Abstract
Secretory clusterin (sCLU) is a stress-activated, cytoprotective chaperone that confers broad-spectrum cancer treatment resistance and its targeted inhibitor (OGX-011) is currently in Phase II trials for prostate, lung, and breast cancer. However, molecular mechanisms by which sCLU inhibits treatment-induced apoptosis in prostate cancer remain incompletely defined. We report that sCLU increases NF-κB nuclear translocation and transcriptional activity by serving as a ubiquitin binding protein that enhances COMMD1 and I-κB proteasomal degradation by interacting with members of the SCF-βTrCP E3 ligase family. Knockdown of sCLU in prostate cancer cells stabilizes COMMD1 and I-κB, thereby sequestrating NF-κB in the cytoplasm and decreasing NF-κB transcriptional activity. Comparative microarray profiling of sCLU over-expressing and knockdown prostate cancer cells confirmed that the expression of many NF-κB regulated genes positively correlate with sCLU levels. We propose that elevated levels of sCLU promote prostate cancer cell survival by facilitating degradation of COMMD1 and I-κB, thereby activating the canonical NF-κB pathway.
Keywords: prostate cancer, clusterin, COMMD1, I-κB, NF-κB
Introduction
Secretory clusterin (sCLU) (NCBI: NM_203339) is a stress-activated, cytoprotective small heat-shock chaperone that potently inhibits protein aggregation at times of cell stress (1). sCLU interacts with and inhibits conformationally-altered Bax in response to cytotoxic stress thereby impeding Bax oligomerization, cytochrome C release, and intrinsic pathway activation (2). sCLU is transcriptionally activated by heath shock factor (HSF-1) (3, 4) and increased levels are associated with a broad range of normal and disease states including aggregopathies in Alzheimer’s Disease (5), mammary gland involution (6) and treatment resistance in cancer. In cancer, sCLU is highly expressed in cells surviving apoptotic stimuli and in advanced and treatment resistant cancers (7), and protects cells against apoptosis in response to hormone-, radiation-, and chemo-therapy (7–9). sCLU increases with androgen withdrawal in hormone-dependent tumors (7, 10). Antisense- or siRNA-induced knockdown of sCLU enhances treatment-induced apoptosis and delays progression in many cancer models (11–13). This identifies sCLU as an anti-cancer target, and a sCLU inhibitor (OGX-011) is currently in phase II clinical trials in prostate, breast, and lung cancer (14, 15).
While sCLU is a multi-functional chaperone that interacts with diverse proteins regulating cell survival, little is known regarding stress-associated pathways regulated by sCLU. In this study, we used two complimentary approaches: a yeast-2 hybrid search for molecules that specifically interact with sCLU, as well as gene profiling to identify genes coordinately altered by sCLU gain- or loss-of-function in prostate cancer cells. COMMD1, an I-κB stabilizing protein, was identified as a sCLU binding protein, and many NF-κB regulated genes were coordinately altered by sCLU over expression- or knockdown. We elucidate a new mechanism whereby sCLU serves as a ubiquitin binding protein to facilitate COMMD1 and I-κB ubiquitination and proteasomal degradation, thereby enhancing NF-κB transcriptional activity and prostate cancer cell survival.
Material and Methods
Cell Culture and siRNA Transfection
LNCaP, PC-3, (ATCC, Manassas, VA), LNsCLU and LNmock were previously generated by clonal selection (7), and Hela-I-kB Fluc were kindly provided by Dr. David Piwnica-Worms (Washington University, St Louis, MO) and maintained in RPMI 1640 and DMEM respectively supplemented with 5% fetal bovine serum. Cells were treated with CLU siRNA or Scr as previously described (16).
Plasmids and Reagents
COMMD1 was amplified from LNCaP tumors and cloned in pcDNA 3.1 TOPO vector (Invitrogen, Life Technologies, Inc.). sCLU, I-κB and βTrCP-1 – 2 plasmids were provided by Dr. Martin Tenniswood (University of Notre Dame, IN), Dr. Piwnica-Worms, and Dr. Tomoki Chiba (University of Tsukuba, Tsukuba, Japan),. Purchased reagents include Ubiquitin (ATCC) and pNF-κB- Luc plasmids (BD Biosciences USA); Protein-G sepharose (Invitrogen); Cyclohexamide and MG-132, fluorogenic substrate N-succinyl-l-leucyl-l-leucyl-l-tyrosine-7-amido-4-methyl-coumarin (Suc-LLVY-AMC) (Calbiochem,San Diego, CA); ubiquitin-Agarose, Flag-Agarose and TNFα (Sigma, St. Louis, MO), antibodies against CLU, I-κB, phospho I-κB, cullin1, ubiquitin, p65 RelA, Flag (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and Vinculin (Sigma, St.Louis, MO). Docetaxel and bortezomib were provided by BC Cancer Agency (Vancouver, BC. Canada).
Yeast two-hybrid screening (Y2H)
Two Y2H screens were performed using the Matchmaker System 3 (BD Biosciences) and Matchmaker library construction & screening Kit (BD Biosciences) according to manufacturer’s protocol and briefly described in MM-S1.
Western Blot and Immunoprecipitation (IP) analysis
Cell pellets were lysed in either RIPA buffer containing 0.1% of SDS, or in 8M Urea buffer as described previously (17). Co-IP or western blotting were described previously (18) and visualized using the Odyssey system (Li-COR Biosciences) according to the manufacture’s protocol.
Transfection and reporter gene assay
LNCaP, LNmock, LNsCLU, or PC-3 cells were transfected with pNF-kB- Luc plasmid using lipofectin (Invitrogen). Luciferase activity was measured using Dual-Luciferase Reporter Assay System (Promega) and a microplate luminometer (EG&G Berthold). Reporter assays were normalized to Renilla and protein concentration for siRNA experiments expressed in arbitrary light units.
Northern Blot Analysis
Total RNA isolated from LNmock or lnsCLU cells +/− sCLU or Scr siRNA using trizol/chloroform extraction (Invitrogen Life Technology Inc.) was subjected to Northern blot analysis using COMMD1 Sema3C (2255bp), NGAL (845bp), sPLA2-IIa (854bp), MIP3α (3000bp) cDNA probes. Loading was normalized to GAPDH as previously described (18).
Statistics
Data was analyzed using GraphPad InStat version 3.00 for Windows 95, (GraphPad Software, San Diego, CA) and were presented as Means ± standard error of the mean (SEM) and p values<0.05 were significant.
Results
sCLU interacts and co-localizes with COMMD1
sCLU was used as bait in Y2H screen of a human normal prostate cDNA library to identify sCLU binding partners. A total of 2 × 106 transformants were screened. Four of nineteen clones contained an insert corresponding to the ORF of COMMD1 gene. Interestingly, a second GAL-4 based Y2H library screen using a cDNA library generated from PC-3 cells stressed with 10nM paclitaxel identified COMMD1 as the only candidate gene for positive interaction with CLU. Identification of COMMD1 as a CLU binding partner under 2 different conditions from 2 different sources, a tissue-based as well as a cell-based cDNA library, prioritized this interaction for further functional studies.
