Abstract
High-level β-globin gene expression is dependent on the presence of the locus control region (LCR), a powerful regulatory element physically characterized by five DNase I-hypersensitive sites (HS), designated HS1–HS5. Of these, HS3 contains seven GT motifs that are essential for its activity. One of the motifs, GT6, has been shown by in vivo footprinting to display the largest difference in signal between fetal and adult globin expressing cells. We assessed the contribution of GT6 on the downstream globin gene expression by mutating this motif in a 248 kb β-globin locus yeast artificial chromosome and measuring the activity of β-globin genes in GT6m β-YAC transgenic mice. Seven transgenic lines were established, three of which contained at least one intact copy of the β-globin locus and were further investigated. The mutation of the GT6 motif reduced the expression of ε- and γ-globin genes during embryonic erythropoiesis. During definitive erythropoiesis, γ-globin gene expression was significantly reduced while β-globin gene expression was virtually indistinguishable from wild-type controls. We conclude that the GT6 motif of hypersensitive site 3 of the LCR is required for normal ε- and γ-globin gene expression during embryonic erythropoiesis and for γ-globin gene expression during definitive erythropoiesis in the fetal liver. Our results provide evidence that mutations of single transcriptional motifs of distant regulatory elements can have profound effects on gene expression.
INTRODUCTION
Five functional genes comprise the human β-globin locus and are arranged in the order of their expression during development (5′ ε–Gγ–Aγ–δ–β 3′). The ε-globin gene is expressed in embryonic yolk sac from the third to the eighth week of gestation. A switch to γ-globin gene expression coincides with a change in the site of erythropoiesis from yolk sac to the fetal liver at around 6–8 weeks of gestation. A second switch, from γ-globin gene expression to β- and δ-globin gene expression takes place late in fetal life. The high levels of expression of the globin genes are dependent on the presence of the locus control region (LCR), a regulatory element located 6–22 kb upstream of the ε-globin gene. The LCR is comprised of five DNase I-hypersensitive sites designated HS1 to HS5 (1,2). Transgenic mice carrying the human globin genes linked to the LCR or to individual HSs, display high-level, copy-number dependent expression (2–7), while fail to express or express low levels the human β-globin genes in the absence of the LCR (8,9). Transgenic mouse studies have been instrumental in the delineation of the molecular control of globin gene switching and the characterization of the functional properties of the LCR (2,10–13).
Most of the activity of the LCR resides in the core elements of the DNAse I-hypersensitive sites (14–17). The core elements range in size from 200–400 bp, and contain binding motifs for erythroid-specific and ubiquitous transcription factors (reviewed in 18,19). Deletion of the core element of a hypersensitive site, in an otherwise intact human β-globin locus, results in severe reduction of globin gene expression (20–23). The deletion of the core element of HS3 results in a characteristic phenotype consisting of absence of ε-globin gene expression in embryonic erythropoiesis, and absence of γ-globin gene expression in definitive erythropoiesis (22). In these mice, human β-globin gene expression is reduced and its levels vary strikingly among transgenic lines, indicating strong position effects by the surrounding mouse chromatin at the site of transgene integration (22,24). A developmental stage-specific phenotype is also characteristic of the deletion of the core element of HS4; this deletion has little effect on ε- and γ-globin gene expression during embryonic erythro-poiesis, but results in a significant decrease in γ-globin gene expression during definitive erythropoiesis in the fetal liver and a decrease in β-globin gene expression in adult erythropoiesis (23).
HS3 has been shown to activate β-globin gene transcription (25) and to open the chromatin of the β-globin locus domain (22,26). A 225 bp core fragment possesses the essential properties of HS3 and contains binding sites for GATA-1 that alternate with GT motifs (14,27,28). GT motifs (5′ GGTGTGGGG 3′) are important cis-regulatory elements required for proper expression of many housekeeping and tissue-specific genes. These G-rich motifs are bound by a family of Sp1- and Kruppel-like transcription factors containing a highly conserved DNA-binding domain consisting of three zinc fingers (29,30). Four GATA-1, and seven GT motifs (GT1–GT7) are contained in the HS3 core; a single NF-E2 motif is located immediately upstream of the 5′ boundary of the core (18,27). In vivo dimethyl sulfate footprinting experiments using human fetal liver erythroid cell hybrids have been used to characterize the protein–DNA interactions during the developmental switch from γ- to β-globin gene expression (31). These hybrids have been formed by fusing mouse erythroleukemia (MEL) cells with human fetal erythroid cells and they retain the unlimited proliferative potential of the MEL cells and the globin gene phenotype of the human parental cells (32). The hybrids initially express almost exclusively human fetal globin but subsequently switch to exclusive adult β-globin gene expression after 20–40 weeks in culture (32). By in vivo footprinting, the major differences between the fetal and the adult globin-expressing hybrids were on the GT motifs of the LCR, particularly the GT1, GT2, GT6 and GT7 motifs of the HS3 core element. Other studies have shown that the Erythoid Kruppel-like factor (EKLF), a transcriptional factor which is necessary for β-globin gene expression, binds to the juxtaposed GT1/GT2 motifs of the HS3 core element and is required for HS3 activity in transgenic mice (33).
To test whether mutation of GT motifs of the LCR affect the expression of the downstream globin genes we produced transgenic mice carrying a human β-globin yeast artificial chromosome (β-YAC), in which the sixth GT motif (GT6) of the HS3 core element was mutated. We established three transgenic mouse lines and determined the effects of the mutation on globin gene expression during development. We found that, as a result of the GT6 mutation, ε- and γ-globin gene expression were significantly reduced in embryonic erythropoiesis. When the erythropoeisis transitioned to the definitive stage of the fetal liver, γ-globin gene expression was significantly reduced, but the levels of β-globin gene expression were normal. These results provide in vivo experimental evidence that mutations of single motifs of distant regulatory elements can affect the expression of the genes these elements control.
RESULTS
Construction of the β-locus YAC containing a mutation of the GT6 motif of HS3
β-Globin yeast artificial chromosomes have been used extensively to study the role of the HS core elements in the control of globin gene expression and globin gene switching. Mice carrying either 150 kb (34) or 248 kb (7) β-locus YACs display normal developmental control of the globin genes. The ε-globin gene is expressed exclusively in the embryonic stage of development. The γ-globin gene is also expressed in the embryonic stage like its murine ortholog, the βh1 gene, but it also continues to be expressed in the definitive erythropoiesis of the fetal liver and switches off around birth. Adult β-globin gene expression begins in the fetal liver stage of definitive erythropoiesis and peaks after birth (7,35). YAC transgenics, therefore, provide an excellent model system to test whether a mutation of a transcriptional motif affects the regulation of globin gene expression during development.
