sadoul K, Pabion M, Khochbin S (2007a) HDAC6, at the crossroads between cytoskeleton and cell signaling by acetylation and ubiquitination. Oncogene 26: 5468–5476 [DOI] [PubMed] [Google Scholar]
- Boyault C, Zhang Y, Fritah S, Caron C, Gilquin B, Kwon SH, Garrido C, Yao TP, Vourc'h C, Matthias P, Khochbin S (2007b) HDAC6 controls major cell response pathways to cytotoxic accumulation of protein aggregates. Genes Dev 21: 2172–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandmeir NJ, Geser F, Kwong LK, Zimmerman E, Qian J, Lee VM-Y, Trojanowski JQ (2008) Severe subcortical TDP-43 pathology in sporadic frontotemporal lobar degeneration with motor neuron disease. Acta Neuropathol 115: 123–131 [DOI] [PubMed] [Google Scholar]
- Buratti E, Baralle FE (2001) Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem 276: 36337–36343 [DOI] [PubMed] [Google Scholar]
- Buratti E, Brindisi A, Giombi M, Tisminetzky S, Ayala YM, Baralle FE (2005) TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J Biol Chem 280: 37572–37584 [DOI] [PubMed] [Google Scholar]
- Buratti E, Dörk T, Zuccato E, Pagani F, Romano M, Baralle FE (2001) Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J 20: 1774–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Wu L, Griffin JD, Paulson HL (2001) The role of protein composition in specifying nuclear inclusion formation in polyglutamine disease. J Biol Chem 276: 44889–44897 [DOI] [PubMed] [Google Scholar]
- Collins LJ, Penny D (2009) The RNA infrastructure: dark matter of the eukaryotic cell? Trends Genet 25: 120–128 [DOI] [PubMed] [Google Scholar]
- D'Ambrogio A, Buratti E, Stuani C, Guarnaccia C, Romano M, Ayala YM, Baralle FE (2009) Functional mapping of the interaction between TDP-43 and hnRNP A2 in vivo. Nucleic Acids Res 37: 4116–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiguin F, Godena VK, Romano G, D'Ambrogio A, Klima R, Baralle FE (2009) Depletion of TDP-43 affects Drosophila motoneurons terminal synapsis and locomotive behavior. FEBS Lett 583: 1586–1592 [DOI] [PubMed] [Google Scholar]
- Giordana MT, Piccinini M, Grifoni S, De Marco G, Vercellino M, Magistrello M, Pellerino A, Buccinna B, Lupino E, Rinaudo MT (2009) TDP-43 redistribution is an early event in sporadic amyotrophic lateral sclerosis. Brain Pathol (advance online publication 17 March 2009; doi:10.1111/j.1750-3639.2009.00284.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggarty SJ, Koeller KM, Wong JC, Grozinger CM, Schreiber SL (2003) Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc Natl Acad Sci USA 100: 4389–4394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang X-F, Yao T-P (2002) HDAC6 is a microtubule-associated deacetylase. Nature 417: 455–458 [DOI] [PubMed] [Google Scholar]
- Ju JS, Miller SE, Hanson PI, Weihl CC (2008) Impaired protein aggregate handling and clearance underlie the pathogenesis of p97/VCP-associated disease. J Biol Chem 283: 30289–30299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Vande Velde C, Bouchard J-P, Lacomblez L, Pochigaeva K, Salachas F, Pradat P-F, Camu W, Meininger V, Dupre N, Rouleau GA (2008) TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet 40: 572–574 [DOI] [PubMed] [Google Scholar]
- Kashima T, Manley JL (2003) A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat Genet 34: 460–463 [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP (2003) The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 115: 727–738 [DOI] [PubMed] [Google Scholar]
- Kumar-Singh S, Van Broeckhoven C (2007) Frontotemporal lobar degeneration: current concepts in the light of recent advances. Brain Pathol 17: 104–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski TJ Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, Valdmanis P, Rouleau GA, Hosler BA, Cortelli P, de Jong PJ, Yoshinaga Y, Haines JL, Pericak-Vance MA, Yan J, Ticozzi N et al. (2009) Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323: 1205–1208 [DOI] [PubMed] [Google Scholar]
- Kwong LK, Neumann M, Sampathu DM, Lee VM-Y, Trojanowski JQ (2007) TDP-43 proteinopathy: the neuropathology underlying major forms of sporadic and familial frontotemporal lobar degeneration and motor neuron disease. Acta Neuropathol 114: 63–70 [DOI] [PubMed] [Google Scholar]
- Lauwers E, Bequé D, Van Laere K, Nuyts J, Bormans G, Mortelmans L, Casteels C, Vercammen L, Bockstael O, Nuttin B, Debyser Z, Baekelandt V (2007) Non-invasive imaging of neuropathology in a rat model of α-synuclein overexpression. Neurobiol Aging 28: 248–257 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lorson CL, Androphy EJ (1998) The domain encoded by exon 2 of the survival motor neuron protein mediates nucleic acid binding. Hum Mol Genet 7: 1269–1275 [DOI] [PubMed] [Google Scholar]
