Abstract
FKLF-2 (KLF13) was cloned from fetal globin-expressing tissues and has been shown to be abundantly expressed in erythroid cells. In this study we examined the transcriptional regulation of the KLF13 gene. A 5.5 kb 5′ flanking region cloned from mouse erythroleukemia (MEL) cell genomic DNA showed that major cis regulatory activities exist in the 550 bp sequence to the unique transcription start site, and that the promoter is more active in K562 cells than in COS-7 cells. The promoter was trans-activated by co-expressed GATA-1 through the sequence containing two CCAAT motifs, suggesting that GATA-1 is involved in the abundant expression of KLF13 mRNA in the erythroid tissue. Dual action, i.e. activating effect in COS-7 and repressive effect in K562 cell, was observed on its own promoter, suggesting a feedback mechanism for the transcriptional control of the KLF13 gene in the erythroid environment. These findings provide an insight on the mechanism of inducible mRNA expression of the KLF13 gene in erythroid cells.
Keywords: Erythroid differentiation, KLF family, Promoter, Transcriptional regulation
1. Introduction
Proteins containing three contiguous Cys2–His2 zinc fingers similar to Sp1 or EKLF constitute a KLF family [1–3]. By using the zinc finger motif, these proteins bind to G-rich (GC or GT/CACCC) motifs that are widely distributed in cis-regulatory DNA sequences for gene expression. Some KLFs are ubiquitously expressed, while others show a tissue-restricted expression pattern. It may be reasonable to speculate that the non-ubiquitous KLFs are involved in cell type specific gene expression. Confirming this notion, EKLF (KLF1), which is specifically expressed in erythroid cells [4], plays a critical role for the expression of the β-globin gene [5,6] by interacting with the proximal CACCC motif of the β gene promoter [7,8]. LKLF (KLF2), which is predominantly expressed in the lung [9], is essential for the normal lung development since LKLF−/− ES cells do not contribute to the lung formation in chimeric mice [10]. KLFs thus play a substantial role in their major expression tissues.
Fetal Krüppel-like factor-2 (FKLF-2: KLF13) was originally cloned from mouse yolk sac and human fetal liver erythroid cells, i.e. fetal globin-expressing tissues [11]. It is predominantly expressed in the bone marrow, striated muscles and a subset of T cells. Other groups, in fact, cloned the same gene from these tissues [12,13]. Of note is that the mRNA expression of KLF13 gene is up-regulated upon the induction of differentiation with chemicals in cell lines with erythroid features [11]. Similarly, the expression of KLF13 protein is up-regulated upon the maturation of T cells [12]. These facts strongly suggest that KLF13 may be involved in the differentiation of the cells in which it is expressed. In addition KLF13 is a powerful activator of a broad range of promoters of erythroid genes in vitro [11]. These results suggest that KLF13 plays a role in the development of erythroid phenotype. The transcriptional regulation for KLF13 gene expression may therefore be a part of molecular events of erythroid cell differentiation.
In this study we have cloned the 5′ flanking region of the mouse KLF13 gene. The promoter activity of the DNA fragment was tested in both erythroid (K562) and non-erythroid (COS-7) cells. Our data show that: the KLF13 gene promoter is more active in the erythroid environment than in the non-erythroid environment; and GATA-1 and KLF13 itself may have substantial effect in the transcriptional regulation of the KLF13 gene expression.
2. Materials and methods
2.1. Isolation of 5′ flanking DNA of the KLF13 gene
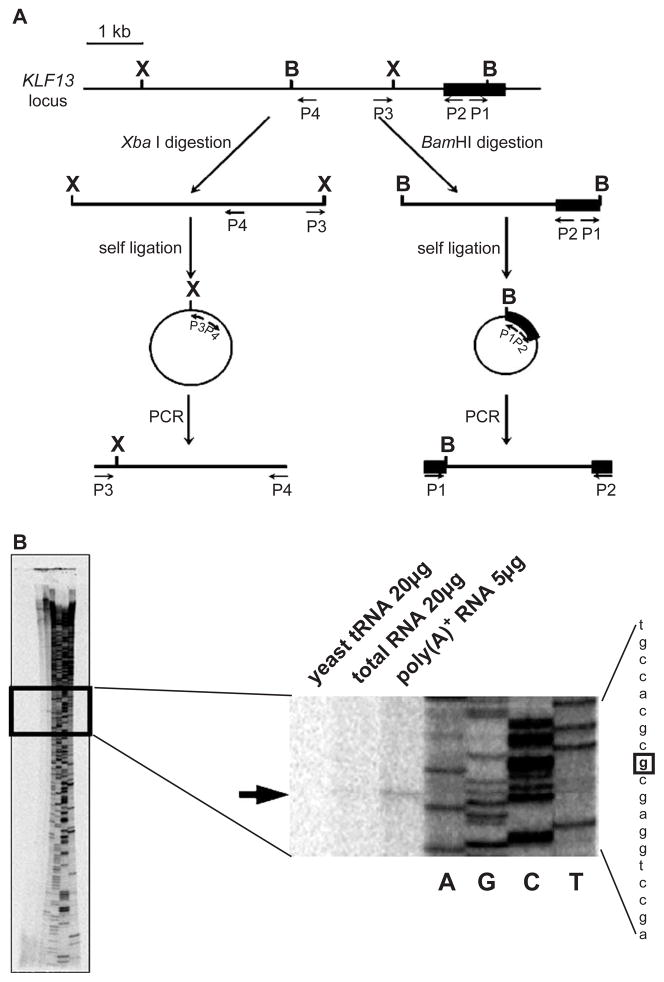
The 5′ flanking region of KLF13 gene was obtained by inverse PCR (Fig. 1A). Four hundred nanograms of genomic DNA extracted from MEL cells was digested with BamHI or XbaI. Subsequently, the DNA was phenol-extracted, ethanol-precipitated and then self-ligated in 100 μL of reaction mixture at 16 °C for 30 min. The self-ligated DNA was dissolved in 8 μL low TE (5 mM Tris–HCl pH 8.0 and 0.1 mM EDTA) and 50 ng of the DNA was subjected to PCR by Expand Long PCR System (Roche) according to manufacture’s instructions. The primer sets used in the PCR are 5′-GCCGCGACGGCAAGGACAGCC-3′ (P1) and 5′-GCATTGTGGCAGGCGGGACAGCC-3′ (P2), and 5′-GGTCTCCCTGACTTGATGGCTG-3′ (P3) and 5′-CAATGCTGAGGGAGTGAGCTAC-3′ (P4) for the BamHI- and XbaI-cut DNA, respectively. The PCR products were cloned into T-vector (Promega) and were fully sequenced. The respective constructs were referred to pBam35 and pXba29.
