Abstract
Sp1/Krüppel-like factor (KLF) family of transcription factors regulates diverse biological processes including cell growth, differentiation, and development through modulation of gene expression. This family of factors regulates transcription positively and negatively by binding to the GC and GT/CACCC boxes in the promoter through their highly conserved three zinc finger domains. Although the molecular mechanism of gene regulation by this family of proteins has been well studied, their exact role in growth and development in vivo remains largely unknown. KLF11 has been implicated in the regulation of cell growth and gene expression. To determine the physiological function of KLF11, we generated KLF11-null mice by gene-targeting technology. Homologous KLF11−/− mice were bred normally and were fertile. Hematopoiesis at all stages of development was normal in the KLF11−/− mice. There was no effect on globin gene expression. These mice lived as long as the wild-type mice without evident pathological defects. Thus, despite its cell growth inhibition and transcriptional regulation functions observed when transiently or stably expressed in cultured cells in vitro, the results from genetic knockout suggest that KLF11 is not absolutely required for hematopoiesis, growth, and development.
Keywords: Sp1/Krüppel-like factor, KLF11, Targeted gene inactivation
Introduction
The Sp1/Sp1/Krüppel-like factor (KLF) proteins are a family of highly conserved transcription factors that are characterized by the presence of three highly homologous Cys2/His2-type zinc fingers near the C-terminus that bind GC/CACCC box, which is one of the most common regulatory elements in promoters of many cellular and viral genes. Amino acid sequences in the transcription activation/repression domains are less conserved among family members. So far, six members in the Sp subgroup and 16 members in the KLF subgroup have been identified in mammalian cells [17]. This family of proteins possesses transcriptional activation, repression, or both functions in a cell context- and gene-specific manner. By regulating gene expression through binding to the GC/CACCC boxes in a target gene promoter, this family of factors regulates diverse cellular processes including cell growth and differentiation and is essential for early embryonic development [3,4,17,27]. Studies on the molecular mechanisms by which they regulate transcription have demonstrated that members of this family form transcriptional activation or repression complexes via direct interactions with coactivators such as CBP/p300, PCAF, and SWI/SNF, as well as corepressors such as CtBP and mSin3A [3,4,17,27]. Currently, however, for most of them, their exact function in vivo and their target specificity remain unknown. This is mainly because they all have similar DNA binding specificity and affinity to the GC, GT, or CACCC boxes at least under in vitro conditions and many of them are ubiquitously expressed.
KLF11 is a member of the Sp1/KLF family of transcription factors and is expressed in many tissues [2,8]. KLF11 was also named FKLF (fetal Krüppel-like factor) and was isolated during a search for factors that may play a role in human γ-globin expression via the CACCC box in its promoter [2]. KLF11 was able to activate ε- and γ-promoter-driven reporter expression, and its overexpression induced endogenous ε- and γ-gene expression in K562 cells [2]. KLF11 is also named TIEG2 [transforming growth factor (TGF) β-inducible early gene-2] on the basis of its homology to KLF10 (also termed TIEG1) and its inducible expression by TGFβ [8,30]. KLF11 was also shown to inhibit transcription through the N-terminal domain [7]. KLF11 interacted with mSin3A through a conserved α-helical repression motif (αHRM) located within the repression domain (R1) of KLF11, and this interaction is disrupted by the epidermal growth factor (EGF)-Ras-MEK1-ERK2 signaling pathway [35]. Therefore, it is possible that KLF11 may have different activity under different conditions.
KLF11 has the highest homology with KLF10 within this family of transcription factors [3], indicating a functional similarity between these two factors. Both proteins are early TGFβ-inducible genes [2,8] and inhibit cell growth by acting as the potential effectors of the TGFβ signaling pathway [9]. Recently, it has been shown that KLF10 enhanced TGFβ stimulation of gene expression by repression of the inhibitory Smad7 expression [15,16]. Overexpression of KLF10 in epithelial cells induced apoptosis [6,28,33]. KLF11 transgenic pancreas also showed an increased rate of apoptosis [9]. KLF10 is highly expressed in normal breast epithelium and completely silenced in invasive carcinoma [31]. These data together suggest that KLF10 and KLF11 may play important roles in the regulation of cell growth and cancer development by functioning as direct downstream factors of growth regulatory signaling pathways.
