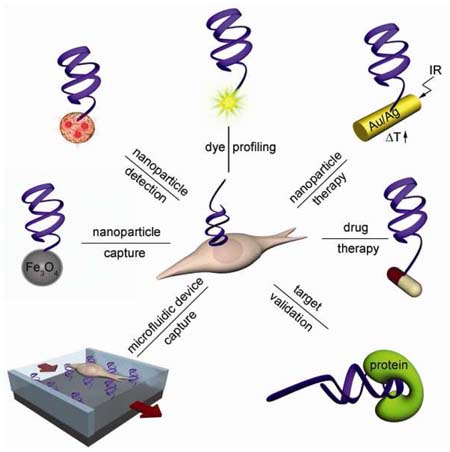
Conspectus
Molecular medicine is an emerging field focused on understanding the molecular basis of diseases and translating this information into strategies for diagnosis and therapy. This approach could lead to personalized medical treatments. Currently, our ability to understand human diseases at the molecular level is limited by the lack of molecular tools to identify and characterize the distinct molecular features of the disease state, especially for diseases such as cancer. Among the new tools being developed by researchers including chemists, engineers and other scientists is a new class of nucleic acid probes called aptamers, which are ssDNA/RNA molecules selected to target a wide range of molecules and even cells. In this Account, we will focus on the use of aptamers, generated from cell-based selections, as a novel molecular tool for cancer research.
Cancers originate from mutations of human genes. These genetic alterations result in molecular changes to diseased cells, which, in turn, lead to changes in cell morphology and physiology. For decades, clinicians have diagnosed cancers primarily based on the morphology of tumor cells or tissues. However, this method does not always give an accurate diagnosis and does not allow clinicians to effectively assess the complex molecular alterations that are predictive of cancer progression. Although genomics and proteomics do not yet allow a full access to this molecular knowledge, aptamer probes represent one effective and practical avenue toward this goal. One special feature of aptamers is that we can isolate them by selection against cancer cells without prior knowledge of the number and arrangement of proteins on the cellular surface. These probes can identify molecular differences between normal and tumor cells and can discriminate among tumor cells of different classifications, at different disease stages, or from different patients.
This Account summarizes our recent efforts to develop aptamers through cell-SELEX for the study of cancer and apply those aptamers in cancer diagnosis and therapy. We first discuss how we select aptamers against live cancer cells. We then describe uses of these aptamers. Aptamers can serve as agents for molecular profiling of specific cancer types. They can also be used to modify therapeutic reagents to develop targeted cancer therapies. Aptamers are also aiding the discovery of new cancer biomarkers through the recognition of membrane protein targets. Importantly, we demonstrate how molecular assemblies can integrate the properties of aptamers and, for example, nanoparticles or microfluidic devices, to improve cancer cell enrichment, detection and therapy.
Introduction
Aptamers, sometimes called chemical antibodies, are antibody-like molecules in that they function primarily in molecular recognition. However, they are very different from antibodies in both structure and properties. Aptamers are short, single-stranded oligonucleotides generated from an in vitro method known as SELEX (Systematic Evolution of Ligands by EXponential enrichment) [1–2]. The SELEX process normally starts with a random library of 1013 ~ 1016 ssDNA or ssRNA molecules and is followed by an iterative process that utilizes PCR to specifically amplify sequences having high binding affinity to a target. The targets of aptamers range from small organic molecules and metal ions to proteins, biological cells and tissues [3–6]. By folding into distinct secondary and tertiary structures, aptamers can bind to their targets with high affinity (dissociation constants on the order of uM to pM) and can recognize their targets with specificity that is comparable to antibodies. Moreover, man-made aptamers possess several advantages over natural-made antibodies [3–7]. These include quick and reproducible synthesis, easy and controllable modification to fulfill different diagnostic and therapeutic purposes, long-term stability as dry powder or in solution, ability to sustain reversible denaturation, non-toxicity and lack of immunogenicity, and fast tissue penetration. These chemical properties make aptamers ideal candidates as probes for use in molecular medicine to elucidate the molecular foundation of diseases, particularly cancer.
Cell-based selection of aptamers specific to cancer cells
Aptamers have shown many important applications in bioanalysis and biomedicine [3–6]. Particularly, several aptamers have been developed against cancer-related proteins, such as PDGF, VEGF, HER3, NFkB, tenascin-C, or PMSA [8–10]. Most of them were generated using single purified proteins as targets. Recently, however, aptamer selection against complex targets, especially whole cells, has been demonstrated and increasingly adopted [10–17]. The advantage of aptamer selection against live cells with target proteins expressing on the cell surface is straightforward: the cell-surface proteins keep their native conformations which are critical for their biological functions. For example, aptamers that recognize human receptor tyrosine kinase RET have been obtained with RET-expressing cells as targets [11]. In contrast to protein-based SELEX, we emphasize in this review that cell-based selection can be carried out without prior knowledge of a cell’s molecular signature. In other words, it is not necessary to know the number or types of proteins on the cell membrane surface in order to generate aptamers which will be highly useful for molecular recognition of cancer cells. It is the selection process itself that differtiates different types of cells, thus making it possible to obtain aptamer probes which only bind to a specific type of cancer cells, not normal cells or other types of cancer cells. Moreover, since the selection can be done with whole cells and many receptor proteins exist on the cell membrane surfaces, a panel of aptamer probes can be selected to profile the molecular characteristics of the target cancer type, which is a main objective in the development of cell-based aptamers for use in cancer diagnosis and treatment.
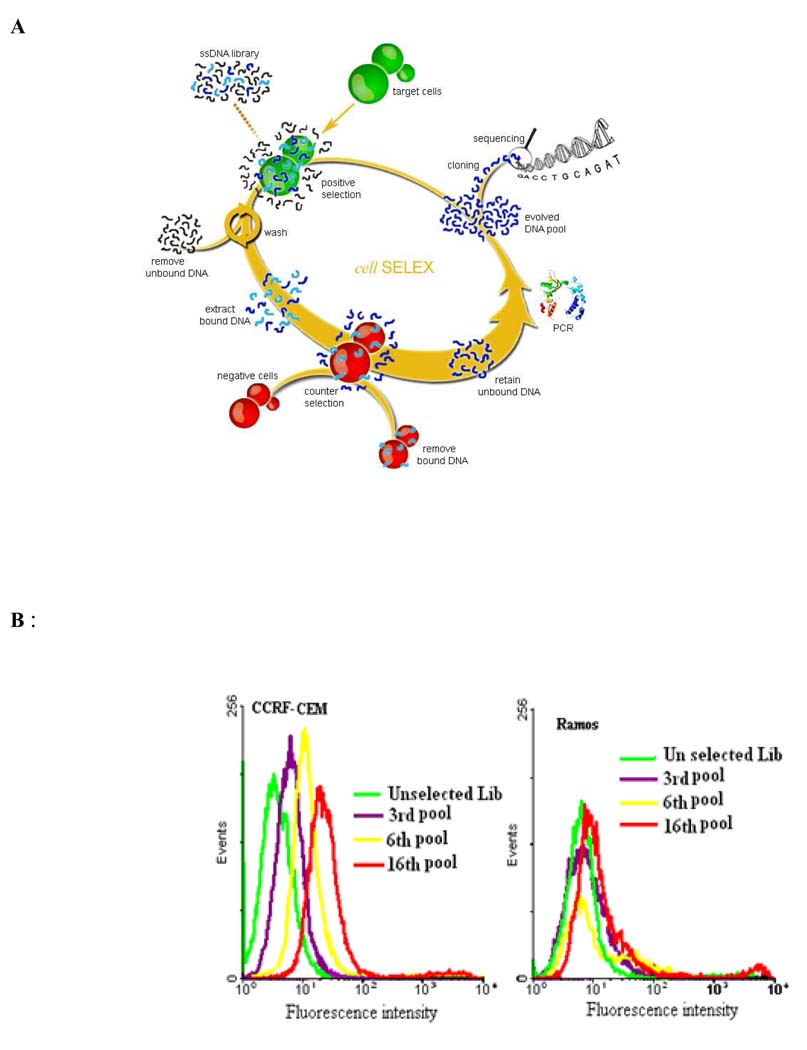
To establish the method for cancer cell-based selection of aptamers, cultured precursor T cell acute lymphoblastic leukemia (ALL) cell line, CCRF-CEM, was used as the target [17]. A counter-selection using a B-cell line from human Burkitt's lymphoma, Ramos, as the negative control, was added in the selection process to exclude DNA sequences that could bind to common molecules on the leukemia cell surface. The typical cell-based selection process is shown schematically in Figure 1A. Briefly, the CCRF-CEM cells were incubated with the ssDNAs library on ice. After washing, the cell-surface-bound sequences were eluted by heating. The eluted sequences were then allowed to interact with excess Ramos cells. Only the sequences that remained free in the supernatant were collected and amplified by PCR to form the starting pool for the next round of selection.