Fig. 1.
Promoter of the KLF13 gene. (A) Cloning of the 5′ flanking region of KLF13 gene by inverse PCRs. The first exon is shown by a solid rectangle. B and X represent the BamHI and XbaI sites, respectively. Primers used in inverse PCRs are indicated by arrows (P1–P4). Note that the positions of upstream BamHI and XbaI sites are unknown prior to the experiments. (B) Primer extension analysis of KLF13 mRNA. RNAs used in the analysis are indicated above. Products of the sequencing reaction using the same probe were run on gel together, of which reading is shown at right. An arrow indicates the position of the extension product. The nucleotide in the sequence is highlighted by a square.
2.2. Plasmid constructions
The 5.5 kb 5′ flanking DNA of the KLF13 gene was reconstituted in pBluescript SK− vector (Stratagene). The NotI–BamHI fragment from pBam35 was subcloned into the same sites of the vector (referred to pBS28). Subsequently, the BamHI–XbaI (blunted) fragment of pXba29 was inserted at the BamHI and EcoRV sites of pBS28 (referred to pBS55). The 5.5 kb DNA was cut as a NotI (blunted)–KpnI fragment and inserted into pGL2-Basic vector (Promega) at the HindIII (blunted) and KpnI sites (referred to pGL55). The truncation of the DNA was performed by either (1) blunting and self-ligation after double digestion with KpnI and a desired enzyme or (2) cutting with XbaI and a desired enzyme (blunted), and subsequent re-insertion into the pGL2-Basic vector cut with SmaI/XbaI.
GATA-1 expression vector was constructed using the coding region of GATA-1 amplified by reverse transcription (RT)-PCR using random-hexamer-primed cDNA from MEL cell total RNA. The PCR primers are 5′-CAGGTTCAACCCCAGTGTTCCCA-3′ and 5′-CCTTCAAGAACTGAGTGGGGCG-3′. The cDNA fragment was cloned into the T vector, and correct amplification was verified by sequencing. Subsequently, the open reading frame was cut out as a SphI (blunted)–SacI fragment and subcloned into a eukaryotic expression vector pSG5DD [14] at the EcoRI (blunted) and SacI sites.
Mutations were introduced using a kit (Altered Sites II in vitro Mutagenesis Systems, Promega) following the manufacture’s instructions. The nucleotide substitution of individual motifs is listed in Table 1.
Table 1.
List of nucleotide substitutions of cis elements
| Motif | Sequencea |
|
|---|---|---|
| Wild type | Mutant | |
| −0.37 kb CACCC | GGGTG | CTAGT |
| −0.23 kb CACCC | CACCC | ACTAG |
| Distal CCAAT | ATTGG | CAGAT |
| Proximal CCAAT | CCAAT | ATCTC |
Sense strand.
2.3. RNA extractions
Total RNA was extracted by a standard method [15]. Poly(A)+ RNA was separated using an oligo (dT)-cellulose spun column (Pharmacia Biotech).
2.4. Primer extension and sequencing
Primer extension analysis was performed by a standard method [15]. An oligo DNA (5′-GCATTGTGG-CAGGCGGGACAGCC-3′) was end-labeled with [γ33P]ATP and 50,000 cpm was put into the extension reaction. Sequencing of the pBam35 by the same probe was performed using a kit (Cyclist, Stratagene).
2.5. Semi-quantitative RT-PCR
Semi-quantitative RT-PCR [11] was carried out using RQ1 DNase (Promega)-treated total RNA of COS-7 and K562 cells. The primers used for the amplification of KLF13 cDNA were 5′-TTCGCCTGCAGCTGGCAGGA-3′ and 5′-TGGCCGGGCTGATGGTGGG-3′. The condition of PCR was 95 °C (3 min) followed by variable number of cycles of 95 °C (30 s), 62 °C (30 s) and 72 °C (30 s).