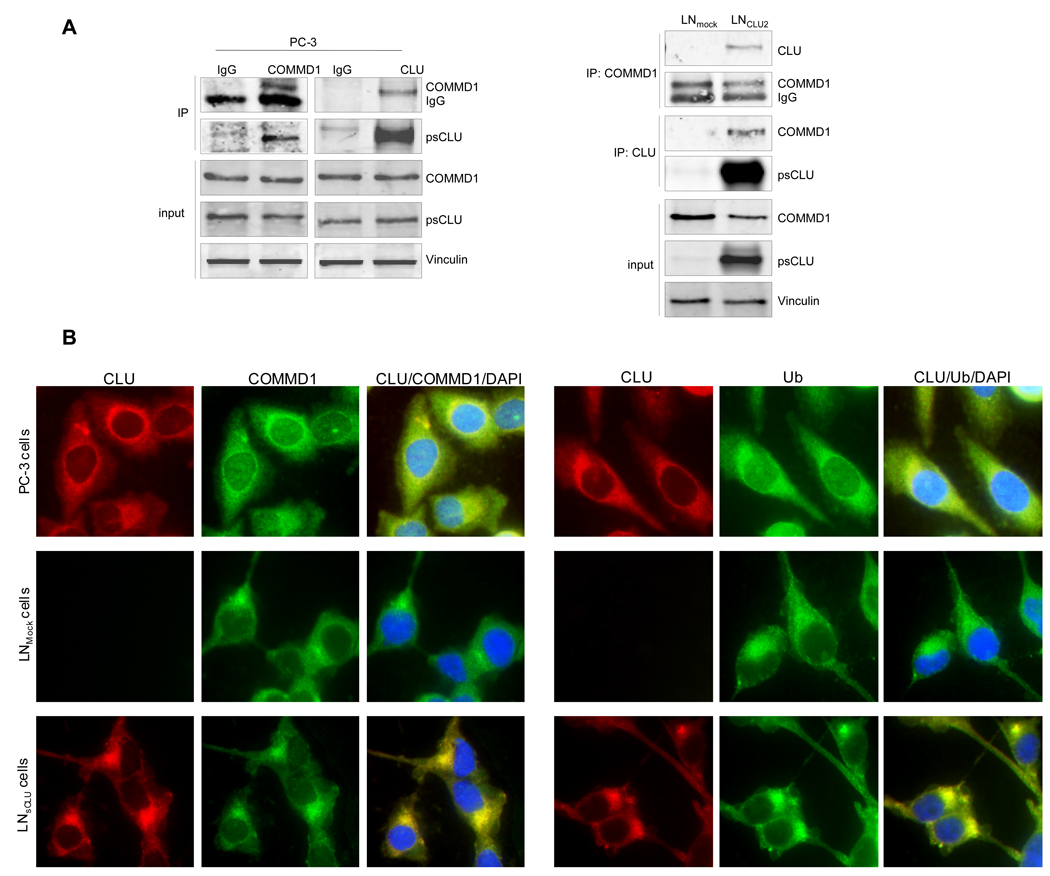
Interaction between COMMD1 and sCLU proteins was confirmed in PC-3 cells by probing reciprocal blots of COMMD1 and sCLU immunoprecipitates with antibodies to detect sCLU and COMMD1, respectively. Parallel experiments were also carried out with corresponding control IgG. Figure 1A (left panel) confirms the existence of specific CLU/COMMD1 complexes in PC-3 cells that express both proteins endogenously but not IgG controls. Specificity of CLU/COMMD1 complexes were corroborated by IP in LNCaP overexpressing sCLU (LNsCLU) versus LNmock cells. Figure 1A (right panel) shows that CLU/COMMD1 complexes are present in cells that express sCLU (LNsCLU) but not in LNmock confirming that sCLU interacts specifically with COMMD1. This interaction was also confirmed using immunofluorescence where both proteins co-localize in the cytoplasm and accumulate in perinuclear foci (Fig. 1B, upper panel) in PC-3 and LNsCLU cells but not in control LNmock. The perinuclear foci identified in Figure 1B may 8 correspond to aggresomes initially described as misfolded, ubiquitinated protein inclusions forming specifically at the centrosome (19). Aggresomes recruit proteasome subunits as well as heat shock proteins, and may be involved in regulating processing in protein folding, aggregation and degradation (20). Since aggresomes frequently immunostain for ubiquitin, we tested for the presence of ubiquitin and observed strong co-localized reactivity in aggresomes (Fig. 1B, lower panel).
Figure 1. sCLU is a COMMD1 and ubiquitin partner in prostate cancer cells.
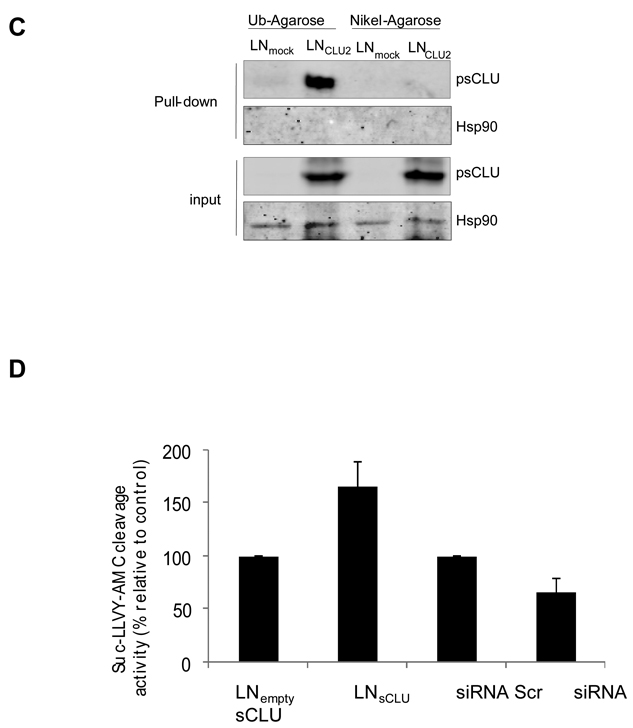
A, sCLU interacts with COMMD1. Total proteins from PC-3, LNmock and LNsCLU were co-IP with anti-CLU or normal IgG followed by western blot using COMMD1 antibody. The inverse experiment was performed using COMMD1 antibody for co-IP. CLU, COMMD1 and vinculin were used for western blot (sCLU is detected as a 60-kDa glycosylated presecretory form of sCLU and is referred as psCLU in all western blot as proposed by Trougakos (43)). B, sCLU co-localizes with COMMD1 and ubiquitin. Immunofluorescence was performed in PC-3, LNmock and LNsCLU using CLU, COMMD1, and ubiquitin antibodies and DAPI for nuclei staining. C, sCLU associates with ubiquitin. Total proteins were incubated with ubiquitin-agarose matrix or Nickel agarose; and bound protein and input were used for western blots with CLU and Hsp90 antibodies. D, sCLU enhances ubiquitinated proteins. Total proteins (50ug) from LNmock and LNsCLU or PC-3 treated with sCLU or Scr siRNA were monitored for cleavage of the Suc-LLVY-AMC. Fluorescence was quantified using a spectrofluorometer (Fluoroskan Ascent FL, Thermo Labsystem). Data is expressed as mean ± SE. *, statistical significance (p<0.05) from 3 biological replicates.
CLU-2 is a ubiquitin binding protein (UBP) that enhances proteasome activity
A pull down assay using ubiquitin agarose matrix (Figure 1C) identifies CLU binding to ubiquitin in LNsCLU but not LNmock cells, and not in cells incubated with nickel-agarose as a pull down assay control. No interaction between ubiquitin and Hsp90 was observed indicating a specific interaction between sCLU and ubiquitin and suggesting that CLU may function as a UBP. The same result was observed using CLU-GST for pull down assay (S1). We next tested the ability of sCLU to modulate proteasome activity and found compared to LNmock, LNsCLU cells exhibited increased rates in Suc-LLVY-AMC cleavage, consistent with enhanced degradation of ubiquitinated proteins (S2). In contrast, sCLU knockdown decreased Suc-LLVY-AMC cleavage (Figure 1D), further supporting a role for sCLU in proteasome activity.
sCLU promotes degradation of COMMD1
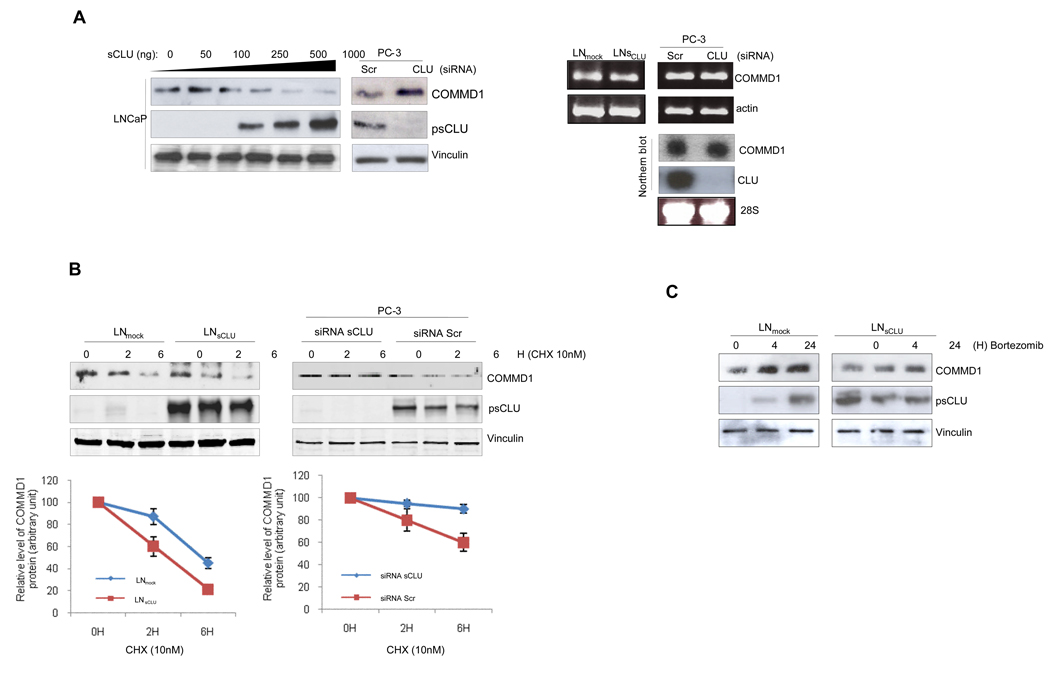
We next tested whether sCLU facilitates degradation of client proteins. Figure 2A shows that COMMD1 protein (left panel) but not mRNA (right panel) levels inversely correlate with sCLU protein levels, suggesting that sCLU negatively regulates COMMD1 levels post-translationally. Figure 2B illustrates the 9 effect of sCLU on COMMD1 protein stability using cyclohexamide, where COMMD1 protein levels decrease rapidly with sCLU overexpression (left panel), while sCLU knockdown prolongs COMMD1 half-life (right panel). Confirmatory data were observed using metabolic labeling (S3). After proteasome inhibition using bortezomib, COMMD1 levels increase in LNmock compared to LNsCLU cells (Fig. 2C) without any effect on cell viability (S4). These results suggest that under basal conditions, sCLU negatively regulates COMMD1 protein levels by facilitating its proteasomal degradation.