To understand the in vivo role of KLF11 in growth and development and gene regulation, we have inactivated the mouse KLF11 gene by gene knockout technology. KLF11−/− mice were normal in growth, bred according to the normal mendelian genetics, and lived as long as the KLF11+/+ mice. Hematological analyses also revealed no abnormalities in the KLF−/− mice. The finding that KLF11 is not absolutely essential for the normal development of mice suggests that there is functional redundancy among this highly conserved family of transcription factors.
Materials and methods
Construction of targeting vector
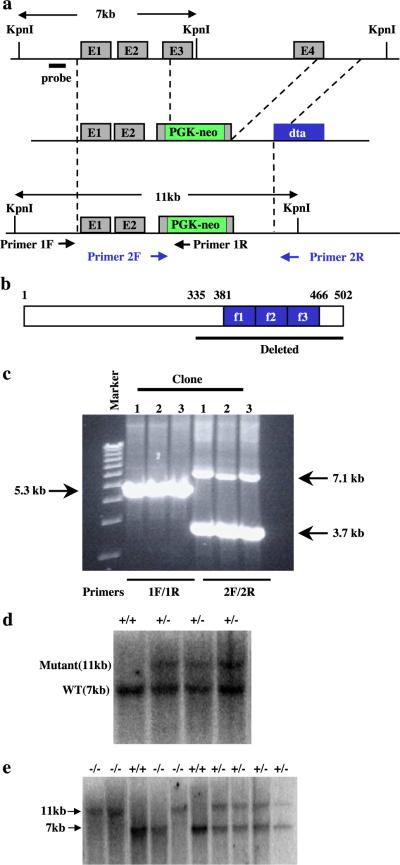
We have isolated a genomic clone that contains the full mouse KLF11 gene from a mouse 129SV genome library (Stratagene). Sequence analysis revealed that KLF11 is encoded by four exons with three introns (Fig. 1a). We intended to make gene-targeting construct that replace the zinc finger-coding region with the Neor gene. The 4.2-kb NotI (from phage vector)–KpnI (intron 3) 5′ homologous fragment and the 4.7-kb HindIII (intron 3 to downstream of exon 4) 3′ homologous fragment were subcloned into pBluescript SK—vector at the NotI and KpnI, and HindIII sites, respectively. An EcoRV fragment was removed from the 3′ HindIII fragment, and a PGK-Neor cassette (blunted EcoRI–SalI fragment) was inserted into the cleaved p3′ H3. Subsequently, the plasmid was digested with HindIII (blunted) and PmlI, and self-ligated to generate pNeo-3′. The NotI–XmnI fragment of p5′ NK was inserted into the NotI/EcoRI (blunted) sites of pNeo-3′ to produce the p5′Neo-3′. The 5′-Neo-3′ sequence was cut by NotI (blunted) and XhoI (blunted), and inserted into diphtheria toxin A gene (DTA)-containing vector at the SmaI site to generate the gene-targeting vector p5′-Neo-3′-DTA. KLF11 targeting vectors were illustrated in Fig. 1a. Homologous recombination of KLF11 will result in the deletion of the C-terminal portion of the protein including the entire DNA-binding zinc finger domain. The DTA gene is cloned outside the region of homologous recombination, thus allowing a negative selection against cells derived from nonhomologous recombination events.
Fig. 1.
Targeted disruption of the KLF11 gene. (a) Schematic representations of the wild-type KLF11 locus, targeting vector, and the targeted allele. (b) Sequences that are deleted from the KLF11 gene. (c) PCR analysis of KLF11 targeted ES cells. The primer set 1F and 1R detects a 5.3-kb band for the targeted allele and no band for the wild-type allele. The primer set 2F and 2R detects a 3.7-kb band for the targeted allele and a 7.1-kb band for the wild-type allele. (d) Southern blot analysis of targeted ES cells. DNA isolated from ES cells was digested with KpnI. The wild-type allele is detected as a 7-kb fragment. The targeted allele is detected as an 11-kb fragment. (e) Southern blot analysis of mice generated from a KLF11+/− cross. DNA isolated from mice tail was digested with KpnI. The wild-type allele is detected as a 7-kb fragment. The targeted allele is detected as an 11-kb fragment.
Isolation of KLF11+/− ES cell clones
Mouse R1 ES cells were cultured as described [1]. R1 ES cells were transfected with NotI-digested targeting vectors of KLF11 by electroporation using a BioRad Gene Pulser II at 250 KV, 500 μF. One day following electroporation, cells were selected with 200 μg/ml G418 for 10 days. Colonies were picked into medium without G418 and were split into duplicate plates, one with gamma-irradiated primary mouse embryonic fibroblast feeder for freeze storage and the other without a feeder layer for analysis. G418-resistant clones were first screened for homologous recombination by PCR using two pairs of primers flanking the 5′ and 3′ integration sites and producing distinct bands of the wild-type and targeted alleles homologous recombination sites (Fig. 1a). Positive clones were further analyzed by Southern blotting using a probe outside the region of the homologous recombination.