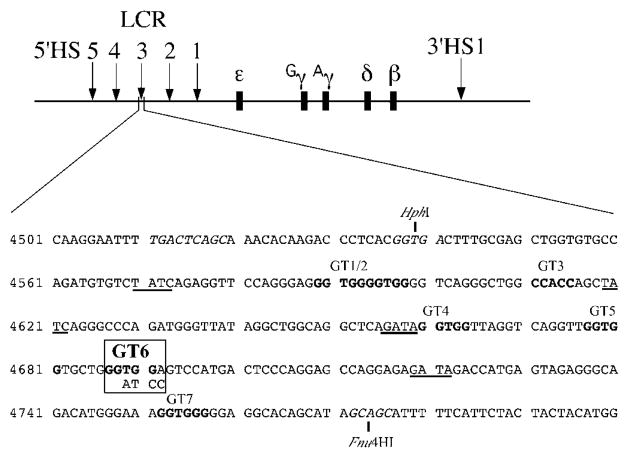
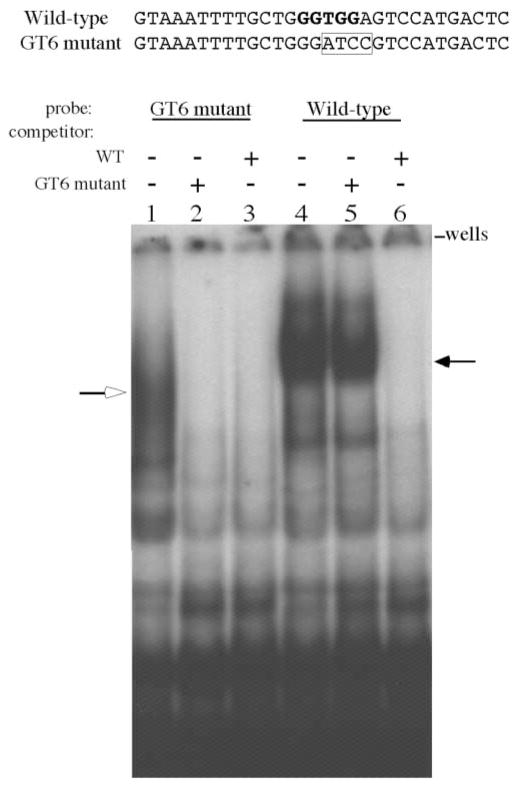
To study the functional effect of the GT6 mutation, we introduced this mutation in the core element of the HS3 of the 248 kb β-locus YAC. The HS3 core element is defined as the 225 bp HphI–Fnu4HI fragment (Fig. 1) (14). Figure 1 shows the transcriptional motifs of the HS3 core; they include four GATA-1 motifs, seven GT motifs and a single NF-E2 motif residing just upstream of the HphI restriction site, at the 5′ boundary of the core element. We performed in vitro mutagenesis to alter the consensus 5′GGTGG3′ core sequence to 5′GATCC3′, effectively mutating the GT6 motif while preserving the size of the HS3 core element of the β-YAC and simultaneously creating a BamHI restriction site which could be used to identify the mutation (Fig. 1). The disruption of protein binding to the mutant GT6 motif was determined by performing electromobility shift assays (EMSA). MEL cell nuclear extracts were incubated with 32P-labeled double stranded oligonucleotides containing either the wild-type GT6 motif or the mutated GT6 motif (GT6m) and the resultant protein–probe complexes were subjected to gel electrophoresis as described in Materials and Methods. As shown by the EMSA autoradiogram in Figure 2, a major complex bound to the wild-type GT6 motif (lane 4) was absent using the GT6m probe (lane 1). A faster migrating complex was identified using the GT6m probe, but was effectively competed by an excess of both the unlabeled GT6m probe (lane 2) and the wild-type probe (lane 3), suggesting that this complex was not specific for the GT6 motif. In contrast, the GT6m probe was unable to compete for the major slow migrating complex bound to the wild-type GT probe (lane 5). Notice the equal signal intensity between lanes 4 (no competitor) and 5 (GT6m competitor) indicating that the four nucleotide substitutions in the GT6 motif completely disrupt protein binding in vitro.
Figure 1.
Mutation of the GT6 motif in the HS3 core element. A diagram of the human β-globin locus is shown in the upper panel. The five DNase I-hypersensitive sites (HS1–HS5) that comprise the LCR are indicated by arrows upstream of the five β-like globin genes, which are represented as dark boxes. 3′HS1 is shown by the arrow located downstream of the β-globin gene. The core element resides on a 225 bp HphI–Fnu4HI fragment. The wild-type sequence is shown with the GT motifs (GGTGG) in bold letters; each GT site is labeled above the sequence (GT1–GT7). GATA-1 sites are underlined and the single NF-E2 motif just upstream of the HphI restriction site is shown in italics. The base substitutions used to mutate the GT6 motif (boxed) are shown underneath the wild-type sequence. The mutation results in the introduction of a BamHI restriction site. Genbank humhbb coordinates (accession number U01317) are shown on the left.
Figure 2.
Electrophoretic mobility shift assay (EMSA) of MEL nuclear extracts with probes derived from the GT6 motif of the HS3 core element. Sequences of the wild-type GT6 motif (WT) and mutant GT6 motif (GT6m) are shown above the autroradiogram. The WT sequence has the GT motif shown in bold and the GT6m sequence shows the four nucleotide substitutions (boxed). A 32P-labeled double stranded oligonucleotide encompassing the WT or mutant GT6 motif was used in the electrophoretic mobility shift assay. Nuclear extracts were prepared from MEL cells as described in the Materials and Methods. Lanes 1–3 used the GT6m probe: lane 1 no competitor; lane 2, competed with unlabeled GT6m probe; lane 3, competed with unlabeled WT probe. Unlabeled competitor DNA was added at 200-fold molar excess. Notice the binding of a complex designated by an open arrow that was effectively competed by both the GT6m and WT probes. Lanes 4–6 used the WT sequence as the probe; lane 4, no competitor; lane 5, competed with unlabeled GT6m probe; lane 6, competed with unlabeled WT probe. A slow migrating complex (filled arrow) is shown in lane 4 and in lane 5 where unlabeled GT6m was used as a competitor. Unlabeled WT probe easily competed for this complex as seen in lane 6.