Figure 2. sCLU induces ubiquitination and proteasome-dependent degradation of COMMD1.
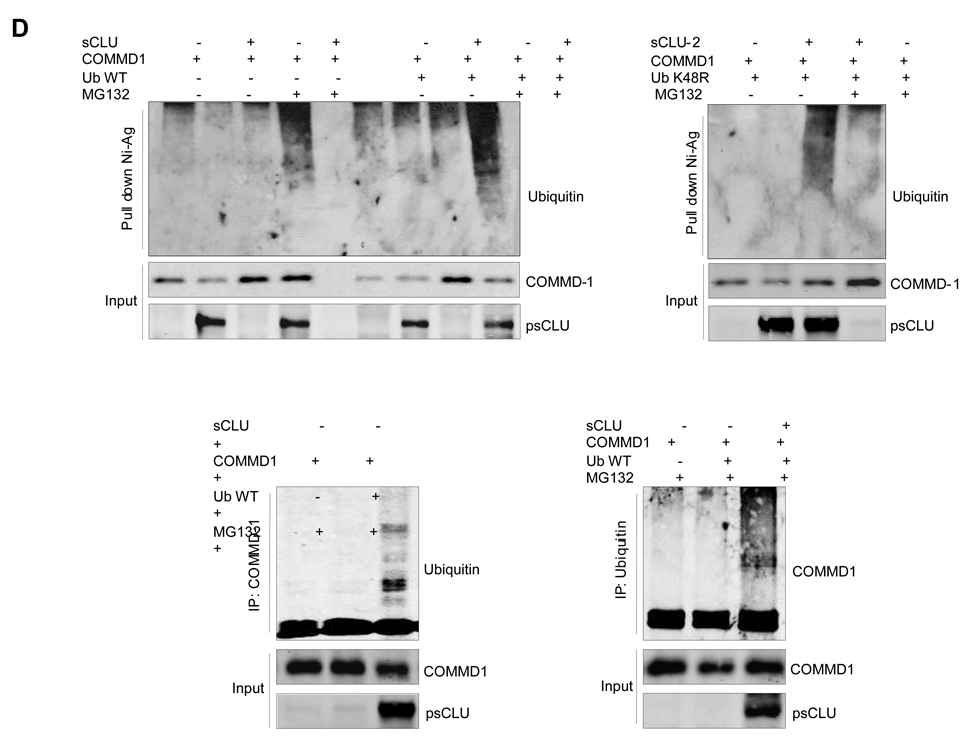
A, COMMD1 levels are negatively regulated by sCLU. Left Panel - LNCaP cells were transiently transfected with increasing amounts of sCLU cDNA as indicated. After 48h, expression levels of COMMD1 were determined by western blot. PC-3 cells were treated with 10nM sCLU or Scr siRNA and western blot was performed using COMMD1, CLU. Right Panel - COMMD1 mRNA levels are not affected by sCLU. RNA was extracted from LNmock, and LNsCLU or PC-3 cells treated sCLU or Scr siRNA. COMMD1 mRNA levels were analyzed using RT-PCR (MM S3) with actin as a control (upper panel) and by northern blot with CLU and 28S as a controls (lower panel). B, sCLU levels affect COMMD1 stability. LNmock, LNsCLU (left panel) or PC-3 cells were treated with 10nM sCLU or Scr siRNA (right panel) followed by 10 µmol/L cycloheximide with DMSO as a control. COMMD1 protein levels were measured by Western blot analysis. C, COMMD1 protein levels are regulated by proteasomal degradation. LNmock or LNsCLU cells were treated with proteasome inhibitor (bortezomib) and COMMD1 levels assessed by western blot using COMMD1 antibodies. D, Effect of sCLU on COMMD1 ubiquitination. LNCaP cells were co-transfected with either empty vector or sCLU in parallel with COMMD1, ubiquitin WT (upper left panel), or mutated ubiquitin (K48R) (upper right panel). After 48 h, cells were treated +/− MG132 for 6 hrs, proteins extracted in RIPA buffer, and pull down assay using Nickel-agarose was performed followed by western blot using ubiquitin, COMMD1 and CLU (input) antibodies. LNCaP cells were co-transfected with ubiquitin +/− sCLU and COMMD1 as indicated. After 48h, cells were treated with MG132 for 6 h. Cells lysates were prepared with 8M Urea buffer; co-IP using COMMD1 (lower left panel) or ubiquitin (lower right panel) antibodies; and western blots (co-IP and Input) were performed using ubiquitin COMMD1 and CLU antibodies.
sCLU promotes COMMD1 ubiquitination
To determine if sCLU promotes ubiquitination of COMMD1, LNCaP cells were transiently co-transfected with COMMD1 and sCLU or empty vector and treated +/− with proteasome inhibitor MG132. sCLU significantly increased COMMD1 ubiquitination levels in the presence MG 132 (Fig. 2D). When COMMD1 and sCLU were co-transfected with wild-type (WT) ubiquitin, increased levels of ubiquitinated COMMD1 were seen in sCLU over-expressing cells in the presence of MG 132 (Fig. 2D) suggesting that sCLU enhances COMMD1 ubiquitination and its subsequent degradation. The same result was observed when COMMD1 and mutated ubiquitin (K48R) +/− sCLU were co-transfected in LNCaP cells (Fig. 2E). These data suggest that, like XIAP (21), sCLU-mediated ubiquitination of COMMD1 can occur independent of K48, a lysine often coupled to degradation of proteins by the 26S proteasome system (22). In LNCaP cells, COMMD1 decreased NF-κB transcriptional activity (S5a. b), while COMMD1 knockdown increased NF-κB transcription activity (S5c). Interestingly, this inhibitory effect of COMMD-1 was abrogated in LNsCLU cells (S5d), further supporting an inhibitory effect of sCLU on COMMD-1 function.