2.6. Cell culture and transfections
K562 and COS-7 cells were cultured in RPMI1640 and DMEM, respectively, supplemented with 10% fetal calf serum. Transient transfections were performed using FuGENE 6 Reagent (Roche) according to the manufacture’s instructions (reagent: DNA=3:1). Briefly, 1 μg plasmid DNA mixture (0.45 μg activator, 0.45 μg reporter and 0.1 μg pSVβ-Gal, or 0.9 μg reporter and 0.1 μg pSVβ-Gal) was transfected into 1×105 COS-7 and 5×105 K562 cells in 1 mL culture medium. COS-7 cells had been plated on a 12-well culture dish 24 h prior to the transfection. K562 cells were plated on the 12-well dish just before the transfection. After, the transfection cells were incubated at 37 °C for 24 h. Then, the cells were harvested, washed once with PBS and lysed in 200 μL Reporter Lysis Buffer (Promega). The cell lysate was subjected to luciferase and β-Gal assays as described elsewhere [8]. All transfections were performed three to five times in triplicate.
2.7. Data analysis and statistics
Data expressed as relative values to the average of the standard group were stored, analyzed and reported with the packages STATISTICA (StatSoft). Tukey’s HSD (honestly significant difference) procedure (two-way ANOVA) was used to evaluate the probability of significant differences. P values less than 0.05 were considered statistically significant.
3. Results
3.1. Promoter of the KLF13 gene
To examine the transcriptional regulation of KLF13 gene, it was required to clone the 5′-flanking DNA. When we started this project the DNA sequence was still uncovered. To determine the unknown genomic DNA sequence we carried out two sequential inverse PCRs using MEL cell genomic DNA (Fig. 1A). Each reaction gave rise to a single band (data not shown). Subsequently, these DNA fragments were fully sequenced, which revealed 6183 nucleotides. (GenBank accession no. AY601638). Using the DNA we first determined the transcription start site of the KLF13 gene by the primer extension analysis. As illustrated in Fig. 1B the poly(A)+ RNA generated a unique band that is at the nucleotide position 5471. A band at the same position, although faint, is formed from the total RNA while no bands were observed in the yeast tRNA. These results indicate that: the transcription of the KLF13 gene starts at a unique site, and the DNA obtained by inverse PCRs contains the promoter region of the KLF13 gene. The presence of the promoter activity was confirmed by the luciferase assay using K562 cells. The DNA fragment clearly raised the luciferase activity (pGL55 in Table 2). The average luciferase count driven by the KLF13 genomic DNA was about 314 times higher compared with that obtained from the control vector (pGL2-Basic). The same experiment was performed using COS-7, i.e. non-erythroid, cells. The pGL55 construct generated approximately 164-fold more luciferase counts compared with the pGL2-Basic vector (Table 2). The fold increase, i.e. 314 in K562 cells vs. 164 in COS-7 cells, was significantly different (P<0.001). These results show that: there exists a promoter activity in the DNA fragment that we cloned, and the promoter is more active in erythroid cells than in non-erythroid cells.
Table 2.
Promoter activity of the 5.5 kb DNA fragment
| COS-7 | K562 | |
|---|---|---|
| pGL2-Basic | 1 ± 0.17 | 1 ± 0.23 |
| pGL55 | 163.5 ± 27.7 | 313.8 ± 54.2 |
Luciferase counts were corrected by the β-Gal activities. The average luciferase activities obtained from pGL2-Basic vector was considered as 1. Data are expressed as mean ±1 S.D. of three independent transfections in triplicate.
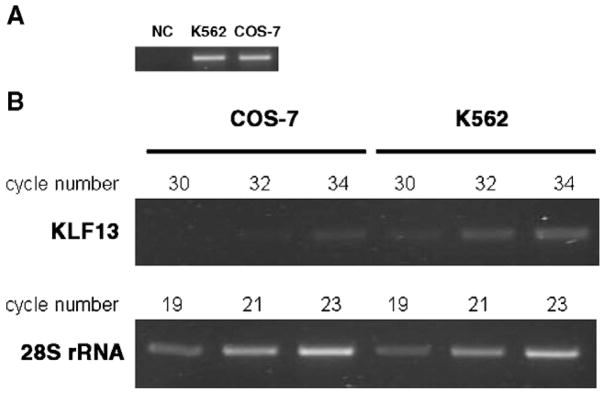
To test whether the different promoter activities were reflected to the gene expression, the expression of KLF13 mRNA was analyzed by semi-quantitative RT-PCR. Since the nucleotide sequence of the KLF13 gene of COS-7 cells was unknown, primers were set in the sequence common between mouse and human KLF13 cDNAs. The primer set amplified the COS-7 and K562 genomic DNA at a comparable efficiency (Fig. 2A). Subsequently, cDNAs of the two cell lines that were diluted to generate similar band patterns on amplification of 28S ribosomal RNA (rRNA) were subjected to PCR of the KLF13 gene. As shown in Fig. 2B cDNA of K562 cells yielded more intense bands in all cycles tested than that of COS-7 cells, demonstrating that higher expression of KLF13 mRNA in K562 cells than in COS-7 cells.
Fig. 2.
Expression of KLF13 mRNAs in COS-7 and K562 cells. (A) PCR using the same amount of genomic DNA. One hundred nanograms of genomic DNA was amplified by 33 cycles of PCR in a 50 μL reaction volume. Subsequently, 10 μL was run on gel. Note that similar bands were generated in COS-7 and K562 cells, indicating comparable efficiencies of amplification between the two cell lines. NC means negative control in which TE buffer instead of DNA was put in the PCR reaction. (B) Semi-quantitative RT-PCR using COS-7 and K562 cell cDNAs. The numbers shown above indicate PCR cycles. Note the similar amplification of 28S rRNA gene between the two cells, and obviously more intense bands on the amplification of KLF13 gene in K562 cells than in COS-7 cells.