Figure 1.
(A) Schematics of the cell-based aptamer selection (B) Flow cytometry assay to monitor the binding of selected pools with CCRF-CEM cells (target cells) and Ramos cells (negative cells).
For the suspension cells, like leukemia cells, it was found that flow cytometry assay is the best way to monitor the enrichment process by virtue of its quantitative nature, high statistical precision, high speed and reproducibility. The DNA products collected after each selection round were sampled for labeling with a fluorescence dye and incubated with CCRF-CEM cells as well as Ramos cells. As the number of selection cycles increased, a corresponding steady increase in fluorescence intensity of the CCRF-CEM cells was observed (Figure 1B), indicating that DNA sequences with higher binding affinity to the target cells were enriched. In contrast, there was no significant change in fluorescence intensity of the Ramos cells. Normally, enriched pools are achieved after about 20 rounds; these can then be cloned and sequenced. Using this method, several high affinity aptamers for CCRF-CEM cells have been obtained with calculated Kds in the nM to pM range. Because they could specifically recognize CCRF-CEM leukemia cells mixed with normal human bone marrow aspirates, the specificity of these aptamers was demonstrated. We then further expanded our cell-based SELEX for solid tumor adherent cells [18]. A nonenzymatic cell dissociation solution containing EDTA was used to detach the adherent cells before incubating them with the labeled DNA pool. Flow cytometry could then be applied to the cell suspension to monitor the enrichment process.
Panels of new aptamer probes have been successfully selected from cell-SELEX for several types of cancer cells, including lymphocytic leukemia, myeloid leukemia, liver cancer, small cell lung cancer, and non-small cell lung cancer [19–21]. This again highlights the usefulness of cell-SELEX in selecting cancer cell-specific probes without the advantage of known targets. These aptamers all showed high affinity and excellent specificity. However, for these aptamer panels, it should be noted that more selection rounds and longer selection times are required compared to SELEX with single compounds and that any damage to fragile cells during the process does incur the risk of selection failure.
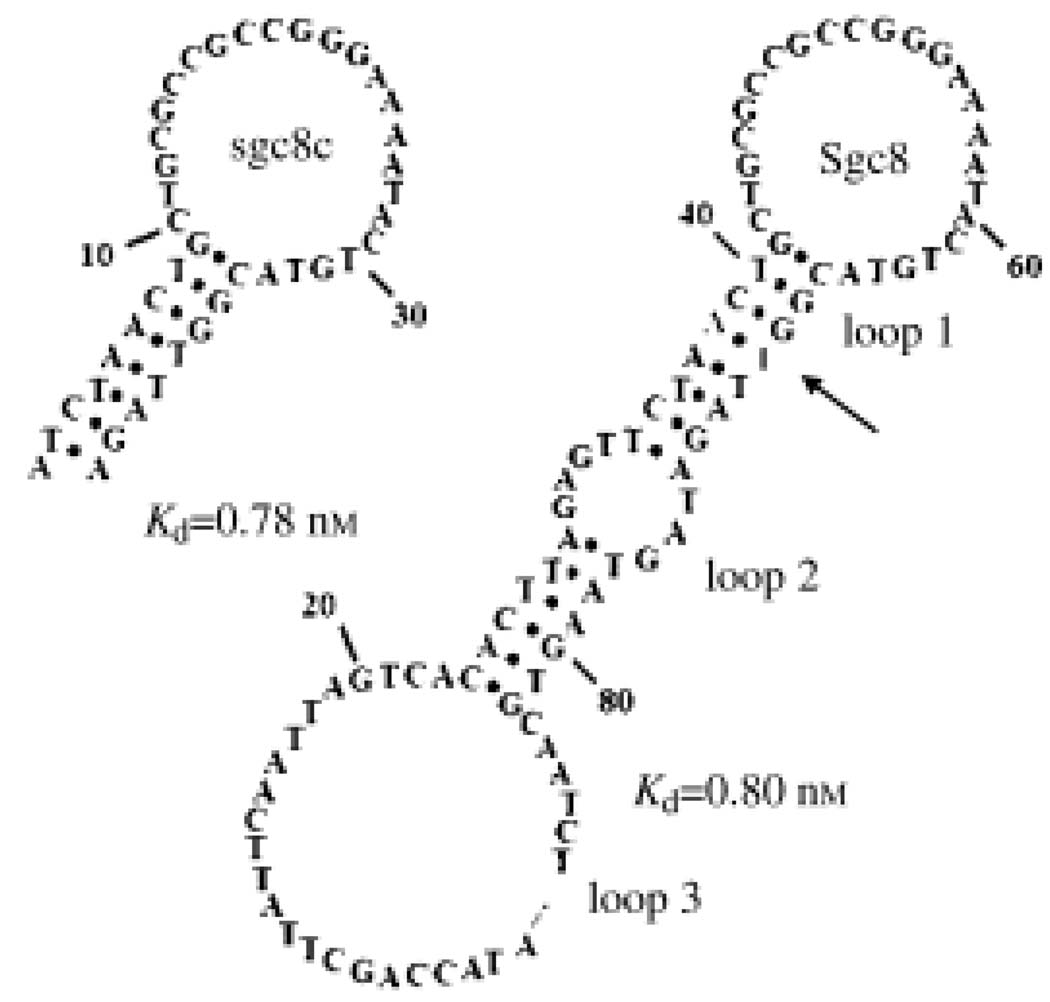
Similar to aptamers selected against purified proteins, aptamers from cell-SELEX can also be analyzed to obtain the binding motif and be post-engineered to enhance their affinity or functionality [22–24]. For example, Figure 2 shows a minimized aptamer, Sgc8c, which was cut from the sequence, Sgc8, obtained in cell-SELEX. Further modification of Sgc8c with LNA bases and PEG spacers resulted in its biostability, as demonstrated by increase in serum, and two-fold higher affinity than the original aptamers [24]. This showed that the aptamers selected from cancer cells are ready for biomedical applications.
Figure 2.
Secondary structures of a selected aptamer and the truncated one.
In the following part of the review, we will explore the chemical biology approaches for these new aptamers and their applications for cancer diagnosis and therapy.
Aptamer-based cancer cell detection
Molecular profiling
Molecular-level differentiation of cancer cells is essential for accurate and early diagnosis [25]. Given the complexity and diversity of cancers, even within similar cancer categories, multiple specific molecular probes are usually needed to delineate unique molecular fingerprints of tumor cells. The systematic production of a panel of antibodies for molecular profiling of cancer cells with unknown biomarkers has proven to be very difficult. As alternative strategies, the phage display peptide library and one-bead one-compound combinatory peptide library have been applied to discover tumor cell-specific peptides [26]. Cell-based selection of aptamers is, however, superior to all of these methods by its use of positive and negative cells to generate multiple probe molecules for cancer cell identification and subcategorization. Moreover, compared to the peptide library (~108), the DNA/RNA library for aptamers is usually much larger (~1016). The cell-SELEX method is also a simple, fast, and economical way to generate high affinity and specific aptamers for molecular recognition.