Generation of FKLF−/− mice
All mice were housed in a University of Washington, Seattle, SPF facility, were fed standard diet and treated according to IAAAC standards. Targeted clones were injected into blastocysts from C57BL/6 mice and transferred to embryonic day (E)2.5 recipients to produce chimeric mice. Chimeric male offspring were mated to C57BL/6 females to test for germ-line transmission of the KLF11 knockout allele. The KLF11+/− mice were then interbred to produce homozygous mutant mice (KLF11−/−).
Analysis of KLF11 expression in wild-type and KLF11−/− mice
To examine the expression of KLF11 in mouse tissues and further confirm that there is no KLF11 expression in KLF11−/− mice, reverse transcription (RT)–PCR was carried out using RNA isolated from different tissues from KLF11+/+ and KLF11−/− mice. Total RNA was isolated from brain, heart, kidney, liver, lung, muscle, spleen, testes, and thymus using TRIZOL reagent (Invitrogen). RT–CR was carried out using the primers that are specific for KLF11 and KLF13.
Expression of potential target genes of KLF11
Total RNA was isolated from yolk sac of day 10 embryos and fetal liver from day 16 embryos. Expression of mouse εy and βH1-globin in the yolk sac and βmajor in the fetal liver was analyzed using RNase protection assay as described [21].
Hematological analysis KLF11−/− mice
Peripheral blood was obtained through retroorbital sinus bleeding. Red blood cells (RBCs), white blood cells (WBCs), platelets (PLTs), hemoglobin (Hb), mean corpuscular volume (MCV), and mean corpuscular hemoglobin (MCH) were analyzed by complete blood counts (CBC) using a Cell-DYN 3500 hematology analyzer (Abbott Diagnostics, Santa Clara, CA). Reticulocytes were counted on a blood smear after staining with brilliant cresyl blue. Peripheral blood and reticulocyte smears were studied under a Zeiss optical microscope, with 10×, 40×, and100} optical lenses.
Bone marrow was collected from KLF11+/+, KLF11+/−, and KLF−/− mice. Cytospin smears were made by centrifugation at 7000 rpm for 10′ and 0.5 } 10 E4 cells were plated in methylcellulose medium containing rhEpo 3 U/ml, rmIL-3 10 ng/ml, rhIL-6 10 ng/ml, and 50 ng/ml rmSCF (MethoCult™, StemCell Technologies, Vancouver, BC).
Irradiation
Mice were exposed to three weekly doses of 2.5 Gy (7.5 Gy total) of total body irradiation in a 137Cs dual source animal irradiator (Gammacell 40, Nordion, Inc., Ottawa, Canada) and were followed up for 12 months.
Results and discussion
Targeted disruption of KLF11 gene
KLF11 sequences including the three zinc finger DNA-binding domains, the sequences N-terminal of the zinc fingers, and some residues N-terminal of the zinc fingers were replaced with the PGK-Neo cassette (Figs. 1a and b). R1 ES cells were transfected with PacI-digested targeting vector. G418-resistant clones were screened for homologous recombination by PCR using primers franking the 5′ and 3′ integration sites and producing distinct bands of the wild-type and targeted alleles PCR (Figs. 1a, c). Homologous recombination was further confirmed by Southern blot analysis using DNA from the targeted ES cells (Fig. 1d). A total of three correctly targeted clones were identified after screening more than 100 ES cell clones transfected with the KLF11 targeting vector.
KLF11+/− ES cell clones were injected into C57BL/6 blastocysts to produce chimeric mice. High percentage chimeras were obtained as judged from coat color. However, only one of the KLF11+/− ES cell clones resulted in germ-line transmission of the KLF11 knockout allele after mating of chimeric males with C57BL/6 female mice. The KLF11+/− mice were interbred to produce KLF11−/− mice (Fig. 1e). Genotyping the tail DNA of 72 offspring from KLF11+/− interbred revealed that the ratio of KLF11+/+–KLF11+/−–KLF11−/− mice is 17:38:17 which is the normal mendelian ratio of 1:2:1.