The GT6 mutation was introduced into the 248 kb β-globin yeast artificial chromosome (β-YAC) by homologous recombination as described in Materials and Methods. The resultant mutant GT6m β-YAC was microinjected into fertilized murine oocytes and ten transgenic founders were produced.
Structural studies of GT6m β-YAC transgenic mice
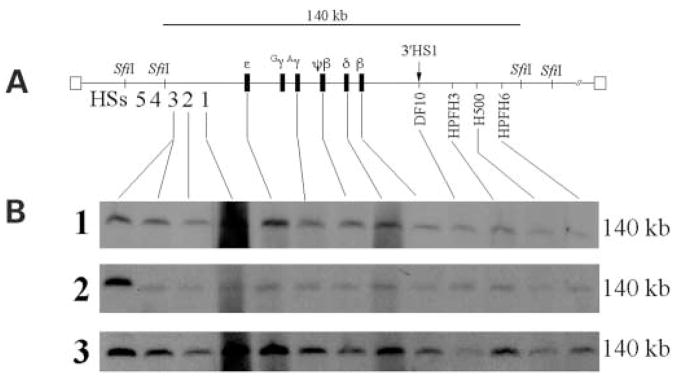
Owing to the large size of the YAC transgene there is a propensity for damage to the DNA during isolation, manipulation microinjection and/or during integration into the mouse genome (22–24,35). Therefore, it was necessary to determine the relative intactness of the β-YAC globin locus in the transgenic mice prior to the analysis of the effects of the GT6 mutation on globin gene expression during development. Pulse-field gel electrophoresis (PFGE) followed by Southern blot hybridization was used to perform detailed structural analysis of the integrated copies of the β-YAC. The majority of the β-globin locus resides on a 140 kb fragment (Fig. 3A). The 5′ SfiI boundary is between HS4 and HS3 of the LCR and the 3′ boundary is located ~60 kb downstream of the β-globin gene, just downstream of the breakpoint of hereditary persistence of fetal hemoglobin (HPFH) 6 (36). β-YAC transgenic mouse DNA embedded in agarose was digested with SfiI, and the DNA fragments were fractionated by PFGE, followed by Southern hybridization analysis. The integrity of an individual β-YAC and the continuity of the globin locus sequences were determined by hybridization using twelve probes distributed along the entire β-globin locus. The probes used are listed in Materials and Methods and are shown on the schematic drawing of the β-globin locus of Figure 3A.
Figure 3.
Structural analysis of GT6m β-YAC transgenic lines. A major problem using YACs as trangenes to produce transgenic mice is the high frequency of deletions within the transferred YACs. We utilize pulsed-field gel electrophoresis followed by Southern blot hybridization analysis to determine the structural integrity of the integrated β-YAC transgene. (A) The figure shows the 140 kb SfiI fragment encompassing most of the β-globin locus from 5′HS3 to the breakpoint of HPFH6 353 kb downstream of the β-globin gene. This various probes used are indicated on the diagram of the β-YAC and in Materials and Methods. (B) Autoradiograms of transgenic lines 1–3 carrying the GT6m β-YAC. Each line contains a single 140 kb SfiI fragment signifying an intact β-globin locus is integrated into the mouse genome. The first lane in each autoradiogram contains DNA from MEL cell line containing a single intact β-YAC, as determined previously by structural analysis and fluorescent in situ hybridization.
Ten transgenic founders were identified, of which three failed to transmit. Seven lines were established from the transmitting founders and liver cell preparations from F2 offspring of each line were used for structural analysis of the β-YAC integrity. Four lines contained β-YACs with deleted sequences of the β locus and were not used for further studies. The remaining three GT6m β-YAC transgenic lines harboring at least one intact β-YAC copy were used for our functional studies. As seen in Figure 3B, each GT6m β-YAC transgenic line has a positive Southern blot hybridization signal of identical size, for DNA sequences from HS3 of the LCR to the HPFH6 breakpoint. The migration of the DNA fragments was identical to the control, a mouse erythroleukemia cell line containing an intact β-globin locus as previously determined by structural analysis and in situ hybridization (Fig. 3B, lane 1) (37). It has been previously shown that when the wild-type β-globin locus is intact from 5′HS4 of the β-globin LCR to the 3′ β-globin gene enhancer, the developmental expression of the globin genes is normal both temporally and spatially (35).
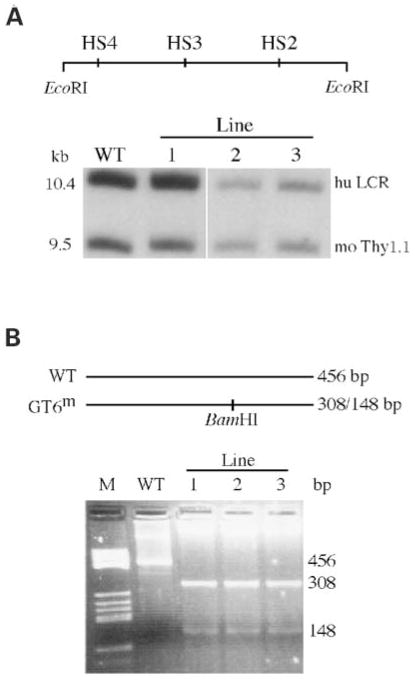
Transgene copy numbers were determined by conventional Southern blot hybridization analyses by comparing hybridization signals between human globin sequences and the endogenous single copy (per haploid genome) murine Thy1.1 gene (Fig. 4A). Line 1 contained two intact copies of the β-globin locus while lines 2 and 3 each contain a single copy of the β-globin locus. The presence of HS4 cannot be identified by the structural analysis of the 140 kb SfiI β-globin locus fragment since it lies upstream of the 5′ most SfiI site used in the assay. However, the presence of a 10.4 kb EcoRI fragment containing HS4, HS3 and HS2 can be confirmed by Southern hybridization analysis (Fig. 4A). Each trangenic line was shown to contain this 10.4 kb EcoRI fragment, indicating that the LCR is intact.
Figure 4.