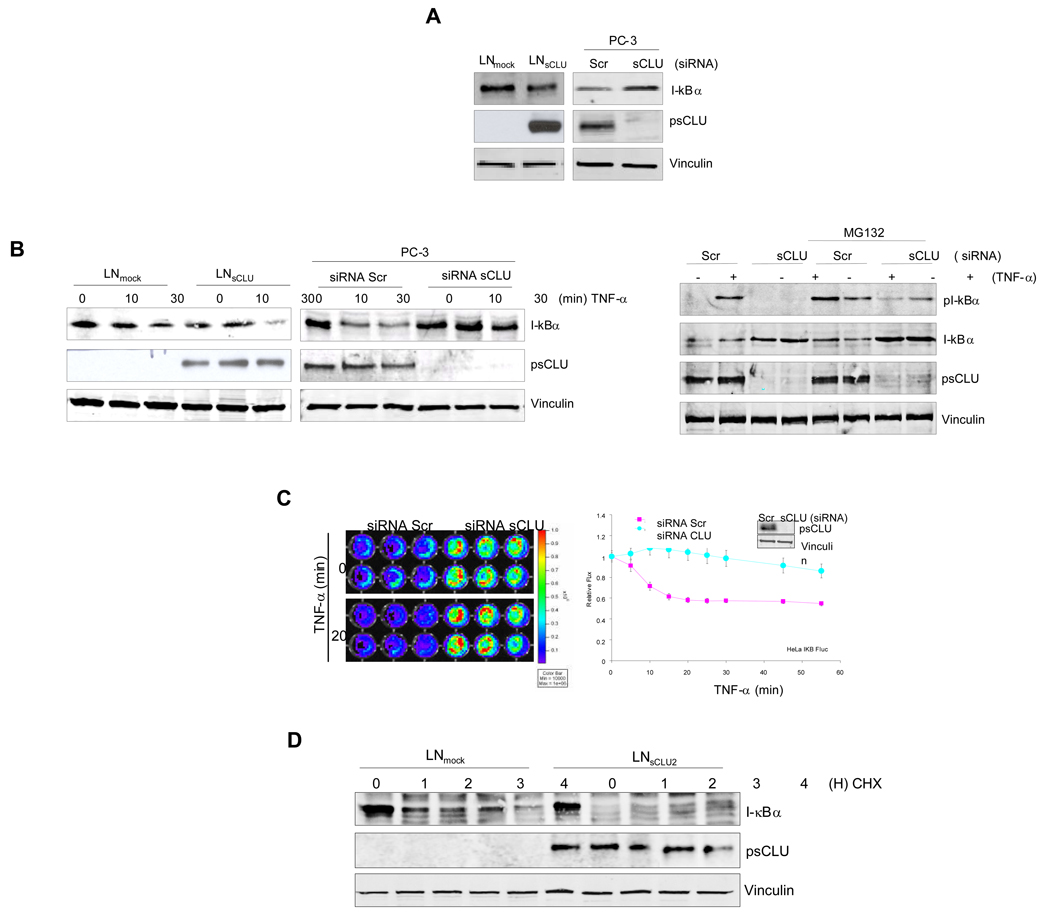
sCLU modulates I-κB stability in vitro and in cellulo
Since COMMD1 inhibits I-κB proteasomal degradation (23), we tested whether sCLU affects I-κB degradation rates. LNsCLU express lower protein levels of I-κB compared with LNmock cells, a result also observed when sCLU is up-regulated endogenously with taxol treatment (S6). Conversely, sCLU knockdown increased total I-κB protein levels (Fig. 3A, left panel). I-κB was more rapidly degraded in LNsCLU compared to LNmock cells after TNF-α stimulation (Fig. 3B, left panel), while sCLU knockdown increased total I-κB in PC-3 cells and decreases pI-κB (Fig. 3B, middle panel) by affecting the ability of IKK to phosphotylates I-κB (S7). Next, Hela cells stably transfected with I-κBα-Fluc fusion reporter were used to monitor the direct effect of sCLU knockdown on I-κB stability in cellulo. Significant increases in I-κB bioluminescence were observed after sCLU knockdown under basal and TNF-α stimulated conditions (Fig. 3C, left panel). Within 20 min after TNF-α stimulation, Hela-IκB-Fluc treated with scramble-siRNA showed rapid decreases in normalized bioluminescence while those treated with sCLU siRNA showed no significant change (Fig. 3C, right panel). This in cellulo data supports previous data that sCLU knockdown stabilizes I-κB. To further corroborate this effect on I-κB, we treated LNsCLU or LNmock cells with cyclohexamide and found I-κB degradation rate increased in LNCLU2 cells compared with controls (Fig. 3D). Collectively, this data indicates that sCLU decreases I-κB levels by enhancing its rate of degradation.
Figure 3. Clusterin facilitates I-kB degradation.
A, sCLU modulates total I-κB protein levels. Total protein from LNmock or LNsCLU (left panel) were extracted and I-κB protein levels assessed by western blot. PC-3 cells (right panel) were treated with 10nM sCLU or Scr siRNA and western blotting was performed using COMMD1, sCLU and vinculin antibodies (left panel). B, sCLU modulates I-κBα degradation. Left panels-LNmock or LNsCLU and PC-3 treated with 10nM sCLU or Scr siRNA were treated with 20ng/ml of TNF-α as indicated. Right Panel- PC-3 treated with 10nM sCLU or Scr siRNA and treated with either 20ng/ml of TNF-α for 30 min or MG132 for 3 hours followed by 30 min TNF-α treatment. Western blots were performed using I-κBα pI-κBα, sCLU and vinculin antibodies. C, sCLU knockdown stabilizes I-κB in cellulo. Hela -I-κB-Fluc cells were treated with 10 nM sCLU- or Scr-siRNA for 48hr. Cells were then treated with 20ng/ml TNF-α and imaged using an IVIS system with luciferin as a substrate (left panel). Photon count was plotted as a function of time after addition of TNF-α or vehicle and normalized at given time point as a fold of TNF α untreated cells (MM S4); values were expressed as a fold of initial value (right panel). D, sCLU regulates steady-state turnover of I-κBα. LNmock or LNsCLU were stimulated by a 10 min pulse of TNF-α and medium containing cyclohexamide (30uM) was added as indicated. Proteins were western blotted using I-κBα, CLU and vinculin antibodies.
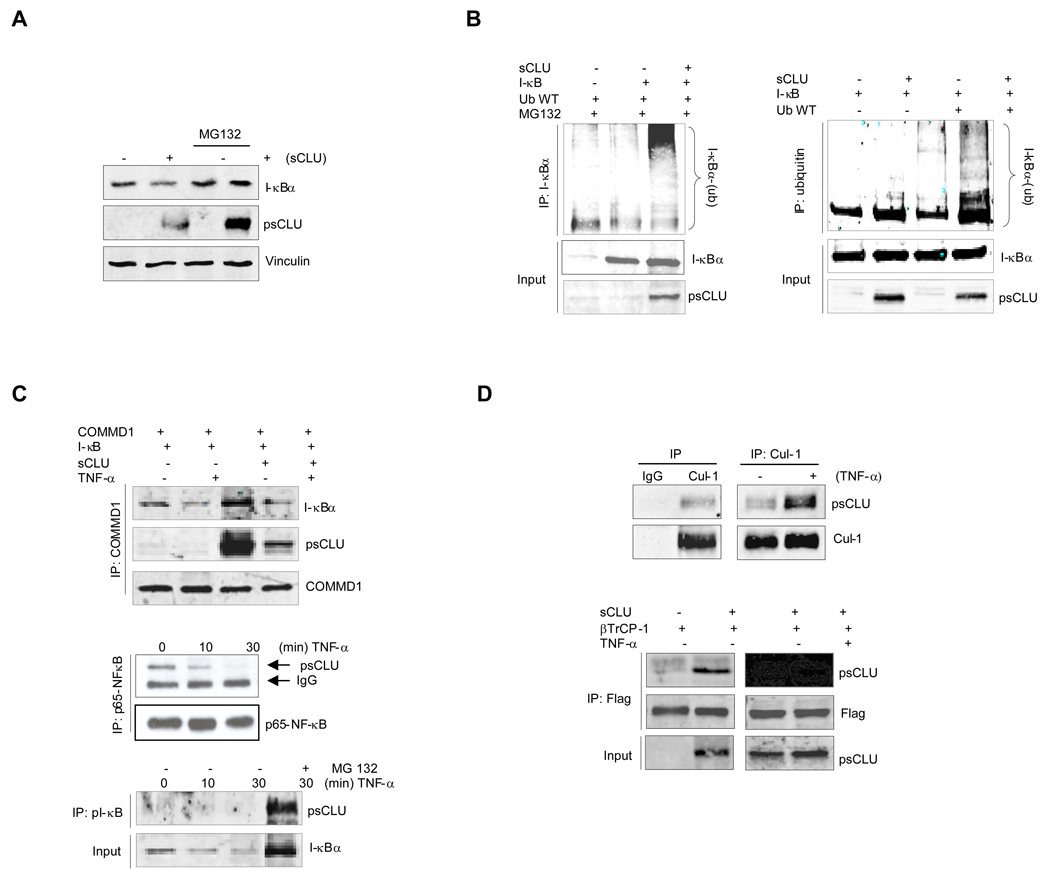
sCLU facilitates proteasome-dependent I-κB degradation
Since sCLU-induced decreases in total I-κB levels was abrogated by the proteasome inhibitor, MG132 (Fig. 4A), we next examined the effects of sCLU on ubiquitinated I-κB levels. Ubiquitination of ectopic I-κB was first evaluated by precipitating I-κB and blotting the recovered material for poly-ubiquitin. Heavily ubiquitinated I-κB was recovered after MG132 treatment only in the presence of sCLU (Fig. 4B, left panel), while precipitated ubiquitinated proteins blotted for I-κB (Fig. 4B, right panel) indicates that sCLU overexpression accelerates I-κB ubiquitination. Interestingly, under basal conditions, sCLU is found in a specific complex with COMMD1, I-κB (Fig. 4C, upper panel) and p65-NF-κB (Fig. 4C, middle panel), but not with control IgG (S8). After TNF-α treatment, sCLU dissociates from p65 NF-κB and associates with phospho-I-κB (Fig. 4C, lower panel). Collectively, these data suggest that TNF-α-induced stress shifts sCLU from a complex with NF-κB to one with COMMD1 and pI-κB which facilitates pI-κB ubiquitination and degradation.
Figure 4. sCLU regulates proteasomal-dependent degradation rates of I-κB.