Characterization of KLF11 knockout mice
We next determined the expression of KLF11 in different mouse tissues and their absence in the KLF−/− mice. Analysis of total RNA by RT-PCR showed that KLF11 is expressed in all the tissues examined in the KLF11+/+ mouse (Fig. 2). This is in agreement with the reports that KLF11 is expressed ubiquitously in human tissues [2,8]. As expected, KLF11 mRNA is absent in all the tissues in the KLF11−/− mouse. The expression of KLF13 (another member of the KLF family that is ubiquitously expressed) was detected in these tissues of the KLF11−/− mice indicating a specific absence of KLF11 expression in the KLF11−/− mice. These results demonstrated that KLF11 is expressed ubiquitously, and KLF11 is completely disrupted in the KLF11−/− mice.
Fig. 2.
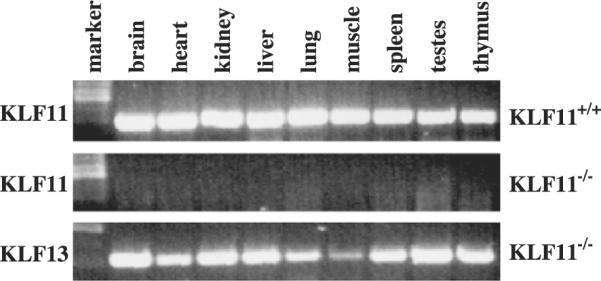
Expression of KLF11 in mouse tissues. RT–PCR analysis for KLF11 expression in tissues from wild-type and KLF11 knockout mice. The PCR primers for KLF11 anneal to the exon 4 region of KLF11, thus they will not be able to amplify the targeted allele.
Homozygous mutant KLF11−/− mice appeared to grow normally and were fertile without any evident defect. We next determined whether KLF11 has an effect on the overall growth of mice since KLF11 has been reported to be able to negatively regulate cell growth in vitro [2,8]. We weighed the knockout offspring at two time points of their development. The average weight at 4 weeks for male knockouts was 17 ± 3.6 g (n = 4) vs. 18.2 ± 1.8 g in the controls (n = 8) and at 6 weeks 22.3 ± 1.3 g (n = 5) vs. 23.5 ± 2.25 g (n = 6), while females were 15.75 ± 6 g (n = 4) and 20.3 ± 0.5 g (n = 3) vs. 14.6 ± 0.9 g (n = 4) and 19.3 ± 1.15 g (n = 3) (Fig. 3). The differences were not statistically significant. These results, as is the case in hematopoietic tissue, indicate that KLF11 is not indispensable for the normal growth and development of mice.
Fig. 3.
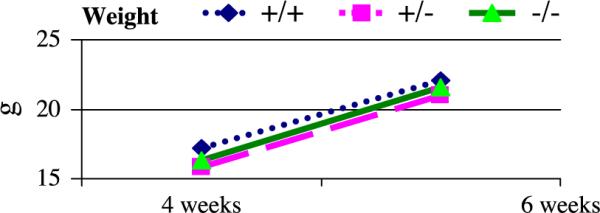
Weight of KLF11 knockout mice. KLF11 knockout mice have similar weight to their heterozygous and wild-type controls (male KLF11+/+ n = 6, KLF11+/− n = 7, KLF11−/− n = 5; female KLF11+/+ n = 3, KLF11+/− n = 8, KLF11−/− n = 3).
KLF11 has been reported to have a suppressive role in tumorigenesis. Mice (three KLF11+/+, three KLF11+/−, and four KLF11−/−) were followed to term. No abnormalities were noted during this long follow-up, and their overall survival was not significantly affected by the lack of KLF11 (Fig. 4).
Fig. 4.
Long-term follow-up of KLF11 knockout mice. Ten littermates were followed to term (3 KLF11+/+, 3 KLF11+/−, and 4 KLF11−/−). No abnormal disease traits were noted and no tumorigenesis as reflected in the similar overall survival.
In addition, eight mice (two KLF11+/+, two KLF11+/−, and four KLF11−/−) were exposed to fractionated sublethal total body g irradiation (7.5 Gy) and were followed up for 12 months. Similar doses of γ irradiation have been shown to induce tumor formation or marrow aplasia in susceptible knockout mouse strains [12–14,19,23]. However, all KLF11−/− mice survived that period showing no evidence of tumor formation, anemia, bleeding diathesis, or susceptibility to infections (data not shown) as did their wild-type and heterozygous littermates. This result indicates that KLF11 is not necessary for radioprotection and suppression of tumor formation under oncogenic stimuli.