3.2. Erythroid cell specific regulation of the KLF13 gene promoter
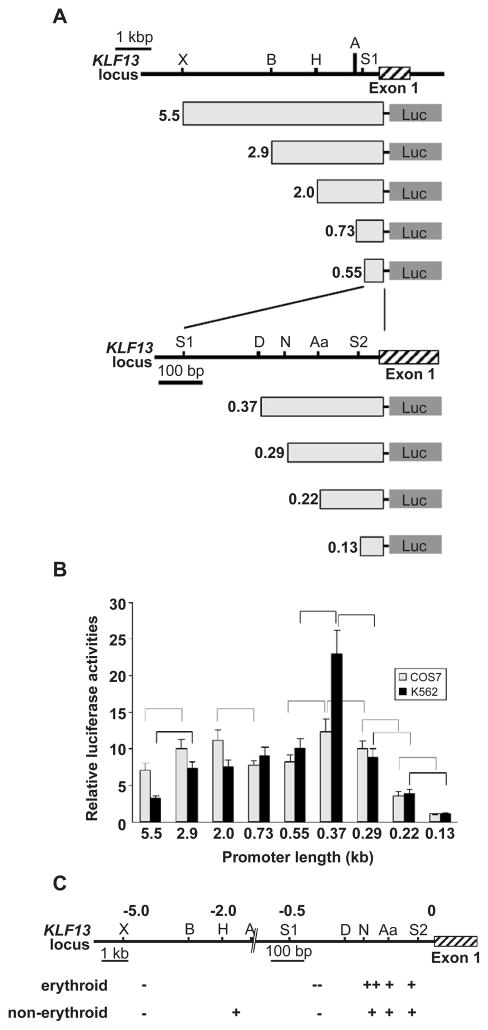
To elucidate the cis regulatory elements of the KLF13 gene promoter, a series of truncated DNA fragments were prepared (Fig. 3A) and their promoter activities were analyzed in COS-7 and K562 cells. Results are expressed as promoter activities relative to those obtained from the 0.13 kb DNA fragment (considered as 1). As shown in Fig. 3B, in COS-7 cells promoter activities were significantly different between 5.5 and 2.9 kb (P<0.001), 2.0 and 0.73 kb (P<0.001), 0.55 and 0.37 kb (P<0.001), 0.37 and 0.29 kb (P<0.001), 0.29 and 0.22 kb (P<0.001), and 0.22 and 0.13 kb (P<0.001), suggesting that two DNA regions, i.e. −0.13 to −0.37 kb and −0.73 to −2.0 kb, function as positive regulatory elements while −0.37 to −0.55 kb and −2.9 to −5.5 kb DNA regions function as negative regulatory elements in non-erythroid cells (Fig. 3C). Since the 0.37 kb fragment exhibited the most powerful promoter activity, which is 12.4-fold higher relative to that of 0.13 kb fragment, among the 9 truncated promoters, major promoter activities exist in this DNA sequence. The highest promoter activity of the 0.37 kb fragment was also observed in K562 cells (Fig. 3B). However, it is of note that the activity is 22.9-fold higher than that of the 0.13 kb fragment, which should be contrasted to the 12.4-fold increase observed in COS-7 cells. The relative promoter activities of DNA fragments containing the upstream sequence (5.5, 2.9 and 2.0 kb) were not as high as those of COS-7 cells (Fig. 3B). The relative activities of 0.73 and 2.0 kb promoters were 9.0 and 7.5, respectively, which was not statistically significant (P=0.46). Therefore the positive regulatory effect of the −0.73 to −2.0 kb region observed in COS-7 cells was absent in K562 cells (Fig. 3C). Together, it is suggested that: the 80 bp sequence between −0.29 and −0.37 kb is more powerful in an erythroid environment than in a non-erythroid environment (expressed as ++ in Fig. 3C), and among the negative regulatory elements the −0.37 to −0.55 kb sequence has a strong effect (expressed as - in Fig. 3C) since the powerful activity of the −0.29 to −0.37 kb region disappeared in the presence of the 180 bp, i.e. −0.37 to −0.55 kb, DNA.
Fig. 3.
Activities of KLF13 gene promoter in erythroid and non-erythroid cells. (A) Reporter constructs and KLF13 locus. Luciferase gene is driven by truncated promoters indicated by light gray rectangles. Restriction sites used for the truncation are indicated above. Abbreviations for restriction sites are X, XbaI; B, BamHI; H, HindIII; A, ApaI; S1, SacI; D, DraIII; N, NaeI; Aa, AatII; and S2, SacII. The length of the promoters is shown at left in kb. (B) Relative luciferase activities obtained from COS-7 (gray bars) and K562 (black bars) cells are shown. Luciferase counts were corrected by the β-Gal activity, and the average count obtained from the 0.13 kb promoter was considered as 1. Error bars indicate one S.D. Results were obtained from three independent transfections in triplicate. Promoter pairs showing significantly different activities are indicated by broken (COS-7) and solid (K562) lines. (C) Positive (+) and negative (−) regulatory regions of KLF13 gene promoter. KLF13 locus is shown above and numbers are the distance to the transcription start site in kb. Note that the scale is altered at −0.7 kb position. Regions having a potent effect are indicated by ++ and −−.