A, sCLU enhances proteasomal degradation of endogenous I-κBα. LNCaP cells were transfected with sCLU or empty vector, and MG132 added 3h before harvesting cells. Total lysates were analyzed by western blot using I-κBα CLU and vinculin antibodies. B, sCLU induces I-κBα ubiquitination. LNCaP cells were co-transfected with ubiquitin +/− sCLU and I-κB as indicated. After 48h, cells were treated with MG132. Total proteins were co-IP with I-κBα antibody. Western blots (co-IP and Input) were performed using ubiquitin or I-κBα and CLU antibodies (left panel). The inverse experiment was performed using ubiquitin antibody for co-IP and I-κB, CLU for western blot (right panel). C, CLU is found in a complex with I-κB, NF-κB and COMMD1. LNCaP cells were co-transfected with COMMD1, I-κB +/− sCLU as indicated. After 48h, cells were treated with TNF-α for 30 min. Co-IP was performed using COMMD1 antibody and western blot with I-κB, CLU and COMMD1 antibodies (upper panel). sCLU interacts with p65-NF-kB. LNsCLU were treated with 20ng/ml TNF-α as indicated and proteins were Co-IP with p65-NF-κB antibody. A western blot was performed using sCLU or p65-NF-κB (middle panel). sCLU interacts with pI-κB. LNsCLU cells were treated with or without MG132 for 3h prior to TNF-α as indicated. Proteins were co-IP with pI-κB antibody and western blot performed using CLU antibody (lower panel). D, sCLU interacts with cullin1: upper panel - LNCLU2 cells were treated with or without TNF-α for 30 min and co-IP with Cul1 or IgG antibodies followed by western blot with CLU and Cul-1 antibodies. Lower panel - LNCaP cells were transfected with βTrCP-1 and sCLU and treated with TNF-α as indicated. Proteins were co-IP with Flag antibody and western blot performed using CLU and Flag antibodies.
Given that sCLU does not possess any known E3 ligase activity, we examined whether sCLU interacts with SCF-βTrCP (Skp1-Cul1-F-box ligase containing the F-box-βTrCP), a known E3 ligase reported to induce I-κB and COMMD1 proteasomal degradation (23–26). Cullin1 co-immunoprecipitated with sCLU and this interaction was enhanced after TNF-α treatment (Fig. 4D, upper panel). To determine whether other components of SCF-βTrCP bind with sCLU, βTrCP-1 or -2 were co-expressed with sCLU or empty vector constructs and cell proteins co-precipitated. CLU also interacts with βTrCP-1 (Fig. 4D, lower panel) indicating that sCLU interacts with the SCF-βTrCP E3 ligase complex, serving as a scaffolding chaperone to form a multimeric protein complex that facilitates COMMD1 and I-κB ubiquitination and proteasomal degradation.
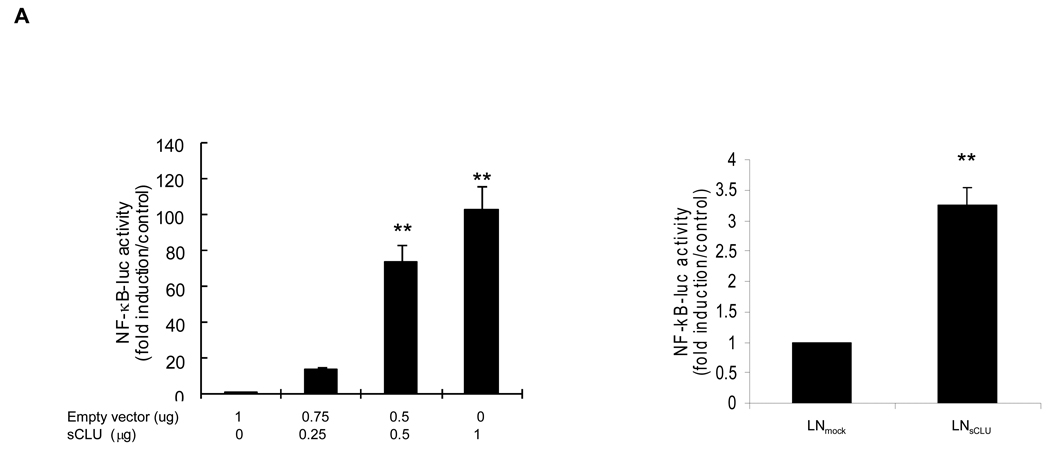
sCLU enhances NF-κB transcription activity
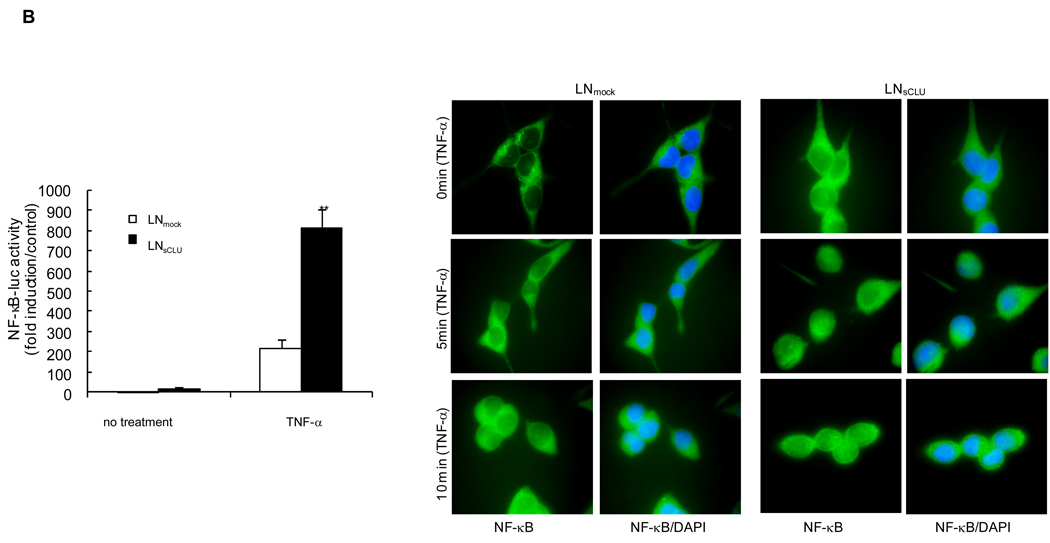
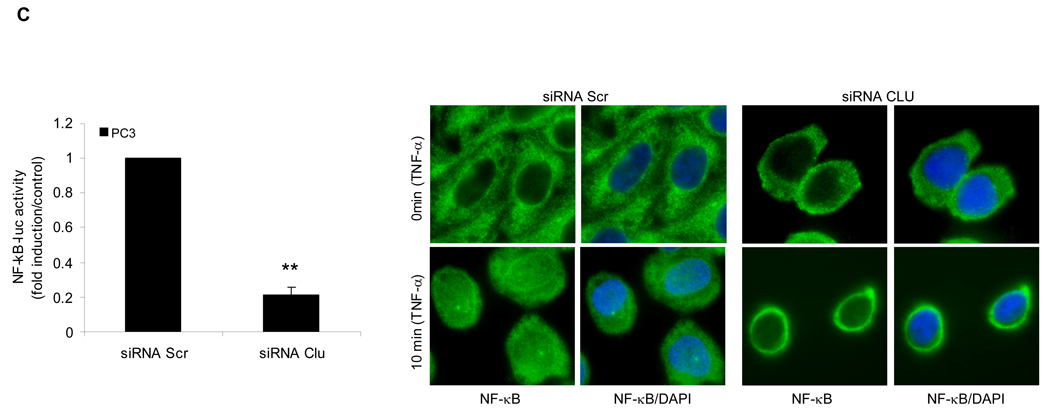
Based on its known anti-apoptotic activity, and the preceding data, NF-κB transactivation assays were performed in LNCaP cells transiently transfected with NF-κB luciferase reporter plasmid, and increasing amounts of exogenous sCLU (Fig. 5A, left panel). NF-κB transcriptional activity increased dose dependently with increasing sCLU levels, reaching 100-fold higher (p<0.001) compared to empty vector controls. NF-κB transcriptional activity was higher in stably transfected LNsCLU compared to LNmock cells (Fig. 5A, right panel), an effect enhanced by TNF-α (p<0.001) (Fig. 5B, left panel). Interestingly, the same results were observed when cells were treated with LPS or IL-1 suggesting that sCLU enhances NF-kB activation (S9). The cellular localization of endogenous p65-NF-κB was exclusively cytoplasmic in LNmock, with slight nuclear staining in LNsCLU cells (Fig. 5B, right panel). After TNF-α treatment, p65-NF-κB translocated to the nucleus more rapidly and completely in LNsCLU compared with LNmock cells. As anticipated, sCLU knockdown using sCLU antisense (OGX-011, S10a) or siRNA (S10b) in PC-3 cells led to a 78% dose-dependent decrease in NF-κB transcriptional activity (n=3, p<0.0001, unpaired t-test) (Fig 5C, left panel) and reduced p65-NF-κB nuclear localization (Fig. 5C, right panel). TNF-α stimulation induced a rapid and complete nuclear translocation of p65-NF-κB in PC-3 cells treated with Scr siRNA, while only cytoplasmic staining was seen in both unstressed and stressed conditions with sCLU siRNA. Together, these results suggest that sCLU enhances NF-κB transcriptional activity by increasing the nuclear accumulation of p65-NF-κB.