The Sp/KLF family of transcription factors contains a highly conserved three C2H2 types of zinc finger DNA binding domain and is capable of binding the GC/CACCC boxes. However, studies have shown that they have distinct functions. For example, SP1, Sp3, and Sp4 knockout mice have different phenotypes. Sp1−/− mice are retarded in development, show a broad range of abnormalities, and die around day 11 of gestation [22]. Sp3 knockout mice showed defects in tooth formation and die at birth due to respiratory failure [5]. Sp4 gene targeting showed that Sp4 is required for normal growth, viability, and male fertility [10,32]. Mice null for KLF1/EKLF, an erythroid-specific KLF family member, die of a lethal anemia due to a specific and substantial decrease in expression of the fetal/adult stage-specific β-globin gene [25,26]. Inactivation of KLF2/LKLF is embryonic-lethal, and developing embryos are growth-retarded [20,34]. KLF4/GKLF knockout mice are normal at birth but die soon thereafter from defects in skin barrier function [29]. KLF4 has also been shown to play a crucial role in colonic epithelial cell differentiation [18]. On the other hand, some functional redundancy among the highly conserved members of this family may exist. For example, mice null of Sp5 [11] and KLF9 [24] showed no overt phenotype. In this study, we also showed that KLF11 is not absolutely essential for normal growth and development.
Hematological analysis KLF11−/− mice
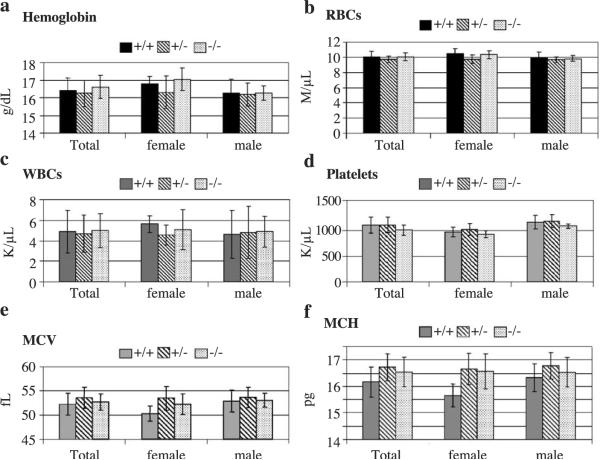
We next examined whether KLF11 plays a role in hematopoiesis. Blood was collected from the retroorbital sinus and analyzed for red blood cells (RBCs), white blood cells (WBCs), platelets (PLTs), hemoglobin (Hb), mean corpuscular volume (MCV), and mean corpuscular hemoglobin (MCH). Hemoglobin of KLF11−/− and KLF11+/+ mice is 16.6 ± 0.7 and 16.4 ± 0.7 g/dl respectively (Fig. 5a). RBCs of KLF11−/− and KLF11+/+ mice are 10 ± 0.5 and 10 ± 0.7 M/μl, respectively (Fig. 5b). WBCs of KLF11−/− and KLF11+/+ mice are 4.97 ± 1.6 and 4.87 ± 2.07 K/μ/l, respectively (Fig. 5c). Platelets of KLF11−/− and KLF11+/+ mice are 1.014 ± 0.102 and 1.11 ± 0.15 M/μl, respectively (Fig. 5d). MCV of KLF11−/− and KLF11+/+ mice is 52.7 ± 1.7 vs. 52.2 ± 2.2, respectively (Fig. 5e). MCH of KLF11−/− and KLF11+/+ mice is 16.5 ± 0.6 vs. 16.15 ± 0.6, respectively (Fig. 5f. Therefore, there is no difference among the KLF11−/− offspring, their wild-type, and heterozygous littermates in RBC, WBC, Hb, MCV, and MCH. In accordance with these results, similar numbers of hematopoietic progenitor colonies were noted on marrow cultures on methylcellulose (Fig. 6) (per 10E4 cells KLF11−/−: 111.5 ± 12 CFU-granulocytic-macrophage (GM), 8.5 ± 07 CFU-erythroid (E), and 2.5 ± 0.7 CFU-megakaryocytic (Meg); KLF11+/+: 115 ± 5.6 CFU-GM, 10.5 ± 2.6 CFU-E, and 1.5 ± 0.7 CFU-Meg). Moreover, no difference was noted on peripheral blood and bone marrow smears (data not shown and Fig. 5). These results indicate that KLF11 is either not required for or functionally redundant in hematopoietic tissue homeostasis.
Fig. 5.