To test whether aptamers derived from cell-SELEX could be used for cancer cell recognition, 6 aptamers, which were respectively generated from two types of leukemia cells, Toledo cells (B-cell ALL) and CCRF-CEM cells (T-ALL), were selected [27]. With more than 20 related leukemia cell lines and the cells from normal human bone marrow aspirates, all of the aptamers demonstrated excellent recognition of their targets, and combinations of selected aptamers produced distinct patterns for different tumor cells. Therefore, these aptamers were next tested for their ability to characterize real samples from leukemia patients. As shown in Table 1, three aptamers (sgc8, sgc3, and sgd3) bound selectively to T-ALL patient samples. Moreover, the differential binding of these three aptamers among the T-ALL patient samples showed the ability of aptamers to distinguish molecular differences among patients with the same diagnosis by current technology. Two aptamers (Sgc4 and sgd2) bound to a high percentage of cells in one leukemia (AML) patient sample, indicating that this particular patient’s cells were expressing surface markers similar to those from patients with T-ALL and B-ALL. These results demonstrate the ability of these aptamers to specifically recognize molecular differences between patients. Correlations between the differential aptamer-binding and the specific phenotypes and prognosis of these patients are being investigated.
Table 1.
Aptamer profiling of cancer cells in patients’ samples
| sgc8 | sgc3 | sgc4 | sgd2 | sgd3 | sgd5 | ||
|---|---|---|---|---|---|---|---|
| T ALL 1 | ++ | +++ | +++ | +++ | +++ | ND | |
| T ALL 2 | ++ | + | +++ | ++ | + | 0 | |
| T ALL 3 | + | + | ++++ | +++ | + | 0 | |
| T ALL 4 | + | + | ++ | +++ | + | 0 | |
| T ALL 5 | + | + | ++ | + | + | 0 | |
| T ALL 6 | 0 | 0 | + | + | 0 | 0 | |
| T ALL 7 | 0 | 0 | ++ | ++ | 0 | 0 | |
| TALL 8 | + | + | ++ | ++ | + | 0 | |
| TALL 9 | + | 0 | + | + | 0 | 0 | |
| TALL10 | 0 | + | + | 0 | + | 0 | |
| B ALL 1 | 0 | 0 | ++ | ++ | 0 | 0 | |
| B ALL 2 | 0 | 0 | ++ | ++ | 0 | + | |
| B ALL 3 | ++ | 0 | ++ | ++ | 0 | + | |
| B-ALL 4 | 0 | 0 | + | + | 0 | 0 | |
| AML 1 | + | + | ++ | + | 0 | 0 | |
| AML 2 | + | 0 | ++ | + | 0 | 0 | |
| AML 3 | + | 0 | + | + | 0 | 0 | |
| AML 4 | 0 | 0 | ++++ | ++++ | 0 | 0 | |
| AML 5 | 0 | 0 | + | 0 | 0 | 0 | |
| AML 6 | + | 0 | 0 | 0 | 0 | 0 | |
| AML 7 | + | 0 | 0 | 0 | 0 | 0 | |
| AML 8 | + | 0 | +++ | +++ | 0 | 0 |
Note: In the flow cytometry analysis, a threshold based on fluorescence intensity of FITC was chosen so that 99 percent of cells incubated with the FITC-labeled unselected DNA library would have fluorescence intensity below it. When FITC-labeled aptamer was allowed to interact with the cells, the percentage of the cells with fluorescence above the set threshold was used to evaluate the binding capacity of the aptamer to the cells. 0:<10%, +:10–35%, ++:35–60%, +++:60–85%, ++++:>85%
Aptamer-nanoparticle conjugation to enhance detection
Early diagnosis of cancer not only relies on the specificity of the molecular probes, but also the sensitivity of their detection. Being DNA molecules with predicable structures and easy site-specific chemical modification, aptamers can readily link to advanced signaling mechanisms. We have attempted such a conjugation of aptamer with nanomaterials to enhance cancer cell detection [28–32]. To explain, some cancer cells, especially those in the early stages of disease development, may have a very low density of target on the cell surface available for aptamer detection. Therefore, to increase the signal and enhance binding affinity, multivalent binding, instead of single-aptamer binding, is usually considered to be an effective approach. Nanomaterials are particularly advantageous as multivalent ligand scaffolds by virtue of their large surface area and variable sizes. For example, we have synthesized Au-Ag nanorods (NRs) 12 nm × 56 nm in size as a nano-platform for multivalent binding by multiple aptamers on the rod [28]. Up to 80 fluorophore-labeled sgc8 aptamers could be attached on one NR, leading to a 26-fold higher affinity and over 300-fold higher fluorescence signal. The molecular assembly of aptamers on nanomaterials clearly shows the potential of multivalency in the case of low binding probes and/or low density binding sites. This also illustrates how molecular assemblies can integrate the properties of aptamers and nanomaterials to improve cancer cell targeting.
We have also developed a colorimetric assay for sensitive cancer cell detection using aptamers conjugated with 50 nm gold nanoparticles [29]. The assembly of aptamer-gold nanoparticle conjugates on the target cell surface resulted in a significant color change because the gold nanoparticles were in close proximity which allowed their surface plasmon resonances to overlap. The non-target cells did not display any change in color, and even a few nonspecific-binding nanoparticles would not have been enough to drastically alter the spectral property of the gold particles. The assay was quite sensitive, as 1,000 target cells could be readily detected by the naked eye, and the detection limit based on simple absorbance measurements was 90 cells. Such a method, which allows us to distinguish cancer cells in a quick and simple way, would therefore be advantageous in point-of-care diagnostics and large-scale cancer screening.
Cancer cell enrichment and detection
Detecting cancer at its earliest stage often requires the testing of body fluids, such as circulating blood and sputum, where the malignant cells are present at very low concentrations. Thus, developing an effective method for enriching and detecting cancer cells in low abundance is critically important. We have developed two such strategies for cancer cell enrichment and detection.