3.3. The erythroid factor GATA-1 and KLF13 are potential transcriptional regulators of the KLF13 gene promoter
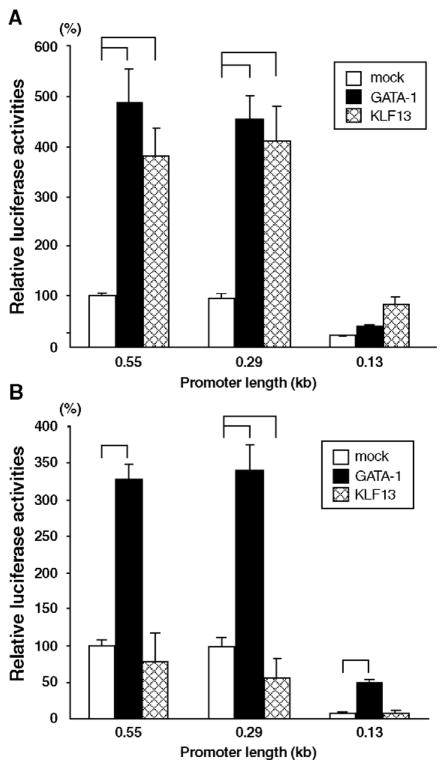
The results shown above indicate that: the regulation of KLF13 gene promoter is distinct in erythroid cells, and major regulatory elements of the promoter are located in the proximal 550 bp region. Inspection and computer database search (TRANSFAC, http://www.motif.genome.jp/) of the DNA sequence revealed a number of transcription factor binding sites. Of remark is the presence of multiple GC, CACCC and GATA-1-binding motifs (Fig. 4), suggesting that Sp1/KLF and GATA factors participate in the regulation of KLF13 gene promoter. To test how the transcription of KLF13 gene is regulated in erythroid cells we analyzed the effects of GATA-1 and KLF13, both of which are expressed in erythroid cells, on the KLF13 gene promoter. The expression vectors of these factors and luciferase reporter constructs were co-transfected into COS-7 and K562 cells. To focus the cis sequence corresponding to the effects, if any, of these factors we utilized three promoters with different length, i.e. 0.55, 0.29 and 0.13 kb fragments. As shown in Fig. 5A GATA-1 activated the 0.55 and 0.29, but not 0.13, kb promoters in COS-7 cells. The respective fold increases compared with its absence were 4.9 (P<0.001), 4.7 (P<0.01) and 1.9 (P=0.99). The activation of the KLF13 gene promoter by GATA-1 was also observed in K562 cells (Fig. 5B). In the presence of GATA-1 the respective promoter activities were 3.3, 3.5 and 6.5 times higher compared with those without GATA-1 (P<0.001 in all promoters). These results show that GATA-1 is a potential trans-activator of the KLF13 gene promoter, and the proximal 130 bp DNA sequence is capable of mediating the GATA-1 effects specifically in the erythroid environment. In addition it is noteworthy that the 0.55 and 0.29 kb promoters showed essentially the same activities, which is similar than the result shown in Fig. 3B, in any experimental condition, suggesting that neither GATA-1 nor KLF13 is responsible for the erythroid specificity in the sequence between −0.55 and −0.29 kb (Figs. 3B and C).
Fig. 4.
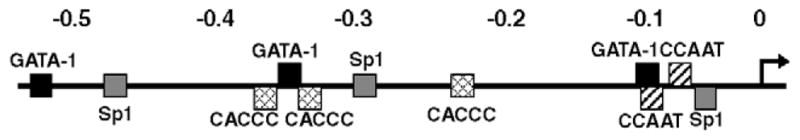
cis elements of the 0.55 kb promoter that are potentially recognized by Sp1/KLFs and GATA-1. Numbers shown above are the distance to the transcription start site in kb. Motifs are indicated by black (GATA-1), gray (Sp1), hatched (CACCC) and striped (CCAAT) squares. The transcription start site is indicated by an arrow.
Fig. 5.
(A and B) Effects of GATA-1 and KLF13 itself on the KLF13 gene promoter. Relative luciferase activities derived from 0.55, 0.29 and 0.13 kb promoters are shown. Reporter constructs were co-transfected with either mock, GATA-1 or KLF13 into COS-7 (A) and K562 (B) cells. Luciferase counts were corrected by the β-Gal activity, and the average count of the 0.55 kb promoter obtained from mock-transfected cells was considered as 100%. Error bars indicate one S.D. Solid lines shown above indicate that the promoter activities were significantly different. Results were obtained from three (COS-7) and four (K562) independent transfections in triplicate.
KLF13 activates its own promoter in COS-7 cells (Fig. 5A). The activities of the 0.55, 0.29 and 0.13 kb promoters in the presence of KLF13 were 3.8 (P<0.01), 4.2 (P<0.01) and 4.0 (P=0.05) times higher compared with its absence. In K562 cells, however, this was not the case — i.e. the respective promoter activities relative to those mock-trans-fected were 0.78 (P=0.27), 0.56 (P<0.001) and 1.04 (P=1.0) (Fig. 5B). KLF13 thus failed to activate its own promoter in K562 cells. Rather, KLF13 tended to repress the 0.55 and 0.29 kb promoters.