Hematological analysis of KLF11 knockout offspring and their heterozygous and wild-type littermates. Males and females compared separately and as a total. (Male KLF11+/+ n = 7, KLF11+/− n = 6, KLF11−/− n = 6, female KLF11+/+ n = 4, KLF11+/− n = 5, KLF11−/− n = 5).
Fig. 6.
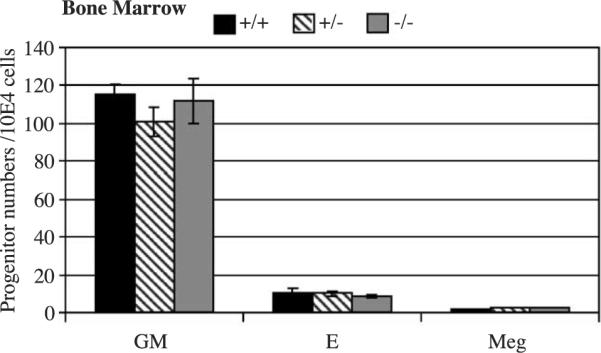
Bone marrow progenitor assay. Progenitor colony numbers were assessed in semisolid media. As expected from the peripheral blood counts and smears, no difference was noted in erythroid (BFU-E, CFU-E), granulocytic-macrophage (CFU-GM), and megakaryocytic progenitors (CFU-Meg). E: erythroid, GM: granulocytic-macrophage, Meg: megakaryocytic.
Expression of globin genes in KLF11−/− mice
KLF11 has been shown to positively and negatively regulate transcription via the CACCC or GC box using either artificial promoter or human hemoglobin promoter-driven reporter expression in transient assays [2,8]. Currently, however, the in vivo target genes of KLF11 are unknown. KLF11 has been shown to activate endogenous human ε- and γ-globin expression when stably overexpressed in K562 cells [2]. Therefore, we first analyzed the effect of KLF11 inactivation on the expression of mouse globin genes. These analyses showed no difference in the expression of all the mouse globin genes between the KLF+/+ and KLF−/− mice (data not shown). It is possible that KLF11 plays a role in globin gene expression, but other members of this family are capable of substituting KLF11 function when it is knocked out. To further test the role of KLF11 in human g-globin expression, we crossed the KLF−/− mice with mice that express the human γ-globin gene. The KLF−/−,γ-globin mice expressed the same level of γ-globin as the KLF +/+,γ-globin mice, indicating that KLF11 is not required for expression of γ-globin transgene in mice (data not shown).
Acknowledgments
We thank Qiliang. Li for analysis of globin gene expression. This work was supported by grants from National Heart, Lung, and Blood Institute, National Institute of Health.
References
- [1].Abbondanzo SJ, Gadi I, Stewart CL. Derivation of embryonic stem cell lines. Methods Enzymol. 1993;225:803–823. doi: 10.1016/0076-6879(93)25052-4. [DOI] [PubMed] [Google Scholar]
- [2].Asano H, Li XS, Stamatoyannopoulos G. FKLF, a novel Krüppel-like factor that activates human embryonic and fetal beta-like globin genes. Mol. Cell. Biol. 1999;19:3571–3579. doi: 10.1128/mcb.19.5.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bieker JJ. Krüppel-like factors: three fingers in many pies. J. Biol. Chem. 2001;276:34355–34358. doi: 10.1074/jbc.R100043200. [DOI] [PubMed] [Google Scholar]
- [4].Black AR, Black JD, Azizkhan-Clifford J. Sp1 and Krüppel-like factor family of transcription factors in cell growth regulation and cancer. J. Cell Physiol. 2001;188:143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- [5].Bouwman P, Gollner H, Elsasser HP, Eckhoff G, Karis A, Grosveld F, Philipsen S, Suske G. Transcription factor Sp3 is essential for post-natal survival and late tooth development. EMBO J. 2000;19:655–661. doi: 10.1093/emboj/19.4.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chalaux E, Lopez-Rovira T, Rosa JL, Pons G, Boxer LM, Bartrons R, Ventura F. A zinc-finger transcription factor induced by TGF-beta promotes apoptotic cell death in epithelial Mv1Lu cells. FEBS Lett. 1999;457:478–482. doi: 10.1016/s0014-5793(99)01051-0. [DOI] [PubMed] [Google Scholar]
- [7].Cook T, Gebelein B, Belal M, Mesa K, Urrutia R. Three conserved transcriptional repressor domains are a defining feature of the TIEG subfamily of Sp1-like zinc finger proteins. J. Biol. Chem. 1999;274:29500–29504. doi: 10.1074/jbc.274.41.29500. [DOI] [PubMed] [Google Scholar]