Southern blot analyses to detect HS4 and PCR analyses to con-firm the presence of the GT6m mutation in the GT6m β-YAC transgenic lines. (A) Southern blot hybridization analysis showing that HS4 resides on a 10.4 kb EcoRI fragment along with HS3 and HS2. Each line produces a fragment that migrates at the same position as the wild-type control. The 9.5 kb fragment is of the murine thy1.1 gene used for copy number analysis. (B) PCR analysis followed by gel electrophoresis to detect BamHI site created by mutating the GT6 motif. As seen in the ethidium bromide stained agarose gel, PCR ampli-fication followed by digestion with BamHI results in a single 456 fragment in the wild-type lane (BamHI resistant) and two fragments of 308 and 148 bp in the GT6m transgenic lines.
The GT6m mutation was identified by polymerase chain reaction (PCR) amplification of the HS3 core element of liver DNA isolated from the GT6m and wild-type transgenic lines. The primers were designed to amplify a 456 bp amplicon (Fig. 4B). The PCR DNA product from the GT6m lines was digested with BamHI, which results in two fragments of 308 and 148 bp in size. The two fragments were detected in each of the GT6m lines, confirming that the mutant GT motif was present (lanes 3–5). Wild-type β-YAC PCR DNA was resistant to BamHI digestion (lane 2).
The GT6 mutation reduces human ε- and γ-globin gene expression during embryonic erythropoiesis
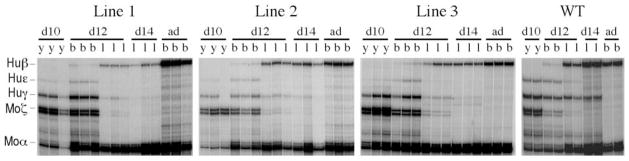
To determine the level of expression of the human globin genes, total RNA was prepared from yolk sac, liver and blood samples of transgenic F2 embryos at 10- and 12-day postconception and subjected to RNase protection analysis. In wild-type β-YAC transgenic mouse lines, human ε- and γ-globin mRNA are detected as early as 9 days post-conception and persist in the circulating blood through day 14 of development (7,35). Anti-sense RNA probes specific for human ε-, γ- and β-globin and mouse α- and ζ-globin were hybridized to total RNA isolated from the erythropoietic tissues. RNase protections were done separately with each embryo of each litter to minimize experimental error and to determine variation in globin gene expression. The amount of human messenger RNA was measured by phosphorimaging analysis and the expression levels were calculated as a percentage of endogenous murine α + ζ globin per gene copy. As shown in Figure 5, and summarized in Table 1, the average expression of the ε-globin gene among the three GT6m β-YAC transgenic lines in day 10 yolk sac and day 12 blood was 3.1±1.2% and 6.2±2.5, respectively, compared with 7.8±1.3 and 16.5±2.3%, respectively, in wild-type β-YAC lines. The average decrease in ε-globin gene expression during embryonic erythropoiesis was 2.4- and 2.9-fold in day 10 yolk sac (P <0.01) and day 12 blood (P <0.01), respectively, relative to the wild-type controls.
Figure 5.
Analysis of human globin mRNA levels in F2 progeny of GT6m β-YAC transgenic lines. Total RNA was isolated from day 10 yolk sac (d10; y), day 12 liver, (d12; l), day 12 blood (d12; b), day 14 liver (d14; l) and adult blood (ad; b) of three F2 littermates from each line and subjected to RNase protection analysis. The migration of the protected fragments is shown on the left of the first autoradiogram; human β-globin, hu β; human γ-globin, hu γ; human ε-globin, hu ε; murine ζ-globin; mo ζ; murine α, mo α-globin. Notice the decrease in ε-globin mRNA in day 10 yolk sac and in γ-globin mRNA levels in day 12 and day 14 fetal liver compared to the wild-type control (last panel). mRNA was quantitated by phosphorimaging and expression levels of the individual human globin genes were calculated as a percentage of murine α-plus ζ-globin per gene copy. Results are presented in Table 1.
Table 1.
Human globin mRNA levels per copy of transgene and copy of endogenous murine α- and ζ-globin in GT6m transgenic mice and wild-type β-YAC control mice
| β-YAC | Line | Percentage of murine α- plus ζ-globin mRNA (mean±SD) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ε-globin mRNA during embryonic erythropoiesis | γ-globin mRNA during: Embryonic erythropoiesis | Definitive erythropoiesis | β-globin mRNA during definitive erythropoiesis | |||||||
| Day 10 yolk sac | Day 12 blood | Day 10 yolk sac | Day 12 blood | Day 12 liver | Day 14 liver | Day 12 liver | Day 14 liver | Adult blood | ||
| 1 | 1.9±0.9 | 3.7±1.4 | 10.9± 3.4 | 12.1±2.1 | 2.6± 1.1 | 1.4±0.1 | 27.3±8.4 | 16.4± 1.8 | 114±10.1 | |
| 2 | 4.3±1.2 | 8.6±0.3 | 14.4± 3.3 | 12.3±2.6 | 6.3± 2.4 | 3.6±0.3 | 36.1±5.2 | 48.8± 19.6 | 113.8±15.3 | |
| 3 | 3.1±0.3 | 6.2±0.2 | 9.6± 0.5 | 13.2±0.2 | 5.6± 0.7 | 2.9±1.8 | 24.1±4.5 | 34.2± 4.4 | 73.9±5.2 | |
| Mean of GT6m lines | 3.1±1.2 | 6.2±2.5 | 11.6± 2.5 | 12.5±0.6 | 4.8± 1.9 | 2.6±1.1 | 29.2±6.2 | 33.1± 16.2 | 100.3±22.8 | |
| Mean of wild-type lines | 7.8±1.3 | 16.5±2.3 | 20.6± 2.4 | 28.7±1.6 | 21.0± 3.7 | 9.4±0.7 | 43.2±5.9 | 43.1± 7.6 | 122.1±18.9 | |
γ-Globin gene expression in day 10 yolk sac was 11.6±2.5%, significantly different (P <0.01) from the mean mRNA level of 20.6±2.4% in the wild-type β-YAC controls. γ-Globin gene expression during embryonic erythropoiesis peaks at day 12 blood, which consists mostly of nucleated erythrocytes from the yolk sac. The mean γ-globin mRNA level in day 12 blood of wild-type β-globin locus YAC mice was 28.7±1.6%. The GT6 mutation resulted in a 2.3-fold reduction of γ-globin gene expression in day 12 blood, the mean mRNA level being 12.5±0.6% (P <0.01) among the GT6m β-YAC transgenic lines (Fig. 5 and Table 1). The significant decrease in ε- and γ-globin gene expression suggests that the GT6 motif is required for normal ε- and γ-globin gene expression during embryonic erythropoiesis.