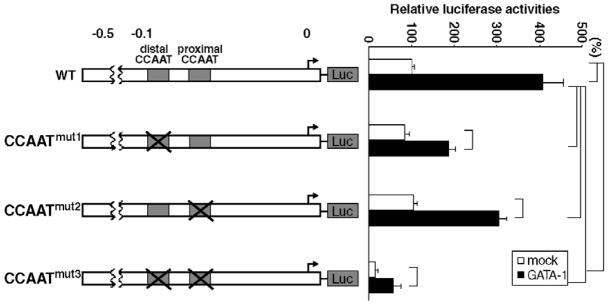
3.4. GATA-1 activates KLF13 gene promoter through the sequence containing CCAAT motifs
The data shown above indicate that erythroid specific GATA-1-responsive element(s) of the KLF13 promoter are present in the very proximal 0.13 kb sequence. There is a non-canonical GATA-1-binding site within the sequence (Fig. 4). In addition two CCAAT boxes could be a potential GATA-1-binding site [16,17]. We, however, failed to demonstrate GATA-1-binding to these sequences by gel shift experiments, in which there was a distinct GATA-1-binding to the canonical GATA sequence (data not shown). Therefore, we focused on the role of activation by GATA-1 of the major cis elements in the 0.13 kb sequence, i.e. two CCAAT boxes. Mutations were introduced into individual CCAAT motifs of the 0.55 kb KLF13 gene promoter. We analyzed how these mutations affect the activity of GATA-1 on the promoter in K562 cells by transient transfection experiments. Results are shown in percentage luciferase counts relative to that obtained from mock-transfected wild type (WT) promoter (considered as 100%). As illustrated in Fig. 6 mutation of the single CCAAT motifs failed to affect the basal promoter activity: 83% in CCAATmut1 (P=0.73), and 105% in CCAATmut2 (P=1.0). In contrast to these single mutations, double mutation (CCAATmut3) brought about remarkable decline of the promoter activity (15%, P<0.001). GATA-1 activated these CCAAT-mutated promoters, however the relative luciferase activities were significantly lower in CCAATmut1 (185%, P<0.001), CCAATmut2 (303%, P<0.001) and CCAATmut3 (56%, P<0.001) compared with that of WT (406%). Thus GATA-1 less effectively activated CCAAT-mutated KLF13 gene promoters.
Fig. 6.
Role of CCAAT motifs for the activity of GATA-1 on the KLF13 gene promoter. They were disrupted by point mutations listed in Table 1. Promoters lacking the distal, proximal and both CCAAT motifs are referred to CCAATmut1, CCAATmut2 and CCAATmut3, respectively, which are shown in the left panel. The numbers shown above are the distance to the transcription start site in kb. The right panel shows the relative luciferase activities tested in K562 cells. Luciferase counts were corrected by the β-Gal activity, and the average count of wild type (WT) promoter without co-expressed GATA-1 was considered as 100%. Error bars indicate one S.D. Solid lines shown at the right indicate that the promoter activities were significantly different. Results were obtained from three independent transfections in triplicate.
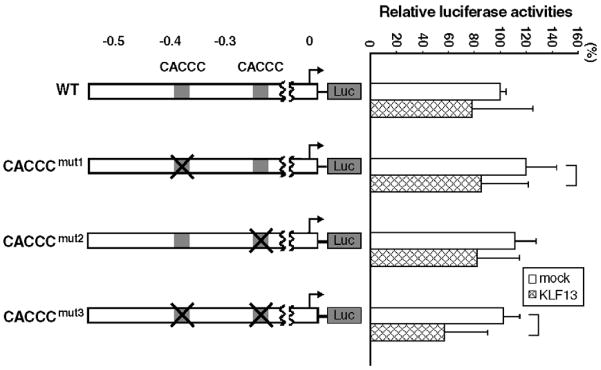
3.5. KLF13 represses its own promoter in K562 cells
As shown in Fig. 5B KLF13 has a tendency to repress its own promoter in K562 cells. Since this was observed in 0.55 and 0.29 kb promoters, and not in 0.13 kb promoter, we focused on two CACCC motifs located at the −0.37 and −0.23 kb positions, potential target sequences of the KLF13 protein. Mutations were introduced into individual motifs of the 0.55 kb promoter (CACCCmut1, CACCCmut2 and CACCCmut3 in Fig. 7). We tested whether the mutations affect the suppressive activity of KLF13 on its own promoter in K562 cells by the transient transfection assay. Results are shown in percentage luciferase counts of the basal WT promoter (Fig. 7). Mutations did not reduce the promoter strength at all: 119% for CACCCmut1 (P=0.61), 111% for CACCCmut2 (P=0.96) and 102% for CACCCmut3 (P=1.0). In the presence of KLF13 the average luciferase counts were low: 78% (WT), 85% (CACCCmut1), 82% (CACCCmut2) and 57% (CACCCmut3). The respective fold increases from the basal promoter activity were 0.78 (P=0.46), 0.71 (P=0.03), 0.74 (P=0.11) and 0.56 (P=0.001). It was thus consistently observed that KLF13, to some extent, represses its own promoter. The repressive effect was strengthened by the mutations of the two CACCC boxes.
Fig. 7.