- [8].Cook T, Gebelein B, Mesa K, Mladek A, Urrutia R. Molecular cloning and characterization of TIEG2 reveals a new subfamily of transforming growth factor-beta-inducible Sp1-like zinc finger-encoding genes involved in the regulation of cell growth. J. Biol. Chem. 1998;273:25929–25936. doi: 10.1074/jbc.273.40.25929. [DOI] [PubMed] [Google Scholar]
- [9].Cook T, Urrutia R. TIEG proteins join the Smads as TGF-beta-regulated transcription factors that control pancreatic cell growth. Am. J. Physiol.: Gastrointest. Liver Physiol. 2000;278:G513–G521. doi: 10.1152/ajpgi.2000.278.4.G513. [DOI] [PubMed] [Google Scholar]
- [10].Gollner H, Bouwman P, Mangold M, Karis A, Braun H, Rohner I, Del Rey A, Besedovsky HO, Meinhardt A, van den Broek M, Cutforth T, Grosveld F, Philipsen S, Suske G. Complex phenotype of mice homozygous for a null mutation in the Sp4 transcription factor gene. Genes Cells. 2001;6:689–697. doi: 10.1046/j.1365-2443.2001.00455.x. [DOI] [PubMed] [Google Scholar]
- [11].Harrison SM, Houzelstein D, Dunwoodie SL, Beddington RS. Sp5, a new member of the Sp1 family, is dynamically expressed during development and genetically interacts with brachyury. Dev. Biol. 2000;227:358–372. doi: 10.1006/dbio.2000.9878. [DOI] [PubMed] [Google Scholar]
- [12].Hollander MC, Sheikh MS, Bulavin DV, Lundgren K, Augeri-Henmueller L, Shehee R, Molinaro TA, Kim KE, Tolosa E, Ashwell JD, Rosenberg MP, Zhan Q, Fernandez-Salguero PM, Morgan WF, Deng CX, Fornace AJ., Jr. Genomic instability in Gadd45a-deficient mice. Nat. Genet. 1999;23:176–184. doi: 10.1038/13802. [DOI] [PubMed] [Google Scholar]
- [13].Hunt CR, Dix DJ, Sharma GG, Pandita RK, Gupta A, Funk M, Pandita TK. Genomic instability and enhanced radiosensitivity in Hsp70.1- and Hsp70.3-deficient mice. Mol. Cell. Biol. 2004;24:899–911. doi: 10.1128/MCB.24.2.899-911.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Inoue A, Seidel MG, Wu W, Kamizono S, Ferrando AA, Bronson RT, Iwasaki H, Akashi K, Morimoto A, Hitzler JK, Pestina TI, Jackson CW, Tanaka R, Chong MJ, McKinnon PJ, Inukai T, Grosveld GC, Look AT. Slug, a highly conserved zinc finger transcriptional repressor, protects hematopoietic progenitor cells from radiation-induced apoptosis in vivo. Cancer Cells. 2002;2:279–288. doi: 10.1016/s1535-6108(02)00155-1. [DOI] [PubMed] [Google Scholar]
- [15].Johnsen SA, Subramaniam M, Janknecht R, Spelsberg TC. TGFbeta inducible early gene enhances TGFbeta/Smad-dependent transcriptional responses. Oncogene. 2002;21:5783–5790. doi: 10.1038/sj.onc.1205681. [DOI] [PubMed] [Google Scholar]
- [16].Johnsen SA, Subramaniam M, Katagiri T, Janknecht R, Spelsberg TC. Transcriptional regulation of Smad2 is required for enhancement of TGFbeta/Smad signaling by TGFbeta inducible early gene. J. Cell. Biochem. 2002;87:233–241. doi: 10.1002/jcb.10299. [DOI] [PubMed] [Google Scholar]
- [17].Kaczynski J, Cook T, Urrutia R. Sp1- and Krüppel-like transcription factors. Genome Biol. 2003;4:206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Katz JP, Perreault N, Goldstein BG, Lee CS, Labosky PA, Yang VW, Kaestner KH. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development. 2002;129:2619–2628. doi: 10.1242/dev.129.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kosugi S, Miyazawa T, Chou D, Saito Y, Shinbo T, Matsuki A, Okano H, Miyaji C, Watanabe H, Hatakeyama K, Niwa O, Kominami R. Mutations in the p53 and scid genes do not cooperate in lymphomagenesis in doubly heterozygous mice. Biochem. Biophys. Res. Commun. 1999;255:99–103. doi: 10.1006/bbrc.1999.0142. [DOI] [PubMed] [Google Scholar]