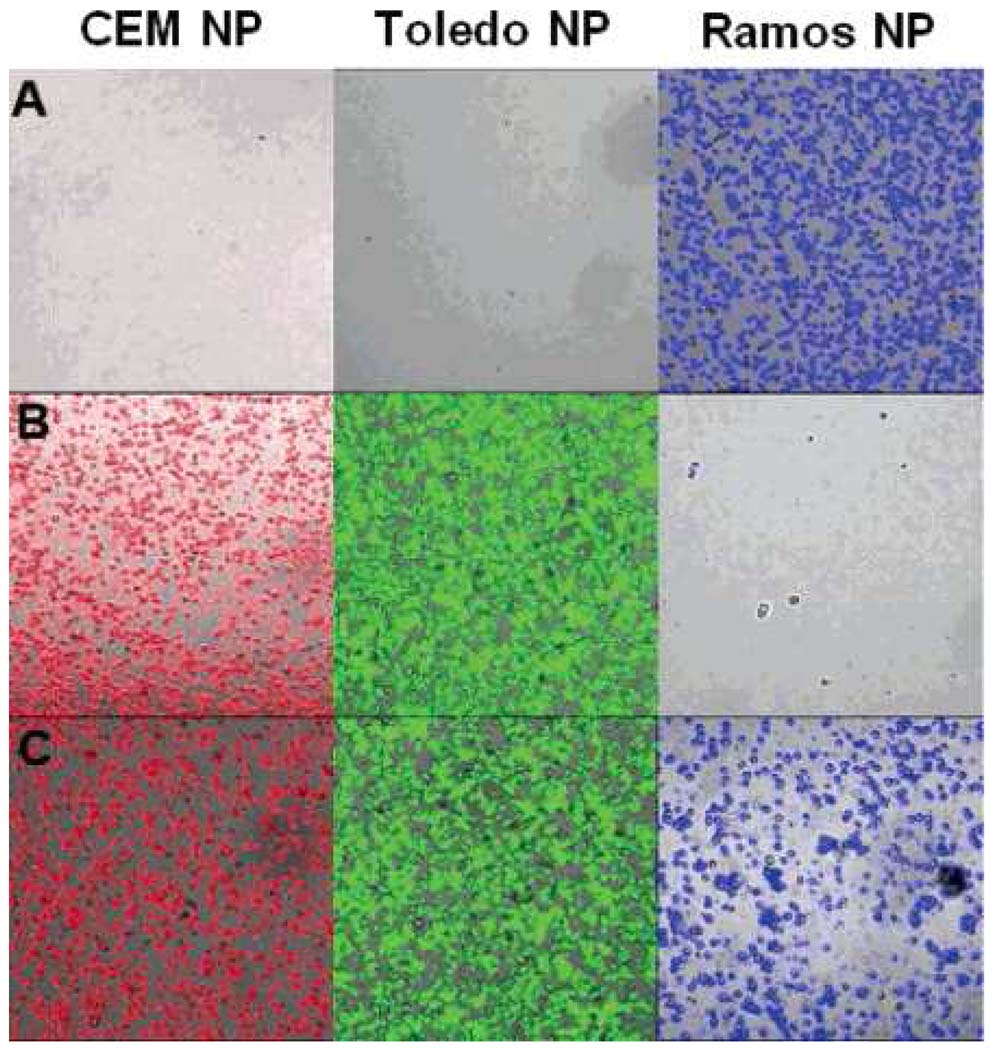
First, a novel two-nanoparticle assay was developed for the rapid collection and detection of leukemia cells [30]. In this assay, aptamer-modified magnetic nanoparticles were used for target cell extraction and enrichment, while aptamer-modified fluorescent dye-doped nanoparticles were simultaneously added for sensitive cell detection. The small size and relatively high surface area of the iron oxide-doped silica nanoparticles provide enhanced extraction capabilities exceeding those of the commonly used micrometer-sized particles [33]. The dye-doped nanoparticles enhance sensitivity by amplifying the signal intensity corresponding to each aptamer binding event. Compared to immunophenotypic- and PCR-based analyses, which take hours to complete, the two-particle assay was very fast, taking less than 1 h to complete. The reproducible extraction of target cells from whole blood samples was successfully carried out. We have extended this method for the collection and detection of multiple cancer cells [31]. As shown in Figure 3, three different cell samples, CEM, Ramos, and Toledo cells, were stepwise extracted and detected by fluorescent imaging using magnetic nanoparticles and dye-doped nanoparticles modified by three aptamers.
Figure 3.
Fluorescence images of the extracted mixed cell samples using the multiple extraction procedure: (A) contains only Ramos cells; (B) contains CEM and Toledo cells; (C) contains all three: CEM, Toledo, and Ramos cells.
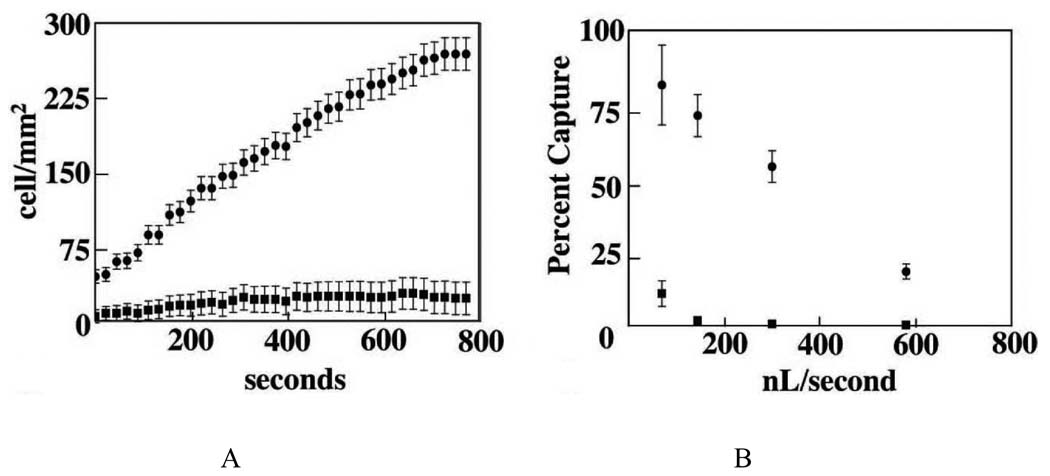
Although the two-nanoparticle assay is efficient, one concern is that the nanoparticle labeling may influence the analysis of extracted cells. In addition, the enrichment is limited as there is no flow system allowing samples to flow through the binding agents. To address these limitations, we have recently developed an aptamer-modified microfluidic device to capture rare cells from a large amount of background cells without the need for much sample pretreatment [32]. As a result of their small physical size and high surface area-to-volume ratio, microfluidic devices are promising platforms to create inexpensive and fast biomedical assays. Even though antibody-coated microfluidic devices have been demonstrated for the enrichment of cancer cells [34], aptamers are a better choice for immobilization on microfluidic devices. DNA-based devices can be stored for a long period of time and can be easily handled in clinical settings. By covalently binding scg8 to a prototype microfabricated poly(dimethylsiloxane) (PDMS) device (Figure 4), >80% capture efficiency with 97% purity for the target CEM cells was obtained. It was found that a significant increase in cell capture efficiency could be achieved by reducing the channel depth, and the pulsing of cell solutions instead of using a constant fluid flow is proposed. To further enhance capture surface and, hence, capture efficiency, micropillars or other microfeatures can also be adapted [34]. After enrichment, the cancer cells can be detected by any sensitive detection strategy discussed above. The aptamer-based microfluidic enrichment scheme is fast and economical and would therefore be a useful tool for early diagnosis of cancer.
Figure 4.
Cell-surface density (A) and cell capture efficiency (B) for target cells (dots) and control cells (squares) in the PDMS device under different conditions.
Aptamer-based target therapy
The goal of molecular medicine is to change the current trial-and-error treatment to evidence-based guided treatment, i.e., treating diseases with knowledge of their molecular origin and molecular changes. Targeted therapy at the molecular level will greatly improve cancer treatment with more efficacy, less toxicity, and fewer side effects. To create effective molecular platforms that target cancer cells, but not normal cells, several strategies, such as immuno-conjugates and peptide or protein conjugates, have been used [35–36]. However, with the advantage of cell-SELEX in generating multiple cancer cell-specific aptamers, which have better chemical properties than antibodies, we made significant strides in developing three types of targeted therapy applications: intracellular delivery, targeted chemotherapy and targeted phototherapy.
Targeted intracellular delivery
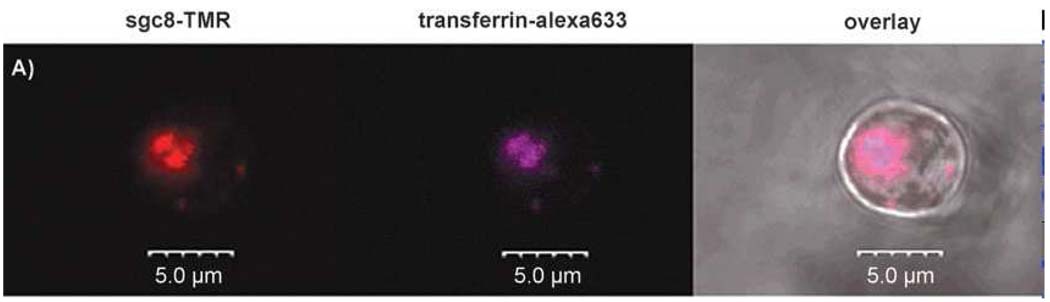
Considerable efforts have been made to incorporate aptamers into liposome vesicles or other delivery vector systems (e.g., siRNA) for possible targeted intracellular drug delivery [36–37]. While most of the existing aptamers cannot be directly taken up by cells, it is highly desirable to generate new aptamers that can be internalized into cells to facilitate their simple delivery. Up to now, the only successful example is the anti-PSMA (prostate-specific membrane antigen) aptamers [37–39]. However, our cell-based SELEX generated many aptamers available for evaluation. For example, we have studied the cellular internalization of one particular aptamer, sgc8, against CEM cells [40]. The results showed that the aptamers were specifically internalized to the target cells. Colocalization of the internalized aptamers and transferrin indicated that the aptamer was taken up into the endosome (Figure 5), a key intracellular compartment where the delivery agents release their cargos. No cytotoxicity was observed when the cells were treated with sgc8, indicating that the aptamer is a promising agent for cell type-specific intracellular delivery.