Role of CACCC motifs for the activity of KLF13 on its own promoter. They were disrupted by the point mutations listed in Table 1. Promoters lacking the −0.37 kb, −0.23 kb and both CACCC motifs are referred to CACCCmut1, CACCCTmut2 and CACCCmut3, respectively, which are shown in the left panel. The numbers shown above are the distance to the transcription start site in kb. Right panel shows relative luciferase activities tested in K562 cells. Luciferase counts were corrected by the β-Gal activity, and the average count of wild type (WT) promoter without co-expressed KLF13 was considered as 100%. Error bars indicate one S.D. The solid lines shown at the right indicate that the promoter activities were significantly different. Results were obtained from five independent transfections in triplicate.
4. Discussion
KLF13 gene is highly expressed in erythroid cells, and its mRNA expression is up-regulated upon the induction of differentiation in erythroid lines [11], which raises the possibility that KLF13 participates in molecular events of erythroid cell differentiation. In view of this it is of interest to investigate how the transcription of the KLF13 gene is regulated in erythroid cells. In this study we cloned and characterized the 5′ flanking regulatory region of the KLF13 gene.
We identified a unique transcription start point of KLF13 gene. To reveal how the transcription is regulated in erythroid cells we analyzed the promoter activity of the 5.5 kb flanking DNA sequence in K562 and COS-7 cells. Our data show that the KLF13 gene promoter in the erythroid environment is more powerful than in the non-erythroid environment, which may be related to the higher KLF13 gene expression in K562 cells than in COS-7 cells (Fig. 2B). In addition positive and negative regulatory regions were at least in part different between the two environments. Of particular interest is the DNA sequence with a potent positive activity between −0.29 and −0.37 kb, which was distinctive in erythroid cells. However, the potent promoter activity disappeared when the −0.55 kb DNA was used, which in turn suggests the presence of a potent negative regulatory activity in the DNA sequence between −0.37 and −0.55 kb. The identification of cis elements responsible for the positive and negative regulation of these DNA sequences may be the next step to uncover the mechanism of erythroid cell specific transcriptional control of the KLF13 gene.
It is interesting that the KLF13 gene promoter was activated by KLF13 itself in COS-7 cells, whereas this was not the case in K562 cells. Rather, the promoter tended to be repressed in the presence of KLF13, suggesting that there exists a feedback mechanism in the control of KLF13 gene transcription in erythroid cells. cis Element(s) responsible for the repressive action of KLF13 remain to be elucidated. Regarding this issue we explored the role of two CACCC boxes of the KLF13 gene promoter. Mutations of the two CACCC motifs, however, made the repressive effect of KLF13 more evident. This might be relevant to the fact that these CACCC motifs were likely involved in the trans-activation of the promoter by KLF13 itself in COS-7 cells since KLF13 failed to sufficiently activate the promoter (data not shown). Therefore, it is suggested that the CACCC boxes may be key cis elements corresponding to the trans-activating effect of KLF13 on its own promoter.
We have shown that the KLF13 promoter is trans-activated by GATA-1, indicating that KLF13 is potentially a downstream gene of GATA-1. The mechanism by which GATA-1 activates the KLF13 gene promoter needs to be determined. The fact that the promoter with distal CCAAT box mutation was less effectively activated by GATA-1 (CCAATmut1 in Fig. 6) should be taken into account, which suggests that the distal CCAAT box or the overlapping non-canonical GATA-1-binding site may play a role in the KLF13 promoter activation by GATA-1. This may be supported by the observation that the reduction of the activation of the CCAATmut1 by GATA-1 was significant, P<0.01, when the same experiment was performed using COS-7 (data not shown). The overlapping CCAAT box and GATA-1-binding site of the KLF13 promoter is structurally similar to the Aγ globin gene promoter [16]. In contrast to the Aγ promoter where GATA-1 binding could be demonstrated by gel shift assays, we have failed to demonstrate GATA-1 binding to the KLF13 promoter containing CCAAT motifs. How does GATA-1 activate the KLF13 promoter in such a situation? There are two possibilities: either GATA-1 binds to the promoter region but the binding is too weak to be detected; or the activation of the promoter by GATA-1 is indirect, a case such as transcription factor(s) regulated by GATA-1 activate the KLF13 gene promoter through the CCAAT or neighboring sequences. The former possibility may be supported by the evidence that the binding to CCAAT element is very unstable [17]. In addition, the non-canonical GATA-1 binding sequence, GATT that overlaps with the distal CCAAT motif of the KLF13 gene promoter (Fig. 4), is supposedly recognized by the GATA-1 carboxyl finger [18]. Therefore, the binding of GATA-1 to the promoter region in vivo may not necessarily be ruled out, even though we failed to demonstrate it in vitro. It should also be mentioned that a number of factors including CP1/NF-Y and CP2 which are expressed in a wide variety of tissues interact with the CCAAT motif [19]. It is therefore possible that GATA-1 activates KLF13 through enhancements of expression of such ubiquitously expressed factors. GATA-1 is also known to influence the expression of other erythroid transcription factors, e.g. EKLF [20], KLFD [21] and GATA-1 itself [22,23]. It has been shown that NF-E4 is a co-factor of CP2 [24], a transcription factor abundantly expressed in erythroid cells and involved in γ globin gene regulation. It is conceivable that GATA-1 activates NF-E4 which may bind to the KLF13 CCAAT box through CP2 and enhance KLF13 expression. It remains to be, however, tested whether the expression of NF-E4 is under the control of GATA-1. The mechanism of action of GATA-1 on the KLF13 promoter remains therefore unknown.
Acknowledgments
This work was supported by NIH grant HL20899 (subcontract 225053).