- [20].Kuo CT, Veselits ML, Barton KP, Lu MM, Clendenin C, Leiden JM. The LKLF transcription factor is required for normal tunica media formation and blood vessel stabilization during murine embryogenesis. Genes Dev. 1997;11:2996–3006. doi: 10.1101/gad.11.22.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Li Q, Duan ZJ, Stamatoyannopoulos G. Analysis of the mechanism of action of non-deletion hereditary persistence of fetal hemoglobin mutants in transgenic mice. EMBO J. 2001;20:157–164. doi: 10.1093/emboj/20.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Marin M, Karis A, Visser P, Grosveld F, Philipsen S. Transcription factor Sp1 is essential for early embryonic development but dispensable for cell growth and differentiation. Cell. 1997;89:619–628. doi: 10.1016/s0092-8674(00)80243-3. [DOI] [PubMed] [Google Scholar]
- [23].Medina D, Ullrich R, Meyn R, Wiseman R, Donehower L. Environmental carcinogens and p53 tumor-suppressor gene interactions in a transgenic mouse model for mammary carcinogenesis. Environ. Mol. Mutagen. 2002;39:178–183. doi: 10.1002/em.10064. [DOI] [PubMed] [Google Scholar]
- [24].Morita M, Kobayashi A, Yamashita T, Shimanuki T, Nakajima O, Takahashi S, Ikegami S, Inokuchi K, Yamashita K, Yamamoto M, Fujii-Kuriyama Y. Functional analysis of basic transcription element binding protein by gene targeting technology. Mol. Cell. Biol. 2003;23:2489–2500. doi: 10.1128/MCB.23.7.2489-2500.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature. 1995;375:316–318. doi: 10.1038/375316a0. [DOI] [PubMed] [Google Scholar]
- [26].Perkins AC, Sharpe AH, Orkin SH. Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature. 1995;375:318–322. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- [27].Philipsen S, Suske G. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 1999;27:2991–3000. doi: 10.1093/nar/27.15.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ribeiro A, Bronk SF, Roberts PJ, Urrutia R, Gores GJ. The transforming growth factor beta(1)-inducible transcription factor TIEG1, mediates apoptosis through oxidative stress. Hepatology. 1999;30:1490–1497. doi: 10.1002/hep.510300620. [DOI] [PubMed] [Google Scholar]
- [29].Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat. Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- [30].Subramaniam M, Harris SA, Oursler MJ, Rasmussen K, Riggs BL, Spelsberg TC. Identification of a novel TGF-beta-regulated gene encoding a putative zinc finger protein in human osteoblasts. Nucleic Acids Res. 1995;23:4907–4912. doi: 10.1093/nar/23.23.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Subramaniam M, Hefferan TE, Tau K, Peus D, Pittelkow M, Jalal S, Riggs BL, Roche P, Spelsberg TC. Tissue, cell type, and breast cancer stage-specific expression of a TGF-beta inducible early transcription factor gene. J. Cell. Biochem. 1998;68:226–236. [PubMed] [Google Scholar]
- [32].Supp DM, Witte DP, Branford WW, Smith EP, Potter SS. Sp4, a member of the Sp1-family of zinc finger transcription factors, is required for normal murine growth, viability, and male fertility. Dev. Biol. 1996;176:284–299. doi: 10.1006/dbio.1996.0134. [DOI] [PubMed] [Google Scholar]
- [33].Tachibana I, Imoto M, Adjei PN, Gores GJ, Subramaniam M, Spelsberg TC, Urrutia R. Overexpression of the TGFbeta-regulated zinc finger encoding gene, TIEG, induces apoptosis in pancreatic epithelial cells. J. Clin. Invest. 1997;99:2365–2374. doi: 10.1172/JCI119418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wani MA, Means RT, Jr., Lingrel JB. Loss of LKLF function results in embryonic lethality in mice. Transgenic Res. 1998;7:229–238. doi: 10.1023/a:1008809809843. [DOI] [PubMed] [Google Scholar]
- [35].Zhang JS, Moncrieffe MC, Kaczynski J, Ellenrieder V, Prendergast FG, Urrutia R. A conserved alpha-helical motif mediates the interaction of Sp1-like transcriptional repressors with the corepressor mSin3A. Mol. Cell. Biol. 2001;21:5041–5049. doi: 10.1128/MCB.21.15.5041-5049.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]