Figure 5.
Co-localization of sgc8 and transferrin in endosomes.
Targeted chemotherapy
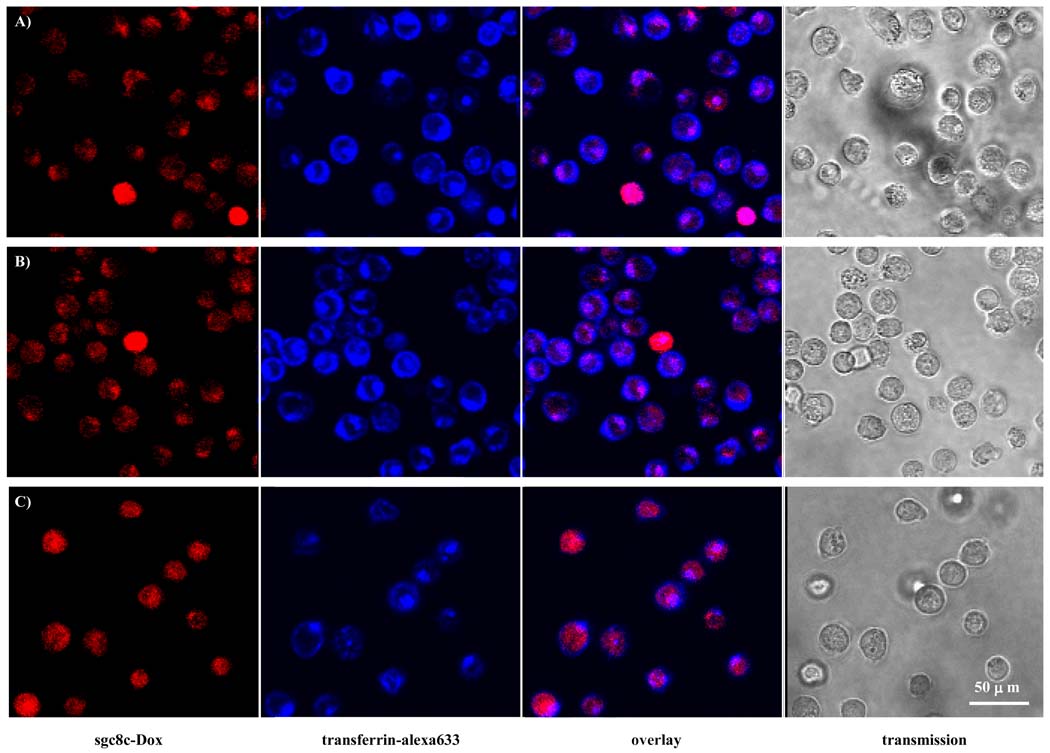
The conjugation of antitumor chemicals to targeting reagents, such as aptamers or antibodies, is a promising method that can increase the efficacy of chemotherapy and reduce its overall toxicity [41,42]. Docetaxel-encapsulated nanoparticles formed by biodegradable polymer and modified with anti-PMSA RNA aptamers have been successfully applied to a model animal, significantly enhancing efficacy and reducing toxicity [42]. We covalently linked the antitumor chemical doxorubicin (Dox) to the DNA aptamer sgc8c [43]. The sgc8c-Dox conjugate possesses many of the properties of the sgc8c aptamer, including high binding affinity and the capability to be efficiently internalized to endosome by target cells (Figure 6). Moreover, as a result of the specific conjugation method, the acid-labile linkage connecting the sgc8c-Dox conjugate can be cleaved inside the acidic endosome environment. Cell viability tests demonstrate that the sgc8c-Dox conjugates not only possess potency similar to the unconjugated Dox, but also have the required molecular specificity which is lacking in most current targeted drug delivery strategies. Nonspecific uptake of membrane-permeable Dox to non-target cell lines could also be inhibited by linking the drug with the aptamer. Compared to the less effective reported Dox-immunoconjugates, this sgc8c-Dox conjugate makes targeted chemotherapy more feasible with drugs having various potencies. This drug-aptamer conjugation method will have broad implications for targeted drug delivery.
Figure 6.
The distribution of sgc8c-Dox conjugates inside CCRF-CEM cells after incubation with cells for 30 min (A), 1 h (B), and 2 h (C), respectively. From left to right, the fluorescence confocal images were monitored for sgc8c-Dox, transferrin-alexa633, overlay of these two channels, and bright field channel, respectively.
Targeted phototherapy
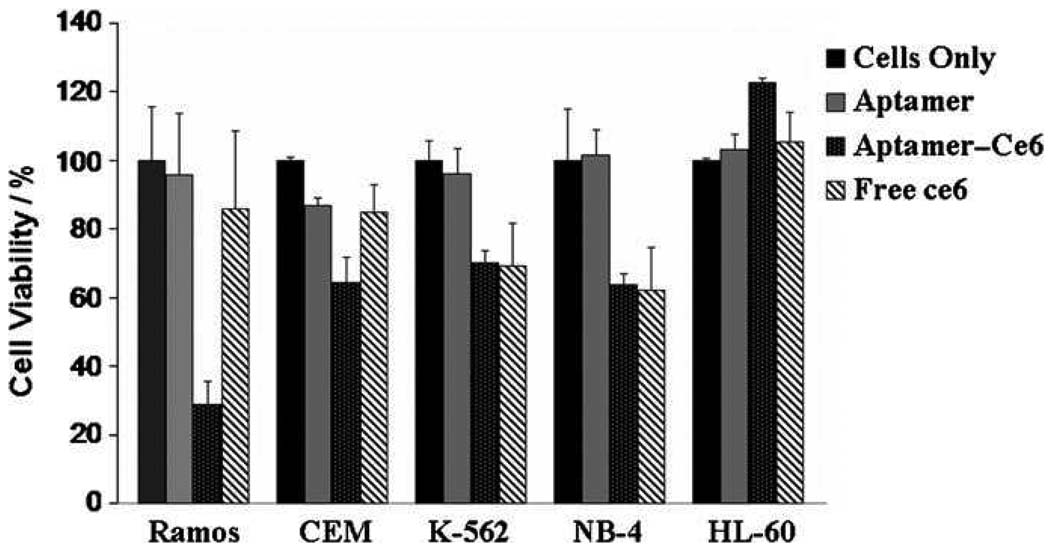
In recent years, photodynamic therapy (PDT), which takes advantage of the ability of a photosensitizer to kill cells by generating reactive oxygen species upon irradiation, has received much attention [44,45]. To overcome the current challenge in localization of photosensitizer at the diseased site, we have engineered a molecular construct with a photosensitizer and a highly selective aptamer for targeted tumor cell phototherapy [46]. An aptamer, TD05, which only recognizes Burkitt’s lymphoma Ramos cells, was used to chemically bind to an efficient phototherapy reagent, Ce6. Introduction of TD05-ce6 followed by light irradiation effectively destroyed the target Ramos cells, but not ALL cells or myeloid leukemia cell lines. As shown in Figure 7, the cell death induced by TD05-Ce6 toxicity in Ramos cells was high, while the toxicity observed in control CEM cells and other cell lines was over 50% less than the targeted cell lines.
Figure 7.
Cell toxicity assay results for Ramos cells (P <0.05) after 30 min incubation followed by irradiation of light for 4 h and subsequent growth for 36 h.
