EMBO J 28 24, 3785–3798 (2009); published online 19 November 2009
In his groundbreaking work, ‘The Fractal Geometry of Nature' (Mandelbrot, 1982), Benoit Mandelbrot described how fractals could account for a long list of natural forms ranging from trees to coastlines. Two recent studies in Science and The EMBO Journal have suggested that chromatin and the nucleoplasmic space surrounding it should be added to the fractal list.
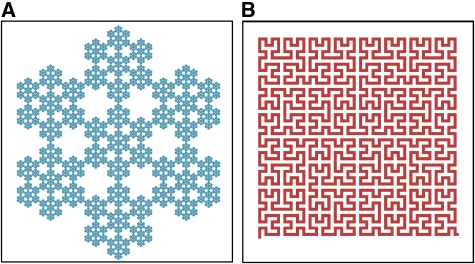
Fractals are ‘self-similar', meaning that they exhibit similar fine-scale features at many magnifications (Figure 1). Over 20 years ago it was argued that folded polymers, including chromatin, should be fractals (Grosberg et al, 1988). The rationale was that as a polymer condenses it is repeatedly subject to the same constraints. Specifically, polymer strands as well as partially folded clumps of the polymer are all impenetrable. Thus, through the self-similar process of crumpling, the resultant condensed polymer becomes a fractal. This ‘fractal globule' structure, as it is called, has advantages because it provides an efficient means to package a long polymer in a small volume without entanglements. This facilitates unravelling the polymer when necessary, for example to gain access to specific DNA segments for transcription, replication or repair.
Figure 1.
(A) The hexa-flake is a fractal that is built recursively from smaller and smaller hexagonal patterns. Four levels of self-similarity are present in this illustration. (B) The Hilbert curve is a fractal that is built recursively from a bilobed pattern. The self-similarity is only evident in the construction of this fractal (see the second website below). In the limit, the hexa-flake has a fractal dimension of d=1.77, indicating that it partially fills the plane, while the Hilbert curve has d=2, indicating that it completely fills the plane. These curves were constructed from the Wolfram demonstration project. http://demonstrations.wolfram.com/NFlakes/; http://demonstrations.wolfram.com/HilbertAndMooreFractalCurves/).
A test of this fractal globule model for chromatin structure has recently been performed using an improved form of 3C called Hi-C (Lieberman-Aiden et al, 2009). Hi-C allows a complete genome-wide identification of chromatin–chromatin interactions. This permits calculating how frequently any pair of genomic loci on the same chromosome make contact as a function of the distance in base pairs (s) separating them. For human chromatin over the range of 500 kb–7 Mb, the average contact frequency decreased at a rate given by 1/s, which is precisely the rate predicted for a fractal globule.
Close on the heels of this Hi-C study is the work of Bancaud et al (2009). These authors measured the diffusion rates of both quantum dots and GFP multimers comprised of 1, 2, 5 or 10 copies of GFP. In all cases they found that the diffusive spreading was slower than conventional diffusion. This is diagnostic of anomalous sub-diffusion, which can occur when obstacles constrain freely diffusive motion. The authors measured an anomaly parameter and found that it was the same for all of the particle sizes they tested. This suggests that the ‘pore' sizes in between chromatin could be similar on different spatial scales, consistent with a fractal structure of the nucleoplasmic space.
The authors found that within heterochromatin the fractal structure of the nucleoplasmic space leads to increased binding association rates for shorter time scales. This results in a process known as compact exploration, in which regulatory factors preferentially sample neighbouring sites. The authors point out that this could aid in the establishment and maintenance of heterochromatin by favouring binding to nearby nucleosomes. The authors did not detect this effect in euchromatin.
Using the measured effects on binding rates and the measured anomaly parameter, the authors were able to predict fractal dimensions of d=2.2 for the open space within heterochromatin and d=2.6 for the open space within euchromatin. The lower fractal dimension of the heterochromatic nucleoplasmic space is consistent with the more condensed nature of heterochromatin, which should have less open space (d=2.2) than euchromatin (d=2.6).
Thus, the two recent studies predict fractal structures for either chromatin or the nucleoplasmic space surrounding chromatin. These results may be consistent, as it is known that if a certain structure is fractal, then its complement may at least resemble a fractal (Crawford and Matsui, 1996). However, in theory, a fractal globule has a fractal dimension of d=3 and should fill the 3D space (Grosberg et al, 1988), leaving no room for a surrounding nucleoplasmic space. In practice, of course no real structure can be packaged so tightly, including the models developed for chromatin fractal globules by Lieberman-Aiden et al (2009). A test of whether the two current studies are consistent could be done by determining first whether the nucleoplasmic space surrounding a chromatin fractal globule itself resembles a fractal, and second, whether under any condition the apparent fractal dimension of this surrounding space would correspond to the predicted values of d=2.2 or d=2.6.
Of course, to know whether chromatin or its surrounding nucleoplasmic space is truly a fractal, we would need to visualize the structures directly. As the two recent studies provide indirect evidence for fractals, it is conceivable that non-fractal structures could also account for the observations. Furthermore, the observations are based on relatively new techniques such as Hi-C and quantitative live cell diffusion and binding analysis. As these tools mature and other more direct techniques evolve, it is likely that our notions about fractal structure in the nucleus will be revised. Regardless, these recent studies with their creative approaches have injected a new life into the longstanding problem of chromatin organization.
Acknowledgments
We thank Tim Stasevich for help with Figure 1.
References
- Bancaud A, Huet S, Daigle N, Mozziconacci J, Beaudouin J, Ellenberg J (2009) Molecular crowding affects diffusion and binding of nuclear proteins in heterochromatin and reveals the fractal organization of chromatin. EMBO J 28: 3785–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford J, Matsui N (1996) Heterogeneity of the pore and solid volume of soil: distinguishing a fractal space from its non-fractal complement. Geoderma 73: 183–195 [Google Scholar]
- Grosberg AY, Nechaev S, Shakhnovich E (1988) The role of topological constraints in the kinetics of collapse of macromolecules. J Phys France 49: 2095–2100 [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelbrot BB (1982) The Fractal Geometry of Nature New York: WH Freeman and Co. [Google Scholar]