The GT6 mutation reduces γ-globin gene expression in the cells of definitive erythropoiesis
The definitive stage of erythropoiesis begins in the murine fetal liver at ~10.5 day post-conception and is characterized by the transcription of two adult globins, β-major and β-minor. In wild-type β-globin locus YAC transgenic mice, the γ-globin gene is also expressed during the definitive erythropoiesis in the fetal liver (7,32). In the wild-type β-YAC transgenic mice the mean γ-globin mRNA level in day-12 fetal livers was 21.0±3.7%; it was 4.6±1.9% (P <0.01) in the 12 day fetal liver cells of the GT6 mutant mice (Table 1 and Fig. 5). In the wild-type β-YAC mice, γ gene expression declined to 9.4± 0.7% by fetal day 14; expression had declined to 2.3± 1.5% (P <0.01) in the 14 day fetal liver cells of the GT6 mutant mice (Table 1 and Fig. 5). Apparently, the GT6 motif of the HS3 core is required for normal γ-globin gene expression in the definitive erythroid cells of the fetal liver.
β-globin gene expression is normal in adult GT6m β-YAC transgenic mice
β-Globin gene expression levels in GT6m β-YAC transgenic mice were similar to those measured in the wild-type controls. Mean β-globin mRNA expression in adult blood was 100.3±22.8%, while in wild-type controls of 122.1±18.9% (P ≈ 0.236). Deletion of the HS3 core element results in striking variation of β-globin gene expression levels among trangenics, indicating that the β-globin gene is not protected from the negative effects of the surrounding chromatin. In contrast, the GT6 mutation resulted in less than two fold variation in β-globin mRNA levels between transgenic lines (114±10.1, 113.8±15.3 and 73.9±5.2% in adult blood collected from lines 1–3, respectively). In general, less than two-fold variation in transgene expression is not considered to reflect effects of position of integration. We conclude that the GT6 motif is not required for normal β-globin gene expression and does not contribute to the function of the HS3, which protects the β-globin genes from position effects. Other motifs of the HS3 core element should be responsible for controlling the levels of β-gene expression in adults.
DISCUSSION
Our data show that a mutation of the sixth GT motif residing in the HS3 core element of the LCR results in a decrease in ε- and γ-globin gene expression in embryonic cells of yolk sac origin, a decrease in γ-globin gene expression in cells of definitive erythropoiesis in the fetal liver, but normal β-globin gene expression during the adult stage of definitive erythropoiesis. These results provide evidence that this motif is an important contributor to the control of globin gene expression during embryonic and early fetal life. This conclusion is consistent with our previous observations, showing that the deletion of 234 bp encompassing the entire core element of HS3 abolishes ε-globin gene expression during the embryonic stage and γ-globin gene expression during the fetal stage of development (22). The GT6 mutation reduced ε-globin gene expression to ~40% of the wild-type level, suggesting that a significant portion of the ε-globin gene enhancement by the HS3 core element is controlled by the GT6 motif. Similarly, the GT6m mutation resulted in a decrease of γ-globin gene expression to less than 43.5% of wild-type levels in day 12 embryonic blood cells, suggesting that the GT6 motif is also required for normal γ-globin gene expression during primitive erythropoiesis in transgenic mice.
In wild-type β-YAC transgenic mice, γ-globin expression continues in the fetal liver stage of definitive erythropoiesis; it declines as development advances and switches off around birth (7,35). Deletion of the HS3 core element totally abolishes γ-globin gene expression in the definitive erythroid cells of the fetal liver, indicating that this core element is necessary for γ-gene expression in definitive erythropoiesis. As we have shown here, mutation of the GT6 motif greatly reduced γ-globin gene expression in definitive cells of the fetal liver indicating that this motif is required for normal γ-globin gene expression during fetal life. Our data suggest that the GT6 motif accounts for ~70% of the enhancement of γ-globin gene expression by HS3 during definitive erythropoiesis in the fetal liver. The remaining level of enhancement should be contributed by the other transcriptional motifs of the HS3 core of the LCR.
Deletion of the HS3 core element reduced the capacity of the LCR to protect the β-globin gene from the repressive effects of the neighboring murine chromatin. β-Globin gene expression among transgenic mice carrying the HS3 core deletion ranged from 3% to 60% of wild-type levels (22,24), indicating the presence of strong position effects. Similar phenotypes were observed by other investigators using either a 72 kb β-locus cosmid transgene (6) or a 155 kb β-YAC (20) in which the HS3 core was deleted. This phenotype is not HS3 specific, since deletions of core elements of HS2 and HS4 resulted in a similar reduction and variable levels of β-globin gene expression in adult cells (12,20,21,23). In contrast, there was essentially no decline in β-gene expression and no significant variation in expression between β-YAC transgenic lines carrying the GT6 motif mutation. Apparently, the GT6 motif is dispensable with regard to β-globin gene expression during definitive erythropoiesis, consistent with the minor in vivo footprints of this motif in human fetal erythroid ×MEL cell hybrids expressing the adult globin program, indicating that this motif is not binding proteins during β-globin gene expression (31). Thus, while HS3 is an important contributor to the function of the LCR in opening the globin locus domain in definitive cells, the GT6 motif is not a necessary component of this function.
Quantitative variation in gene expression presumably underlies several quantitative traits and contributes to the pathogenesis of common diseases in humans (38–40). This quantitative variation most likely reflects structural variation in the regulatory elements controlling the genes involved. Here we show that mutations of a single motif of a distant and highly complex regulatory element can also contribute to the quantitative variation of downstream located genes. Identification of regulatory elements controlling structural genes will represent an important step towards the detection of regulatory mutants affecting specific motifs involved in the control of gene expression.