Another strategy we designed employs aptamer-nanorod conjugates for photothermal therapy [47]. This was achieved by combining the high absorption efficiency of Au-Ag nanorods with the target specificity of molecular aptamers to develop an efficient and selective therapeutic agent for targeted cancer cell photothermal destruction. As mentioned before, aptamer Scg8 conjugated with Au-Ag nanorods showed excellent recognition ability to CEM cells with an enhanced affinity exceeding that of the original aptamer probes. The high absorption characteristic of our Au-Ag nanorods requires up to a relative 6 orders of lower laser exposure than the other materials. When tested with both cell suspensions and solid tumor samples, these aptamer conjugates showed excellent hyperthermia efficiency and selectivity.
Aptamer-directed cancer biomarker discovery
Biomarker discovery is a pressing task in molecular medicine [14, 25]. Especially for many types of cancers, the lack of effective biomarkers greatly hinders early diagnosis and effective therapy. Although the combination of mass spectrometry (MS) and two-dimensional gel electrophoresis (2D-GE) enables the analysis of the whole cell proteome and identification cell-specific proteins, the main limitation of current biomarker discovery schemes is under-representation of membrane proteins in proteome analysis, which mainly results from intensive sample processing. It is estimated that 30% of proteins consists of membrane proteins and that less than 5% of this total is recognized by 2D-GE-MS [48]. Differential selection of aptamers from cell-SELEX facilitates biomarker discovery for membrane proteins. For example, the surface marker that distinguishes mature from immature dendritic cells has been discovered by this strategy [14]. In this discussion, we have shown how molecular probes generated by the cell-based SELEX method can bind to a specific type of cancer cells with high specificity. However, it should also be noted that these aptamers are expected to bind to some as yet unidentified membrane proteins, or biomarkers, on the cell-surface membrane. We used aptamer sgc8, which showed the highest affinity and specificity among the aptamers selected for the target CEM cells, for its binding molecule elucidation [49].
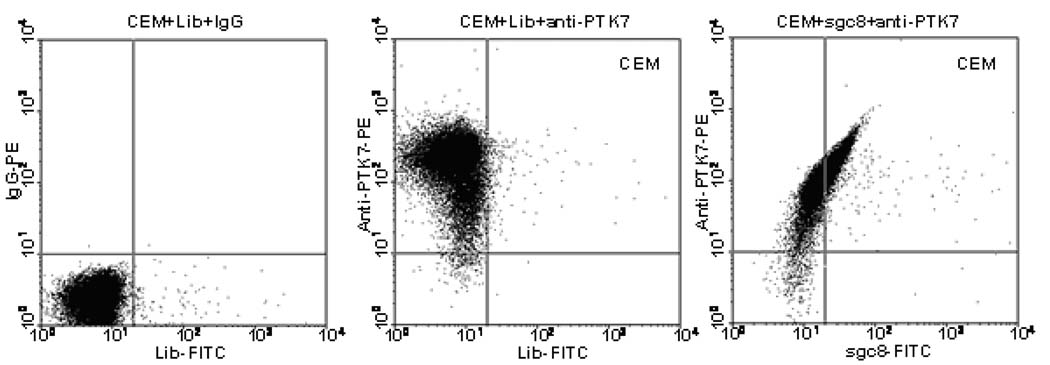
The procedure for target membrane protein identification mainly includes the following steps: 1) preparation of a cell lysate from the target cells, 2) separation of the membrane fraction from the soluble proteins, 3) extraction of the aptamer-protein complex, 4) separation of the target protein by SDS-PAGE, 5) sequencing the protein by mass spectrometry and 6) validation of the target protein. In our first try, sgc8 was labeled with a biotin tag, and its binding complex with the target was extracted using streptavidin-coated magnetic beads. By comparing with the control experiments using random DNA, characteristic protein bands on the SDS-PAGE captured by sgc8 were digested and subjected to LC-MS/MS analysis. A transmembrane receptor, PTK7 (protein tyrosine kinase 7), was identified and verified to be the aptamer target. It is interesting to find the co-binding of sgc8 and anti-PTK7 antibody on PTK7, instead of competitive binding, as evident by a linear relationship shown on CEM cells stained with anti-PTK7-PE and sgc8-FITC in the flow cytometry results (Figure 8). Although PTK7 is also known as colon carcinoma kinase-4 (CCK-4) and is reported to have a high expression level in several tumors [50], its exact function is not yet clear. Our results suggested for the first time a link between PTK7 over-expression and T-ALL cells.
Figure 8.
Flow cytometry assay of CEM cells stained with anti-PTK7-PE and sgc8-FITC.
We have also identified the target of another aptamer, TD05, which is specific to Ramos cells [51]. By chemical modification of TD05 with a photoactive group to covalently crosslink the aptamers with its target, we improved upon the previous method to allow more efficient capture and enrichment of the target receptors. Immunoglobulin Heavy mu chain (IGHM) was found to be the target of TD05. It has been reported that IGHM expression level on premature B-lymphocytes is closely related to Burkitt’s lymphoma development [52]. Therefore, using the aptamer approach is effective in identifying cell membrane receptors that have altered expression levels in tumor cells.
By developing high-quality aptamer probes through cell-SELEX and using them to identify their target proteins, these studies demonstrate that new disease-related potential markers can be discovered. This has major clinical implications in that the technique promises to improve the overall effectiveness of biomarker discovery.
Conclusion and future perspective
Cell-SELEX provides an effective approach to generate a large number of aptamer probes that specifically target a variety of cancer cells whose molecular makers are unknown. These aptamers can be applied to many areas of cancer cell biology, generating a wide variety of directions for future work. For example, the use of aptamers to stain patient samples and detect cancer using flow cytometry or microscopy can be easily transferred to the clinic. Also, using existing biotechnologies and analytical methods, we can functionalize aptamers by integrating them into microfluidic devices and nanobiosensors in order to create inexpensive early detection schemes. Furthermore, aptamers highly specific to cancer cells can be used as drug targeting agents. This may result in reducing toxicity while increasing the efficacy of current therapeutic drugs.
Another exciting new research focus will study the interactions between aptamers and the cells they bind. Using cell-SELEX aptamers to fish for target proteins, new cancer biomarkers will be discovered. Aptamers that are internalized by cells through their surface protein-binding partners will be used to study internalization pathways. They can also function as regulators to inhibit or activate the signaling pathways for the study of cancer development.
All these efforts will lead to an improved understanding of the biochemistry and molecular basis of cancer, which will, in turn, spur exciting new technologies for detection, diagnosis, and treatment of cancer. This aptamer-based chemical biology approach can also be applied to the investigation of other diseases. With a large number of aptamers specific to different types of disease cells, personalized medicine can become a practical reality. For instance, the molecular profiling of blood or other easily obtainable bodily fluids will give a virtual picture of an individual’s current state of health, as well as provide the potential prognosis factors.
Acknowledgements
The authors would like to acknowledge many of our co-workers whose work is reported here. This work has been supported over the years by NIH, NSF and CAS grants.
Biographies
Xiaohong Fang is a professor in the Institute of Chemistry at the Chinese Academy of Sciences. She received her BS at Wuhan University and her PhD in Chemistry in 1996 at Peking University. From 1998 to 2001, she worked as a research associate at the University of Florida, where she started developing aptamer-based analytical methods in Dr. Weihong Tan’s group. Her current research interests include nanobiotechnology, real-time biomolecule detection and biomolecular interaction study at the single-molecule level.
Weihong Tan is V. T. and Louis Jackson Professor of Chemistry and Physiology at the University of Florida. He obtained his BS from Hunan Normal University, his MS from the Chinese Academy of Sciences, and his PhD in physical chemistry in 1993 from the University of Michigan. His research interests include chemical biology, bioanalytical chemistry, bionanotechnology and molecular engineering. Recently, his group has developed cell-SELEX for the generation and application of aptamers as molecular tools to elucidate the molecular foundation of biological and pathological processes.