Figure 5. Effect of sCLU on NF-kB transcription activity.
A, sCLU enhances NF-κB transactivation. Left panel - LNCaP cells were transiently co-transfected with NF-κB-Luciferase and Renilla plasmids and sCLU plasmid. Total amount of plasmid DNA transfected was normalized to 1.65 ug/well by addition of empty vector. Right panel - LNmock and LNCLU2 were transiently transfected with NF-κB-Luciferase and Renilla plasmids. After 48 h, cells were harvested and luciferase activity determined. B, Left panel - TNF-α enhances the effect of sCLU on NF-κB activation: LNmock and LNsCLU were transfected with NF-κB-Luciferase plasmid. After 24 h, cells were treated with 20ng/ml of TNF-α for 24h and luciferase activity determined. Data represents the mean of at least 3 independent experiments performed in triplicate. Fold is measured relative to NF-κB activation in LNmock without treatment. Right panel - sCLU accelerates NF-κB nuclear translocation after TNF-α. LNmock and LNsCLU were treated with +/− 20ng/ml TNF-α and immunofluorescence performed using p65-NF-κB antibody. C. sCLU knockdown decreases NF-κB activation. Left - PC3 cells were transfected simultaneously with NF-κB-Luciferase plasmid and sCLU or Scr siRNA. After 48 h, luciferase activity was determined. Triplicate luciferase assays repeated 3 times and reported as mean + SE. **, statistical significance, (p<0.001). Right - sCLU knockdown inhibits p65-NF-kB nuclear localization. PC-3 cells were transfected with 10nM sCLU or Scr siRNA, for 48h followed by +/− 20ng/ml TNF-α Immunofluorescence was performed using p65 NF-kB antibody and DAPI for nuclei staining.
sCLU modulates NF-κB regulated transcriptome
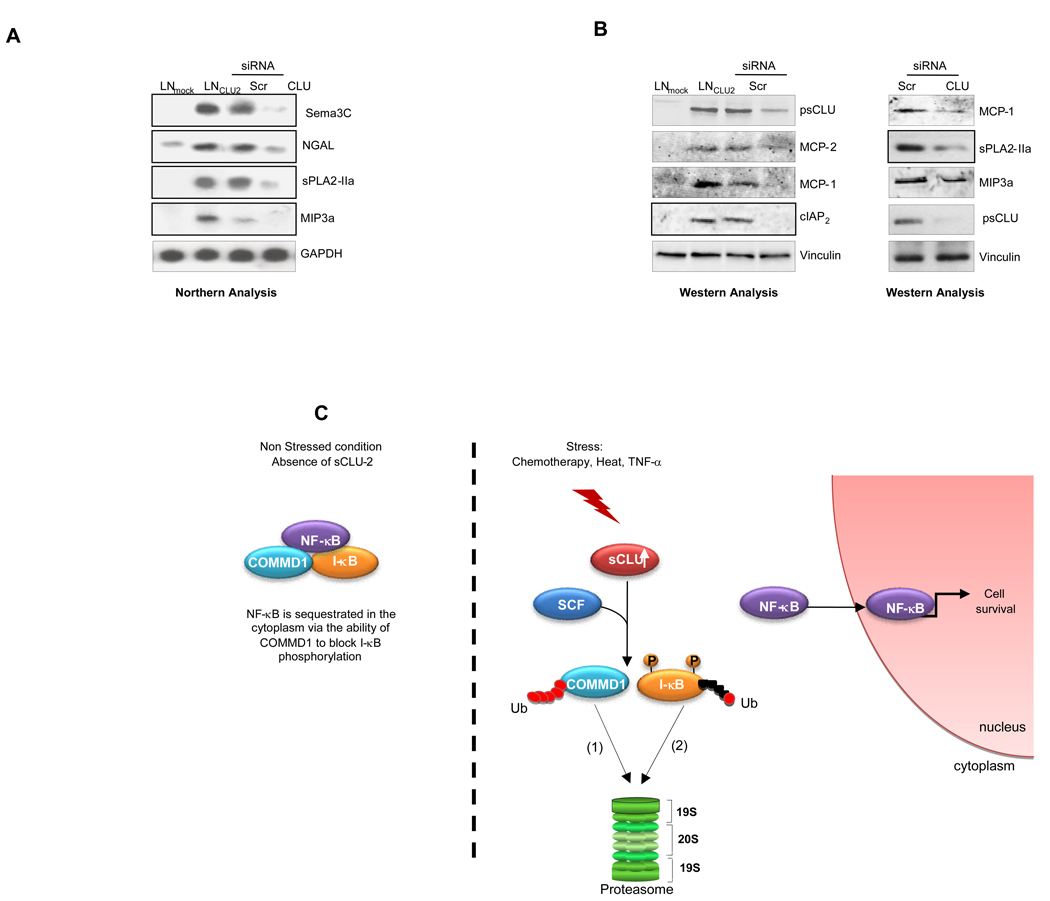
A genome-wide, unbiased analysis of comparative microarrays with sCLU over-expression versus knockdown in LNCaP cells also identified a link between sCLU levels and NF-κB activity (S11). mRNA levels of several NF-κB-regulated genes (Sema3C, NGAL, sPLA2-IIa, MIP3a) were upregulated in LNsCLU cells and downregulated after sCLU knock-down (Fig. 6A). Protein levels several more NF-κB-regulated genes (MCP-1, MCP-2, cIAP2) were upregulated in LNsCLU cells and downregulated after sCLU knock-down (Fig. 6B left panel). In addition a western blot of PC-3 cells following sCLU knockdown confirmed that sCLU expression correlates with levels of MCP-1, MIP3a and sPLA2-IIa (Fig. 6B right panel).
Figure 6. sCLU modulates NF-κB dependent genes.
A, sCLU expression correlates with NF-kB dependent genes. Total RNA were extracted from LNmock and LNsCLU, and also from LNsCLU cells treated with Scr- or CLU-siRNA. Northern analysis was performed using Sema3C, NGAL, sPLA2-IIa, and MIP3a probes. B, Validation of gene array at the protein level. Total proteins were extracted from LNmock and LNsCLU, and also from LNsCLU (left panel) and PC-3 cells (right panel) treated with Scr- or CLU-siRNA, western blots were performed using CLU, MCP-1, MCP-2, cIAP2, sPLA2-IIa, MIP-3 and vinculin antibodies. C, Schema illustrating ligand-independent mode of NF-κB transactivation involving the molecular chaperone, sCLU.
Discussion
sCLU is a stress-activated, cytoprotective glycoprotein implicated in numerous physiologic processes. While its multifunctional roles link this versatile chaperone to the activity of many pathways, the downstream effectors and mechanism(s) of sCLU action remain poorly defined. Proposed mechanisms for its anti-apoptotic activity include decreased caspase 3 activation (27) and inhibition of activated Bax (2). We describe a novel property of sCLU, namely its ability to regulate ubiquitination and degradation rates of COMMD1 and I-κB to enhance NF-κB nuclear translocation and transcriptional activity. Accordingly, sCLU knockdown stabilizes COMMD1 and I-κB, sequesters NF-κB in the cytoplasm, and decreases transcription of many NF-κB regulated genes, including NGAL, lipocalin2, sPLA2-IIa, semaphorin3C, apoptosis inhibitor2, and syndecan4.
The links between sCLU and NF-κB began when a two-hybrid approach identified COMMD1 as a sCLU binding protein. COMMD1 is a ubiquitously expressed protein implicated initially in regulation of copper homeostasis (28). COMMD1 also inhibits HIV replication in unstimulated CD4+ T cells, through its ability to suppress basal and cytokine-stimulated NF-κB activity (23). COMMD1 knockdown enhances phosphorylation and proteasomal degradation of I-κBα decreasing total I-κBα and increasing NF-κB activity (23). Moreover, COMMD1 represses NF-κB-dependent transcription by decreasing the duration of NF-κB-chromatin interaction (21). COMMD1 also accelerates ubiquitination and degradation of NF-κB subunits through its interaction with multimeric ubiquitin ligase containing Elongins B and C, Cul2 and SOCS1 (17). Consequently, COMMD1 is an important negative regulator of NF-κB (17, 23, 29).