MATERIALS AND METHODS
Construction of the GT6m β-YAC transgene
Plasmid pRSIII1.8 (HpaI–HindIII) is a yeast-integrating-plasmid (YIP) containing the 1.8 kb HpaI–HindIII fragment encompassing the 225 bp HS3 core element and 1572 bp of flanking DNA sequence (GenBank humhbb, accesssion number U01317; coordinates 3375–5172). Complimentary 33-mer primers were synthesized with the DNA sequence of 5′GTTGGTGGTGCTGGATCCAGTCCATGACTCCCC3′ (nucleotide substitutions are shown in italics and the BamHI site is underlined). This set of complimentary primers was used to mutate the GT6 motif by in vitro site-directed mutagenesis using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla CA, USA) following the manufacturer’s protocol. The mutation in the resultant plasmid, pRSHS3-GT6m, was confirmed by restriction enzyme digestion and DNA sequence analysis. pRSHS3-GT6m was linearized with the restriction enzyme SpeI and transformed into spheroplasted Saccharomyces cerevisiae strain AB1380 containing the 248 kb β-YAC (7). Transformants were selected for uracil prototrophy on complete medium and correct recombinants were identified by Southern blot analysis. Spontaneous excision of the YIP was induced by overnight growth in non-selective rich medium (yeast–peptone–dextrose). The yeast cells were plated on 5-fluoroorotic acid plates to select for loss of the URA3 gene residing on the YIP vector, which results in 5-fluoroorotic acid resistance. The presence of the GT6 mutation was confirmed by Southern blot analysis.
Electromobility shift assay
Nuclear extracts were prepared from murine erythroleukemia cells (MEL). MEL cells were harvested and washed twice with cold phosphate-buffered saline (PBS) followed by a wash in a 5-fold volume related to the packed cell volume (PCV) of hypotonic buffer [10 mM HEPES–KOH, pH 7.9; 1.5 mM MgCl2; 10 mM KCl; 0.5 mM dithiothreitol (DTT); 0.2 mM phenylmethylsulfonyl fluoride]. The MEL cells were incubated in a 3-fold PVC volume of hypotonic buffer for 10 min at 4°C, followed by homogenization with a Dounce tissue grinder (Wheaton Science, Millville, NJ, USA). The homogenized tissue was centrifuged at 3300g for 15 min at 4°C. The supernatant was discarded and the pellet comprised primarily of nuclei was resuspended in 1× volume of low salt buffer (10 mM HEPES–KOH, pH 7.9; 1.5 mM MgCl2; 10 mM KCl; 0.2 mM phenylmethylsulfonyl fluoride; 0.5 mM DTT), and high salt buffer (20 mM HEPES–KOH, pH 7.9; 25% of glycerol; 1.5 mM MgCl2; 1.2 M KCl; 0.2 M EDTA; 0.2 mM phenylmethylsulfonyl fluoride; 0.5 mM DTT) was added to obtain a final salt concentration of 0.3 M. The suspension was incubated for 30 min at 4°C. The cellular debris was removed by centrifugation at 12 000g for 15 min at 4°C and the nuclear extract supernant was collected and stored at −70°C. The wild-type GT6 sequence (WT) of the double stranded oligonucleotide used as a probe was 5′GTAAATTTTGCTGGGTGGAGTCC-ATGACTC3′, the GT motif is underlined and five nucleotides in italics were substituted to eliminate the GT5 motif that resides 5 bp upstream of GT6. The GT6 mutant (GT6m) sequence of the double stranded oligonucleotide used as probe was 5′GTAAATTTTGCTGGGATCCGTCCATGACTC3′, the BamHI site shown in bold. Radiolabeled WT and GT6m double-stranded oligonucleotides (2 ×104 cpm) were incubated with 3 μg of MEL cell nuclear extract for 20 min at room temperature in binding buffer [20 mM Tris–HCl, pH 7.5; 100 mM NaCl; 1 mM DTT; 10% glycerol; 0.5 μg of poly(dI-dC); 0.05% NP-40] and 200-fold excess of competitor, when used. Samples were subjected to electrophoresis in a 5% polyacrylamide gel in 1× Tris–borate/EDTA (TBE) buffer containing 5 mM Mg2+ at 4°C, followed by autoradiography.
YAC purification and production of transgenic mice
The yeast strain containing the GT6m β-YAC was grown and YAC DNA isolated as previously described (22). The purified and filtered YAC was injected into fertilized mouse eggs (B6/C3F1) and then transferred to pseudopregnant foster mothers (B6/D2F1). Transgenic founder animals were identified by Southern hybridization slot blot of tail DNA. The transgenic founders were bred with non-transgenic mice (B6/D2F1) to produce F1 progeny, and the F1 males were subsequently bred for staged pregnancies that were interrupted at post-conception days 10, 12 and 14 to collect fetuses or bred to produce F2 adults.
Determination of transgene copy number
Agarose plugs containing high molecular weight liver DNA were digested with restriction enzymes overnight. The resultant fragments were fractionated by agarose gel electrophoresis and the gel was blotted onto Zeta-probe positive-charged nylon membrane (Bio-Rad, Hercules, CA, USA). Transgene copy number was determined by comparing HS4, HS3, HS2, ε-, Aγ- and β-globin gene hybridization signals to the endogenous murine Thy1.1 signal as previously described (22). The radioactive signals were measured using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA, USA) and the ratio of human globin sequences to Thy1.1 (corrected for a haploid genome) was calculated to determine transgene copy number.
Structural analysis of GT6m β-YAC transgenic mice
High molecular weight liver DNA embedded in agarose was prepared as previously described (23). The probes used in the Southern hybridization analyses were radiolabeled using a Decaprime II random probe labeling kit following the manufacturer’s instructions (Ambion, Austin, TX, USA). The radiolabeled fragments used as probes for the structural analyses are as follows: 0.7 kb PstI HS3, 1.9 kb HindIII HS2, 1.8 kb XbaI HS1, 3.7 kb EcoRI ε-globin gene, 2.4 kb EcoRI fragment 3′ of the Aγ-globin gene, 1.0 kb EcoRV ψβ region, 2.1 kb PstI fragment upstream of the δ-globin gene, 0.9 kb EcoRI–BamHI fragment 3′ of the β-globin gene, 1.4 kb XbaI DF10 (3′HS1), 1.9 kb BglII HPFH3, 0.5 kb HindIII H500 and 1.5 kb EcoRI–BglII HPFH6.