References
- 1.Tuerk C, Gold L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 2.Ellington AD, Szostak JW. In vitro Selection of RNA Molecules that Bind Specific Ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 3.Osborne SE, Matsumura I, Ellington AD. Aptamers as Therapeutic and Diagnostic Reagents: Problems and Prospects. Curr. Opin. Chem. Biol. 1997;1:5–9. doi: 10.1016/s1367-5931(97)80102-0. [DOI] [PubMed] [Google Scholar]
- 4.Famulok M, Hartig JS, Mayer G. Functional Aptamers and Aptazymes in Biotechnology, Diagnostics, and Therapy. Chem. Rev. 2007;107:3715–3743. doi: 10.1021/cr0306743. [DOI] [PubMed] [Google Scholar]
- 5.Navani NK, Li Y. Nucleic Acid Aptamers and Enzymes as Sensors. Curr. Opin. Chem. Biol. 2006;10:272–281. doi: 10.1016/j.cbpa.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 6.Bunka DH, Stockley PG. Aptamers Come of Age-at Last. Nat. Rev. Microbiol. 2006;4:588–596. doi: 10.1038/nrmicro1458. [DOI] [PubMed] [Google Scholar]
- 7.Jiang Y, Zhu C, Ling L, Wan L, Fang X, Bai C. Specific Aptamer-Protein Interaction Studied by Atomic Force Microscopy. Anal. Chem. 2003;75:2112–2116. doi: 10.1021/ac026182s. [DOI] [PubMed] [Google Scholar]
- 8.Ireson CR, Kelland LR. Discovery and Development of Anticancer Aptamers. Mol. Cancer Ther. 2006;5:2957–2962. doi: 10.1158/1535-7163.MCT-06-0172. [DOI] [PubMed] [Google Scholar]
- 9.Lupold SE, Hicke BJ, Liny Y, Coffey DS. Identification and Characterization of Nuclease-Stabilized RNA Molecules that Bind Human Prostate Cancer Cells via the Prostate-Specific Membrane Antigen. Cancer Res. 2002;62:4029–4033. [PubMed] [Google Scholar]
- 10.Daniels DA, Chen H, Hicke BJ, Swiderek KM, Gold L. A Tenascin-C Aptamer Identified by Tumor Cell SELEX: Systematic Evolution of Ligands by Exponential Enrichment. Proc. Natl. Acad. Sci. U.S.A. 2003;100:15416–15421. doi: 10.1073/pnas.2136683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cerchia L, Ducongé F, Pestourie C, Boulay J, Aissouni Y, Gombert K, Tavitian B, Franciscis V, Libri D. Neutralizing Aptamers from Whole-Cell SELEX Inhibit the RET Receptor Tyrosine Kinase. PLoS. Biol. 2005;3:e123. doi: 10.1371/journal.pbio.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Shamah SM, Healy JM, Cload ST. Complex Target SELEX. Acc. Chem. Res. 2008;41:130–138. doi: 10.1021/ar700142z. [DOI] [PubMed] [Google Scholar]
- 13.Blank M, Weinschenk T, Priemer M, Schluesener H. Systematic Evolution of a DNA Aptamer Binding to Rat Brain Tumor Microvessels. Selective Targeting of Endothelial Regulatory Protein Pigpen. J. Biol. Chem. 2001;276:16464–16468. doi: 10.1074/jbc.M100347200. [DOI] [PubMed] [Google Scholar]
- 14.Berezovski MV, Lechmann M, Musheev MU, Mak TW, Krylov SN. Aptamer-Facilitated Biomarker Discovery (AptaBiD) J. Am. Chem. Soc. 2008;130:9137–9143. doi: 10.1021/ja801951p. [DOI] [PubMed] [Google Scholar]
- 15.Guo K, Schafer R, Paul A, Gerber A, Ziemer G, Wendel HP. A New Technique for the Isolation and Surface Immobilization of Mesenchymal Stem Cells from Whole Bone Marrow Using High-Specific DNA Aptamers. Stem Cells. 2006;24:2220–2231. doi: 10.1634/stemcells.2006-0015. [DOI] [PubMed] [Google Scholar]
- 16.Raddatz ML, Dolf A, Endl E, Knolle P, Famulok M, Mayer G. Enrichment of Cell-Targeting and Population-Specific Aptamers by Fluorescence-Activated Cell Sorting. Angew. Chem. Int. Ed. 2008;47:5190–5193. doi: 10.1002/anie.200800216. [DOI] [PubMed] [Google Scholar]
- 17.Shangguan D, Li Y, Tang Z, Cao Z, Chen H, Mallikaratchy P, Sefah K, Yang C, Tan W. Aptamers Evolved from Live Cells as Effective Molecular Probes for Cancer Study. Proc. Natl. Acad. Sci. U.S.A. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shangguan D, Meng L, Cao Z, Xiao Z, Fang X, Li Y, Cardona D, Witek R, Liu C, Tan W. Identification of Liver Cancer-Specific Aptamers Using Whole Live Cells. Anal. Chem. 2008;80:721–728. doi: 10.1021/ac701962v. [DOI] [PubMed] [Google Scholar]
- 19.Sefah K, Tang Z, Shangguan D, Chen H, Lopez-Colon D, Li Y, Parekh P, Martin J, Meng L, Phillips JA, Kim YM, Tan W. Molecular Recognition of Acute Myeloid Leukemia Using Aptamers. Leukemia. 2009;23:235–244. doi: 10.1038/leu.2008.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen H, Medley C, Sefah K, Shangguan D, Tang Z, Meng L, Smith J, Tan W. Molecular Recognition of Small-Cell Lung Cancer Cells Using Aptamers. ChemMedChem. 2008;3:991–1001. doi: 10.1002/cmdc.200800030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang Z, Shangguan D, Wang K, Shi H, Sefah K, Mallikratchy P, Chen H, Li Y, Tan W. Selection of Aptamers for Molecular Recognition and Characterization of Cancer Cells. Anal. Chem. 2007;79:4900–4907. doi: 10.1021/ac070189y. [DOI] [PubMed] [Google Scholar]
- 22.Hasegawa H, Taira KI, Sode K, Ikebukuro K. Improvement of Aptamer Affinity by Dimerization. Sensors. 2008;8:1090–1098. doi: 10.3390/s8021090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Müller J, Wulffen B, Pötzsch B, Mayer G. Multidomain Targeting Generates a High-Affinity Thrombin-Inhibiting Bivalent Aptamer. ChemBioChem. 2007;8:2223–2226. doi: 10.1002/cbic.200700535. [DOI] [PubMed] [Google Scholar]
- 24.Kim Y, Cao Z, Tan W. Molecular Assembly for High-Performance Bivalent Nucleic Acid Inhibitor. Proc. Natl. Acad. Sci. U.S.A. 2008;105:5664–5669. doi: 10.1073/pnas.0711803105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Negm RS, Verma M, Srivastava S. The Promise of Biomarkers in Cancer Screening and Detection. Trends Mol. Med. 2002;8:288–293. doi: 10.1016/s1471-4914(02)02353-5. [DOI] [PubMed] [Google Scholar]
- 26.Aina OH, Liu R, Sutcliffe JL, Marik J, Pan CX, Lam KS. From Combinatorial Chemistry to Cancer-Targeting Peptides. Mol Pharm. 2007;4:631–651. doi: 10.1021/mp700073y. [DOI] [PubMed] [Google Scholar]
- 27.Shangguan D, Cao Z, Li Y, Tan W. Aptamers Evolved from Cultured Cancer Cells Reveal Molecular Differences of Cancer Cells in Patient Samples. Clin. Chem. 2007;53:1153–1155. doi: 10.1373/clinchem.2006.083246. [DOI] [PubMed] [Google Scholar]
- 28.Huang Y, Chang H, Tan W. Cancer Cell Targeting Using Multiple Aptamers Conjugated on Nanorods. Anal. Chem. 2008;80:567–572. doi: 10.1021/ac702322j. [DOI] [PubMed] [Google Scholar]