References
- 1.Philipsen S, Suske G. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 1999;27:2991–3000. doi: 10.1093/nar/27.15.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bieker JJ. Krüppel-like factors: three fingers in many pies. J Biol Chem. 2001;276:34355–34358. doi: 10.1074/jbc.R100043200. [DOI] [PubMed] [Google Scholar]
- 3.Kaczynski J, Cook T, Urrutia R. Sp1- and Krüppel-like transcription factors. Genome Biol. 2003;4:206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller IJ, Bieker JJ. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Krüppel family of nuclear proteins. Mol Cell Biol. 1993;13:2776–2786. doi: 10.1128/mcb.13.5.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature. 1995;375:316–318. doi: 10.1038/375316a0. [DOI] [PubMed] [Google Scholar]
- 6.Perkins AC, Sharpe AH, Orkin SH. Lethal β-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature. 1995;375:318–322. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- 7.Donze D, Townes TM, Bieker JJ. Role of erythroid Kruppel-like factor in human γ- to β-globin gene switching. J Biol Chem. 1995;270:1955–1959. doi: 10.1074/jbc.270.4.1955. [DOI] [PubMed] [Google Scholar]
- 8.Asano H, Stamatoyannopoulos G. Activation of β-globin promoter by erythroid Krüppel-like factor. Mol Cell Biol. 1998;18:102–109. doi: 10.1128/mcb.18.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson KP, Kern CB, Crable SC, Lingrel JB. Isolation of a gene encoding a functional zinc finger protein homologous to erythroid Krüppel-like factor: identification of a new multigene family. Mol Cell Biol. 1995;15:5957–5965. doi: 10.1128/mcb.15.11.5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wani MA, Wert SE, Lingrel JB. Lung Krüppel-like factor, a zinc finger transcription factor, is essential for normal lung development. J Biol Chem. 1999;274:21180–21185. doi: 10.1074/jbc.274.30.21180. [DOI] [PubMed] [Google Scholar]
- 11.Asano H, Li XS, Stamatoyannopoulos G. FKLF-2: a novel Krüppel-like transcriptional factor that activates globin and other erythroid lineage genes. Blood. 2000;95:3578–3584. [PubMed] [Google Scholar]
- 12.Song A, Chen YF, Thamatrakoln K, Storm TA, Krensky AM. Transcriptional regulation of RANTES expression in T lymphocytes. Immunity. 1999;10:93–103. doi: 10.1016/s1074-7613(00)80010-2. [DOI] [PubMed] [Google Scholar]
- 13.Martin KM, Cooper WN, Metcalfe JC, Kemp PR. Mouse BTEB3, a new member of the basic transcription element binding protein (BTEB) family, activates expression from GC-rich minimal promoter regions. Biochem J. 2000;345:529–533. [PMC free article] [PubMed] [Google Scholar]
- 14.Asano H, Li XS, Stamatoyannopoulos G. FKLF, a novel Krüppel-like factor that activates human embryonic and fetal β-like globin genes. Mol Cell Biol. 1999;19:3571–3579. doi: 10.1128/mcb.19.5.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3. Cold Spring Harbor Laboratory Press; New York: 2001. [Google Scholar]
- 16.Berry M, Grosveld F, Dillon N. A single point mutation is the cause of the Greek form of hereditary persistence of fetal haemoglobin. Nature. 1992;358:499–502. doi: 10.1038/358499a0. [DOI] [PubMed] [Google Scholar]
- 17.Li Q, Duan ZJ, Stamatoyannopoulos G. Analysis of the mechanism of action of non-deletion hereditary persistence of fetal hemoglobin mutants in transgenic mice. EMBO J. 2001;20:157–164. doi: 10.1093/emboj/20.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whyatt DJ, deBoer E, Grosveld F. The two zinc finger-like domains of GATA-1 have different DNA binding specificities. EMBO J. 1993;12:4993–5005. doi: 10.1002/j.1460-2075.1993.tb06193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- 20.Crossley M, Tsang AP, Bieker JJ, Orkin SH. Regulation of the erythroid Krüppel-like factor (EKLF) gene promoter by the erythroid transcription factor GATA-1. J Biol Chem. 1994;269:15440–15444. [PubMed] [Google Scholar]
- 21.Oates AC, Pratt SJ, Vail B, Yan Y, Ho RK, Johnson SL, Postlethwait JH, Zon LI. The zebrafish klf gene family. Blood. 2001;98:1792–1801. doi: 10.1182/blood.v98.6.1792. [DOI] [PubMed] [Google Scholar]
- 22.Tsai SF, Strauss E, Orkin SH. Functional analysis and in vivo footprinting implicate the erythroid transcription factor GATA-1 as a positive regulator of its own promoter. Genes Dev. 1991;5:919–931. doi: 10.1101/gad.5.6.919. [DOI] [PubMed] [Google Scholar]
- 23.Nishikawa K, Kobayashi M, Masumi A, Lyons SE, Weinstein BM, Liu PP, Yamamoto M. Self-association of gata1 enhances transcriptional activity in vivo in zebra fish embryos. Mol Cell Biol. 2003;23:8295–8305. doi: 10.1128/MCB.23.22.8295-8305.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou W, Clouston DR, Wang X, Cerruti L, Cunningham JM, Jane SM. Induction of human fetal globin gene expression by a novel erythroid factor, NF-E4. Mol Cell Biol. 2000;20:7662–7672. doi: 10.1128/mcb.20.20.7662-7672.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]