Measurement of globin mRNA synthesis
Total RNA was isolated from yolk sac, liver and blood from F2 transgenic embryos, fetuses, and adults using the RNAgents total RNA Isolation System following the manufacturer’s instructions (Promega, Madison, WI, USA). Human and murine globin mRNAs were detected by RNase protection analysis and quantitated using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA, USA). Template DNAs used to prepare riboprobes to measure human ε-, γ- and β-globin mRNA were pT7Huε(188), pT7Aγm(170) and pT7βm, respectively; templates to measure endogenous murine α- and ζ-globin were pT7 Moα and pT7 Moζ, respectively. The source and amount (in parentheses) of isolated RNAs used in RNase protection assays were as follows: day 10 yolk sac (1000 ng), day 12 liver (500 ng), day 12 blood (80 ng), day 14 liver (500 ng), and adult blood (50 ng).
Polymerase chain reactions
The HS3 core region was amplified using the following HS3 specific primers: 5′ HS3 core (proximal) 5′GAGGAGGATCA-GATGGATGGGGC3′ and 3′HS3 core (distal) 5′GGCACTT-GCCCCTAGCTGG3′. 100 ng of genomic DNA were PCR amplified using Taq DNA polymerase in storage buffer B following the manufacturer’s instructions (Promega, Madison, WI, USA). The PCR conditions were the following: 30 s at 95 °C, 45 s at 55°C, 40 s at 72°C for 30 cycles.
Acknowledgments
We thank Alex Rohde, Julianne Roy, Julie Stewart and Betty Mastropaolo for excellent technical support. This work was supported by the National Institutes of Health grants DK45365 and HL20899.
References
- 1.Tuan D, Solomon W, Li Q, London IM. The ‘beta-like-globin’ gene domain in human erythroid cells. Proc Natl Acad Sci USA. 1985;82:6384–6388. doi: 10.1073/pnas.82.19.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grosveld F, van Assendelft GB, Greaves DR, Kollias G. Position-independent, high-level expression of the human β-globin gene in transgenic mice. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 3.Blom van Assendelft G, Hanscombe O, Grosveld F, Greaves DR. The β-globin dominant control region activates homologous and heterologous promoters in a tissue-specific manner. Cell. 1989;56:969–977. doi: 10.1016/0092-8674(89)90630-2. [DOI] [PubMed] [Google Scholar]
- 4.Talbot D, Collis P, Antoniou M, Vidal M, Grosveld F, Greaves DR. A dominant control region from the human β-globin locus conferring integration site-independent gene expression. Nature. 1989;338:352–355. doi: 10.1038/338352a0. [DOI] [PubMed] [Google Scholar]
- 5.Fraser P, Hurst J, Collis P, Grosveld F. DNaseI hypersensitive sites 1, 2 and 3 of the human β-globin dominant control region direct position-independent expression. Nucl Acids Res. 1990;18:3503–3508. doi: 10.1093/nar/18.12.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strouboulis J, Dillon N, Grosveld F. Developmental regulation of a complete 70-kb human β-globin locus in transgenic mice. Genes Dev. 1992;6:1857–1864. doi: 10.1101/gad.6.10.1857. [DOI] [PubMed] [Google Scholar]
- 7.Peterson KR, Clegg CH, Huxley C, Josephson BM, Haugen HS, Furukawa T, Stamatoyannopoulos G. Transgenic mice containing a 248-kb yeast artificial chromosome carrying the human β-globin locus display proper developmental control of human globin genes. Proc Natl Acad Sci USA. 1993;90:7593–7597. doi: 10.1073/pnas.90.16.7593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Townes TM, Lingrel JB, Chen HY, Brinster RL, Palmiter RD. Erythroid-specific expression of human β-globin genes in transgenic mice. EMBO J. 1985;4:1715–1723. doi: 10.1002/j.1460-2075.1985.tb03841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chada K, Magram J, Costantini F. An embryonic pattern of expression of a human fetal globin gene in transgenic mice. Nature. 1986;319:685–689. doi: 10.1038/319685a0. [DOI] [PubMed] [Google Scholar]
- 10.Forrester WC, Novak U, Gelinas R, Groudine M. Molecular analysis of the human β-globin locus activation region. Proc Natl Acad Sci USA. 1989;86:5439–5443. doi: 10.1073/pnas.86.14.5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Q, Stamatoyannopoulos JA. Position independence and proper developmental control of γ-globin gene expression require both a 5′ locus control region and a downstream sequence element. Mol Cell Biol. 1994;14:6087–6096. doi: 10.1128/mcb.14.9.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milot E, Strouboulis J, Trimborn T, Wijgerde M, de Boer E, Langeveld A, Tan-Un K, Vergeer W, Yannoutsos N, Grosveld F, et al. Heterochromatin effects on the frequency and duration of LCR-mediated gene transcription. Cell. 1996;87:105–114. doi: 10.1016/s0092-8674(00)81327-6. [DOI] [PubMed] [Google Scholar]
- 13.Ryan TM, Behringer RR, Martin NC, Townes TM, Palmiter RD, Brinster RL. A single erythroid-specific DNase I super-hypersensitive site activates high levels of human β-globin gene expression in transgenic mice. Genes Dev. 1989;3:314–323. doi: 10.1101/gad.3.3.314. [DOI] [PubMed] [Google Scholar]
- 14.Philipsen S, Talbot D, Fraser P, Grosveld F. The β-globin dominant control region: hypersensitive site 2. EMBO J. 1990;9:2159–2167. doi: 10.1002/j.1460-2075.1990.tb07385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Talbot D, Philipsen S, Fraser P, Grosveld F. Detailed analysis of the site 3 region of the human β-globin dominant control region. EMBO J. 1990;9:2169–2177. doi: 10.1002/j.1460-2075.1990.tb07386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pruzina S, Hanscombe O, Whyatt D, Grosveld F, Philipsen S. Hypersensitive site 4 of the human β globin locus control region. Nucl Acids Res. 1991;19:1413–1419. doi: 10.1093/nar/19.7.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu D, Chang JC, Moi P, Liu W, Kan YW, Curtin PT. Dissection of the enhancer activity of β-globin 5′ DNase I-hypersensitive site 2 in transgenic mice. Proc Natl Acad Sci USA. 1992;89:3899–3903. doi: 10.1073/pnas.89.9.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardison R, Slightom JL, Gumucio DL, Goodman M, Stojanovic N, Miller W. Locus control regions of mammalian β-globin gene clusters: combining phylogenetic analyses and experimental results to gain functional insights. Gene. 1997;205:73–94. doi: 10.1016/s0378-1119(97)00474-5. [DOI] [PubMed] [Google Scholar]