- 29.Medley C, Smith J, Tang Z, Wu Y, Bamrungsap S, Tan W. Gold Nanoparticle-Based Colorimetric Assay for the Direct Detection of Cancerous Cells. Anal. Chem. 2008;80:1067–1072. doi: 10.1021/ac702037y. [DOI] [PubMed] [Google Scholar]
- 30.Herr J, Smith J, Medley C, Shangguan D, Tan W. Aptamer-Conjugated Nanoparticles for Selective Collection and Detection of Cancer Cells. Anal. Chem. 2006;78:2918–2924. doi: 10.1021/ac052015r. [DOI] [PubMed] [Google Scholar]
- 31.Smith J, Medley C, Tang Z, Shangguan D, Lofton C, Tan W. Aptamer-Conjugated Nanoparticles for the Collection and Detection of Multiple Cancer Cells. Anal. Chem. 2007;79:3075–3082. doi: 10.1021/ac062151b. [DOI] [PubMed] [Google Scholar]
- 32.Phillips J, Xu Y, Xia Z, Fan Z, Tan W. Enrichment of Cancer Cells Using Aptamers Immobilized on a Microfluidic Channel. Anal. Chem. 2009;81:1033–1039. doi: 10.1021/ac802092j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iinuma H, Okimaga K, Adachi M, Suda K, Sekine T, Sakagawa K, Baba Y, Tamura J, Kumagai H, Ida A. Detection of Tumor Cells in Blood Using CD45 Magnetic Cell Separation Followed by Nested Mutant Allele-Specific Amplification of p53 and K-ras Genes in Patients with Colorectal Cancer. Int. J. Cancer. 2000;89:337–344. doi: 10.1002/1097-0215(20000720)89:4<337::aid-ijc4>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 34.Nagrath S, Sequist L, Maheswaran S, Bell D, Irimia1 D, Ulkus L, Smith M, Kwak E, Digumarthy S, Muzikansky A, Ryan P, Balis U, Tompkins R, Haber D, Toner M. Isolation of Rare Circulating Tumor Cells in Cancer Patients by Microchip Technology. Nature. 2007;450:1235–1241. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morishita R, Gibbons GH, Horiuchi M, Ellison KE, Nakajima M, Zhang L, Kaneda Y, Ogihara T, Dzau VJ. A Gene Therapy Strategy Using a Transcription Factor Decoy of the E2F Binding Site Inhibits Smooth Muscle Proliferation in vivo. Proc. Natl. Acad. Sci. U.S.A. 1995;92:5855–5859. doi: 10.1073/pnas.92.13.5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaloin L, Lehmann MJ, Georg S, Tobias R. Endogenous Expression of a High-Affinity Pseudoknot RNA Aptamer Suppresses Replication of HIV-1. Nucleic Acids Res. 2002;30:4001–4008. doi: 10.1093/nar/gkf522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farokhzad OC, Jon S, Khademhosseini A, Tran TT, LaVan DA, Langer R. Nanoparticle-Aptamer Bioconjugates: A new Approach for Targeting Prostate Cancer Cells. Cancer Res. 2004;64:7668–7672. doi: 10.1158/0008-5472.CAN-04-2550. [DOI] [PubMed] [Google Scholar]
- 38.Gu F, Zhang L, Teply BA, Mann N, Wang A, Radovic-Moreno AF, Langer R, Farokhzad OC. Precise Engineering of Targeted Nanoparticles by Using Self-Assembled Biointegrated Block Copolymers. Proc. Natl. Acad. Sci. U.S.A. 2008;105:2586–2591. doi: 10.1073/pnas.0711714105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chu TC, Twu KY, Ellington AD, Levy M. Aptamer Mediated siRNA Delivery. Nucleic Acids Res. 2006;34:e73. doi: 10.1093/nar/gkl388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao Z, Shangguan D, Cao Z, Fang X, Tan W. Cell-Specific Internalization Study of an Aptamer from Whole Cell Selection. Chemistry, A Eoropean Journal. 2008;14:1769–1775. doi: 10.1002/chem.200701330. [DOI] [PubMed] [Google Scholar]
- 41.Chu TC, Marks JW, Lavery LA, Faulkner S, Rosenblum MG, Ellington AD, Levy M. Aptamer: Toxin Conjugates that Specifically Target Prostate Tumor Cells. Cancer Res. 2006;66:5989–5992. doi: 10.1158/0008-5472.CAN-05-4583. [DOI] [PubMed] [Google Scholar]
- 42.Farokhzad OC, Cheng JJ, Teply BA, Sherifi I, Jon SY, Kantoff PW, Richie JP, Langer R. Targeted Nanoparticle-Aptamer Bioconjugates for Cancer Chemotherapy in vivo. Proc. Natl. Acad. Sci. U.S.A. 2006;103:6315–6320. doi: 10.1073/pnas.0601755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang Y, Shangguan D, Liu H, Phillips JA, Zhang X, Chen Y, Tan W. Molecular Assembly of an Aptamer-Drug Conjugate for Targeted Drug Delivery to Tumor Cells. ChemBioChem. 2009;10:862–868. doi: 10.1002/cbic.200800805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dolmans DE, Fukumura D, Jain RK. Photodynamic Therapy for Cancer. Nat. Rev. Cancer. 2003;3:380–387. doi: 10.1038/nrc1071. [DOI] [PubMed] [Google Scholar]
- 45.Oseroff AR, Ohuoha D, Hasan T, Bommer JC, Yarmush ML. Antibody-Targeted Photolysis: Selective Photodestruction of Human T-cell Leukemia Cells Using Monoclonal Antibody-Chlorin e6 Conjugates. Proc. Natl. Acad. Sci. U.S.A. 1986;83:8744–8748. doi: 10.1073/pnas.83.22.8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mallikaratchy P, Tang Z, Tan W. Cell Specific Aptamer-Photosensitizer Conjugates as a Molecular Tool in Photodynamic Therapy. ChemMedChem. 2008;3:425–428. doi: 10.1002/cmdc.200700260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang Y, Sefah K, Bamrungsap S, Chang H, Tan W. Selective Photothermal Therapy for Mixed Cancer Cells Using Aptamer-Conjugated Nanorods. Langmuir. 2008;24:11860–11865. doi: 10.1021/la801969c. [DOI] [PubMed] [Google Scholar]
- 48.Wu C, Yates JR., III The Application of Mass Spectrometry to Membrane Proteomics. Nat. Biotechnol. 2003;21:262–267. doi: 10.1038/nbt0303-262. [DOI] [PubMed] [Google Scholar]
- 49.Shangguan D, Cao Z, Meng L, Mallikaratchy P, Sefah K, Wang H, Li Y, Tan W. Cell-Specific Aptamer Probes for Membrane Protein Elucidation in Cancer Cells. J. Proteome Res. 2008;7:2133–2139. doi: 10.1021/pr700894d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Müller-Tidow C, Schwäble J, Steffen B, Tidow N, Brandt B, Becker K, Schulze-Bahr E, Halfter H, Vogt U, Metzger R, Schneider PM, Büchner T, Brandts C, Berdel WE, Serve H. High-Throughput Analysis of Genome-Wide Receptor Tyrosine Kinase Expression in Human Cancers Identifies Potential Novel Drug Targets. Clin. Cancer Res. 2004;10:1241–1249. doi: 10.1158/1078-0432.ccr-0954-03. [DOI] [PubMed] [Google Scholar]
- 51.Mallikaratchy P, Tang Z, Kwame S, Meng L, Shangguan D, Tan W. Aptamer Directly Evolved from Live Cells Recognizes Membrane Bound Immunoglobin Heavy Mu Chain in Burkitt's lymphoma cells. Mol. Cell Proteomics. 2007;6:2230–2238. doi: 10.1074/mcp.M700026-MCP200. [DOI] [PubMed] [Google Scholar]
- 52.Thomas MD, Srivastava B, Allman D. Regulation of Peripheral B-Cell Maturation. Cell. Immun. 2006;239:92–102. doi: 10.1016/j.cellimm.2006.04.007. [DOI] [PubMed] [Google Scholar]