- Mackenzie IR, Neumann M, Bigio EH, Cairns NJ, Alafuzoff I, Kril J, Kovacs GG, Ghetti B, Halliday G, Holm IE, Ince PG, Kamphorst W, Revesz T, Rozemuller AJ, Kumar-Singh S, Akiyama H, Baborie A, Spina S, Dickson DW, Trojanowski JQ et al. (2009) Nomenclature for neuropathologic subtypes of frontotemporal lobar degeneration: consensus recommendations. Acta Neuropathol 117: 15–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado PA, Ayala YM, Romano M, Buratti E, Baralle FE (2005) Depletion of TDP 43 overrides the need for exonic and intronic splicing enhancers in the human apoA-II gene. Nucleic Acids Res 33: 6000–6010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Mackenzie IR, Cairns NJ, Boyer PJ, Markesbery WR, Smith CD, Taylor JP, Kretzschmar HA, Kimonis VE, Forman MS (2007) TDP-43 in the ubiquitin pathology of frontotemporal dementia with VCP gene mutations. J Neuropathol Exp Neurol 66: 152–157 [DOI] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM-Y (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314: 130–133 [DOI] [PubMed] [Google Scholar]
- Nonaka T, Kametani F, Arai T, Akiyama H, Hasegawa M (2009) Truncation and pathogenic mutations facilitate the formation of intracellular aggregates of TDP-43. Hum Mol Genet 18: 3353–3364 [DOI] [PubMed] [Google Scholar]
- Olivé M, Janue A, Moreno D, Gamez J, Torrejon-Escribano B, Ferrer I (2009) TAR DNA-Binding protein 43 accumulation in protein aggregate myopathies. J Neuropathol Exp Neurol 68: 262–273 [DOI] [PubMed] [Google Scholar]
- Ou S-HI, Wu F, Harrich D, García-Martínez LF, Gaynor RB (1995) Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J Virol 69: 3584–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, Padmanabhan R, Hild M, Berry DL, Garza D, Hubbert CC, Yao T-P, Baehrecke EH, Taylor JP (2007) HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature 447: 859–863 [DOI] [PubMed] [Google Scholar]
- Salajegheh M, Pinkus JL, Taylor JP, Amato AA, Nazareno R, Baloh RH, Greenberg SA (2009) Sarcoplasmic redistribution of nuclear TDP-43 in inclusion body myositis. Muscle Nerve 40: 19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T, Landwehrmeyer GB, Schmitt I, Trottier Y, Auburger G, Laccone F, Klockgether T, Volpel M, Epplen JT, Schols L, Riess O (1998) An isoform of ataxin-3 accumulates in the nucleus of neuronal cells in affected brain regions of SCA3 patients. Brain Pathol 8: 669–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Xiao Z, Nagashima K, Li CC, Lockett SJ, Dai RM, Cho EH, Conrads TP, Veenstra TD, Colburn NH, Wang Q, Wang JM (2008) The heavy metal cadmium induces valosin-containing protein (VCP)-mediated aggresome formation. Toxicol Appl Pharmacol 228: 351–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong MJ, Volkening K, Hammond R, Yang W, Strong W, Leystra-Lantz C, Shoesmith C (2007) TDP43 is a human low molecular weight neurofilament (hNFL) mRNA-binding protein. Mol Cell Neurosci 35: 320–327 [DOI] [PubMed] [Google Scholar]
- Valenzuela-Fernandez A, Cabrero JR, Serrador JM, Sanchez-Madrid F (2008) HDAC6: a key regulator of cytoskeleton, cell migration and cell-cell interactions. Trends Cell Biol 18: 291–297 [DOI] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, Ganesalingam J, Williams KL, Tripathi V, Al-Saraj S, Al-Chalabi A, Leigh PN, Blair IP, Nicholson G, de Belleroche J, Gallo JM et al. (2009) Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323: 1208–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt A, Pflanz R, Schafer U, Jackle H (2002) Perlecan participates in proliferation activation of quiescent Drosophila neuroblasts. Dev Dyn 224: 403–412 [DOI] [PubMed] [Google Scholar]
- Vumbaca F, Phoenix KN, Rodriguez-Pinto D, Han DK, Claffey KP (2008) Double-stranded RNA-binding protein regulates vascular endothelial growth factor mRNA stability, translation, and breast cancer angiogenesis. Mol Cell Biol 28: 772–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H-Y, Wang I-F, Bose J, Shen C-KJ (2004) Structural diversity and functional implications of the eukaryotic TDP gene family. Genomics 83: 130–139 [DOI] [PubMed] [Google Scholar]
- Wang I-F, Reddy NM, Shen C-KJ (2002) Higher order arrangement of the eukaryotic nuclear bodies. Proc Natl Acad Sci USA 99: 13583–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang I-F, Wu L-S, Chang H-Y, Shen C-KJ (2008) TDP-43, the signature protein of FTLD-U, is a neuronal activity-responsive factor. J Neurochem 105: 797–806 [DOI] [PubMed] [Google Scholar]
- Watts GDJ, Wymer J, Kovach MJ, Mehta SG, Mumm S, Darvish D, Pestronk A, Whyte MP, Kimonis VE (2004) Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet 36: 377–381 [DOI] [PubMed] [Google Scholar]
- Weihl CC, Temiz P, Miller SE, Watts G, Smith C, Forman M, Hanson PI, Kimonis V, Pestronk A (2008) TDP-43 accumulation in inclusion body myopathy muscle suggests a common pathogenic mechanism with frontotemporal dementia. J Neurol Neurosurg Psychiatry 79: 1186–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winton MJ, Igaz LM, Wong MM, Kwong LK, Trojanowski JQ, Lee VM-Y (2008) Disturbance of nuclear and cytoplasmic TAR DNA-binding protein (TDP-43) induces disease-like redistribution, sequestration, and aggregate formation. J Biol Chem 283: 13302–13309 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Movie S1
Supplementary Figure S3
Supplementary Figure S4
Supplementary Table S1
Supplementary Information
Review Process File