NF-κB, a stress-regulated transcription factor belonging to the Rel family, plays a pivotal role in the control of inflammatory and innate immune responses. N F-κB activation is also associated with oncogenesis, through control of cell proliferation, migration, cell cycle progression, and apoptosis. Inactive NF-κB is sequestered in the cytoplasm by interaction with I-κB family members inhibiting its nuclear translocation and transcriptional activity. In the canonical pathway, IKK complex phosphorylates I-κB leading to ubiquitination and degradation in the 26S proteosome, thereby exposing nuclear localization signals on NF-κB subunits with nuclear translocation and transactivation of NF-κB-regulated genes (30). NF-κB is activated in cancer cells by chemo- and radiation therapy and is associated with acquired anti-cancer treatment resistance, including androgen independent prostate cancer (31–33). sCLU functions like a small heat shock protein, potently inhibiting stress-induced protein misfolding and aggregation (34). sCLU is highly expressed in cancer, particularly post-treatment (1), where increased levels of heat shock proteins and sCLU manage the stress-increased unfolded protein burden by facilitating degradation of terminally misfolded proteins by the ubiquitin-proteasomal degradation system. Here, we report that sCLU enhances the catalytic activity of the proteasome and degradation of ubiquitinated proteins, while sCLU knockdown leads to accumulation of ubiquitinated proteins.
We found that sCLU acts as a UBP, a role which accounts for recent findings that the sCLU N-terminus co-localizes with ubiquitin in aggregosomes (35). These inclusions contain misfolded, ubiquitinated proteins sheathed in a cage of intermediate filaments. Interestingly, the N-terminus of sCLU co-localizes with the MSS1 subunit of the 19S proteasome, which led us to test whether sCLU interacts with ubiquitinated proteins. A well characterized function of the ubiquitin-proteasome complex is regulation of the NF-κB pathway. However, the ability of sCLU to affect this pathway remains controversial (36, 37). Interestingly, NF-κB has been reported to induce sCLU expression (38) suggesting a possible feed-forward loop regulating the shared anti-apoptotic effects of sCLU and NF-κB. In this study we introduce a new role for sCLU, the regulation of ligand-independent NF-κB activation by facilitating the ubiquitination and degradation of COMMD1 and I-κB via its interaction with a member of the E3 ligase complex SCF-βTrCP. We demonstrate that sCLU is part of a multimeric complex with NF-κB-COMMD1 and I-κB. sCLU reduces COMMD1 and I-κB protein levels, a process mediated by the proteasome as bortezomib and MG132 inhibit this effect and enhance NF-kB nuclear translocation. In contrast, sCLU knockdown decreases rates of COMMD1 and I-κB ubiquitination and degradation, sequestering NF-κB in the cytoplasm and identifying a mechanism by which OGX-011, an antisense inhibitor of sCLU, may enhance treatment-induced apoptosis. COMMD1 reportedly binds and inhibits the E3 ligase SCF-βTrCP-1, thereby preventing proteasomal degradation of I-κB. Knockdown of COMMD1 increased phospho-I-κB levels (23), an effect also observed with sCLU over-expression. Taken together, these data suggest that sCLU interacts with and inhibits COMMD1 stabilization of I-κB, exposing I-κB for phosphorylation and ubiquitination.
sCLU is a chaperone with no known intrinsic E3 ligase activity. Both COMMD1 and I-κB interact with the E3 ligase complex SCF-βTrCP, and we demonstrate here that sCLU also interacts with two components of this complex - Cul1 and βTrCP. SCF-βTrCP ubiquitinates and facilitates degradation of I-κB and COMMD1 (23, 24, 39), leading to activation of NF-κB. Elevated expression of β-TrCP1 has been reported in metastatic colon (40) and pancreatic (41) cancer, and correlated with constitutive NF-κB activation and acquired resistance. Accordingly, targeting β-TrCP1 through siRNA or expression of a dominant-negative mutant suppressed growth and survival of human breast cancer cells and sensitized them to apoptosis induced by anticancer agents (42). Interestingly, sCLU binds to β-TrCP1–2 and Cul1 members of SCF-βTrCP complex. This interaction between sCLU, β-TrCP and Cul1 are enhanced by TNF-α stimulation and correlates with increased I-κB phosphorylation, ubiquitination and proteasomal degradation. This data identify and define a novel scaffolding role for sCLU in regulating the NF-κB signalosome, including its ability to recruit E3 ligases, chaperone and stabilize I-κB-E3 and COMMD1-E3 ligase complexes, or act as a ubiquitin binding protein to facilitate proteasomal degradation of ubiquitinated proteins. Stress-induced increases in sCLU enhance rates of COMMD1 and I-κB ubiquitination and proteasomal degradation through recruitment of SCF-βTrCP E3 ligase complex, and lead to increased NF-κB nuclear translocation and transcription of NF-κB regulated genes. We also observed that sCLU knockdown stabilizes COMMD1 and I-κB, and abrogates NF-κB nuclear translocation in the presence of TNF-α. sCLU knockdown inhibits NF-κB transcriptional activity as shown by luciferase assay and confirmed by gene profiling, induces apoptosis (S12), and identify a set of putative pharmacodynamic markers that may prove clinically useful as inhibitors of sCLU like OGX-011 move through clinical development.
Collectively, these studies provide the first evidence that sCLU functions as a UBP, enhancing COMMD1 and I-κB ubiquitination and proteasomal degradation through recruitment of SCF-βTrCP E3 ligase complex, increasing NF-κB nuclear translocation with transcription of NF-κB regulated genes, and defining a novel mechanism whereby stress-induced increases in sCLU activates the NF-κB signalosome (Fig. 6C).
Supplementary Material
Acknowledgements
This work was supported by grants from the Terry Fox Foundation of the National Cancer Institute of Canada and the NIH Pacific Northwest Prostate Specialized Programs of Research Excellence.
References
- 1.Shannan B, Seifert M, Leskov K, et al. Challenge and promise: roles for clusterin in pathogenesis, progression and therapy of cancer. Cell Death Differ. 2006;13:12–19. doi: 10.1038/sj.cdd.4401779. [DOI] [PubMed] [Google Scholar]
- 2.Zhang H, Kim JK, Edwards CA, Xu Z, Taichman R, Wang CY. Clusterin inhibits apoptosis by interacting with activated Bax. Nat Cell Biol. 2005;7:909–915. doi: 10.1038/ncb1291. [DOI] [PubMed] [Google Scholar]
- 3.Bayon Y, Ortiz MA, Lopez-Hernandez FJ, Howe PH, Piedrafita FJ. The retinoid antagonist MX781 induces clusterin expression in prostate cancer cells via heat shock factor-1 and activator protein-1 transcription factors. Cancer Res. 2004;64:5905–5912. doi: 10.1158/0008-5472.CAN-03-3657. [DOI] [PubMed] [Google Scholar]
- 4.Loison F, Debure L, Nizard P, le Goff P, Michel D, le Drean Y. Up-regulation of the clusterin gene after proteotoxic stress: implication of HSF1–HSF2 heterocomplexes. Biochem J. 2006;395:223–231. doi: 10.1042/BJ20051190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lidstrom AM, Bogdanovic N, Hesse C, Volkman I, Davidsson P, Blennow K. Clusterin (apolipoprotein J) protein levels are increased in hippocampus and in frontal cortex in Alzheimer's disease. Exp Neurol. 1998;154:511–521. doi: 10.1006/exnr.1998.6892. [DOI] [PubMed] [Google Scholar]
- 6.French LE, Soriano JV, Montesano R, Pepper MS. Modulation of clusterin gene expression in the rat mammary gland during pregnancy, lactation, and involution. Biol Reprod. 1996;55:1213–1220. doi: 10.1095/biolreprod55.6.1213. [DOI] [PubMed] [Google Scholar]
- 7.Miyake H, Nelson C, Rennie PS, Gleave ME. Testosterone-repressed prostate message-2 is an antiapoptotic gene involved in progression to androgen independence in prostate cancer. Cancer Res. 2000;60:170–176. [PubMed] [Google Scholar]
- 8.Kalka K, Ahmad N, Criswell T, Boothman D, Mukhtar H. Up-regulation of clusterin during phthalocyanine 4 photodynamic therapy-mediated apoptosis of tumor cells and ablation of mouse skin tumors. Cancer Res. 2000;60:5984–5987. [PubMed] [Google Scholar]
- 9.Klokov D, Criswell T, Leskov KS, Araki S, Mayo L, Boothman DA. IR-inducible clusterin gene expression: a protein with potential roles in ionizing radiation-induced adaptive responses, genomic instability, and bystander effects. Mutat Res. 2004;568:97–110. doi: 10.1016/j.mrfmmm.2004.06.049. [DOI] [PubMed] [Google Scholar]