- 19.Stamatoyannopoulos G, Grosveld F. Hemoglobin switching. In: Stamatoyannopoulos G, Majerus PW, Perlmutter RM, Varmus H, editors. Molecular Basis of Blood Diseases. 3. WB Saunders; Philadephia, PA: 2001. pp. 135–182. [Google Scholar]
- 20.Bungert J, Dave U, Lim KC, Lieuw KH, Shavit JA, Liu Q, Engel JD. Synergistic regulation of human β-globin gene switching by locus control region elements HS3 and HS4. Genes Dev. 1995;9:3083–3096. doi: 10.1101/gad.9.24.3083. [DOI] [PubMed] [Google Scholar]
- 21.Bungert J, Tanimoto K, Patel S, Liu Q, Fear M, Engel JD. Hypersensitive site 2 specifies a unique function within the human β-globinlocus control region to stimulate globin gene transcription. Mol Cell Biol. 1999;19:3062–3072. doi: 10.1128/mcb.19.4.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Navas PA, Peterson KR, Li Q, Skarpidi E, Rohde A, Shaw SE, Clegg CH, Asano H, Stamatoyannopoulos G. Developmental specificity of the interaction between the locus control region and embryonic or fetal globin genes in transgenic mice with an HS3 core deletion. Mol Cell Biol. 1998;18:4188–4196. doi: 10.1128/mcb.18.7.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Navas PA, Peterson KR, Li Q, McArthur M, Stamatoyannopoulos G. The 5′ HS4 core element of the human β-globin locus control region is required for high-level globin gene expression in definitive but not in primitive erythropoiesis. J Mol Biol. 2001;312:17–26. doi: 10.1006/jmbi.2001.4939. [DOI] [PubMed] [Google Scholar]
- 24.Navas PA, Li Q, Peterson KR, Swank RA, Rohde A, Roy J, Stamatoyannopoulos G. Activation of the β-like globin genes in transgenic mice is dependent on the presence of the β-locus control region. Hum Mol Genet. 2002;11:893–903. doi: 10.1093/hmg/11.8.893. [DOI] [PubMed] [Google Scholar]
- 25.Fraser P, Pruzina S, Antoniou M, Grosveld F. Each hypersensitive site of the human β-globin locus control region confers a different developmental pattern of expression on the globin genes. Genes Dev. 1993;7:106–113. doi: 10.1101/gad.7.1.106. [DOI] [PubMed] [Google Scholar]
- 26.Ellis J, Tan-Un KC, Harper A, Michalovich D, Yannoutsos N, Philipsen S, Grosveld F. A dominant chromatin-opening activity in 5′ hypersensitive site 3 of the human β-globin locus control region. EMBO J. 1996;15:562–568. [PMC free article] [PubMed] [Google Scholar]
- 27.Philipsen S, Pruzina S, Grosveld F. The minimal requirements for activity in transgenic mice of hypersensitive site 3 of the β globin locus control region. EMBO J. 1993;12:1077–1085. doi: 10.1002/j.1460-2075.1993.tb05749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strauss EC, Orkin SH. In vivo protein-DNA interactions at hypersensitive site 3 of the human β-globin locus control region. Proc Natl Acad Sci USA. 1992;89:5809–5813. doi: 10.1073/pnas.89.13.5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Philipsen S, Suske G. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucl Acids Res. 1999;27:2991–3000. doi: 10.1093/nar/27.15.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaczynski J, Cook T, Urrutia R. Sp1- and Kruppel-like transcription factors. Genome Biol. 2003;4:206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ikuta T, Papayannopoulou T, Stamatoyannopoulos G, Kan YW. Globin gene switching. In vivo protein-DNA interactions of the human β-globin locus in erythroid cells expressing the fetal or the adult globin gene program. J Biol Chem. 1996;271:14082–14091. doi: 10.1074/jbc.271.24.14082. [DOI] [PubMed] [Google Scholar]
- 32.Papayannopoulou T, Brice M, Stamatoyannopoulos G. Analysis of human hemoglobin switching in MEL × human fetal erythroid cell hybrids. Cell. 1986;46:469–476. doi: 10.1016/0092-8674(86)90667-7. [DOI] [PubMed] [Google Scholar]
- 33.Gillemans N, Tewari R, Lindeboom F, Rottier R, de Wit T, Wijgerde M, Grosveld F, Philipsen S. Altered DNA-binding specificity mutants of EKLF and Sp1 show that EKLF is an activator of the β-globin locus control region in vivo. Genes Dev. 1998;12:2863–2873. doi: 10.1101/gad.12.18.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaensler KM, Kitamura M, Kan YW. Germ-line transmission and developmental regulation of a 150-kb yeast artificial chromosome containing the human β-globin locus in transgenic mice. Proc Natl Acad Sci USA. 1993;90:11381–11385. doi: 10.1073/pnas.90.23.11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peterson KR, Navas PA, Li Q, Stamatoyannopoulos G. LCR-dependent gene expression in β-globin YAC transgenics: detailed structural studies validate functional analysis even in the presence of fragmented YACs. Hum Mol Genet. 1998;7:2079–2088. doi: 10.1093/hmg/7.13.2079. [DOI] [PubMed] [Google Scholar]
- 36.Kosteas T, Palena A, Anagnou NP. Molecular cloning of the breakpoints of the hereditary persistence of fetal hemoglobin type-6 (HPFH-6) deletion and sequence analysis of the novel juxtaposed region from the 3′ end of the β-globin gene cluster. Hum Genet. 1997;100:441–445. doi: 10.1007/s004390050530. [DOI] [PubMed] [Google Scholar]
- 37.Peterson KR, Zitnik G, Huxley C, Lowrey CH, Gnirke A, Leppig KA, Papayannopoulou T, Stamatoyannopoulos G. Use of yeast artificial chromosomes (YACs) for studying control of gene expression: correct regulation of the genes of a human β-globin locus YAC following transfer to mouse erythroleukemia cell lines. Proc Natl Acad Sci USA. 1993;90:11207–11211. doi: 10.1073/pnas.90.23.11207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheung VG, Spielman RS. The genetics of variation in gene expression. Nat Genet. 2002;32 (suppl):522–525. doi: 10.1038/ng1036. [DOI] [PubMed] [Google Scholar]
- 39.Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW. Allelic variation in human gene expression. Science. 2002;297:1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
- 40.Cheung VG, Conlin LK, Weber TM, Arcaro M, Jen KY, Morley M, Spielman RS. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat Genet. 2003;33:422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]