- 10.Miyake H, Tolcher A, Gleave ME. Antisense Bcl-2 oligodeoxynucleotides inhibit progression to androgen-independence after castration in the Shionogi tumor model. Cancer Res. 1999;59:4030–4034. [PubMed] [Google Scholar]
- 11.Miyake H, Hara I, Gleave ME. Antisense oligodeoxynucleotide therapy targeting clusterin gene for prostate cancer: Vancouver experience from discovery to clinic. Int J Urol. 2005;12:785–794. doi: 10.1111/j.1442-2042.2005.01173.x. [DOI] [PubMed] [Google Scholar]
- 12.Yamanaka K, Gleave M, Muramaki M, Hara I, Miyake H. Enhanced radiosensitivity by inhibition of the anti-apoptotic gene clusterin using antisense oligodeoxynucleotide in a human bladder cancer model. Oncol Rep. 2005;13:885–890. [PubMed] [Google Scholar]
- 13.So A, Sinnemann S, Huntsman D, Fazli L, Gleave M. Knockdown of the cytoprotective chaperone, clusterin, chemosensitizes human breast cancer cells both in vitro and in vivo. Mol Cancer Ther. 2005;4:1837–1849. doi: 10.1158/1535-7163.MCT-05-0178. [DOI] [PubMed] [Google Scholar]
- 14.Chi KN, Eisenhauer E, Fazli L, et al. A phase I pharmacokinetic and pharmacodynamic study of OGX-011, a 2'-methoxyethyl antisense oligonucleotide to clusterin, in patients with localized prostate cancer. J Natl Cancer Inst. 2005;97:1287–1296. doi: 10.1093/jnci/dji252. [DOI] [PubMed] [Google Scholar]
- 15.Chi KN, Siu LL, Hirte H, et al. A phase I study of OGX-011, a 2'-methoxyethyl phosphorothioate antisense to clusterin, in combination with docetaxel in patients with advanced cancer. Clin Cancer Res. 2008;14:833–839. doi: 10.1158/1078-0432.CCR-07-1310. [DOI] [PubMed] [Google Scholar]
- 16.Rocchi P, Beraldi E, Ettinger S, et al. Increased Hsp27 after androgen ablation facilitates androgen-independent progression in prostate cancer via signal transducers and activators of transcription 3-mediated suppression of apoptosis. Cancer Res. 2005;65:11083–11093. doi: 10.1158/0008-5472.CAN-05-1840. [DOI] [PubMed] [Google Scholar]
- 17.Maine GN, Mao X, Komarck CM, Burstein E. COMMD1 promotes the ubiquitination of NF-kappaB subunits through a cullin-containing ubiquitin ligase. Embo J. 2007;26:436–447. doi: 10.1038/sj.emboj.7601489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zoubeidi A, Zardan A, Beraldi E, et al. Cooperative interactions between androgen receptor (AR) and heat-shock protein 27 facilitate AR transcriptional activity. Cancer Res. 2007;67:10455–10465. doi: 10.1158/0008-5472.CAN-07-2057. [DOI] [PubMed] [Google Scholar]
- 19.Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fabunmi RP, Wigley WC, Thomas PJ, DeMartino GN. Activity and regulation of the centrosome-associated proteasome. J Biol Chem. 2000;275:409–413. doi: 10.1074/jbc.275.1.409. [DOI] [PubMed] [Google Scholar]
- 21.Burstein E, Ganesh L, Dick RD, et al. A novel role for XIAP in copper homeostasis through regulation of MURR1. Embo J. 2004;23:244–254. doi: 10.1038/sj.emboj.7600031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doskeland AP. A method for simple identification of signature peptides derived from polyUb-K48 and K63 by MALDI-TOF MS and chemically assisted MS/MS fragmentation. Amino Acids. 2006;30:99–103. doi: 10.1007/s00726-005-0227-4. [DOI] [PubMed] [Google Scholar]
- 23.Ganesh L, Burstein E, Guha-Niyogi A, et al. The gene product Murr1 restricts HIV-1 replication in resting CD4+ lymphocytes. Nature. 2003;426:853–857. doi: 10.1038/nature02171. [DOI] [PubMed] [Google Scholar]
- 24.Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, Harper JW. The SCFbeta-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IkappaBalpha and beta-catenin and stimulates IkappaBalpha ubiquitination in vitro. Genes Dev. 1999;13:270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang CY, Mayo MW, Baldwin AS., Jr TNF-and cancer therapy-induced apoptosis: potentiation by inhibition of NF-kappaB. Science. 1996;274:784–787. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- 26.Spencer E, Jiang J, Chen ZJ. Signal-induced ubiquitination of IkappaBalpha by the F-box protein Slimb/beta-TrCP. Genes Dev. 1999;13:284–294. doi: 10.1101/gad.13.3.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Savkovic V, Gantzer H, Reiser U, et al. Clusterin is protective in pancreatitis through anti-apoptotic and anti-inflammatory properties. Biochem Biophys Res Commun. 2007;356:431–437. doi: 10.1016/j.bbrc.2007.02.148. [DOI] [PubMed] [Google Scholar]
- 28.Mercer JF. The molecular basis of copper-transport diseases. Trends Mol Med. 2001;7:64–69. doi: 10.1016/s1471-4914(01)01920-7. [DOI] [PubMed] [Google Scholar]
- 29.Maine GN, Burstein E. COMMD Proteins and the Control of the NFkappaB Pathway. Cell Cycle. 2007:6. doi: 10.4161/cc.6.6.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shishodia S, Aggarwal BB. Nuclear factor-kappaB: a friend or a foe in cancer? Biochem Pharmacol. 2004;68:1071–1080. doi: 10.1016/j.bcp.2004.04.026. [DOI] [PubMed] [Google Scholar]
- 31.Nakanishi C, Toi M. Nuclear factor-kappaB inhibitors as sensitizers to anticancer drugs. Nat Rev Cancer. 2005;5:297–309. doi: 10.1038/nrc1588. [DOI] [PubMed] [Google Scholar]
- 32.Beg AA, Baltimore D. An essential role for NF-kappaB in preventing TNF-alpha-induced cell death. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 33.Jin RJ, Lho Y, Connelly L, et al. The nuclear factor-kappaB pathway controls the progression of prostate cancer to androgen-independent growth. Cancer Res. 2008;68:6762–6769. doi: 10.1158/0008-5472.CAN-08-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carver JA, Rekas A, Thorn DC, Wilson MR. Small heat-shock proteins and clusterin: intra- and extracellular molecular chaperones with a common mechanism of action and function? IUBMB Life. 2003;55:661–668. doi: 10.1080/15216540310001640498. [DOI] [PubMed] [Google Scholar]
- 35.Debure L, Vayssiere JL, Rincheval V, Loison F, Le Drean Y, Michel D. Intracellular clusterin causes juxtanuclear aggregate formation and mitochondrial alteration. J Cell Sci. 2003;116:3109–3121. doi: 10.1242/jcs.00619. [DOI] [PubMed] [Google Scholar]
- 36.Santilli G, Aronow BJ, Sala A. Essential requirement of apolipoprotein J (clusterin) signaling for IkappaB expression and regulation of NF-kappaB activity. J Biol Chem. 2003;278:38214–38219. doi: 10.1074/jbc.C300252200. [DOI] [PubMed] [Google Scholar]
- 37.Devauchelle V, Essabbani A, De Pinieux G, et al. Characterization and functional consequences of underexpression of clusterin in rheumatoid arthritis. J Immunol. 2006;177:6471–6479. doi: 10.4049/jimmunol.177.9.6471. [DOI] [PubMed] [Google Scholar]
- 38.Li X, Massa PE, Hanidu A, et al. IKKalpha, IKKbeta, and NEMO/IKKgamma are each required for the NF-kappa B-mediated inflammatory response program. J Biol Chem. 2002;277:45129–45140. doi: 10.1074/jbc.M205165200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yaron A, Hatzubai A, Davis M, et al. Identification of the receptor component of the IkappaBalpha-ubiquitin ligase. Nature. 1998;396:590–594. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]
- 40.Ougolkov A, Zhang B, Yamashita K, et al. Associations among beta-TrCP, an E3 ubiquitin ligase receptor, beta-catenin, and NF-kappaB in colorectal cancer. J Natl Cancer Inst. 2004;96:1161–1170. doi: 10.1093/jnci/djh219. [DOI] [PubMed] [Google Scholar]
- 41.Muerkoster S, Arlt A, Sipos B, et al. Increased expression of the E3-ubiquitin ligase receptor subunit betaTRCP1 relates to constitutive nuclear factor-kappaB activation and chemoresistance in pancreatic carcinoma cells. Cancer Res. 2005;65:1316–1324. doi: 10.1158/0008-5472.CAN-04-1626. [DOI] [PubMed] [Google Scholar]
- 42.Tang W, Li Y, Yu D, Thomas-Tikhonenko A, Spiegelman VS, Fuchs SY. Targeting beta-transducin repeat-containing protein E3 ubiquitin ligase augments the effects of antitumor drugs on breast cancer cells. Cancer Res. 2005;65:1904–1908. doi: 10.1158/0008-5472.CAN-04-2597. [DOI] [PubMed] [Google Scholar]
- 43.Trougakos IP, Djeu JY, Gonos ES, Boothman DA. Advances and challenges in basic and translational research on clusterin. Cancer Res. 2009;69:403–406. doi: 10.1158/0008-5472.CAN-08-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.